Abstract
We describe a case of carbapenem resistant E. coli isolated from urine in an 87-year-old woman with recurrent urinary tract infections. Using whole genome sequencing (WGS), we identified the carbapenem resistance mechanism to be a combination of ompC porin loss and plasmid-mediated AmpC gene blaCMY-2, which was not detected by routine molecular and phenotypic carbapenemase assays. Our case raises a concern for the limitation of current CRE screening tools for emerging resistance mechanisms and demonstrates the utility of WGS as a better tool for characterization of CRE in the clinical setting.
Abbreviations: AMR, Antimicrobial resistance; CRE, Carbapenem resistant Enterobacterales; mCIM, Modified carbapenem inactivation method; MHT, Modified Hodge test; Omp, Outer membrane porin; pAmpC, Plasmid-mediated AmpC; WGS, Whole genome sequencing
Keywords: Carbapenem resistant, Enterobacterales, E. coli, plasmid-mediated AmpC, Porin loss, OmpC, blaCMY-2
Introduction
Carbapenem resistant Enterobacterales (CRE) is a global concern in both the hospital and community setting. Patients with CRE infections are three times more likely to die compared to patients with non-CRE infections [1], with crude mortality estimated to be up to 70% [2]. Rapid and accurate identification of CRE is critical for infection prevention as well as guiding appropriate treatment. Once a patient has a confirmed CRE, additional precautions, including patient isolation, screening other at-risk patients, and tracking the spread of the CRE are warranted [2].
There are multiple resistance mechanisms for CRE including carbapenemase or β-lactamase with residual carbapenemase activity combined with concurrent porin deficiency [3,4]. The AmpC gene is mainly encoded in the chromosomal DNA of the many enteric bacteria including Serratia, Citrobacter, and Enterobacter, and its expression requires induction [5,6]. However, AmpC has increasingly been found constitutively expressed on plasmids in Klebsiella pneumoniae and Escherichia coli [5], contributing to carbapenem resistance [7,8]. While modified Hodge test (MHT), modified carbapenem inactivation method (mCIM) [9], and PCR targeting specific carbapenemase genes are commonly used to characterize carbapenem resistance mechanisms [10], there are currently no commercially available tests for organisms with resistance due to plasmid-mediated AmpC (pAmpC). In the U.S., pAmpC is predominately found in K. pneumoniae and E. coli. One study found pAmpC-producing K. pneumoniae in 42% of hospitals surveyed between 1996 and 2000 [11]. Another study found that 17.85% of cefoxitin-nonsusceptible [12] and 8.5% of oxyimino-β-lactam resistant K. pneumoniae [13] were AmpC producers. One children’s hospital in Washington State noted an increase in the incidence of pAmpC-producing strains of Enterobacterales between 1999 and 2007, with 39.2% of broad-spectrum β-lactam resistant E. coli isolates producing AmpC [14]. However, limited larger-scale surveillance data is available regarding pAmpC-producing E. coli in the U.S. Here we describe a case of carbapenem resistant E. coli isolated from urine where whole genome sequencing (WGS) identified a pAmpC.
Case Report
An 87-year-old woman presented to the emergency department with a complicated medical history including dementia, chronic constipation, paroxysmal atrial fibrillation, hypertension, persistent atrial flutter, recurrent urinary tract infections, and urinary incontinence resulting in intermittent catherization. Her current complaint was worsening abdominal pain and nausea. Upon examination, she was found to have cecal volvulus and underwent emergent exploratory laparotomy and large bowel resection. Her urine culture was positive for Pseudomonas aeruginosa and E. coli on hospital day 17. The patient was treated with nitrofurantoin and was discharged on hospital day 21 with improvement of symptoms.
The susceptibility of the E. coli isolate (designated EC01) was performed by broth microdilution method according to the Clinical and Laboratory Standards Institute (CLSI) reference using panels prepared in house. EC01 showed resistance to imipenem, ertapenem, and meropenem (Table 1). However, Xpert Carba-R (Cepheid, Sunnyvalle, CA) did not detect IMP, VIM, NDM, KPC, or OXA-48 carbapenemase genes. Moreover, carbapenemase activity was not detected by routine phenotypic carbapenemase assays MHT or mCIM, suggesting alternative carbapenem resistance mechanisms.
Table 1.
Antimicrobial Susceptibility Profile of EC01. Broth microdilution was used to determine susceptibility of EC01 to the antibiotics. The interpretive criteria were based on CLSI 2019 breakpoints. Cefazolin was used to predict susceptibility to oral cephalosporins.
| EC01 |
||
|---|---|---|
| Antimicrobial Agent | MIC (mcg/mL) | Interpretation |
| Amikacin | < = 4 | S |
| Ampicillin | >256 | R |
| Cefepime | 16 | R |
| Ceftazidime | >32 | R |
| Ceftazidime/Avibactam | 4 | S |
| Ceftolozane/Tazobactam | >32 | R |
| Ceftriaxone | >64 | R |
| Ciprofloxacin | >4 | R |
| Ertapenem | >4 | R |
| Gentamicin | < = 1 | S |
| Imipenem | >16 | R |
| Levofloxacin | >8 | R |
| Meropenem | 8 | R |
| Minocycline | 1 | S |
| Nitrofurantoin | < = 16 | S |
| Oral Cephalosporins | R | |
| Piperacillin + Tazobactam | >128 | R |
| Tobramycin | < = 1 | S |
| Trimethoprim/ Sulfamethoxazole | < = 1/20 | S |
WGS was performed using MiSeq (Illumina, San Diego, CA). Plasmid replicons, closely related strains, and antimicrobial resistance (AMR) genes were identified using Center for Genomic Epidemiology (CGE) tools (http://www.genomicepidemiology.org/). A Kmer tree was constructed using CLC Genomics Workbench (Qiagen, Valencia, CA, USA). Geneious (Biomatters, New Zealand) was used to perform sequence alignment analysis.
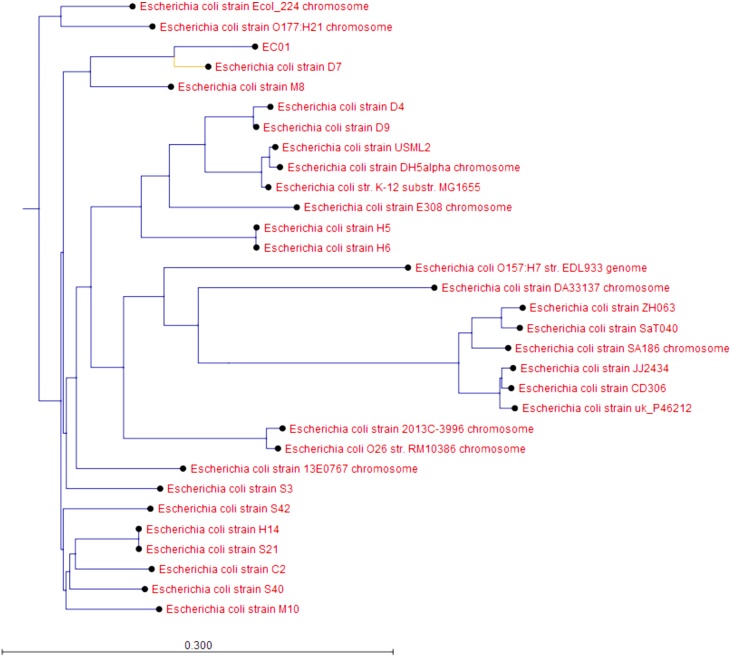
WGS analysis revealed that EC01 harbored multiple chromosomal point mutations (parC p.S80I, parE p.S458A, gyrA p.S83L, gyrA p.D87N) conferring resistance to fluoroquinolones. EC01 contained an average of 2.42 copies of IncI1-type plasmids per bacteria, which carried a pAmpC gene blaCMY-2. The most closely related strain was E. coli strain D7 (NZ_CP010150.1), with 99.4% pairwise identity and 97.1% coverage (Fig. 1).
Fig. 1.
EC01 is most closely related to E. coli strain D7. A Kmer tree was constructed using the most closely related strains as well as a large sampling of E. coli strains from around the world and derived from different sources.
To examine changes in the porins, the sequences of EC01 were aligned to wild-type E. coli strain K-12, which revealed that ompC in EC01 had a 12-bp deletion at position 524, a 12-bp insertion sequence (GGTACCATCGCT) at position 918, and a total of 44 point mutations throughout the gene. However, no point mutations, insertions, or deletions were identified in ompF.
Discussion
WGS was instrumental in characterizing a urinary E. coli isolate that was carbapenem resistant, but not detected by traditional carbapenemase assays. The mechanism for resistance was found to be a combination of a β-lactamase and outer membrane porin mutation. The AmpC β-lactamase, blaCMY-2, was found in EC01, which has been previously implicated in neonatal meningitis [15] and was associated with ceftriaxone resistance [16].
The most closely related strain, E. coli strain D7 (NZ_CP010150.1), was isolated from dog faeces in China. Interestingly, multiple studies have demonstrated shared plasmid-mediated blaCMY-2 between humans and animals. One study found that humans and poultry meat shared blaCMY-2, suggesting a potential food-borne exposure [17]. Plasmid-mediated blaCMY-2 was also identified in humans and their companion pets, with IncI1 as the most prominent plasmid type [18]. This plasmid was also found in our E. coli isolate, however, in our case, it is unclear whether the patient had any animal contact.
Outer membrane porin (Omp) mutations that decrease or obliterate the Omp protein level are often accompanied by AmpC production resulting in enhanced resistance to carbapenems [4,19]. For instance, decreased levels of OmpC and OmpF were associated with AmpC-mediated carbapenem resistant Enterobacter aerogenes [20]. We found multiple mutations and deletions in ompC. This highly mutated ompC is predicted to result in porin loss, which in conjunction with the plasmid-mediated blaCMY-2, explains the carbapenem resistance in our case. Notably this resistance mechanism (i.e., pAmpC + porin loss) has already been identified in both K. pneumoniae and E. coli previously [3,21]. However, the current prevalence of carbapenem resistant E. coli with this mechanism in the U.S. is still unknown.
Taken together, we identified an unusual carbapenem resistant E. coli with plasmid-mediated AmpC and porin loss, which was not detected by routine molecular and phenotypic carbapenemase assays currently used in clinical microbiology laboratories. As pAmpC can exhibit atypical susceptibility patterns and lead to confusion for clinicians, its timely and accurate detection is important for guiding appropriate treatment. In addition, close monitoring of pAmpC-mediated CRE is of great epidemiological value. Our case raises a concern for the limitation of current CRE screening tools and demonstrates the utility of WGS as a better method for characterization of CRE in the clinical setting.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Paige M.K. Larkin: Conceptualization, Methodology, Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Susan Realegeno: Investigation, Methodology, Writing - review & editing. Kevin W. Ward: Investigation, Methodology, Writing - review & editing. Omai B. Garner: Writing - review & editing. Shangxin Yang: Conceptualization, Methodology, Investigation, Formal analysis, Supervision, Writing - review & editing.
Declaration of Competing Interest
All authors declared no conflict of interest.
Acknowledgements
We would like to thank the UCLA Clinical Microbiology Bacteriology & Antimicrobial Bench staff for processing and working up this urine specimen and for recognizing the unique susceptibility profile of this isolate.
References
- 1.Martin A., Fahrbach K., Zhao Q., Lodise T. Association Between Carbapenem Resistance and Mortality Among Adult, Hospitalized Patients With Serious Infections Due to Enterobacteriaceae: Results of a Systematic Literature Review and Meta-analysis. Open Forum Infect Dis. 2018;5 doi: 10.1093/ofid/ofy150. ofy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman N.D., Carmeli Y., Walton A.L., Schwaber M.J. Carbapenem-Resistant Enterobacteriaceae: A Strategic Roadmap for Infection Control. Infect Control Hosp Epidemiol. 2017;38:580–594. doi: 10.1017/ice.2017.42. [DOI] [PubMed] [Google Scholar]
- 3.van Boxtel R., Wattel A.A., Arenas J., Goessens W.H., Tommassen J. Acquisition of Carbapenem Resistance by Plasmid-Encoded-AmpC-Expressing Escherichia coli. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.01413-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye Y., Xu L., Han Y., Chen Z., Liu C., Ming L. Mechanism for carbapenem resistance of clinical Enterobacteriaceae isolates. Exp Ther Med. 2018;15:1143–1149. doi: 10.3892/etm.2017.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philippon A., Arlet G., Jacoby G.A. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemother. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomson K.S. Extended-spectrum-beta-lactamase, AmpC, and Carbapenemase issues. J Clin Microbiol. 2010;48:1019–1025. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacoby G.A. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meini S., Tascini C., Cei M., Sozio E., Rossolini G.M. AmpC beta-lactamase-producing Enterobacterales: what a clinician should know. Infection. 2019;47:363–375. doi: 10.1007/s15010-019-01291-9. [DOI] [PubMed] [Google Scholar]
- 9.Pierce V.M., Simner P.J., Lonsway D.R., Roe-Carpenter D.E., Johnson J.K., Brasso W.B., Bobenchik A.M., Lockett Z.C., Charnot-Katsikas A., Ferraro M.J., Thomson R.B., Jr., Jenkins S.G., Limbago B.M., Das S. Modified Carbapenem Inactivation Method for Phenotypic Detection of Carbapenemase Production among Enterobacteriaceae. J Clin Microbiol. 2017;55:2321–2333. doi: 10.1128/JCM.00193-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Codjoe F.S., Donkor E.S. Carbapenem Resistance: A Review. Med Sci (Basel) 2017;6 doi: 10.3390/medsci6010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moland E.S., Black J.A., Ourada J., Reisbig M.D., Hanson N.D., Thomson K.S. Occurrence of newer beta-lactamases in Klebsiella pneumoniae isolates from 24 U.S. hospitals. Antimicrob Agents Chemother. 2002;46:3837–3842. doi: 10.1128/AAC.46.12.3837-3842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coudron P.E., Hanson N.D., Climo M.W. Occurrence of extended-spectrum and AmpC beta-lactamases in bloodstream isolates of Klebsiella pneumoniae: isolates harbor plasmid-mediated FOX-5 and ACT-1 AmpC beta-lactamases. J Clin Microbiol. 2003;41:772–777. doi: 10.1128/JCM.41.2.772-777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez M., Tran J.H., Chow N., Jacoby G.A. Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother. 2004;48:533–537. doi: 10.1128/AAC.48.2.533-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qin X., Zerr D.M., Weissman S.J., Englund J.A., Denno D.M., Klein E.J., Tarr P.I., Kwong J., Stapp J.R., Tulloch L.G., Galanakis E. Prevalence and mechanisms of broad-spectrum beta-lactam resistance in Enterobacteriaceae: a children’s hospital experience. Antimicrob Agents Chemother. 2008;52:3909–3914. doi: 10.1128/AAC.00622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fakioglu E., Queenan A.M., Bush K., Jenkins S.G., Herold B.C. Amp C beta-lactamase-producing Escherichia coli in neonatal meningitis: diagnostic and therapeutic challenge. J Perinatol. 2006;26:515–517. doi: 10.1038/sj.jp.7211550. [DOI] [PubMed] [Google Scholar]
- 16.Tamma P.D., Shahara S.L., Pana Z.D., Amoah J., Fisher S.L., Tekle T., Doi Y., Simner P.J. Molecular Epidemiology of Ceftriaxone Non-Susceptible Enterobacterales Isolates in an Academic Medical Center in the United States. Open Forum Infect Dis. 2019;6 doi: 10.1093/ofid/ofz353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voets G.M., Fluit A.C., Scharringa J., Schapendonk C., van den Munckhof T., Leverstein-van Hall M.A., Stuart J.C. Identical plasmid AmpC beta-lactamase genes and plasmid types in E. coli isolates from patients and poultry meat in the Netherlands. Int J Food Microbiol. 2013;167:359–362. doi: 10.1016/j.ijfoodmicro.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Bortolaia V., Hansen K.H., Nielsen C.A., Fritsche T.R., Guardabassi L. High diversity of plasmids harbouring blaCMY-2 among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J Antimicrob Chemother. 2014;69:1492–1496. doi: 10.1093/jac/dku011. [DOI] [PubMed] [Google Scholar]
- 19.Logan L.K., Weinstein R.A. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu J., Taga T., Sato T., Eguchi Y. Sepsis in a 4-Month-Old Boy Due to Carbapenem-Resistant Enterobacteriaceae Characterized by AmpC beta-Lactamase with Porin Loss. Int J Appl Basic Med Res. 2018;8:263–265. doi: 10.4103/ijabmr.IJABMR_383_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee K., Yong D., Choi Y.S., Yum J.H., Kim J.M., Woodford N., Livermore D.M., Chong Y. Reduced imipenem susceptibility in Klebsiella pneumoniae clinical isolates with plasmid-mediated CMY-2 and DHA-1 beta-lactamases co-mediated by porin loss. Int J Antimicrob Agents. 2007;29:201–206. doi: 10.1016/j.ijantimicag.2006.09.006. [DOI] [PubMed] [Google Scholar]