Abstract
Most literature describing pharmacogenetic implementations are within academic medical centers and use single‐gene tests. Our objective was to describe the results and lessons learned from a multisite pharmacogenetic pilot that utilized panel‐based testing in academic and nonacademic settings. This was a retrospective analysis of 667 patients from a pilot in 4 perioperative and 5 outpatient cardiology clinics. Recommendations related to 12 genes and 65 drugs were classified as actionable or not actionable. They were ascertained from Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines and US Food and Drug Administration (FDA) labeling. Patients displayed a high prevalence of actionable results (88%, 99%) and use of medications (28%, 46%) with FDA or CPIC recommendations, respectively. Sixteen percent of patients had an actionable result for a current medication per CPIC compared with 5% per FDA labeling. A systematic approach by a health system may be beneficial given the quantity and diversity of patients affected.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Most patients possess at least one potentially actionable result.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ What is the clinical relevance of panel‐based testing using 12 genes with Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines in outpatient cardiology or perioperative clinics?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ One in six patients tested in an outpatient cardiology clinic or perioperative clinic possessed an actionable result per CPIC for a medication that was currently prescribed. That number changed to 1 in 20 patients if using pharmacogenetic recommendations in the US Food and Drug Administration label.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ This study provides further evidence that panel‐based testing is a reasonable approach; however, panel‐based testing can be accompanied by considerations (e.g., source of pharmacogenetic recommendations) that should be taken seriously. Future studies assessing the change in medication therapy related to this intervention are needed.
Guidelines from the Clinical Pharmacogenetics Implementation Consortium (CPIC) facilitate the translation of pharmacogenetic results to prescribing decisions in clinical practice.1 As of September 2019, the CPIC has published guidelines for 19 genes and 47 drugs.2 In addition to CPIC guidelines, pharmacogenetic data are available from numerous sources including the US Food and Drug Administration (FDA) labeling, the Pharmacogenomics Knowledgebase (PharmGKB), and the Dutch Pharmacogenetics Working Group (DPWG).3, 4 As of August 2019, PharmGKB noted that 37 of the 47 (79%) drugs with CPIC guidelines have descriptions of drug‐gene relationships in the FDA labeling.5
Despite recent advances, fewer health systems have clinically implemented pharmacogenetics than initially projected.6, 7 A number of barriers have been identified, including the lack of uptake in clinical guidelines.8 Several institutions, often at academic medical centers, have overcome these barriers and clinical implementations are underway.9, 10, 11 These institutions contribute to expanding evidence supporting clinical utility of pharmacogenetics and provide lessons learned, which can facilitate additional clinical implementations.12, 13, 14, 15
Many institutions have started with cardiology, largely in the inpatient setting, when establishing a pharmacogenetic program.11 Cardiology services are a reasonable target for pharmacogenetic implementations as CPIC guidelines and FDA labeling provide pharmacogenetic recommendations for multiple cardiac medications (e.g., clopidogrel and warfarin). Likewise, CPIC guidelines for medications used for conditions important in the perioperative setting (e.g., pain management, nausea/vomiting, and identification of malignant hyperthermia susceptibility (MHS)) are available. The perioperative setting exposes patients to a high volume of new medications in a condensed period. Furthermore, the planned nature of elective procedures represents a feasible opportunity to incorporate preemptive pharmacogenetic testing into a clinical workflow.
In an effort to expand the uptake of CPIC guidelines and to make pharmacogenetics more generalizable to patient care, there is a need to expand clinical implementation efforts to community and academic centers, particularly when multiple pharmacogenes are tested simultaneously (i.e., panel‐based testing). Our objective is to describe the results and lessons learned from a large multisite pilot in academic and nonacademic settings within one regional health system in the Washington‐Baltimore metropolitan area, MedStar Health. We also examine how the use of pharmacogenetic data can be applied in clinical specialties using either CPIC or FDA guidance.
Materials And Methods
Design
We formed an internal governing body, the MedStar Health Pharmacogenomics (PGx) Steering Committee, which has met monthly since May 2017. It aims to operationalize pharmacogenetic testing across MedStar Health, emphasizing improved outcomes, research opportunities, and education. Membership includes physician leadership from specialties relevant to where pharmacogenetics may be applied (e.g., primary care and anesthesiology), pharmacists, nursing, an education specialist, the Chief Medical Information Officer, a genetic counselor, a representative from a payor, legal, an ethicist, human factors research specialists, the MedStar Health Research Institute, and regulatory compliance.
We report the results of a retrospective analysis of a de‐identified data set from a clinical pharmacogenetics pilot. Patients were approached and offered pharmacogenetic testing in perioperative (n = 4) and outpatient cardiology (n = 5) clinics. In addition to clinical evidence, physician demand was a principle driver in selection of clinical sites and specialties for initial deployment.
Study population
Patients were adults who received clinical pharmacogenetic testing in a multisite (n = 9) pilot over a 4‐month period (December 2017–April 2018). Patients were approached at their provider’s discretion with no formal inclusion/exclusion criteria. Patients provided written informed consent for this clinical pharmacogenetic test. Although there is no consensus about whether the use of a clinical pharmacogenetic panel warrants obtaining written informed consent,16 the MedStar Health PGx Steering Committee decided that written informed consent for this test would help set appropriate patient expectations and ensure that patients received education about the potential benefits, limitations, and risks of the testing.
Pharmacogenetic testing
A buccal sample was collected from patients and mailed to a commercial laboratory to perform targeted next‐generation sequencing on 41 genes (Table S1 ). For cytochrome P450 family 2 subfamily D member 6 (CYP2D6), a PCR‐based assay assessed copy number variation. The polymerase chain reaction assay was able to detect which allele was duplicated but not the number of duplications. Samples were processed in accordance with College of American Pathologists (CAP)‐accredited, Clinical Laboratory Improvement Amendments (CLIA)‐certified laboratory procedures. Among 41 genes on the panel, 12 genes (i.e., calcium voltage‐gated channel subunit alpha1 S (CACNA1S), cytochrome P450 family 2 subfamily B member 6 (CYP2B6), cytochrome P450 family 2 subfamily C member 19 (CYP2C19), cytochrome P450 family 2 subfamily C member 9 (CYP2C9), CYP2D6, cytochrome P450 family 3 subfamily A member 5 (CYP3A5), dihydropyrimidine dehydrogenase (DPYD), interferon lambda 3 (IFNL3), solute carrier organic anion transporter family member 1B1 (SLCO1B1), ryanodine receptor 1 (RYR1), thiopurine S‐methyltransferase (TPMT), and vitamin K epoxide reductase complex subunit 1 (VKORC1)) have published CPIC guidelines available.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31
Reporting of results
For select genes (i.e., CYP2C19, CYP2C9, CYP2D6, CYP3A4, CYP3A5, DPYD, and TPMT), the laboratory translated genotype data to star alleles and phenotypes. These results were available as discrete, structured data in a PGx tab within the electronic health record (EHR). Additionally, results for the full panel were reported as a PDF available in the PGx tab and on the laboratory portal. Patients and the ordering provider had access to results on the laboratory portal.
Phenotype assignment and definition of actionable results
For the purposes of this report, all genetic results were translated (by author D.M.S.) to phenotypes in alignment with CPIC guidelines. CYP2D6 phenotype translation is a dynamic area.32 The CPIC recently completed the CYP2D6 Genotype to Phenotype Standardization Project, which proposes several changes to CYP2D6 phenotype translation.33 Analyses within this report were conducted using the new activity score definitions and phenotype assignments.
The definition of an actionable result aimed to take the perspective of a practicing clinician making a prescribing decision for an adult patient. Thus, phenotypes with a recommended change in prescribing (i.e., dose or drug selection) were defined as actionable results. However, this definition did not account for clinical factors outside of the drug and genotype. Other factors (e.g., indication, dose, and drug interactions) may have a significant role in how or whether pharmacogenetic results are applied, but are beyond the scope of this article. The assessment of recommendations at the level of a drug‐gene‐phenotype trio instead of a drug‐gene pair facilitates identification of differences in recommendations at the phenotype level. For example, codeine‐CYP2D6 is an actionable drug‐gene pair per the CPIC and the FDA. However, codeine‐CYP2D6‐ultrarapid metabolizer (UM) is actionable per the CPIC and the FDA, whereas codeine‐CYP2D6‐poor metabolizer is only actionable per CPIC.
Recommendations were collected from CPIC guidelines and FDA labeling and classified as actionable or not actionable for each drug‐gene‐phenotype trio (Tables S2‐S5 ).17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 34 Drugs were included if a CPIC guideline or FDA labeling had a recommendation or comment related to a gene of interest. The 12 genes of interest (i.e., CACNA1S, CYP2B6, CYP2C19, CYP2C9, CYP2D6, CYP3A5, DPYD, IFNL3, SLCO1B1, RYR1, TPMT, and VKORC1) were selected as they were on the test used in this pilot and have CPIC guidelines. The FDA recommendations were collected via Drug Label Annotations in PharmGKB then confirmed using the FDA labeling.5 It was not required for a drug to have a CPIC guideline. If a recommendation lacked a specific prescribing action, the pharmacogenetic result was considered “not actionable.” Evaluation of CPIC guidelines and FDA labeling for 12 genes led to categorization of actionability for 65 drugs, which included 72 drug‐gene pairs and 130 drug‐gene‐phenotype trios (Tables S2‐S5 ). This methodology identified 86 and 29 drug‐gene‐phenotype trios with an actionable recommendation from the CPIC and FDA, respectively. Figure 1 displays how each drug‐gene‐phenotype trio was categorized per the CPIC and FDA.
Figure 1.
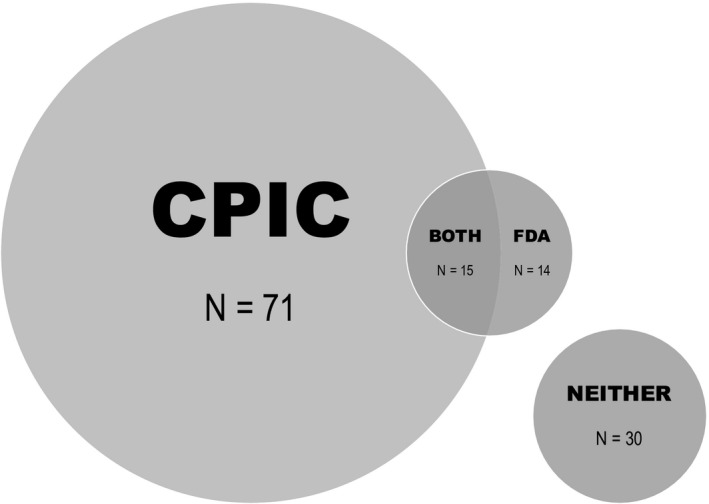
Classification of actionable recommendations for each drug‐gene‐phenotype trio. Both represent drug‐gene‐phenotype trios that had actionable recommendations from both organizations. The area covered by either Clinical Pharmacogenetics Implementation Consortium (CPIC) or US Food and Drug Administration (FDA) represents a drug‐gene‐phenotype trio with an actionable recommendation only provided by that organization. Neither represents drug‐gene‐phenotype trios that had a comment by at least one of the resources but was not deemed actionable by either organization.
Secondary findings
Secondary findings were defined as genes (i) in which variants may be associated with non‐drug‐induced phenotypic manifestations, (ii) recommended by the American College of Medical Genetics and Genomics (ACMG) for return of secondary findings,35 or (iii) in which variants are associated with potential reproductive implications. Genes were assessed for secondary findings via an evidence review of multiple sources (e.g., ACMG, ClinGen, CPIC, FDA labeling, Online Mendelian Inheritance in Man, and primary literature though PubMed searches). We evaluated each of the genes tested (Table S1 ) for secondary findings. The genes that may possess secondary findings were CACNA1S, DPYD, F2, F5, and RYR1. Table S6 displays the specific results classified as secondary findings and the rationale for each gene. The goal of this process was to connect patients with secondary findings to experts trained to manage such results. Upon secondary finding detection, the ordering provider was notified via the EHR. Providers were also given education, conversation guides, contact information for a genetic counselor, and descriptive letters that may be forwarded to their patient.
Data collection, outcome measures, and data analysis
Data in the de‐identified data set included age, sex, race, clinic site, provider, list of medications in the EHR at the time of sample collection, and genetic results. Outcomes included the prevalence of actionable results (e.g., CYP2D6‐UM), current use of a medication with CPIC guideline or FDA recommendations (e.g., codeine), and actionable results relevant to a current medication (e.g., codeine prescribed to a patient with CYP2D6‐UM). Physician champions and program leaders anecdotally provided lessons learned at the MedStar Health PGx Steering Committee meeting. Descriptive statistics were used. This study was deemed to be exempt from review by the Georgetown University Institutional Review Board.
Normality was assessed through histograms and the ShapiroWilks test with P < 0.05 translating to non‐normal distribution. One sample t‐test and Wilcoxon were used to assess continuous variables normally and not normally distributed, respectively. Chi‐square and Fisher’s exact test were used for categorical variables, as appropriate.
Results
Patient characteristics
Six hundred sixty‐eight patients underwent clinical pharmacogenetic testing via a panel consisting of 41 genes. One patient had incomplete genetic results (58 of 169 single nucleotide polymorphisms) and is excluded from the remainder of this report. Patient characteristics are presented in Table 1. Most patients (64%) received testing at outpatient cardiology clinics. Compared with patients tested at perioperative clinics, these patients tended to be older (P < 0.0001) and male (P < 0.01). Self‐reported race differed between clinics (P < 0.0001).
Table 1.
Patient characteristics
| Characteristic |
All patients N = 667 (%) |
Perioperative N = 237 (%) |
Cardiology N = 430 (%) |
|---|---|---|---|
| Age, yearsa | 70 (61, 76) | 67 (57, 74) | 72 (63, 78) |
| Sex, female | 339 (51) | 138 (58) | 201 (47) |
| Race | |||
| White | 424 (64) | 123 (52) | 301 (70) |
| African American | 87 (13) | 45 (19) | 42 (10) |
| Other | 67 (10) | 20 (8) | 47 (11) |
| Unknown | 89 (13) | 49 (21) | 40 (9) |
Data for age are displayed as median (interquartile range‐1, interquartile range‐3).
Actionability per CPIC and the FDA
Phenotype results for the genes tested with CPIC guidelines are shown in Table 2, with genotype and diplotype data available in Table S7 . Actionable results per CPIC guidelines were present in 660 of 667 patients (99%) tested. In other words, 99% of patients had a result that may be actionable if exposed to a relevant medication. Of the 12 genes with CPIC guidelines tested, patients had a mean of 3.4 ± 1.4 and a maximum of 8 actionable results per the CPIC. The genes most commonly found to be actionable per the CPIC were CYP2C19 (395, 59%), VKORC1 (393, 59%), CYP2B6 (312, 47%), and CYP2D6 (301, 45%). Actionable results per FDA labeling were present in 584 of 667 patients (88%) tested, which was lower than CPIC recommendations (P < 0.0001). Of the same 12 genes reported above, patients had a mean of 1.5 ± 0.90 and a maximum of 4 actionable results per the FDA. The genes most commonly found to be actionable per the FDA were VKORC1 (393, 59%), CYP2D6 (301, 45%), CYP2C9 (233, 35%), and TPMT (60, 9%).
Table 2.
Phenotype results for genes with CPIC guidelines
| Gene | Phenotypea | No. (%) |
|---|---|---|
| CACNA1S | Uncertain susceptibilityc | 667 (100) |
| CYP2B6 | Normal metabolism | 355 (53) |
| Intermediate metabolism | 247 (37) | |
| Poor metabolism | 65 (10) | |
| CYP2C19 | Ultrarapid metabolism | 27 (4) |
| Rapid metabolism | 160 (24) | |
| Normal metabolism | 269 (40) | |
| Intermediate metabolism | 186 (29) | |
| Poor metabolism | 22 (3) | |
| Uncertain functiond | 2 (< 1) | |
| Unknown resulte | 1 (< 1) | |
| CYP2C9 | Normal metabolism | 434 (65) |
| Intermediate metabolism | 218 (33) | |
| Poor metabolism | 15 (2) | |
| CYP2D6 | Ultrarapid metabolism | 19 (3) |
| Normal to ultrarapid metabolism | 12 (2) | |
| Normal metabolism | 351 (53) | |
| Intermediate metabolism | 248 (37) | |
| Poor metabolism | 34 (5) | |
| Unknown resulte | 3 (< 1) | |
| CYP3A5 | Normal metabolism | 46 (7) |
| Intermediate metabolism | 125 (19) | |
| Poor metabolism | 496 (74) | |
| DPYD | Normal metabolism | 659 (99) |
| Intermediate metabolism | 8 (1) | |
| IFNL3 | Favorable response | 306 (46) |
| Unfavorable response | 360 (54) | |
| Unknown resulte | 1 (< 1) | |
| RYR1 | Malignant hyperthermia susceptible | 5 (1) |
| Uncertain susceptibilityc | 662 (99) | |
| SLCO1B1 | Normal function | 495 (74) |
| Decreased function | 158 (24) | |
| Poor function | 14 (2) | |
| TPMT | Normal metabolism | 607 (91) |
| Intermediate metabolism | 59 (9) | |
| Poor metabolism | 1 (< 1) | |
| VKORC1 b | Wild type | 274 (41) |
| Heterozygous | 305 (46) | |
| Homozygous | 88 (13) |
CPIC, Clinical Pharmacogenetics Implementation Consortium. CACNA1S, calcium voltage‐gated channel subunit alpha1 S; CYP2B6, cytochrome P450 family 2 subfamily B member 6; CYP2C19, cytochrome P450 family 2 subfamily C member 19; CYP2C9, cytochrome P450 family 2 subfamily C member 9; CYP2D6, cytochrome P450 family 2 subfamily D member 6; CYP3A5, cytochrome P450 family 3 subfamily A member 5; DPYD, dihydropyrimidine dehydrogenase; IFNL3, interferon lambda 3; SLCO1B1, solute carrier organic anion transporter family member 1B1; RYR1, ryanodine receptor 1; TPMT, thiopurine S‐methyltransferase; VKORC1, vitamin K epoxide reductase complex subunit 1.
Data may not total 100% due to rounding.
CPIC guidelines were used as the standard for phenotype nomenclature.
Rather than a phenotype, CPIC recommendations related to VKORC1 are for a specific single nucleotide polymorphism (c.‐1639G > A, rs9923231).24
Uncertain susceptibility: The test did not find the patient to possess malignant hyperthermia susceptibility but the results do not eliminate the chance that a patient is susceptible to malignant hyperthermia. Patients may be susceptible to malignant hyperthermia due to genetic variants not tested or for nongenetic reasons.
Uncertain function: Definitive allele function has not been assigned by CPIC.
Unknown result: The laboratory did not generate a result.
The remaining analyses report on the 600 patients for which medication data were available. The prevalence of actionable results, current use of an affected medication, and actionable results relevant to a current medication are shown per CPIC guidelines and FDA labeling (Table 3), CPIC (Figure 2), and the FDA (Figure 3). The most commonly prescribed medications with CPIC guidelines included clopidogrel (102, 17%), simvastatin (62, 10%), warfarin (31, 5%), tramadol (28, 5%), and escitalopram (27, 5%). The most commonly prescribed medications with FDA pharmacogenetic recommendations were clopidogrel (102, 17%), warfarin (31, 5%), tramadol (28, 5%), celecoxib (8, 1%), and citalopram (7, 1%). Thirteen of 23 medications with actionable pharmacogenetic information in the FDA label were not prescribed. Per CPIC guidelines, clopidogrel‐CYP2C19 (37, 6%), warfarin‐VKORC1/CYP2C9 (22, 4%), and simvastatin‐SLCO1B1 (18, 3%) were the most prevalent actionable drug‐gene pairs. Per FDA labeling, warfarin‐VKORC1/CYP2C9 (22, 4%) and clopidogrel‐CYP2C19 (5, 1%) were the only actionable drug‐gene pairs.
Table 3.
Prevalence of actionability using CPIC guidelines and FDA labeling
| Medication data available (N = 600) | CPIC | FDA |
|---|---|---|
| Genetic resultsa | ||
| Actionable result (i.e., may be actionable now or in the future) | 595 | 523 |
| No actionable result | 5 | 77 |
| Current prescriptions affected by PGx results | ||
| Current prescription for an affected drugb | 277 | 168 |
| Recommendation: modify usec | 97 | 27 |
| Recommendation: regular use | 180 | 141 |
| No current prescription for an affected drug | 323 | 432 |
CPIC, Clinical Pharmacogenetics Implementation Consortium; FDA, US Food and Drug Administration; PGx, pharmacogenomics.
More patients possessed an actionable results per CPIC than the FDA (P < 0.0001).
More patients possessed a prescription for drug affected by PGx results per CPIC than the FDA (P < 0.0001).
More patients possessed an actionable result for a medication they were currently prescribed per CPIC than the FDA (P < 0.0001).
Figure 2.
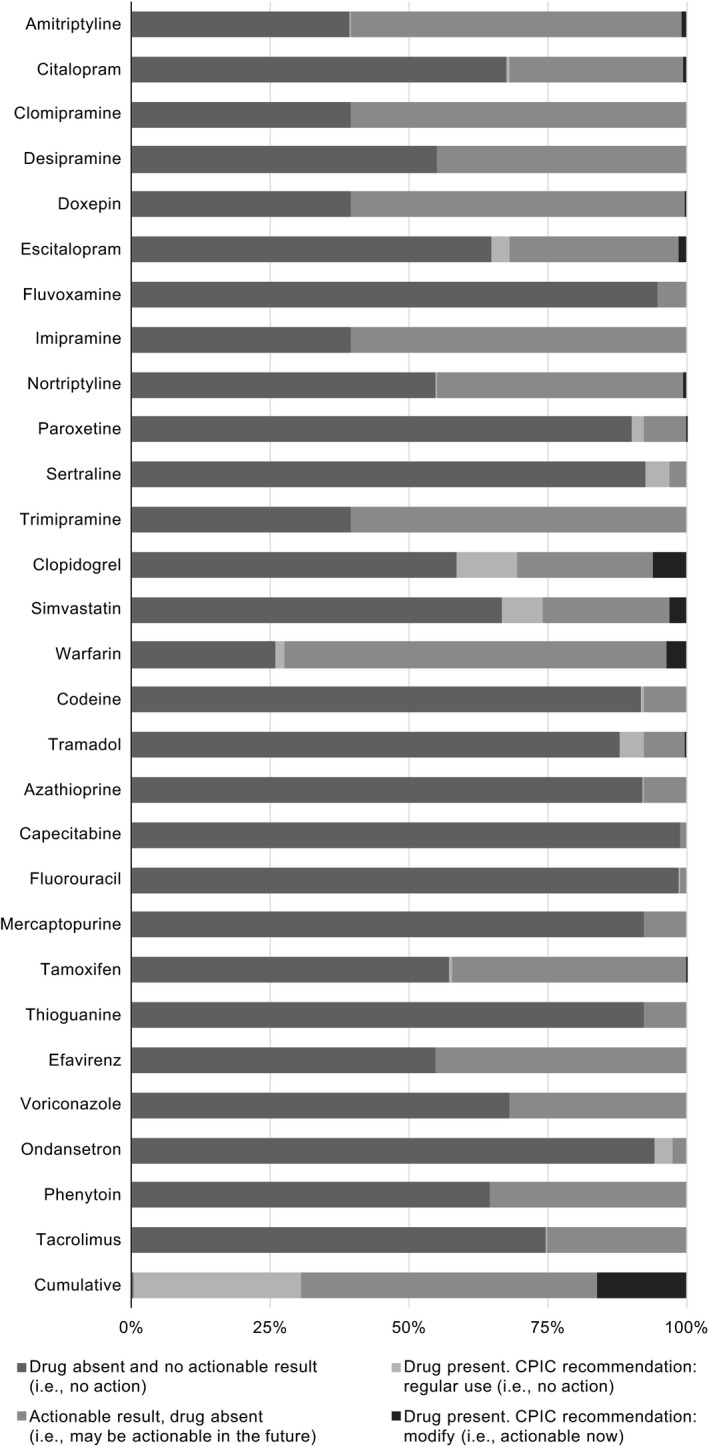
Actionable results per Clinical Pharmacogenetics Implementation Consortium (CPIC) by medication. Percentages are calculated using N = 600. Exact numbers are available in the supplement (Table S8 ).
Figure 3.
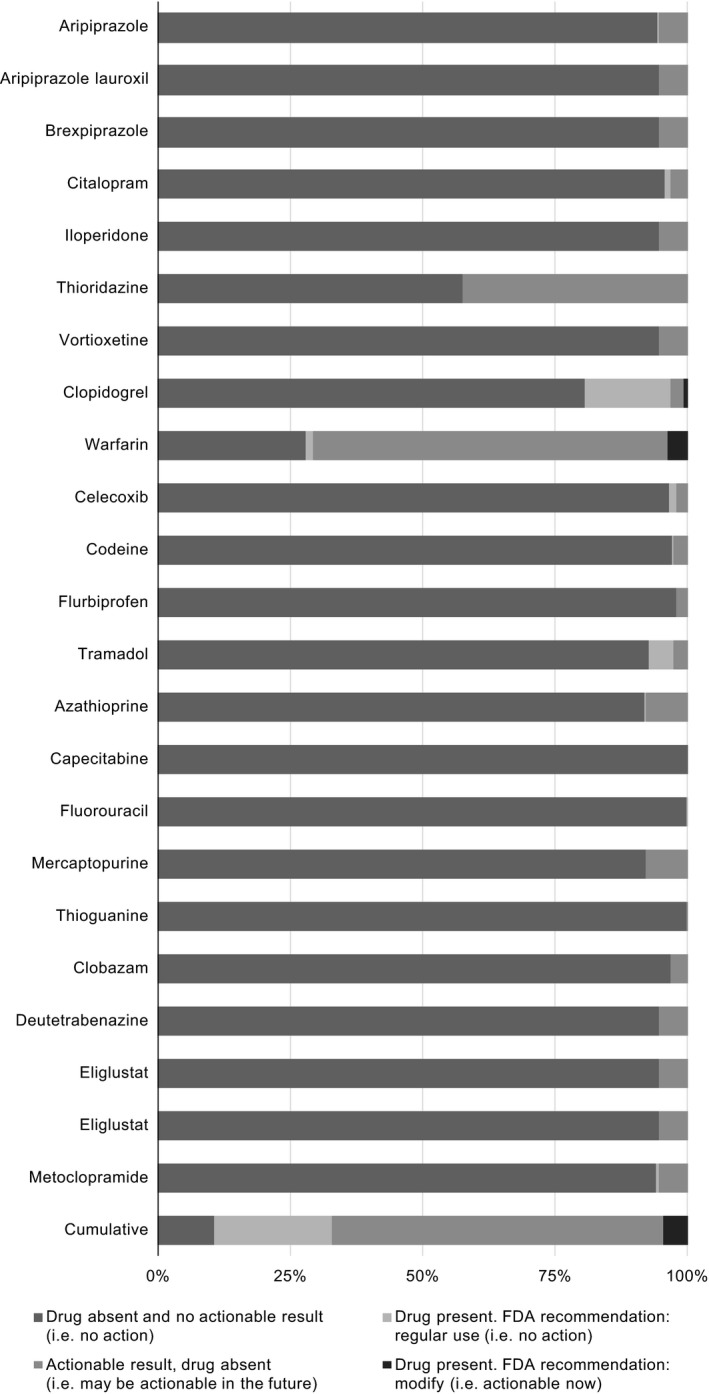
Actionable results per US Food and Drug Administration (FDA) by medication. Percentages are calculated using N = 600. Exact numbers are available in the supplement (Table S9 ).
Actionable results per CPIC by clinic site
Orders for at least one medication with CPIC guidelines were found for 195 of 399 patients (49%) compared with 82 of 201 patients (41%) tested at cardiology and perioperative clinics, respectively. The most commonly prescribed medications with CPIC guidelines for patients tested at cardiology clinics were clopidogrel (90, 23%), simvastatin (43, 11%), warfarin (28, 7%), tramadol (18, 5%), and escitalopram (17, 4%). The most commonly prescribed medications with CPIC guidelines for patients tested at perioperative clinics were simvastatin (19, 9%), ondansetron (14, 7%), clopidogrel (12, 6%), tramadol (10, 5%), and escitalopram (10, 5%).
The most prevalent actionable drug‐gene pairs for patients tested at cardiology clinics were clopidogrel‐CYP2C19 (31, 8%), warfarin‐VKORC1/CYP2C9 (19, 5%), and simvastatin‐SLCO1B1 (9, 2%). The most prevalent actionable drug‐gene pairs for patients tested at perioperative clinics were simvastatin‐SLCO1B1 (9, 5%), clopidogrel‐CYP2C19 (6, 3%), and warfarin‐VKORC1/CYP2C9 (3, 1%).
Secondary findings
Forty‐nine secondary findings were identified in 48 of 600 patients (7%). One patient had two secondary findings. These secondary findings were associated with variants in: F5 (21, 3%), F2 (20, 3%), RYR1 (5, 1%), and DPYD (3, < 1%). No patient was found to carry a variant in CACNA1S.
Lessons learned
Several lessons learned were identified by the MedStar Health PGx Steering Committee. First, expanded clinical expertise is necessary to guide ordering providers in the interpretation and application of pharmacogenetic data. To address this need, inpatient and outpatient pharmacists were identified and trained to serve as PGx consultants to clinical providers. These volunteers completed a PGx certificate from a national pharmacy organization, receive weekly “PGx Pearls” email updates, and meet monthly for continued education and other purposes. Moving forward, PGx consultants will serve as an expert resource for clinical consults, routine questions, and education delivery. A PGx subcommittee of the MedStar Health Pharmacy and Therapeutics committee was also formed. This subcommittee uses an evidence‐based approach in the clinical application of pharmacogenetics and influences the creation of related in‐house professional educational materials.
As commonly seen in practice,36 laboratory phenotype translation differed from the CPIC in some instances. The translation from diplotype to phenotype differed between the laboratory and the CPIC for select CYP2C9 alleles (i.e., CYP2C9*5, *6, *8, and *11) and CYP2C19 diplotypes (e.g., CYP2C19*2/*17). CYP2C19 diplotypes with a loss of function allele and an increased function allele are currently conditionally classified as intermediate metabolizers (IMs) per the CPIC whereas the laboratory took a different approach of classifying these results as IM/normal metabolizers.25
Nineteen patients possessed at least one of the CYP2C9*5, *6, *8, and *11 alleles, which were tested but not initially translated to star alleles, and, thus, the laboratory classified these results as CYP2C9*1. Upon translating these results per CPIC guidelines, the CYP2C9 phenotype was changed from normal metabolizer to IM for 18 patients and from IM to poor metabolizer for 1 patient. This issue predominantly affected patients who did not identify as white (15 of 19 (79%)). The rapidly growing literature is challenging to maintain alignment with CPIC guidelines as the CYP2C9 discrepancy is a result of a CPIC guideline update earlier in that year.24
Providers were receptive to the pilot as measured by test order volume (668 tests ordered in ~ 4 months); however, they reported translation of results to clinical practice to be a significant challenge. Although the laboratory‐generated PDF delivered genotype‐guided recommendations, providers noted multiple limitations (e.g., difficulty finding relevant recommendations and report length). Providers expressed preference for electronic clinical decision support alerts at the time of medication order entry and access to pharmacist consultations. Due to the timing of an EHR software upgrade, patients tested at two sites did not initially have results available in the EHR. Accessing results for these patients via the laboratory’s patient portal was feasible for the ordering provider but was a barrier to access for other clinicians. Moving forward, all implementation sites will be on the same version of the EHR.
The last lesson learned was that many providers did not expect to be contacted about secondary findings. However, they appreciated the clinical support to aid in connecting patients to the appropriate expertise (e.g., hematologists and clinical geneticist). Providers relayed several comments, including: a need for more patient and clinician education regarding secondary findings at the time of testing and a preference for pharmacogenetic testing that excludes genes with known secondary findings.
Discussion
This pilot identified 88–99% patients to have an actionable result, depending on the source used for the calculation. These data are similar to those reported by others.37, 38, 39, 40 Patients are likely to carry a potentially actionable result with clinical utility depending on exposure to a medication relevant to that result. This pilot identified 16% of patients who had an actionable result per CPIC guidelines for a currently prescribed medication. This is a snapshot in time, which likely underestimates the total utility of these results. However, just 5% of patients possessed an actionable result per FDA labeling for a currently prescribed medication.
The FDA has a similar but different scope than the CPIC for clinical pharmacogenetic recommendations, which may partially explain the differences in clinical pharmacogenetic recommendations between the two. CPIC guidelines are written under the assumption that the genetic test result is known.1 The FDA evaluates multiple variables when considering drug label modifications that address pharmacogenetic information.41 In the case of warfarin, leading experts in pharmacogenetics at the FDA stated one concern was related to “being overly prescriptive in labeling” due to clinician access to an analytically validated assay. Access to testing is certainly a valid concern, but many health systems have access to an analytically validated (CAP/CLIA‐certified) laboratory.42 Interestingly, warfarin is one example where CPIC and FDA recommendations are largely similar, although there are subtle yet clinically relevant differences (e.g., additional CYP2C9 alleles in CPIC guideline, dosing algorithm in CPIC guideline vs. dosing table in FDA label).
One example of discordance between CPIC guidelines and FDA labeling are recommendations related to RYR1 and CACNA1S for identification of MHS with select anesthetics (Tables S2‐S5 ). CPIC states these agents are relatively contraindicated in persons with MHS,22 whereas FDA labeling for several anesthetics states that they are “contraindicated in individuals with known, or suspected, genetic predisposition to malignant hyperthermia.” Notably, the label does not name genes associated with MHS and, thus, was classified as “not actionable” for the purposes of this study.
Differences between recommendations from the CPIC and the FDA have increased importance. A laboratory marketing a panel‐based pharmacogenetic test recently received a warning letter from the FDA that cited several concerns, including the clinical validity of the test.43 For example, the FDA stated that two drugs (i.e., escitalopram‐CYP2C19 and sertraline‐CYP2C19) had an unestablished relationship between genotype and drug response, which contradicts CPIC guidelines. The CPIC has since responded to the FDA and indicated CPIC recommendations are analogous to adjustments routinely made based upon pharmacokinetic data.44 Although a lack of randomized controlled trials is often cited as a barrier to implementation of pharmacogenetics, randomized controlled trials may not be feasible or ethical.45 In practice, clinical pharmacogenetics is the utilization of additional evidence in clinical decisions that would have otherwise been left to clinical judgment. Differences between CPIC and FDA recommendations are an example of a problem that health systems could address through an interdisciplinary committee, like our PGx Steering Committee, which can provide guidance on the interpretation of pharmacogenetic data into the practice of medicine.
Numerous institutions have implemented pharmacogenetics, some of which are part of the Implementing GeNomics In pracTicE (IGNITE) Network. The IGNITE Pharmacogenetics Working Group has reported strategies for two of the most widely tested pharmacogenes, CYP2C19 and CYP2D6.9, 11 Our pilot differs from other pharmacogenomic implementations, with the largest difference being the target population. Another difference was our identification and management of secondary findings. The ACMG published a list of 59 medically actionable high‐risk genes, which recommends that laboratories offer to return these results, regardless of the indication for whole exome/genome sequencing.22, 35 RYR1 and CACNA1S are currently the only genes with CPIC guidelines, which are also on this ACMG list. Secondary findings associated with RYR1 and CACNA1S may have broad implications. For instance, identifying a carrier of a pathogenic variant in RYR1 or CACNA1S may result in military discharge or ineligibility as the US Department of Defense defines MHS as a disqualifying condition.46 Including this information in a consent prior to testing could give patients the option to opt out of testing for MHS. This process speaks to a bigger issue where our program is unique, as we are addressing concerns regarding genetic discrimination in subsequent consent. Our PGx Steering Committee will discuss the inclusion of RYR1 and CACNA1S before testing resumes.
During this pilot, any positive finding for a variant reported for RYR1 and CACNA1S was treated as diagnostic for MHS, and, thus, avoidance of triggering agents (i.e., succinylcholine and potent volatile anesthetics) was recommended. An important consideration related to the pre‐operative identification of patients with MHS is that it causes workflow changes in the operating room. Because extensive cleaning of equipment is required prior to anesthetizing a patient with MHS, it is optimal for an affected patient to be the first case of the day. Additionally, although the anesthesiologists were the physician champions actively seeking pharmacogenetic testing to guide perioperative care, anesthesiologists often had to rely on the orthopedic surgeon to order the test. Reasons for this collaboration included constraints due to test turnaround time, workflow, and reimbursement (e.g., test ordered inpatient vs. outpatient). Follow‐up with anesthesiologists after surgery is also less common for patients and, thus, having orthopedic surgeons order pharmacogenetic testing helped to maximize continuity of care and appropriate follow‐up. Our study had several strengths: (i) we provided pharmacogenetic testing to outpatient cardiology and perioperative clinics, which are uncommonly reported in the literature, (ii) this was a large pilot, with 668 patients undergoing clinical pharmacogenetic testing, (iii) we comprehensively reviewed and categorized the actionability of all CPIC guidelines and FDA labels for medications affected by the 12 genes of interest from the perspective of a prescribing clinician, (iv) we coordinated an interdisciplinary team possessing the expertise to address pharmacogenetic implications as well as secondary findings, and (v) we provide a measure of clinical relevance by assessing the number of patients with actionable results relevant to a current medication using both CPIC guideline and FDA recommendations.
There are several limitations to our work. (i) Our analyses were limited to 12 genes with CPIC guidelines that were included on the panel and, therefore, do not account for other drug‐gene pairs with a high level of evidence (e.g., abacavir‐HLA‐B). (ii) The number of patients prescribed a medication with CPIC guidelines (46%) or FDA recommendations (28%) is likely an underestimate as it only includes medications prescribed at the time pharmacogenetic testing was ordered. It does not include medications being considered by prescribers (e.g., patients at perioperative clinics were tested to guide pharmacotherapy decisions at a future procedure). (iii) Insurance reimbursement and the cost of testing is a widely cited barrier to implementation of pharmacogenetic testing; however, we did not have reimbursement data available.47 (iv) This was a retrospective cohort created from a de‐identified data set; therefore, we could not assess clinical outcomes or changes in medication. However, we hypothesize results were underutilized due to reliance on laboratory‐generated PDF reports to drive changes in clinical practice and that we currently lack, but are building clinical decision support and a clinical consult service. Anecdotally, the majority of providers indicated they could not easily use the PDF report to make changes in care, whereas the minority of providers found the PDF report useful. (v) The definition of actionable results used in this paper does not account for clinical factors outside of drug and genotype. Inclusion of other factors (e.g., patient history, indication, and result availability at initiation of therapy) would provide a more global assessment of whether the result is actionable.
Given the diversity of medications with pharmacogenetic recommendations in the FDA labeling and CPIC guidelines, a systematic approach across a health system may be beneficial. Testing for 12 genes with CPIC recommendations found 16% of patients had an actionable result for a current medication compared with just 5% of patients per FDA labeling. The prevalence of actionable results (88–99%) and use of medications (28–46%) with FDA or CPIC recommendations was high. Future studies will assess the clinical utility and outcomes of pharmacogenetics.
Funding
Institutional support was received from MedStar Health.
Conflict of Interest
Ms. Peshkin reports personal fees from Clear Genetics, outside the submitted work. Dr. Shapiro reports non‐financial support from CQuentia NGS, LLC, during the conduct of the study. Dr. Swain reports personal fees and non‐financial support from Athenex, personal fees and non‐financial support from Daiichi‐Sankyo, personal fees and non‐financial support from Eli Lilly and Company, grants, personal fees, non‐financial support and other from Genentech/Roche, personal fees from Genomic Health, personal fees and non‐financial support from Inivata, Ltd., personal fees and non‐financial support from Novartis, grants from Pfizer, personal fees and non‐financial support from Pieris Pharmaceuticals, personal fees from Tocagen, non‐financial support from Caris Life Sciences, non‐financial support from NanoString Technologies, grants from Merrimack, non‐financial support and other from AstraZeneca, non‐financial support from Bristol‐Myers Squibb, outside the submitted work. All other authors declared no competing interests for this work.
Author Contributions
D.M.S., B.N.P., T.B.S., R.P.B., E.H., and S.M.S. wrote the manuscript. D.M.S. and S.M.S. designed the research. D.M.S., B.N.P., T.B.S., R.P.B., E.H., S.K., R.S., Z.E., C.L., J.M., B.L., and S.M.S. performed the research. D.M.S. and S.M.S. analyzed the data.
Supporting information
Table S1. Genes tested.
Table S2. CPIC guidelines and clinical actionability.
Table S3. FDA labeling and clinical actionability.
Table S4. FDA labeling and translation to actionability.
Table S5. Pharmacogenetic evidence for each drug‐gene pair.
Table S6. Secondary findings (SFs).
Table S7. Diplotype results.
Table S8. Actionable results per CPIC by medication.
Table S9. Actionable results per FDA by medication.
Acknowledgments
The authors thank the following current or former members of the PGx Steering Committee: Alex Walker, PhD; Allison Reschovsky, Esq; Amber Stumpf, MBA; Amy Will; Daniel Feeley, CPA; Eric Slechter; Erin Agelakopoulos, MBA; Geoffrey Cox, PharmD; Gregory Dohmeier, DO; Inna Kats, MD; James Welsh, MD, MBA, MPH; Jennifer Wilkerson, MHSA; Jessica Howe, MA; Jim McMahon, MBA; Joan Bardsley, MBA; Katie Carlin, MBA; Kevin FitzGerald, SJ, PhD; Kumudhini Hendrix, MD; Lisa Speight, MD; Mark Call; Mark Hoeflich; Mary Anne Hinkson, MBA; Mitch Herbert, MS; Moira Larsen, MD, MBA; Mutanu Mutuvi‐Thomas, MSIT; Neil J. Weissman, MD; Patryce Toye, MD; Paul Scherz, PhD; Pete Celano, MBA; Rachel Wynn, PhD; Raj Ratwani, PhD; Stephen R.T. Evans, MD; Subha Madhavan, PhD; Terry Fairbanks, MD, MS; and Zoey Pruitt. The authors thank Dr. Reem Saadeh‐Haddad, Dr. Adrian Hui, and Mr. Joseph Welsh for their contributions to this work.
References
- 1. Caudle, K.E. et al Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 15, 209–217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clinical Pharmacogenetics Implementation Consortium (CPIC) . Guidelines <https://cpicpgx.org/guidelines/>. Accessed September 30, 2019.
- 3. Swen, J.J. et al Pharmacogenetics: from bench to byte–an update of guidelines. Clin. Pharmacol. Ther. 89, 662–673 (2011). [DOI] [PubMed] [Google Scholar]
- 4. Caudle, K.E. et al Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am. J. Health Syst. Pharm. 73, 1977–1985 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whirl‐Carrillo, M. et al Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKinnon, R.A. , Ward, M.B. & Sorich, M.J. A critical analysis of barriers to the clinical implementation of pharmacogenomics. Ther. Clin. Risk Manag. 3, 751–759 (2007). [PMC free article] [PubMed] [Google Scholar]
- 7. Owusu Obeng, A. et al Physician‐reported benefits and barriers to clinical implementation of genomic medicine: a multi‐site IGNITE‐network survey. J. Pers. Med. 8, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frigon, M.P. et al Pharmacogenetic testing in primary care practice: opinions of physicians, pharmacists and patients. Pharmacogenomics 20, 589–598 (2019). [DOI] [PubMed] [Google Scholar]
- 9. Cavallari, L.H. et al Multi‐site investigation of strategies for the clinical implementation of CYP2D6 genotyping to guide drug prescribing. Genet. Med. 21, 2255–2263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunnenberger, H.M. et al Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Empey, P.E. et al Multisite investigation of strategies for the implementation of CYP2C19 genotype‐guided antiplatelet therapy. Clin. Pharmacol. Ther. 104, 664–674 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cicali, E.J. et al Challenges and lessons learned from clinical pharmacogenetic implementation of multiple gene‐drug pairs across ambulatory care settings. Genet. Med. 21, 2264–2274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith, D.M. et al CYP2D6‐guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet. Med. 21, 1842–1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cavallari, L.H. et al Multisite investigation of outcomes with implementation of CYP2C19 genotype‐guided antiplatelet therapy after percutaneous coronary intervention. JACC Cardiovasc. Interv. 11, 181–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weitzel, K.W. et al Implementation of standardized clinical processes for TPMT testing in a diverse multidisciplinary population: challenges and lessons learned. Clin. Transl. Sci. 11, 175–181 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haga, S.B. & Mills, R. A review of consent practices and perspectives for pharmacogenetic testing. Pharmacogenomics 17, 1595–1605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amstutz, U. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin. Pharmacol. Ther. 103, 210–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bell, G.C. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 102, 213–218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Birdwell, K.A. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin. Pharmacol. Ther. 98, 19–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Caudle, K.E. et al Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA‐B genotypes and phenytoin dosing. Clin. Pharmacol. Ther. 96, 542–548 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goetz, M.P. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and tamoxifen therapy. Clin. Pharmacol. Ther. 103, 770–777 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonsalves, S.G. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for the use of potent volatile anesthetic agents and succinylcholine in the context of RYR1 or CACNA1S genotypes. Clin. Pharmacol. Ther. 105, 1338–1344 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Johnson, J.A. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics‐guided warfarin dosing: 2017 update. Clin. Pharmacol. Ther. 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moriyama, B. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C19 and voriconazole therapy. Clin. Pharmacol. Ther. 102, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muir, A.J. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon‐alpha‐based regimens. Clin. Pharmacol. Ther. 95, 141–146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Relling, M.V. et al Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 93, 324–325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hicks, J.K. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramsey, L.B. et al The Clinical Pharmacogenetics Implementation Consortium guideline for SLCO1B1 and simvastatin‐induced myopathy: 2014 update. Clin. Pharmacol. Ther. 96, 423–428 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott, S.A. et al Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin. Pharmacol. Ther. 94, 317–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Desta, Z. et al Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2B6 and efavirenz‐containing antiretroviral therapy. Clin. Pharmacol. Ther. 106, 726–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gaedigk, A. , Dinh, J.C. , Jeong, H. , Prasad, B. & Leeder, J.S. Ten years' experience with the CYP2D6 activity score: a perspective on future investigations to improve clinical predictions for precision therapeutics. J. Pers. Med. 8, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Caudle, K.E. et al Standardizing CYP2D6 genotype to phenotype translation: consensus recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clin. Transl. Sci. 13, 116–124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luzum, J.A. et al The pharmacogenomics research network translational pharmacogenetics program: outcomes and metrics of pharmacogenetic implementations across diverse healthcare systems. Clin. Pharmacol. Ther. 102, 502–510 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kalia, S.S. et al Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2. 0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 19, 249–255 (2017). [DOI] [PubMed] [Google Scholar]
- 36. Bousman, C.A. & Dunlop, B.W. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic‐based decision support tools. Pharmacogenomics J. 18, 613–622 (2018). [DOI] [PubMed] [Google Scholar]
- 37. Van Driest, S.L. et al Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin. Pharmacol. Ther. 95, 423–431 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ji, Y. et al Preemptive pharmacogenomic testing for precision medicine: a comprehensive analysis of five actionable pharmacogenomic genes using next‐generation DNA sequencing and a customized CYP2D6 genotyping cascade. J. Mol. Diagn. 18, 438–445 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bush, W.S. et al Genetic variation among 82 pharmacogenes: the PGRNseq data from the eMERGE network. Clin. Pharmacol. Ther. 100, 160–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Samwald, M. et al Incidence of exposure of patients in the United States to multiple drugs for which pharmacogenomic guidelines are available. PLoS One 11, e0164972 (2016).27764192 [Google Scholar]
- 41. Drozda, K. , Pacanowski, M.A. , Grimstein, C. & Zineh, I. Pharmacogenetic labeling of FDA‐approved drugs: a regulatory retrospective. JACC Basic Transl. Sci. 3, 545–549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haga, S.B. & Kantor, A. Horizon scan of clinical laboratories offering pharmacogenetic testing. Health Aff. 37, 717–723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. US Food and Drug Administration . Letter to Inova Genomics Laboratory. FDA Warning <https://www.fda.gov/inspections-compliance-enforcement-and-criminal-investigations/warning-letters/inova-genomics-laboratory-577422-04042019?utm_campaign=040419_PR_FDA%2520issues%2520warning%2520letter%2520to%2520Inova%2520Genomics%2520Laboratory%26utm_medium=email%26utm_source=Eloqua> Accessed May 15, 2019.
- 44. Klein, T.E. & Relling, M.V. CYP2C19 testing may assist with prescribing sertraline and escitalopram. The PharmGKB Blog (2019).
- 45. Huddart, R. , Sangkuhl, K. , Whirl‐Carrillo, M. & Klein, T.E. Are randomized controlled trials necessary to establish the value of implementing pharmacogenomics in the clinic? Clin. Pharmacol. Ther. 106, 284–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee, M.A. , McGlinch, E.B. , McGlinch, M.C. & Capacchione, J.F. Malignant hyperthermia susceptibility and fitness for duty. Mil. Med. 182, e1854–e1857 (2017). [DOI] [PubMed] [Google Scholar]
- 47. Phillips, K.A. et al Payer coverage policies for multigene tests. Nat Biotechnol. 35, 614–617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Genes tested.
Table S2. CPIC guidelines and clinical actionability.
Table S3. FDA labeling and clinical actionability.
Table S4. FDA labeling and translation to actionability.
Table S5. Pharmacogenetic evidence for each drug‐gene pair.
Table S6. Secondary findings (SFs).
Table S7. Diplotype results.
Table S8. Actionable results per CPIC by medication.
Table S9. Actionable results per FDA by medication.


