Abstract
The hepatic influx transporter OATP1B1 (SLCO1B1) plays an important role in the disposition of endogenous substrates and drugs prescribed to children. Alternative splicing increases the diversity of protein products from > 90% of human genes and may be triggered by developmental signals. As concentrations of several endogenous OATP1B1 substrates change during growth and development, with this exploratory study we investigated age‐dependent alternative splicing of SLCO1B1 mRNA in 97 postmortem livers (fetus‐adolescents). Twenty‐seven splice variants were detected; 10 were confirmed by additional bioinformatic analyses and verified by quantitative polymerase chain reaction, and selected for detailed analysis based on relative abundance, association with age, and overlap with an adjacent gene. Two splice variants code for reference OATP1B1 protein, and eight code for truncated proteins. The expression of eight isoforms was associated with age. We conclude that alternative splicing of SLCO1B1 occurs frequently in children; although the functional consequences remain unknown, the data raise the possibility of a regulatory role for alternative splicing in mediating developmental changes in drug disposition.
Transporters are membrane‐bound proteins that are present in the apical and basolateral membranes of organs, such as the liver.1 Their biological role is the trafficking of substrates across membranes, making them critical determinants of tissue and cellular substrate disposition. Moreover, they act in concert with drug‐metabolizing enzymes (DMEs) to maintain homeostatic balance for endogenous substrates and to facilitate the detoxification and elimination of exogenous substrates, such as drugs and food toxins.2
This latter is important for newborns, as after birth they become dependent on exogenous food sources for nutrition, and the diet expands as they grow into infanthood. During all these changes in food exposure, the child must defend itself against potentially toxic dietary constituents, recruiting pathways not or differentially expressed during fetal life. Hence, ontogeny of DMEs and transporters occurs, influencing the disposition of their endogenous and exogenous substrates over age. Moreover, ontogeny may well be driven by developmental homeostatic changes in endogenous substrates.3
A classic example of age‐related changes in a DME that plays an important physiological role is hepatic cytochrome P450 (CYP)3A7. CYP3A7 is highly expressed in fetal liver but steadily declines throughout the last trimester of pregnancy and the first year of postnatal life to low levels characteristic of an adult liver.4, 5 From a functional perspective, CYP3A7 catalyzes the 16α‐hydroxylation of dehydroepiandosterone 3‐sulfate (DHEA‐S), to form 16α‐DHEA‐S, a precursor for estriol synthesis by placental syncytiotrophoblasts.6 DHEA‐S concentrations are high during the fetal and neonatal periods and decline postnatally.7, 8 DHEA‐S has been reported to activate CYP3A7 activity, which explains the high expression of CYP3A7 in the fetal liver.9 DHEA‐S also provides an example of the interplay between DMEs and transporters in a developmental context, as prior to biotransformation by CYP3A7 in fetal liver DHEA‐S needs to cross the hepatocyte membrane, using the solute carrier organic anion transporter (gene name SLCO1B1, protein name OATP1B1) located on the basolateral membrane.10 Consistent with CYP3A7, the OATP1B1 expression also has been demonstrated to decline directly after birth,11 followed by age‐dependent increases in mRNA expression throughout childhood.12 Data are conflicting regarding developmental patterns of OATP1B1 protein, with expression increased around age 8 years, compared with younger children in one study, using immunoblotting techniques,13 and no apparent statistically significant relationship with age, using a quantitative proteomic approach.14
Whereas the contribution of CYP3A7 to drug clearance postnatally is relatively minor, OATP1B1 plays an important role in the clearance of potentially toxic endogenous molecules. One example illustrating the importance of transporter function early after birth involves bilirubin; an association has been demonstrated between the SLCO1B1 388G>A allele, a variant associated with reduced function of the transporter, and unconjugated hyperbilirubinemia in newborns, which is associated with neurotoxicity.15, 16 Moreover, OATP1B1 is not only involved in the disposition of endogenous substrates but also of drugs that are used in pediatrics, such as statins, methotrexate and bosentan.10 Malfunctioning of the OATP1B1 transporter may put children at risk of toxic or subtherapeutic effects of these drugs. Thus, understanding the regulatory mechanisms of the gene SLCO1B1 in response to developmental signals is critical to understand physiological changes in endogenous substrates and to provide safe and effective drug therapy in children.
To date, ontogeny studies have generally focused on mRNA expression and, more recently, have expanded to protein abundance targeting specific regions of the reference gene and/or protein sequence. Recently, it has become increasingly apparent that alternative splicing, a process that increases the diversity of products from a single gene, may have functional consequences. Due to alternative splicing events, > 90% of our genes give rise to more than one mRNA transcript, varying with respect to numbers of exons, different length of exons, and varying lengths of untranslated regions.17 Not all products of alternative splicing necessarily result in the production of functional proteins. Alternative splicing may be the result of developmental signals expressed during the course of growth and development.17, 18, 19 For example, developmentally regulated alternative splicing has been demonstrated for neuronal sodium channels genes SCN1A, SCN2A, SCN3A, SCN8A, and SCN9A in brain tissue; the alternative exon 5N predominates in the neonatal period whereas 5A predominates in the adult.20, 21, 22, 23, 24, 25 In the case of SCN1A, a gene implicated in the pathogenesis of febrile seizures in newborns,26 an allelic variant SCN1A IVS5–91 G>A disrupts the 5' splice donor site of exon 5N and potentially influences the relative expression of exons 5N and 5A. Although the SCN1A IVS5–91 G>A variant does not seem to be associated with febrile seizures per se,27 studies suggest that presence of the variant allele may affect dose requirements for phenytoin and carbamazepine.25, 28 Thus, alternative splicing and genetic variants affecting alternative splicing may have therapeutic consequences.
The SLCO1B1 gene, consisting of 14 coding and one non‐coding exons, codes for the protein OATP1B1 that is composed of 691 amino acids, and consists of 12 transmembrane (TM) regions.10 It is part of the SLCO1B family, for which splice variants have been described. For example, five mRNA transcripts for SLCO1B3 have been deposited in Ensembl, of which four represent full‐length or truncated protein‐coding sequences.29 Furthermore, the splice variant LST‐3TM12 is a hybrid transcript with sequences derived from SLCO1B3 and SLCO1B7, and has functional transporter properties.30 In contrast, for SLCO1B1, there is only one reported mRNA transcript, ENST00000256958.2, encoding the functional 691 amino acid OATP1B1 protein; referred to hereafter as the “reference isoform.”10
Given these considerations, the purpose of this exploratory study was to investigate alternative splicing of SLCO1B1 in postmortem pediatric liver tissue over a wide age range from fetal to adolescent ages. Using RNA sequencing (RNA‐seq) data, we created a process involving computational software integrating our bioinformatics pipelines and an in‐house developed RNA‐seq database query system to perform a structured analysis of the RNA‐seq data in silico. Using this data analysis pipeline, we aimed (1) to predict splice variants for SLCO1B1, (2) to identify potential hybrid splice variants overlapping SLCO1B1 and SCLO1B7 (another member of the SLCO1B family), and (3) to study age‐related changes in expression of SLCO1B1 splice variants.
MATERIALS AND METHODS
See Figure 1 for the workflow of the methods, and the explanation underneath.
Figure 1.
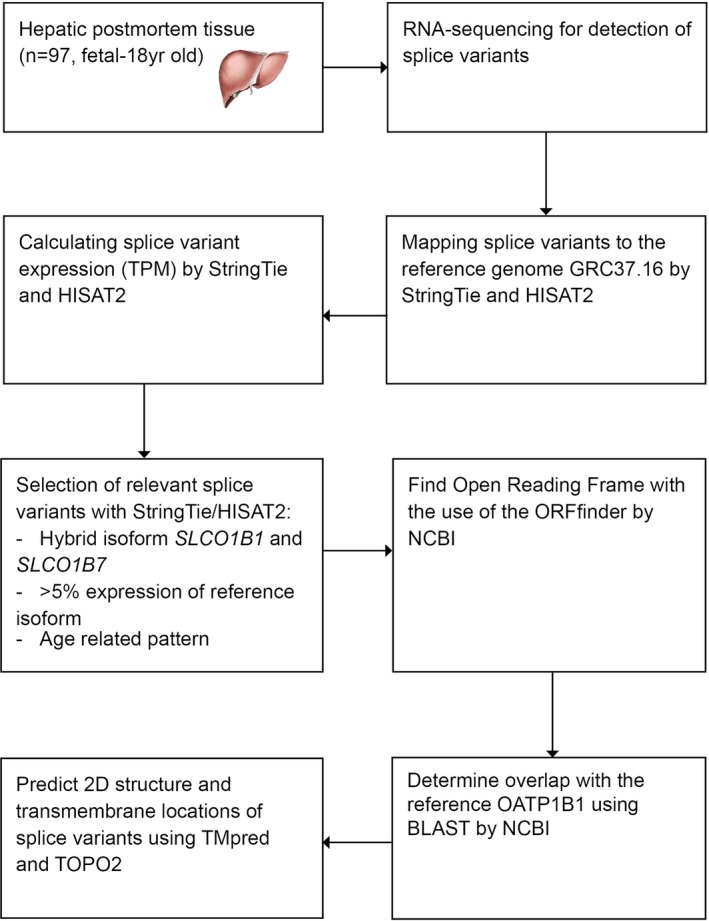
Flow of methods predicting splice variants of SLCO1B1. NCBI, National Center for Biotechnology Information; ORF, open reading frame; TM, transmembrane; TPM, transcripts per million.
Tissue samples
Postmortem liver tissue samples from autopsies of fetuses (therapeutic abortions or stillbirths) and infants were provided by the Erasmus MC Tissue Bank (Rotterdam, The Netherlands) and the repository of the Division of Clinical Pharmacology, Toxicology, and Therapeutic Innovation at Children’s Mercy Kansas City (Kansas City, MO). Tissue was procured at the time of autopsy within 24 hours after death and snap‐frozen at −80°C for later research use. For the tissues provided by the Erasmus MC Tissue Bank, the Erasmus MC Research Ethics Board waived the need for formal ethics approval according to the Dutch Law on Medical Research in Humans. Tissue was collected when parental written informed consent for both autopsy and the explicit use of the tissue for research was present. The samples were selected when the clinical diagnosis of the patient was not related to hepatic problems and the tissue was histologically normal (as estimated by a pathologist based on hematoxylin and eosin staining). Postmortem pediatric liver tissues in the repository of the Division of Clinical Pharmacology, Toxicology, and Therapeutic Innovation at Children’s Mercy Kansas City (Kansas City, MO) were obtained from multiple sources, including the Brain and Tissue Bank for Developmental Disorders at the University of Maryland (Baltimore, MD), the Liver Tissue Cell Distribution System (University of Pittsburgh, and University of Minnesota), University of Washington Center for Birth Defects Research (Seattle, WA), and XenoTech LLC (Lenexa, KS). The use of these tissues was declared nonhuman subject research by the Children's Mercy Hospital Pediatric Institutional Review Board.
RNA‐seq
The mRNA expression of SLCO1B1 was determined using RNA‐seq. RNA was isolated from hepatic tissue according to the manufacturer’s instructions using the RNeasy Mini kit (Qiagen, Valencia, CA). Samples with an RNA integrity number of < 5 were excluded. The RNA‐seq experiments were performed according to the Illumina RNA‐seq protocol (San Diego, CA). In brief, a population of poly(A)+ mRNA was selected and converted to a library of cDNA fragments (220–450 bp) with adaptors attached to both ends, using an Illumina mRNA‐Seq sample preparation kit. The quality of the library preparation was confirmed by analysis on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cDNA fragments were then sequenced on an Illumina HiSeq 2000 to obtain 100‐bp sequences from both ends (paired end). The resulting reads were mapped by Bowtie2 and StringTie31, 32, 33 to the transcriptome constructed through reference genes/transcripts, according to the reference human genome GRCh37.61/hg19.34 The mapped reads were then assigned to transcripts from which the abundance of the reference transcript is estimated by RSEM35 and for the splice variants with HISAT2.36 The counts of RNA‐seq fragments were used to indicate the amount of identified mRNA transcripts, presented in transcripts per million (TPM).35
Validation data set
To validate the RNA‐seq results, the presence of the reference transcript was confirmed (Ensembl transcript ID ENST00000256958.3). Moreover, to further validate the results, the presence of the alternatively spliced transcript LST‐3TM1230 was detected using the algorithm RSEM combined with Bowtie2 and the assembly GRCh37.
Protein prediction
Sequence alignment and overlap of splice variants with consensus coding sequences37 for SLCO1B1 and the adjacent gene SLCO1B7 was performed using Basic Local Alignment Search Tool (BLAST).38 Splice variants were prioritized for further investigation when one of the following criteria was met and when the presence of the splice variant was verified with real‐time polymerase chain reaction (RT‐PCR; see section Verification of splice variants by RT‐PCR and sequencing):
The expression of the splice variant was > 5% of the expression of the reference isoform,
The splice variant was a SLCO1B7 and SLCO1B1 hybrid transcript, or
The expression of the splice variant was associated with age (see section Data and statistical analysis for statistical analysis).
Next, the open reading frame (ORF) of > 600 nucleotides (nts) of the relevant splice variants was predicted with ORF‐Finder by the National Center for Biotechnology Information.39 Prediction of TM regions and orientation was done with TMpred based on the TMbase database.40 Two‐dimensional protein structures were generated with TOPO2.41
To provide additional bioinformatic confirmation that candidate novel alternatively spliced products represent coding transcripts, sequencing data were analyzed using two additional tools: the Coding Potential Calculator Algorithm (CPC2) and the Coding‐Potential Assessment Tool (CPAT).42, 43 These algorithms both use logistic regression to distinguish between coding and noncoding transcripts based on four intrinsic features: the Fickett testcode score (both), ORF length (both), ORF integrity (CPC2), isoelectric point (CPC2), ORF coverage defined as the ratio of ORF to transcript lengths (CPAT), and hexamer usage bias (CPAT).
Verification of splice variants by RT‐PCR and sequencing
The presence of the splice variants selected for further investigation was verified by RT‐PCR and sequencing. Primers were designed to be specific for each splice variant (Figure S1 and Table S1 ). In addition, a universal primer pair was designed to amplify all splice variants and to function as a positive control. Due to the low abundance of some of the variants, a nested forward primer was also designed.
RNA was extracted from frozen liver tissue, utilizing the Qiagen RNeasy Mini Kit (Qiagen, Valencia, CA). One µg of total RNA was DNase treated and reverse transcribed with the Maxima H Minus First Strand cDNA Synthesis Kit, following the manufacturer’s instructions (Thermo Scientific, Waltham, MA). The cDNA equivalent to 10 ng of total RNA was used per polymerase chain reaction (PCR; 2G Fast ReadyMix, KAPA Biosystems, Wilmington, MA). The cycling conditions were: 94°C, 3 minutes, followed by 40 cycles of 94°C for 15 seconds, 60°C for 15 seconds, and 72°C for 5 seconds. The primary PCR amplicons were diluted 1:4,000 and a nested PCR was performed with the same KAPA mix and the same cycling conditions. The PCRs were column purified up with the QIAquick PCR Purification Kit (Qiagen). One ng was used in subsequent sequencing reactions with BigDye version 3.1 and run on a 3,730xl DNA Analyzer (Thermo Scientific). The results were analyzed with Sequencher software (Gene Codes, Ann Arbor, MI).
Data and statistical analysis
Because of non‐normal distribution, the data are presented as median (range). First, the relative abundance of the expression of each transcript compared with the reference isoform was calculated. In addition, the relationship of age with expression levels (TPM) was studied by comparing expression levels between age groups. Samples were assigned to one of five age groups: fetal, 0–1.5, 1.5–6, 6–12, and 12–18 years. KruskalWallis test with Dunn's post hoc test were used for multiple comparisons of expression levels between age groups. For Dunn’s post hoc test for multiple comparisons, the adjusted P values are reported, in which a correction for multiple testing for age groups is applied. Second, Spearman correlations between age (on a continuous scale) and expression levels of splice variants were examined. To control for the number of correlations tested (54), P values were considered statistically significant only if their corresponding q value was < .05 after BenjaminiHochberg adjustment to control the false discovery rate. Statistical analyses were performed using IBM SPSS Statistics software (SPSS Statistics for Windows, version 21.0; IBM, Armonk, NY) and graphical exploration was done with GraphPad Prism. We explored negative binomial and zero‐inflated negative binomial models in SAS 9.4, but the former did not fit well, and we were unable to identify predictors of excess zeros for the latter.
RESULTS
Descriptive results
The mRNA expression of the reference isoform and splice variants of SLCO1B1 was quantified in 97 postmortem liver tissues of humans of various ages, of which the age distribution can be found in Table 1. The reference isoform of SLCO1B1 was detected in all but one sample with a median expression of 33.4 (range 0–134.2) TPM. The transcript consisted of 2,791 nts of which 95 nts comprise the 5'‐UTR and 621 nts the '3‐UTR, resulting in a protein of 691 amino acids. This is in accordance with Niemi et al.10 In Figure 2 a the expression of this transcript in various age groups is presented and did not show any age‐related changes when binned in age groups. On the other hand, on a continuous scale, postnatal age was related to transcript expression (ρ = 0.316, P = 0.002; Figure 2 d). Twenty‐seven splice variants of SLCO1B1 were identified using RNA‐seq, representing a total expression of 18.8 (0.2–105.0) TPM (Figure 2 b). These are numbered randomly between 21 and 55. The ratio of the total expression of the splice variants vs. the reference isoform is presented in Figure 2 c and was not different between age groups. Thirteen splice variants met the selection criteria for further analysis, and 10 of these were subsequently verified by RT‐PCR (Table 2), as described below.
Table 1.
Median (range) age by group for postmortem liver samples
| Age groups | Number of samples | Gestational age (weeks) | Postnatal age (years) |
|---|---|---|---|
| Fetus | 22 | 16.4 (14.7–41.3) | – |
| 0–1.5 years | 35 | – | 0.1 (0–1.2) |
| 1.5–6 years | 16 | – | 3 (1.8–6) |
| 6–12 years | 15 | – | 9 (7–12) |
| 12–18 years | 9 | – | 15 (13–17) |
| Total | 97 |
Figure 2.
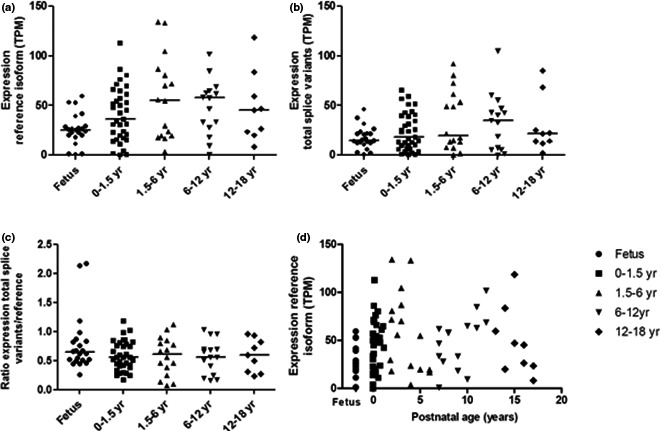
Transcripts per million (TPM) expression of (a) the reference isoform of SLCO1B1 (b) the total TPM values of splice variants and (c) the ratio of total TPM values of splice variants to TPM values of the reference isoform in various age groups; and (d) the relationship of the reference isoform of SLCO1B1 with postnatal age (ρ = 0.316, P = 0.002).
Table 2.
Relevant splice variants of SLCO1B1 in 97 pediatric liver samples for which the presence in the samples is confirmed by RT‐PCR
| Splice variant | Abundance of all novel isoforms (%) | Abundance compared with reference isoform (%) | Found in number of samples | Number of exons | Length (nt) | ORF (n AA) | Overlapping number of AA with locus: | Number of TM helices | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF SLCO1B1 (% of reference SLCO1B1) | Intron SLCO1B1 | ORF SLCO1B7 | In between SLCO1B1 and SLCO1A2 a , b | SLCO‐1A2 a , b | ||||||||
| 46 a , b | 26.55 | 16.48 | 63 | 10 | 4,638 | 284 | 274 (40%) | 10 | – | – | – | 6 |
| 50 a , b | 14.26 | 8.85 | 93 | 17 | 34,388 | 453 | 444 (65%) | 9 | – | – | – | 10 |
| 34 a , b | 9.24 | 5.73 | 46 | 18 | 4,156 | 625 | 622 (90%) | – | – | 3 | – | 11 |
| 24 a , b | 0.32 | 0.20 | 77 | 25 | 3,151 | 659 | 622 (90%) | – | 0 | – | 37 | 11 |
| 26 a , b | 1.80 | 1.12 | 81 | 24 | 13,158 | 691 | 691 (100%) | – | 0 | – | – | 12 |
| 28 a , b | 1.21 | 0.75 | 96 | 20 | 14,577 | 691 | 691 (100%) | – | 0 | – | – | 12 |
| 210 | – | – | – | – | 210 | 0 | ||||||
| 881 | – | – | – | – | 881 | 1 | ||||||
| 30 a , b | 0.34 | 0.21 | 54 | 20 | 5,684 | 484 | 453 (66%) | – | 0 | – | 31 | 8 |
| 38 a , b | 0.52 | 0.33 | 95 | 15 | 22,310 | 453 | 444 (65%) | 9 | – | – | – | 10 |
| 44 a , b | 7.86 | 4.88 | 65 | 3 | 7,364 | 98 | 98 (14%) | – | – | – | – | 2 |
| 51 a , b | 0.73 | 0.46 | 76 | 17 | 15,407 | 522 | 522 (75%) | – | – | – | – | 8 |
AA, amino acid; nt, nucleotides; ORF, open reading frame; RT‐PCR, real‐time polymerase chain reaction; TM, transmembrane.
SLCO1A2 is on the reverse strand. bAbundant splice variant. cHybrid SLCO1B1 and ‐1B7. dAge‐related changes in expression.
Verification of splice variants by RT‐PCR and sequencing
The presence of the splice variants meeting one or more criteria of (i) expression level > 5% of the expression of the reference isoform, (ii) a hybrid SLCO1B7 and SLCO1B1 transcript, or (iii) expression was associated with age, were verified for 10 of 13 samples; splice variants 21, 37, and 48 could not be verified by RT‐PCR (see Figure S2 ). Splice variants 21 and 37 had sizes different than expected. Splice variant 48 could not be amplified. Further analysis of variants 21, 37, and 48 by sequencing was also unable to confirm the presence of the 21, 37, and 48 splice variants, and, thus, these transcripts were excluded from further analysis. The splice junctions of variant 46 were not sufficiently unique to design primers that would amplify only this variant or to generate a product with a size that could be resolved on an agarose gel from amplicons generated from other transcripts as templates. The results for splice variant 46 should, therefore, be interpreted with caution.
Validation data set
To further validate our RNA‐seq results, we confirmed the presence of the LST‐3TM12 transcript30 in our samples. The transcript emerged in 3 of the 97 samples with a low abundance of 0.11, 0.18, and 0.30 TPM.
Splice variants meeting the abundance criterion
Three splice variants had an abundance of > 5% of the expression of the reference isoform (Table 2). They had 40–90% overlap with the ORF from the reference amino acid sequence for OATP1B1, resulting in a prediction of a number of TM helices ranging from 6 to 11. These three splice variants are, therefore, predicted to result in truncated versions of OATP1B1.
Splice variants overlapping with SLCO1B7 and SLCO1B1
Four splice variants were identified with exons overlapping the SLCO1B1 as well as the SLCO1B7 gene region (Table 2). They all had an ORF overlapping > 40% of the amino acid sequence of OATP1B1. However, the ORF of none of them was overlapping the SLCO1B7 region. Two of these isoforms are predicted to translate into similar protein versions of OATP1B1, as they have 100% overlap with the reference isoform. All four splice variants have longer untranslated regions than the reference isoform.
One isoform, sv28, overlapped with SLCO1A2. This hybrid isoform contains an intron‐less complete coding sequence, which is officially located in intron 13 of SLCO1A2.
Splice variants with age‐related expression
Age groups
The splice variants 26 and 46 showed age‐related changes in their expression with lower expression in fetal tissue than in tissue from older children (see Figure 3 a for specific changes and Table 2 for splice variant information). When analyzed as a ratio to the expression of the reference isoform, for one splice variant (26) the ratio variant/reference isoform increased with age, whereas three splice variants decreased with age: isoforms 24, 28, and 44 (see Figure 3 b for specific changes and Table 2 for splice variant information). This latter observation reflects that for the individual samples either the expression of the splice variant was lower, or the expression of reference isoform was higher. As the expression of the reference isoform was shown to be similar when binned in age groups (Figure 3 a), it is, therefore, likely that the expression of the splice variants that decreased with age was lower.
Figure 3.
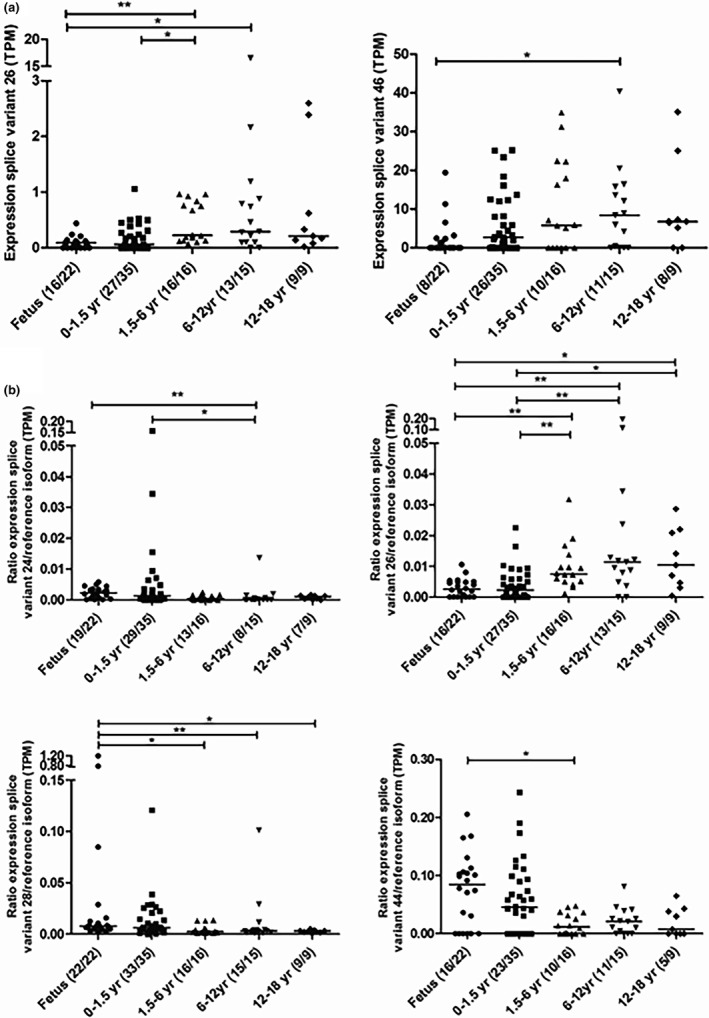
Expression (a) and expression in relation to the reference isoform (b) of developmentally regulated splice variants of SLCO1B1 in various age groups. Counts of tissues with isoform expressed out of total counts by age group are provided in parentheses. *P < 0.05; **P < 0.01. TPM, transcripts per million.
Age on continuous scale
The expression of four of the five hybrid SLCO1B7 and SLCO1B1 splice variants (24, 26, 28, and 30) and the abundant splice variant 46 are significantly correlated with age (see Table 3). More specifically, the expression of the splice variants 24 and 28 decreased with increasing age, and the expression of 26, 30, and 46 increased with increasing age. When splice variant expression is analyzed as a ratio to the expression of the reference isoform, correlation with age was found for the same and for four additional splice variants (see Table 3). The expression of splice variant 48 was correlated with age, but was excluded for further analysis as their presence was not verified by RT‐PCR (see section Verification splice variants by RT‐PCR and sequencing).
Table 3.
Spearman correlations expression splice variant vs. postnatal age
| Splice variant | Expression splice variant (TPM) vs. postnatal age (weeks) | Ratio expression splice variant/reference isoform vs. postnatal age (weeks) | ||
|---|---|---|---|---|
| rs | P value | rs | P value | |
| 21 | 0.231 | 0.023 | 0.194 | 0.057 |
| 24 | −0.330 | 0.001 a , b | –0.392 | 0.000 a , b |
| 26 | 0.489 | 0.000 a , b | 0.518 | 0.000 a , b |
| 28 | –0.263 | 0.009 a , b | –0.433 | 0.000 a , b |
| 30 | 0.290 | 0.004 a , b | 0.296 | 0.003 a , b |
| 33 | 0.142 | 0.166 | 0.143 | 0.163 |
| 34 | 0.133 | 0.193 | 0.110 | 0.285 |
| 35 | –0.035 | 0.734 | –0.222 | 0.029 |
| 36 | 0.063 | 0.539 | 0.058 | 0.575 |
| 37 | 0.069 | 0.501 | 0.006 | 0.955 |
| 38 | –0.070 | 0.496 | –0.271 | 0.007 a , b |
| 39 | –0.141 | 0.167 | –0.188 | 0.065 |
| 40 | –0.017 | 0.869 | –0.092 | 0.371 |
| 41 | 0.003 | 0.977 | –0.001 | 0.990 |
| 42 | 0.055 | 0.591 | –0.059 | 0.566 |
| 43 | 0.065 | 0.528 | 0.063 | 0.542 |
| 44 | –0.121 | 0.240 | –0.259 | 0.010 a , b |
| 45 | –0.017 | 0.867 | –0.020 | 0.843 |
| 46 | 0.386 | 0.000 a , b | 0.343 | 0.001 a , b |
| 47 | –0.023 | 0.825 | –0.106 | 0.299 |
| 48a | –0.243 | 0.016 | –0.463 | 0.000 a , b |
| 49 | –0.117 | 0.253 | –0.146 | 0.152 |
| 50 | 0.204 | 0.045 | –0.202 | 0.047 |
| 51 | –0.177 | 0.083 | –0.308 | 0.002 a , b |
| 53 | –0.029 | 0.779 | –0.043 | 0.678 |
| 54 | –0.155 | 0.129 | –0.169 | 0.097 |
| 55 | –0.173 | 0.091 | –0.203 | 0.046 |
TPM, transcripts per million.
Excluded from further analysis because the presence was not confirmed by real‐time polymerase chain reaction (RT‐PCR; see section Verification splice variants by RT‐PCR and sequencing).
Bold and * indicate significant after adjustment to control false discovery rate at 0.05.
Predicted protein structure
In Table S3 , the coding‐potential prediction results using the CPAT and CPC2 tool are depicted. Splice variant 44 has a low coding probability, hence will likely not result in a protein product. This can be explained by the fact that the ORF length was small compared with the size of the splice variant or because of a high hexamer‐score.42, 43 The hexamer‐score is a feature dependent on adjacent amino acids in proteins and is based on a log‐likelihood ratio to measure differential hexamer usage between coding and noncoding sequences.44 All other splice variants have a very high probability to be translated into a protein product. In Figure 4 a, the 2D structure of OATP1B1 is presented, consisting of 12 TM helices. In Figure 4 b, the predicted 2D structure of the splice variants with an ORF with high probability to be translated in a protein product are presented.
Figure 4.
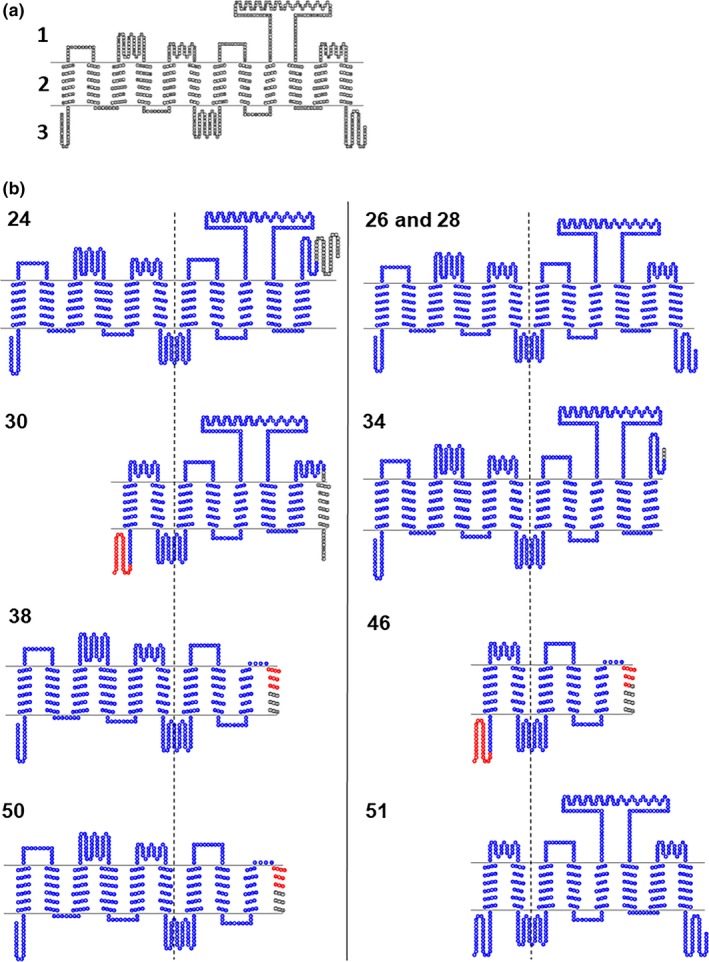
(a) Predicted 2D structure of reference OATP1B1 (1: extracellular, 2: transmembrane, 3: intracellular) and (b) the predicted 2D structure of splice variants of OATP1B1, centered on the fourth intracellular loop (dashed line) of the reference structure for OATP1B1. The number of the splice variant is presented in the upper left corner of each structure. Red and blue: overlapping amino acid sequence with OATP1B1. Blue: overlapping structure OATP1B1.
DISCUSSION
In the current study, we have developed a data analysis pipeline that allowed a structured analysis of a large amount of RNA‐seq data generated from pediatric liver samples and used this to investigate alternative splicing of the SLCO1B1 gene that could potentially translate into functional OATP1B1 proteins. More specifically, we report three major findings from the 10 relevant splice variants that we identified: (i) 2 splice variants are predicted to translate into the same amino acid sequence as the reference isoform for OATP1B1; (ii) 8‐splice variants may translate into truncated versions of the OATP1B1 protein because of an altered length of amino acid sequence, and (iii) the expression of 8 splice variants was associated with age. None of the splice variants had an ORF that covered the SLCO1B7 region.
Our results show that the SLCO1B1 gene locus is subject to alternative splicing, as supported by the major findings presented above. More specifically, the fact that eight splice variants of SLCO1B1 showed a developmental pattern is consistent with developmentally regulated alternative splicing as a mechanism for altered SLCO1B1/OATP1B1 expression during growth and maturation. This finding may have implications for the functionality of the transporter in children, and with that the disposition of its endogenous and exogenous substrates, as most of these splice variants were predicted to result in truncated OATP1B1 isoforms with fewer TM regions compared with the reference OATP1B1 protein. Available evidence suggests that SLCO1B7 is a pseudogene resulting in a nonfunctional protein product with only 11 TM regions, whereas the SLCO1B1 gene, the SLCO1B3 gene, and the hybrid transcript LST‐3TM12 give rise to at least one mRNA transcript that translates into functional transporters with 12 TM regions.30 Moreover, the truncated proteins encoded by the transcripts we observed may lack one or more of the N‐glycosylation sites Asn134, Asn503, and Asn516, located at the extracellular loops 2 and 5 of OATP1B1.45 Glycosylation is a post‐translational modification that is suggested to be essential for the proper function of OATP1B1. Disruption of all these sites led to lower protein stability with reduced total protein levels, and nonglycosylated OATP1B1 was retained within the endoplasmic reticulum (e.g., it was not present on the cell membrane).45
Some of the alternative proteins of OATP1B1 reported in this study, therefore, are likely to result in nonfunctional protein products incapable of cellular transport, but could possibly possess alternative functional properties, such as regulating the activity of the functional OATP1B1 transporter protein. A precedent for this type of regulatory role is illustrated by the DME UGT1A, a complex gene with three major mRNA variants created by splicing events involving an alternative 5a or 5b exon. The classic variant (i1) with exon 5a has transferase activity, whereas the alternative proteins (i2), with either exon 5b or with both exon 5a and 5b, lack transferase activity.46 The relative glucuronidation of SN‐38, a substrate for UGT1A, was decreased in the presence of i2 proteins despite the same amount of i1 enzyme.47 This phenomenon is explained by oligomerization of UGT1A enzyme; i2 proteins can form dimers with i1 enzymes, inhibiting the activity of i1 enzymes.48 Interestingly, OATP1B transporters not only form homo‐oligomers, but can also form hetero‐oligomers, even with transporters from another family (e.g., with Na+/taurocholate co‐transporting polypeptide (NTCP)).49, 50 It has been demonstrated that a nonfunctional unit of OATP1B3, containing a lysine at position 41 in place of the wild‐type cysteine in the homodimer, did not affect normal substrate transport by the functional, cysteine‐containing OATP1B3 component of the homodimer, suggesting that each unit within the dimer works as an independent functional unit.49 However, each splice variant may have its own function and so we hypothesize that those resulting in truncated proteins may influence the transporter activity of the reference protein OATP1B1 and other transporters.
The specific SLCO1B1 region is part of the wider SLCO1B‐family region, which gives rise to the SLCO1B3/SLCO1B7 hybrid transcript LST‐3TM12 that result in a functional transporter.30 The four SLCO1B1 splice variants found in this study that contained exons covering the region of SLCO1B7 did not contain the start codon from the SLCO1B7 locus, thus we conclude that the SLCO1B1 gene is not subject to hybridization with adjacent genes. However, the length of the untranslated region (UTR) of these and other transcripts could well be influencing the function of the transporter, even when the ORF of the splice variant is the same as the reference sequence. We note that a transcript of CYP3A4 with a shorter 3'‐UTR than the canonical transcript due to an additional polyadenylation site was more stable and generated more protein51 than an alternative transcript with a longer 3'‐UTR. Interestingly, this shorter transcript showed developmental regulation as it was higher expressed in adult livers than in pediatric livers. Nevertheless, it remains to be seen whether this is also the case for the splice variants presented in this study.
Another consequence of these truncated versions of OATP1B1 is that they may interfere with the estimation of OATP1B1 content using liquid chromatography‐tandem mass spectrometry‐based proteomic methods. This quantitative proteomic approach utilizes short peptides to target the protein of interest. All truncated versions of OATP1B1 presented in this paper contained the amino acid sequences NVTGFFQSFK14 and of LNTVGIAK11 that have been used in studies presenting results on expression of OATP1B1 protein in pediatric liver tissue. The latter refers to our previous study, where a poor correlation was seen between total mRNA expression of SLCO1B1 as measured with RNA‐seq and protein abundance of OATP1B1 in a subset of the samples presented in the current study.11 This lack of correlation can be explained by the fact that not all mRNA transcripts translate into protein. Moreover, potential translation of splice variants into nonfunctional proteins that are nevertheless detected by the peptide sequences used to quantitate OATP1B1 content could also result in poor correlations between abundance and activity. Thus, care may be needed when extrapolating mRNA expression to protein abundance, protein abundance to actual activity, and ultimately the prediction of disposition of transporter substrates.
We recognize that a limitation of our study is that our results are based on in silico predictions, and the presence of the corresponding truncated proteins must be confirmed by protein abundance studies before any of the implications we propose above can be assessed, including investigations of a dominant‐negative regulatory role analogous to the UGT1A situation. Developmental regulation of alternative splicing is a commonly recognized phenomenon during tissue development and cell differentiation. In fact, level of expression, localization within the cell, mRNA stability, translation efficiency, and splicing of specific RNA binding proteins (RBPs) is finely regulated. RBPs bind to cis‐elements and promote or inhibit splice site recognition, and, therefore, RBP expression coordinates alternative splicing networks during development.18 Further work is necessary to characterize the specific developmental signals responsible for the observed changes in expression of the SLCO1B1 splice variants.
These exploratory data imply that the complexity of processes involved with growth and development throughout childhood may have influences on transporter expression and subsequent substrate disposition, as yet unrecognized. The observed age‐related changes in expression of splice variants in the context of age‐related changes in concentrations of endogenous OATP1B1 substrates, such as DHEA‐S and 16alpha‐hydroxylated metabolites, makes it tempting to speculate that additional regulatory mechanisms may be in play, with implications for the disposition of exogenous substrates used in pediatrics. Most importantly, the data analysis pipeline we have developed allows the analyses described in this paper for SLCO1B1 to be applied to any other gene of interest and will be repeated for other transporters and genes involved in drug disposition or growth of children in the future.
In conclusion, we have shown that SLCO1B1 splice variants with an ORF could potentially translate into proteins with unknown function; they are unlikely to code for functional transporters but may have other roles, such as regulatory activity. Moreover, as the expression of particular SLCO1B1 splice variants showed age‐related changes, the data raise the possibility of a regulatory role for alternative splicing in mediating developmental changes of SLCO1B1/OATP1B1 in drug disposition. These data can contribute to improved understanding of age‐related changes in expression of SLCO1B1, and possibly other enzymes and transporters involved in the disposition of endogenous and exogenous substrates throughout growth and development.
Funding
B.G. was supported, in part, by the Ter Meulen Grant 2018 of the Royal Netherlands Academy of Arts and Sciences. Bioinformatic and statistical support was provided by the Administrative Core of grant U54 HD090258 (C.B., V.S., and J.S.L.). The National Institute of Child Health and Human Development Brain and Tissue Bank for Developmental Disorders at the University of Maryland is funded by the National Institutes of Health (NIH) contract HHSN275200900011C, reference number, N01‐HD‐9‐0011 and the Liver Tissue Cell Distribution System is funded by NIH contract number N01‐DK‐7‐0004/HHSN267200700004C.
Conflict of Interest
All other authors declared no competing interests for this work.
Author Contributions
B.G., C.B., R.G., V.S., D.T., S.W., and J.L. wrote the manuscript. B.G., C.B., R.G., V.S., D.T., S.W., and J.L. designed the research. B.G., C.B., V.S., R.G., and J.L. performed the research. B.G., C.B., R.G., V.S., and J.L. analyzed the data.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ The hepatic influx transporter OATP1B1 (SLCO1B1) transports substrates that include drugs that are prescribed to children, and endogenous substrates that are involved in growth and development. Alternative splicing may be triggered by developmental signals; no data regarding alternative splicing of SLCO1B1 are available.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ Given that concentrations of several endogenous OATP1B1 substrates change during growth and development, this study addressed the question of whether SLCO1B1 undergoes alternative splicing and the potential relationship with age.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ Alternative splicing of SLCO1B1 occurred commonly in pediatric liver tissue, and the expression of several splice variants was associated with postnatal age. Most alternative transcripts were predicted to code for truncated forms that may lack transporter activity.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ Interpretation of these data raises the possibility of a regulatory role for alternative splicing in mediating developmental changes in drug disposition pathways during growth and maturation and can stimulate further research to better understand age‐related changes in the expression of OATP1B1.
Supporting information
Figures S1–S2. Tables S1–S3
Acknowledgments
We thank the Department of Pathology at Erasmus MC for sample collection for the Erasmus MC Tissue Biobank, and Prof. G.M.M. Groothuis and Prof. P. Artursson for the adult samples.
References
- 1. Brouwer, K.L. et al Human ontogeny of drug transporters: review and recommendations of the pediatric transporter working group. Clin. Pharmacol. Ther. 98, 266–287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nigam, S.K. What do drug transporters really do? Nat. Rev. Drug Discov. 14, 29–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mooij, M.G. et al Development of human membrane transporters: drug disposition and pharmacogenetics. Clin. Pharmacokinet. 55, 507–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stevens, J.C. et al Developmental expression of the major human hepatic CYP3A enzymes. J. Pharmacol. Exp. Ther. 307, 573–582 (2003). [DOI] [PubMed] [Google Scholar]
- 5. Leeder, J.S. et al. Variability of CYP3A7 expression in human fetal liver. J. Pharmacol. Exp. Ther. 314, 626–635 (2005). [DOI] [PubMed] [Google Scholar]
- 6. Oshiro, C. , Mangravite, L. , Klein, T. & Altman, R. PharmGKB very important pharmacogene: SLCO1B1. Pharmacogenet. Genomics 20, 211–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kojima, S. , Yanaihara, T. & Nakayama, T. Serum steroid levels in children at birth and in early neonatal period. Am. J. Obstet. Gynecol. 140, 961–965 (1981). [DOI] [PubMed] [Google Scholar]
- 8. France, J.T. Levels of 16‐alpha‐hydroxydehydroepiandrosterone, dehydroepiandrosterone and pregnenolone in cord plasma of human normal and anencephalic fetuses. Steroids 17, 697–719 (1971). [DOI] [PubMed] [Google Scholar]
- 9. Nakamura, H. et al CYP3A4 and CYP3A7‐mediated carbamazepine 10,11‐epoxidation are activated by differential endogenous steroids. Drug Metab. Dispos. 31, 432–438 (2003). [DOI] [PubMed] [Google Scholar]
- 10. Niemi, M. , Pasanen, M.K. & Neuvonen, P.J. Organic anion transporting polypeptide 1B1: a genetically polymorphic transporter of major importance for hepatic drug uptake. Pharmacol. Rev. 63, 157–181 (2011). [DOI] [PubMed] [Google Scholar]
- 11. van Groen, B.D. et al Proteomics of human liver membrane transporters: a focus on fetuses and newborn infants. Eur. J. Pharm. Sci. 124, 217–227 (2018). [DOI] [PubMed] [Google Scholar]
- 12. Mooij, M.G. et al Ontogeny of human hepatic and intestinal transporter gene expression during childhood: age matters. Drug Metab. Dispos. 42, 1268–1274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thomson, M.M. , Hines, R.N. , Schuetz, E.G. & Meibohm, B. Expression patterns of organic anion transporting polypeptides 1B1 and 1B3 protein in human pediatric liver. Drug Metab. Dispos. 44, 999–1004 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prasad, B. et al Ontogeny of hepatic drug transporters as quantified by LC‐MS/MS proteomics. Clin. Pharmacol. Ther. 100, 362–370 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dennery, P.A. , Seidman, D.S. & Stevenson, D.K. Neonatal hyperbilirubinemia. N. Engl. J. Med. 344, 581–590 (2001). [DOI] [PubMed] [Google Scholar]
- 16. Huang, M.J. , Kua, K.E. , Teng, H.C. , Tang, K.S. , Weng, H.W. & Huang, C.S. Risk factors for severe hyperbilirubinemia in neonates. Pediatr. Res. 56, 682–689 (2004). [DOI] [PubMed] [Google Scholar]
- 17. Wang, E.T. et al Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baralle, F.E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Castle, J.C. et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 40, 1416–1425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plummer, N.W. & Meisler, M.H. Evolution and diversity of mammalian sodium channel genes. Genomics 57, 323–331 (1999). [DOI] [PubMed] [Google Scholar]
- 21. Plummer, N.W. , McBurney, M.W. & Meisler, M.H. Alternative splicing of the sodium channel SCN8A predicts a truncated two‐domain protein in fetal brain and non‐neuronal cells. J. Biol. Chem. 272, 24008–24015 (1997). [DOI] [PubMed] [Google Scholar]
- 22. Gustafson, T.A. , Clevinger, E.C. , O'Neill, T.J. , Yarowsky, P.J. & Krueger, B.K. Mutually exclusive exon splicing of type III brain sodium channel alpha subunit RNA generates developmentally regulated isoforms in rat brain. J. Biol. Chem. 268, 18648–18653 (1993). [PubMed] [Google Scholar]
- 23. Sarao, R. , Gupta, S.K. , Auld, V.J. & Dunn, R.J. Developmentally regulated alternative RNA splicing of rat brain sodium channel mRNAs. Nucleic Acids Res. 19, 5673–5679 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Belcher, S.M. , Zerillo, C.A. , Levenson, R. , Ritchie, J.M. & Howe, J.R. Cloning of a sodium channel alpha subunit from rabbit Schwann cells. Proc. Natl. Acad. Sci. USA 92, 11034–11038 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tate, S.K. et al Genetic predictors of the maximum doses patients receive during clinical use of the anti‐epileptic drugs carbamazepine and phenytoin. Proc. Natl. Acad. Sci. USA 102, 5507–5512 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mulley, J.C. , Scheffer, I.E. , Petrou, S. , Dibbens, L.M. , Berkovic, S.F. & Harkin, L.A. SCN1A mutations and epilepsy. Hum. Mutat. 25, 535–542 (2005). [DOI] [PubMed] [Google Scholar]
- 27. Petrovski, S. , Scheffer, I.E. , Sisodiya, S.M. , O'Brien, T.J. & Berkovic, S.F . Lack of replication of association between scn1a SNP and febrile seizures. Neurology 73, 1928–1930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tate, S.K. et al A common polymorphism in the SCN1A gene associates with phenytoin serum levels at maintenance dose. Pharmacogenet. Genomics 16, 721–726 (2006). [DOI] [PubMed] [Google Scholar]
- 29. Hunt, S.E. et al Ensembl variation resources. Database (2018). 10.1093/database/bay119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Malagnino, V. , Hussner, J. , Seibert, I. , Stolzenburg, A. , Sager, C.P. & Meyer Zu Schwabedissen, H.E. LST‐3TM12 is a member of the OATP1B family and a functional transporter. Biochem. Pharmacol. 148, 75–87 (2018). [DOI] [PubMed] [Google Scholar]
- 31. Pertea, M. , Kim, D. , Pertea, G.M. , Leek, J.T. & Salzberg, S.L. Transcript‐level expression analysis of RNA‐seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Langmead, B. & Salzberg, S.L. Fast gapped‐read alignment with Bowtie 2. Nat. Meth. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pertea, M. , Pertea, G.M. , Antonescu, C.M. , Chang, T.‐C. , Mendell, J.T. & Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nat. Biotechnol. 33, 290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Genome Reference Consortium . Genome Reference Consortium Human Build 37. <https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13/>. Accessed May 2018.
- 35. Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA‐Seq data with or without a reference genome. BMC Bioinformat. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim, D. , Langmead, B. & Salzberg, S.L. HISAT: a fast spliced aligner with low memory requirements. Nat. Meth. 12, 357 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Farrell, C.M. et al Current status and new features of the Consensus Coding Sequence database. Nucleic Acids Res. 42, D865–D872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boratyn, G.M. , Thierry‐Mieg, J. , Thierry‐Mieg, D. , Busby, B. & Madden, T.L. Magic‐BLAST, an accurate DNA and RNA‐seq aligner for long and short reads. bioRxiv (2018). doi: 10.1101/390013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. NCBI . ORFfinder. <https://www.ncbi.nlm.nih.gov/orffinder/>. Accessed May 2018.
- 40. ExPaSy . TMpred. <https://embnet.vital-it.ch/software/TMPRED_form.html>. Accessed May 2018.
- 41. UCSF . TOPO2. <http://www.sacs.ucsf.edu/cgi-bin/open-topo2.py>. Accessed May 2018.
- 42. Kang, Y.J. et al CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 45, W12–W16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang, L. , Park, H.J. , Dasari, S. , Wang, S. , Kocher, J.P. & Li, W. CPAT: Coding‐Potential Assessment Tool using an alignment‐free logistic regression model. Nucleic Acids Res. 41, e74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fickett, J.W. & Tung, C.S. Assessment of protein coding measures. Nucleic Acids Res. 20, 6441–6450 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yao, J. , Hong, W. , Huang, J. , Zhan, K. , Huang, H. & Hong, M. N‐Glycosylation dictates proper processing of organic anion transporting polypeptide 1B1. PLoS One 7, e52563 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Girard, H. , Levesque, E. , Bellemare, J. , Journault, K. , Caillier, B. & Guillemette, C. Genetic diversity at the UGT1 locus is amplified by a novel 3' alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet. Genomics 17, 1077–1089 (2007). [DOI] [PubMed] [Google Scholar]
- 47. Rouleau, M. , Roberge, J. , Bellemare, J. & Guillemette, C. Dual roles for splice variants of the glucuronidation pathway as regulators of cellular metabolism. Mol. Pharmacol. 85, 29–36 (2014). [DOI] [PubMed] [Google Scholar]
- 48. Bellemare, J. , Rouleau, M. , Harvey, M. & Guillemette, C. Modulation of the human glucuronosyltransferase UGT1A pathway by splice isoform polypeptides is mediated through protein‐protein interactions. J. Biol. Chem. 285, 3600–3607 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang, Y. , Boxberger, K.H. & Hagenbuch, B. Organic anion transporting polypeptide 1B3 can form homo‐ and hetero‐oligomers. PLoS One 12, e0180257 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ni, C. , Yu, X. , Fang, Z. , Huang, J. & Hong, M. Oligomerization study of human organic anion transporting polypeptide 1B1. Mol. Pharm. 14, 359–367 (2017). [DOI] [PubMed] [Google Scholar]
- 51. Li, D. , Gaedigk, R. , Hart, S.N. , Leeder, J.S. & Zhong, X.B. The role of CYP3A4 mRNA transcript with shortened 3'‐untranslated region in hepatocyte differentiation, liver development, and response to drug induction. Mol. Pharmacol. 81, 86–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S2. Tables S1–S3


