Summary
Slowing down translation in either the cytosol or the mitochondria are conserved longevity mechanisms. Here, we found a non-interventional natural correlation of mitochondrial and cytosolic ribosomal proteins (RPs) in mouse population genetics, suggesting a translational balance. Inhibiting mitochondrial translation in C. elegans through mrps-5 RNAi repressed cytosolic translation. Transcriptomics integrated with proteomics revealed that this inhibition specifically reduced translational efficiency of mRNAs required in growth pathways while increasing stress response mRNAs. The repression of cytosolic translation and extension of lifespan from mrps-5 RNAi were dependent on atf-5/ATF4, and independent from metabolic phenotypes. We found the translational balance to be conserved in mammalian cells upon inhibiting mitochondrial translation pharmacologically with doxycycline. Lastly, extending this in vivo, doxycycline repressed cytosolic translation in the livers of germfree mice. These data demonstrate that inhibiting mitochondrial translation initiates an atf-5/ATF4-dependent cascade leading to coordinated repression of cytosolic translation, which could be targeted to promote longevity.
Keywords: longevity, ribosomes, mitochondrial translation, cytosolic translation, translational balance
Introduction
The translation of mRNA into protein is essential for the growth and survival of every cell. Translation is carried out by small and large ribosomal subunits associating with mRNA assisted by eukaryotic initiation factors (eIFs) and elongation factors (eEFs) to ensure its speed and accuracy. The biogenesis of ribosomes coupled to mRNA translation has been estimated to be the most energy-consuming process in the cell (Buttgereit and Brand, 1995; Lane and Martin, 2010; Wieser and Krumschnabel, 2001). Slowing down translation by reducing expression of ribosomal proteins (RPs), eIFs/eEFs, or ribosomal RNA (rRNA) results in lifespan extension in yeast, worms, and flies (Chiocchetti et al., 2007; Hansen et al., 2007; Pan et al., 2007). This suggests a universal mechanism of lifespan extension lies in conserving the energy normally expended on cytosolic mRNA translation.
Distinct from the ribosomes in the cytosol, ribosomes reside in the mitochondria that are dedicated to translating the mRNAs encoded by the mitochondrial DNA. Due to the endosymbiotic ‘bacterial’ origin of the mitochondria, mitochondrial ribosomes are structurally more similar to bacterial ribosomes than to their cytosolic counterparts (Greber et al., 2015; Greber et al., 2014; Smits et al., 2007). Compared to cytosolic ribosomes, mitochondrial ribosomes only need to translate a relatively small set of 13 mRNAs coding for subunits of the oxidative phosphorylation (OXPHOS) complexes that are required for the generation of ATP. Remarkably, slowing down mitochondrial translation also extends lifespan (Houtkooper et al., 2013). Inhibiting mitochondrial translation genetically (e.g. by knocking down mitochondrial ribosomal genes) or pharmacologically (e.g. by using the antibiotic doxycycline) results in mitonuclear protein imbalance and activates the mitochondrial unfolded protein response (UPRmt) in a process conserved from worms to mammals (Houtkooper et al., 2013; Jovaisaite and Auwerx, 2015). The UPRmt includes the upregulated expression of cytoprotective genes, such as those coding for mitochondrial heat shock proteins 6 and 60 (hsp-6 and hsp-60) (Haynes and Ron, 2010). Other aspects of this response initiated by stressed mitochondria are mediated by Atf4 (C. elegans atf-5) (Quiros et al., 2017), a cyclic AMP-dependent transcription factor that has been implicated in a variety of mouse longevity models (Li et al., 2014).
Although mitochondrial and cytosolic ribosomes are separate translational apparatuses they depend on each other to coordinate mitochondrial function, in particular the synthesis of OXPHOS proteins. A regulatory signaling route from cytosolic translation to mitochondrial translation has been reported in yeast, which allegedly is strictly unidirectional (Couvillion et al., 2016). However, since mitochondria produce cellular energy and translation in the cytosol is a vast consumer of this energy, we hypothesized that there may be bidirectional translational control.
Here, we investigated the correlation of abundances between mitochondrial and cytosolic RPs in the BXD genetic reference population of mice (Wang et al., 2016). To address the direct link between mitochondrial and cytosolic translation, we have used a worm model of genetically impaired mitochondrial translation (RNAi mediated knockdown of mrps-5) and observed a decrease in cytosolic translation using polysome profiling. By integrating ‘omics’ data on three levels of biology [transcriptomics of the whole worm, transcriptomics of the polysomal fraction of the worm (highly translated transcripts), and proteomics], we explored an integrated, system-level view of the molecular changes induced via knockdown of mitochondrial translation that result in longevity. To show that key elements of this process are evolutionarily conserved, we used the antibiotic doxycycline as a means to reduce mitochondrial translation in human K562 cells and in vivo in mice. The findings of this study indicate that a bidirectional regulation exists between the two translation machineries across species, regulating longevity.
Results
Mitochondrial and cytosolic ribosomal proteins co-correlate in a natural population of mice
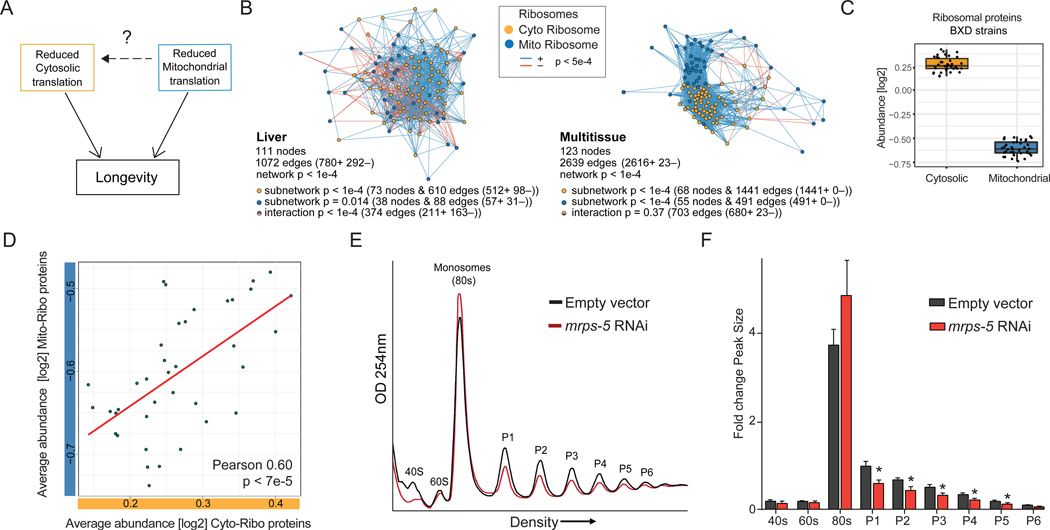
The degree to which the mitochondrial and cytosolic ribosomes longevity pathways regulate one another is not well understood (illustrated in Figure 1A). However, it seems likely that cross-communication between these systems exists allowing for the suppression of translation should either machinery become compromised in order to maintain equal rates of production. Therefore, one would expect the natural abundances of these two translation machineries to match one another in varying cell types, tissues, or individuals in a population. In order to evaluate the possible natural balance in abundances that occurs between elements of the mitochondrial and cytosolic translational machinery, we turned to the population of BXD mouse strains. These segregate for around 5 million sequence variants, which in turn lead to expression variation in the transcriptome and proteome with a complexity similar to human populations (Wang et al., 2016). To resolve the relationships between the mitochondrial and cytosolic translation machineries in the mouse BXD liver proteome data, we reconstructed the protein correlation network between the individual elements of these protein complexes (Figure 1B, Table S1). Using a fairly permissive cutoff criteria (p < 5e-4), we observed robust correlations both within the subunits of the cytoribosome and mitoribosome (p < 1e-4, p = 0.014, respectively), and importantly, strong interactions between these translation machineries (p < 1e-4). To confirm the robustness of this observation, we turned to a separate mouse study (Williams et al., 2018) examining the regulation of proteins across eight BXD strains and across four different tissues (Figure 1B). Indeed, we found RPs from both ribosome types clustered and co-varied as observed in the liver-specific data, suggesting a general mechanism is in place ensuring mito-cytosolic translational balance. As expected, taking the average abundance of either cytosolic or mitochondrial RPs for each BXD strain showed variation to exist within each ribosome type (Figure 1C), and evaluating average mitochondrial ribosome abundances in relation to the average levels of cytosolic ribosomes within individual BXD strains, confirmed the strong correlation between these two translation machineries (Pearson R = 0.60, p < 0.05) (Figure 1D). Taken together, these findings demonstrate that mitochondrial ribosome and cytosolic ribosome abundances are tightly regulated to stay in natural stoichiometric ratios.
Figure 1. A conserved mito-cytosolic translational balance in mice and worms.
(A) Model showing possible links between two branches of translation regulation in lifespan control; cytosolic (yellow) and mitochondrial (blue).
(B) Correlation networks in BXD mice show strong relationships between mitochondrial and cytosolic ribosomal protein levels both in liver (left) and across multiple tissues (right), suggesting that the subunits within ribosomes and across ribosomes are coregulated. Both networks show enriched connectivity. Liver: 73 nodes for the cytoribosome subnetwork have 610 edges (p < 1e-4, 512 are positively correlated), 38 nodes for the mitoribosome subnetwork have 88 edges (p = 0.014, 57 are positively correlated), 374 nodes are involved in the interaction between mitoribosome and cytoribosome subnetworks (p < 1e-4, 211 are positively correlated). Multitissue: 68 nodes for the cytoribosome subnetwork have 1441 edges (p < 1e-4, 1441 are positively correlated), 55 nodes for the mitoribosome subnetwork have 491 edges (p <1e-4, 491 are positively correlated), 703 nodes are involved in the interaction between mitoribosome and cytoribosome subnetworks (p < 1e-4, 680 are positively correlated). For significance testing, 10,000 random networks were permuted using the same input data and gene set size for each data set and subset with gene sets of the same size selected at random out of the full proteomic datasets. A reported p < 1e-4 means that none of the 10,000 random networks were as significant as the selected ribosome gene sets taken from literature.
(C) BXD mouse strains showing natural variation in cytosolic (yellow) or mitochondrial (blue) ribosome abundances in liver proteome. Each dot represents one BXD strain’s average abundance of either the cytosolic or mitochondrial ribosomes.
(D) Averaged abundances from mitochondrial and cytosolic ribosomes in the BXD liver proteome (from (C)) show correlation between mitochondrial and cytosolic abundances (Pearson 0.60, p < 7e-5).
(E) Representative polysome profiles showing decreased cytosolic polysome abundances in worms with impaired mitochondrial ribosomal biogenesis (mrps-5 RNAi). Lysate is normalized to protein levels. The subunits (40S and 60S), monosomal peak (80S), and polysomal peak numbers are indicated (P1–P6).
(F) Quantification of polysome peak sizes of representative experiment with N=4 per condition normalized to P1 peak of empty vector control worms. Error bars represent mean ± SD and significance was tested with Student’s t-test, and p-values were adjusted to correct for multiple testing using the Holm-Šídák method, with α = 0.05.
Previous work in yeast has suggested a unidirectional relationship between these two translation machineries, where disruption of cytosolic ribosomes can concomitantly reduce abundances of mitochondrial ribosomes but not the reverse (Couvillion et al., 2016). To test the hypothesis that regulation could also occur in the opposite direction, namely mitochondrial ribosomes influencing the cytosolic ribosomes, we turned to C. elegans with reduced levels of mitochondrial small ribosomal protein 5 (mrps-5). In addition to inhibiting mitochondrial translation, RNAi of mrps-5 induces the mitochondrial unfolded protein response (UPRmt), alters mitochondrial function, prevents aging-associated functional decline, and extends lifespan in C. elegans (Houtkooper et al., 2013). To assess the status of cytosolic translation in mrps-5 worms, we used a polysome profiling technique where free ribosomal subunits, monosomes (mRNA with one ribosome associated), and polysomes (mRNA with two or more ribosomes associated), are separated over a sucrose density gradient and quantified by optical density. Remarkably, in mrps-5 RNAi treated worms we observed a shift from polysomes to monosomes, suggesting that more cytosolic mRNAs are being translated by a single ribosome instead of multiple following mitochondrial translation inhibition (Figure 1E). Indeed, quantification of the profiles confirmed a significant reduction of polysomal peaks in the mrps-5 worms, revealing a suppression of cytosolic translation occurring in worms with an impaired mitochondrial ribosome (Figure 1F). To establish if this is a universal effect caused by mitochondrial disturbances, we performed polysome profiling in worms treated with either eat-3 or fzo-1 RNAi, which do not target the mito-ribosome but mitochondrial fusion. Both these conditions activated the UPRmt and decreased respiration (Liu et al., 2019), but did not repress cytosolic translation (Figure S1A–B).
Together, these observations demonstrate that in addition to the established communication from the cytosol and mitochondria, a second and novel relationship exists whereby the mitochondrial ribosome communicates with the cytosolic translational machinery.
mrps-5 RNAi treated worms display reduced translational efficiency (TE) of cytosolic ribosomal proteins
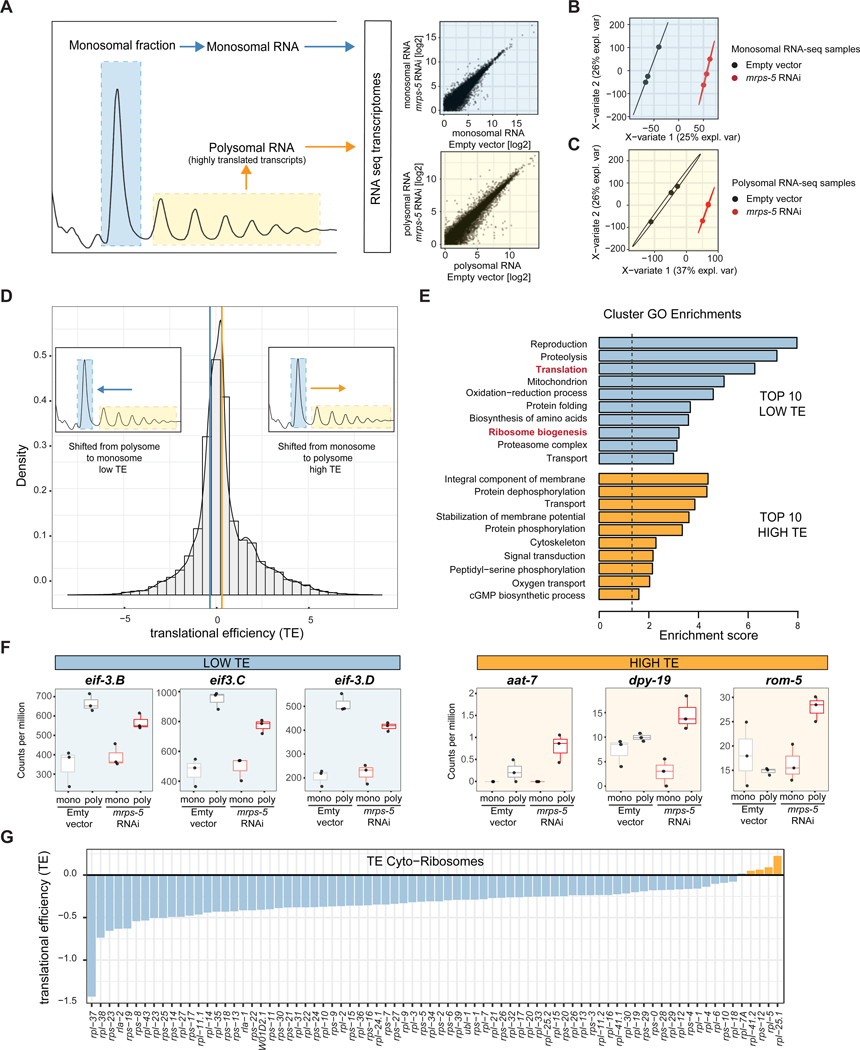
Having observed a decrease in global translation occurring in mrps-5 RNAi treated worms, we next inquired as to what genes are specifically repressed at the translational level, i.e. in our polysomal fractions. In order to do so, we generated RNA-seq libraries of polysomal and monosomal fractions of RNA from mrps-5 and control worms for comparison. The RNA-seq transcriptomes provided reliable quantification of over 16000 genes covering the majority of the worm genome, allowing for a global description of the differences existing between monosome and polysomal RNA fractions (Figure 2A, Table S2).
Figure 2. mrps-5 RNAi-treated worms show reduced translational efficiencies for cytosolic ribosomal genes.
(A) Schematic showing RNA isolated from polysome profiles, from highly translated fractions (yellow, polysome), and lesser translated fractions (blue, monosome). RNA-seq transcriptomics was performed on polysomal and monosomal fractions for mrps-5 RNAi and control worms. Transcripts (log2 transformed normalized counts) are depicted (scatterplots). Biological triplicates were used for each condition.
(B-C) Partial least squares discriminant analysis (PLS-DA) for monosomal (B) and polysomal (C) RNA libraries showing clear distinction between mrps-5 RNAi and control samples. Analysis was used to derive VIP scores, for genes contributing most to the group separation.
(D) Translational efficiencies (TE) of transcripts, defined as the log2 ratio of polysomal vs monosomal differences between mrps-5 RNAi and control gene expression, shows shifts in transcripts from either the monosome to the polysome (high TE, yellow) or the polysome to the monosome (low TE, blue). Insets schematically depict the shifts observed in the histogram. Cutoff lines and VIP scores were used to remove noise for calculating enrichment of biological processes in low or high TE mRNAs.
(E) Top 10 biological processes with highest enrichments scores in low TE (blue) or high TE (yellow) transcripts using DAVID analysis clusters enrichments in Gene Ontologies (GO). Decreased TE was observed in pathways such as ribosome biogenesis and translation. Clusters with an ‘Enrichment Score’ above 1.3 (p < 0.05, indicated with dotted line) were considered significantly enriched.
(F) Selection of mRNAs with either low TE (blue) or high TE (yellow), which contributed to the biological enrichments (‘translation’ or ‘integral component of membrane,’ respectively). Boxplots depict abundances (counts per million) of mRNA in the monosome and polysome for control and mrps-5 RNAi treated worms, used to calculate TEs.
(G) Individual cytosolic ribosomal proteins have a low TE in mrps-5 RNAi compared to control worms.
To capture the broad-spectrum set of changes that discriminate between mrps-5 and control worms at either the monosomal or polysomal levels, we performed a partial least squares discriminant analysis (PLS-DA) on the samples for each condition. We found this to effectively distinguish mrps-5 from control worms (Figure 2B–C, Table S2). Notably, this method is effective as a feature selector in addition to a sample classifier and is therefore useful in that it provides a variable of importance (VIP) ranking for each gene corresponding to its contribution in group discrimination (Boulesteix and Strimmer, 2007). This VIP score correlates highly to traditional p-value ranking (Figure S2A–B), though allows for a broader set of changes to be considered, which is more representative of the multifactorial nature of complex biological phenotypes. To explore the biological differences between groups in this way, we assessed genes with a high VIP score (>1) and turned to the DAVID bioinformatics resource (Huang da et al., 2009). DAVID analysis clusters enrichments in Gene Ontologies (GO) and various pathway databases, including KEGG and Interprot, based on recurrence and similarities of genes in enriched pathways. With this method we noted a wide variety of changes between mrps-5 RNAi and control worms (Table S2). For instance, changes in the mrps-5 monosomal RNA included an upregulation of RNAs coding for mitochondrial transit peptides (e.g. genes asg-2, atad-3, cyc-2.1) and a suppression of RNAs important for cuticle formation and other developmental pathways (e.g. genes bli-6, col-81, rol-1). A parallel upregulation of RNAs coding for mitochondrial transit peptides in the polysomes of mrps-5 (i.e. genes aco-2, coq-8, phb-2) was also found along with a similar suppression of RNAs coding for proteins important for reproduction and growth (e.g. genes acn-1, epi-1, ptr-4) (Table S2).
In order to identify what transcripts were shifting from polysomal to monosomal fractions, and vice versa, we calculated the translational efficiency (TE) of each gene. The TE is defined as the ratio of the polysomal RNA to the monosomal RNA fraction, for mrps-5 RNAi relative to control worm. Upon log2 transformation, a positive TE value then corresponds to transcripts that shift from the monosome to the polysome in mrps-5 RNAi relative to control, while negative TE values correspond to a shift in the opposite direction (Figure 2D, Table S2). Setting a low minimal fold change TE cutoff of 1.25 with a significant VIP score in at least one of the two group comparisons, we assessed genes with altered TE using DAVID GO term clustering. This analysis revealed that upon mrps-5 RNAi, genes with a high TE were coding for membrane components, proteins involved in stabilization of membrane potential, and proteins used in oxygen transport that include genes such as aat-7, dpy-19, and rom-5 (Figure 2E–F, Table S2). This observation is likely reflecting the worm’s need to adapt to its altered mitochondrial biology. Strikingly, genes with a low TE were by and large coding for proteins involved in translation and ribosome biogenesis (Figure 2E, Table S2), suggesting that mrps-5 RNAi worms suppress the translation of cytosolic ribosome components. These genes included translation elongation factors such as eif-3.B, eif-3.C, and eif-3.D (Figure 2F), in addition to those coding for the vast majority of RPs. Remarkably, visualizing the TE of the ribosome-associated proteins individually highlights what amounts to be almost a complete repression of the genes coding for the RPs that assemble the cytosolic ribosome (Figure 2G). These findings demonstrate the mrps-5-deficient mitochondrial translation machinery wields a strong regulatory influence over the cytosolic ribosomal translation machinery.
mrps-5 worms suppress growth pathways and upregulate stress responses at both the proteome and transcriptome levels
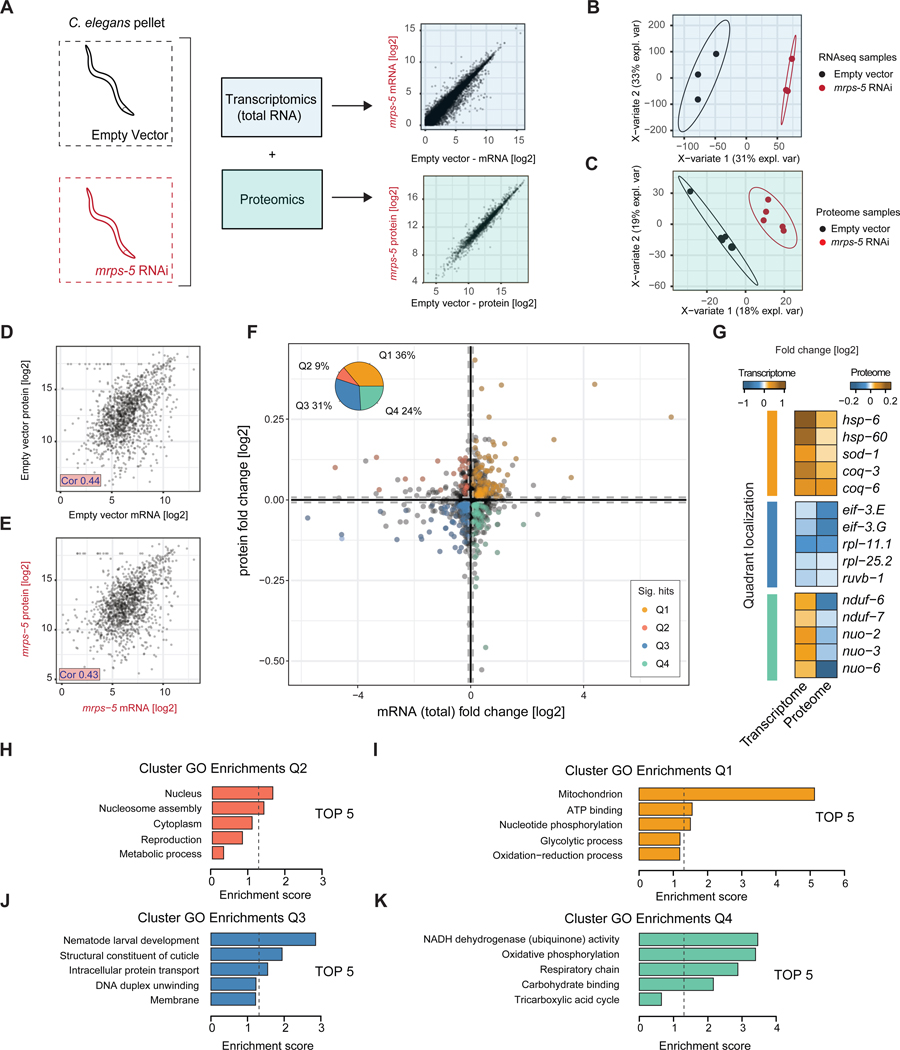
To better understand how the transcriptional responses ensuing from mrps-5 RNAi impact the proteome, we next performed both whole worm RNA-seq, and SWATH mass spectrometry proteomics of mrps-5 RNAi versus empty vector control worms. Our whole worm RNA profiling tracked the abundances of over 16000 transcripts (Table S3), while proteomics was able to quantify over 1700 proteins (Table S4). Again, the worm samples were readily distinguished through PLS-DA (Figure 3A–C), attributable to the large differences inherent between mrps-5 RNAi worms and controls reflected at both the transcript and protein levels. Using the VIP scores from the PLS-DA classification, we found major changes in the mrps-5 RNAi worms. These are characterized by an upregulation of oxidation-reduction and glycolysis-related genes and a downregulation of development, growth, and reproduction-related genes at the transcript level (Table S3). At the protein level we found an upregulation of mitochondrial (transit peptides) and nucleosome assembly proteins and a downregulation of the mitochondrial respiratory chain (complex I) subunits along with proteins involved in development and reproduction (Table S4).
Figure 3. Proteome-transcriptome cross-comparison of mrps-5 RNAi worms reveals transcriptional suppression of growth pathways and upregulation of stress responses.
(A) Schematic showing RNA (light blue) and proteins (light green) isolated from whole worms for RNA-seq or SWATH proteomics. Transcripts or proteins (Log2-transformed normalized counts) are depicted (scatterplots). RNA-seq was performed in biological triplicates and proteomics performed in biological quintuple per condition.
(B-C) Partial least squares discriminant analysis (PLS-DA) for transcriptome (B) and proteome (C) shows clear distinction between mrps-5 RNAi and control samples. PLS-DA was used to derive VIP scores, for genes contributing most to the group separation.
(D-E) Significant correlations of 0.44 and 0.43 were observed in the fold changes between the 1407 transcript-protein pairs overlapping the mRNA-protein datasets for control (D) and mrps-5 RNAi (E) samples respectively.
(F) Co-regulation plot of transcript and protein fold changes of mrps-5 RNAi vs control samples. Quadrants are defined as Q1 co-upregulated, Q2, post-transcriptionally increased, Q3 co-downregulated, and Q4, post-transcriptionally suppressed. Dashed lines indicate cutoff used to eliminate noise of near zero fold change (mRNA and protein cutoffs each relative to the standard deviation of their distributions), and a VIP score > 1 for both mRNA and protein levels were considered as significant hits. Pie chart inset shows percentage regulated, revealing a majority of changes being co-regulated.
(G) Examples of co-upregulated, co-downregulated, or post-transcriptionally suppressed genes, derived from the significant hits of panel (F).
(H-K) Top 5 biological processes enriched in each quadrant of the co-regulation plot, derived from the significant hits of panel (F).
To better understand if the changes detected at the transcript and protein levels were representative of one another, we assessed the 1407 protein-transcript pairs overlapping between both ‘omics’ datasets (Table S5). Given their good correlation in abundance (Figure 3D–E), we proceeded to check their differences in a co-regulation analysis. This was achieved by investigating fold changes occurring at the transcript level versus those occurring at the protein level for each individual gene and protein (Figure 3F). To reduce noise, we omitted genes with near zero-fold changes (those within dashed lines in Figure 3F) and again used the PLS-DA derived (Figure 3B–C) VIP scores for selection criteria, whereby a VIP score >1 identified genes contributing to the difference between control and mrps-5 RNAi treated worms. Our co-regulation analysis showed that the majority (67%) of genes were similarly regulated at the protein and transcript levels in mrps-5 RNAi worms (Figure 3F, pie chart inset and scatterplot, whereby 36% were co-upregulated [Q1] and 31% were co-downregulated [Q3]). Manual inspection of co-upregulated (Q1) genes revealed an increase antioxidant-related defense genes such as sod-1, coq-3, and coq-6 as well as the UPRmt proteins hsp-6 and hsp-60, the latter of which are in line with what we previously reported (Houtkooper et al., 2013). Co-downregulated genes (Q3) included growth- and translation-related genes such as eif-3.E, eif-3.G, rpl-11.1 and rpl-25.2 (Figure 3G). While only a minority (9%) of genes were downregulated at the transcript level and upregulated at the proteins level (Q2), our co-regulation analysis demonstrated that a sizeable proportion of genes (24%) exhibited an upregulation at the transcript level and a downregulation at the protein level (Q4). Manual inspection showed these genes to be predominantly related to cellular respiration, such as the NADH:ubiquinone oxidoreductase core subunit genes nduf-6, nduf-7, nuo-2, nuo-3, nuo-6 (Figure 3G). While the upregulation at mRNA level likely reflects the worm’s need to adapt to a loss of respiratory capacity from mrps-5 RNAi, the OXPHOS complexes cannot be assembled properly due to the lack of their mitochondrial translated partners and are downregulated on the protein level.
To better understand changes present in each quadrant of our co-regulation plot, we performed GO term enrichment analyses (Figure 3H–K, Table S5). While the genes associated with upregulated proteins and downregulated transcripts had relatively few enriched GO terms (Figure 3H), the remaining quadrants showed strong enrichments. Co-upregulated (Q1) genes enriched for mitochondrial-related processes likely reflecting an active adaptation in mitochondria to the knockdown of mrps-5 (Figure 3I). Co-downregulated (Q3) gene enrichments supported a role for decreased growth and development, with such terms as “nematode larval development” and “structural constituent of cuticle,” being most prominent (Figure 3J). Meanwhile, as observed in the individual gene analysis, genes with upregulated transcripts and downregulated proteins showed enrichments for OXPHOS (Figure 3K). Taken together, co-suppression of the individual translation-related genes in Q2 (Figure 3G) with the suppression of developmental related processes (Figure 3J) supports the idea that mitochondrial ribosomes crosstalk with cytosolic growth pathways to adapt the worm to a low energy state. Indeed, this adaptation is reflected in protein components of the worm’s respiratory chain, whereby OXPHOS proteins were suppressed despite their transcriptional upregulation (Figure 3G, K).
The mitochondrial to cytosolic translation response and lifespan are dependent on the atf-5/Atf4 transcription factor but can be decoupled from metabolic phenotypes
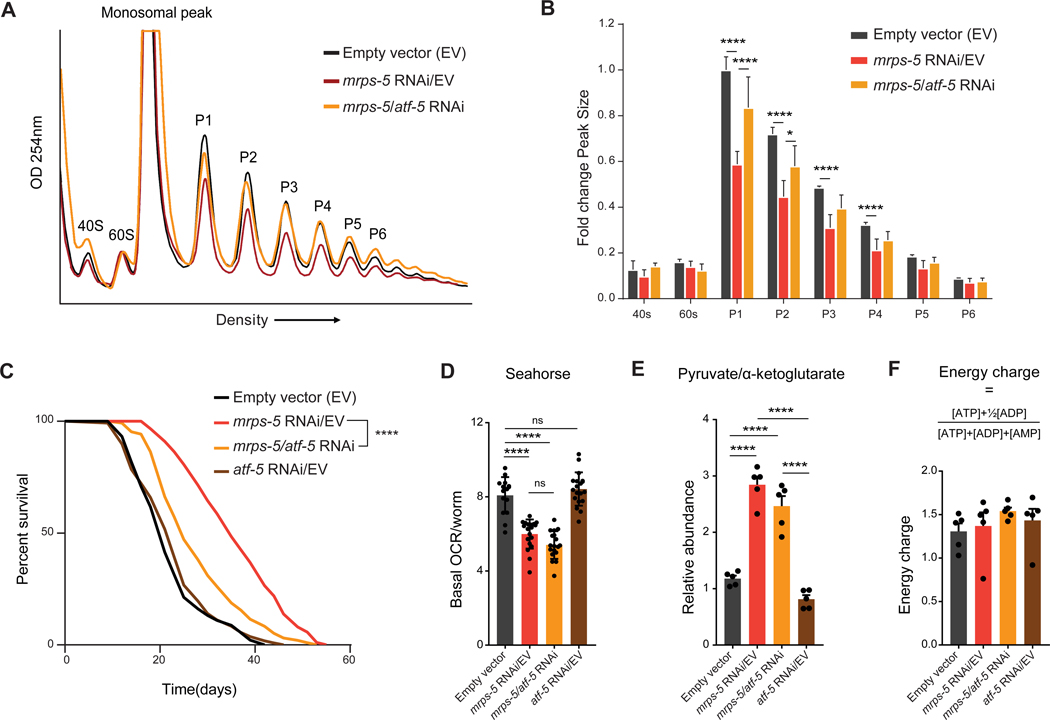
Our proteome-transcriptome co-regulation analysis of mrps-5 RNAi worms indicates that the upregulation of cytoprotective genes and downregulation of growth and biogenesis-related genes is to a significant extent a transcriptionally-regulated phenotype. This suggests that transcription factors may be core mediators of this process. Recent studies identified the transcription factor Atf4 (atf-5 in C. elegans) as a key player in communication from mitochondria towards the cytosol during mitochondrial stress induced by inhibition of either mitochondrial import, OXPHOS, membrane potential, or mitochondrial translation (Quiros et al., 2017). Elevated levels of ATF4 were also observed in many mouse longevity models that involve the suppression of cytosolic translation (Li et al., 2014). Given these reports, we investigated a potential role of C. elegans atf-5 coordinating the balance between cytosolic and mitochondrial translation. Using polysome profiling, we observed that the decreased cytosolic translation occurring in mrps-5 RNAi worms is almost fully restored to control levels upon additional feeding of atf-5 RNAi to worms (Figure 4A, Figure S3A–B). Indeed, quantification of the peaks in the polysome profiles showed significant rescue of the repressed polysomal peaks in mrps-5 worms when atf-5 was also knocked down (Figure 4B, Figure S3A–B). This epistasis experiment implicated atf-5 as a mediator of the suppression of cytosolic translation that is induced by the suppression of mitochondrial translation via mrps-5 knockdown.
Figure 4. Atf-5 mediates the balance between mitochondrial and ribosomal protein translation and lifespan extension, but not the metabolic phenotype in mrps-5 RNAi worms.
(A) Representative polysome profiles of control (black), mrps-5 RNAi/EV (red), and mrps-5 RNAi + atf-5 RNAi (yellow) worms shows mrps-5 mediated cytosolic translational suppression is dependent on the atf-5 transcription factor. Lysate is normalized to protein levels. The subunits (40S and 60S), monosomal peak (80S), and polysomal peak numbers are indicated (P1–P6).
(B) Quantification of polysome peak sizes of representative experiment with N=4 per condition normalized to P1 peak of empty vector control worms. Bars represent mean ± SD and significance was tested with Student’s t-test, and p-values were adjusted to correct for multiple testing using the Holm-Šídák method, with α = 0.05.
(C) Lifespan curves of control (black), mrps-5 RNAi (red), and mrps-5/atf-5 RNAi (yellow) and atf-5 RNAi (brown) worms shows mrps-5-mediated lifespan extension is dependent on the atf-5 transcription factor. N= 120 worms per condition.
(D) Seahorse respirometry measurements of control, mrps-5 RNAi/EV, mrps-5/atf-5 double RNAi, and atf-5 RNAi/EV worms show reduced basal OCR in both mrps-5 RNAi/EV and mrps-5/atf-5 RNAi-treated worms compared to controls. Data points represent average OCR per worm per well.
(E) Ratio between the relative abundances of pyruvate (glycolysis) and α-ketoglutarate (TCA cycle) of control, mrps-5 RNAi/EV, mrps-5/atf-5 double RNAi, and atf-5 RNAi/EV worms show a similar metabolic shift toward glycolysis in both mrps-5 RNAi/EV and mrps-5/atf-5 RNAi treated worms compared to control worms as measured by mass spectrometry.
(F) No difference was observed in energy charge measured by mass spectrometry. Bars represent mean ± SD with N=5 per condition and significance was tested with Student’s t-test, and p-values were adjusted to correct for multiple testing using the Holm-Šídák method, with α = 0.05.
In order to test if atf-5 also plays a role in the lifespan extension of mrps-5 RNAi worms, we performed lifespan experiments in which we silenced both mrps-5 and atf-5 through double RNAi (Figure 4C). We observed significant lifespan extension in the mrps-5 RNAi treated worms as reported before (Houtkooper et al., 2013). This lifespan extension was dependent on atf-5 since the mrps-5/atf-5 double RNAi condition led to a marked reduction of lifespan compared to mrps-5 RNAi alone, although not a complete reversal of the lifespan of controls (Figure 4C; Table S6).
In addition to repressed cytosolic translation, mrps-5 RNAi worms also have metabolic phenotypes, including reduced respiration (Houtkooper et al., 2013). We next aimed to elucidate whether the changes in translation can be decoupled from changes in metabolism. Since both the cytosolic translation and lifespan phenotype were dependent on atf-5, we explored if atf-5 RNAi also rescued the metabolic phenotypes. As described before, we found lower basal respiration in mrps-5 RNAi treated worms compared to the control worms (Houtkooper et al., 2013) (Figure 4D). When these worms were simultaneously treated with atf-5 RNAi (to block the cross-organellar translational feedback), basal respiration remained similar to the mrps-5 RNAi treated worms. This indicates that atf-5 RNAi does not rescue the reduced mitochondrial respiration in the mrps-5 RNAi worms. To substantiate these observations, we performed metabolomics on the worms subjected to these various RNAi conditions. In line with the reduced mitochondrial activity, we observed lower amounts of TCA cycle intermediates in the mrps-5 worms compared to control worms (Figure S3C). This was accompanied by increased pyruvate levels (Figure S3C), suggesting increased glycolysis as a likely means to maintain energy homeostasis in the mrps-5 RNAi treated worms. Taking the ratio between a TCA cycle intermediate (α-ketoglutarate) and a glycolysis intermediate (pyruvate), we see a significant shift from mitochondrial metabolism towards glycolysis in the mrps-5 worms (Figure 4E). Again, when these worms were simultaneously treated with both mrps-5 and atf-5 RNAi, a similar metabolic shift was observed, just like mrps-5 RNAi alone (Figure 4E). Finally, from our metabolomics data we calculated the energy charge, an index used to measure the energy status ([ATP]+½[ADP])/([ATP]+[ADP]+[AMP]). We observed no changes between worms subjected to any of the different RNAi conditions (Figure 4F). These data again support the notion that metabolic changes imposed by mrps-5 RNAi are not causing the changes in cytosolic translation.
Moreover, we found that the mito-cytosolic translational balance does not depend on the UPRmt. In the atfs-1(cmh15) mutant, which is unable to mount a UPRmt response (Deng et al., 2019), we found that the expression of cytosolic translation genes eif-3.c, eif3-g and rpl-14 was still downregulated upon mrps-5 RNAi (Figure S3D). Furthermore, although it was recently suggested that ATF4/ATF-5 displays cross-talk with the oxidative stress response transcription factor NRF2/SKN-1 (Kasai et al., 2019), ROS was not induced following either mrps-5 RNAi or doxycycline treatment and supplementation with the antioxidant N-acetylcysteine did not suppress the lifespan extension in these conditions, suggesting indepdendence from oxidative stress signaling (Houtkooper et al., 2013). In line with this, double knockdown of both mrps-5 and skn-1 still repressed the expression of eif-3.c, eif-3.g and rpl-14, similar to mrps-5 RNAi alone, suggesting that this repression is independent of skn-1 (Figure S3E).
In conclusion, we show that atf-5 is specifically regulating the mito-cytosolic translational balance and lifespan extension in mrps-5 worms, while our respiration and metabolomics measurements allow us to decouple mitochondrial respiration from cytosolic translation in our model with altered mitochondrial translation.
The mitochondrial to cytosolic translation response is conserved in mammalian cells
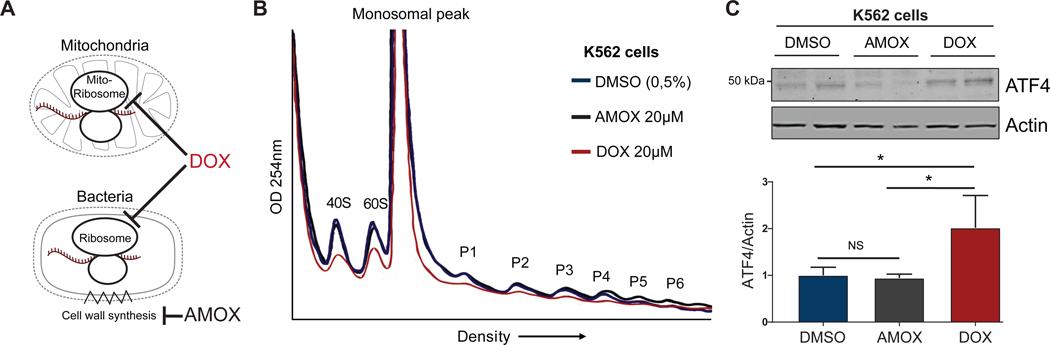
In order to see if this communication between mitochondrial translation machinery and cytosolic translation machinery is conserved in mammalian cells, we turned to a pharmacological model of reducing mitochondrial translation with the antibiotic doxycycline. Doxycycline is in the tetracycline family of antibiotics that inhibits mitochondrial translation by binding the 30S mitochondrial ribosomal subunit, while in contrast other antibiotics such as amoxicillin target bacterial cell wall synthesis. Amoxicillin is used here as a control in line with our previous work (Houtkooper et al., 2013; Moullan et al., 2015) (Figure 5A). The human K562 leukemia cell line was treated with DMSO, amoxicillin, or doxycycline and subjected to polysome profiling. We found that doxycycline treatment resulted in an overall repression of all peaks, including the monosomal and polysomal fractions, revealing a global repression of cytosolic translation as a result of mitochondrial translation inhibition (Figure 5B). In line with our observations that the C. elegans atf-5 is involved in this response, we observed that protein expression of ATF4, the mammalian homolog of atf-5, is upregulated in doxycycline- but not amoxicillin-treated K562 cells (Figure 5C). Taken together, these findings suggest that ATF4/atf-5 is mediating the communication between mitochondrial and cytosolic translation machineries in a manner that is conserved from nematodes to mammals.
Figure 5. The balance between mitochondrial and ribosomal protein translation is conserved in human cells.
(A) Schematic of doxycycline (DOX) mediated inhibition of mitochondrial ribosomes with amoxicillin not interfering with mitochondria as control antibiotic.
(B) Polysome profiles of DMSO and amoxicillin (AMOX) controls and doxycycline (DOX)-treated K562 cells shows doxycycline-induced suppression of cytosolic translation.
(C) Western blot analyses of amoxicillin control and doxycycline-treated K562 cells shows doxycycline-induced expression of ATF4. Bars represent mean ± SD and significance was tested with 1way ANOVA with multiple comparisons, with α = 0.05
The mitochondrial to cytosolic translation response is conserved in vivo in mice
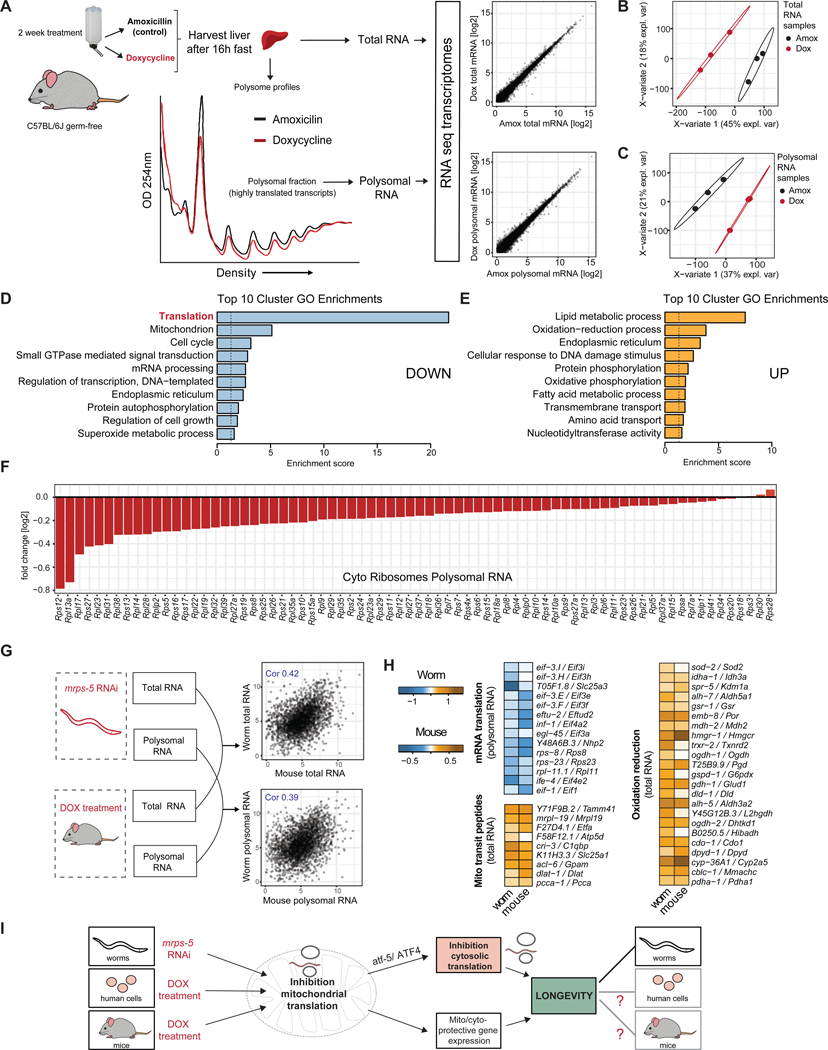
Having observed a conserved repression of cytosolic translation upon inhibition of mitochondrial translation in mammalian cells with doxycycline treatment, we next investigated these effects in vivo in mice. We treated mice for two weeks with doxycycline or amoxicillin in their drinking water, using doses that were previously reported to induce a mitonuclear protein imbalance (Moullan et al., 2015). To eliminate potential indirect effects of the antibiotics through the microbiome, we performed the antibiotic treatments on germ-free C57BL/6J mice. We found that doxycycline treatment resulted in suppression of global translation in liver in a dose-dependent manner (Figure S4A–B). Following this, we performed RNA-seq of the polysomal and total RNAs of liver tissue harvested from the antibiotic-treated mice (Figure 6A, Table S7). Similar as to what was observed in worms, we found the samples were readily classified through PLS-DA (Figure 6B–C) and again used VIP scores to distinguish differentially expressed genes (VIP > 1). DAVID analysis of the RNA-seq results showed a striking downregulation of translation-related mRNAs in the polysomal RNA fractions of doxycycline-treated mice (Figure 6D, Table S7), and an upregulation of various cellular defense processes including oxidation-reduction (Figure 6E) that is in line with our observations in mrps-5 deficient C. elegans. This suppression of cytosolic ribosomes was greater in the polysomal RNA fraction (Figure 6F) than in total RNA (Figure S4C), together resulting in a suppression of the TE of ribosomal genes occurring in doxycycline-treated mice (Figure S4D, Table S7). Mitochondrial ribosomes were most strongly suppressed at the total RNA level (Figure S4E–F). In line with our findings in C. elegans, DAVID enrichment analyses showed cytoprotective processes to have high TEs, developmental processes to have low TEs, and visual inspection confirmed the near total suppression of RP genes’ TEs (Figure S4D, Table S7). Taken together, these findings indicate that in vivo pharmacological inhibition of mitochondrial translation co-suppresses cytosolic translation, directly reducing cytosolic RP transcript TE. This confirms that the mechanism for balancing mitochondrial and cytosolic translation is conserved from nematodes to mammals.
Figure 6. Suppressing mitochondrial ribosomes by doxycycline treatment suppresses cytosolic ribosome abundance in vivo in mice liver, resulting in a conserved transcriptional signature for longevity assurance.
(A) Schematic showing RNA isolated from either the polysomal fraction or total RNA, from livers of germ-free mice treated with either doxycycline (DOX, red) or amoxicillin control (AMOX, black). Transcripts (Log2 transformed normalized counts) are depicted (scatterplots). For RNA-seq, biological triplicates were used for each condition.
(B-C) Partial least squares discriminant analysis (PLS-DA) for total (B) and polysomal (C) RNA libraries showing clear distinction between amoxicillin- and doxycycline-treated samples. Analysis was used to derive VIP scores, for genes contributing most to the group separation.
(D-E) Downregulated (D) or upregulated (E) top 10 biological processes enriched in the polysomal RNA of doxycycline-treated mice compared to amoxicillin, shows a suppression of translation-related transcripts after doxycycline treatment. A VIP score > 1 was used to identify differentially expressed genes.
(F) Individual cytosolic ribosomal proteins are downregulated in the polysomal RNA in doxycycline-treated mouse livers.
(G-H) Mouse-worm cross species comparison of total and polysomal RNA-seq libraries reveals a conserved transcriptional response for a polysomal downregulation of ‘mRNA translation’-related proteins and total RNA upregulation of ‘mito transit peptides’ and ‘oxidation reduction’ related genes.
(I) Model depicting how downregulating mitochondrial translation results in a coordinated downregulation of cytosolic translation via Atf4/atf-5, along with an upregulation of oxidation-reduction genes pathways ultimately leading towards longevity in C. elegans and possibly mammals.
Having observed a strong conservation of the molecular phenotypes in mrps-5 RNAi treated worms and doxycycline-treated mouse livers, we next asked what molecular signatures were specifically conserved between the two models. To do so, we performed a mouse-worm cross-species comparison of both total and polysomal RNA-seq libraries for orthologous genes (Figure 6G). Total and polysomal RNA abundances showed interspecies correlations of 0.42 and 0.39, respectively. Assessing the overlap of differentially expressed murine and C. elegans genes (Figure 6H) showed a conserved upregulation of genes coding for mitochondrial transit peptides and oxidation reduction in the total RNA and a downregulation of genes involved in mRNA translation in the polysomal RNA (Table S8).
Taken all together, our findings suggest that a conserved program for mito-cytosolic translational balance is activated across species when mitochondrial translation is inhibited by distinct and diverse methods. Our findings demonstrate that this leads to ATF4/atf-5 mediated signaling to slow down cytosolic translation while upregulating cyto-protective genes. Considering the remarkable longevity induced by mrps-5 and doxycycline treatments (Houtkooper et al., 2013), we propose that mito-cytosolic translational balance can ultimately be used to assure longevity in C. elegans, and possibly also other organisms (Figure 6I).
Discussion
In this study we used a systems biology approach operating on multiple levels of biology to elucidate the relationship between mitochondrial and cytosolic translation. This included analyzing mouse population transcriptomics, worm proteome-transcriptome regulation, and polysome fractioned transcriptomics from both worms and mice with impaired mitochondrial ribosomes. Through this approach, we identified a mito-cytosolic translational balance whereby the functional ability and abundance levels of mitochondrial ribosomes are communicated to the cytosolic ribosomes to suppress cytosolic translation. This was found conserved across species and in different genetic backgrounds. Furthermore, we have shown evidence that this communication is dependent on the transcription factor ATF4. Taken together, our work establishes the presence of a retrograde communication process between mitochondrial and cytosolic ribosomes.
Communication occurring from the cytosolic ribosomes regulates mitochondrial protein abundance (Couvillion et al., 2016; Fujiki and Verner, 1993; Surguchov et al., 1983). Indeed, mitochondria are dependent on cytosolic translation, as the vast majority of the ~1200 mitochondrial proteins, including mitochondrial RPs, are synthesized by cytosolic ribosomes (Attimonelli et al., 2002). However, given the energy requirement for cytosolic ribosome biogenesis and function coupled to the mitochondrial function as the main source of cellular energy, a clear need exists for the cytosolic and mitochondrial ribosomes to communicate their operational state to each other. Indeed, evidence for other mechanisms of communication from the mitochondria to the nucleus has been well documented. When a mitonuclear protein imbalance occurs due to inhibition of mitochondrial translation, the UPRmt is activated which triggers a nuclear transcriptional response to ultimately restore mitochondrial proteostasis (Haynes and Ron, 2010; Houtkooper et al., 2013). However, the effect on cytosolic proteostasis in a situation where mitochondrial translation was inhibited was still unclear. Also, a cytosolic proteostatic response occurs when mitochondrial proteins are not properly targeted to mitochondria and erroneously accumulate in the cytosol (Wrobel et al., 2015). Similarly, it has been shown that defective mitochondrial tRNA taurine modifications can also result in global cellular proteostasis activation mechanisms (Fakruddin et al., 2018), which again points to an intrinsic protein homeostasis network between the mitochondria and cytosol. Indeed, our work builds upon these prior observations of inter-organelle communication and establishes the ribosomes as cross-communicating complexes between subcellular organelles. Our findings provide a framework to better understand prior observations in mitochondrial biology. For example, dysfunction in mitochondrial ribosomes was shown to block cell proliferation prior to any loss of mitochondrial respiration (Richter et al., 2013). Our findings suggest that the mito-to-cytosolic translation balance could play a role in this anti-proliferation signal from the mitochondrial ribosomes by slowing down cytosolic translation. Similarly, our findings can help explain the anti-carcinogenic activity of antibiotics such as doxycycline and tigecycline (Kuntz et al., 2017; Skrtic et al., 2011). With cytosolic translation already established as a target for cancer therapy (Bhat et al., 2015; Faller et al., 2015; Pelletier et al., 2018), our work points to the possibility that this could be achieved through the cellular mechanisms in place to achieve mito-cytosolic translation balance.
The loss of respiration is one of the downstream effects of impaired mitochondrial translation in the mrps-5 worms (Houtkooper et al., 2013). Although we observed a strong upregulation of transcripts coding for mitochondrial OXPHOS proteins, the levels of these proteins were in fact downregulated in a way that likely explains the observed loss of respiration. OXPHOS in the mitochondria is carried out by five different protein complexes, of which four are dependent on mtDNA-encoded subunits. Interestingly, many of the genes that were up at the transcript level and down at the protein level (such as nuo−6, nuo−3, nuo−2, nduf−7 and nduf−6) were mainly nuclear genes coding for OXPHOS complex I proteins. The vast majority of OXPHOS complex I proteins are transcribed from nuclear DNA (38 genes) compared to mtDNA (7 genes) (Bar-Yaacov et al., 2012), suggesting the mrps-5 worms may be trying to redress their loss of respiration capacity by upregulating translation of complex proteins. However, the translation impairment of mtDNA-encoded components of OXPHOS complexes and the resulting stochiometric imbalance makes it seem likely that the unassembled nuclear-encoded complex proteins are degraded. This, in turn, may aggravate mitochondrial proteotoxic stress. Interestingly, this protein-specific degradation of members of stoichiometric complexes was observed before in human fibroblast (Liu et al., 2017). We suggest that mitochondrial (OXPHOS) proteins are degraded when they are not properly integrated in their preferred structures, i.e. the OXPHOS complexes. This has been previously reported in patients with mutations in the complex IV protein COA6, where the authors observed not only a complete absence of COA6 but also reduced levels of other complex IV proteins such as COX1, COX2, and COX4 (Baertling et al., 2015).
Inhibiting mitochondrial translation obviously leads to numerous downstream metabolic effects. Surprisingly, we showed that while atf-5 is required for the mito-cytosolic translational balance and lifespan extension in mrps-5 worms, it appears dispensable for the metabolic phenotypes such as mitochondrial respiration and metabolite profiles. Metabolic reprogramming is an important consequence of inhibiting mitochondrial translation, but our results suggest that this is not the driving force behind the mito-cytosolic translational balance. Rather we propose it goes hand in hand with the mito-cytosolic translational balance regulated by atf-5 in order to activate cytoprotection and maintain cellular homeostasis. This concept is supported by previous work showing that mrps-5 RNAi still extends lifespan in the AMPK (aak-2) mutant, demonstrating that it is not exclusively metabolic effects that are important for the observed lifespan extension (Houtkooper et al., 2013).
We found that Atf4 plays a role in orchestrating the communication from the mitoribosome to the cytoribosome. Interestingly, upregulation of this transcription factor has been linked to longevity in mice (Li et al., 2014) and overexpression of the orthologous transcription factor in yeast (GCN4) extended lifespan (Mittal et al., 2017), both in line with the longevity of mrps-5 RNAi-treated worms (Houtkooper et al., 2013) which was dependent on atf-5. In the yeast overexpression study, it was observed that Gcn4 acts as a repressor of protein synthesis (Mittal et al., 2017), in line with our observations in nematode and mammalian models. We did not observe changes in atf-5 mRNA in our mrps-5 RNAi treated worm transcriptomics, nor did we find changes in Atf4 mRNA in our doxycycline treated mice transcriptomics. While the regulation of Atf4 and its orthologues Gcn4 and atf-5 is not fully understood, it is known that differential contribution of two upstream ORFs (uORFs) in the 5’ leader of the mouse Atf4 mRNA regulates Atf4 expression (Vattem and Wek, 2004). Translation of ATF4 can be increased by phospho-eIF2α, phosphorylated by one of the four eIF2α kinases (PERK, GCN2, PKR, HRI) (Harding et al., 2000). However, the activated ATF4 induced by mitochondrial stress (e.g. doxycycline treatment), was not dependent on any of the individual eIF2α kinases in mammalian cells (Quirós et al., 2017). More recently, it was shown that there are also GCN2- and EIF2α-independent ways to activate ATF4, for instance in the case of methionine restriction (Mazor and Stipanuk, 2016). Understanding how ATF4 activation is regulated after inhibition of mitochondrial ribosomes may be valuable to target the aging process.
Slowing down cytosolic mRNA translation itself can extend lifespan (Hansen et al., 2007; Pan et al., 2007). Also other longevity mechanism are also known to be marked by reduced cytosolic translation such as the reduction of mTORC1 (Genolet et al., 2008) or insulin signaling (Stout et al., 2013) which both also extends lifespan in many organisms (Bluher et al., 2003; Harrison et al., 2009; Jia et al., 2004; Kaeberlein et al., 2005; Kapahi et al., 2004; Tissenbaum and Ruvkun, 1998; Vellai et al., 2003). In all likelihood, the mechanism leading to the observed reduction of cytosolic translation in the context of longevity is multifactorial. Since C. elegans mRNAs are known to by and large be lacking TOP 5’UTR sequences (Thoreen et al., 2012), it seems unlikely that the 5’UTRs are playing an important role in the regulation of mRNA translation. Therefore, we suggest an alternative mechanism may be at play involving mRNA binding proteins, many of which we found downregulated in the proteome of mrps-5 RNAi worms. One of these downregulated proteins is polyA binding protein 2 (pab-2) which is associates with several ribosomal proteins and localizes to polysomes; its knockdown also results in a reduction of polysomes (Lemieux and Bachand, 2009). Exploring this and other putative mechanisms that are responsible for the repression of mRNA translation in our models of longevity will be a very exciting area of future work.
Our cross-species analyses resulted in a transcriptional signature that was changing similarly in both worms and mice upon inhibition of mitochondrial translation. Most of the upregulated hits in our signature were mitochondrial proteins suggesting a mitochondrial quality control system. This signature involved cytoprotective transcriptional changes, including an upregulation of oxidation-reduction processes, and was characterized by a reduction of translation EFs. These findings are in line with the observation that inhibiting mitochondrial translation is a conserved longevity mechanism (Houtkooper et al., 2013). Our work here shows that this inhibition can also induce a previously unknown mito-cytosolic translational balance whereby cytosolic translation is also lowered, linking to another longevity mechanism (MacInnes, 2016). Finally, we identified a cyto-protective transcriptional signature characteristic of this response, and together these changes have the potential to drive healthy aging (Figure 5I).
Limitations of Study
While inhibiting mitochondrial ribosomes in C. elegans extends lifespan and its dependence on atf-5 was robustly demonstrated in our study, it remains to be determined whether inhibition of mitochondrial translation and activation of ATF4 can extend lifespan in mice and humans. Furthermore, due to the fact that our pharmacological intervention was an antibiotic, our work in vivo was carried out in germ-free mice and therefore may not fully recapitulate what may occur in wildtype mice. Likewise, our in vivo study focused on liver tissue, and it remains to be seen if these observations are also conserved in other tissues.
STAR methods
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Riekelt H. Houtkooper (r.h.houtkooper@amsterdamumc.nl).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Nematode growing conditions and RNAi experiments
Worms were cultured at 20°C on nematode growth medium (NGM) agar plates seeded with OP50 strain Escherichia coli. In brief, synchronized N2 worms were harvested and snap frozen at L4 larval stage for either total mRNA isolation, polysome profiling, proteomics, or metabolomics. For RNAi knock down experiments, synchronized L1 worms were plated on NGMi (containing 2 mM IPTG) with a bacterial lawn of either E. coli HT115 (RNAi control strain, containing an empty vector) or mrps-5, atf-5, eat-3, fzo-1, or skn-1 RNAi bacteria from the Ahringer C. elegans RNAi library. For double RNAi experiments bacteria were mixed in a 50/50 ratio and the control RNAi conditions were mixed with the empty vector (EV) RNAi bacteria HT115 in a 50/50 ratio. Lifespan tests were performed at 20°C as described (Houtkooper et al., 2013). In brief, synchronized worms were grown on NGM plates seeded with OP50 until reaching the L4 stage and then transferred to RNAi plates (F0). F0 adults were synchronized after 24h and transferred to NGMi RNAi plates to generate F1 offspring. At the L4 stage, F1 worms were transferred to plates containing 10 μM 5-Fluorouracil (5-FU, Sigma) to prevent growth of progeny. Worms were transferred to fresh RNAi plates once a week and after two transfers no 5-FU was added to plates.
K562 cell culture
K562 cells (female origin) were cultured at 37°C and in 5% CO2 in antibiotic-free RPMI 1640 medium (Thermo Fischer Scientific) supplemented with 10% FBS (Bodinco) and 25 mM HEPES (VWR Life Science). After 48 treatment with either 20 μM amoxicillin (Sigma), 20 μM doxycycline (Sigma), or DMSO (Sigma) as a vehicle control (0.05% in media), cells were harvested for western blotting or polysome profiling.
C57BL/6J germ-free mice
C57BL/6J germ-free male mice were housed 4 animals per cage and randomly assigned to the 3 experimental groups with N=8 mice per treatment. The germ-free mice were housed in sterile flexible-film isolators (Getinge France, Les Ulis, France). They were given free access to autoclaved tap water and a gamma-irradiated (45 kGy) standard diet (R03; Scientific Animal Food and Engineering, Augy, France). The isolators were maintained at 20–24°C and on a 12-h light/dark cycle (lights on at 07:30am). The germ-free status was monitored weekly by microscopic examination and aerobic and anaerobic cultures of freshly voided feces. At the age of 9 weeks mice were treated for two weeks with either amoxicillin (50 mg/kg/d), or one of two doses of doxycycline (50 mg/kg/d or 500 mg/kg/d) in the drinking water supplemented with 5% sucrose. Mice were sacrificed after 12 hours of fasting, and the livers were harvested and snap frozen in liquid N2. Liver tissue (~20 mg) was ground in liquid N2 in a mortar and pestle and used for either mRNA isolation or polysome profiling. Procedures were carried out in accordance with the European guidelines for the care and use of laboratory animals; they were approved by the Ethics Committee of AgroParisTech and the INRA Research Center of Jouy-en-Josas (approval reference: 14–40).
METHOD DETAILS
BXD strains
The BXD mouse liver proteome data used for network analyses were used from a prior publication (Williams et al., 2016) that consists of 73 cohorts of BXD mice—38 strains on chow diet and 35 strains on high fat diet for proteomics, and were combined. Note that the raw liver proteomics data from PXD003266 from 2016 were re-analyzed using the newer SWATH search library from PXD005044 from 2018, which was designed to improve coverage for mitochondrial proteins. These re-processed and normalized and processed mouse data are also available as Supplemental table 1. BXD mouse multi-tissue data were used from a prior publication (Williams et al., 2018) using datasets available in the Gene Expression Omnibus (GEO; GSE60149, GSE60150, GSE60151, and GSE60489). BXD mouse liver proteome data for average abundance quantification (Fig 1C–D) were downloaded from www.genenetwork.org (GN540).
Network analysis
Correlation network graphs were calculated using the above-referenced BXD proteome data using the iGraph v1.2.1 package in R. In the liver proteomics data, 77 cohorts were measured. 73 distinct cytosolic ribosome genes were quantified while 38 distinct mitochondrial ribosome genes were quantified. In the cross-tissue data, 40 samples from 5 tissues were measured. The cytosolic ribosome had 68 proteins quantified, while the mitochondrial ribosome had 55 proteins quantified. A fixed p-value cutoff for both networks was used (p < 5e-4), with varying corresponding rho due to varying n. Note that protein expression is generally more highly variable across 40 samples from 5 tissues than it is across 77 samples just from liver. Nodes that have 0 or 1 edge connection to the rest of the network were visually suppressed, but are still counted in the edge and node counts reported. For significance testing, 10,000 random networks were permuted using the same input data and gene set size for each data set and subset with gene sets of the same size selected at random out of the full proteomic datasets. A reported p < 1e-4 means that none of the 10,000 random networks were as significant as the selected ribosome gene sets taken from literature. Testing was only performed for overall edge counts.
Polysome profiling
Gradients of 17–50% sucrose (11 ml) in gradient buffer (110 mM KAc, 20 mM MgAc2 and 10 mM HEPES pH 7.6) were prepared on the day before use in thin-walled, 13.2 ml polypropylene (14 × 89 mm) centrifuge tubes (Beckman-Coulter, USA). Approximately 5000 nematodes were lysed in 500 ml polysome lysis buffer (gradient buffer containing 100 mM KCl, 10 mM MgCl2, 0.1% NP-40, 2 mM DTT, 0.5 mM cycloheximide and RNaseOUT™ (Thermo Fischer Scientific) using a Dounce homogenizer (Wheaton, USA) as described before (Molenaars et al., 2018). Mouse livers and K562 cells were lysed similarly with polysome lysis buffer but containing 5 mM cycloheximide. The samples were centrifuged at 1200 g for 10 min to remove debris and the supernatant was subjected to BCA protein assay (company). In all, 500 μg of total protein for each sample was loaded on top of the sucrose gradients. The gradients were ultra-centrifuged for 2 h at 40,000 g in a SW41Ti rotor (Beckman-Coulter, USA). The gradients were displaced into a UA6 absorbance reader (Teledyne ISCO, USA) using a syringe pump (Brandel, MD, USA) containing 60% sucrose. Absorbance was recorded at an OD of 254 nm. All steps of this assay were performed at 4°C or on ice and all chemicals came from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. Polysome peaks were quantified by measuring area under the curve (AUC) in ImageJ. Significance was tested with Student’s t-test, and p-values were adjusted to correct for multiple testing using the Holm-Šídák method, with alpha = 0.05.
Seahorse respirometry in C. elegans
Seahorse measurements were performed as described (Koopman et al., 2016). In brief, synchronized N2 worms grown on different RNAi conditions were collected at L4 larval stage, washed three times with M9 buffer, and ± 20 worms were transferred to 200 μl M9 buffer in a Seahorse XF96 microplate to measure respiration. Prior to and during the measurement the heater of the Seahorse respirometer was turned off. The chemical uncoupler FCCP (Abcam) and the complex IV inhibitor sodium azide (Sigma) treatments were used at a final concentration of 10 μM and 40 mM, respectively.
Metabolomics in C. elegans
A 75 μL mixture of the following internal standards in water was added to each freeze-dried sample: adenosine-15N5-monophosphate (100 μM), adenosine-15N5-triphosphate (1 mM), D4-alanine (100 μM), D7-arginine (100 μM), D3-aspartic acid (100 μM), D4-citric acid (100 μM), 13C1-citrulline (100 μM), 13C6-fructose-1,6-diphosphate (100 μM), guanosine-15N5-monophosphate (100 μM), guanosine-15N5-triphosphate (1 mM), 13C6-glucose (1 mM), 13C6-glucose-6-phosphate (100 μM), D3-glutamic acid (100 μM), D5-glutamine (100 μM), 13C6-isoleucine (100 μM), D3-leucine (100 μM), D4-lysine (100 μM), D3-methionine (100 μM), D6-ornithine (100 μM), D5-phenylalanine (100 μM), D7-proline (100 μM), 13C3-pyruvate (100 μM), D3-serine (100 μM), D5-tryptophan (100 μM), D4-tyrosine (100 μM), D8-valine (100 μM). Subsequently, 425 μL water, 500 μL methanol was added and worms were homogenized with a 5 mm steel bead using a TissueLyser II (Qiagen) for 5 min at frequency of 30 times/sec, and 1 mL chloroform was added to the same 2 mL tube before thorough mixing and centrifugation for 10 min at 14.000 rpm. The top layer, containing the polar phase, was transferred to a new 1.5 mL tube and dried using a vacuum concentrator at 60°C. Dried samples were reconstituted in 100 μL methanol/water (6/4; v/v). Metabolites were analyzed using a Waters Acquity ultra-high performance liquid chromatography system coupled to a Bruker Impact II™ Ultra-High Resolution Qq-Time-Of-Flight mass spectrometer. Samples were kept at 15°C during analysis and 5 μL of each sample was injected. Chromatographic separation was achieved using a Merck Millipore SeQuant ZIC-cHILIC column (PEEK 100 × 2.1 mm, 3 μm particle size). Column temperature was held at 30°C. Mobile phase consisted of (A) 1:9 acetonitrile:water and (B) 9:1 acetonitrile:water, both containing 5 mM ammonium acetate. Using a flow rate of 0.25 mL/min, the LC gradient consisted of: 100% B for 0–2 min, ramp to 0% B at 28 min, 0% B for 28–30 min, ramp to 100% B at 31 min, 100% B for 31–35 min. MS data were acquired using negative ionization in full scan mode over the range of m/z 50–1200. Data were analyzed using Bruker TASQ software version 2.1.22.3. All reported metabolite intensities were normalized to the number of worms in each sample, as well as to internal standards with comparable retention times and response in the MS. Metabolite identification has been based on a combination of accurate mass, (relative) retention times and fragmentation spectra, compared to the analysis of a library of standards.
Western blotting
For extraction of protein, K562 pellets were resuspended in RIPA buffer with cOmplete Mini Protease Inhibitor Cocktail (Sigma-Aldrich). Lysates were incubated on ice for 10 minutes and then centrifuged at 16,000 × g for 10 minutes in a pre-chilled centrifuge to remove debris and quantification was carried out with BCA protein assay. Equal amounts of protein (25–50 μg) were loaded onto NuPAGE® Novex 4–12% Bis-Tris Gel 1.5 mm, 10 Well (Invitrogen) and subsequently transferred to nitrocellulose membranes using iBlot™ 2 Transfer Stacks (Invitrogen) for 7 minutes at 14 V in iBlot™ 2 Dry Blotting System (Invitrogen). Western blotting was performed with antibodies against ATF4 (Cell Signaling Technology, #11815), β-actin (Sigma-Aldrich, #A5441), IRDye® 680RD Donkey anti-Rabbit secondary antibody (Li-Cor, #925–68073), and IRDye® 800CW Goat anti-Mouse secondary antibody (Li-Cor, #925–32210) and visualized using the Odyssey® Imaging System.
Isolation of mRNA (Total, Monosomal, and Polysomal)
For isolation of total mRNA, whole worms or liver tissue were homogenized with a 5 mm steel bead using a TissueLyser II (Qiagen) for 5 min at frequency of 30 times/sec in the presence of TRIzol (Invitrogen), then the isolation was continued according to manufacturer’s protocol. Polysomal or monosomal fractions from two experiments were pooled and mRNA was extracted using TRIzol Liquid Sample (LS) (Invitrogen) according to the manufacturer’s protocol. For RNAseq, contaminating genomic DNA was removed using RNase-Free DNase (Qiagen) and samples were cleaned up with the RNeasy MinElute Cleanup Kit (Qiagen).
Library Preparation
RNA libraries were prepared and sequenced with the Illumina platform by Genome Scan (Leiden, Netherlands). Libraries were prepared using the NEBNext Ultra Directional RNA Library Prep Kit (NEB #E7420) according to manufacturer’s protocols. Briefly, rRNA was depleted from total RNA using the rRNA depletion kit (NEB# E6310). After fragmentation of the rRNA-reduced RNA, a cDNA synthesis was performed in order to ligate with the sequencing adapters and PCR amplification of the resulting product. Quality and yields after sample preparation were measured with the Fragment Analyzer (Agilent). Sizes of the resulting products were consistent with the expected size distribution (a broad peak between 300–500 bp). Clustering and DNA sequencing using the Illumina cBot and HiSeq 4000 was performed according to manufacturer’s protocol with a concentration of 3.0 nM of DNA. HiSeq control software HCS v3.4.0, image analysis, base calling, and quality check was performed with the Illumina data analysis pipeline RTA v2.7.7 and Bcl2fastq v2.17.
Read mapping, statistical analyses, and data visualization
Reads were subjected to quality control FastQC (Andrews, 2010) trimmed using Trimmomatic v0.32 (Bolger et al., 2014) and aligned to either the C. elegans (worm) or M. musculus (mouse) genomes obtained from Ensembl, wbcel235.v91 and GRCm38v87, respectively, using HISAT2 v2.0.4 (Kim et al., 2015). Worm and mouse samples were analyzed separately; different fractions (total, monosomal, polysomal) within each species were analyzed together. Counts were obtained using HTSeq (v0.6.1, default parameters) (Anders et al., 2015) using the corresponding GTF taking into account the directions of the reads. Statistical analyses were performed using the edgeR (Robinson et al., 2010) and limma/voom (Ritchie et al., 2015) R packages. All genes with more than 2 counts in at least 4 of the samples were kept. Count data were transformed to log2-counts per million (logCPM), normalized by applying the trimmed mean of M-values method (Robinson et al., 2010) and precision weighted using voom (Law et al., 2014). All data analyses and visualizations were done using transcript data with a count value greater than 0. Differential expression was assessed using either an empirical Bayes moderated t-test within limma’s linear model framework including the precision weights estimated by voom (Ritchie et al 2015, Law et al 2014), or a Partial least squares discriminant analysis (PLS-DA) using mixomics (Rohart et al., 2017) setting a variable of importance (VIP) score of greater than 1 as significant. Resulting p values (where applicable) were corrected for multiple testing using the Benjamini-Hochberg false discovery rate. Genes were re-annotated using biomaRt using the Ensembl genome databases (v91). Data visualization was performed using gplots (Warnes et al., 2016) and ggplot2 (Wickham, 2009) selecting colors from RcolorBrewer (Neuwirth, 2014). Data processing and visualization was performed using R v3.4.3 and Bioconductor v3.5. The RNA-seq data are available on GEO under the ID GSE122097.
Translational efficiency calculations
Translational efficiencies (TEs) were acquired by considering the ratio of a highly translated ribosome-mRNA fraction (i.e. polysomal RNA) over a lower translated ribosome-mRNA fraction (i.e. either monosomal RNA for C. elegans or total RNA for M. musculus). Monosomal RNA was used for C. elegans as it showed an increase in abundance in our polysome profiles. The monosomal fraction did not show an increase in abundance in M. musculus and therefore total RNA was used. This ratio (highly translated ribosome-mRNA fraction / lower translated ribosome-mRNA fraction) was then compared between treated samples (i.e. mrps-5 for C. elegans or doxycycline treatment for M. musculus) and control samples (i.e. empty vector for C. elegans or Amoxicilin treatment for M. musculus). This resulted in TE being defined as: log2[ (treatedpolysome / treatedmonosome) / (controlpolysome / controlmonosome)] in the case of C. elegans and log2[ (treatedpolysome / treatedtotal) / (controlpolysome / controltotal)] in the case of M. musculus. Means from each set of replicates were used to calculate the ratios. A TE value was defined as significant if the gene in question had a PLS-DA VIP score above 1, in either the polysomal RNA (for both C. elegans and M. musculus) or monosomal RNA (for C. elegans) or total RNA (for M. musculus), and changed at least 1.25 fold in its TE (to eliminate noise of near zero TEs).
Reverse Transcription and qPCR
For qPRC, 1 μg of extracted RNA was pre-treated with gDNA Wipeout Buffer and reverse transcribed into cDNA according to the manufacturer’s instructions using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative gene expression analysis was performed using the LightCycler® 480 SYBR Green I Master (Roche) and measured using the LightCycler® 480 Instrument (Roche). Relative expression was calculated with the linRegPCR method ((Ruijter et al., 2009) and normalized to reference genes (F35G12.2 and pmp-3). A complete list of primers is available in Key Resources Table.
Key resources
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-ATF4 (1:1000) | Cell Signaling Technology | Cat# 11815 RRID: AB_2616025 |
| Mouse monoclonal anti-β-Actin (1:5000) | Sigma-Aldrich | Cat# A5441 RRID: AB_476744 |
| Donkey anti-Rabbit IgG (1:10000) | LI-COR Biosciences | Cat# 925-68073 RRID: AB_2716687 |
| Goat anti-mouse IgG (1:10000) | LI-COR Biosciences | Cat# 925-32210 RRID: AB_2687825 |
| Bacterial and Virus Strains | ||
| Escherichia coli:OP50 | Caenorhabditis Genetics Center | RRID:WB-STRAIN:OP50 |
| Escherichia coli: HT115 (DE3) | Caenorhabditis Genetics Center | RRID:WB-STRAIN:HT115(DE3) |
| Ahringer C. elegans RNAi library | Source BioScience | http://www.sourcebioscience.com/products/life-science-research/clones/rnai-resources/c-elegans-rnai-collection-ahringer/ |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cycloheximide | Sigma-Aldrich | Cat# C6255 |
| Doxycycline | Sigma-Aldrich | Cat# D9891 |
| Amoxicillin | Sigma-Aldrich | Cat# A8523 |
| DMSO | Sigma -Aldrich | Cat# D8418 |
| 5-FU | Sigma -Aldrich | Cat# f6627 |
| Sodium azide | Sigma-Aldrich | Cat# s8032 |
| FCCP | Abcam | Cat# ab120081 |
| Critical Commercial Assays | ||
| BCA protein assay Reagent A | Thermo Fisher Scientific | Cat# 23223 |
| TRIzol (LS) | Invitrogen | Cat# 15596026 (#10296010) |
| RNeasy MinElute Cleanup Kit | Qiagen | Cat# 74204 |
| NEBNext Ultra Directional RNA Library Prep Kit | NEB | Cat# E7420 |
| rRNA depletion kit | NEB | Cat# E6310 |
| QuantiTect Reverse Transcription Kit | Qiagen | Cat# 205314 |
| LightCycler® 480 SYBR Green I Master | Roche | Cat# 04887352001 |
| Deposited Data | ||
| C. elegans and Mouse RNA-seq data | This paper | GEO: GSE122097 |
| C. elegans Proteomics data | This paper | PRIDE: PXD009223 |
| BXD mouse liver proteome data (reprocessed) | This paper | Table S1 |
| BXD mouse liver proteome data | Genenetwork.org | GN540 |
| BXD mouse multi tissue proteome data | Molecular and Cellular Proteomics Journal | DOI: 10.1074/mcp.RA118.000554 |
| Experimental Models: Cell Lines | ||
| Human: K562 cells | ATCC | CCL-243 RRID:CVCL_0004 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: N2 Bristol | Caenorhabditis Genetics Center (CGC); http://cbs.umn.edu/cgc/home | CGC:10570 RRID:WB-STRAIN: N2_(ancestral) |
| C. elegans: atfs-1(cmh15) | (Deng et al., 2019) | NA |
| Mouse: C57BL/6J germ-free | Micalis Institute (INRA Research Center of Jouy-en-Josas) | NA |
| Oligonucleotides | ||
| F35G12.2 forward primer: 5’- ACTGCGTTCATCCGTGCCGC-3’ | Sigma-Aldrich | NA |
| F35G12.2 reverse primer: 5’-TGCGGTCCTCGAGCTCCTTC-3’ | Sigma-Aldrich | NA |
| pmp-3 forward primer: 5’-GCTGGAGTCACTCATCGTGTTA -3’ | Sigma-Aldrich | NA |
| pmp-3 reverse primer: 5’- CCGGCCAATCATCCTCTTGA -3’ | Sigma-Aldrich | NA |
| mrps-5 forward primer: 5’-ACTGGCCGAACGAAAAGGTCT -3’ | Sigma-Aldrich | NA |
| mrps-5 reverse primer: 5’-AGTGGAAATCGGTGACGCCACAA -3’ | Sigma-Aldrich | NA |
| atf-5 forward primer: 5’- GTGCGAAGAAATCGAGCGTC -3’ | Sigma-Aldrich | NA |
| atf-5 reverse primer: 5’- GCTCCAACTCGGATACCTGAT -3’ | Sigma-Aldrich | NA |
| skn-1 forward primer: 5’- TCAATTTATGGAGTGTCGTCCA -3’ | Sigma-Aldrich | NA |
| skn-1 reverse primer: 5’- CCGTTGATCATCACGCCAAC -3’ | Sigma-Aldrich | NA |
| eif-3.c forward primer: 5’- TTTCCAGGAAGGAACGAGCG -3’ | Sigma-Aldrich | NA |
| eif-3.c reverse primer: 5’- CCGTCTTGTCCTTGTCCTCC -3’ | Sigma-Aldrich | NA |
| eif-3.g forward primer: 5’- GCATTAACGCAATGGCTCCC -3’ | Sigma-Aldrich | NA |
| eif-3.g reverse primer: 5’- TCGGTACGTGTTCCATCAGC -3’ | Sigma-Aldrich | NA |
| rpl-14 forward primer: 5’- CAAGCTCACCGACTTCGAGA -3’ | Sigma-Aldrich | NA |
| rpl-14 reverse primer: 5’- GAGCTCCACTCGGACGATTC -3’ | Sigma-Aldrich | NA |
| Recombinant DNA | ||
| Software and Algorithms | ||
| R (v3.4.3) | R | https://www.r-project.org |
| FastQC | (Andrews, 2010) | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| iGraph v1.2.1 package | iGraph | https://CRAN.R-project.org/package=igraph |
| RTA v2.7.7 | Illumina, Real-Time Analysis | Illumina.com |
| Bcl2fastq v2.17 | Illumina | Illumina.com |
| Trimmomatic v0.32 | (Bolger et al., 2014) | http://www.usadellab.org/ |
| HISAT2 v2.0.4 | (Kim et al., 2015) | https://ccb.jhu.edu/software/hisat2/index.shtml |
| HTSeq v0.6.1 | (Anders et al., 2015) | https://htseq.readthedocs.io/en/release_0.10.0/ |
| Comet v2016.01 r3 | doi: 10.1002/pmic.201200439 | v2016.01 r3 |
| OpenSWATH v2.1.0 | http://www.openswath.org/ | |
| MSstats v3.12.0 | http://msstats.org/ | |
| Limma/voom | (Ritchie et al., 2015)/(Law et al.,2014) | https://bioconductor.org/packages/release/bioc/html/limma.html |
| edgeR | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| HiSeq control software HCS v3.4.0 | Illumina | https://support.illumina.com/downloads/hiseq-x-hcs-v3-4-0.html |
| Bioconductor v3.5 | Bioconductor | https://www.bioconductor.org/install/ |
| GraphPad Prism v7.03 | GraphPad Software, Inc. | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Bruker TASQ software v2.1.22.3 | Bruker | https://www.bruker.com/products/mass-spectrometry-and-separations/ms-software/tasq/tasq-software.html |
| Other | ||
Protein-Transcript cross comparison
Fold change (log2 transformed) of the genes in common between the transcriptome and proteome data were plotted in a co-regulation plot, with mRNA or protein data on the X or Y axis, respectively. A cutoff was used to eliminate noise of near zero-fold change when identifying genes differentially expressed in the plot. Selecting cutoffs relative to each dataset allowed us to address the large difference in fold changes observed between the different mRNA and protein datasets, and we therefore used a quarter of the standard deviation of each log2-transformed dataset’s distribution as a standard threshold for noise (dashed line in plot). This corresponded to a value of 1.13 (0.179 in log2 scale) for the transcriptome and 1.01 (0.016 in log2 scale) for the proteome. Values above these cutoffs in each respective dataset that had VIP scores above 1 were considered as significant in the quadrants of the co-regulation plot.
Worm-Mouse cross comparison
Worm and mouse orthologous genes were acquired using the R package annotationTools (Kuhn et al., 2008) and HomoloGene v68 species annotation.
Ribosomal gene lists
Cytosolic and mitochondrial ribosomal gene lists for both C. elegans and M. musculus were downloaded from the Ribosomal Protein Gene Database (http://ribosome.med.miyazaki-u.ac.jp/).
Functional Annotation of gene sets
Gene sets were analysed for functional enrichments using the DAVID bioinformatics resource version 6.8 (Huang da et al., 2009). Functional annotation clustering was performed using DAVID defined default settings incorporating gene sets from Gene Ontologies (biological process, cellular component, and molecular function), functional categories (including Clusters of Orthologous Groups (COG) ontologies, UP keywords, UP seq features), pathways (including KEGG), and protein domains (including INTERPRO PIR superfamily and SMART). Background datasets to check enrichments against included the measured transcriptome (for enrichments at mRNA level within a single species), measured proteome (for enrichments at the protein level), gene overlap between the measured transcriptome and proteome (for enrichments of the co-regulation analysis), or genes in common between mouse and worms (for cross-species enrichment). Resulting clusters were manually annotated for purposes of visualization and summarization. Clusters with an ‘Enrichment Score’ above 1.3 (p < 0.05) were considered significantly enriched.
Protein isolation for SWATH proteomics
Worm pellets were freeze-dried overnight and stored at room temperature until use. C. elegans proteins were isolated using the standard protocol used for tissue preparation (Wu et al., 2017) with the exception that, to ensure that the cuticle was fully ruptured, during the protein isolation from total cells in 8M urea, the samples were sonicated at a high power (150 W) for 10×10 cycles of 10 seconds (UP200St-G sonicator, Hielsher, Germany). In brief (see above source for more details): proteins were extracted in an 8M urea buffer, 300 μg were aliquoted and precipitated by acetone overnight, then reduced with DTT, alkylated with IAA, and digested with trypsin at a 1:25 ratio. C18 spin columns (The Nest Group) were used for cleanup.
SWATH proteomics – library acquisition
For library acquisition, 150 μg of digested peptide from three sets of pooled N2 wildtype adult C. elegans were pH fractionated (protocol #84868, Thermo Fisher Scientific) into 10 fractions. The resulting 30 fractions were separated on an Eksigent liquid chromatography machine coupled with a 20 cm PicoFrit emitter injected on an AbSciex 5600+ TripleTOF using a 120 minute gradient going from 2% to 35% acetonitrile at 300 nL/minute. At the MS1 level, the 20 most intense precursors were selected in the range of 350 – 1460 m/z with a 500 ms survey scan. At the MS2 level, spectra were acquired at 150 ms survey scans between 50 – 2000 m/z. These samples were used for preparing a spectral library to support the DIA/SWATH data (Rost et al., 2014). iRT peptides (Biognosys) were added to all samples, for both DDA injections (library acquisition) and DIA injections (for quantification).
SWATH proteomics – sample acquisition
For all samples measured for quantitative DIA (SWATH) acquisition, 1 μg of non-fractionated sample were injected in the same AbSciex 5600+ TripleTOF using the same parameters except using a 60 minute gradient and in data-independent acquisition mode using 64 m/z windows. The output .wiff files from both DIA and DDA acquisition mode were converted to centroided mzXML files using FileConverter v2.2.0. For library generation from the DDA mzXML files, samples were searched against the canonical UniProtKB C. elegans proteome database containing 27481 proteins and searched with Comet v2016.01 r3. Reverse decoy proteins were generated and up to 2 tryptic missed cleavages were allowed with a precursor mass error of 50 ppm and fragment error of 0.1 Da. Cysteine carbamidomethylation was used as the fixed modification and methionine oxidation as the variable modification. PeptideProphet was used to search the data and scored with iProphet. A 1% protein FDR was used for significance cutoff. The in-house results from the DDA runs were combined with extensive published C. elegans DDA data (PXD004584) and (Narayan et al., 2016). The light channel from these SILAC-labeled samples was also searched with Comet as above, using the same parameters, using CiRT peptides instead of iRT peptides (Parker et al., 2015). The results from the in-house and downloaded DDA files were combined, and peptides with retention time variance of ≥ 150 were removed. For peptides identified in-house and in the downloaded dataset, the in-house peptide was retained and the other peptide discarded. The final assembled library contains 67’612 peptides (of which 18’072 from the in-house DDA runs and 49’540 from the downloaded DDA runs), corresponding to 9’438 unique proteins. This library was then used as the reference library for OpenSWATH v2.1.0. OpenSWATH was run on all DIA acquisitions, followed by mProphet scoring using the msproteomicstools package available on GitHub (September 2017). 10750 peptides corresponding to 1715 proteotypic proteins were quantified. Protein data were generated from the peptide matrix using MSstats v3.12.0. Differential expression was determined using either a Student’s t-test (with p values corrected for multiple testing using the Benjamini-Hochberg false discovery rate, presented for reference) or partial least squares discriminant analysis (PLS-DA) with mixomics (Rohart et al., 2017) setting a variable of importance (VIP) score of greater than 1 as significant. The DIA data and the assembled library file are available on PRIDE under the ID PXD009223.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details and tests used, the number and representation of n and any other forms of quantification present are specified in the respective figure legends and results section. Statistical analyses for quantified polysome profiles and western blotting (WB) were performed using the Prism 7 software (GraphPad Software, La Jolla, CA, USA). Significance was tested with Student’s t-test, and p-values were adjusted to correct for multiple testing using the Holm-Šídák method. All other statistics were performed as described in each respective methods section and unless otherwise noted, were performed using R v3.4.3. Gene expression was considered differential relative to control using the variable of importance (VIP) score from a partial least squares discriminant analysis (PLSA-DA) greater than 1. In all supplemental tables where relevant, p-values (Bayes moderated t-test for transcriptomics or Student’s t-test for proteomics), and adjusted p-values (Benjamini-Hochberg method) are presented in addition to the VIP scores (from PLS-DA). For lifespan studies, survival curves were calculated using the log-rank (Mantel-Cox) method.
DATA AND SOFTWARE AVAILABILITY
The accession number for the C. elegans and mice RNA sequencing data reported in this paper is GEO: GSE122097. The proteomics DIA data and the assembled library file are available on PRIDE under the ID PXD009223. The reference liver mouse transcriptomics and proteomics data examined in Figure 1 are available under GEO at GSE60149 and PRIDE at PXD003266. The reference multi-tissue mouse transcriptome data are available under GEO GSE60149, GSE60150, GSE60151, and GSE60489 and are from the same mice despite the non-continuous and separate entries. The multi-tissue proteomics data are from PXD005044. Note that the raw liver proteomics data from PXD003266 from 2016 were re-analyzed using the newer SWATH search library from PXD005044 from 2018, which was designed to improve coverage for mitochondrial proteins. These re-processed and normalized and processed mouse data are also available as Supplemental Table S1.
Supplementary Material
Table S1. Re-analyzed liver proteomics data from PXD003266, related to Figure 1
Table S2. Monosomal/polysomal RNA-seq and translational efficiency worm, related to Figure 2.
Table S3. Total RNA-seq worm, Related to Figure 3
Table S4. Proteome worm, Related to Figure 3
Table S7. RNA-seq mice, Related to Figure 6
Table S8. Cross species RNA-seq comparison, Related to Figure 6
Acknowledgements
Work in the Houtkooper group is financially supported by an ERC Starting grant (no. 638290), a VIDI grant from ZonMw (no. 91715305), and a grant from the Velux Stiftung (no. 1063). AWM is supported by E-Rare-2, the ERA-Net for Research on Rare Diseases (ZonMW #40-44000-98-1008). GEJ is supported by a Federation of European Biochemical Society (FEBS, https://www.febs.org) long-term fellowship and a VENI grant from ZonMw (no. 09150161810014, https://www.zonmw.nl) EGW was supported by an NIH F32 Ruth Kirchstein Fellowship (F32GM119190). RA was supported by the European Research Council advanced grant ERC-20140AdG 670821. MAM is supported by NIH P20GM121176 and a Glenn Foundation for Medical Research and American Federation for Aging Research Junior Faculty Grant.
Footnotes
Conflict of interest
RHH is an inventor on a patent related to mitochondrial ribosomal proteins as aging regulators. The other authors declare that they have no conflict of interest.
References
- Anders S, Pyl PT, and Huber W (2015). HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S (2010). FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. [Google Scholar]
- Attimonelli M, Catalano D, Gissi C, Grillo G, Licciulli F, Liuni S, Santamaria M, Pesole G, and Saccone C (2002). MitoNuc: a database of nuclear genes coding for mitochondrial proteins. Update 2002. Nucleic acids research 30, 172–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baertling F, M A.M.v.d.B.., Hertecant JL, Al-Shamsi A, L P.v.d.H., Distelmaier F, Mayatepek E, Smeitink JA, Nijtmans LG, and Rodenburg RJ (2015). Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Human mutation 36, 34–38. [DOI] [PubMed] [Google Scholar]
- Bar-Yaacov D, Blumberg A, and Mishmar D (2012). Mitochondrial-nuclear co-evolution and its effects on OXPHOS activity and regulation. Biochimica et biophysica acta 1819, 1107–1111. [DOI] [PubMed] [Google Scholar]
- Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, and Topisirovic I (2015). Targeting the translation machinery in cancer. Nature reviews Drug discovery 14, 261–278. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, and Kahn CR (2003). Extended longevity in mice lacking the insulin receptor in adipose tissue. Science (New York, NY) 299, 572–574. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulesteix AL, and Strimmer K (2007). Partial least squares: a versatile tool for the analysis of high-dimensional genomic data. Briefings in bioinformatics 8, 32–44. [DOI] [PubMed] [Google Scholar]
- Buttgereit F, and Brand MD (1995). A hierarchy of ATP-consuming processes in mammalian cells. The Biochemical journal 312 (Pt 1), 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiocchetti A, Zhou J, Zhu H, Karl T, Haubenreisser O, Rinnerthaler M, Heeren G, Oender K, Bauer J, Hintner H, et al. (2007). Ribosomal proteins Rpl10 and Rps6 are potent regulators of yeast replicative life span. Experimental gerontology 42, 275–286. [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Soto IC, Shipkovenska G, and Churchman LS (2016). Synchronized mitochondrial and cytosolic translation programs. Nature 533, 499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P, Uma Naresh N, Du Y, Lamech LT, Yu J, Zhu LJ, Pukkila-Worley R, and Haynes CM (2019). Mitochondrial UPR repression during Pseudomonas aeruginosa infection requires the bZIP protein ZIP-3. Proceedings of the National Academy of Sciences of the United States of America 116, 6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakruddin M, Wei FY, Suzuki T, Asano K, Kaieda T, Omori A, Izumi R, Fujimura A, Kaitsuka T, Miyata K, et al. (2018). Defective Mitochondrial tRNA Taurine Modification Activates Global Proteostress and Leads to Mitochondrial Disease. Cell reports 22, 482–496. [DOI] [PubMed] [Google Scholar]
- Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, et al. (2015). mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517, 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki M, and Verner K (1993). Coupling of cytosolic protein synthesis and mitochondrial protein import in yeast. Evidence for cotranslational import in vivo. The Journal of biological chemistry 268, 1914–1920. [PubMed] [Google Scholar]
- Genolet R, Araud T, Maillard L, Jaquier-Gubler P, and Curran J (2008). An approach to analyse the specific impact of rapamycin on mRNA-ribosome association. BMC medical genomics 1, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, and Ban N (2015). Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science (New York, NY) 348, 303–308. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, and Ban N (2014). The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515, 283–286. [DOI] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, and Kenyon C (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging cell 6, 95–110. [DOI] [PubMed] [Google Scholar]
- Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, and Ron D (2000). Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6, 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. (2009). Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460, 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes CM, and Ron D (2010). The mitochondrial UPR - protecting organelle protein homeostasis. Journal of cell science 123, 3849–3855. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, and Auwerx J (2013). Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Jia K, Chen D, and Riddle DL (2004). The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development (Cambridge, England) 131, 3897–3906. [DOI] [PubMed] [Google Scholar]
- Jovaisaite V, and Auwerx J (2015). The mitochondrial unfolded protein response-synchronizing genomes. Current opinion in cell biology 33, 74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, and Kennedy BK (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science (New York, NY) 310, 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, and Benzer S (2004). Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Current biology : CB 14, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai S, Yamazaki H, Tanji K, Engler MJ, Matsumiya T, and Itoh K (2019). Role of the ISR-ATF4 pathway and its cross talk with Nrf2 in mitochondrial quality control. Journal of clinical biochemistry and nutrition 64, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nature methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman M, Michels H, and Dancy BM (2016). A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. 11, 1798–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Luthi-Carter R, and Delorenzi M (2008). Cross-species and cross-platform gene expression studies with the Bioconductor-compliant R package ‘annotationTools’. BMC bioinformatics 9, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, Helgason GV, and Gottlieb E (2017). Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nature medicine 23, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N, and Martin W (2010). The energetics of genome complexity. Nature 467, 929–934. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, and Bachand F (2009). Cotranscriptional recruitment of the nuclear poly(A)-binding protein Pab2 to nascent transcripts and association with translating mRNPs. Nucleic acids research 37, 3418–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Li X, and Miller RA (2014). ATF4 activity: a common feature shared by many kinds of slow-aging mice. Aging cell 13, 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Borel C, Li L, Muller T, Williams EG, Germain PL, Buljan M, Sajic T, Boersema PJ, Shao W, et al. (2017). Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. 8, 1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, McIntyre R, Janssens G, Williams E, Lan J, van Veen H, van der Wel N, Mair WB, Aebersold R, MacInnes A, et al. (2019). Mitochondrial translation and dynamics synergistically extend lifespan in C. elegans through HLH-30. bioRxiv, 871079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes AW (2016). The role of the ribosome in the regulation of longevity and lifespan extension. Wiley interdisciplinary reviews RNA 7, 198–212. [DOI] [PubMed] [Google Scholar]
- Mazor KM, and Stipanuk MH (2016). GCN2- and eIF2alpha-phosphorylation-independent, but ATF4-dependent, induction of CARE-containing genes in methionine-deficient cells. Amino acids 48, 2831–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, and Zavolan M (2017). The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nature communications 8, 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaars M, Janssens GE, Santermans T, Lezzerini M, Jelier R, MacInnes AW, and Houtkooper RH (2018). Mitochondrial ubiquinone-mediated longevity is marked by reduced cytoplasmic mRNA translation. Life science alliance 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, et al. (2015). Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell reports. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan V, Ly T, Pourkarimi E, Murillo AB, Gartner A, Lamond AI, and Kenyon C (2016). Deep Proteome Analysis Identifies Age-Related Processes in C. elegans. Cell systems 3, 144–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwirth E (2014). RColorBrewer: ColorBrewer Palettes. R package version 1.1–2. https://CRAN.R-project.org/package=RColorBrewer.
- Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, and Kapahi P (2007). Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging cell 6, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SJ, Rost H, Rosenberger G, Collins BC, Malmstrom L, Amodei D, Venkatraman V, Raedschelders K, Van Eyk JE, and Aebersold R (2015). Identification of a Set of Conserved Eukaryotic Internal Retention Time Standards for Data-independent Acquisition Mass Spectrometry. Molecular & cellular proteomics : MCP 14, 2800–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Thomas G, and Volarevic S (2018). Ribosome biogenesis in cancer: new players and therapeutic avenues. Nature reviews Cancer 18, 51–63. [DOI] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirós PM, Prado MA, Zamboni N, D’amico D, Williams RW, Finley D, Gygi SP, and Auwerx J (2017). Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology 216, 2027–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Lahtinen T, Marttinen P, Myohanen M, Greco D, Cannino G, Jacobs HT, Lietzen N, Nyman TA, and Battersby BJ (2013). A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Current biology : CB 23, 535–541. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (Oxford, England) 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Gautier B, Singh A, and Le Cao KA (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS computational biology 13, e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost HL, Rosenberger G, Navarro P, Gillet L, Miladinovic SM, Schubert OT, Wolski W, Collins BC, Malmstrom J, Malmstrom L, et al. (2014). OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nature biotechnology 32, 219–223. [DOI] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, and Moorman AF (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic acids research 37, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. (2011). Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer cell 20, 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, and Ettema TJ (2007). Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic acids research 35, 4686–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout GJ, Stigter EC, Essers PB, Mulder KW, Kolkman A, Snijders DS, van den Broek NJ, Betist MC, Korswagen HC, Macinnes AW, et al. (2013). Insulin/IGF-1-mediated longevity is marked by reduced protein metabolism. Molecular systems biology 9, 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surguchov AP, Sudarickov AB, Telckov MV, Smirnov VN, Ter-Avanesyan MD, and Inge-Vechtomov SG (1983). Relationship between cytoplasmic and mitochondrial apparatus of protein synthesis in yeast Saccharomyces cerevisiae. Molecular & general genetics : MGG 189, 172–174. [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, and Sabatini DM (2012). A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, and Ruvkun G (1998). An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics 148, 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattem KM, and Wek RC (2004). Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 101, 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, and Muller F (2003). Genetics: influence of TOR kinase on lifespan in C. elegans. Nature 426, 620. [DOI] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, et al. (2016). Joint mouse-human phenome-wide association to test gene function and disease risk. Nature communications 7, 10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnes GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S, et al. (2016). gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1. https://CRAN.R-project.org/package=gplots. [Google Scholar]
- Wickham H (2009). ggplot2: Elegant Graphics for Data Analysis Springer-Verlag New York. [Google Scholar]
- Wieser W, and Krumschnabel G (2001). Hierarchies of ATP-consuming processes: direct compared with indirect measurements, and comparative aspects. The Biochemical journal 355, 389–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Wu Y, Jha P, Dubuis S, Blattmann P, Argmann CA, Houten SM, Amariuta T, Wolski W, Zamboni N, et al. (2016). Systems proteomics of liver mitochondria function. Science (New York, NY) 352, aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Wu Y, Wolski W, Kim JY, Lan J, Hasan M, Halter C, Jha P, Ryu D, Auwerx J, et al. (2018). Quantifying and Localizing the Mitochondrial Proteome Across Five Tissues in A Mouse Population. Molecular & cellular proteomics : MCP 17, 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel L, Topf U, Bragoszewski P, Wiese S, Sztolsztener ME, Oeljeklaus S, Varabyova A, Lirski M, Chroscicki P, Mroczek S, et al. (2015). Mistargeted mitochondrial proteins activate a proteostatic response in the cytosol. Nature 524, 485–488. [DOI] [PubMed] [Google Scholar]
- Wu Y, Williams EG, and Aebersold R (2017). Application of SWATH Proteomics to Mouse Biology. Current protocols in mouse biology 7, 130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Re-analyzed liver proteomics data from PXD003266, related to Figure 1
Table S2. Monosomal/polysomal RNA-seq and translational efficiency worm, related to Figure 2.
Table S3. Total RNA-seq worm, Related to Figure 3
Table S4. Proteome worm, Related to Figure 3
Table S7. RNA-seq mice, Related to Figure 6
Table S8. Cross species RNA-seq comparison, Related to Figure 6
Data Availability Statement
The accession number for the C. elegans and mice RNA sequencing data reported in this paper is GEO: GSE122097. The proteomics DIA data and the assembled library file are available on PRIDE under the ID PXD009223. The reference liver mouse transcriptomics and proteomics data examined in Figure 1 are available under GEO at GSE60149 and PRIDE at PXD003266. The reference multi-tissue mouse transcriptome data are available under GEO GSE60149, GSE60150, GSE60151, and GSE60489 and are from the same mice despite the non-continuous and separate entries. The multi-tissue proteomics data are from PXD005044. Note that the raw liver proteomics data from PXD003266 from 2016 were re-analyzed using the newer SWATH search library from PXD005044 from 2018, which was designed to improve coverage for mitochondrial proteins. These re-processed and normalized and processed mouse data are also available as Supplemental Table S1.