Abstract
Background & Aims
Nonalcoholic steatohepatitis (NASH) occurs in the context of aberrant metabolism. Glutaminolysis is required for metabolic reprograming of hepatic stellate cells (HSCs) and liver fibrogenesis in mice. However, it is unclear how changes in HSC glutamine metabolism contribute to net changes in hepatic glutaminolytic activity during fibrosis progression, or whether this could be used to track fibrogenic activity in NASH. We postulated that increased HSC glutaminolysis marks active scarring in NASH.
Methods
Glutaminolysis was assessed in mouse NASH fibrosis models and in NASH patients. Serum and liver levels of glutamine and glutamate and hepatic expression of glutamine transporter/metabolic enzymes were correlated with each other and with fibrosis severity. Glutaminolysis was disrupted in HSCs to examine if this directly influenced fibrogenesis. 18F-fluoroglutamine positron emission tomography was used to determine how liver glutamine assimilation tracked with hepatic fibrogenic activity in situ.
Results
The serum glutamate/glutamine ratio increased and correlated with its hepatic ratio, myofibroblast content, and fibrosis severity. Healthy livers almost exclusively expressed liver-type glutaminase (Gls2); Gls2 protein localized in zone 1 hepatocytes, whereas glutamine synthase was restricted to zone 3 hepatocytes. In fibrotic livers, Gls2 levels reduced and glutamine synthase zonality was lost, but both Slc1a5 (glutamine transporter) and kidney-type Gls1 were up-regulated; Gls1 protein was restricted to stromal cells and accumulated in fibrotic septa. Hepatocytes did not compensate for decreased Gls2 by inducing Gls1. Limiting glutamine or directly inhibiting GLS1 inhibited growth and fibrogenic activity in cultured human HSCs. Compared with healthy livers, fibrotic livers were 18F-fluoroglutamine-avid by positron emission tomography, suggesting that glutamine-addicted myofibroblasts drive increased hepatic utilization of glutamine as fibrosis progresses.
Conclusions
Glutaminolysis is a potential diagnostic marker and therapeutic target during NASH fibrosis progression.
Keywords: Liver Diseases, Biomarker, Metabolomics, Amino Acid
Abbreviations used in this paper: α-KG, α-ketoglutarate; αSMA, α-smooth muscle actin; BMI, body mass index; CDAA-HFD, choline-deficient, L-amino acid–defined high-fat diet; CT, computed tomography; GLS, glutaminase; GLS1, kidney-type glutaminase; GLS2, liver-type glutaminase; GS, glutamine synthase; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HSC, hepatic stellate cell; IHC, immunohistochemistry; IRB, Institutional Review Board; MCDE, methionine/choline-deficient diet supplemented with 0.15% ethionine; MF, myofibroblast; mRNA, messenger RNA; MS, mass spectrometry; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; PET, positron emission tomography; siRNA, small interfering RNA; TCA, tricarboxylic acid; UHPLC, ultrahigh performance liquid chromatography
Summary.
Serum glutamate/glutamine increased and correlated with its hepatic ratio and fibrosis severity in nonalcoholic steatohepatitis fibrosis. These changes reflect increased glutaminolysis in myofibroblasts and are shown by noninvasive positron emission tomography. Increased glutaminolysis is a potential diagnostic marker and therapeutic target during nonalcoholic steatohepatitis fibrosis progression.
Nonalcoholic fatty liver disease (NAFLD) is the hepatic manifestation of the metabolic syndrome, a systemic disorder of energy homeostasis.1 However, relatively little is known about how dysregulated energy homeostasis in the metabolic syndrome impacts metabolic reprogramming responses that are needed for wound healing in injured livers, or how wound healing–related metabolic reprogramming contributes to the disordered systemic energy homeostasis that defines the metabolic syndrome. Because effective wound healing responses are required to fully recover from NAFLD, clarification of the key processes may show new diagnostic and therapeutic targets in NAFLD and progressive liver fibrosis.
Liver disease–related morbidity and mortality in NAFLD are largely restricted to individuals who develop progressive fibrosis that results in cirrhosis.2 Hence, current efforts to reduce NAFLD mortality focus on improving early identification of patients at high risk for progressive fibrosis, and on developing effective therapeutics to prevent and reverse liver fibrosis. Whether or not repair-related shifts in metabolism might be targeted to improve fibrosis detection or constrain fibrosis progression is still largely unknown. However, this concept is plausible given that NAFLD cirrhosis is characterized by the excessive accumulation of fibrogenic myofibroblasts derived from hepatic stellate cells (HSCs).3 Liver injury stimulates HSCs to be reprogrammed (ie, to transdifferentiate) from their quiescent state to become proliferative, migratory, and fibrogenic myofibroblasts (MFs). Metabolic reprogramming must be maintained to satisfy the high energy demands of such cells.4,5 Otherwise, MF HSCs die or revert back to a less fibrogenic, more quiescent state. Transient accumulation of MF HSCs is necessary for effective liver regeneration, but excessive accumulation of such cells causes progressive fibrosis, defective repair, and, ultimately, cirrhosis.2 Because liver injury outcome is dictated by factors that control the size of MF-HSC populations, the mechanisms that orchestrate HSC reprogramming are attractive therapeutic targets.
We recently reported that murine MF HSCs are addicted to glutamine,5 the most abundant amino acid in mammalian plasma. Glutamine carries both nitrogen and carbon moieties and plays a critical role in various biological functions including protein and lipid biosynthesis, acid–base balance, and cellular energy homeostasis. Increased glutamine catabolism (glutaminolysis) is a key metabolic characteristic of rapidly proliferating cells, including cancer cells, and glutaminolysis inhibition has been proposed as a cancer treatment.6 Glutamine metabolism is controlled primarily by 2 enzymes: glutaminase (Gls; for glutamine catabolism) and glutamine synthetase (GS; for glutamine anabolism). The 2 tissue-specific glutaminase isoforms: kidney-type glutaminase (known as Gls1) and liver-type glutaminase (known as Gls2), are the products of distinct genes on different chromosomes and show significant differences in their respective affinities for glutamine (Gls1 > Gls2).7 High-affinity Gls (Gls1) are not detectable in healthy adult livers, whereas low-affinity Gls (Gls2) are strongly expressed and active in periportal hepatocytes, where they generate glutamate and release ammonia for urea synthesis.7,8 In contrast, GS is strictly localized to pericentral hepatocytes, where it condenses residual glutamate and ammonia to regenerate glutamine.9 Altered expression of either Gls or GS impacts hepatic glutamate–glutamine homeostasis. A recent study showed that plasma glutamate is increased in NAFLD patients, and reported that the glutamate/(serine + glycine) index positively correlated with hepatocellular ballooning and inflammation, and could discriminate mild from severe fibrosis.10 However, the sample size was relatively small in that cross-sectional study, and whether the findings can be confirmed in a prospective study with larger sample size is still unknown. The underlying mechanisms for altered glutamate levels and associated correlations also are unclear, but may be informed by evidence that Gls1 is induced and required for liver fibrosis in murine models of liver fibrosis.5,11 Because myofibroblastic HSCs require glutaminolysis and selectively up-regulate Gls1 expression to satisfy their bioenergetic/biosynthetic demands,5,11 we hypothesize that Gls1 induction critically modifies glutamine utilization in fibrosing nonalcoholic steatohepatitis (NASH) progression and that the associated change in glutamine metabolism is a potential diagnostic marker and therapeutic target for NASH.
Results
Increased Serum Glutamate/Glutamine Ratio Reflects Increased Glutaminolytic Activity and Fibrogenic Activity in Injured Livers
Induction of glutaminolysis is a critical component of the metabolic reprogramming that is necessary to fuel the growth of highly proliferative cells during tissue repair and regeneration because the conversion of glutamine into α ketoglutarate (α-KG) (ie, glutaminolysis) enhances tricarboxylic acid (TCA) cycle activity, enabling adenosine triphosphate production while providing precursors for new biomass synthesis.6 Although it was reported that serum glutamate is increased in NASH fibrosis patients,10,12 it is not known if/how hepatic glutaminolytic activity is altered in fibrosis and NASH progression.
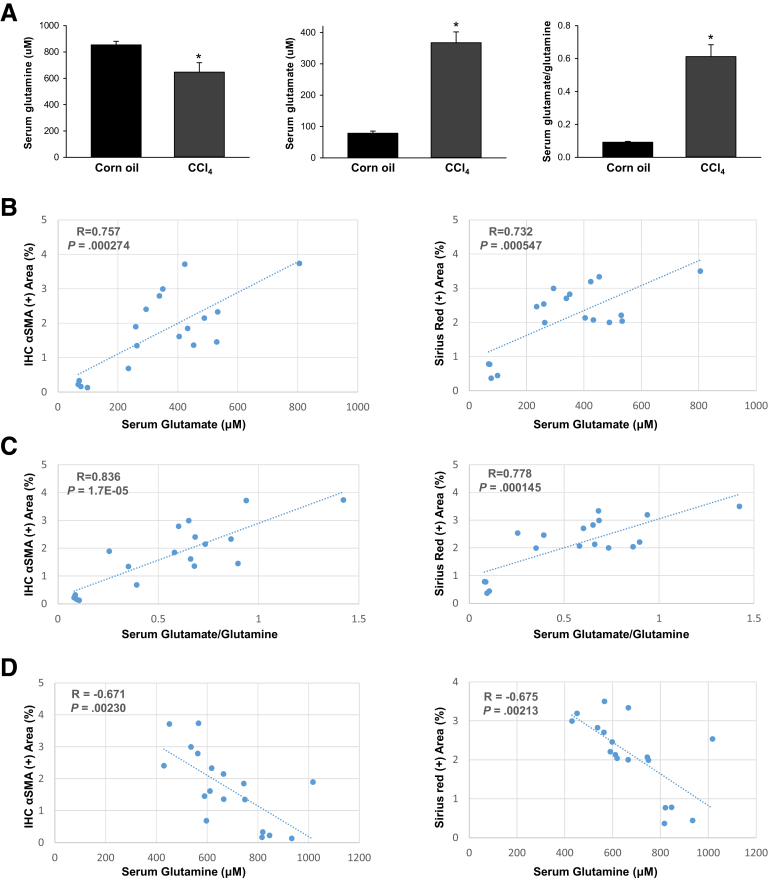
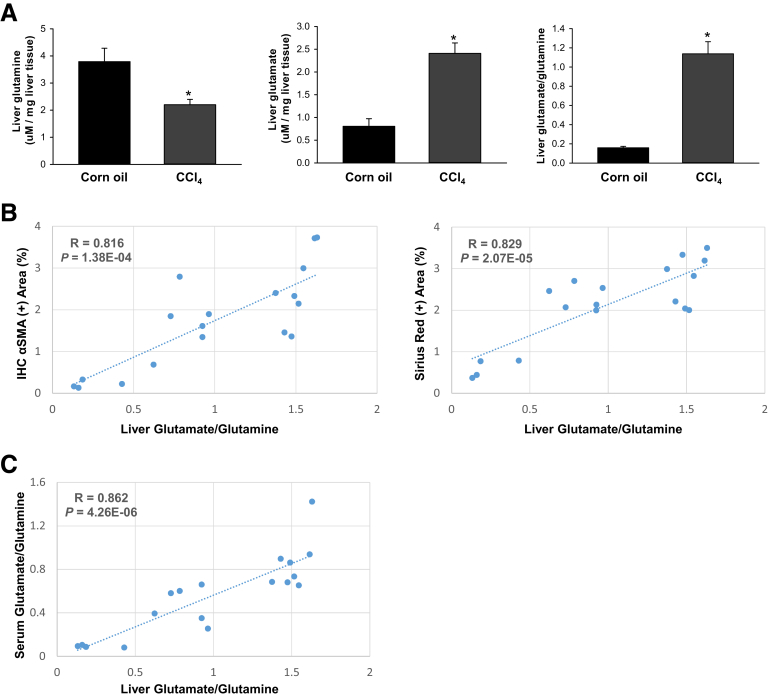
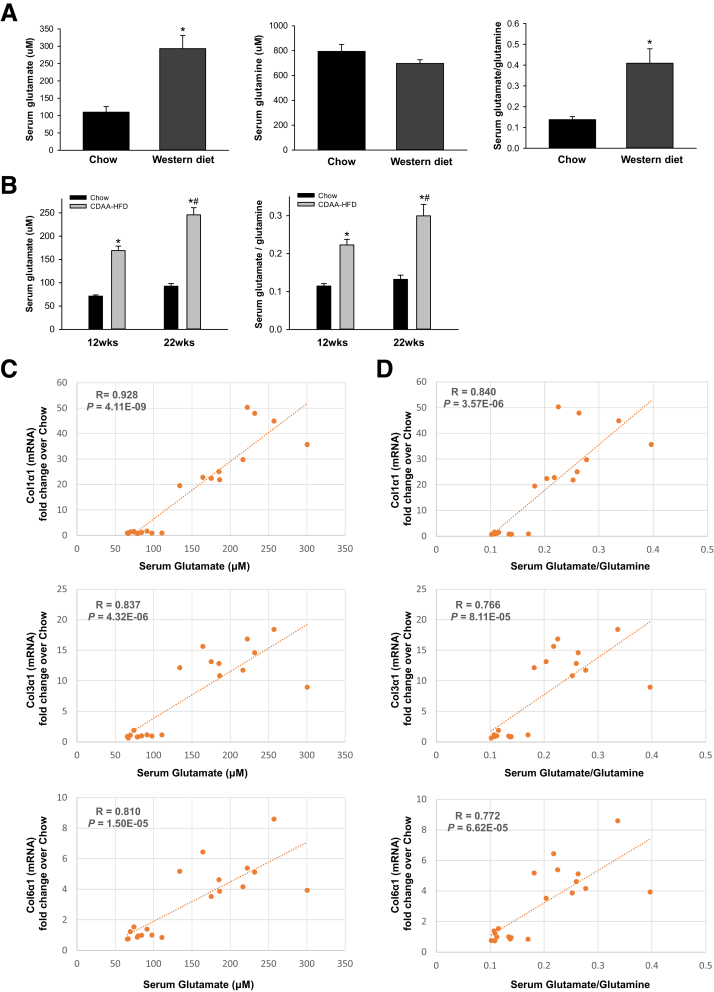
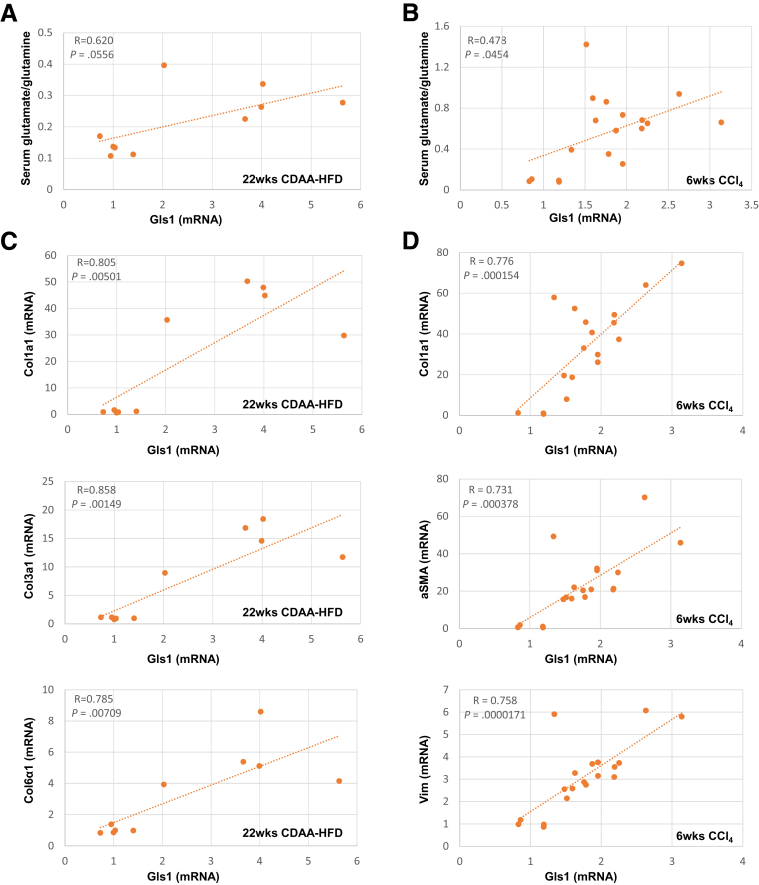
To address this question, we first compared serum glutamate and glutamine levels in healthy mice and mice that were chronically treated with CCl4, a direct hepatotoxin that is known to evoke robust fibrotic responses in the liver.5 Serum glutamine levels slightly decreased although glutamate levels increased, and, thus, the ratio of glutamate/glutamine increased significantly in CCl4-treated mice (Figure 1A). We then evaluated how glutamate levels and the glutamate/glutamine ratio in sera correlate with markers of fibrosis in injured livers. We found that the serum glutamate concentration (Figure 1B) and glutamate/glutamine ratio (Figure 1C) correlated strongly with hepatic accumulation of myofibroblasts and fibrosis severity, as assessed by α smooth muscle actin (αSMA) immunohistochemistry (IHC) and Sirius red staining. Conversely, serum glutamine levels were correlated negatively with these fibrosis markers (Figure 1D). Fibrotic livers themselves had lower glutamine concentrations, higher glutamate concentrations, and higher glutamate/glutamine ratios than nonfibrotic livers (Figure 2A). Furthermore, the liver glutamate/glutamine ratio strongly correlated with the severity of myofibroblast accumulation and fibrosis (Figure 2B), as well as the glutamate/glutamine ratio in serum (Figure 2C). Thus, the aggregate data in this mouse model of CCl4-induced liver fibrosis suggest that changes in liver glutamine homeostasis may have contributed to the altered circulating levels of glutamate and glutamine that were reported to occur in patients with NASH fibrosis.10,12 To address this issue more directly, we compared serum glutamate/glutamine levels in obese, insulin-resistant mice with NASH caused by chronic consumption of either a Western diet for 16 weeks (Figure 3A) or a choline-deficient, L-amino acid–defined high-fat diet (CDAA-HFD) for 12 or 22 weeks (Figure 3B). Serum glutamine levels were decreased slightly, whereas glutamate levels and glutamate/glutamine ratios were approximately 2- to 3-fold higher in these mouse models of NASH than in the respective control groups (Figure 3A and B). As in the CCl4 liver fibrosis model, changes in circulating levels of glutamate and glutamine reflected changes in liver fibrosis severity in the NASH models. For example, in mice fed CDAA-HFD, serum levels of glutamate and the serum glutamate/glutamine ratio increased in parallel with hepatic messenger RNA (mRNA) expression of Col1α1, Col3α1, and Col6α1, and were highly correlated with these fibrosis markers (Figure 3C and D). Collectively, these results show that glutamate–glutamine homeostasis is altered in fibrosis induced by NASH and other types of chronic liver injury, justifying further investigation to determine the mechanism for, and role of, glutaminolysis in NASH-related liver fibrosis.
Figure 1.
Serum levels of glutamate, glutamine, and glutamate/glutamine change and correlation with fibrosis in CCl4-injured livers. Mice were treated with corn oil vehicle or CCl4 for 6 weeks. (A) Serum levels of glutamine, glutamate, and glutamate/glutamine. Bars represent means ± SEM of n = 4–7 mice/group. *P < .05 vs corn oil vehicle. (B–D) Correlative analysis of serum level of glutamate, glutamate/glutamine, and glutamine with αSMA IHC positively stained area (in percentages) or Sirius red positively stained area (in percentages) in liver sections. R, correlation coefficient.
Figure 2.
Increased serum glutamate/glutamine ratio reflects increased glutaminolytic activity and fibrosis in CCl4-injured livers. Mice were treated with corn oil vehicle or CCl4 for 6 weeks. (A) Hepatic levels of glutamine, glutamate, and glutamate/glutamine. Bars represent means ± SEM of n = 4–7 mice/group. *P < .05 vs corn oil vehicle. (B) Correlative analysis of hepatic levels of glutamate/glutamine with αSMA IHC positively stained area (in percentages) or Sirius red positively stained area (in percentages) in liver sections. (C) Correlative analysis of liver vs serum glutamate/glutamine levels. R, correlation coefficient.
Figure 3.
Increased serum glutamate/glutamine ratio correlates with fibrosis severity in NASH-injured livers. Mice were treated with chow diet or Western diet for 16 weeks, or CDAA-HFD for 12 or 22 weeks. (A and B) Serum levels of glutamine, glutamate, and glutamate/glutamine were measured in these mice. Bars represent means ± SEM of n = 4–8 mice/group. *P < .05 vs chow diet or corn oil vehicle. #P < .05 vs CDAA-HFD group at 12 weeks. (C and D) Correlative analysis of serum levels of glutamate or glutamate/glutamine with fibrogenic marker Col1α1, Col3α1, or Col6α1 mRNA expression in the liver. R, correlation coefficient.
Glutamine Metabolism Is Differentially Regulated in Hepatocytes and Myofibroblasts During Liver Fibrosis
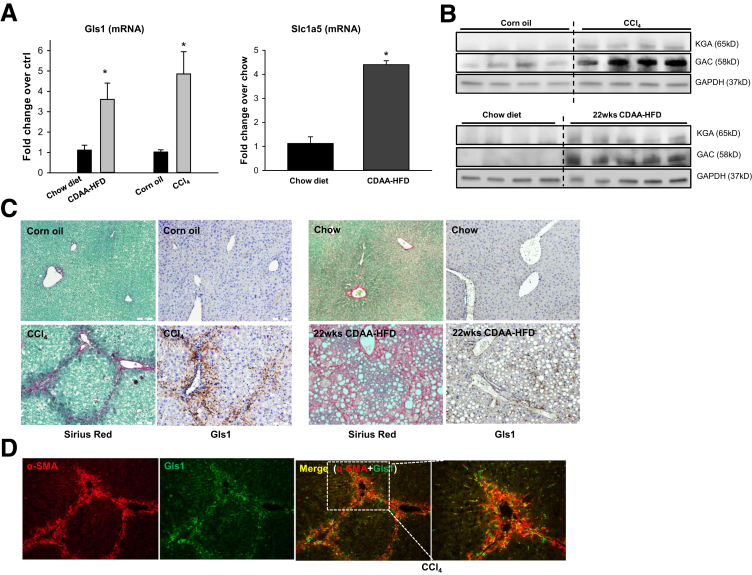
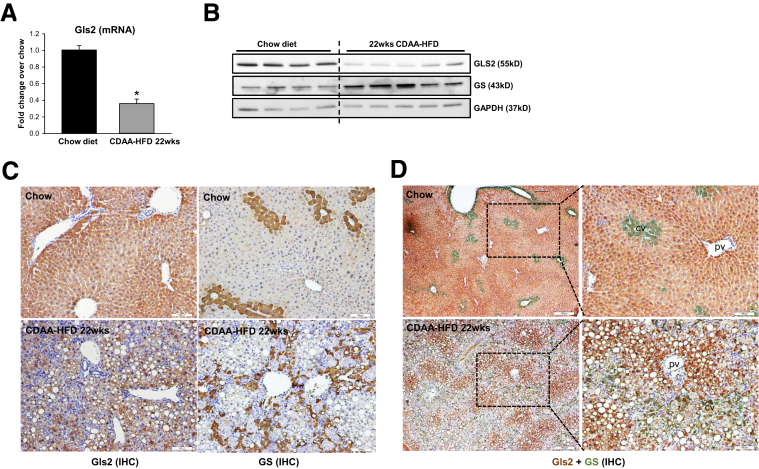
The conversion of glutamine to glutamate can be catalyzed by a high-affinity glutaminase (Gls1) or a low-affinity glutaminase (Gls2). We previously showed that mRNA levels of Gls1 doubled, whereas Gls2 mRNAs decreased by more than 50%, in livers of mice treated with a methionine/choline-deficient diet to induce steatohepatitis.5 Interestingly, we also observed that the glutamine transporter Slc1a5 increased in these mice (data not shown), suggesting that glutamine uptake increases in chronically injured livers. Mice that developed mild liver fibrosis during Western diet feeding also showed mild (approximately 1.7-fold) but significant (P = .042) increases in Gls1 mRNA levels relative to chow-fed controls. Here, we show that net mRNA expression of Slc1a5 and Gls1 also increased in models of liver fibrosis caused by feeding mice CDAA-HFD, or by treating mice chronically with CCl4 (Figure 4A). More importantly, immunoblot analysis showed that the proteins encoded by both Gls1 mRNA splice variants, kidney-type glutaminase and Glutaminase C, increased in fibrotic livers (Figure 4B). Immunohistochemistry confirmed these results, showing that Gls1 is barely detected in control mice but robustly accumulates in fibrotic areas during both NASH- and CCl4-induced liver fibrosis (Figure 4C). Furthermore, we found that Gls1 strongly colocalized with αSMA in fibrotic septa (Figure 4D), supporting previous evidence that the Gls1 isoenzyme is expressed strongly by fibrogenic myofibroblasts.5,11 Importantly, we found that Gls1 mRNA levels correlated with the serum glutamate/glutamine ratio (Figure 5A and B) and with hepatic fibrosis markers (Figure 5C and D) in both CCl4- and CDAA-HFD–damaged livers, complementing the other evidence that serum and liver glutamate/glutamine ratios parallel measures of liver fibrosis (Figure 1, Figure 2, Figure 3). Conversely, we found that net hepatic expression of both Gls2 mRNA and protein decreased in mice with NASH fibrosis (Figure 6A and B). Immunohistochemistry confirmed that Gls2 localizes to zone 1 (periportal) hepatocytes in healthy livers and showed that its expression decreased and became more diffuse in fibrotic livers (Figure 6C and D). Hence, because Gls1 has a much higher affinity for glutamine than Gls2,7,13 induction of Gls1 most likely is responsible for increasing glutaminolytic activity during liver repair. Expression of GS, the enzyme that catalyzes resynthesis of glutamine from glutamate and ammonia, also changed in injured livers, but this response was more variable (Figure 6B). In healthy livers, GS expression was restricted to hepatocytes surrounding terminal hepatic venules (Figure 6C and D). This zonal pattern of expression was lost during NASH-induced liver fibrosis, as evidenced by more diffuse GS staining (Figure 6C and D).
Figure 4.
Gls1 is increased in myofibroblasts and correlates with liver fibrosis during liver injury. Mice were treated with chow diet or CDAA-HFD for 12 or 22 weeks, or corn oil vehicle or CCl4 for 4–6 weeks. (A) Liver Gls1 and Slc1a5 mRNA was quantified by reverse-transcription polymerase chain reaction. Bars represent means ± SEM of n = 4–5 mice/group. *P < .05 vs chow diet or corn oil control. (B) Protein expression of Gls1 (both variants: KGA and GAC) measured by Western blot. (C) Sirius red staining and IHC staining of Gls1 in livers of mice treated with corn oil vehicle or CCl4, or chow diet or CDAA-HFD for 22 weeks. (D) αSMA and Gls1 co-immunofluorescent staining in livers from mice treated with CCl4. GAC, Glutaminase C; KGA, kidney-type glutaminase.
Figure 5.
Increased Gls1 mRNA correlates with serum glutamate/glutamine ratios and liver fibrosis during liver injury. Mice were treated with chow diet or CDAA-HFD for 12 or 22 weeks, or corn oil vehicle or CCl4 for 6 weeks. (A and B) mRNA of Gls1 was correlated with serum glutamate/glutamine ratios. (C and D) mRNA of Gls1 was correlated with the expression level of fibrosis markers (αSMA, Col1α1, Col3α1, Col6α1, and vimentin). R, correlation coefficient.
Figure 6.
Gls2 is decreased while GS becomes more diffuse in hepatocytes in NASH-injured livers. Mice were treated with chow diet or CDAA-HFD for 22 weeks. (A) Liver Gls2 mRNA was quantified by reverse-transcription polymerase chain reaction. Bars represent means ± SEM of n = 5 mice/group. *P < .05 vs chow diet. (B) Protein expression of Gls2 and GS was measured by Western blot. (C) IHC staining of Gls2 and GS in chow or CDAA-HFD diet–treated mice. (D) Representative co-IHC staining of Gls2 (brown) and GS (green) in mice treated with chow diet or CDAA-HFD for 22 weeks. cv, central vein; pv, portal vein.
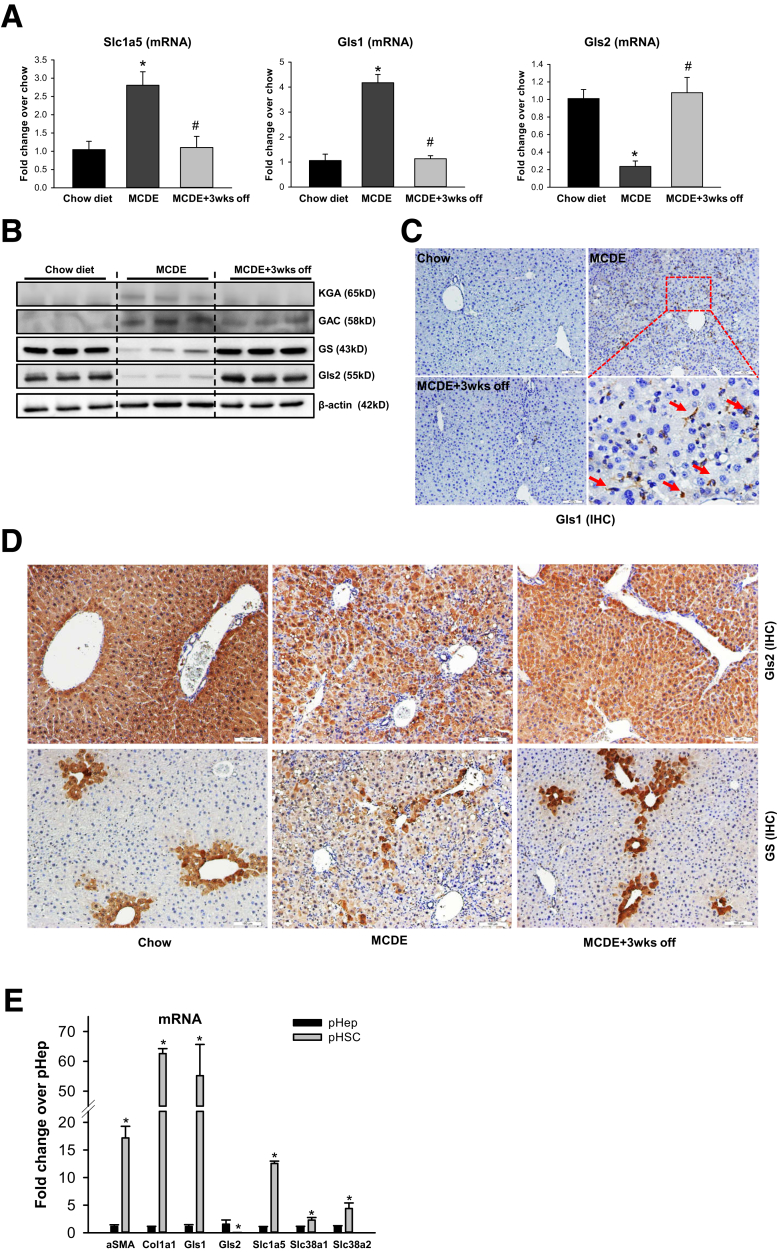
To further clarify how changes in hepatic glutamine catalysis and resynthesis relate to net changes in fibrogenic activity, we studied mice that were fed methionine-choline–deficient diets supplemented with ethionine (MCDE). The MCDE diet rapidly increases hepatic fibrogenesis, but this quickly abates when the diet is withdrawn, such that very little fibrosis remains 3 weeks after mice have been returned to regular chow.14 As shown in Figure 7A–D, changes in Slc1a5, Gls1, Gls2, and GS tightly track with changes in fibrogenic activity. When fibrogenic activity is high, Slc1a5 and Gls1 expression is increased, and expression of Gls2 and GS is decreased. Conversely, as fibrogenic activity decreases and fibrosis dissipates, levels of Slc1a5 and Gls1 decrease and Gls2 and GS recover back to their baseline expression levels. These results show that the dysregulation of glutamine metabolism is dynamic and reversible in liver fibrosis.
Figure 7.
Dysregulation of glutamine metabolism is dynamic and reversible in liver fibrosis. Mice were treated with chow diet or MCDE diet for 3 weeks. The MCDE diet was removed in some mice for 3 weeks after 3 weeks of treatment. (A) mRNA of Slc1a5, Gls1, and Gls2 was measured by reverse-transcription polymerase chain reaction. Bars represent means ± SEM of n = 3 mice/group. *P < .05 vs chow diet; #P < .05 vs MCDE group. (B) Protein expression of Gls1 (both variants: KGA and GAC), GS, and Gls2 was measured by Western blot. (C) Representative IHC staining of Gls1 in mice treated with chow or MCDE diet for 3 weeks. Red arrows indicate positively stained cells. (D) Representative IHC staining of GS and Gls2 in mice treated with chow or MCDE diet for 3 weeks. (E) mRNA expression was measured by reverse-transcription polymerase chain reaction in freshly isolated primary mouse hepatocytes and hepatic stellate cells. Bars represent means ± SEM of n = 3–4 assays. *P < .05 vs hepatocyte. GAC, Glutaminase C; KGA, kidney-type glutaminase; pHep, primary hepatocyte; pHSC, primary hepatic stellate cell.
Collectively, these strong links between Gls1 induction, Gls2 reduction, increased glutaminolysis, and fibrogenesis suggest that fibrogenic myofibroblasts become major glutamine consumers (and glutamate producers) in damaged livers. This conclusion also is supported by comparative analysis of glutaminase and glutamine transporter gene expression in freshly isolated primary stellate cells (the main source of fibrogenic MFs in NASH) and hepatoctyes (the main target of lipotoxicity in NASH). Stellate cells express higher levels of several glutamine transporters, including Slc1a5, and 50 times more Gls1 mRNA than hepatocytes (Figure 7E), but do not express Gls2 either when quiescent or myofibroblastic (Figures 6C and D, and 7D). In contrast, hepatocytes express mainly Gls2 mRNA (Figure 7E), and immunohistochemistry localizes Gls2 protein to hepatocytes in healthy livers (Figures 6C and D, and 7D). During liver repair, net hepatic Gls2 expression decreases (Figures 6, and 7B and D) and it is not evident that hepatocytes compensate for this by inducing Gls1 (Figures 4C and D, and 7C). Thus, although Gls2-expressing hepatocytes may be major glutamine consumers in healthy livers where fibrogenic MFs are rare, hepatocyte glutamine utilization seems to be compromised in injured livers by suppression of Gls2 and accumulation of MFs that express Gls1, the more efficient glutaminolytic enzyme.7,13
Glutaminolytic Activity Is Increased in NASH Patients and Correlates With Fibrosis Severity
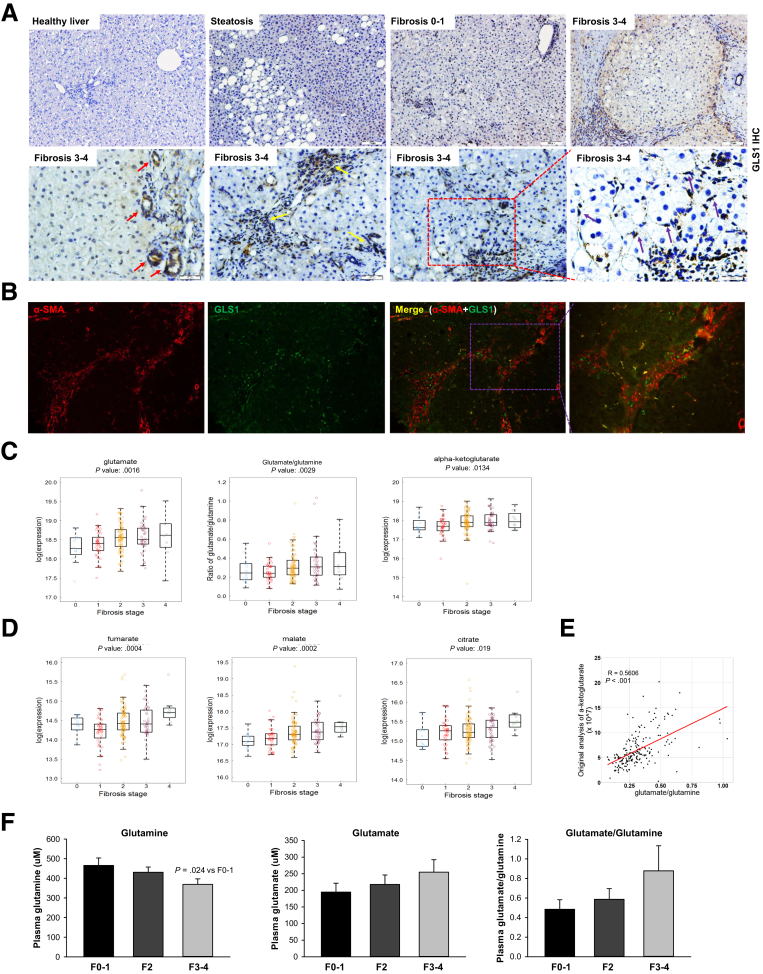
To clarify whether findings in our murine models might be relevant to human beings, we re-examined our published gene expression microarray data from more than 70 biopsy-proven NASH patients with varying degrees of liver fibrosis.15 The data showed that GLS1 expression is 1.6-fold higher (P = 1.10E-07) in the group of patients with advanced (F3–4) liver fibrosis (n = 32) than in those with mild (F0–1) liver fibrosis (n = 40). Previously, we showed that Gls1 mRNAs increase during NAFLD progression.5 Here, our immunohistochemistry shows that GLS1 protein also increases with the severity of liver fibrosis (Figure 8A), and shows that the increased GLS1 localizes mostly to areas of scarring (Figure 8A). Double-immunofluorescence staining for GLS1 and αSMA show that GLS1 localizes to myofibroblasts within fibroductular septa in human NASH (Figure 8B).
Figure 8.
Glutaminolytic activity is increased in fibrotic patients and correlates with fibrosis severity. (A) GLS1 expression was assessed by IHC in NAFLD patients with varying degrees of liver fibrosis. Red arrows indicate the positively stained ductal cells, yellow arrows indicate the positively stained immune cells, and purple arrows indicate the positively stained stromal cells such as MF-HSCs. (B) GLS1 and αSMA co-stained liver sections from fibrosing NASH patients. (C and D) Distribution of glutamate, glutamate/glutamine, α-KG, fumarate, malate, and citrate in serum samples of staged fibrotic NAFLD patients (F0, 12; F1, 38; F2, 100; F3, 42; and F4, 8). The top and bottom of the box are the third and first quartiles, respectively. The middle line of the box is the median. P values of univariate ordinal logistic regression analyses are shown on the top. (E) Pearson correlation analysis of glutamate/glutamine and α-KG in NAFLD patients. (F) Plasma levels of glutamine, glutamate, and glutamate/glutamine were analyzed in an independent cohort of 40 biopsy-proved fibrotic patients with HIV/HCV infections (F0–1, 11; F2, 15; and F3–4, 14). Bars represent means ± SEM. P value was calculated using an unpaired Student t test.
We also performed metabolomics analysis of serum samples from a separate cohort of 200 biopsy-proven NAFLD patients. By using univariate ordinal logistic regression analysis, we showed that glutamate, glutamate/glutamine, α-KG (Figure 8C), and their downstream metabolites in the TCA cycle (eg, fumarate, malate, citrate) (Figure 8D) all increased as fibrosis severity worsened. Furthermore, the serum glutamate/glutamine ratio strongly correlated with α-KG levels in these NAFLD patients (Figure 8E), suggesting that glutaminolysis fuels α-KG production for the TCA cycle. These increases remain significant even after regression analysis to control for multiple known confounders, such as age, sex, body mass index (BMI), diabetes, hypertension, past/current alcohol, and past/current smoking (Table 1). Interestingly, as we noted in NAFLD patients, plasma glutamine decreased, whereas glutamate and the glutamate/glutamine ratio tended to increase, with fibrosis progression in an independent cohort of 40 human immunodeficiency virus (HIV)/hepatitis C virus (HCV)-infected patients with biopsy-proven liver fibrosis (Figure 8F). Thus, data in human beings replicate data in mice, which together indicate that increased glutaminolytic activity is a conserved response that accompanies liver fibrosis progression.
Table 1.
Association Analysis Between Amino Acids and Fibrosis Stage Using Ordinal Logistic Model, With or Without Controlling for Age, Sex, BMI, Diabetes, Hypertension, Past/Current Alcohol, and Past/Current Smoking
| Outcomes | Amino acid | Covariates | β | SE | P value | FDR |
|---|---|---|---|---|---|---|
| Fibrosis stage, F0, F1, F2, F3, F4 | Glutamate | Univariate | 1.1119 | 0.3531 | .0016 | 0.0043 |
| Glutamine | Univariate | -0.4065 | 0.8139 | .6175 | 0.6175 | |
| α-KG | Univariate | 0.6251 | 0.2528 | .0134 | 0.0214 | |
| Fumarate | Univariate | 1.2280 | 0.3487 | .0004 | 0.0016 | |
| Malate | Univariate | 1.3767 | 0.3640 | .0002 | 0.0016 | |
| Citrate | Univariate | 0.9224 | 0.3934 | .0190 | 0.0253 | |
| Glutamate/glutamine | Univariate | 2.6480 | 0.8876 | .0029 | 0.0058 | |
| Glutamate/(Ser + Gly) | Univariate | 0.0284 | 0.0145 | .0502 | 0.0574 | |
| Glutamate | Age + sex + BMI + DM | 0.9090 | 0.3588 | .0113 | 0.0301 | |
| Glutamine | Age + sex + BMI + DM | 0.1635 | 0.8416 | .8459 | 1.8459 | |
| α-KG | Age + sex + BMI + DM | 0.5783 | 0.2551 | .0234 | 0.0356 | |
| Fumarate | Age + sex + BMI + DM | 1.0235 | 0.3639 | .0049 | 0.0196 | |
| Malate | Age + sex + BMI + DM | 1.1960 | 0.3764 | .0015 | 0.0120 | |
| Citrate | Age + sex + BMI + DM | 0.9729 | 0.4194 | .0202 | 0.0356 | |
| Glutamate/glutamine | Age + sex + BMI + DM | 2.0170 | 0.9102 | .0267 | 0.0356 | |
| Glutamate/(Ser + Gly) | Age + sex + BMI + DM | 0.0165 | 0.0151 | .2733 | 0.3123 | |
| Glutamate | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.9639 | 0.3728 | .0097 | 0.0259 | |
| Glutamine | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.0620 | 0.8559 | .9423 | 0.9423 | |
| α-KG | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.5944 | 0.2607 | .0226 | 0.0382 | |
| Fumarate | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.9954 | 0.3700 | .0071 | 0.0259 | |
| Malate | Age + sex + BMI + DM + HPT + smoking + alcohol | 1.1914 | 0.3871 | .0021 | 0.0168 | |
| Citrate | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.9526 | 0.4376 | .0295 | 0.0393 | |
| Glutamate/glutamine | Age + sex + BMI + DM + HPT + smoking + alcohol | 2.1033 | 0.9312 | .0239 | 0.0382 | |
| Glutamate/(Ser + Gly) | Age + sex + BMI + DM + HPT + smoking + alcohol | 0.0163 | 0.0156 | .2966 | 0.3390 |
DM, diabetes mellitus; Gly, glycine; HPT, hypertension; Ser, serine.
Increased Glutaminolysis Is a Novel Biomarker for Active Scarring
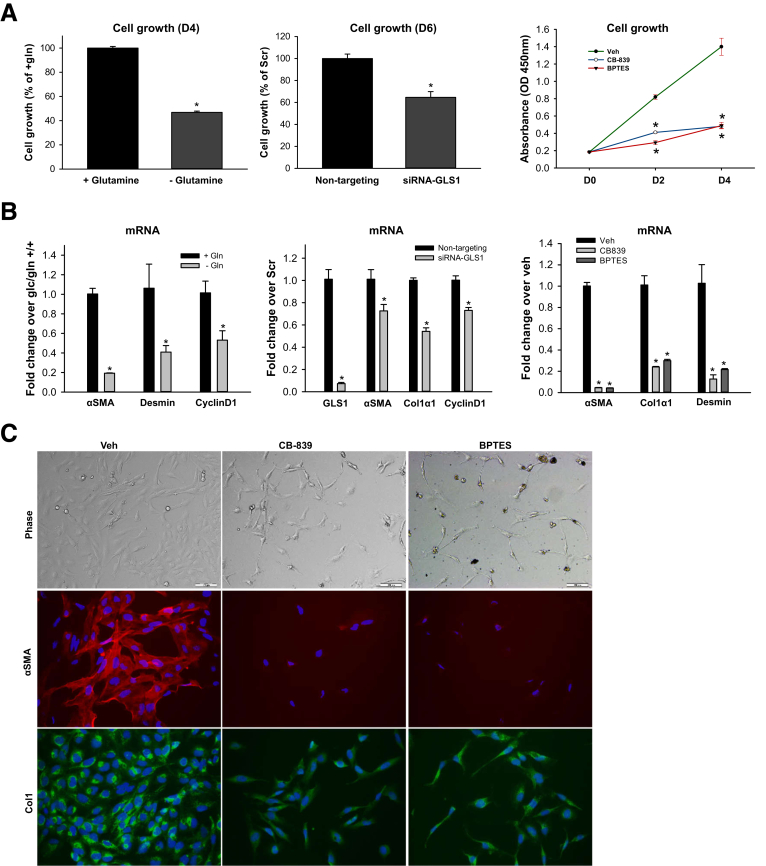
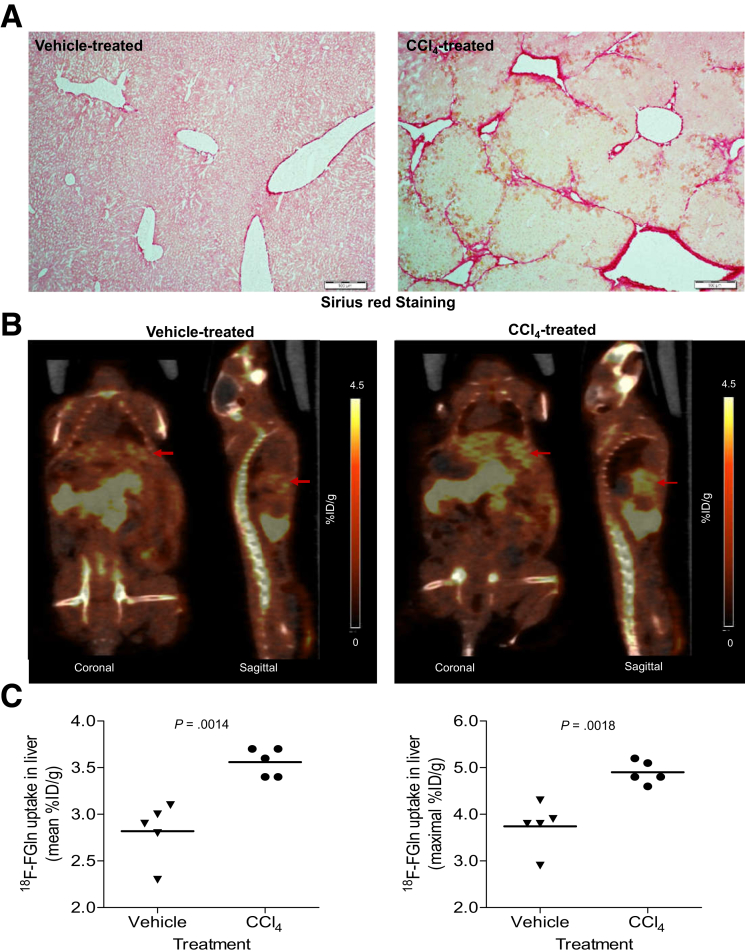
Previously, we and others have shown that glutamine transport and glutaminolysis increase during the transdifferentiation of rodent HSCs.5,11 Hence, avid glutamine uptake may reflect a stage of fibrosis in which the myofibroblasts are highly proliferative and fibrogenic. To assess the direct role of glutaminolysis in HSC growth and fibrogenesis further, we disrupted the glutaminolytic pathway in human myofibroblastic HSCs by glutamine deprivation, GLS1 small interfering RNA (siRNA), or pharmaceutical inhibitors of GLS1. Depleting glutamine from the culture medium suppressed MF-HSC growth (Figure 9A) and decreased expression of the cell-cycle marker, cyclinD1, and fibrogenic markers such as αSMA and desmin (Figure 9B). Knocking-down GLS1 or treating with the GLS1-specific inhibitors Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide or CB-839 also suppressed the growth of MF HSCs (Figure 9A) and decreased expression of cyclinD1, αSMA, Col1α1, and desmin (Figure 9B and C). All these data indicate that GLS1-dependent glutaminolysis is critical for growth and fibrogenic activity of human MF-HSCs, as it is in rodent MF HSCs.5,11 Evidence that increased glutamine catabolism generally is required to expand populations of MF-HSCs and parallels hepatic accumulation of fibrogenic myofibroblasts suggested that positron emission tomography (PET) might be able to track changes in liver fibrogenic activity based on its ability to detect increased rates of tissue glutamine assimilation.16,17 To evaluate this tenet we treated mice with CCl4 for 10 weeks to increase liver fibrogenesis (Figure 10A), gave them an intravenous injection of 18F-fluoroglutamine (18F-FGln), and monitored accumulation and clearance of the labeled substrate by PET. Compared with control mice that had received vehicle for 10 weeks, CCl4-treated mice showed significantly higher hepatic uptake of 18F-FGln, enabling PET to distinguish the CCl4-treated mice from controls (Figure 10B and C). This finding indicates that some of the changes in cell metabolism that mark highly proliferative cancer cells (eg, increased glutaminolysis) occur relatively selectively in nonmalignant fibrogenic myofibroblasts during liver injury. Further research is necessary to determine how metabolic differences among cells in injured livers might be exploited to track (or treat) liver fibrogenesis in NASH and other chronic liver diseases.
Figure 9.
Inhibition of glutaminolysis suppressed growth and fibrogenic activity of human MF-HSCs. Human MF-HSCs (LX2 cells) were grown in glutamine-containing or glutamine-free medium, or treated with 20 nmol/L nontargeting siRNA or GLS1 siRNA, or GLS1 inhibitor CB-839 (1 μmol/L), BPTES (10 μmol/L), or vehicle (0.1% dimethyl sulfoxide). (A) Cell growth was determined by CCK8 assay. (B) mRNA was quantified by quantitative reverse-transcription polymerase chain reaction. (C) Representative phase pictures and immunocytochemistry of αSMA and Col1. Bars represent means ± SEM of n = 3–5 assays. *P < .05 vs glutamine-free medium, nontargeting RNA, or vehicle group. BPTES, Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide; gln, glutamine; Veh, vehicle.
Figure 10.
Noninvasive imaging of increased glutaminolysis in fibrotic livers with 18F-FGln PET. Mice treated with vehicle (corn oil) or CCl4 (10 weeks) were imaged with 18F-FGln at 60 minutes after injection of the radiotracer by small-animal PET/CT (n = 5 per group). (A) Sirius red–stained sections from livers of these mice. (B) Examples of PET and co-registered PET/CT images of mice treated with vehicle or CCl4 and imaged at 60 minutes after injection of 18F-FGln. Representative pictures of coronal and sagittal sections are shown, arrows indicate the liver tissue. (C) 18F-Fluoroglutamine mean or maximal uptake is presented as the percentage of injected dose per gram (% ID/g) liver for individual mice from the 2 treatment groups. Bars represent means ± SEM. P value was calculated using an unpaired Student t test.
Discussion
Emerging human data from epidemiologic analyses, preclinical studies, and clinical trials support dysregulated metabolism as a critical driver of NASH pathogenesis, and, thus, targeting metabolic pathways is a putative therapeutic approach for NASH.18 However, although numerous efforts have focused on correcting the lipid metabolic defects in hepatocytes, effective pharmacotherapy for NASH is still lacking and relatively little is known about how metabolism in multiple other cell types might impact NASH progression.
Glutamine is the most abundant amino acid in the circulation and liver is one of the key organs involved in its metabolism. In healthy livers, gut-derived glutamine in portal blood is converted immediately to glutamate and ammonia in periportal hepatocytes that express Gls2, a low-affinity glutaminase. Hepatocytes in zones 1–2 also show high urea cycle activity, and, thus, readily convert glutamine-derived ammonia into urea. Perivenous hepatocytes in zone 3, in turn, condense residual ammonia with glutamate to regenerate glutamine and limit ammonia release into the systemic circulation. Because none of the cells in healthy livers are very proliferative, Gls1 (the high-affinity glutaminase expressed in rapidly growing cells) is barely detectable. In contrast, proliferative cells in many cancers, including some hepatocellular carcinomas, are glutamine addicted, and must induce Gls1 to increase glutamine catabolism to fuel their growth.6 Hence, Gls1 inhibitors are being evaluated as cancer therapeutics.6 Increased glutaminase activity was reported in cirrhotic human livers more than 20 years ago,19 prompting speculation that glutamine metabolism and signal transduction may be therapeutic targets for liver diseases.20 However, the role of glutaminolysis in NAFLD and its utility as a potential diagnostic biomarker or therapeutic target remain unclear. Metabolomics analysis of plasma from a small cohort of NAFLD patients (11 with hepatic steatosis and 24 with NASH vs 25 healthy controls) showed increased glutamate in patients with steatosis or NASH.12 In addition, measurements in the hepatic artery and the hepatic vein of 8 NAFLD and 8 healthy subjects showed uptake of glutamine and net release of glutamate and α-KG in NAFLD patients.21 Furthermore, random forest analysis of the plasma biochemical profile showed glutamate as the most effective metabolite for separating NASH patients from healthy subjects.12 Another recent small study (44 NAFLD patients) also showed increased plasma glutamate in NASH and reported that the glutamate/(serine + glycine) index correlated positively with liver ballooning and inflammation and distinguished advanced from mild fibrosis.10 Collectively, these studies suggest that glutaminolysis increases during NASH progression, but the mechanisms involved have not been investigated.
The present study includes serum metabolomics data from a large cohort of NAFLD patients (n = 200) with varying degrees of clinically occult liver fibrosis. The results not only confirm that circulating glutamate levels are increased in NASH but show that serum levels of glutamate generally parallel fibrosis severity. We also observed a positive association between serum glutamate concentrations and liver fibrosis stage in a separate cohort of patients with chronic hepatitis C (and in mice with CCl4-induced liver fibrosis), and, thus, concluded that glutamate levels in the systemic circulation typically increase during liver fibrogenesis. Glutamine catabolism is a potential source of glutamate. We found that hepatic concentrations of glutamate and glutamine are regulated reciprocally in multiple mouse models of liver fibrogenesis and showed that these changes parallel changes in the glutamate/glutamine ratio in both liver and sera. Consistent with the strong preclinical evidence that liver glutaminolytic activity increases during fibrogenesis, we observed that several glutaminolytic by-products, including α-KG, fumarate, malate, and citrate, all are increased in human NASH sera and correlate positively with liver fibrosis severity. The glutamate/glutamine ratio significantly correlates with α-KG levels in NAFLD patients, suggesting that glutaminolysis is induced to fuel TCA cycle activity and increase production of energy and biomass during fibrogenic repair. The correlations between liver fibrosis and glutaminolytic metabolites remain significant in human beings even after controlling for multiple known confounding factors (eg, age and diabetes). However, the strength of the associations is somewhat attenuated and this may explain why serum glutamate per se is an imperfect fibrosis biomarker in NASH.10 Surprisingly, we failed to replicate an earlier report that showed that the glutamate/(serine + glycine) index is associated significantly with hepatocellular ballooning and inflammation, and discriminates severe from mild fibrosis in NASH patients10 (data not shown). Although the exact reason for this discrepancy is unclear, it may reflect in part the fact that neither serum serine nor glycine decreases with NASH fibrosis progression in our cohort.
One of the most significant results of our study was the discovery that glutamine is consumed preferentially by different cell types in healthy livers compared with injured livers that are undergoing fibrogenic repair. This finding has both diagnostic and therapeutic implications. We confirmed previous reports showing that only rare cells in healthy livers express Gls1 (the glutaminase that has a high affinity for glutamine),13 and that Gls2 (the low-affinity so-called “liver-type glutaminase”) is expressed predominately in periportal hepatocytes.22 Furthermore, we showed that Gls2 and Gls1 are regulated reciprocally in injured livers: Gls2 decreases and Gls1 increases significantly. We also showed that Gls1 induction is accompanied by increased expression of glutamine transporters, likely helping injured livers to up-regulate net glutaminolytic activity significantly. We found that stromal cells (eg, myofibroblasts) are the main Gls1-expressing cells in injured livers. Hepatocytes suppress their typical expression of Gls2 and do not compensate for this by up-regulating Gls1. Although unexpected initially, this result is consistent with evidence that Gls1 is the glutaminase that is induced in rapidly growing cells6 and others have reported that NASH livers are enriched with senescent hepatocytes.23,24 Senescent hepatocytes release factors that stimulate transdifferentiation of hepatic stellate cells into proliferative myofibroblasts, driving fibrosis progression in NASH.23,24 We and other investigators recently reported that murine HSCs require glutamine to become and remain proliferative myofibroblasts,5,11 and here we confirm that human MF HSCs are similarly glutamine-addicted. Hence, the cumulative data suggest that the increased glutaminolytic activity observed in human NASH stems at least in part from the increased glutamine requirements of the fibrogenic stromal compartment. This tenet is supported further by re-examination of our published liver transcriptomic data from 70 NASH patients,15 which showed that GLS1 expression is significantly higher in human livers with advanced fibrosis than in those with no or mild fibrosis. Interestingly, our new immunoblot/immunohistochemistry results suggest that although glutamine consumption by fibrogenic stroma increases in NASH livers, hepatocytes may switch from consuming to producing glutamine as Gls2 expression is suppressed consistently, and GS expression is changed variably, during fibrogenic repair. It is important to point out that our studies have focused on glutaminolysis and simply show that cell type–specific changes in glutaminolytic gene expression parallel net changes in the glutamate/glutamine ratio and fibrosis severity in NASH. Because multiple pathways interact to maintain glutamine homeostasis,20 future studies that include detailed metabolic flux assays will be necessary to clarify the relative contribution of different cell types to injury-related changes in glutamine metabolism.
We suggest that these changes in hepatic glutamine metabolism might be exploited to develop novel noninvasive approaches to track dynamic changes in hepatic fibrogenic activity in NASH patients. Fibrogenic MFs drive fibrosis progression in NASH and we found that suppressing Gls1 activity directly reverted human MF HSCs to a more quiescent, less-fibrogenic state. In addition, we showed that liver glutaminolytic activity increases when fibrogenesis is active during NASH, and then promptly subsides as NASH resolves and fibrogenesis regresses in mice. Finally, we used PET to show that liver accumulation of 18F-FGln readily was detected, and increased significantly in a non-NASH preclinical model of relatively advanced liver fibrosis. Studies in models of more transient and less-severe fibrosis will be necessary to map the sensitivity and specificity of this PET-based approach for tracking changes in hepatic fibrogenesis. Those results will determine if PET (or approaches with less radiation exposure) might be developed to monitor active liver scarring in NASH, overcoming an inherent limitation of current fibrosis biomarkers that mainly reflect fibrosis status at the time of measurement, rather than predicting whether it is progressing or regressing.25,26 This success would improve identification of NASH patients with progressive fibrosis at an early stage, enable their appropriate stratification for antifibrotic interventions, facilitate real-time monitoring of treatment responses, and provide mechanistic insight into therapeutic successes/failures.
In summary, we deployed an array of complementary tools (human stellate cell cultures, mouse models of NASH and liver fibrosis, and biosamples from large cohorts of NAFLD patients with staged liver fibrosis) to confirm and extend previous reports that have suggested that glutaminolytic activity increases in NASH. We discovered that the observed changes in circulating levels of glutamine and its metabolites tightly parallel changes in the hepatic expression of genes that encode glutaminolytic enzymes, and, thus, likely reflect increased glutaminolysis in liver. Importantly, we mapped the increased hepatic glutaminolytic activity to fibrogenic stromal cells and showed that these cells significantly up-regulate their expression of Gls1 (a high-affinity glutaminase). In contrast, we found that hepatocytes in NASH livers retain expression of GS, an enzyme that generates glutamine, but significantly suppress their basal expression of a low-affinity glutaminase (Gls2) without inducing Gls1. Thus, the gene expression changes suggest that the liver switches from relatively low glutamine catabolism by healthy hepatocytes to significantly higher glutamine consumption by fibrogenic stromal cells as fibrosis progresses in NASH. These changes in liver glutamine metabolism are dynamic, track with the severity of liver fibrosis, and may be shown by PET. Thus, glutaminolysis is a potential target for future biomarker development and pharmacotherapy in NASH.
Methods and Materials
Metabolomics Analysis
Sample handling and processing
All samples were maintained at -80ºC until processed on the metabolomics platform at Metabolon, Inc (Morrisvile, NC). Samples were extracted and split into equal parts for analysis on the gas chromatography/mass spectrometry (GC/MS) and liquid chromatography/tandem mass spectrometry (MS/MS) platforms. A combination of 3 platforms was used for metabolic profiling: ultrahigh performance liquid chromatography (UHPLC/MS/MS) optimized for basic species, UHPLC/MS/MS optimized for acidic species, and GC/MS as previously described.27
GC/MS and UHPLC/MS analysis
UHPLC/MS was performed using a Waters Acquity UHPLC (Waters Corporation, Milford, MA) coupled to a linear trap quadrupole mass spectrometer (Thermo Fisher Scientific, Inc, Waltham, MA) equipped with an electrospray ionization source. Two separate UHPLC/MS injections were performed on each sample: 1 optimized for positive ions and 1 for negative ions. Samples for GC/MS were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole MS (Thermo Fisher Scientific, Inc, Waltham, MA) operated at unit mass resolving power. Chromatographic separation followed by full-scan mass spectra were performed to record retention time, molecular weight (m/z), and MS/MS of all detectable ions presented in the samples.
Quality assurance and control
Several types of controls were analyzed in concert with the experimental samples. Specifically, a pooled matrix sample generated by taking a small volume of each experimental sample served as a technical replicate throughout the data set. Extracted water samples served as process blanks, and a cocktail of quality control standards chosen not to interfere with the measurement of endogenous compounds were spiked into every analyzed sample to allow for instrument performance monitoring and aided chromatographic alignment. Instrument variability was determined by calculating the median relative SD for the standards that were added to each sample before injection into the mass spectrometers. Process variability was determined by calculating the median relative SD for all endogenous metabolites (ie, noninstrument standards) present in 100% of the pooled matrix samples. Experimental samples were randomized across the platform run with quality control samples spaced evenly among the injections.
Cell Culture Studies
Human MF HSCs (LX2 cells) were maintained in Gibco Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. To test the role of glutaminolysis, cells were grown in glutamine-containing or glutamine-free medium, or treated with 20 nmol/L nontargeting siRNA or GLS1 siRNA (Dharmacon, Lafayette, CO), GLS1 inhibitor CB-839 (1 μmol/L), Bis-2-(5-phenylacetamido-1,3,4-thiadiazol-2-yl)ethyl sulfide (10 μmol/L), or vehicle (0.1% dimethyl sulfoxide). Cell growth was monitored with the Cell Counting Kit-8 (CCK8; Dojindo Molecular Technologies, Inc, Rockville, MD).
Glutamine and Glutamate Measurement
The Glutamine/Glutamate-Glo Assay kit (J8022; Promega, Madison, WI) was used to measure glutamine and glutamate levels according to the manufacturer’s instructions.
Quantitative Real Time Reverse-Transcription Polymerase Chain Reaction
Total RNA was isolated from whole liver or cultured cells using TRIzol (Thermo Fisher Scientific) reagent and complementary DNAs were generated using Superscript II Reverse Transcriptase (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. Each complementary DNA sample was assayed by quantitative reverse-transcription polymerase chain reaction using SYBR Green Super-mix (Thermo Fisher Scientific) using the primers listed in Table 2. Results were normalized to the housekeeping gene S9 based on the threshold cycle, and relative fold change was determined using the 2-ΔΔCt method.
Table 2.
Primer Used for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Gene symbol | Primer forward | Primer reverse | |
|---|---|---|---|
| Human | S9 | GACTCCGGAACAAACGTGAGGT | CTTCATCTTGCCCTCGTCCA |
| Gls1 | GGTCTCCTCCTCTGGATAAGATGG | CCCGTTGTCAGAATCTCCTTGAGG | |
| Acta2 (αSma) | GGAGATCACGGCCCTAGCAC | AGGCCCGGCTTCATCGTAT | |
| Col1a1 (Col1α1) | CGGTGTGACTCGTGCAGC | ACAGCCGCTTCACCTACAGC | |
| Desmin | CAGTGGCTACCAGGACAACA | GCTGGTTTCTCGGAAGTTGA | |
| CyclinD1 | GGCCTCTAAGATGAAGGAGA | GGTTCCACTTGAGCTTGTT | |
| Mouse | S9 | GGGCCTGAAGATTGAGGATT | CGGGCATGGTGAATAGATTT |
| Gls1 | GCAAGTTCTTGCTGGAGACTCTCAT | AGTTGTCCCCAACGTCATGGGC | |
| Gls2 | TTTGCTGCATATAGTGGAGATGTC | GTTGAACTGCACAGCATCGTCCAG | |
| Slc1a5 | TCGCTATCGTCTTTGGTGTG | ATGGTGGCATCATTGAAGGAG | |
| Slc38a1 | GGACGGAGATAAAGGCACTC | CAGAGGGATGCTGATCAAGG | |
| Slc38a2 | GACACAGTAAGTGAGTGACG | CTCTCTTTGGATACCTGACC | |
| Acta2 (αSma) | GCCAGTCGCTGTCAGGAACCC | AGCCGGCCTTACAGAGCCCA | |
| Col1a1 (Col1α1) | GAGCGGAGAGTACTGGATCG | GCTTCTTTTCCTTGGGGTTC | |
| Vim (Vimentin) | GCCGAAAGCACCCTGCAGTCA | TGGGCCTGCAGCTCCTGGAT |
Immunoblotting, IHC, and Immunocytochemistry
Protein extracts were prepared from total liver tissue using RIPA buffer with protease inhibitors (Roche, Madison, WI). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis on 4%–20% Criterion gels (BioRad, Hercules, CA), and then transferred to polyvinylidene difluoride membranes, and incubated with the kidney-type glutaminase (20170-1-AP), Glutaminase C (19958-1-AP), Gls2(ab113509), and GS(ab73593) primary antibody overnight at 4°C. Blots were visualized with horseradish-peroxidase–conjugated secondary antibodies. β-actin or glyceraldehyde-3-phosphate dehydrogenase was used as an internal loading control for immunoblot normalization. For IHC, liver tissue was fixed in formalin, embedded in paraffin, and cut into sections, dewaxed, hydrated, and incubated for 10 minutes in 3% hydrogen peroxide to block endogenous peroxidase. Antigen retrieval was performed by heating in 10 mmol/L sodium citrate buffer (pH 6.0) for 10 minutes. Sections were blocked in Dako protein block solution for 1 hour and incubated overnight at 4°C with indicated primary antibodies: αSMA (M0851; Dako, Santa Clara, CA), Gls1 (ab156876), Gls2 (ab113509), and GS (ab73593). Polymer–horseradish peroxidase secondary antibodies were applied for 45 minutes at room temperature, and the 3,3′-diaminobenzidine substrate chromogen system (Dako) was used for detection. To see whether gls1 co-localizes with the MF-HSC marker αSMA, in our co-immunofluorescence staining we used primary monoclonal mouse anti-αSMA antibody (M0851; Dako, Santa Clara, CA) and secondary goat anti-mouse antibody (Alexa Fluor 647; Invitrogen, Waltham, MA), and primary rabbit monoclonal anti-Gls1 antibody (ab156876) and secondary goat anti-rabbit antibody (Alexa Fluor 488; Invitrogen). Zonal distribution of Gls2 and GS was identified by double IHC with 3,3′-diaminobenzidine tetra hydrochloride and Vina Green chromogens (Biocare Medical, Pacheco, CA), according to the manufacturer’s instructions. Picrosirius red (#365548; Sigma-Aldrich, St. Louis, MO) staining was performed according to the manufacturer’s instructions. For immunocytochemistry, cells were fixed in 4% paraformaldehyde, permeabilized, blocked with normal goat serum, and incubated overnight with αSMA (M0851; Dako) or Col1α1 (ab34710; Abcam, Cambridge, MA). Cells were washed in phosphate-buffered saline and incubated with Texas Red goat anti-mouse IgG or Alexa Fluor 488 goat anti-rabbit IgG (Heavy + Light chains; ThermoFisher Scientific) for 45 minutes at room temperature. 4',6-diamidino-2-phenylindole was used to visualize nuclei. Images were acquired and processed using a Zeiss LSM710 (Zeiss Microscopy, Oberkochen, Germany) inverted confocal microscope system.
Animal Studies
To generate NASH fibrosis mouse models, C57BL/6 mice were fed with a chow diet (Picolab [St. Louis, MO] Rodent diet 20, #5053), a Western diet (TD.120330 22% hydrotreated vegetable oil + 0.2% cholesterol diet; Teklad Research [Indianapolis, IN]; supplemented with high–corn fructose syrup equivalents in the drinking water; n = 8 mice) for 16 weeks, a CDAA-HFD diet (A06071302, choline-deficient, L-amino acid defined diet with 60 kcal% fat; Research Diets, Inc, New Brunswick, NJ; n = 10 mice) for 12 or 22 weeks, or a MCDE (supplemented with 0.15% ethionine) for 3 weeks (n = 3 mice), or treated with 0.6 mL/kg CCl4 or corn oil by intraperitoneal injection, twice a week, for 4–6 weeks (n = 26 mice). The MCDE diet was removed in some mice for 3 weeks after 3 weeks of treatment (n = 3 mice). Liver tissue was fixed in phosphate-buffered formalin for histology, or flash-frozen in liquid nitrogen and stored at −80°C. All studies were approved by the Duke University Institutional Animal Care and Use Committee as set forth in the Guide for the Care and Use of Laboratory Animals, published by the National Research Council.
18F-FGln PET/Computed Tomography Imaging in Mice With Fibrotic Livers
The radiosynthesis of [18F]-(2S, 4R)-4-fluoroglutamine (18F-FGln) was performed following the methods reported in the literature, and the final product was dissolved in saline and sterile-filtered (0.22 μm) for use in the imaging studies.28,29 For small animal PET/computed tomography (CT) imaging, CCl4 or vehicle-treated C57BL/6 mice (n = 5 mice/group) were injected with 18F-FGln via the tail vein and were imaged on an Inveon PET/CT scanner (Siemens, Knoxville, TN) 60 minutes after injection under isoflurane anesthesia (1%–1.5% in O2). Images were reconstructed and analyzed by drawing regions of interest over the liver on the co-registered PET/CT images for each of the animals using Inveon Research Workplace software (Siemens).30 The animals were killed at the end of the study and livers were collected and processed for histology and Sirius red staining.
Human Subjects
Metabolomic analysis was performed on de-identified serum samples from 200 biopsy-proven NAFLD patients across different stages of fibrosis (F0–1, 50; F2, 100; and F3–4, 50) obtained from the Duke University Health System NAFLD Clinical Database and Biorepository following nonexempt Institutional Review Board (IRB) approval. The Duke University Health System NAFLD Clinical Database is linked to tissue repositories in which biosamples (eg, blood, liver tissues) were collected at the time of a clinically indicated liver biopsy. All participants provided written informed consent before study enrollment. Patient characteristics are summarized in Table 3. Expression of GLS1 was assessed in a subset of patients with available paraffin-embedded slides (healthy controls, 1; NAFLD patients: simple steatosis, 9; mild fibrosis, 10; and advanced fibrosis, 9) following exempt Duke IRB approval (Pro00087196). Microarray data from our previously published study in 72 NASH patients (n = 32 with fibrosis stage F3–4 and n = 40 with F0–1 fibrosis) also were re-analyzed for GLS1 expression.15 In addition, sera were assayed in 40 HIV/HCV-infected patients with varying degrees of biopsy-proven liver fibrosis (F0–1, 11; F2, 15; and F3–4, 14) from an independent cohort. Subjects with HIV and HCV infections, aged 18 years of age and older, were identified from the Duke HIV Research Database and Biorepository, which was opened in 1998. All subjects either provided informed consent for the use of their personal health information and plasma samples for future research purposes or samples were used under IRB approved waiver of informed consent. The proposed research project was approved by the Duke Institutional Review Board. Subjects were eligible if they had a liver biopsy at Duke University Medical Center as part of standard of care and an available plasma specimen in the biorepository within 6 months of the liver biopsy. In all subjects, serum was prepared by centrifugation (2500 g, 15 min). Supernatants were placed into cryovials and stored at –80°C until analysis.
Table 3.
Patient Characteristics in Metabolomics Analysis
| Fibrosis stage 0 | Fibrosis stage 1 | Fibrosis stage 2 | Fibrosis stage 3 | Fibrosis stage 4 | |
|---|---|---|---|---|---|
| Subjects, n | 12 | 38 | 100 | 42 | 8 |
| Age, y | 53 ± 9 | 49 ± 11 | 50 ± 11 | 51 ± 8 | 59 ± 5 |
| Sex, males/females | 5/7 | 16/22 | 50/50 | 22/20 | 2/6 |
| BMI, kg/m2 | 34 ± 8 | 35 ± 7 | 35 ± 7 | 35 ± 7 | 38 ± 14 |
| Diabetes subjects, n (%) | 2 (17) | 8 (21) | 36 (36) | 23 (55) | 6 (75) |
| Steatosis | 1.5 ± 0.9 | 1.9 ± 0.8 | 1.8 ± 0.8 | 1.8 ± 0.8 | 1.6 ± 0.9 |
| Lobular inflammation | 0.8 ± 0.4 | 1.3 ± 0.6 | 1.4 ± 0.6 | 1.6 ± 0.7 | 1.4 ± 0.5 |
| Portal inflammation | 0.1 ± 0.3 | 0.2 ± 0.4 | 0.4 ± 0.5 | 0.5 ± 0.5 | 1.0 ± 0.0 |
| Ballooning | 0.4 ± 0.5 | 1.1 ± 0.7 | 1.2 ± 0.6 | 1.5 ± 0.6 | 1.5 ± 0.8 |
| NASH activity score | 2.8 ± 1.3 | 4.2 ± 1.5 | 4.5 ± 1.4 | 5.0 ± 1.3 | 4.3 ± 1.6 |
| Glucose, mg/dL | 79 ± 19 | 93 ± 42 | 100 ± 33 | 103 ± 18 | 107 ± 22 |
| Aspartate aminotransferase, IU/L | 34 ± 19 | 62 ± 48 | 71 ± 54 | 74 ± 41 | 67 ± 23 |
| Alanine aminotransferase, IU/L | 48 ± 38 | 85 ± 75 | 92 ± 62 | 88 ± 56 | 73 ± 32 |
| Triglycerides, mg/dL | 237 ± 173 | 151 ± 64 | 184 ± 167 | 208 ± 163 | 164 ± 60 |
| Hemoglobin A1c, % | 5.7 ± 0.6 | 6.2 ± 1.1 | 6.2 ± 1.3 | 6.9 ± 1.6 | 6.8 ± 0.9 |
| Insulin, mIU/L | 17 ± 70 | 18 ± 7 | 27 ± 23 | 22 ± 13 | 60.1 |
NOTE. Patient information pertaining to the Duke cohort. Subjects are shown according to different fibrosis stages, ranging from absent (stage 0) to cirrhosis (stage 4).
Statistics
Data were expressed as means ± SEM. Except for metabolomics analysis, statistical significance between 2 groups was evaluated using the Student t test, whereas comparisons of multiple groups were assessed by 1-way analysis of variance, followed by the Student–Newman–Keul test. A P value less than .05 was considered statistically significant. For metabolomics data from human NAFLD patients, amino acid values were log-transformed before statistical analyses. An ordinal logistic regression model was used to assess the association between amino acids and fibrosis stage. A proportional odds assumption of the model was tested. A multiple ordinal logistic regression model was used to assess amino acid association with fibrosis stage while controlling for age, sex, BMI, diabetes, hypertension, smoking, and alcohol consumption. A P value less than .05 was considered significant. The Benjamini–Hochberg procedure was used to correct for multiple testing with a false-discovery rate of 5%, considering data relevant to the scope of this study. Statistical analyses of the data were performed using SAS software (version 9.4; Cary, NC).
Acknowledgments
The authors thank Metabolon, Inc, for performing the metabolomics study, the Duke NAFLD program for metabolomics analysis of human serum samples, the Duke Department of Pathology for providing paraffin-embedded slides from human liver biopsy specimens, Dr Michael R. Zalutsky and Dr Jason S. Lewis (Memorial Sloan Kettering Cancer Center) for providing the 18F-FGln precursor, the staff of the preclinical PET/CT molecular imaging center for help with the imaging studies, and the Charles E. Putman MD Vision Award, Department of Radiology, Duke University School of Medicine, for additional support.
Footnotes
Author contributions Kuo Du, Satish K. Chitneni, Mustafa R. Bashir, Manal F. Abdelmalek, and Anna Mae Diehl conceived the experiments; Kuo Du, Satish K. Chitneni, Jeongeun Hyun, and Richard T. Premont performed the experiments; Kuo Du, Satish K. Chitneni, Ayako Suzuki, Ying Wang, Ricardo Henao, Cynthia A. Moylan, Mustafa R. Bashir, and Anna Mae Diehl analyzed data; Ying Wang, Susanna Naggie, and Manal F. Abdelmalek provided clinical biosamples, and Kuo Du and Anna Mae Diehl wrote the manuscript. All authors reviewed and approved the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants R37 AA010154, R01 DK077794, and R56 DK106633 (A.M.D.). Additional funding was provided by the American Association for the Study of Liver Diseases Afdhal/McHutchison Liver Institute and Foundation for Education and Research Award (K.D.).
References
- 1.Diehl A.M., Day C. Cause, pathogenesis, and treatment of nonalcoholic steatohepatitis. N Engl J Med. 2017;377:2063–2072. doi: 10.1056/NEJMra1503519. [DOI] [PubMed] [Google Scholar]
- 2.Diehl A.M., Chute J. Underlying potential: cellular and molecular determinants of adult liver repair. J Clin Invest. 2013;123:1858–1860. doi: 10.1172/JCI69966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujii H., Kawada N. Inflammation and fibrogenesis in steatohepatitis. J Gastroenterol. 2012;47:215–225. doi: 10.1007/s00535-012-0527-x. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y., Choi S.S., Michelotti G.A., Chan I.S., Swiderska-Syn M., Karaca G.F., Xie G., Moylan C.A., Garibaldi F., Premont R., Suliman H.B., Piantadosi C.A., Diehl A.M. Hedgehog controls hepatic stellate cell fate by regulating metabolism. Gastroenterology. 2012;143:1319–1329 e11. doi: 10.1053/j.gastro.2012.07.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du K., Hyun J., Premont R.T., Choi S.S., Michelotti G.A., Swiderska-Syn M., Dalton G.D., Thelen E., Rizi B.S., Jung Y., Diehl A.M. Hedgehog-YAP signaling pathway regulates glutaminolysis to control activation of hepatic stellate cells. Gastroenterology. 2018;154:1465–1479 e13. doi: 10.1053/j.gastro.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang L., Venneti S., Nagrath D. Glutaminolysis: a hallmark of cancer metabolism. Annu Rev Biomed Eng. 2017;19:163–194. doi: 10.1146/annurev-bioeng-071516-044546. [DOI] [PubMed] [Google Scholar]
- 7.Katt W.P., Lukey M.J., Cerione R.A. A tale of two glutaminases: homologous enzymes with distinct roles in tumorigenesis. Future Med Chem. 2017;9:223–243. doi: 10.4155/fmc-2016-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curthoys N.P., Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu Rev Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 9.Brosnan M.E., Brosnan J.T. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr. 2009;90:857S–861S. doi: 10.3945/ajcn.2009.27462Z. [DOI] [PubMed] [Google Scholar]
- 10.Gaggini M., Carli F., Rosso C., Buzzigoli E., Marietti M., Della Latta V., Ciociaro D., Abate M.L., Gambino R., Cassader M., Bugianesi E., Gastaldelli A. Altered amino acid concentrations in NAFLD: impact of obesity and insulin resistance. Hepatology. 2018;67:145–158. doi: 10.1002/hep.29465. [DOI] [PubMed] [Google Scholar]
- 11.Li J., Ghazwani M., Liu K., Huang Y., Chang N., Fan J., He F., Li L., Bu S., Xie W., Ma X., Li S. Regulation of hepatic stellate cell proliferation and activation by glutamine metabolism. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalhan S.C., Guo L., Edmison J., Dasarathy S., McCullough A.J., Hanson R.W., Milburn M. Plasma metabolomic profile in nonalcoholic fatty liver disease. Metabolism. 2011;60:404–413. doi: 10.1016/j.metabol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Botman D., Tigchelaar W., Van Noorden C.J. Determination of phosphate-activated glutaminase activity and its kinetics in mouse tissues using metabolic mapping (quantitative enzyme histochemistry) J Histochem Cytochem. 2014;62:813–826. doi: 10.1369/0022155414551177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syn W.K., Jung Y., Omenetti A., Abdelmalek M., Guy C.D., Yang L., Wang J., Witek R.P., Fearing C.M., Pereira T.A., Teaberry V., Choi S.S., Conde-Vancells J., Karaca G.F., Diehl A.M. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478–1488 e8. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moylan C.A., Pang H., Dellinger A., Suzuki A., Garrett M.E., Guy C.D., Murphy S.K., Ashley-Koch A.E., Choi S.S., Michelotti G.A., Hampton D.D., Chen Y., Tillmann H.L., Hauser M.A., Abdelmalek M.F., Diehl A.M. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology. 2014;59:471–482. doi: 10.1002/hep.26661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou R., Pantel A.R., Li S., Lieberman B.P., Ploessl K., Choi H., Blankemeyer E., Lee H., Kung H.F., Mach R.H., Mankoff D.A. [(18)F](2S,4R)4-fluoroglutamine PET detects glutamine pool size changes in triple-negative breast cancer in response to glutaminase inhibition. Cancer Res. 2017;77:1476–1484. doi: 10.1158/0008-5472.CAN-16-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu H., Liu F., Zhang Y., Yang J., Xu X., Guo X., Liu T., Li N., Zhu L., Kung H.F., Yang Z. (2S,4R)-4-[(18)F]Fluoroglutamine as a PET indicator for bone marrow metabolism dysfunctional: from animal experiments to clinical application. Mol Imaging Biol. 2019;21:945–953. doi: 10.1007/s11307-019-01319-4. [DOI] [PubMed] [Google Scholar]
- 18.Esler W.P., Bence K.K. Metabolic targets in nonalcoholic fatty liver disease. Cell Mol Gastroenterol Hepatol. 2019;8:247–267. doi: 10.1016/j.jcmgh.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuno T., Goto I. Glutaminase and glutamine synthetase activities in human cirrhotic liver and hepatocellular carcinoma. Cancer Res. 1992;52:1192–1194. [PubMed] [Google Scholar]
- 20.Haussinger D., Schliess F. Glutamine metabolism and signaling in the liver. Front Biosci. 2007;12:371–391. doi: 10.2741/2070. [DOI] [PubMed] [Google Scholar]
- 21.Hyotylainen T., Jerby L., Petaja E.M., Mattila I., Jantti S., Auvinen P., Gastaldelli A., Yki-Jarvinen H., Ruppin E., Oresic M. Genome-scale study reveals reduced metabolic adaptability in patients with non-alcoholic fatty liver disease. Nat Commun. 2016;7:8994. doi: 10.1038/ncomms9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D., Shi X., Meng G., Chen J., Yan C., Jiang Y., Wei J., Ding Y. Kidney-type glutaminase (GLS1) is a biomarker for pathologic diagnosis and prognosis of hepatocellular carcinoma. Oncotarget. 2015;6:7619–7631. doi: 10.18632/oncotarget.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravinthan A., Scarpini C., Tachtatzis P., Verma S., Penrhyn-Lowe S., Harvey R., Davies S.E., Allison M., Coleman N., Alexander G. Hepatocyte senescence predicts progression in non-alcohol-related fatty liver disease. J Hepatol. 2013;58:549–556. doi: 10.1016/j.jhep.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 24.Ogrodnik M., Miwa S., Tchkonia T., Tiniakos D., Wilson C.L., Lahat A., Day C.P., Burt A., Palmer A., Anstee Q.M., Grellscheid S.N., Hoeijmakers J.H.J., Barnhoorn S., Mann D.A., Bird T.G., Vermeij W.P., Kirkland J.L., Passos J.F., von Zglinicki T., Jurk D. Cellular senescence drives age-dependent hepatic steatosis. Nat Commun. 2017;8:15691. doi: 10.1038/ncomms15691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong V.W., Adams L.A., de Ledinghen V., Wong G.L., Sookoian S. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat Rev Gastroenterol Hepatol. 2018;15:461–478. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 26.Decaris M.L., Li K.W., Emson C.L., Gatmaitan M., Liu S., Wang Y., Nyangau E., Colangelo M., Angel T.E., Beysen C., Cui J., Hernandez C., Lazaro L., Brenner D.A., Turner S.M., Hellerstein M.K., Loomba R. Identifying nonalcoholic fatty liver disease patients with active fibrosis by measuring extracellular matrix remodeling rates in tissue and blood. Hepatology. 2017;65:78–88. doi: 10.1002/hep.28860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evans A.M., DeHaven C.D., Barrett T., Mitchell M., Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 28.Qu W., Zha Z., Ploessl K., Lieberman B.P., Zhu L., Wise D.R., Thompson C.B., Kung H.F. Synthesis of optically pure 4-fluoro-glutamines as potential metabolic imaging agents for tumors. J Am Chem Soc. 2011;133:1122–1133. doi: 10.1021/ja109203d. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Basuli F., Shi Z.D., Xu B., Blackman B., Choyke P.L., Swenson R.E. Automated synthesis of [(18)F](2S,4R)-4-fluoroglutamine on a GE TRACERlab FX-N Pro module. Appl Radiat Isot. 2016;112:110–114. doi: 10.1016/j.apradiso.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chitneni S.K., Yan H., Zalutsky M.R. Synthesis and evaluation of a (18)F-labeled triazinediamine analogue for imaging mutant IDH1 expression in gliomas by PET. ACS Med Chem Lett. 2018;9:606–611. doi: 10.1021/acsmedchemlett.7b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]