Abstract
In this study, a series of new indole derivatives of ursolic acid bearing different N-(aminoalkyl)carboxamide side chains were designed, synthesized, and evaluated for their in vitro cytotoxic activities against two human hepatocarcinoma cell lines (SMMC-7721 and HepG2) and normal hepatocyte cell line (LO2) via MTT assay. Among them, compound 5f exhibited the most potent activity against SMMC-7721 and HepG2 cells with IC50 values of 0.56 ± 0.08 μM and 0.91 ± 0.13 μM, respectively, and substantially lower cytotoxicity to LO2 cells. A follow-up enzyme inhibition assay and molecular docking study indicated that compound 5f can significantly inhibit the activity of Topoisomerase IIα. Further mechanistic studies performed in SMMC-7721 cells revealed that compound 5f can elevate the intracellular ROS levels, decrease mitochondrial membrane potential, and finally lead to the apoptosis of SMMC-7721 cells. Collectively, compound 5f is a promising Topoisomerase II (Topo II) inhibitor, which exhibited the potential as a lead compound for the discovery of novel anticancer agents.
Keywords: ursolic acid, indole derivatives, anticancer activity, topoisomerase II, apoptosis
1. Introduction
Cancer has become a major cause of death in human beings. In 2018, there were estimated 18.1 million new cancer cases and 9.6 million cancer deaths worldwide [1]. In China, about 3.8 million new cancer cases and 2.29 million cancer deaths occurred in 2018, both ranked first among countries worldwide [1]. Despite their indispensable role in the treatment of cancer, chemotherapies often cause less satisfactory results due to severe side effects and rapidly occurring drug resistance [2]. Therefore, efforts to develop low-toxic and high-efficacy anticancer agents have still been the primary task of medicinal chemists.
Topoisomerase, which are generally classified as types I and II, are critical nuclear enzymes essential for cell proliferation by solving the topological hurdles in the process of DNA replication. Topoisomerase I (Topo I) handles the breaking and rejoining of only a single strand of a DNA duplex to relieve the topological tension. In the case of topoisomerase II (Topo II), it relaxes the DNA double helices by scissoring and religating both strands [3,4,5]. The monomeric enzyme Topo I does not require Mg(II) for DNA relaxation, while homodimeric enzyme Topo II requires Mg(II) and ATP hydrolysis for the enzyme’s function and rapid kinetics [3]. The Topo II family has two isozymes—Topo IIα and IIβ. Only the level of Topo IIα protein among human topo isozymes (Topo I, Iiα, and IIβ) increases in proliferative stage dependent manner. In addition, Topo II is frequently overexpressed in different types of tumors, so the expression level of Topo IIα has been used as a useful diagnostic marker for chemotherapy in diverse cancers [6,7]. These findings provide insight that targeting Topo II, especially Topo IIα, can be a good strategy for aiming at actively proliferating cancer cells rather than normal cells. A number of Topo II-targeted chemotherapy drugs, such as etoposide (VP-16), doxorubicin (DOX), and mitoxantrone have been approved by the FDA for clinical use [8]. However, most Topo II inhibitors exhibited rebarbative side effects including cardiotoxicity, development of secondary malignancies and multidrug resistance [9]. Hence, it is still a necessity to develop novel Topo II inhibitors with high efficacy and selectivity.
Natural products have long been used to treat various diseases due to their wide range of pharmacological activities [10]. Since the structural modified derivatives of natural products with anticancer activities generally have fewer side effects, a simple and effective method to obtain anticancer agents is the structural modification of bioactive natural products [11]. Ursolic acid (UA, 1) is a natural pentacyclic triterpenoid abundant in many vegetables, fruits, and herbs. The compound and its derivatives exhibit a wide variety of biological activities, such as antibacterial [12], anticancer [13], antiviral [14], anti-inflammatory [15], antidiabetic [16], antiulcer [17], antiarrhythmic [18], anti-osteoporosis [19], anti-atherosclerotic [20], and anti-neurodegenerative activities [21]. In addition, some pentacyclic triterpenoids including UA were reported to inhibit Topo I and IIα by contending with DNA for topoisomerase binding sites, thus preventing topoisomerase-DNA cleavable complex formation [22,23,24]. Therefore, UA could be a promising starting material for the discovery of new topoisomerase inhibitors as anticancer agents.
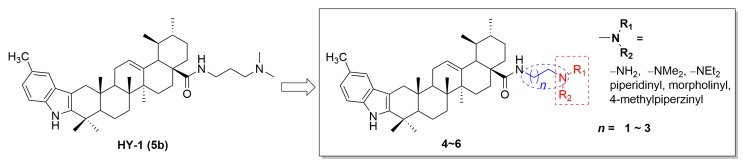
In an effort to discover novel anticancer agents, we have previously designed and synthesized a series of indole derivatives of UA bearing N-aminoalkyl side chains. In the in vitro cytotoxic assay, some derivatives showed significant cytotoxic activity against two human hepatocarcinoma cell lines (SMMC-7721 and HepG2). Among them, HY-1 (compound 5b) exhibited the strongest activity with IC50 values of about 1 μM [25]. Moreover, in a preliminary screening, HY-1 showed considerable inhibitory activity to Topo I and IIα. Based on these findings, this compound was chosen as the starting material for further structure modifications. Since, in our preliminary study, the carbon chain and N-containing moiety of C-28 side chain played an important role in the cytotoxic activity of derivatives, we focused on the modification of the side chain in this work (Figure 1). As a result, a series of new UA derivatives were designed and synthesized in order to find derivatives with better anticancer activities. Furthermore, the anticancer mechanisms of the active compounds were also extensively explored.
Figure 1.
The structural modification of HY-1 (5b).
2. Results
2.1. Chemistry
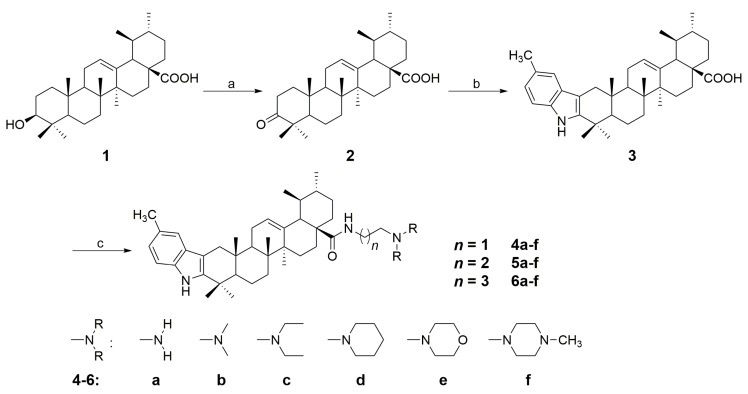
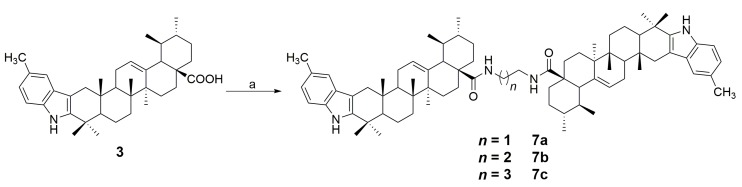
Based on the method reported earlier [25], a series of new indole derivatives of ursolic acid with different amine side chains were synthesized. Synthetic routes for the target compounds were summarized in Scheme 1 and Scheme 2. Briefly, the starting material UA (1) was oxidized by Jones reagent to afford 3-oxo-ursolic acid (2) in 76% yield. Subsequently, compound 2 was converted to the indole derivative (3) in 69% yield by reacting with p-tolylhydrazine hydrochloride through Fisher indole reaction. Further, compound 3 was reacted with different N1,N1-dialkyl-1,2-ethanediamine, N1,N1-dialkyl-1,3-propanediamine, or N1,N1-dialkyl-1,4-butanediamine to afford the corresponding amide derivatives 4a–f, 5a–f, and 6a–f in 72–93% yields (Scheme 1). Interestingly, when compound 3 was reacted with 1,2-ethanediamine, 1,3-propanediamine, or 1,4-butanediamine in the presence of HOBt and DCC, the dimeric byproducts (7a–c) could always be detected. However, if we treat compound 3 with 0.5 eq of 1,2-ethanediamine, 1,3-propanediamine or 1,4-butanediamine, the dimeric derivatives 7a–c became the main products with the yield of 61%, 65%, 63%, respectively (Scheme 2). All the synthesized compounds were purified by recrystallization or silica gel column chromatography, and the structures of these compounds were characterized through their HRMS, IR, 1H NMR and 13C NMR spectra (1H and 13C NMR spectra: Figure S1–S42 in the Supplementary Materials).
Scheme 1.
Synthetic route for the indole derivatives (4a–f, 5a–f, 6a–f) from ursolic acid. Reagents and conditions: (a) Jones reagent, acetone, 0 °C, 5 h; (b) p-methyl phenylhydrazine hydrochloride, EtOH, conc. HCl, reflux for 3 h; (c) N,N-dialkylethane-1,2-diamine, N,N-dialkylpropane-1,3-diamine, or N,N-dialkylbutane-1,4-diamine (1.2 eq), HOBt, DCC, CH2Cl2, rt, 12 h.
Scheme 2.
Synthesis of compounds 7a–c from 3. Reagents and conditions: (a) 1,2-ethylenediamine, 1,3-propanediamine and 1,4-butanediamine (0.5 eq), HOBt, DCC, CH2Cl2, rt, 12 h.
2.2. Cytotoxic Assay
The in vitro cytotoxic activities of synthesized compounds were evaluated against two human hepatocarcinoma cell lines (SMMC-7721 and HepG2) and the human normal hepatocyte cell line (LO2) via MTT assay. All tested compounds were dissolved in DMSO and the stock solutions were diluted by DMEM medium before treatment of the cultured cells. The IC50 values of the tested compounds against four cell lines were shown in Table 1.
Table 1.
The cytotoxicity Topo I and II inhibitory activities of the target compounds.
| IC50 Values (μM) | Topo I Inhibition (%) | Topo II Inhibition (%) | |||||
|---|---|---|---|---|---|---|---|
| SMMC-7721 | HepG2 | LO2 | 100 μM | 20 μM | 100 μM | 20 μM | |
| 4a | 3.50 ± 0.61 | 3.31 ± 0.35 | 29.59 ± 2.12 | 15.1 | 5.6 | 69.7 | 25.9 |
| 4b | 4.42 ± 0.71 | 4.11 ± 0.55 | 31.06 ± 2.63 | 3.7 | -1 | 68.0 | 36.9 |
| 4c | 2.10 ± 0.21 | 2.24 ± 0.32 | 26.39 ± 1.32 | 0 | - | 68.7 | 38.3 |
| 4d | 1.75 ± 0.11 | 1.47 ± 0.12 | 16.03 ± 1.07 | 13.0 | 0 | 71.7 | 32.7 |
| 4e | 7.82 ± 0.78 | 10.72 ± 2.09 | >50 | 0 | - | 63.3 | 26.8 |
| 4f | 1.24 ± 0.16 | 1.35 ± 0.17 | 18.27 ± 0.78 | 0 | - | 73.7 | 31.3 |
| 5a | 2.02 ± 0.21 | 2.45 ± 0.38 | 21.32 ± 1.59 | 24.5 | 12.4 | 72.3 | 31.2 |
| 5b | 1.08 ± 0.22 | 1.26 ± 0.17 | 20.65 ± 1.73 | 34.2 | 9.0 | 72.5 | 38.2 |
| 5c | 1.15 ± 0.14 | 1.20 ± 0.24 | 15.85 ± 1.06 | 0 | - | 75.8 | 38.9 |
| 5d | 0.89 ± 0.11 | 1.04 ± 0.09 | 12.82 ± 0.83 | 0 | - | 81.1 | 41.3 |
| 5e | 3.75 ± 0.37 | 5.27 ± 0.67 | 28.62 ± 0.76 | 0 | - | 70.8 | 31.5 |
| 5f | 0.56 ± 0.08 | 0.91 ± 0.13 | 10.58 ± 0.52 | 0 | - | 83.5 | 43.2 |
| 6a | 1.21 ± 0.19 | 1.32 ± 0.21 | 15.23 ± 1.15 | 28.8 | 13.7 | 71.1 | 27.0 |
| 6b | 2.43 ± 0.21 | 2.91 ± 0.18 | 18.19 ± 1.64 | 15.1 | 0 | 69.0 | 31.7 |
| 6c | 3.62 ± 0.23 | 3.45 ± 0.25 | 28.42 ± 2.98 | 16.8 | 0 | 70.0 | 35.2 |
| 6d | 2.72 ± 0.31 | 2.31 ± 0.21 | 18.65 ± 1.36 | 0 | - | 72.5 | 32.8 |
| 6e | 4.55 ± 1.56 | 7.84 ± 2.31 | 35.39 ± 3.13 | 0 | - | 65.6 | 28.2 |
| 6f | 0.65 ± 0.07 | 1.01 ± 0.12 | 13.72 ± 1.28 | 0 | - | 81.8 | 39.8 |
| 7a | >50 | >50 | >50 | 0 | - | 23.8 | 9.7 |
| 7b | >50 | >50 | >50 | 2.0 | - | 17.8 | 9.8 |
| 7c | >50 | >50 | >50 | 0 | - | 16.9 | 8.1 |
| CPT 2 | 0.56 ± 0.18 | 1.12 ± 0.26 | 8.93 ± 0.52 | 75.1 | 38.2 | - | - |
| VP-16 | 0.36 ± 0.10 | 0.57 ± 0.16 | 5.92 ± 0.29 | - | - | 88.5 | 47.8 |
| DOX | 0.68 ± 0.11 | 0.81 ± 0.19 | 5.32 ± 0.67 | - | - | - | - |
1 -: Not tested. 2 CPT: camptothecin; VP-16: etoposide; DOX: doxorubicin.
As shown in Table 1, most synthesized compounds showed considerable cytotoxic activity against two cancer cell lines. Among them, compounds 4d, 4f, 5b–d, 5f, 6a and 6f displayed strong inhibition with IC50 values of around 1 μM. Especially, compound 5f exhibited the most potent anticancer activity with IC50 values of 0.56 ± 0.08 and 0.91 ± 0.13 μM, respectively, which were comparable to those of reference drugs. Notably, compound 5f showed significantly lower cytotoxicity to human normal hepatocyte cell line LO2 (IC50: 10.58 ± 0.52 μM), indicating that the compound had good selectivity between hepatocarcinoma and noncancerous hepatocyte cells. Compounds 5d and 6f also exhibited significant activities against SMMC-7721 cells with IC50 values of 0.89 ± 0.11 and 0.65 ± 0.07 μM, respectively. In addition, compounds 4a–c, 4e, 5a, 5e, and 6b–e showed good inhibitory activity to the two cancer cell lines. On the contrary, compounds 7a–c were inactive to all three cell lines (IC50 > 50 μM).
The results indicated that the anticancer activities of the synthesized compounds were affected by the N-containing moieties anchored on the side chain. For Series 4a–f, 5a–f, and 6a–f, compounds 4f, 5f, and 6f with N-methylpiperazine moieties all exhibited the strongest activities in their own series. Compounds 4a–d, 5a–d, and 6a–d containing NH2, N(Me)2, N(Et)2, and piperidine moieties generally showed strong activities. However, compounds 4e, 5e, and 6e with morpholine rings displayed relatively weaker inhibition to cancer cells compared with their analogs in the same series. In general, the order of effects to the cytotoxicity of these derivatives could be summarized as N-methylpiperazine > NH2, N(Me)2, N(Et)2 and piperidine > morpholine. On the other hand, the length of amide side chain also affected the cytotoxicity to some extent. Generally, compounds 5af exhibited stronger cytotoxic activities than compounds 4a–f and 6a–f, which suggested that the 3C alkyl linker could be more beneficial to the cytotoxicity than 2C and 4C alkyl linkers. In addition, three dimeric derivatives 7a–c were all inactive to tested cancer cells (IC50 > 50 μM), which indicated that such dimerization by alkyl linkers might hinder the interaction between these compounds and their potential targets, thus significantly decreasing their anticancer activities.
2.3. Topoisomerase Inhibition Assay
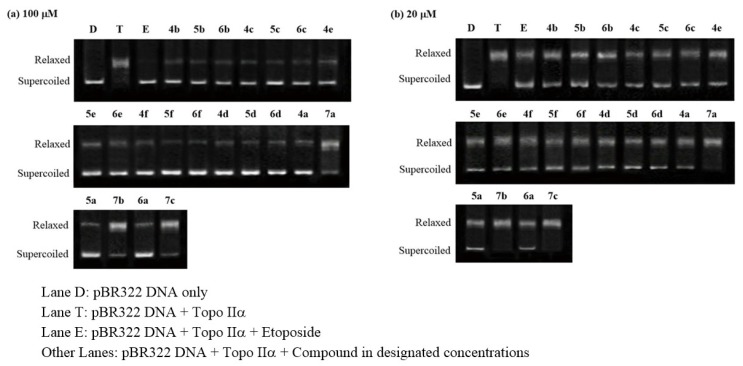
The conversion of supercoiled pBR322 plasmid DNA to relaxed DNA by Topo I and II was examined in the presence of the prepared compounds 4a–f, 5a–f, 6a–f, and 7a–c. Camptothecin and etoposide, well-known Topo I and II inhibitors, respectively, were used as positive controls. The effect of the tested compounds on Topo I and II activities were evaluated by Topo I/II relaxation assays. The reaction products, relaxed DNA could be distinguished from supercoiled DNA by gel electrophoresis and visualized using gel imaging system.
As shown in Figure 2 and Table 1, compounds 5a, 5b, and 6a displayed moderate and weak inhibitory activities against Topo I at concentrations of 100 μM and 20 μM, respectively. In addition, compounds 4a, 4b, 4d, 6b, and 6c also showed mild Topo I inhibitory activities at 100 μM. However, the other compounds, especially compounds with potent anticancer activity such as 5d, 5f, and 6f, were not inhibitory to Topo I even at 100 μM. Therefore, it is evident that the Topo I inhibition of the title compounds only have minor correlations with their anticancer activities.
Figure 2.
Topoisomerase I inhibitory activity of the prepared compounds 4a–f, 5a–f, 6a–f, and 7a–c. (a) The DNA relaxation assay of the prepared compounds at 100 μM; (b) The DNA relaxation assay of the selected active compounds at 20 μM.
Concerning Topo II inhibitory activity, it was observed that most compounds exhibited considerable inhibitory activity at concentrations of 100 and 20 μM (Figure 3 and Table 1). Among them, compound 5f with the strongest cytotoxic activity exhibited the most potent inhibitory activity with inhibition rates of 83.5% and 43.2% at concentrations of 100 and 20 μM, respectively. Compounds 5d and 6f also showed significant activities comparable to positive control etoposide. On the contrary, three dimer derivatives 7a–c with poor cytotoxic activities only showed weak inhibitory activities at 100 μM and 20 μM. The close correlations between the Topo II inhibitory activity and cytotoxic activity suggested that Topo II inhibition of the title compounds probably played an important role in their anticancer activity.
Figure 3.
Topoisomerase II inhibitory activity of the prepared compounds 4a–f, 5a–f, 6a–f, and 7a–c. (a) The DNA relaxation assay of the prepared compounds at 100 μM; (b) The DNA relaxation assay of the prepared compounds at 20 μM.
2.4. Molecular Docking
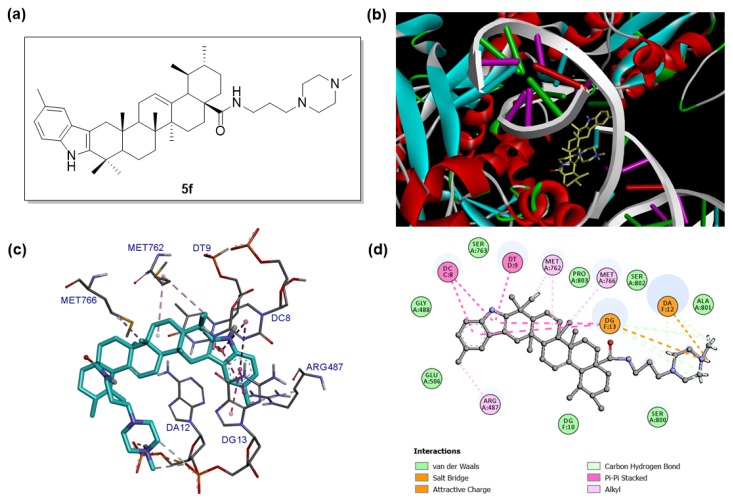
To gain more understanding between target compounds and Topo II, the binding modes of compound 5f with Topo II were explored by molecular docking based on the reported Topo IIα/DNA/inhibitor complex structure (PDB: 5GWK). The docking studies were performed by using the GLIDE docking module of Schrodinger suite 2015-1 and the docking results were analyzed and visualized by Discovery Studio 2016 Client. The binding modes of compound 5f with Topo IIα/DNA complex are shown in Figure 4.
Figure 4.
Binding mode of compound 5f with human topoisomerase IIα in complex with DNA (PDB: 5GWK). (a) Molecular structure of compound 5f. (b) Binding pose of compound 5f within the active site of Topo IIα. Ligand is presented as stick models and colored by atom type, whereas the protein is presented as ribbons and DNA is presented as arrows (for backbone) and ladders (for base pairs). (c) Detailed docked view of compound 5f in the active site. The amino acid residues and base pairs are presented as sticks and colored by atom type, while the interactions are presented as dash lines. (d) Two-dimensional projection drawing of compound 5f docked into the active site of Topo IIα.
It was observed that compound 5f could be suitably docked into the binding site of Topo IIα (Figure 4b,c), affording a significant docking score (−8.240). Specifically, compound 5f was deeply embedded into the major groove of DNA. The planar 5-methyl-indole moiety of 5f was intercalated into the gap of DNA double strand and formed π–π stacking interactions with Cytosine DC8, Thymine DT9 and Guanine DG13. The methyl group on the indole ring formed hydrophobic alkyl interactions with Arg487. Other hydrophobic alkyl interactions were also detected between the methyl group (C40), methylene groups (C7 and C8) and MET762, methyl group (C32) and MET766. The N-(3-(4-methylpiperazin-1-yl)propyl)carboxamide side chain of compound 5f also played an important role in the molecular docking. The nitrogen atom (N4) on the piperazine ring formed an electrostatic attractive charge interaction with the phosphate group of guanosine monophosphate residue (DG13), and the hydrogen on N4 formed a salt bridge with the phosphate group of adenosine monophosphate residue (DA12). In addition, compound 5f also formed van der Waals interactions with residues Gly488, Glu506, Ser763, Pro803, Ser802, Ala801, Ser800, and Guanine DG10 (Figure 4d). Taken together, the molecular docking results in combination with topoisomerase inhibition assay data indicated that compound 5f could be a promising Topo IIα inhibitor worthy of further investigations.
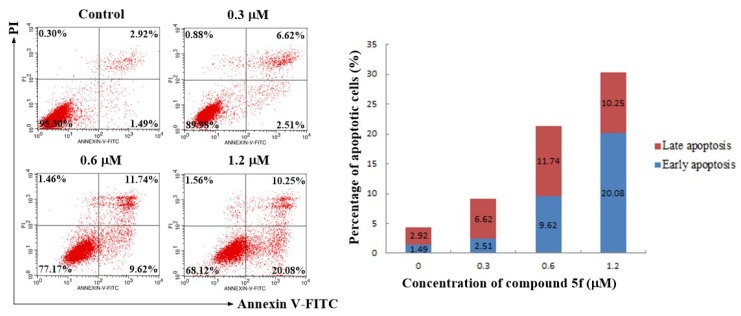
2.5. Annexin V-FITC/PI Staining Assay
To further investigate the role of apoptosis in the antitumor activity of compound 5f, SMMC-7721 cells were treated with different concentrations of compound 5f (0, 0.3, 0.6, and 1.2 μM) for 48 h. Then the treated cells were stained with Annexin V-FITC/PI followed by flow cytometry analysis. As shown in Figure 5, the percentage of early and late apoptotic cells (early apoptosis: Lower right quadrant, AV+/PI−; late apoptosis: Upper right quadrant, AV+/PI+) considerably increased from 4.41% (0 μM) to 9.13% (0.3 μM), 21.36% (0.6 μM), and 30.33% (1.2 μM), respectively. These results indicated that compound 5f could trigger the apoptosis of SMMC-7721 cells in a dose-dependent manner.
Figure 5.
Apoptotic assay by flow cytometry. SMMC-7721 cells were treated with compound 5f at 0, 0.3, 0.6, and 1.2 μM for 48 h. The cells were stained with Annexin V-FITC/PI and were detected by flow cytometry.
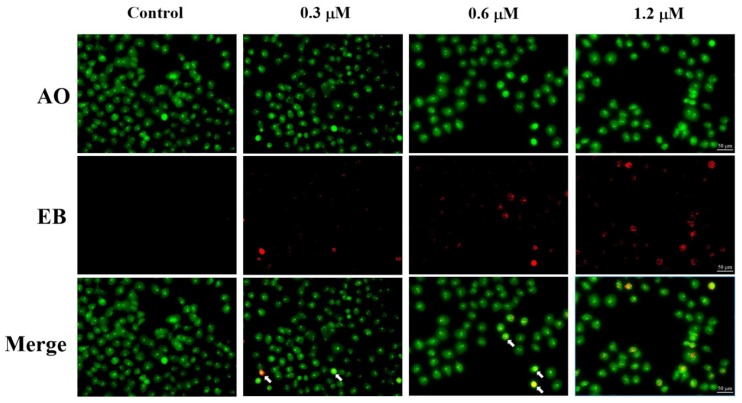
2.6. AO/EB Staining Assay
Apoptosis induced by compound 5f was further evaluated using acridine orange / ethidium bromide (AO/EB) double staining. AO, as a vital dye showing green fluorescence, can pass through cell membrane of living or dead cells, while EB only stains apoptotic or necrotic cells that have lost their integrity and appear red fluorescence. Therefore, the normal cells will be stained only by AO with bright green while apoptotic cells will be stained by AO and EB with red-orange [26,27]. As shown in Figure 6, the cell population in control appeared in green color with usual morphology. Fluorescence images of cells treated with various concentrations of 5f displayed the increase of cells with red-orange fluorescence, the reduced number of normal cells and also displayed distorted structural characteristics such as chromatin condensation, cell shrinkage and membrane blebbing, which further indicated that compound 5f induced apoptosis in SMMC-7721 cells.
Figure 6.
AO/EB staining of compound 5f in SMMC-7721 cells. SMMC-7721 cells were treated with compound 5f at 0, 0.3, 0.6, and 1.2 μM for 48 h. The cells were stained with AO/EB and were detected by fluorescence microscope. White arrow: apoptotic cells.
2.7. ROS Generation Assay
Reactive oxygen species (ROS), which can exert oxidative stress to cells and lead to disturbances in mitochondrial membrane potential, are important mediators for the induction of apoptosis in many cancer cells [28]. To determine whether the apoptosis induced by compound 5f was associated with intracellular ROS level, SMMC-7721 cells treated with different concentrations of 5f were stained with the fluorescent probe DCFH-DA and analyzed by flow cytometry. As shown in Figure 7, the cells with elevated ROS levels increased from 3.97% (Control) to 9.89% (0.3 μM), 19.84% (0.6 μM) and 34.61% (1.2 μM), which indicated that compound 5f could induce ROS generation in SMMC-7721 cells in a dose-dependent manner.
Figure 7.
Effects of compound 5f on the intracellur reactive oxygen species (ROS) level in SMMC-7721 cells. SMMC-7721 cells were treated with different concentrations of compound 5f (0, 0.3, 0.6, and 1.2 μM) for 48 h, stained with DCFH-DA and analyzed using flow cytometry.
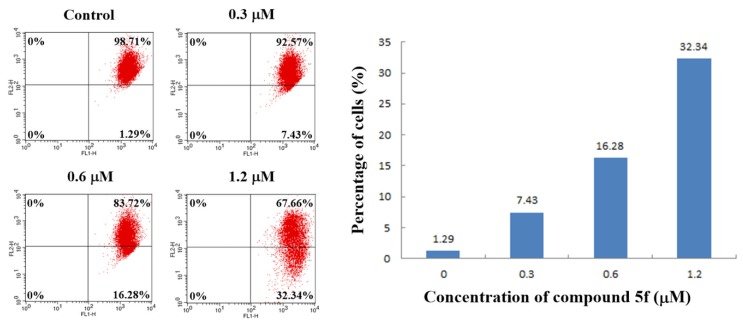
2.8. Mitochondrial Membrane Potential Assay
It has been widely believed that intracellular ROS accumulation could decrease mitochondrial membrane potential (ΔΨm), which is a characteristic phenomenon of early apoptosis [29]. To investigate the correlation between ΔΨm and apoptosis induced by compound 5f, the measurement of cells with decreased ΔΨm were carried out by JC-1 assay kit on the instructions of the manual. As shown in Figure 8, the percentage of cells with low ΔΨm (Lower right quadrant) increased from 1.29% (Control) to 7.43% (0.3 μM), 16.28% (0.6 μM), and 32.34% (1.2 μM). Therefore, the observations clearly showed that compound 5f could decrease mitochondrial membrane potential (ΔΨm) of SMMC-7721 cells in a dose-dependent manner, and the compound probably induced the apoptosis through ROS-mediated mitochondrial pathway.
Figure 8.
Compound 5f-induced loss of the mitochondrial membrane potential (ΔΨm). SMMC-7721 cells were treated with compound 5f (0, 0.3, 0.6, and 1.2 μM) for 24 h, incubated with JC-1, and analyzed using flow cytometry.
3. Discussion
A series of new indole derivatives of ursolic acid bearing N-(aminoalkyl)carboxamide side chains were designed, synthesized, and screened for their in vitro cytotoxic activity against two human hepatocarcinoma cell lines (SMMC-7721, HepG2) and a normal human hepatocyte cell line (LO2). A number of derivatives showed considerable cytotoxic activities in MTT assay. Especially, compound 5f exhibited the most potent inhibitory activity against SMMC-7721 and HepG2 cells among all derivatives and significantly lower cytotoxicity to LO2 cells. Concerning structure-activity relationships, the order of effects of the N-containing moieties to the cytotoxicity could be summarized as N-methylpiperazine > NH2, N(Me)2, N(Et)2, and piperidine > morpholine. In addition, 3C linker on the side chain could be more beneficial to the cytotoxicity than 2C and 4C alkyl linkers. In vitro pharmacological studies showed that compound 5f could significantly inhibit Topoisomerase II activity and thus hinder the relaxation of supercoiled DNA. In addition, it could induce intracellular ROS generation, decrease mitochondrial membrane potential, and finally lead to the apoptosis of SMMC-7721 cells. Due to its strong Topoisomerase II inhibitory activity and anticancer activity, this compound exhibited potential as an anticancer drug candidate. In addition, this class of derivatives could expand the structural diversity of anticancer agents and affirm the perspectives of further studies. More investigations on the structural modification and the structure–activity relationship of this scaffold will be carried out in the future.
4. Materials and Methods
4.1. General
Melting points were measured on an XT-4 apparatus (Taike Corp., Beijing, China) and were uncorrected. IR spetra were measured on a Nexus 870 FT-IR spectrometer (Thermo Fisher Scientific, Waltham, MA, USA), and the absorption bands were expressed in cm−1. The HRMS spectra were recorded on a high-resolution mass spectrometer equipped with electrospray (ESI) and nanospray sources, and a quadrupole-time of flight hybrid analyzer (Q-TOF Premier/nanoAquity, Waters, Milford, MA, USA). 1H and 13C NMR spectra were obtained in CDCl3 on a Bruker DRX-600 NMR spectrometer (Billerica, MA, USA) using TMS as internal standard. Reactions and the resulted products were monitored by TLC which was carried out on TLC Silica gel 60 F254 Aluminum sheets from Merck KGaA, Darmstadt, Germany, and visualized in UV light (254 nm). Silica gel (300–400 mesh) for column chromatography was purchased from Qingdao Marine Chemical Factory (Qingdao, China). The reagents (chemicals), all being of A.R. grade, were purchased from Shanghai Chemical Reagent Company (Shanghai, China) and Energy Chemical (Shanghai, China). Ursolic acid (95%) was bought from Jingzhu Biological Technology Co., Ltd. (Nanjing, China).
4.2. Procedure for the Synthesis of Compound 3
To a solution of ursolic acid (1.0 g, 2.2 mmol) in acetone (150 mL) was added dropwise to 1 mL of Jones reagent (0.27 g CrO3 dissolved in 0.23 mL concentrated H2SO4 and 0.73 mL H2O) at 0 °C. The mixture was then stirred at room temperature for 5 h (TLC control). Subsequently, isopropanol (45 mL) was added and the mixture was stirred for additional 0.5 h. Then the mixture was filtered, the filtrate was concentrated in vacuo to remove solvent. The residue was then recrystallized in MeOH to afford compound 2 as a white solid (0.56 g, yield 76%). The spectral data of compound 2 were in accordance with the records of previous literature [30].
To a solution of compound 2 (0.44 g, 1 mmol) in 10 mL EtOH was added 0.33 g (2.1 mmol) of p-tolylhydrazine hydrochloride and 0.5 mL concentrated HCl. The mixture was refluxed at 85 °C for 3 h. After cooling, the mixture was poured into 100 mL of ice-cold water and extracted with CH2Cl2 (3 × 50 mL). The organic layer was combined, washed with water, saturated NaHCO3 solution and brine, dried over anhydrous Na2SO4 and concentrated to give a crude product, which was purified by silica gel column chromatography (petroleum ether–acetone 15:1, v/v) to afford compound 3 as a white solid (69%, 0.37 g) [25].
4.3. General Procedure for the Synthesis of Compounds 4a–f, 5a–f, and 6a–f
To a solution of compound 3 (54 mg, 0.1 mmol) in CH2Cl2 (5 mL) were added HOBt (19 mg, 0.12 mmol) and DCC (24 mg, 0.12 mmol). The reaction mixture was stirred at room temperature for 0.5 h and different amine (0.12 mmol) was added. The mixture was stirred for 12 h at room temperature. At the end of reaction, the mixture was filtered, and the filtrate concentrated in vacuo. The obtained residue was purified by silica gel column chromatography (petroleum ether–acetone 30:1, v/v) to afford the corresponding product 4a–f, 5a–f, and 6a–f with 2-, 3- and 4-carbon side chains, respectively (Scheme 1).
4.3.1. N-(2-Aminoethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4a)
Yellow solid; Yield: 72%; mp. 228–230 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.93 (d, J = 6.2 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.29–1.90 (m, 15H), 1.90–2.08 (m, 2H), 2.08–2.25 (m, 3H), 2.32 (brs, 2H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 2.83 (brs, 2H), 3.12 (m, 1H), 3.42 (m, 1H), 5.45 (brs, 1H), 6.46 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.21 (s, 1H), 7.87 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.92, 16.95, 17.36, 19.39, 21.35, 21.60, 23.35, 23.37, 23.66, 25.01, 28.07, 31.09, 31.12, 32.46, 34.15, 37.23, 37.50, 38.10, 39.19, 39.80, 39.99, 41.19, 41.77, 42.77, 46.38, 48.07, 53.30, 54.10, 106.32, 110.20, 117.81, 122.44, 126.17, 128.10, 128.54, 134.59, 139.51, 141.19, 178.82; IR (KBr, cm−1): 3311, 2924, 2854, 1635, 1519, 1457, 1378, 1305, 1276, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C39H58N3O: 584.4580; found: 584.4584.
4.3.2. N-(2-(Dimethylamino)ethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4b)
Yellow solid; Yield: 89%; mp. 166–168 °C; 1H NMR (600 MHz, CDCl3): δ 0.90 (s, 3H), 0.93 (d, J = 6.4 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.2 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.35–2.00 (m, 16H), 2.10–2.23 (m, 3H), 2.25 (s, 6H), 2.30–2.42 (m, 3H), 2.43 (s, 3H), 2.78 (d, J = 14.8 Hz, 1H), 3.19 (m, 1H), 3.35 (m, 1H), 5.41 (brs, 1H), 6.59 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.20 (s, 1H), 7.87 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.91, 17.00, 17.34, 19.43, 21.34, 21.59, 23.31, 23.37, 23.69, 25.02, 28.12, 31.08, 31.12, 32.60, 34.18, 36.98, 37.28, 37.41, 38.13, 39.23, 39.86, 40.02, 42.79, 45.35, 46.44, 48.01, 53.40, 54.30, 57.75, 106.33, 110.20, 117.81, 122.41, 126.24, 128.09, 128.59, 134.64, 139.25, 141.22, 178.33; IR (KBr, cm−1): 3298, 2923, 2854, 1633, 1510, 1457, 1380, 1307, 1186, 1054, 793; HRMS (ESI): m/z [M + H]+ calcd. for C41H62N3O: 612.4893; found: 612.4887.
4.3.3. N-(2-(Diethylamino)ethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4c)
Yellow solid; Yield: 88%; mp. 163–165 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.93 (d, J = 6.3 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.3 Hz, 3H), 1.04 (t, J = 6.9 Hz, 6H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.31–2.00 (m, 16H), 2.12 (m, 1H), 2.18–2.25 (m, 2H), 2.43 (s, 3H), 2.53 (m, 4H), 2.56–2.61 (m, 3H), 2.78 (d, J = 14.8 Hz, 1H), 3.10 (m, 1H), 3.42 (m, 1H), 5.41 (brs, 1H), 6.61 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.19 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.96 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 11.79, 15.82, 16.85, 17.42, 19.40, 21.34, 21.59, 23.34, 23.36, 23.55, 24.96, 28.05, 31.08, 31.09, 32.43, 34.14, 36.87, 37.17, 37.51, 38.10, 39.29, 39.77, 39.95, 42.66, 46.38, 46.64, 47.92, 51.40, 53.32, 54.15, 106.25, 110.19, 117.76, 122.34, 125.99, 128.01, 128.53, 134.59, 139.24, 141.23, 178.03; IR (KBr, cm−1): 3393, 3301, 2924, 2854, 1634, 1508, 1457, 1378, 1306, 1185, 1083, 795; HRMS (ESI): m/z [M + H]+ calcd. for C43H66N3O: 640.5206; found: 640.5203.
4.3.4. N-(2-(Piperidin-1-yl)ethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4d)
Yellow solid; Yield: 90%; mp. 195–197 °C; 1H NMR (600 MHz, CDCl3): δ 0.89 (s, 3H), 0.94 (d, J = 6.5 Hz, 3H), 0.95 (s, 3H), 0.97 (d, J = 6.5 Hz, 3H), 1.16 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.35–2.10 (m, 23H), 2.10–2.26 (m, 3H), 2.36–2.42 (m, 6H), 2.43 (s, 3H), 2.78 (d, J = 14.8 Hz, 1H), 3.23 (m, 1H), 3.37 (m, 1H), 5.44 (brs, 1H), 6.62 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.21 (s, 1H), 7.85 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.87, 16.90, 17.45, 19.44, 21.36, 21.59, 23.35, 23.37, 23.63, 24.48, 25.02, 26.18, 28.09, 31.13, 31.15, 32.48, 34.18, 36.08, 37.24, 37.51, 38.15, 39.27, 39.84, 40.03, 42.75, 46.43, 48.00, 53.37, 54.25, 54.44, 57.15, 106.34, 110.20, 117.79, 122.42, 126.04, 128.09, 128.58, 134.64, 139.29, 141.22, 178.08; IR (KBr, cm−1): 3312, 2926, 2853, 1633, 1508, 1456, 1379, 1305, 1127, 794, 736; HRMS (ESI): m/z [M + H]+ calcd. for C44H66N3O: 652.5206; found: 652.5212.
4.3.5. N-(2-Morpholinoethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4e)
Yellow solid; Yield: 82%; mp. 104–105 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.93 (d, J = 6.5 Hz, 3H), 0.94 (s, 3H), 0.98 (d, J = 6.5 Hz, 3H), 1.16 (s, 3H), 1.20 (s, 3H), 1.34 (s, 3H), 1.36–2.35 (m, 22H), 2.43 (s, 3H), 2.50 (brs, 4H), 2.77 (d, J = 14.8 Hz, 1H), 3.26 (m, 1H), 3.43 (m, 1H), 3.76 (brs, 4H), 5.44 (brs, 1H), 6.53 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.76 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.92, 16.97, 17.45, 19.44, 21.36, 21.60, 22.82, 23.41, 23.69, 25.05, 25.79, 28.10, 30.36, 31.16, 31.58, 34.18, 37.24, 37.53, 38.16, 39.30, 39.85, 40.05, 42.83, 46.40, 48.10, 53.36, 53.54, 54.32, 57.14, 67.01, 106.37, 110.19, 117.87, 122.51, 125.89, 128.21, 128.59, 134.64, 139.63, 141.14, 178.22; IR (KBr, cm−1): 3313, 2954, 2923, 2852, 1635, 1498, 1458, 1379, 1303, 1266, 1187, 1119, 1081, 967, 792, 739; HRMS (ESI): m/z [M + H]+ calcd. for C43H64N3O2: 654.4999; found: 654.4995.
4.3.6. N-(2-(4-Methylpiperazin-1-yl)ethyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (4f)
Yellow solid; Yield: 87%; mp. 188–190 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.94 (d, J = 6.5 Hz, 3H), 0.95 (s, 3H), 0.97 (d, J = 6.4 Hz, 3H), 1.15 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.31–2.05 (m, 16H), 2.10–2.25 (m, 3H), 2.31 (s, 3H), 2.43 (s, 3H), 2.48–2.60 (m, 12H), 2.77 (d, J = 14.8 Hz, 1H), 3.23 (m, 1H), 3.37 (m, 1H), 5.43 (brs, 1H), 6.53 (brs, 1H), 6.92 (d, J = 8.2 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.97 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.96, 16.87, 17.43, 19.37, 21.34, 21.58, 23.27, 23.32, 23.66, 25.01, 28.01, 31.08, 31.09, 32.39, 34.13, 36.00, 37.22, 37.43, 38.09, 39.22, 39.75, 39.98, 42.75, 46.13, 46.34, 47.99, 52.88, 53.27, 54.21, 55.28, 56.46, 106.17, 110.21, 117.70, 122.37, 125.91, 128.01, 128.46, 134.58, 139.44, 141.20, 178.16; IR (KBr, cm−1): 3404, 3311, 2924, 2868, 2853, 2791, 1635, 1507, 1457, 1377, 1301, 1165, 1094, 1013, 795, 736; HRMS (ESI): m/z [M + H]+ calcd. for C44H67N4O: 667.5315; found: 667.5309.
4.3.7. N-(3-Aminopropyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5a)
Yellow solid; Yield: 73%; mp. 205–207 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.91 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.1 Hz, 3H), 1.14 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.32–2.10 (m, 19H), 2.10–2.30 (m, 3H), 2.42 (s, 3H), 2.62 (brs, 2H), 2.76 (d, J = 14.6 Hz, 1H), 2.78 (m, 2H), 3.13 (m, 1H), 3.44 (m, 1H), 5.42 (brs, 1H), 6.39 (brs, 1H), 6.93 (d, J = 8.2 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.20 (s, 1H), 7.82 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.91, 16.91, 17.39, 19.40, 21.35, 21.60, 23.36, 23.39, 23.65, 25.02, 26.67, 28.04, 31.08, 31.14, 32.42, 34.16, 37.22, 37.56, 38.12, 39.23, 39.65, 39.79, 40.00, 42.79, 46.38, 47.99, 53.30, 54.07, 55.34, 106.34, 110.19, 117.82, 122.46, 126.05, 128.15, 128.55, 134.59, 139.73, 141.19, 178.87; IR (KBr, cm−1): 3294, 2924, 2854, 1637, 1524, 1457, 1379, 1306, 1187, 1078, 963, 794, 738; HRMS (ESI): m/z [M + H]+ calcd. for C40H60N3O: 598.4736; found: 598.4730.
4.3.8. N-(3-(Dimethylamino)propyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5b)
Yellow powder solid; Yield: 81%; mp. 186–188 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.92 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 7.4 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.35–2.00 (m, 18H), 2.10–2.25 (m, 3H), 2.26 (s, 6H), 2.30–2.40 (m, 3H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 3.07 (m, 1H), 3.48 (m, 1H), 5.39 (t, J = 3.2 Hz, 1H), 6.93 (d, J = 8.2 Hz, 1H), 6.96 (brs, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.20 (s, 1H), 7.81 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.86, 16.94, 17.49, 19.44, 21.36, 21.59, 23.38, 23.49, 23.61, 24.92, 26.35, 28.08, 31.06, 31.14, 32.51, 34.17, 37.22, 37.70, 38.15, 39.21, 39.39, 39.83, 39.94, 42.69, 45.68, 46.44, 47.82, 53.38, 54.08, 58.76, 106.41, 110.19, 117.83, 122.44, 125.91, 128.13, 128.60, 134.65, 139.45, 141.24, 177.93; IR (KBr, cm−1): 3411, 3298, 2946, 2924, 2866, 1636, 1522, 1459, 1380, 1305, 1185, 1050, 794; HRMS (ESI): m/z [M + H]+ calcd. for C42H64N3O: 626.5049; found: 626.5053.
4.3.9. N-(3-(Diethylamino)propyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5c)
Yellow solid; Yield: 87%; mp. 147–149 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.93 (d, J = 6.4 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.5 Hz, 3H), 1.06 (t, J = 7.1 Hz, 6H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.39–1.85 (m, 16H), 1.95–2.05 (m, 3H), 2.13 (m, 1H), 2.15–2.25 (m, 2H), 2.43 (s, 3H), 2.51 (t, J = 6.4 Hz, 2H), 2.58 (q, J = 7.1 Hz, 4H), 2.77 (d, J = 14.8 Hz, 1H), 3.10 (m, 1H), 3.42 (m, 1H), 5.41 (brs, 1H), 6.65 (brs, 1H), 6.93 (d, J = 8.1 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.85 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 11.28, 15.85, 16.95, 17.39, 19.44, 21.37, 21.59, 23.37, 23.46, 23.61, 24.92, 26.29, 28.10, 31.11, 31.13, 32.51, 34.17, 37.22, 37.60, 38.15, 39.00, 39.25, 39.82, 39.93, 42.70, 46.44, 46.86, 47.78, 51.54, 53.38, 53.97, 106.36, 110.20, 117.81, 122.41, 125.93, 128.09, 128.58, 134.64, 139.56, 141.21, 177.97; IR (KBr, cm−1): 3411, 3304, 2962, 2925, 2855, 1636, 1520, 1456, 1380, 1306, 1188, 1084, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C44H68N3O: 654.5362; found: 654.5358.
4.3.10. N-(3-(Piperidin-1-yl)propyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5d)
Yellow solid; Yield: 88%; mp. 183–185 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.93 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.29 (s, 3H), 1.30–1.90 (m, 23H), 1.90–2.06 (m, 2H), 2.07–2.25 (m, 3H), 2.30–2.40 (m, 6H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 3.10 (m, 1H), 3.41 (m, 1H), 5.42 (brs, 1H), 6.66 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.20 (s, 1H), 7.82 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.87, 17.01, 17.38, 19.45, 21.38, 21.59, 23.39, 23.52, 23.67, 24.37, 24.87, 25.64, 25.88, 28.07, 31.09, 31.15, 32.54, 34.18, 37.23, 37.62, 38.16, 39.12, 39.23, 39.84, 39.92, 42.69, 45.40, 46.45, 47.78, 53.38, 53.97, 54.87, 106.41, 110.19, 117.83, 122.44, 125.90, 128.13, 128.60, 134.64, 139.58, 141.21, 178.01; IR (KBr, cm−1): 3300, 2925, 2854, 1635, 1522, 1455, 1379, 1306, 1188, 1125, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C45H68N3O: 666.5362; found: 666.5369.
4.3.11. N-(3-Morpholinopropyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5e)
Yellow solid; Yield: 90%; mp. 108–110 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.93 (d, J = 6.3 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.20 (s, 3H), 1.33 (s, 3H), 1.36–2.35 (m, 22H), 2.42 (s, 3H), 2.43–2.50 (m, 2H), 2.51 (brs, 4H), 2.77 (d, J = 14.8 Hz, 1H), 3.09 (m, 1H), 3.44 (m, 1H), 3.77 (brs, 4H), 5.41 (brs, 1H), 6.45 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.20 (s, 1H), 7.89 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.87, 16.97, 17.39, 19.40, 21.35, 21.59, 23.37, 23.43, 23.65, 24.93, 25.47, 28.03, 31.04, 31.11, 32.46, 34.15, 37.20, 37.58, 38.11, 38.49, 39.25, 39.78, 39.93, 42.73, 46.37, 47.85, 53.30, 53.79, 54.08, 57.37, 66.71, 106.27, 110.19, 117.79, 122.41, 125.89, 128.09, 128.51, 134.59, 139.72, 141.17, 178.21; IR (KBr, cm−1): 3313, 2954, 2923, 2853, 1638, 1520, 1459, 1378, 1305, 1186, 1119, 1082, 968, 795, 739; HRMS (ESI): m/z [M + H]+ calcd. for C44H66N3O2: 668.5155; found: 668.5161.
4.3.12. N-(3-(4-Methylpiperazin-1-yl)propyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (5f)
Yellow solid; Yield: 88%; mp. 171–173 °C; 1H NMR (600 MHz, CDCl3): δ 0.86 (s, 3H), 0.93 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.4 Hz, 3H), 1.14 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.30–1.90 (m, 17H), 1.95–2.25 (m, 5H), 2.30 (s, 3H), 2.40 (m, 4H), 2.42 (s, 3H), 2.42–2.52 (m, 6H), 2.77 (d, J = 14.8 Hz, 1H), 3.07 (m, 1H), 3.45 (m, 1H), 5.41 (brs, 1H), 6.51 (brs, 1H), 6.92 (d, J = 8.0 Hz, 1H), 7.17 (d, J = 8.1 Hz, 1H), 7.20 (s, 1H), 7.95 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.85, 16.92, 17.37, 19.38, 21.32, 21.58, 23.34, 23.44, 23.63, 24.86, 25.77, 28.01, 31.03, 31.09, 32.44, 34.13, 37.18, 37.53, 38.09, 38.89, 39.21, 39.76, 39.91, 42.68, 46.04, 46.36, 47.78, 53.29, 53.36, 54.02, 55.07, 57.24, 106.23, 110.18, 117.75, 122.36, 125.86, 128.02, 128.49, 134.58, 139.68, 141.19, 178.02; IR (KBr, cm−1): 3295, 2924, 2870, 2847, 2796, 1637, 1520, 1458, 1376, 1303, 1283, 1186, 1014, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C45H69N4O: 681.5471; found: 681.5475.
4.3.13. N-(4-Aminobutyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6a)
Yellow solid; Yield: 72%; mp. 256–258 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.92 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.4 Hz, 3H), 1.15 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.30–2.10 (m, 21H), 2.10–2.30 (m, 3H), 2.43 (s, 3H), 2.45 (brs, 2H), 2.76 (t, J = 5.2 Hz, 2H), 2.77 (d, J = 15.2 Hz, 1H), 3.03 (m, 1H), 3.36 (m, 1H), 5.43 (brs, 1H), 6.09 (t, J = 5.3 Hz, 1H), 6.93 (dd, J = 8.2, 1.1 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.20 (s, 1H), 7.86 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.93, 16.93, 17.39, 19.39, 21.36, 21.61, 23.31, 23.39, 23.66, 25.08, 26.87, 28.05, 31.09, 31.13, 31.56, 32.44, 34.16, 37.23, 37.40, 38.12, 39.23, 39.37, 39.78, 40.03, 41.52, 42.84, 46.38, 47.95, 53.31, 54.21, 106.30, 110.19, 117.80, 122.44, 125.98, 128.11, 128.53, 134.58, 139.96, 141.18, 178.41; IR (KBr, cm−1): 3309, 2924, 2855, 1638, 1525, 1456, 1381, 1305, 1185, 1078, 967, 792, 738; HRMS (ESI): m/z [M + H]+ calcd. for C41H62N3O: 612.4893; found: 612.4887.
4.3.14. N-(4-(Dimethylamino)butyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6b)
Yellow solid; Yield: 90%; mp. 163–165 °C; 1H NMR (600 MHz, CDCl3): δ 0.86 (s, 3H), 0.91 (d, J = 6.4 Hz, 3H), 0.95 (s, 3H), 0.96 (d, J = 6.0 Hz, 3H), 1.14 (s, 3H), 1.20 (s, 3H), 1.29 (s, 3H), 1.35–2.00 (m, 20H), 2.10–2.25 (m, 3H), 2.32 (s, 6H), 2.30–2.42 (m, 3H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 3.03 (m, 1H), 3.35 (m, 1H), 5.41 (t, J = 3.1 Hz, 1H), 6.15 (t, J = 5.3 Hz, 1H), 6.92 (d, J = 8.1 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.20 (s, 1H), 7.99 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.89, 16.87, 17.36, 19.38, 21.34, 21.60, 23.32, 23.35, 23.63, 24.49, 24.99, 27.15, 28.02, 31.05, 31.11, 32.41, 34.15, 37.21, 37.42, 38.09, 39.16, 39.20, 39.76, 39.98, 42.78, 44.93, 46.36, 47.89, 53.30, 54.09, 58.86, 106.22, 110.22, 117.76, 122.38, 125.95, 128.04, 128.50, 134.59, 139.84, 141.22, 178.35; IR (KBr, cm−1): 3287, 2924, 2855, 1637, 1523, 1459, 1306, 1276, 1187, 1083, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C43H66N3O: 640.5206; found: 640.5201.
4.3.15. N-(4-(Diethylamino)butyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6c)
Yellow solid; Yield: 88%; mp. 132–134 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.92 (d, J = 6.4 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.4 Hz, 3H), 1.02 (t, J = 7.1 Hz, 6H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.36–2.05 (m, 21H), 2.13 (m, 1H), 2.18–2.25 (m, 2H), 2.43 (s, 3H), 2.45 (m, 2H), 2.53 (q, J = 7.1 Hz, 4H), 2.77 (d, J = 14.8 Hz, 1H), 3.04 (m, 1H), 3.37 (m, 1H), 5.43 (brs, 1H), 6.01 (brs, 1H), 6.93 (d, J = 8.2 Hz, 1H), 7.18 (d, J = 8.2 Hz, 1H), 7.21 (s, 1H), 7.88 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 11.61, 15.90, 16.91, 17.38, 19.40, 21.34, 21.58, 23.31, 23.34, 23.66, 24.73, 25.08, 27.54, 28.07, 31.09, 31.11, 32.45, 34.17, 37.26, 37.37, 38.12, 39.26, 39.53, 39.80, 40.04, 42.86, 46.41, 46.93, 47.94, 52.60, 53.35, 54.27, 106.26, 110.21, 117.77, 122.41, 125.97, 128.07, 128.54, 134.64, 140.03, 141.21, 178.22; IR (KBr, cm−1): 3304, 2964, 2926, 2869, 1637, 1524, 1456, 1379, 1306, 1262, 1086, 1051, 796; HRMS (ESI): m/z [M + H]+ calcd. for C45H70N3O: 668.5519; found: 668.5523.
4.3.16. N-(4-(Piperidin-1-yl)butyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6d)
Yellow solid; Yield: 90%; mp. 121–123 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.93 (d, J = 6.4 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.30 (s, 3H), 1.37–2.08 (m, 27H), 2.08–2.25 (m, 3H), 2.25–2.42 (m, 6H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 3.03 (m, 1H), 3.35 (m, 1H), 5.43 (brs, 1H), 6.02 (brs, 1H), 6.93 (d, J = 8.1 Hz, 1H), 7.18 (d, J = 8.1 Hz, 1H), 7.21 (s, 1H), 7.85 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.91, 16.93, 17.38, 19.41, 21.35, 21.59, 23.33, 23.37, 23.67, 24.18, 24.30, 25.08, 25.64, 27.51, 28.08, 31.11, 31.14, 32.47, 34.17, 37.27, 37.41, 38.13, 39.25, 39.45, 39.82, 40.04, 42.86, 46.43, 47.94, 53.35, 54.22, 54.46, 58.76, 106.34, 110.21, 117.81, 122.46, 125.99, 128.12, 128.57, 134.65, 140.01, 141.22, 178.28; IR (KBr, cm−1): 3298, 2924, 2853, 1637, 1526, 1456, 1379, 1306, 1188, 1082, 963, 793, 738; HRMS (ESI): m/z [M + H]+ calcd. for C46H70N3O: 680.5519; found: 680.5516.
4.3.17. N-(4-Morpholinobutyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6e)
Yellow solid; Yield: 86%; mp. 221–223 °C; 1H NMR (600 MHz, CDCl3): δ 0.88 (s, 3H), 0.92 (d, J = 6.4 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.29 (s, 3H), 1.30–2.05 (m, 23H), 2.13 (m, 1H), 2.22 (m, 2H), 2.35 (brs, 4H), 2.43 (s, 3H), 2.77 (d, J = 14.8 Hz, 1H), 3.04 (m, 1H), 3.37 (m, 1H), 3.71 (m, 4H), 5.43 (brs, 1H), 5.99 (t, J = 5.5 Hz, 1H), 6.93 (d, J = 8.2 Hz, 1H), 7.18 (d, J = 8.0 Hz, 1H), 7.21 (s, 1H), 7.93 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.91, 16.90, 17.39, 19.37, 21.34, 21.59, 23.29, 23.35, 23.64, 24.18, 25.08, 27.39, 28.04, 31.07, 31.10, 32.42, 34.14, 37.23, 37.37, 38.09, 39.26, 39.48, 39.77, 40.01, 42.85, 46.37, 47.93, 53.28, 53.81, 54.27, 58.65, 67.01, 106.22, 110.20, 117.78, 122.42, 125.93, 128.07, 128.50, 134.60, 140.08, 141.17, 178.27; IR (KBr, cm−1): 3314, 2953, 2925, 2853, 1635, 1530, 1456, 1376, 1305, 1274, 1118, 1084, 788, 732; HRMS (ESI): m/z [M + H]+ calcd. for C45H68N3O2: 682.5312; found: 682.5317.
4.3.18. N-(4-(4-Methylpiperazin-1-yl)butyl)-5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide (6f)
Yellow solid; Yield: 91%; mp. 146–148 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 3H), 0.92 (d, J = 6.1 Hz, 3H), 0.96 (s, 3H), 0.97 (d, J = 6.5 Hz, 3H), 1.15 (s, 3H), 1.21 (s, 3H), 1.29 (s, 3H), 1.30–2.10 (m, 20H), 2.10–2.25 (m, 3H), 2.28 (s, 3H), 2.35 (brs, 4H), 2.43 (s, 3H), 2.44–2.51 (m, 7H), 2.77 (d, J = 14.8 Hz, 1H), 3.04 (m, 1H), 3.35 (m, 1H), 5.42 (brs, 1H), 5.99 (brs, 1H), 6.93 (d, J = 8.0 Hz, 1H), 7.18 (d, J = 8.0 Hz, 1H), 7.20 (s, 1H), 7.88 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.94, 16.91, 17.38, 19.39, 21.33, 21.57, 23.30, 23.35, 23.66, 24.51, 25.08, 27.43, 28.06, 31.08, 31.11, 32.44, 34.16, 37.26, 37.37, 38.11, 39.25, 39.53, 39.80, 40.03, 42.86, 46.01, 46.40, 47.94, 53.13, 53.33, 54.27, 55.07, 58.16, 106.27, 110.20, 117.78, 122.42, 125.95, 128.08, 128.54, 134.62, 140.04, 141.19, 178.26; IR (KBr, cm−1): 3289, 2925, 2854, 2799, 1638, 1522, 1457, 1375, 1305, 1282, 1165, 1013, 794, 737; HRMS (ESI): m/z [M + H]+ calcd. for C46H71N4O: 695.5628; found: 695.5620.
4.4. General Procedure for the Synthesis of Compounds 7a–c
To a solution of compound 3 (98 mg, 0.18 mmol) in CH2Cl2 (5 mL) were added HOBt (36 mg, 0.24 mmol) and DCC (48 mg, 0.24 mmol). The reaction mixture was stirred at room temperature for 0.5 h and then ethane-1,2-diamine, propane-1,3-diamine, or butane-1,4-diamine (0.09 mmol) was added. The mixture was stirred for 12 h at room temperature. At the end of reaction, the mixture was worked up as above to afford the corresponding dimer product 7a–c with 2-, 3- and 4-carbon chain, respectively (Scheme 2).
4.4.1. N,N′-(ethane-1,2-diyl)-bis(5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide) (7a)
Yellow solid; Yield: 61%; mp. 301–303 °C; 1H NMR (600 MHz, CDCl3): δ 0.86 (s, 6H), 0.93 (d, J = 6.2 Hz, 6H), 0.95 (s, 6H), 0.98 (s, 6H), 1.16 (s, 6H), 1.19 (s, 6H), 1.29 (s, 6H), 1.30–2.10 (m, 34H), 2.10–2.28 (m, 6H), 2.44 (s, 6H), 2.77 (d, J = 14.8 Hz, 2H), 3.14 (m, 2H), 3.48 (m, 2H), 5.48 (brs, 2H), 6.58 (brs, 2H), 6.93 (d, J = 7.9 Hz, 2H), 7.18 (d, J = 8.0 Hz, 2H), 7.21 (s, 2H), 7.86 (brs, 1H), 7.92 (brs, 1H); 13C NMR (150 MHz, CDCl3): δ 15.88, 16.85, 17.39, 19.38, 21.39, 21.61, 23.35, 23.39, 23.64, 25.02, 25.07, 28.08, 31.11, 32.37, 34.07, 34.14, 37.19, 37.65, 38.11, 39.22, 39.77, 39.97, 42.68, 46.38, 47.86, 53.30, 53.73, 106.30, 110.18, 117.84, 122.42, 126.33, 128.10, 128.54, 134.60, 139.29, 141.20, 179.39; IR (KBr, cm−1): 3415, 3322, 2923, 2853, 1639, 1522, 1456, 1379, 1304, 1237, 1083, 963, 792, 732; HRMS (ESI): m/z [M + H]+ calcd. for C76H107N4O2: 1107.8394; found: 1107.8387.
4.4.2. N,N′-(propane-1,3-diyl)-bis(5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide) (7b)
Yellow solid; Yield: 65%; mp. 276–278 °C; 1H NMR (600 MHz, CDCl3): δ 0.87 (s, 6H), 0.92–0.98 (m, 18H), 1.15 (s, 6H), 1.16 (s, 3H), 1.21 (s, 3H), 1.28 (s, 3H), 1.33 (s, 3H), 1.30–2.10 (m, 36H), 2.10–2.26 (m, 6H), 2.42 (s, 3H), 2.43 (s, 3H), 2.77 (m, 2H), 3.13 (m, 2H), 3.27 (m, 2H), 5.37 (brs, 1H), 5.48 (brs, 1H), 6.92 (d, J = 7.1 Hz, 1H), 6.93 (d, J = 7.1 Hz, 1H), 7.17 (d, J = 7.7 Hz, 1H), 7.18 (d, J = 7.7 Hz, 1H), 7.20 (s, 2H), 7.93 (brs, 2H); 13C NMR (150 MHz, CDCl3): δ 15.91 (15.94), 17.03 (17.05), 17.34 (17.36), 19.66 (19.68), 21.35 (21.37), 21.57 (21.61), 23.30 (23.33), 23.34 (23.37), 23.63 (23.65), 24.86 (24.99), 26.59 (26.69), 28.00 (28.09), 31.05 (31.11), 32.50, 34.12 (34.15), 34.24 (34.26), 36.38 (36.41), 37.63 (37.66), 38.10 (38.11), 39.09 (39.11), 39.15 (39.16), 39.80 (39.85), 42.63 (42.72), 46.37 (46.38), 46.86 (46.89), 47.98 (48.00), 53.86 (53.91), 55.29 (55.35), 106.24 (106.27), 110.17 (110.21), 117.76 (117.80), 122.36 (122.42), 126.08 (126.11), 128.01 (128.06), 128.50 (128.52), 134.60 (134.61), 139.41 (139.47), 141.15 (141.18), 178.37 (178.57); IR (KBr, cm−1): 3318, 2924, 2853, 1634, 1521, 1455, 1379, 1305, 1083, 963, 796; HRMS (ESI): m/z [M + H]+ calcd. for C77H109N4O2: 1121.8551; found: 1121.8558.
4.4.3. N,N′-(butane-1,4-diyl)-bis(5′-methyl-1′H-ursa-2,12-dieno[3,2-b]indol-28-carboxamide) (7c)
Yellow solid; Yield: 63%; mp. 255–258 °C; 1H NMR (600 MHz, CDCl3): δ 0.86 (s, 3H), 0.87 (s, 3H), 0.89–0.92 (m, 12H), 0.94 (s, 3H), 0.96 (s, 3H), 1.07 (s, 3H), 1.08 (s, 3H), 1.13 (s, 3H), 1.15 (s, 3H), 1.24 (s, 3H), 1.25 (s, 3H), 1.30–2.08 (m, 38H), 2.08–2.25 (m, 6H), 2.31 (s, 3H), 2.32 (s, 3H), 2.68 (d, J = 14.7 Hz, 2H), 2.99 (m, 4H), 5.31 (brs, 2H), 6.78 (d, J = 8.2 Hz, 2H), 7.06 (s, 2H), 7.08–7.15 (m, 4H), 10.50 (d, J = 7.1 Hz, 2H); 13C NMR (150 MHz, CDCl3): δ 15.52, 16.72 (16.74), 17.12, 19.03 (19.10), 21.10, 21.28, 22.75 (22.76), 23.00 (23.01), 23.18 (23.19), 23.52 (23.64), 26.33 (26.41), 27.37 (27.41), 30.55 (30.56), 33.66, 33.80 (33.82), 36.12, 36.82, 37.05, 37.57, 38.45, 41.72 (41.81), 45.76, 46.04, 46.53, 46.65, 52.20, 53.01, 54.19, 104.23, 110.16, 117.08, 121.34, 124.76, 125.91, 127.77, 134.54, 138.45, 141.21, 176.04; IR (KBr, cm−1): 3313, 2925, 2853, 1635, 1523, 1455, 1379, 1306, 1087, 792, 732; HRMS (ESI): m/z [M + H]+ calcd. for C78H111N4O2: 1135.8707; found: 1135.8715.
4.5. Biological Evaluation
4.5.1. In Vitro Anti-Proliferation Assay
Compounds 4a–f, 5a–f, 6a–f, and 7a–c were evaluated for their antitumor activity against two human hepatocarcinoma cells (HepG2 and SMMC-7721) and the human normal hepatocyte cell line (LO2) using the typical MTT assay according to the literature [31]. Cell lines were obtained from American Tissue Culture Collection (ATCC). Briefly, different tumor cells were grown in DMEM medium supplemented with 10% fetal bovine serum, penicillin (100 U/mL), and streptomycin (50 μg/mL). Cells were harvested at log phase of growth and seeded in 96-well plates (100 μL/well at a density of 2 × 105 cells/mL). After 24 h incubation at 37 °C and 5% CO2 to allow cell attachment, cultures were exposed to various concentrations of the isolated compounds for 48 h. Finally, MTT solution (2.5 mg/mL in PBS) was added (40 μL/well). Plates were further incubated for 4 h at 37 °C, and the formazan crystals formed were dissolved by adding 150 μL/well of DMSO. Absorption at 570 nm was measured with an ELISA plate reader. The results were expressed as IC50 values, which was defined as the concentration at which 50% survival of cells was obtained. Doxorubicin, camptothecin, and etoposide (VP-16) were co-assayed as positive controls.
4.5.2. Topoisomerase I Inhibition Assay
The activity of DNA topoisomerase I (TaKaRa, Kyoto, Japan) was determined by measuring the relaxation of supercoiled DNA pBR322 [32]. Camptothecin was used as a positive control. The reaction mixture was prepared according to the manufacture’s protocol, and incubated at 37 °C for 30 min. The reactions were terminated by adding dye solution containing 1% SDS, 0.02% bromophenol blue and 50% glycerol. The mixtures were applied to 1% agarose gel and subjected to electrophoresis for 1 h, in 1× TAE buffer (40 mM Tris-acetate, 1 mM EDTA). Gels were stained for 30 min in 60 mL 1× TAE buffer with ethidium bromide (0.5 μg/mL). DNA bands were visualized by transillumination with UV light and then photographed by a Gel Doc XR+ System (BIO-RAD, Hercules, CA, USA).
4.5.3. Topoisomerase II Inhibition Assay
Human Topoisomerase IIα activity was detected by measuring the relaxation of supercoiled pBR322 plasmid DNA [22]. Reaction mixture contained 10 mM Tris (pH 6.9), 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 1 mM ATP, pBR322 DNA (100 ng), and 2 units of Topo IIα (USB, Affymetrix, Waltham, MA, USA) in a final volume of 20 μL. For inhibition studies, the compounds were pre-incubated with human Topo IIα and DNA for 15 min. Compounds were used at the appropriate concentrations by dissolving in 2% (v/v) DMSO. DMSO didn’t show detrimental effect on the enzyme activity at concentration up to 2% (v/v). Reaction mixture was incubated at 37 °C for 30 min and stopped by the addition of 7 mM EDTA. Reaction product was mixed with DNA loading dye and electrophoresed on 1% TAE-Agarose. The gel was stained with ethidium bromide (0.5 μg/mL) for 30 min, destained twice in 1× TAE buffer, and then visualized using a BIO-RAD Gel Doc XR+ System. Etoposide (VP-16) was co-assayed as a positive control.
4.5.4. Molecular Docking
The molecular modeling of compound 5f was performed with Schrödinger Suite 2015-1 (Schrödinger LLC., New York, NY, USA). The crystal structure of Topo IIα (PDB ID: 5GWK) was downloaded from Protein Data Bank (PDB) and prepared using the Protein Preparation Wizard workflow from Schrödinger Suite, including the optimization of hydrogen bond network and a short energy minimization with position restraints on heavy atoms using OPLS_2005 force field. The docking grid was generated according to the initial ligand etoposide. Then, the target compounds were freely docked into the designated binding site using the standard protocol implemented in Maestro v 10.1 (Schrödinger LLC, Cambridge, MA, USA). Van der Waals (vdW) scaling of 0.8 and partial cut-off of 0.15 were set to soften the potential for non-polar sites, and no constraints were specified. The best docked pose ranked by Glide Score value was recorded and saved for the ligand. The structures of complexes were analyzed for interaction modes, and the binding pose of compound 5f with Topo IIα was displayed using the Discovery Studio 2016 client (Accelrys Software, San Diego, CA, USA).
4.5.5. Cell Apoptosis Analysis
The extent of apoptosis was quantitatively measured using Annexin V-FITC/PI dual staining assay [33]. SMMC-7721 cells were seeded into a six-well plate at 5 × 105 cells per well in 10% fetal calf serum (FBS)-DMEM into six-well plates and treated with different concentrations of the indicated compound 5f for 48 h. The cells were detached with 0.25% trypsin, washed with ice-cold PBS for twice and then resuspended in 1 × Binding buffer (0.1 M Hepes/NaOH (pH 7.4), 1.4 M NaCl, 25 mM CaCl2). The cells were stained with 5 μL of Annexin V-FITC and 5 μL of PI (propidium indole) to each tube. The cells were gently vortexed and incubated in the dark at room temperature for 15 min and then kept at 4 °C. The samples were analyzed by a flow cytometer (Becton-Dickinson FACSCalibur, Franklin Lakes, NJ, USA), and data were analyzed using the FlowJo software (Becton-Dickinson Biosciences, Franklin Lakes, NJ, USA).
4.5.6. Acridine Orange/Ethidium Bromide (AO/EB) Staining
SMMC-7721 cells were seeded into six-well plates and incubated at 37 °C with 5% CO2 for 24 h. Then, the cells were exposed to different concentrations of compound 5f (0, 0.3, 0.6, and 1.2 μM) for 48 h. After that, 10 μL of each fluorescent dye cantaining AO and EB (10 μg/mL) were added to each well in equal volumes respectively and incubated for 5 min in the dark. The cells were visualized through an inverted fluorescence microscope (Olympus 1X71 Inverted System Microscope, Olympus, Tokyo, Japan) [26].
4.5.7. ROS Generation Assay
ROS generation assay was performed by using the reactive oxygen species assay kit (Beyotime Biotech., Shanghai, China). Intracellular ROS generation was tested through dichlorodihydro fluorescein diacetate (DCFH-DA) assay [34]. DCFH-DA is taken up by SMMC-7721 cells, and then activated by esterase-mediated cleavage of acetate to form dichlorodihydro fluorescein (DCFH), which is trapped in the cells. DCFH is converted to fluorescein DCF in the presence of ROS. SMMC-7721 cells were seeded in six-well plates and incubated with different concentrations of compound 5f for 48 h. After removing the compound solution, cells were treated with 10 μM of DCFH-DA at 37 °C for 20 min. Subsequently, the cells were washed with PBS for three times and then exposed to light. Immediately after light exposure, the fluorescence intensity of dichlorofluorescein (DCF) was measured with excitation at 488 nm and emission at 525 nm by a flow cytometry (Becton-Dickinson FACSCalibur, Franklin Lakes, NJ, USA).
4.5.8. JC-1 Mitochondrial Membrane Potential Assay
The JC-1 mitochondrial membrane potential assay kit (Keygene Biotech., Nanjing, China) was employed to measure mitochondrial depolarization in SMMC-7721 cells. The fluorescence probe JC-1 was used to detect the change of mitochondrial membrane potential [35]. Briefly, cells cultured in six-well plates after indicated treatments by compound 5f were incubated with an equal volume of JC-1 staining solution (5 μg/mL) at 37 °C for 20 min and rinsed twice with PBS. Mitochondrial membrane potentials were monitored by determining the relative amounts of dual emissions from mitochondrial JC-1 monomers or aggregates using flow cytometry (Becton-Dickinson FACSCalibur, Franklin Lakes, NJ, USA). Mitochondrial membrane depolarization is indicated by an increase in the percentage of cells with low ΔΨm (green fluorescence, lower right quadrant) compared with cells with high ΔΨm (red fluorescence, upper right quadrant).
Acknowledgments
The authors would thank the Modern Analysis and Testing Center, Nanjing Forestry University, for the measurement of spectroscopic data.
Abbreviations
| Topo | Topoisomerase |
| UA | Ursolic acid |
| HOBt | 1-Hydroxybenzotriazole |
| DCC | N,N′-Dicyclohexylcarbodiimide |
| MTT | 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide |
| PI | Propidium Iodide |
| AO | Acridine orange |
| EB | Ethidium bromide |
| ROS | Reactive oxygen species |
| EDTA | Ethylene diamine tetraacetic acid |
| PBS | Phosphate buffer saline |
| SDS | Sodium dodecyl sulfate |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/8/2876/s1.
Author Contributions
W.G. conceived and designed the experiments and wrote the manuscript; A-L.L. and Y.H. synthesized and characterized the compounds; W.-Y.W. and Y.H. determined the biological activities; A-L.L., Q.-S.L., and Y.S. analyzed the experimental data and wrote the experimental part. All authors gave approval to the final version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31770616), the Natural Science Foundation for Colleges and Universities in Jiangsu Province (17KJA220002), and the Top-Notch Academic Programs Project of Jiangsu Higher Education Institutions (PPZY2015C221).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang S.-Y., Wang L.-J., Jiang B., Wu N., Li X.-Q., Luo J., Wang B.-C., Zhang R.-S., Xu Q., Shi D.-Y. Synthesis and biological evaluation of novel fluorinated anticancer agents incorporating the indolin-2-one moiety. RSC Adv. 2015;5:91795–91801. doi: 10.1039/C5RA19617E. [DOI] [Google Scholar]
- 3.Champoux J.J. DNA Topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 4.Nitiss J. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jun K.Y., Lee E.Y., Jung M.J., Lee O.H., Lee E.S., Park Choo H.Y., Na Y., Kwon Y. Synthesis, biological evaluation, and molecular docking study of 3-(3′-heteroatom substituted-2′-hydroxy-1′-propyloxy) xanthone analogues as novel topoisomerase IIα catalytic inhibitor. Eur. J. Med. Chem. 2011;46:1964–1971. doi: 10.1016/j.ejmech.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Park S.-E., Chang I.-H., Jun K.-Y., Lee E., Lee E.-S., Na Y., Kwon Y. 3-(3-Butylamino-2-hydroxy-propoxy)-1-hydroxy-xanthen-9-one acts as a topoisomerase IIα catalytic inhibitor with low DNA damage. Eur. J. Med. Chem. 2013;69:139–145. doi: 10.1016/j.ejmech.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 7.Byl J.A.W., Cline S.D., Utsugi T., Kobunai T., Yamada Y., Osheroff N. DNA topoisomerase II as the target for the anticancer drug TOP-53: Mechanistic basis for drug action. Biochemistry. 2001;40:712–718. doi: 10.1021/bi0021838. [DOI] [PubMed] [Google Scholar]
- 8.Chikamori K., Grozav A.G., Kozuki T., Grabowski D., Ganapathi R., Ganapathi M.K. DNA topoisomerase II enzymes as molecular targets for cancer chemotherapy. Curr. Cancer Drug Targets. 2010;10:758–771. doi: 10.2174/156800910793605785. [DOI] [PubMed] [Google Scholar]
- 9.Azarova A.M., Lyu Y.L., Lin C.-P., Tsai Y.-C., Lau J.Y.-N., Wang J.C., Liu L. Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies. Proc. Natl. Acad. Sci. USA. 2007;104:11014–11019. doi: 10.1073/pnas.0704002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan H., Ma Q., Ye L., Piao G. The traditional medicine and modern medicine from natural products. Molecules. 2016;21:559. doi: 10.3390/molecules21050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Safe S., Kasiappan R. Natural products as mechanism-based anticancer agents: Sp transcription factors as targets. Phytother. Res. 2016;30:1723–1732. doi: 10.1002/ptr.5669. [DOI] [PubMed] [Google Scholar]
- 12.Zhou T., Li Z., Kang O.-H., Mun S.-H., Seo Y.-S., Kong R., Shin D.-W., Liu X.-Q., Kwon D.-Y. Antimicrobial activity and synergism of ursolic acid 3-O-α-L-arabinopyranoside with oxacillin against methicillin-resistant Staphylococcus aureus. Int. J. Mol. Med. 2017;40:1285–1293. doi: 10.3892/ijmm.2017.3099. [DOI] [PubMed] [Google Scholar]
- 13.Wu J., Zhang Z.-H., Zhang L.-H., Jin X., Ma J., Piao H.-R. Design, synthesis, and screening of novel ursolic acid derivatives as potential anti-cancer agents that target the HIF-1α pathway. Bioorg. Med. Chem. Lett. 2019;29:853–858. doi: 10.1016/j.bmcl.2018.12.060. [DOI] [PubMed] [Google Scholar]
- 14.Tohmé M., Gimenez M., Peralta A., Colombo M., Delgui L.R. Ursolic acid: A novel antiviral compound inhibiting rotavirus infection in vitro. Int. J. Antimicrob. Agents. 2019;54:601–609. doi: 10.1016/j.ijantimicag.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Rai S.N., Zahra W., Singh S.S., Birla H., Keswani C., Dilnashin H., Rathore A.S., Singh R., Singh R.K., Singh S. Anti-inflammatory activity of ursolic acid in MPTP-induced parkinsonian mouse model. Neurotox. Res. 2019;36:452–462. doi: 10.1007/s12640-019-00038-6. [DOI] [PubMed] [Google Scholar]
- 16.Wu P., Huang T., Li D., Hu Q., Cheng A., Jiang Z., Jiao L., Zhao S., Zhang K. Synthesis and evaluation of novel triterpene analogues of ursolic acid as potential antidiabetic agent. PLoS ONE. 2015;10:e0138767. doi: 10.1371/journal.pone.0138767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan M.S.A., Khundmiri S.U.K., Khundmiri S.R., Al-Sanea M.M., Mok P.L. Fruit-derived polysaccharides and terpenoids: Recent update on the gastroprotective effects and mechanisms. Front. Pharmacol. 2018;9:569. doi: 10.3389/fphar.2018.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain H., Green I.R., Ali R.N.I., Khan I., Ali Z., Al-Sadi A., Ahmed I. Ursolic acid derivatives for pharmaceutical use: A patent review (2012–2016) Expert Opin. Ther. Pat. 2017;27:1061–1072. doi: 10.1080/13543776.2017.1344219. [DOI] [PubMed] [Google Scholar]
- 19.Fu H.-J., Zhou Y.-R., Bao B.-H., Jia M.-X., Zhao Y., Zhang L., Li J.-X., He H., Zhou X. Tryptophan hydroxylase 1 (Tph-1)-targeted bone anabolic agents for osteoporosis. J. Med. Chem. 2014;57:4692–4709. doi: 10.1021/jm5002293. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen H.N., Ahn Y.J., Medina E.A., Asmis R. Dietary 23–hydroxy ursolic acid protects against atherosclerosis and obesity by preventing dyslipidemia-induced monocyte priming and dysfunction. Atherosclerosis. 2018;275:333–341. doi: 10.1016/j.atherosclerosis.2018.06.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramos-Hryb A.B., Pazini F.L., Kaster M.P., Rodrigues A.L.S. Therapeutic potential of ursolic acid to manage neurodegenerative and psychiatric diseases. CNS Drugs. 2017;31:1029–1041. doi: 10.1007/s40263-017-0474-4. [DOI] [PubMed] [Google Scholar]
- 22.Mandal A., Ghosh S., Bothra A.K., Nanda A.K., Ghosh P. Synthesis of friedelan triterpenoid analogs with DNA topoisomerase IIα inhibitory activity and their molecular docking studies. Eur. J. Med. Chem. 2012;54:137–143. doi: 10.1016/j.ejmech.2012.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Kim D.K., Baek J.H., Kang C.M., Yoo M.A., Sung J.W., Chung H.Y., Kim N.D., Choi Y.H., Lee S.H., Kim K.W. Apoptotic activity of ursolic acid may correlate with the inhibition of initiation of DNA replication. Int. J. Cancer. 2000;87:629–636. doi: 10.1002/1097-0215(20000901)87:5<629::AID-IJC2>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 24.Mizushina Y., Iida A., Ohta K., Sugawara F., Sakaguchi K. Novel triterpenoids inhibit both DNA polymerase and DNA topoisomerase. Biochem. J. 2000;3:757–763. doi: 10.1042/bj3500757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu W., Hao Y., Zhang G., Wang S., Miao T.-T., Zhang K.-P. Synthesis, in vitro antimicrobial and cytotoxic activities of new carbazole derivatives of ursolic acid. Bioorg. Med. Chem. Lett. 2015;25:554–557. doi: 10.1016/j.bmcl.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 26.Wang R., Yang W., Fan Y., Dehaen W., Li Y., Li H.-J., Wang W., Zheng Q., Huai Q.-Y. Design and synthesis of the novel oleanolic acid-cinnamic acid ester derivatives and glycyrrhetinic acid-cinnamic acid ester derivatives with cytotoxic properties. Bioorg. Chem. 2019;88:102951. doi: 10.1016/j.bioorg.2019.102951. [DOI] [PubMed] [Google Scholar]
- 27.Huang X., Huang R., Liao Z., Pan Y., Gou S., Wang H.-S. Synthesis and pharmacological evaluation of dehydroabietic acid thiourea derivatives containing bisphosphonate moiety as an inducer of apoptosis. Eur. J. Med. Chem. 2016;108:381–391. doi: 10.1016/j.ejmech.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 28.Kumar N.P., Vanjari Y., Thatikonda S., Pooladanda V., Sharma P., Sridhar B., Godugu C., Kamal A., Shankaraiah N., Sridhar B. Synthesis of enamino-2-oxindoles via conjugate addition between α-azido ketones and 3-alkenyl oxindoles: Cytotoxicity evaluation and apoptosis inducing studies. Bioorg. Med. Chem. Lett. 2018;28:3564–3573. doi: 10.1016/j.bmcl.2018.07.038. [DOI] [PubMed] [Google Scholar]
- 29.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Boil. Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gnoatto S.B., Dassonville-Klimpt A., Da Nascimento S., Galéra P., Boumediene K., Gosmann G., Sonnet P., Moslemi S. Evaluation of ursolic acid isolated from Ilex paraguariensis and derivatives on aromatase inhibition. Eur. J. Med. Chem. 2008;43:1865–1877. doi: 10.1016/j.ejmech.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Wang C., Delcros J.-G., Biggerstaff J., Phanstiel O., IV Synthesis and biological evaluation ofN1-(Anthracen-9-ylmethyl)triamines as molecular recognition elements for the polyamine transporter. J. Med. Chem. 2003;46:2663–2671. doi: 10.1021/jm030028w. [DOI] [PubMed] [Google Scholar]
- 32.Wu W.-B., Ou J.-B., Huang Z., Chen S.-B., Ou T.-M., Tan J.-H., Li D., Shen L.-L., Huang S.-L., Gu L.Q., et al. Synthesis and evaluation of mansonone F derivatives as topoisomerase inhibitors. Eur. J. Med. Chem. 2011;46:3339–3347. doi: 10.1016/j.ejmech.2011.04.059. [DOI] [PubMed] [Google Scholar]
- 33.Su C.C., Chen J.Y., Din Z.H., Su J.H., Yang Z.Y., Chen Y.J., Wang R.Y., Wu Y.J. 13-acetoxysarcocrassolide induces apoptosis on human gastric carcinoma cells through mitochondria- related apoptotic pathways: p38/JNK activation and PI3K/AKT suppression. Mar. Drugs. 2014;12:5295–5315. doi: 10.3390/md12105295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Gu Z., Zhang C., Peng Y., Zhang L., Zhou G., Wang S., Zhang J. Synthesis, characterization and ROS-mediated antitumor effects of palladium(II) complexes of curcuminoids. Eur. J. Med. Chem. 2018;144:662–671. doi: 10.1016/j.ejmech.2017.12.027. [DOI] [PubMed] [Google Scholar]
- 35.Donthiboina K., Anchi P., Ramya P.S., Karri S., Srinivasulu G., Godugu C., Shankaraiah N., Kamal A. Synthesis of substituted biphenyl methylene indolinones as apoptosis inducers and tubulin polymerization inhibitors. Bioorg. Chem. 2019;86:210–223. doi: 10.1016/j.bioorg.2019.01.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.