Abstract
Autophagy is a highly conserved intracellular degradation pathway that breaks down damaged macromolecules and/or organelles. It is involved in plant development and senescence, as well as in biotic and abiotic stresses. However, the autophagy process and related genes are largely unknown in citrus. In this study, we identified 35 autophagy-related genes (CsATGs—autophagy-related genes (ATGs) of Citrus sinensis, Cs) in a genome-wide manner from sweet orange (Citrus sinensis). Bioinformatic analysis showed that these CsATGs were highly similar to Arabidopsis ATGs in both sequence and phylogeny. All the CsATGs were randomly distributed on nine known (28 genes) and one unknown (7 genes) chromosomes. Ten CsATGs were predicted to be segmental duplications. Expression patterns suggested that most of the CsATG were significantly up- or down-regulated in response to drought; cold; heat; salt; mannitol; and excess manganese, copper, and cadmium stresses. In addition, two ATG18 members, CsATG18a and CsATG18b, were cloned from sweet orange and ectopically expressed in Arabidopsis. The CsATG18a and CsATG18b transgenic plants showed enhanced tolerance to osmotic stress, salt, as well as drought (CsATG18a) or cold (CsATG18b), compared to wild-type plants. These results highlight the essential roles of CsATG genes in abiotic stresses.
Keywords: citrus, autophagy, ATG18a, ATG18b, drought, heat, cold, heavy metal
1. Introduction
Autophagy is a highly conserved intracellular degradation process that occurs in eukaryotes including yeasts, animals, and plants [1,2]. During this process, damaged macromolecules and/or organelles are sequestered into a double-membrane vesicle (called an autophagosome) and then delivered into a vacuole (yeast and plants) or a lysosome (animals) for breakdown. The cellular cytoplasmic contents are then recycled [1,2,3,4]. Autophagy-related genes (ATGs) are responsible for the regulation and control of the autophagy process. The first ATG gene was identified from yeast, and at least 32 ATGs have been shown to participate in yeast autophagy [5,6]. To date, many homologues of ATGs have been identified from various plant species, including 40 AtATGs in Arabidopsis [1,3] (Arabidopsis thaliana, At), 33 OsATGs in rice [7] (Oryza sativa, Os), 30 NtATGs in tobacco [8] (Nicotiana tabacum, Nt), 45 ZmATGs in maize [9] (Zea mays, Zm), 29 CaATGs in pepper [10] (Capsicum annuum, Ca), 37 SiATGs in foxtail millet [11] (Setaria italic, Si), 32 MaATGs in banana [12] (Musa acuminate, Ma), and 35 VvATGs in grapevine [13] (Vitis vinifera, Vv). According to the reported characterizations of ATGs in yeast and Arabidopsis, these ATGs can be divided into the following functional groups: (1) the ATG1/13 kinase complex consisting of ATG1, ATG13, ATG20, and TOR (target of rapamycin kinase), mainly functioning on autophagy induction and initiation; (2) the PI3K (phosphatidylinositol 3 kinase) complex consisting of ATG6, VPS15 (vacuolar protein sorting-associated protein), and VPS34 that is involved in vesicle nucleation and autophagosome formation; (3) the ATG9/2/18 complex consisting of ATG9, ATG2, and ATG18 that is responsible for the delivery of membranes for autophagosome formation; (4) the ubiquitin-like ATG8-PE (phosphatidylethanolamine) conjugation pathway (including ATG3, ATG4, ATG7, and ATG8) and ATG12–ATG5 conjugation pathway (including ATG5, ATG7, ATG10, ATG12, and ATG16) that are involved in the elongation of autophagic vesicles; and (5) the VTI12 (vesicle transport v-soluble N-ethylmaleimide-sensitive-factor attachment protein receptor (SNARE) protein) family belonging to the SNARE group, which contributes to the fusion of autophagosomes with vacuoles [1,2,3,4,6].
The autophagy process maintains a basal level under normal conditions, but it can be quickly induced by external stimuli such as nutrition starvation, drought, salt, heat, and oxidative and osmotic stresses [3,12,14,15,16]. Many studies have demonstrated the functions of ATGs in alleviating abiotic stresses in plants. For example, an apple MdATG8i (Malus domestica, Md) was significantly up-regulated in response to nitrogen depletion and oxidative stress, and overexpression of MdATG8i in apple callus conferred enhanced tolerance to nutrient-limited conditions [17]. MdATG3a, MdATG3b, MdATG7b, and MdATG18a of apple were shown to confer tolerance to nitrogen/carbon starvation, drought, salt, or osmotic stresses by ectopic-expressing in Arabidopsis and apple [18,19,20,21]. Li et al. reported that a banana MaATG8f improved drought stress tolerance in transgenic Arabidopsis through modulating reactive oxygen species (ROS) metabolism, abscisic acid biosynthesis, and autophagy activity [22]. Chen et al. overexpressed an ATG8 in Arabidopsis that increased autophagic activity and improved nitrogen remobilization efficiency [23]. Moreover, overexpression of a SiATG8a of foxtail millet conferred tolerance to both nitrogen starvation and drought in transgenic Arabidopsis and rice [11,24]. In contrast, silencing of AtATG18a in Arabidopsis reduced drought and salt tolerance [25]. Knockdown of the ATG6 of wheat inhibited autophagy and resulted in accelerated programmed cell death (PCD) under drought stress [26]. Loss of ATG10 and ATG18f significantly reduced HsfA1a-induced drought tolerance and autophagosome formation in tomato [27]. Under salt stress, autophagosome formation was rapidly induced, and the atg2 and atg7 mutants of Arabidopsis showed hypersensitive phenotype [28]. Under heat stress, atg mutants of Arabidopsis displayed visibly impaired pollen development (including atg2-1, atg5-1, atg7-2, and atg10-1) [29] or enhanced growth inhibition (including atg2-1, atg5-1, atg12ab, and atg18a-2) [30]. Recently, Shinozaki et al. proved that autophagy played important role in maintaining zinc bioavailability to avoid ROS accumulation under zinc deficiency in Arabidopsis [31]. Moreover, the potential roles of autophagy in waterlogging and excess aluminum and copper stresses were also reported [13,32,33].
As the largest fruit crop, citrus is widely grown around the world. In 2017, the global citrus growing area and yield were 14.1 million hectares and 146.6 million tons, respectively (FAO statistics, http://faostat.fao.org/default.aspx). However, citrus production is limited by abiotic stresses including drought, heat, cold, and nutrient disorders. Although the underlying physiological and molecular mechanisms triggered in response to these stresses have been well studied, the autophagy process and ATG genes in citrus are largely unknown. In this study, we systematically identified 35 ATGs from citrus by using the genome of sweet orange [34]. Comprehensive analysis including bioinformatic characterizations and expression profiles of these ATGs were conducted and discussed. Moreover, two ATGs of sweet orange, CsATG18a and CsATG18b (Citrus sinensis, Cs), were cloned and overexpressed in Arabidopsis. This further supported their functions in improving tolerance to drought, salt, cold, and osmotic stresses.
2. Results
2.1. Identification of 35 ATGs in the Sweet Orange Genome
After a BLASTP search of the sweet orange genome with known AtATG proteins as queries and confirmation of the existence of the ATG domains in the PFAM database, a total of 35 putative ATG proteins were identified from sweet orange (Table 1). These ATGs showed 47.6% to 90.9% sequence identity with AtATGs, and they were named and numbered as CsATGs according to the highest sequence identity and the closest phylogenic relationship of AtATG homologues (Table 1 and Figure 1). On the basis of known functional classification of AtATGs, all CsATGs were also classified in the categories of the ATG1/13 kinase complex, PI3K complex, ATG9/2/18 complex, ubiquitin-like ATG8 and PE conjugation pathway, ubiquitin-like ATG12 and ATG5 conjugation pathway, and SNARE (Table 1). Among the 35 CsATGs, 15 contained only one member, whereas 4 CsATGs were encoded by gene families (three members for CsATG1, six members for CsATG8, seven members for CsATG18, and four members for CsVTI12). The ORF length of the CsATGs ranged from 315 bp (CsATG12) to 7095 bp (CsTOR), encoding 104 to 2364 amino acids (Table 1). This suggested the existence of significant variations and potential function differentiation. Predicted subcellular localization suggested that most CsATGs express in the cytoplasm, but there are also members located in the mitochondria, plasma membrane, Golgi apparatus, vacuoles, and nucleus (Table 1).
Table 1.
Detailed information of autophagy-related genes (ATGs) in Arabidopsis and sweet orange.
| Gene | Arabidopsis ID | Gene | Citrus ID | Identity to Arabidopsis | ORF (bp) | Protein (aa) |
Predicted Localization 1 | Predicted Function 2 |
|---|---|---|---|---|---|---|---|---|
| ATG1/13 kinase complex | Initiation of autophagy | |||||||
| AtATG1a | At1g49180 | CsATG1a | Cs1g05170 | 56.3% | 894 | 297 | Cytoplasmic | Serine/threonine-protein kinase |
| AtATG1b | At2g37840 | CsATG1b | orange1.1t03071 | 66.5% | 2157 | 718 | Nuclear | Serine/threonine-protein kinase |
| AtATG1c | At3g53930 | NA | NA | NA | NA | NA | NA | NA |
| AtATG1d | At3g61960 | CsATG1d | orange1.1t00581 | 59.1% | 2079 | 692 | Cytoplasmic | Serine/threonine-protein kinase |
| AtATG13a | At3g18770 | CsATG13 | Cs1g05100 | 61.7% | 3222 | 1073 | Cytoplasmic | Autophagocytosis-associated protein |
| AtATG13b | AT3G49590 | NA | NA | NA | NA | NA | NA | NA |
| AtATG20 | At5g06140 | CsATG20 | Cs6g14260 | 83.3% | 1212 | 403 | Mitochondrial | Sorting nexin 2-like protein |
| AtTOR | At1g50030 | CsTOR | Cs8g03630 | 83.6% | 7095 | 2364 | Cytoplasmic | Phosphoinositide 3-kinase |
| PI3K complex | Autophagosome formation | |||||||
| AtVPS15 | At4g29380 | CsVPS15 | Cs4g15580 | 73.7% | 4662 | 1553 | Cytoplasmic | Serine/threonine-protein kinase |
| AtVPS34 | At1g60490 | CsVPS34 | Cs7g08800 | 83.8% | 2469 | 822 | Cytoplasmic | Phosphoinositide 3-kinase |
| AtATG6 | At3g61710 | CsATG6 | orange1.1t00487 | 77.4% | 1551 | 516 | Cytoplasmic | Autophagy protein Apg6 |
| ATG9/2/18 complex | Membrane recruitment to autophagosome | |||||||
| AtATG2 | At3g19190 | CsATG2 | Cs8g05820 | 51.6% | 5985 | 1994 | Plasma membrane | Autophagy-related protein 2 |
| AtATG9 | At2g31260 | CsATG9 | Cs5g29970 | 69.9% | 2625 | 874 | Vacuolar | Autophagy protein Apg9 |
| AtATG18a | At3g62770 | CsATG18a | Cs5g32770 | 76.5% | 1221 | 406 | Cytoplasmic | WD-40 repeat containing protein |
| AtATG18b | At4g30510 | CsATG18b | Cs4g02320 | 74.2% | 1095 | 364 | Mitochondrial | WD-40 repeat containing protein |
| AtATG18c | At2g40810 | CsATG18c | Cs7g29360 | 75.6% | 1248 | 415 | Mitochondrial | WD-40 repeat containing protein |
| AtATG18d | At3g56440 | NA | NA | NA | NA | NA | NA | NA |
| AtATG18e | At5g05150 | CsATG18e | Cs3g14170 | 47.6% | 1083 | 360 | Cytoplasmic | WD-40 repeat containing protein |
| AtATG18f | At5g54730 | CsATG18f | Cs4g11290 | 48.7% | 2340 | 779 | Plasma membrane | Breast carcinoma amplified sequence 3 |
| AtATG18g | At1g03380 | CsATG18g | Cs5g32120 | 57.5% | 2985 | 994 | Plasma membrane | Breast carcinoma amplified sequence 3 |
| AtATG18h | At1g54710 | CsATG18h | Cs5g34870 | 62.0% | 3021 | 1006 | Mitochondrial | Breast carcinoma amplified sequence 3 |
| Ubiquitin-like ATG8 and PE conjugation pathway | Conjugation of ATG8 to PE | |||||||
| AtATG3 | At5g61500 | CsATG3 | Cs9g15060 | 82.1% | 960 | 319 | Cytoplasmic | Autophagocytosis-associated protein 3 |
| AtATG4a | At2g44140 | NA | NA | NA | NA | NA | NA | NA |
| AtATG4b | At3g59950 | CsATG4 | Cs5g20640 | 60.8% | 1332 | 443 | Cytoplasmic | Peptidase family C54 |
| AtATG7 | At5g45900 | CsATG7 | Cs2g04910 | 71.9% | 2148 | 715 | Cytoplasmic | Autophagy protein Apg7 |
| AtATG8a | At4g21980 | CsATG8a | Cs2g03030 | 88.2% | 363 | 120 | Cytoplasmic | Autophagy protein Apg8 |
| AtATG8b | AT4G04620 | NA | NA | NA | NA | NA | NA | NA |
| AtATG8c | At1g62040 | CsATG8c | Cs4g10210 | 90.8% | 363 | 120 | Cytoplasmic | Autophagy protein Apg8 |
| AtATG8d | At2g05630 | CsATG8d | Cs2g21350 | 90.8% | 360 | 119 | Mitochondrial | Autophagy protein Apg8 |
| AtATG8f | At4g16520 | CsATG8f | Cs5g08990 | 90.9% | 369 | 122 | Cytoplasmic | Autophagy protein Apg8 |
| AtATG8g | At3g60640 | CsATG8g | orange1.1t00201 | 84.6% | 354 | 117 | Mitochondrial | Autophagy protein Apg8 |
| AtATG8h | At3g06420 | NA | NA | NA | NA | NA | NA | NA |
| AtATG8i | At3g15580 | CsATG8i | orange1.1t01753 | 79.1% | 378 | 125 | Mitochondrial | Autophagy protein Apg8 |
| Ubiquitin-like ATG12 and ATG5 conjugation pathway | Conjugation of ATG12, ATG5, and ATG16 | |||||||
| AtATG5 | At5g17290 | CsATG5 | Cs9g11540 | 70.2% | 1233 | 410 | Cytoplasmic | Autophagy protein Apg5 |
| AtATG7 | At5g45900 | CsATG7 | Cs2g04910 | 71.9% | 2148 | 715 | Cytoplasmic | Autophagy protein Apg7 |
| AtATG10 | At3g07525 | CsATG10 | Cs2g18190 | 56.1% | 693 | 230 | Plasma membrane | Autophagocytosis-associated protein |
| AtATG12a | At1g54210 | CsATG12 | Cs1g17600 | 81.1% | 315 | 104 | Cytoplasmic | Autophagocytosis-associated protein |
| AtATG12b | At3g13970 | NA | NA | NA | NA | NA | NA | NA |
| AtATG16 | At5g50230 | CsATG16 | Cs5g06560 | 74.1% | 1533 | 510 | Cytoplasmic | WD-40 repeat containing protein |
| SNARE | Fusion of autophagososme with the vacuole | |||||||
| AtVTI12 | At1g26670 | CsVTI12a | orange1.1t04711 | 70.1% | 666 | 221 | Golgi | Vesicle transport v-SNARE protein |
| CsVTI12b | Cs7g03410 | 72.7% | 519 | 172 | Golgi | Vesicle transport v-SNARE protein | ||
| CsVTI12c | Cs6g21610 | 68.3% | 666 | 221 | Golgi | Vesicle transport v-SNARE protein | ||
| CsVTI12d | orange1.1t00788 | 71.7% | 522 | 173 | Golgi | Vesicle transport v-SNARE protein | ||
1 Localizations of CsATGs (Citrus sinensis, Cs) were prediceted online by using the ProtComp. 2 Functions of CsATGs were predicted according to the annotations of AtATGs (Arabidopsis thaliana, At) in the Arabidopsis information resource (TAIR) database.
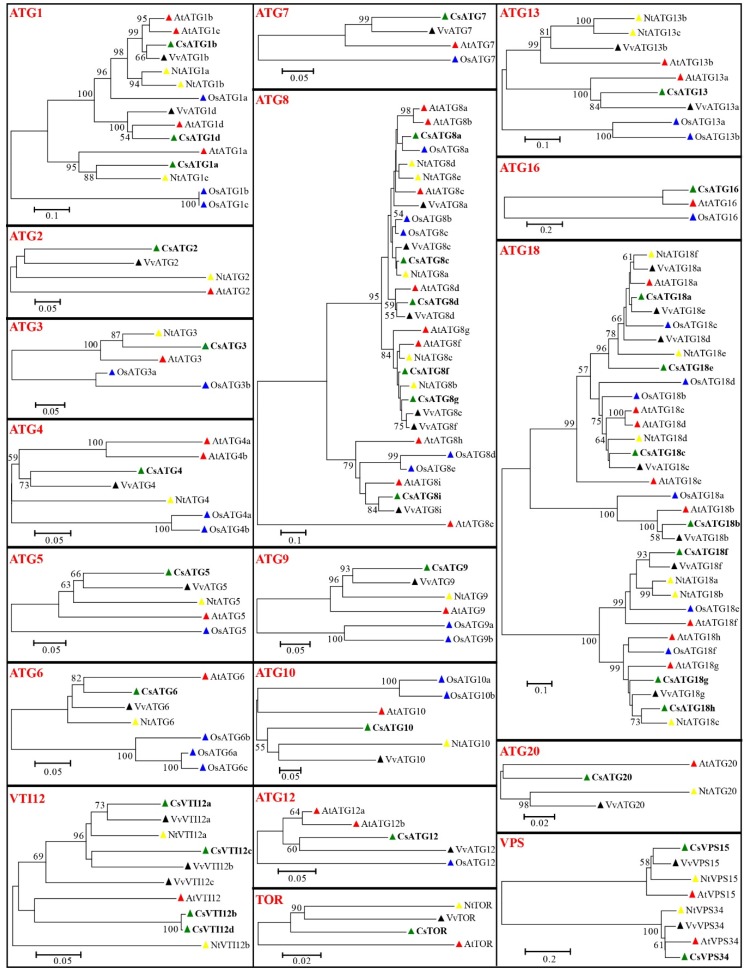
Figure 1.
Phylogenetic analysis of sweet orange and other plant ATG proteins. Thirty-five CsATGs of sweet orange (Citrus sinensis, Cs), 40 AtATGs of Arabidopsis thaliana (At), 30 NtATGs of tobacco (Nicotiana tabacum, Nt), 31 OsATGs of rice (Oryza sativa, Os), and 33 VvATGs of grapevine (Vitis vinifera, Vv) were used to construct the neighbor-joining tree in MEGA 6.0 software, with 1000 bootstrap replicates. The same plant of ATGs are marked with the same color of triangles. The bootstrap values (the number at each node) higher than 50% are shown in the phylogenetic trees.
2.2. Bioinformatic Characterizations of CsATGs
Neighbor-joining phylogenetic trees were constructed with 35 CsATGs, 40 AtATGs, 30 NtATGs, 31 OsATGs, and 33 VvATGs, and the results showed that most of the CsATG proteins closely clustered with their homologues in Arabidopsis and grapevine (Figure 1). For the ATGs containing multiple members (such as CsATG1, CsATG8, CsATG18, CsVTI12, and CsVPS in sweet orange), the different members of same subfamily were clustered in two or three different branches. For example, three CsATG1 members were clustered in two branches (CsATG1b and CsATG1d in one branch, and CsATG1a in another branch), and seven CsATG18 members were clustered in three branches (CsATG18a, CsATG18c, and CsATG18e in one branch; CsATG18f, CsATG18g, and CsATG18h in one branch; and CsATG18b in another branch). This result suggests potential functional differentiations in these subfamilies.
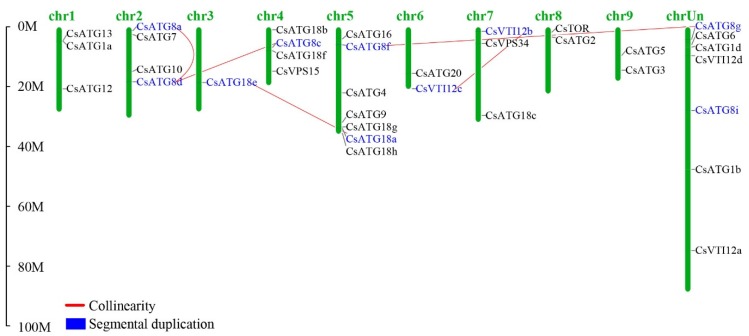
The chromosomal distribution of 35 CsATGs is shown in Figure 2. In total, 28 CsATGs were distributed on 9 chromosomes, whereas the chromosomal location of 7 CsATGs were unknown. Chromosome 5 (chr5) contained the most CsATGs (seven genes), followed by chr2 (four genes) and chr4 (four genes). Gene duplication analysis showed that 10 CsATGs were predicted to be segmental duplicated genes, accounting for about 29% of all CsATGs. In these segmental duplicated genes, five pairs—CsATG8a and CsATG8d, CsATG8c and CsATG8d, CsATG18a and CsATG8e, CsATG8f and CsATG8g, and CsVTI12b and CsVTI12c—were predicted to have collinear correlations.
Figure 2.
Chromosomal distribution and gene duplication of CsATG genes. The genes highlighted with blue color represent segmental duplications. The pairs linked with red lines represent collinear genes.
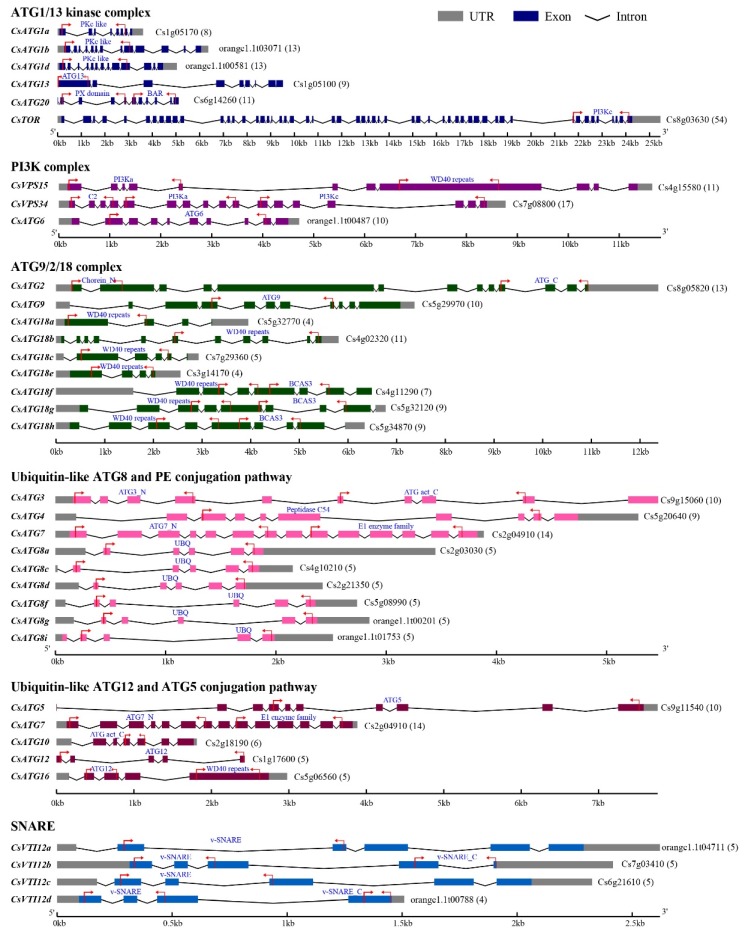
The detailed intron-exon characterizations and conserved domains of CsATGs are shown in Figure 3. We found that 34 of the CsATGs had 4 to 17 exons, but 1 gene (CsTOR) had 54 exons. All the CsATG genes contained introns, but three members (CsATG12, CsATG13, and CsATG20) had no untranslated region (UTR). The main conserved domains of CsATGs were predicted to be protein kinase superfamily (PKc)-like, ATG, phox homology (PX) domain, Bin/amphiphysin/Rvs (BAR), PI3K, WD40, breast carcinoma amplified sequence 3 (BCAS3), chorein N, peptidase C54, ubiquitin homologues (UBQ), E1 enzyme family, and vesicle transport v-SNARE (V-SNARE). In general, the CsATGs in the same functional group or subfamily had similar domains (Figure 3).
Figure 3.
Gene structures and conserved domains of CsATGs. The detailed intron-exon structures and domains of all CsATGs were predicted in the Gene Structure Display Server and the NCBI Conserved Domain Database, respectively. The results are shown and listed according to their functional classifications. Arabic numerals in the parentheses after each gene ID represent exon number. The arrows show the start and end positions of domains. Full names of the conserved domains are listed in the Abbreviations section.
2.3. Expression Patterns of CsATGs Under Abiotic Stresses
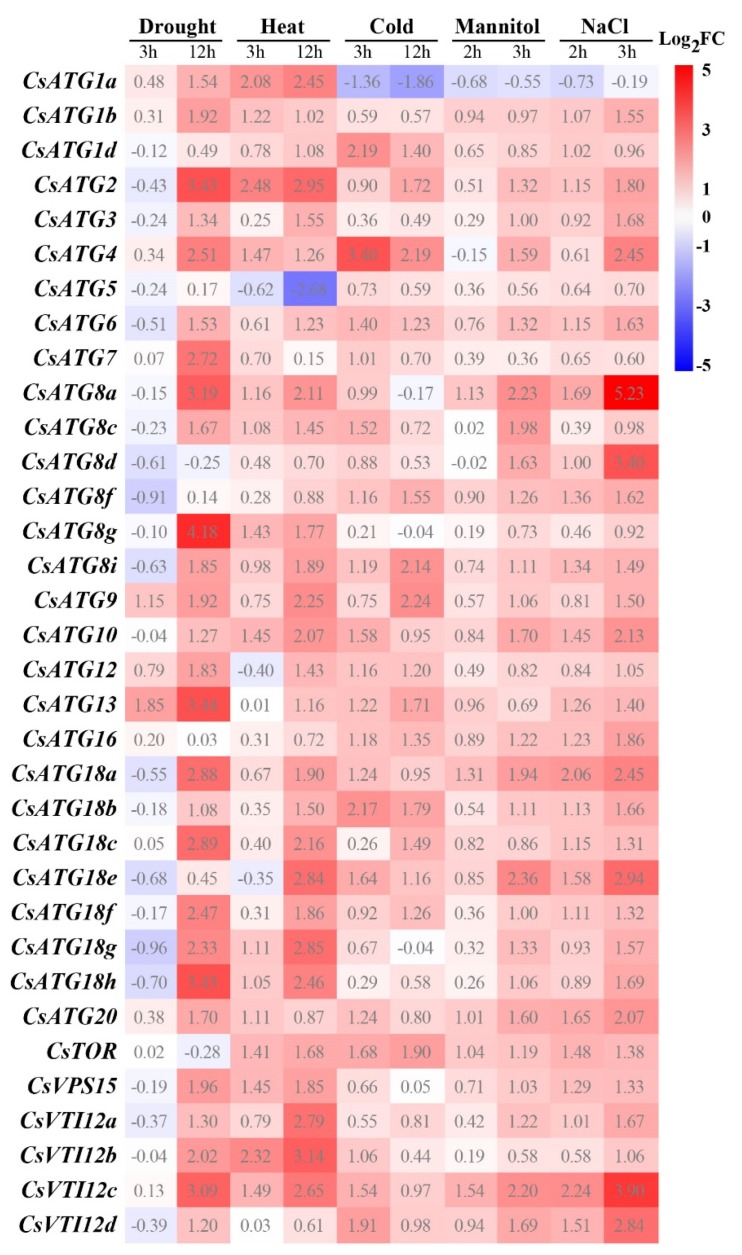
As shown in Figure 4, most of CsATG genes were up-regulated under different stresses. For example, 27, 28, 23, 22, and 29 of CsATGs were significantly up-regulated (log2FC > 1) during at least one time-point of drought, heat, cold, mannitol, or salt treatments, respectively. In particular, CsATG2, CsATG8g, CsATG13, and CsATG18h under drought; CsATG2, CsATG18e, CsATG18g, and CsVTI12b under heat; CsATG4 under cold; and CsATG8a, CsATG18e, and CsVTI12c under mannitol and salt showed high expression levels, suggesting their potential key roles in response to these stresses. Apart from CsATG1d, CsATG5, CsATG7, CsATG8d, CsATG8g, CsATG16, CsATG18e, CsTOR, and CsVTI12d, the other 26 tested genes were significantly up-regulated during both drought and heat stresses. Twenty CsATGs showed significant up-regulation under both mannitol and salt stress. Remarkably, CsATG2, CsATG4, CsATG8i, CsATG9, CsATG10, CsATG18a, CsATG18b, and CsVTI12c were significantly up-regulated under all tested stresses simultaneously, suggesting their important roles in response to these stresses. In addition, some genes were significantly up-regulated at all tested time points of treatment, such as CsATG9 and CsATG13 under drought. Apart from these up-regulated CsATGs, we also noticed that CsATG1a was down-regulated under cold, mannitol, and salt treatments, which differed from the expression of the other two members of the ATG1 subfamily, CsATG1b and CsATG1d. The expression of CsATG5 was not obvious (absolute value of log2FC < 1) under most of the stresses, except for a significant down-regulation under 12 h of heat treatment. Similarly, the expression of CsATG8d was also steady, except for a significant up-regulation under mannitol and salt treatments.
Figure 4.
Expression patterns of CsATG genes under drought, heat, cold, mannitol, and salt stresses. qRT-PCR was used to determine the relative expression levels of CsATGs in the leaves of sweet orange treated with drought, heat (42 °C), cold (4 °C), mannitol (400 mM), and NaCl (200 mM) stresses. Leaves sampled from plants under normal conditions were used as controls (CK). The fold change (FC) was calculated as (relative expression of CsATGs under treatments)/(relative expression of CsATGs in the CK). The R package was used to generate the heat map based on the mean log2FC values of three biological replicates.
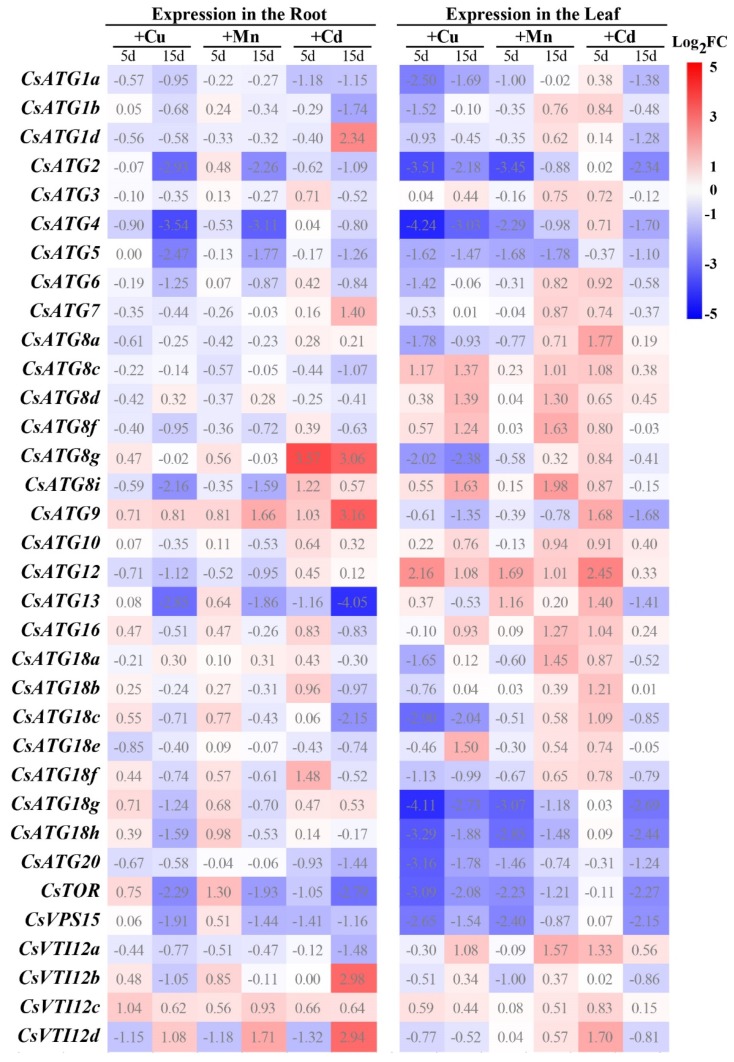
Under excess Cu (+Cu), Mn (+Mn), and Cd (+Cd) treatments, the expression of most CsATGs were down-regulated or not obvious in roots and leaves, and only a few CsATGs were significantly up-regulated at one or two time-points of treatment (Figure 5). Generally, CsATG2, CsATG4, CsATG5, CsTOR, and CsVPS15 were significantly down-regulated (log2FC < -1) in both roots and leaves, CsATG18g, CsATG18h, and CsATG20 were significantly down-regulated in leaves, and CsATG13 was significantly down-regulated in roots under +Cu, +Mn, and +Cd stresses simultaneously. In addition, the expression of CsATG1a in +Cd roots and +Cu leaves, as well as CsATG8g and CsATG18c in +Cu leaves showed extreme down-regulation. Expression of CsATG8g and CsATG9 in +Cd roots, CsATG8c in +Cu leaves, and CsATG12 in both +Cu and +Mn leaves showed obvious up-regulation (Figure 5). These genes may play roles in response to heavy metal stress.
Figure 5.
Expression patterns of CsATG genes under excess Cu, Mn, and Cd stresses. qRT-PCR was used to determine the relative expression levels of CsATGs in the roots and leaves of sweet orange treated with excess Cu, Mn, and Cd for 5 and 15 days. The roots or leaves sampled from plants under normal conditions for 5 and 15 days were used as controls (CK). The fold change (FC) was calculated as (relative expression of CsATGs under treatments)/(relative expression of CsATGs in the CK). The R package was used to generate a heat map based on the mean log2FC values of three biological replicates.
2.4. Overexpression of CsATG18a/b Conferred Osmotic and Salt Tolerance in Arabidopsis
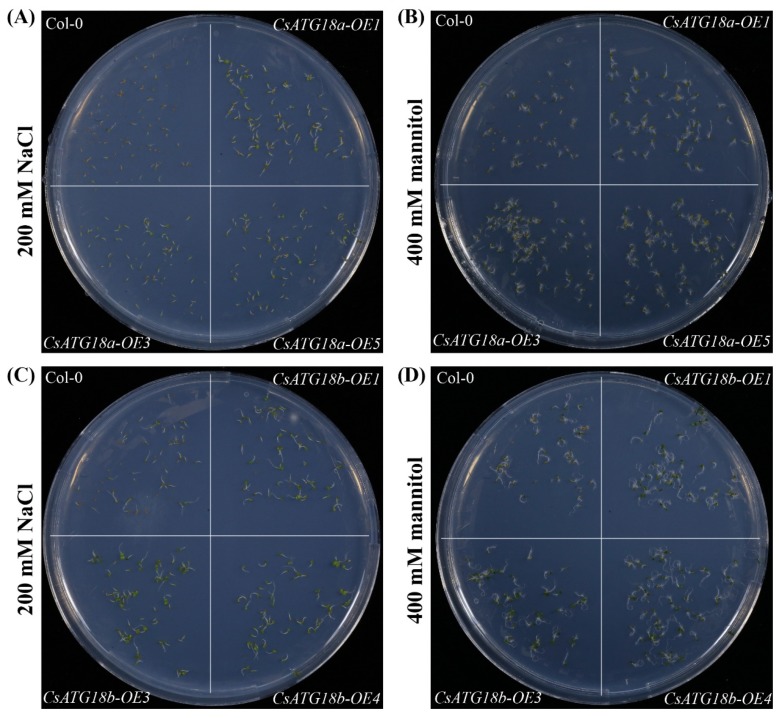
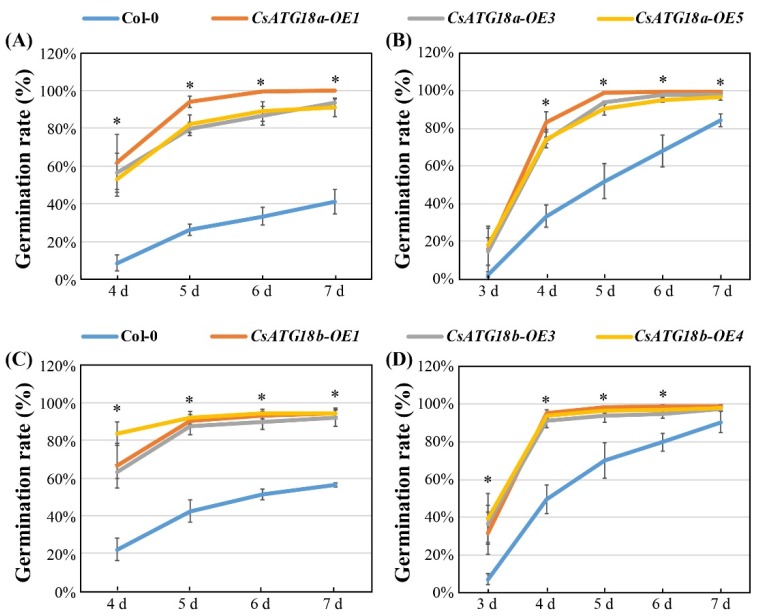
To confirm the functions of CsATGs in abiotic stresses, CsATG18a and CsATG18b were cloned and overexpressed in Arabidopsis. Three homozygous overexpression (OE) lines (T4 generation) and wild-type (WT, Col-0) of Arabidopsis were selected to perform germination experiments. The results showed that the OE lines of CsATG18a (OE1, OE3, and OE5) and CsATG18b (OE1, OE3, and OE4) had significantly higher germination rates than WT on 200 mM NaCl and 400 mM mannitol (Figure 6 and Figure 7). These data suggest that overexpression of both CsATG18a and CsATG18b confer osmotic and salt tolerance.
Figure 6.
Germination phenotype of CsATG18a- and CsATG18b-overexpressed (OE) lines and wild-type (WT) under NaCl and mannitol stresses. (A–D) Representative pictures of three homozygous OE lines (OE1, OE3, and OE5 of CsATG18a, and OE1, OE3, and OE4 of CsATG18b) and WT (Col-0) of Arabidopsis that germinated on 200 mM NaCl (A,C) and 400 mM mannitol (B,D) for 7 days. Three independent experiments were performed and the similar phenotype was observed.
Figure 7.
Germination rate of CsATG18a- and CsATG18b-overexpressed (OE) lines and WT under NaCl and mannitol stresses. (A–D) Germination rate (%) of three OE lines of CsATG18a and CsATG18b and WT (Col-0) calculated after 4, 5, 6, and 7 days of 200 mM NaCl (A,C) treatment and 3, 4, 5, 6, and 7 days of 400 mM mannitol (B,D) treatment. The data are the means ± SE of three independent repeats (n = 60). * indicates a significant difference at p < 0.05 using the least significant difference (LSD) test.
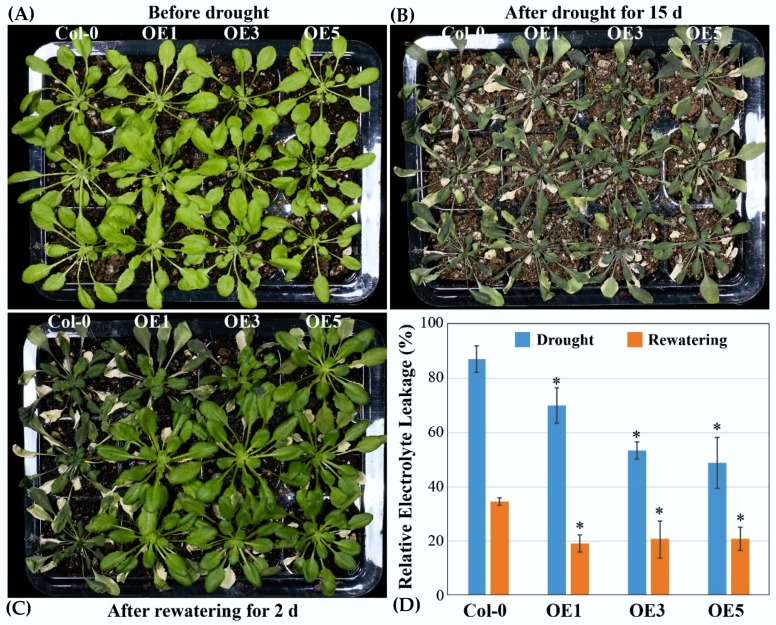
2.5. Overexpression of CsATG18a Conferred Drought Tolerance in Arabidopsis
Figure 8 shows that the OE lines of CsATG18a showed similar wilting phenotype with WT after 15 days of drought treatment. However, after rewatering for 2 days, all of the OE lines recovered to near normal, whereas the WT still exhibited wilting. In addition, the relative electrolyte leakage of OE lines was significantly lower than that of WT after either drought for 15 days or recovery for 2 days. These results suggest that overexpression of CsATG18a conferred drought tolerance in Arabidopsis. For the OE lines of CsATG18b, we also evaluated their tolerance to drought stress but observed no obvious tolerant phenotype (Figure S1).
Figure 8.
Overexpression of CsATG18a enhanced drought tolerance in transgenic Arabidopsis. (A–C) The representative pictures of three homozygous OE lines (OE1, OE3, and OE5) of CsATG18a and WT (Col-0) of Arabidopsis that were treated before drought (A), after drought for 15 days (B), and after rewatering for 2 days (C). (D) Relative electrolyte leakage of WT and OE lines measured after drought for 15 days and after rewatering for 2 days.
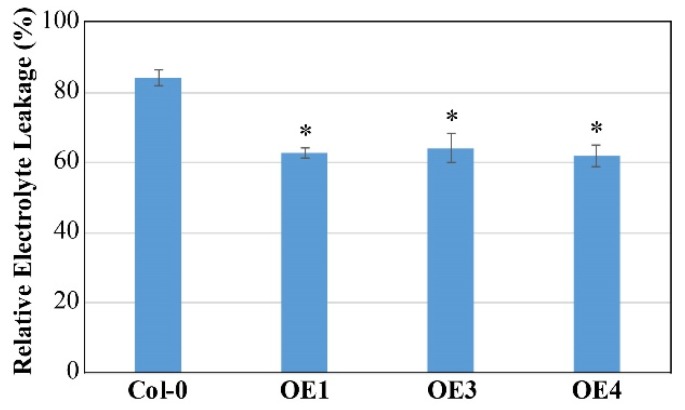
2.6. Overexpression of CsATG18b Conferred Cold Tolerance in Arabidopsis
As shown in Figure 9, the OE lines of CsATG18b had a similar growth phenotype with WT before and after cold treatment (-20 °C). However, after recovery for 2 days, the three OE lines had less leaf damage. In addition, the relative electrolyte leakage of OE lines was significantly lower than that of WT after cold treatment for 5 min (Figure 10). These results suggest that overexpression of CsATG18b conferred cold tolerance in Arabidopsis. For the OE lines of CsATG18a, we did not observe a cold tolerance phenotype (Figure S2).
Figure 9.
Overexpression of CsATG18b reduced leaf damage of transgenic Arabidopsis under cold stress. Representative pictures of WT and OE lines (OE1, OE3, and OE4) of CsATG18b were photographed before treatment, after −20 °C for 5 min, and after recovery (25 °C) for 2 days.
Figure 10.
Overexpression of CsATG18b decreased relative electrolyte leakage of transgenic Arabidopsis under cold stress. Relative electrolyte leakages of WT and OE lines were measured after −20 °C treatment for 5 min. The data are the means ± SE of three independent repeats. * indicates a significant difference at p < 0.05 using an LSD test.
3. Discussion
ATG genes are known to be involved in the autophagy process [1]. Most ATGs were first identified and functionally characterized by mutagenesis studies in yeast [2]. More than 30 ATGs have now been identified in yeast, and 23, including ATG1-10, 12-14, 16-18, 20, 27, 29, 31, TOR, VPS15, and VPS34, are considered to be the core ATGs participating in autophagy [3,4]. In Arabidopsis, around 40 homologues to these core ATGs have been identified, except for ATG14, 17, 27, 29, and 31 with no homologues [1,3]. Similarly, a total of 19 core CsATGs, including 35 members, were identified from the sweet orange genome in this study. The quantity of ATGs in sweet orange is similar to the quantity in grape (35 VvATGs) [13], banana (32 MaATGs) [12], rice (33 OsATGs) [7], and foxtail millet (37 SiATGs) [11], suggesting very conservative evolution of ATGs in these plants. Among the 19 core CsATGs, we found that CsATG1, CsATG8, and CsATG18 contained multiple members, which is similar to that in Arabidopsis and other plants [1,8,13]. However, unlike having two ATG4, ATG12, and ATG13 genes and one VTI12 in Arabidopsis, sweet orange has one member of CsATG4, CsATG12, and CsATG13 and four members of CsVTI12. This indicates a citrus crop-specificity in the number of these ATGs [1,8]. On the basis of gene duplication analysis, a high proportion of segmental duplications was predicted for those multiple members of CsATGs, especially for CsATG8. This suggests that expansion of these CsATG subfamilies was possibly derived from gene duplication during evolution [34].
Before functions were identified via transgenic strategy, the potential functions of unknown genes were often predicted according to their orthologous genes, conserved domains, and expression patterns. In this study, 29 of the CsATG genes showed higher than 60% sequence identity to homologues of Arabidopsis. Most of these high identities of CsATG and AtATG were also closely clustered together in phylogenetic trees. This result indicates that some CsATG genes may have functions similar to their homologues in Arabidopsis. On the basis of analysis of conserved domains, three CsATG1 members have similar serine/threonine-protein kinase domains that function in forming a complex with ATG13 and regulating the initiation of autophagy [35]. The UBQ and v-SNARE domains are conserved in all CsATG8 and CsVTI12 members, respectively. In general, the similar domain containing in the family members indicates the possible functional redundancy of this family [8]. However, the phylogenetic analysis showed that the members of CsATG1, CsATG8, and CsVTI12 were obviously clustered in two different branches, indicating that functional divergences are also existing in these subfamilies. A functional divergence had been reported for ATG8b and ATG8i during the hydrotropic response in Arabidopsis roots [36], which agrees well with their different phylogenetic branches in this study. In the CsATG18 subfamily, all members contain the WD40 domain, except CsATG18f, CsATG18g, and CsATG18h with an extra BCAS3 domain. The phylogenetic analysis also showed that the CsATG18f, CsATG18g, and CsATG18h were clustered in an independent branch, indicating functional divergence of these three CsATGs with the other members in this family [10].
ATG genes have played key roles in plant responses to different abiotic stresses. We found that the expression of most CsATG genes were induced under drought, heat, cold, mannitol, or salt stresses. Our results support the previous findings regarding the involvement of autophagy in abiotic stresses [3,25,28]. Although most CsATGs were induced by these stresses, some CsATGs showed different expression patterns to different stresses. For example, CsATG1a was significantly up-regulated under drought and heat but down-regulated under cold. In contrast, CsATG1d was significantly up-regulated under cold. In the CsATG8 subfamily, CsATG8a, CsATG8c, CsATG8g, and CsATG8i were obviously induced by drought and heat, but this was not true for CsATG8d and CsATG8f. Similar results have been reported for ATGs of other plant species, such as VvATGs of grape [13], NtATGs of tobacco [8], and CaATGs of pepper [10]. These results indicate that there may be different pathways to regulate the autophagy process under different abiotic stresses [10,15,25]. Compared to the above tested abiotic stresses, the roles of autophagy in heavy metals toxicity are less well known in plants. To provide more evidence, expression patterns of CsATGs were analyzed under high levels of Cu, Mn, and Cd. Unlike the predominant up-regulation of CsATGs under drought, heat, cold, and salt stress, more down-regulated CsATGs were found upon exposure to excess Cu, Mn, and Cd. In addition, the expression patterns were different between heavy metal-treated roots and leaves. Although we could not find similar reports to support our result at present, these data provide a new reference for understanding potential roles of ATGs in heavy metal stress.
ATG18 is a WD40 domain containing protein family with multiple members. They can function in vesicle formation in autophagy by binding to the phosphatidylinositol 3-phosphate sites in plants [2]. Among the multiple ATG18 members, ATG18a is the most studied. RNAi-AtATG18a transgenic Arabidopsis plants were autophagy-defective and more sensitive to oxidative, salt, and drought stresses than WT [25,37]. MdATG18a of apple showed significant up-regulation in response to leaf senescence, drought, heat, oxidative stress, and nitrogen starvation, and overexpression of MdATG18a in tomato and apple enhanced drought tolerance [21,38]. In the present study, overexpression of a CsATG18a in Arabidopsis also improved tolerance to osmotic stress, salt, and drought, which is consistent with previous results. Another member, CsATG18b, was shown to confer enhanced tolerance to osmotic stress, salt, and cold. This result provides new evidence regarding the functions of ATG18b in plants. Under abiotic stresses, a common response in cells is ROS accumulation and oxidative damage, including membrane breakage and protein oxidation and aggregation [21,37]. Thus, we speculate that enhanced tolerance in CsATG18a and CsATG18b transgenic plants might be because the damaged proteins and components are more efficiently degraded and recycled by the autophagy pathway, thereby reducing ROS accumulation and maintaining membrane integrity under stress [37,39]. Interestingly, a functional difference between CsATG18a and CsATG18b was noticed under drought and cold. Overexpression of CsATG18a conferred drought but not cold tolerance, whereas overexpression of CsATG18b conferred cold but not drought tolerance. This result appears reasonable and explainable. First, CsATG18a and CsATG18b have very different gene structures and predicted subcellular localization, which indicates their functional divergence (Table 1 and Figure 3). Second, qRT-PCR analysis shows that CsATG18a has a higher expression level under drought but a lower expression level under cold than CsATG18b (Figure 4). Third, CsATG18a and CsATG18b may be regulated by different transcription factors (TFs) under drought and cold. There is evidence that ATG18a interacts with the WRKY33 transcription factor to mediate autophagosome formation and necrotrophic pathogen resistance in Arabidopsis [40]. The HsfA1a transcription factor binds to the ATG18f promoter to regulate drought tolerance in tomato [27]. ATG18a and ATG18f are regulated by two different TFs, indicating that CsATG18a and CsATG18b may be also regulated by different TFs, thereby resulting in their functional differentiations.
4. Materials and Methods
4.1. Plant Materials and Treatments
“Hamlin” sweet orange (Citrus sinensis (L.) Osbeck) was used as the plant material in this study. The seeds of “Hamlin” sweet orange were collected from the Citrus Research Institute, Southwest University, Chongqing, China. The seeds were sterilized and germinated, and the seedlings were hydroponically cultured at 25 °C and 16 h photoperiod (50 µmol m−2 s−1), as described by Fu et al. [41]. After 30 days of growth in hydroponic solution, uniform seedlings were divided into different groups (3 biological repeats and 10 plants per repeat for each group) for the following sets of treatments. For the drought treatment, the seedlings were dehydrated under room temperature (25°C) by removing the hydroponic solution. For the heat and cold treatments, two groups of seedlings were put in a growth chamber under 42 °C and a refrigerator under 4 °C, respectively. For the mannitol and NaCl treatments, two groups of seedlings were transferred to 400 mM mannitol and 200 mM NaCl solutions, respectively. For the excess Cu, Mn, and Cd treatments, the seedlings were transferred to new hydroponic solutions containing 0.01 mM CuSO4 (+Cu), 0.1 mM MnSO4 (+Mn), and 0.038 mM CdSO4 (+Cd), respectively. The seedlings grown under normal conditions were used as controls (CK). The leaves were sampled after 3 h and 12 h of drought, heat, and cold treatments or 2 h and 3 h of mannitol and NaCl treatments, and the leaves and roots were sampled upon 5 days and 15 days of excess Cu, Mn, and Cd treatments. Samples were immediately frozen in liquid nitrogen and stored at −80 °C until use.
4.2. Genome-Wide Identification of ATG Genes in Sweet Orange
To identify all putative ATG proteins of the sweet orange, a BLASTP search of the sweet orange genome database (http://citrus.hzau.edu.cn/orange/) [34] was performed using the known Arabidopsis ATG proteins (AtATGs) as query sequences [1,3]. The results were filtered using a score value of ≥ 100 and an e-value ≤ e−10 [42]. After removal of redundant sequences, all putative ATG genes were submitted to the PFAM database (http://pfam.xfam.org/) to confirm the existence of the ATG domains. All of the identified ATG genes of sweet orange were named as CsATGs.
4.3. Bioinformatic Analysis of CsATGs
The subcellular locations of the CsATG proteins were predicted online by using the ProtComp v. 9.0 (http://www.softberry.com/berry.phtml?topic=protcomppl&group=programs&subgroup=proloc). To construct phylogenetic trees, all putative CsATGs and the ATG proteins from Arabidopsis thaliana [1,3], tobacco (Nicotiana tabacum) [8], rice (Oryza sativa) [7], and grapevine (Vitis vinifera) [13] were first collected (as shown in Table S1) and then submitted to ClustalW for performing multiple sequence alignment [43]. The generated files were used to construct the phylogenetic tree with the neighbor-joining method and 1000 replicates of bootstrap analysis in MEGA 6 software [44]. The bootstrap values lower than 50% are not shown in the phylogenetic trees. The chromosomal positions of the CsATG genes were provided by the sweet orange genome database, and MapInspect software (http://mapinspect.software.informer.com) was used to draw the location images. For the gene structure analysis, the generic feature format (GFF) file of each CsATG was downloaded from the sweet orange genome database and then submitted to the Gene Structure Display Server (GSDS, http://gsds.cbi.pku.edu.cn/) to display intron-exon structures [45]. The GFF files were also inputted into MCScanX software to identify gene duplications and collinear correlations [46]. The conserved domains of CsATG proteins were predicted online in the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd/).
4.4. Total RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA was extracted from the CK and stress-treated leaves or roots of “Hamlin” sweet orange seedlings using a RNAprep pure plant kit (Tiangen Biotech Co., Ltd., Beijing, China), and the RNA concentration and quality were determined with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Then, 1 μg of high-quality RNA was used for cDNA synthesis with an iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. qRT-PCR was performed on the Bio-Rad CFX Connect RealTime system using ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd., Nanjing, China). Each PCR reaction contained 5.0 μL SYBR mix, 0.2 μM primers, and 1.0 μL diluted cDNA in a final volume of 10 μL. Specific primers of CsATG genes were designed using the online primer-blast program in the NCBI website, whereas Actin (Cs1g05000.1) was used as a reference gene to normalize the relative expression levels of the tested genes. Three biological replicates and three technical replicates were performed for each treatment.
4.5. Construction of Transgenic Arabidopsis Expressing 35S-CsATG18a and 35S-CsATG18b
The CDS (coding sequence) fragments of CsATG18a and CsATG18b were first isolated from “Hamlin” sweet orange. After being confirmed by sequencing, the full CDS fragments of CsATG18a and CsATG18b were recombined into a pGBO vector to produce 35S-CsATG18a and 35S-CsATG18b constructs, respectively. The two constructs were then transformed into Agrobacterium tumefaciens strain GV3101 by electroporation. The Arabidopsis thaliana wild-type (WT; ecotype Col-0) plants were used to perform Agrobacterium-mediated transformation by the floral dipping method [47]. The transgenic plants were selected on 1/2 MS medium containing 50 mg L−1 kanamycin until a homozygous T4 generation was obtained. The expression levels of transgenes were measured by qRT-PCR.
4.6. Phenotypic and Physiological Analysis of Transgenic Arabidopsis Lines
To evaluate the tolerance of CsATG18a and CsATG18b transgenic Arabidopsis plants to different abiotic stresses, three independent T4 homozygous lines with relatively high expression levels of CsATG18a and CsATG18b were selected for NaCl, mannitol, drought, and cold treatments, and WT (Col-0) plants were used as controls. For the NaCl and mannitol treatments, the seeds of transgenic lines and WT were sown on 1/2 MS with 200 mM NaCl or 400 mM mannitol, and the germination rate was calculated at 3 to 7 days. For the drought treatment, 30 day old transgenic and WT plants were dewatered for 15 days and then watered for recovery. For the cold treatment, 30 day old transgenic and WT plants were transferred to a −20 °C freezer for 5 min and then back to room temperature for recovery. After drought and cold treatments, the relative electrolytic leakage was measured as described by Shi et al. [48].
5. Conclusions
A total of 35 CsATG genes were identified from the genome of sweet orange. These CsATGs showed high sequence and phylogeny similarities with the AtATGs of Arabidopsis. Most CsATG genes were significantly up- or down-regulated in response to drought, heat, cold, salt, and osmotic stresses, as well as to excess Cu, Mn, and Cd stresses. Overexpression of CsATG18a and CsATG18b enhanced tolerance to both salt and osmotic stresses in Arabidopsis. Overexpression of CsATG18a in Arabidopsis conferred drought but not cold tolerance, whereas overexpression of CsATG18b conferred cold but not drought tolerance. This study analyzed CsATG genes in citrus and preliminarily revealed their important roles in response to abiotic stresses. In future, their functions and the underlying mechanisms will be further uncovered by overexpressing in citrus plants.
Abbreviations
| TOR | target of rapamycin kinase |
| PE | phosphatidylethanolamine |
| ROS | reactive oxygen species |
| PCD | programmed cell death |
| UTR | untranslated region |
| FC | fold change |
| OE | overexpression |
| PKc like | protein kinase superfamily |
| ATG C | autophagy-related protein C-terminal domain |
| ATG N | autophagy-related protein N terminal domain |
| Chorein N | N-terminal region of chorein or VPS13 |
| Autophagy N | autophagocytosis-associated protein N-terminal domain |
| Autophagy C | autophagocytosis-associated protein C-terminal domain |
| Autophagy act C | autophagocytosis-associated protein, active-site domain |
| Peptidase C54 | peptidase family C54 |
| UBQ | ubiquitin homologues |
| WD40 Repeats | WD40 repeats |
| BCAS3 | breast carcinoma amplified sequence 3 |
| PX domain | the phox homology domain, a phosphoinositide binding module |
| BAR | the Bin/amphiphysin/Rvs (BAR) domain |
| PI3Kc | catalytic domain of phosphoinositide 3-kinase |
| PI3Ka | phosphoinositide 3-kinase family, accessory domain |
| C2 | C2 domain |
| SNARE | soluble N-ethylmaleimide-sensitive-factor attachment protein receptor |
| V-SNARE N | vesicle transport v-SNARE protein N-terminus |
| V-SNARE C | vesicle transport v-SNARE protein C-terminus |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/8/2699/s1.
Author Contributions
X.-Z.F. and L.-Z.P. conceived and designed the study; X.Z. performed bioinformatic analysis. Y.-Y.X. and Q.-L.H. performed stress treatments and qRT-PCR. C.-P.C. and L.-L.L. prepared experimental materials. X.-Z.F. prepared all of the figures and tables and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (31772280), the National Key Research and Development Program of China (2018YFD1000300, 2017YFD0202000), the National Citrus Engineering Research Center (NCERC2019001), and the Technological Innovation and Application Program of Chongqing (cstc2018jscx-mszdX0002).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Il Kwon S., Park O.K. Autophagy in plants. J. Plant Biol. 2008;51:313–320. doi: 10.1007/BF03036132. [DOI] [Google Scholar]
- 2.Liu Y., Bassham D.C. Autophagy: Pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 2012;63:215–237. doi: 10.1146/annurev-arplant-042811-105441. [DOI] [PubMed] [Google Scholar]
- 3.Han S., Yu B., Wang Y., Liu Y. Role of plant autophagy in stress response. Protein Cell. 2011;2:784–791. doi: 10.1007/s13238-011-1104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Avila-Ospina L., Moison M., Yoshimoto K., Masclaux-Daubresse C. Autophagy, plant senescence, and nutrient recycling. J. Exp. Bot. 2014;65:3799–3811. doi: 10.1093/jxb/eru039. [DOI] [PubMed] [Google Scholar]
- 5.Ohsumi Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Bio. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 6.Thompson A.R., Vierstra R.D. Autophagic recycling: Lessons from yeast help define the process in plants. Curr. Opin. Plant Biol. 2005;8:165–173. doi: 10.1016/j.pbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Xia K.F., Liu T., Ouyang J., Wang R., Fan T., Zhang M.Y. Genome-wide identification, classification, and expression analysis of autophagy-associated gene homologues in rice (Oryza sativa L.) DNA Res. 2011;18:363–377. doi: 10.1093/dnares/dsr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou X.M., Zhao P., Wang W., Zou J., Cheng T.H., Peng X.B., Sun M.X. A comprehensive, genome-wide analysis of autophagy-related genes identified in tobacco suggests a central role of autophagy in plant response to various environmental cues. DNA Res. 2015;22:245–257. doi: 10.1093/dnares/dsv012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F.Q., Chung T., Pennington J.G., Federico M.L., Kaeppler H.F., Kaeppler S.M., Otegui M.S., Vierstra R.D. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015;27:1389–1408. doi: 10.1105/tpc.15.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Y.F., Guo M., Wang H., Lu J.P., Liu J.H., Zhang C., Gong Z.H., Lu M.H. Autophagy, a conserved mechanism for protein degradation, responds to heat, and other abiotic stresses in Capsicum annuum L. Front. Plant Sci. 2016:7. doi: 10.3389/fpls.2016.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W.W., Chen M., Wang E.H., Hu L.Q., Hawkesford M.J., Zhong L., Chen Z., Xu Z.S., Li L.C., Zhou Y.B., et al. Genome-wide analysis of autophagy-associated genes in foxtail millet (Setaria italica L.) and characterization of the function of SiATG8a in conferring tolerance to nitrogen starvation in rice. BMC Genomics. 2016;17:797. doi: 10.1186/s12864-016-3113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y.X., Liu W., Hu W., Liu G.Y., Wu C.J., Liu W., Zeng H.Q., He C.Z., Shi H.T. Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017;36:1237–1250. doi: 10.1007/s00299-017-2149-5. [DOI] [PubMed] [Google Scholar]
- 13.Shangguan L.F., Fang X., Chen L.D., Cui L.W., Fang J.G. Genome-wide analysis of autophagy-related genes (ARGs) in grapevine and plant tolerance to copper stress. Planta. 2018;247:1449–1463. doi: 10.1007/s00425-018-2864-3. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J., Yu J.Q., Chen Z.X. The perplexing role of autophagy in plant innate immune responses. Mol. Plant Pathol. 2014;15:637–645. doi: 10.1111/mpp.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lv X., Pu X., Qin G., Zhu T., Lin H. The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis. 2014;19:905–921. doi: 10.1007/s10495-014-0981-4. [DOI] [PubMed] [Google Scholar]
- 16.Avin-Wittenberg T., Bajdzienko K., Wittenberg G., Alseekh S., Tohge T., Bock R., Giavalisco P., Fernie A.R. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell. 2015;27:306–322. doi: 10.1105/tpc.114.134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang P., Sun X., Jia X., Wang N., Gong X.Q., Ma F.W. Characterization of an autophagy-related gene MdATG8i from apple. Front. Plant Sci. 2016;7:720. doi: 10.3389/fpls.2016.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P., Sun X., Jia X., Ma F.W. Apple autophagy-related protein MdATG3s afford tolerance to multiple abiotic stresses. Plant Sci. 2017;256:53–64. doi: 10.1016/j.plantsci.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Sun X., Wang N., Jia X., Ma F.W. Ectopic expression of an autophagy-associated MdATG7b gene from apple alters growth and tolerance to nutrient stress in Arabidopsis thaliana. Plant Cell Tiss. Org. 2017;128:9–23. doi: 10.1007/s11240-016-1070-x. [DOI] [Google Scholar]
- 20.Sun X., Jia X., Huo L.Q., Che R.M., Gong X.Q., Wang P., Ma F.W. MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 2018;41:469–480. doi: 10.1111/pce.13110. [DOI] [PubMed] [Google Scholar]
- 21.Sun X., Wang P., Jia X., Huo L.Q., Che R.M., Ma F.W. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018;16:545–557. doi: 10.1111/pbi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B., Liu G.Y., Wang Y.Q., Wei Y.X., Shi H.T. Overexpression of banana ATG8f modulates drought stress resistance in Arabidopsis. Biomolecules. 2019;9:814. doi: 10.3390/biom9120814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Q.W., Soulay F., Saudemont B., Elmayan T., Marmagne A., Masclaux-Daubresse C. Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 2019;60:343–352. doi: 10.1093/pcp/pcy214. [DOI] [PubMed] [Google Scholar]
- 24.Li W.W., Chen M., Zhong L., Liu J.M., Xu Z.S., Li L.C., Zhou Y.B., Guo C.H., Ma Y.Z. Overexpression of the autophagy-related gene SiATG8a from foxtail millet (Setaria italica L.) confers tolerance to both nitrogen starvation and drought stress in Arabidopsis. Biochem. Biophys. Res. Commun. 2015;468:800–806. doi: 10.1016/j.bbrc.2015.11.035. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y.M., Xiong Y., Bassham D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy. 2009;5:954–963. doi: 10.4161/auto.5.7.9290. [DOI] [PubMed] [Google Scholar]
- 26.Li Y.B., Cui D.Z., Sui X.X., Huang C., Huang C.Y., Fan Q.Q., Chu X.S. Autophagic survival precedes programmed cell death in wheat seedlings exposed to drought stress. Int. J. Mol. Sci. 2019;20:5777. doi: 10.3390/ijms20225777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., Cai S.Y., Yin L.L., Shi K., Xia X.J., Zhou Y.H., Yu J.Q., Zhou J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015;11:2033–2047. doi: 10.1080/15548627.2015.1098798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo L.M., Zhang P.P., Zhu R.H., Fu J., Su J., Zheng J., Wang Z.Y., Wang D., Gong Q.Q. Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 2017;8:1459. doi: 10.3389/fpls.2017.01459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dundar G., Shao Z.H., Higashitani N., Kikuta M., Izumi M., Higashitani A. Autophagy mitigates high-temperature injury in pollen development of Arabidopsis thaliana. Dev. Biol. 2019;456:190–200. doi: 10.1016/j.ydbio.2019.08.018. [DOI] [PubMed] [Google Scholar]
- 30.Sedaghatmehr M., Thirumalaikumar V.P., Kamranfar I., Marmagne A., Masclaux-Daubresse C., Balazadeh S. A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 2019;42:1054–1064. doi: 10.1111/pce.13426. [DOI] [PubMed] [Google Scholar]
- 31.Shinozaki D., Merkulova E.A., Naya L., Horie T., Kanno Y., Seo M., Ohsumi Y., Masclaux-Daubresse C., Yoshimoto K. Autophagy increases zinc bioavailability to avoid light-mediated reactive oxygen species production under zinc deficiency. Plant Physiol. 2020;182:1284–1296. doi: 10.1104/pp.19.01522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren H., Li Y.N., Zhao F.F., Pu X.J., Wei L.J., Lv X., Zhu F., Lin H.H. The role of autophagy in alleviating damage of aluminum stress in Arabidopsis thaliana. Plant Growth Regul. 2016;79:167–175. doi: 10.1007/s10725-015-0122-2. [DOI] [Google Scholar]
- 33.Guan B., Lin Z., Liu D.C., Li C.Y., Zhou Z.Q., Mei F.Z., Li J.W., Deng X.Y. Effect of waterlogging-induced autophagy on programmed cell death in Arabidopsis roots. Front. Plant Sci. 2019;10:468. doi: 10.3389/fpls.2019.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Q., Chen L.L., Ruan X., Chen D., Zhu A., Chen C., Bertrand D., Jiao W.B., Hao B.H., Lyon M.P., et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–66. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 35.Cheong H., Nair U., Geng J.F., Klionsky D.J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2008;19:668–681. doi: 10.1091/mbc.e07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jimenez-Nopala G., Salgado-Escobar A.E., Cevallos-Porta D., Cardenas L., Sepulveda-Jimenez G., Cassab G., Porta H. Autophagy mediates hydrotropic response in Arabidopsis thaliana roots. Plant Sci. 2018;272:1–13. doi: 10.1016/j.plantsci.2018.03.026. [DOI] [PubMed] [Google Scholar]
- 37.Xiong Y., Contento A.L., Nguyen P.Q., Bassham D.C. Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang P., Sun X., Yue Z.Y., Liang D., Wang N., Ma F.W. Isolation and characterization of MdATG18 alpha, a WD40-repeat AuTophaGy-related gene responsive to leaf senescence and abiotic stress in Malus. Sci. Hortic. 2014;165:51–61. doi: 10.1016/j.scienta.2013.10.038. [DOI] [Google Scholar]
- 39.Perez-Perez M.E., Lemaire S.D., Crespo J.L. Reactive oxygen species and autophagy in plants and algae. Plant Physiol. 2012;160:156–164. doi: 10.1104/pp.112.199992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai Z.B., Wang F., Zheng Z.Y., Fan B.F., Chen Z.X. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 2011;66:953–968. doi: 10.1111/j.1365-313X.2011.04553.x. [DOI] [PubMed] [Google Scholar]
- 41.Fu X.Z., Tong Y.H., Zhou X., Ling L.L., Chun C.P., Cao L., Zeng M., Peng L.Z. Genome-wide identification of sweet orange (Citrus sinensis) metal tolerance proteins and analysis of their expression patterns under zinc, manganese, copper, and cadmium toxicity. Gene. 2017;629:1–8. doi: 10.1016/j.gene.2017.07.072. [DOI] [PubMed] [Google Scholar]
- 42.Kumar R., Tyagi A.K., Sharma A.K. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics. 2011;285:245–260. doi: 10.1007/s00438-011-0602-7. [DOI] [PubMed] [Google Scholar]
- 43.Thompson J.D., Higgins D.G., Gibson T.J. Clustal-W-improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo A.Y., Zhu Q.H., Chen X., Luo J.C. GSDS: A gene structure display server. Yi Chuan. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y.P., Tang H.B., DeBarry J.D., Tan X., Li J.P., Wang X.Y., Lee T.H., Jin H.Z., Marler B., Guo H., et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic. Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 48.Shi J., Fu X.Z., Peng T., Huang X.S., Fan Q.J., Liu J.H. Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 2010;30:914–922. doi: 10.1093/treephys/tpq030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.