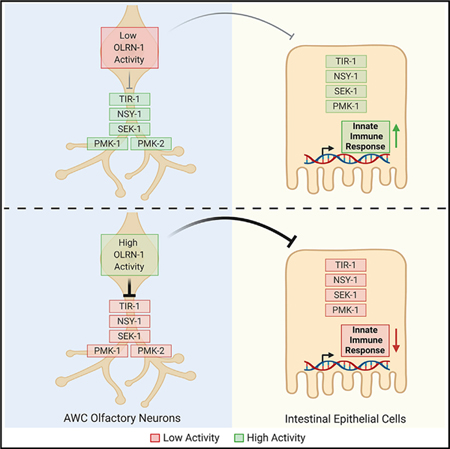
SUMMARY
Olfactory neurons allow animals to discriminate nutritious food sources from potential pathogens. From a forward genetic screen, we uncovered a surprising requirement for the olfactory neuron gene olrn-1 in the regulation of intestinal epithelial immunity in Caenorhabditis elegans. During nematode development, olrn-1 is required to program the expression of odorant receptors in the AWC olfactory neuron pair. Here, we show that olrn-1 also functions in AWC neurons in the cell non-autonomous suppression of the canonical p38 MAPK PMK-1 immune pathway in the intestine. Low activity of OLRN-1, which activates the p38 MAPK signaling cassette in AWC neurons during larval development, also de-represses the p38 MAPK PMK-1 pathway in the intestine to promote immune effector transcription, increased clearance of an intestinal pathogen, and resistance to bacterial infection. These data reveal an unexpected connection between olfactory receptor development and innate immunity and show that anti-pathogen defenses in the intestine are developmentally programmed.
Graphical Abstract
In Brief
During nematode development, the olfactory neuron gene olrn-1 programs odorant receptor expression in AWC neurons. Foster et al. show that olrn-1 also functions in AWC neurons to suppress innate immune defenses in the intestine. These data reveal an unexpected connection between olfactory receptor development and innate immunity.
INTRODUCTION
The expression of a diverse array of olfactory receptors within sensory neurons is essential for metazoans to survive in microbe-rich environments. For example, amphid neurons in the head of the nematode C. elegans sample the environment and program rapid changes in locomotion, which allows nematodes to forage decomposing organic matter for bacterial food sources and avoid potential pathogens. Thus, C. elegans provides an experimental platform to understand how the development of sensory neurons is integrated with the physiology of the organism as a whole.
In addition to learned behavioral aversion responses to bacterial pathogens, innate immune defenses in intestinal epithelial cells allow nematodes to survive challenge from environmental pathogens (Kim and Ewbank, 2018; Pukkila-Worley, 2016). The canonical immune pathway in intestinal cells is anchored by the p38 mitogen-activated protein kinase (MAPK) PMK-1 (Kim et al., 2002; Troemel et al., 2006). p38 MAPK PMK-1 functions as part of a classical MAPK signaling cassette, which is activated by MAPKKK NSY-1 and MAPKK SEK-1, homologs of mammalian ASK1 and MKK3/6, respectively (Kim et al., 2002), and by TIR-1, a toll/interleukin-1 receptor (TIR) domain protein (Liberati et al., 2004). The p38 MAPK PMK-1 pathway ensures the basal expression of immune effectors in the absence of a pathogen (Peterson et al., 2019; Troemel et al., 2006). Thus, mechanisms that adjust basal levels of p38 MAPK PMK-1 pathway activity could act as a rheostat for immune effector expression, functioning both to prime C. elegans intestinal epithelial cells for the induction of anti-pathogen responses and to limit the deleterious effects of immune hyperactivation.
Here, we conducted a forward genetic screen to identify endogenous regulators of the p38 MAPK PMK-1 pathway. Genetic analyses of mutants identified in this screen uncovered a signaling axis between amphid wing C (AWC) sensory neurons and the intestinal epithelium that promotes immune homeostasis by suppressing the canonical p38 MAPK PMK-1 immune pathway. Interestingly, olrn-1, the newly discovered neuronal regulator of this pathway, was previously shown to control the expression of olfactory receptors in AWC sensory neurons (Bauer Huang et al., 2007). During neuronal development, olrn-1 acts cell autonomously in AWC neurons to suppress the TIR-1/NSY-1/SEK-1 cassette, which signals redundantly through the p38 MAPK PMK-1 or PMK-2 (Bauer Huang et al., 2007; Chuang and Bargmann, 2005; Pagano et al., 2015; Troemel et al., 1999). Modulation of p38 MAPK pathway activity by OLRN-1 in AWC neurons leads to differentiation of olfactory receptor expression, a developmental step that is required for C. elegans to detect specific chemoattractive odors (Wes and Bargmann, 2001). We show that neuronal olrn-1 also functions cell non-autonomously to suppress the p38 MAPK PMK-1 innate immune pathway in the intestine. Low olrn-1 activity in AWC neurons, as recapitulated in multiple olrn-1 loss-of-function mutant strains, de-represses the p38 MAPK PMK-1 pathway in the intestine, which promotes immune effector transcription, increased clearance of an intestinal pathogen, and resistance to bacterial infection. Interestingly, olrn-1 and p38 MAPK pmk-1-dependent immune effectors are enriched among genes that are induced during larval development in wild-type nematodes. These data suggest that low activity of neuronal OLRN-1 de-represses the p38 MAPK PMK-1 pathway to prime the immune response in the intestine to handle challenges from bacterial pathogens encountered during larval development.
RESULTS
Loss-of-Function Mutations in olrn-1 Cause Constitutive Immune Activation
We conducted a forward genetic screen to identify endogenous regulators of the p38 MAPK PMK-1 innate immune pathway. The innate immune reporter Pirg-4(F08G5.6)::GFP was used for this experiment to provide a convenient readout of innate immune activation (Pukkila-Worley et al., 2014). irg-4 is a putative C. elegans immune effector whose basal expression is under the control of the p38 MAPK PMK-1 innate immune pathway (Anderson et al., 2019; Peterson et al., 2019; Pukkila-Worley et al., 2012; Troemel et al., 2006). In addition, irg-4 is upregulated during infection by multiple bacterial pathogens and is required for host defense against the bacterial pathogen Pseudomonas aeruginosa (Anderson et al., 2019; Nandakumar and Tan, 2008; Peterson et al., 2019; Shapira et al., 2006; Troemel et al., 2006). Previously, we screened the F1 progeny of mutagenized Pirg-4::GFP animals for dominant mutations that cause constitutive immune activation and identified nsy-1(ums8), a gain-of-function mutation in the MAPKKK that functions upstream of p38 MAPK pmk-1 (Cheesman et al., 2016). Thus, this approach can identify mutations that affect the activity of the p38 MAPK PMK-1 pathway.
To identify regulators of immune activation in C. elegans, we screened the F2 progeny of approximately 40,000 mutagenized Pirg-4::GFP haploid genomes for recessive mutations that caused constitutive Pirg-4::GFP expression, and we identified two mutant alleles: ums9 and ums11 (Figure 1A). Whole-genome sequencing of pooled F2 recombinants, homozygous for the mutant phenotype following two outcrosses to wild-type N2 animals, was performed to identify the mutations that caused constitutive Pirg-4::GFP expression. Sequencing revealed that both ums9 and ums11 contain different recessive mutations in olrn-1, a neuronally expressed protein with putative transmembrane domains (Bauer Huang et al., 2007; Torayama et al., 2007). ums9 is a nonsense mutation (Q497*), and ums11 is a mutation that disrupts an olrn-1 splice acceptor site (Figure 1B).
Figure 1. Loss-of-Function Mutations in olrn-1 Cause Constitutive Immune Activation.
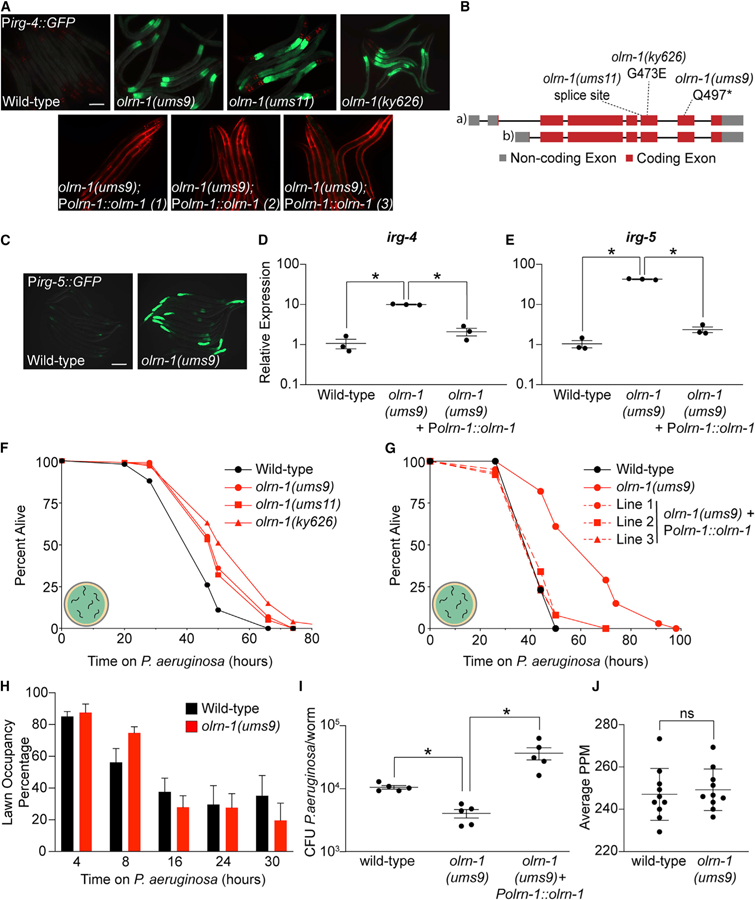
(A) Images of olrn-1 mutants and three independent rescue lines with olrn-1 expressed under the control of its own promoter in the Pirg-4::GFP immune reporter background are shown. Red pharyngeal expression is the Pmyo-2::mCherry co-injection marker, which confirms the presence of the Pirg-4::GFP transgene. The Pmyo-3::mCherry co-injection marker confirms expression of the Polrn-1::olrn-1 construct in the rescue lines.
(B) Schematic of the olrn-1 locus with the locations of the ums9, ums11, and ky626 mutations is shown.
(C) Immune reporter Pirg-5::GFP in the olrn-1(ums9) background is shown.
(D and E) Presence of the Pirg-5::GFP transgene was confirmed by assaying for the Rol phenotype. qRT-PCR data of irg-4 (D) and irg-5 (E) of the indicated genotypes is presented. Data are the average of three independent replicates, each normalized to a control gene, with error bars representing SEM, Data are presented as the value relative to the average expression from all replicates of the indicated gene in wild-type animals. *p < 0.05 by one-way ANOVA for the indicated comparison.
(F and G) C. elegans pathogenesis assay conducted with a large lawn of P. aeruginosa and C. elegans of indicated genotypes at L4 is shown. Data are representative of three trials. The Kaplan-Meier method was used to estimate the survival curves for each group, and the log rank test was used for all statistical comparisons. Sample sizes, mean lifespan, and p values for all trials are shown in Table S2.
(H) Quantification of the propensity of olrn-1(ums9) and wild-type animals to avoid a lawn of P. aeruginosa is shown. Data are presented as the average number of animals that were on a small lawn of P. aeruginosa from three separate replicates, with error bars representing SEM. There is no significant difference by one-way ANOVA between these mutants, except at the 8-h time point.
(I) P. aeruginosa, isolated from the intestines of animals with the indicated genotypes, was quantified after 24 h of bacterial infection. Data are colony-forming units (CFUs) of P. aeruginosa and are presented as the average of five separate replicates, with each replicate containing 10–11 animals. *p < 0.05 by one-way ANOVA for the indicated comparison.
(J) Data are the pharyngeal pumping rates, recorded as pumps per minute (PPM), of 10 individual young adult C. elegans feeding on non-pathogenic OP50 in wild-type and olrn-1(ums9) mutants, with error bars representing SEM. ns indicates no significant difference by one-way ANOVA for the indicated comparison.
Scale bars in (A) and (C) are 100 μm. See also Figure S1.
Several experiments demonstrated that loss-of-function mutations in olrn-1 cause constitutive activation of Pirg-4::GFP. First, expression of olrn-1 under the control of its own promoter in three independent extrachromosomal arrays rescued the constitutive activation of Pirg-4::GFP in the ums9 mutant (Figure 1A). A previously characterized loss-of-function allele, olrn-1(ky626) (a missense mutation [G473E]), also caused constitutive activation of Pirg-4::GFP (Figures 1A and 1B; Bauer Huang et al., 2007). Finally, we outcrossed olrn-1(ums9) to wild type six times and confirmed that Pirg-4::GFP was still constitutively activated (Figure 1A).
A second transcriptional reporter for a different innate immune effector, Pirg-5(F35E12.5)::GFP, is also constitutively activated in the intestine of olrn-1(ums9) mutants (Figure 1C). Like irg-4, irg-5 is an immune effector that is required for host defense during P. aeruginosa infection and is a target of the p38 MAPK PMK-1 pathway (Bolz et al., 2010; Peterson et al., 2019; Troemel et al., 2006). Altogether, irg-4 and irg-5 are useful readouts of innate immune activation in the C. elegans intestine. Endogenous irg-4 and irg-5 were transcriptionally induced to comparable levels in olrn-1(ums9), olrn-1(ums11), and olrn-1(ky626) mutants, and reintroduction of olrn-1 under the control of its own promoter rescued the constitutive activation of these genes in the olrn-1(ums9) mutant background (Figures 1D, 1E, and S1A–S1C).
Consistent with the constitutive activation of innate immune effector transcription in olrn-1 loss-of-function animals, olrn-1 mutants displayed increased resistance to infection by P. aeruginosa (Figure 1F). Importantly, we confirmed that loss of function of olrn-1 modulates the susceptibility of C. elegans to P. aeruginosa infection by testing the pathogen susceptibility of the three independent olrn-1 rescue lines described earlier (Figure 1G). Expression of olrn-1 under the control of its own promoter rescued the pathogen-resistance phenotype of the olrn-1(ums9) mutant (Figure 1G). We considered that olrn-1 mutants may be resistant to P. aeruginosa infection because they are better able to avoid the pathogen than wild-type animals. However, olrn-1(ums9), olrn-1(ums11), and olrn-1(ky626) mutants were resistant to P. aeruginosa infection in a pathogenesis assay conducted with a lawn of bacteria that was spread to the edges of the agar, which negates the contribution of behavioral avoidance to the pathogen-resistance phenotype (Figure 1F; Reddy et al., 2009; Styer et al., 2008; Sun et al., 2011). In addition, the olrn-1(ums9) mutant did not have increased propensity to avoid a small lawn of P. aeruginosa compared with wild type (Figure 1H).
The olrn-1(ums9) mutant animals accumulated significantly less P. aeruginosa in their intestines compared with wild-type animals (Figure 1I). Reintroduction of olrn-1 under the control of its own promoter complemented this olrn-1(ums9) mutant phenotype (Figure 1I). There was no difference in pharyngeal pumping rates between wild-type and olrn-1(ums9) mutants (Figure 1J). In addition, olrn-1(ums9) and olrn-1(ky626) mutants each have a lifespan that is similar to that of wild-type animals (Figure S1D). Altogether, these data demonstrate that olrn-1(ums9) mutants drive a transcriptional response that promotes clearance of bacteria from the intestine and resistance to P. aeruginosa infection, without pleiotropic effects on feeding behavior or nematode lifespan.
olrn-1 Suppresses the p38 MAPK PMK-1 Innate Immune Pathway
To characterize the full spectrum of genes regulated by olrn-1, we performed RNA sequencing (RNA-seq). The transcriptomes of wild-type and olrn-1(ums9) mutant animals growing in standard culture conditions (i.e., in the absence of pathogen challenge) were profiled. In olrn-1(ums9) mutants, 549 genes were upregulated compared with wild type (greater than 2-fold, p < 0.05) (Figure 2A; Table S1). Analysis of these differentially expressed transcripts revealed significant enrichment for innate immune genes (Figure 2B), including irg-4 and irg-5 (Figure 2A). Indeed, 41 of the 236 genes that are induced during pseudomonal infection (Miller et al., 2015) are also constitutively upregulated in the olrn-1(ums9) mutant (hypergeometric p value 3.22e—21) (Figure 2C).
Figure 2. olrn-1 Suppresses Innate Immune Effector Expression.
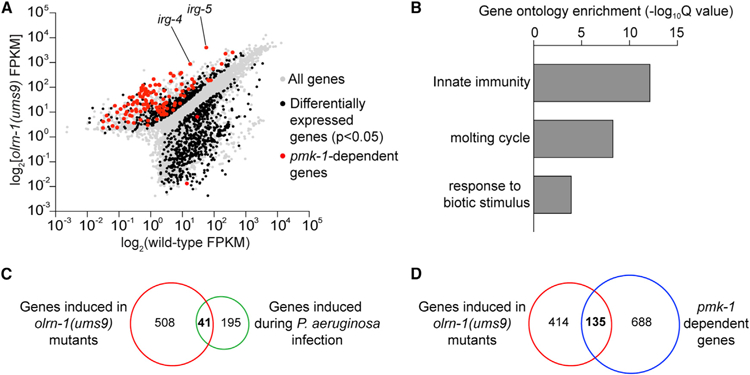
(A) Data from an mRNA sequencing (mRNA-seq) experiment comparing gene expression in olrn-1(ums9) mutants with wild-type animals are shown. All genes are shown in gray. Genes that are differentially expressed in olrn-1(ums9) mutants compared with wild-type animals are shown in black (fold change > 2, p < 0.05). Genes that are known targets of the p38 MAPK pmk-1 pathway are highlighted in red. The locations of the representative genes irg-4 and irg-5, whose expression is examined throughout this manuscript, are shown.
(B) Gene Ontology enrichment analysis for the 549 genes whose transcription was significantly upregulated in olrn-1(ums9) mutants compared with wild type is shown. The three most significantly enriched categories are shown, reported as the —log10 transformation of the Q value for the enrichment of each category.
(C and D) Venn diagrams show the overlap of the 549 genes upregulated in olrn-1 mutants with genes that are known to be induced during P. aeruginosa infection (C) and are targets of the p38 MAPK PMK-1 pathway (D). Hypergeometric p values for the overlap in (C) and (D) are 3.22e–21 and 9.10e–67, respectively.
See also Figure S2.
Loss-of-function olrn-1 mutants demonstrated constitutive activation of Pirg-4::GFP (Figure 1A) and Pirg-5::GFP (Figure 1C) in the intestinal epithelial cells of C. elegans, the tissue that directly interfaces with ingested pathogens. In the RNA-seq experiment, 171 of the 549 olrn-1-repressed genes are expressed in intestinal epithelial cells (hypergeometric p value 1.16e—32) (Haenni et al., 2012). Altogether, these data indicate that olrn-1 suppresses the transcription of genes that are expressed in the intestine, which includes a significant number of immune effectors.
The RNA-seq experiment also revealed that olrn-1 negatively regulates a significant number of genes that are known targets of the p38 MAPK PMK-1 pathway (Bond et al., 2014; Troemel et al., 2006). Of the 549 genes that are upregulated in olrn-1(ums9) mutants, 135 depend on pmk-1 for their basal transcription (hypergeometric p value 9.10e–67) (Figures 2A and 2D; Bond et al., 2014). To determine whether olrn-1 controls the activity of the p38 MAPK PMK-1 pathway, we performed western blot experiments with antibodies that recognize the doubly phosphorylated TGY motif of activated PMK-1 and the total PMK-1 protein. Both the olrn-1(ums9) and the olrn-1(ky626) loss-of-function mutants have an increased ratio of phosphorylated PMK-1 relative to total PMK-1 compared with wild-type controls, as quantified from four biological replicates (Figures 3A and 3B).
Figure 3. olrn-1 Suppresses the p38 MAPK PMK-1 Innate Immune Pathway.
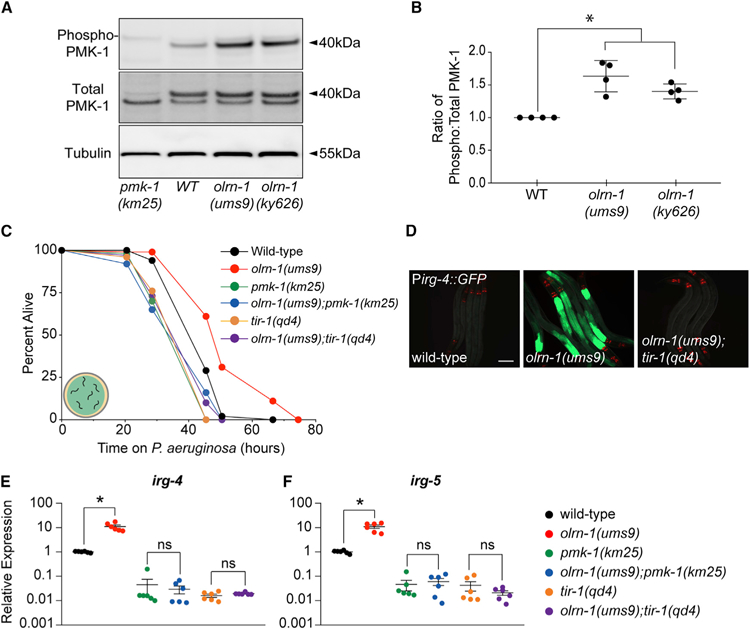
(A) Immunoblot analysis of lysates from animals of the indicated genotype using antibodies that recognize the doubly phosphorylated TGY motif of PMK-1 (phospho-PMK-1), the total PMK-1 protein (total PMK-1), and tubulin is shown. PMK-1 is a 40 kDa protein, and tubulin is a 55 kDa protein.
(B) Band intensities of four biological replicates of the western blot shown in (A) were quantified. The ratio of active to total PMK-1 is shown for each genotype and is presented relative to the ratio in wild-type animals for each replicate. *p < 0.05 by one-way ANOVA for the indicated comparison.
(C) C. elegans pathogenesis assay conducted with a large lawn of P. aeruginosa and C. elegans of indicated genotypes at L4 is shown. Data are representative of three trials. The Kaplan-Meier method was used to estimate the survival curves for each group, and the log rank test was used for all statistical comparisons. Sample sizes, mean lifespan, and p values for all trials are shown in Table S2.
(D) Images of olrn-1(ums9) mutants and olrn-1(ums9);tir-1(qd4) double mutants are shown. Red pharyngeal expression is the Pmyo-2::mCherry co-injection marker, which confirms the presence of the Pirg-4::GFP transgene. Scale bar is 100 μm.
(E and F) qRT-PCR data show irg-4 (E) and irg-5 (F) expression in the indicated genotypes. Data are the average of six independent replicates, each normalized to a control gene, with error bars representing SEM. Data are presented as the value relative to the average expression from all replicates of the indicated gene in wild-type animals. *p < 0.05 by one-way ANOVA for the indicated comparison. ns denotes that the difference between the indicated comparisons was not statistically significant.
Consistent with these data, loss-of-function mutations in p38 MAPK PMK-1 pathway components pmk-1(km25) and tir-1(qd4) suppressed the pathogen-resistance phenotype of the olrn-1(ums9) mutants (Figure 3C). In addition, both tir-1(qd4) (Figure 3D) and sek-1(km4) (Figure 6A) suppressed the hyperactivation of Pirg-4::GFP in the intestinal epithelial cells of the olrn-1(ums9) mutant. Moreover, tir-1(qd4) and pmk-1(km25) mutations each suppressed the constitutive activation of the immune effectors irg-4 (Figure 3E) and irg-5 (Figure 3F) in the olrn-1(ums9) background.
Figure 6. Immune Effectors Regulated by Neuronal olrn-1 Are Dynamically Expressed during Nematode Development.
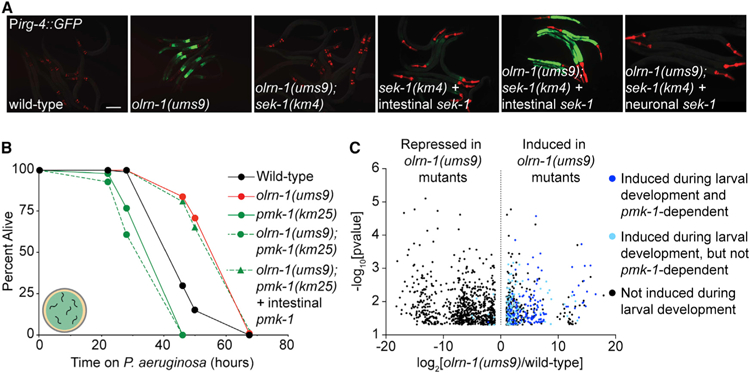
(A) Representative images of animals with the indicated genotypes carrying an integrated Pirg-4::GFP reporter. Red pharyngeal expression is the Pmyo-2::mCherry co-injection marker, which confirms the presence of the Pirg-4::GFP transgene. Bright red pharyngeal expression in C. elegans with intestinal sek-1 (Pges-1::sek-1::GFP) and neuronal sek-1 (Punc-119::sek-1::GFP) is the Pmyo-2:mStrawberry co-injection marker. The presence of the Pirg-4::GFP reporter was confirmed in these animals by Pmyo-2::mCherry expression in siblings that did not contain the indicated extrachromosomal arrays. Scale bar is 100 μm.
(B) C. elegans pathogenesis assay conducted with a large lawn of P. aeruginosa and C. elegans of indicated genotypes at L4 is shown. Data are representative of three trials. Sample sizes, mean lifespan, and p values for all trials are shown in Table S2. ‘‘Intestinal pmk-1’’ indicates that these animals have the Pvha-6::pmk-1 extrachromosomal array.
(C) Volcano plot of the mRNA-seq transcriptome profiling analysis shows all genes that were differentially expressed in olrn-1(ums9) mutants compared with wild-type animals (fold change > 2, p < 0.05), as described in Figure 2A. Highlighted in dark blue are the genes whose transcription are (1) dependent on the p38 MAPK pmk-1 (from the overlap in Figure 2D) and (2) induced in wild-type animals at L1, L2, L3, or L4 compared with wild-type young adult animals. Highlighted in light blue are the genes that are induced in L1, L2, L3, or L4 wild-type nematodes compared with adult animals but whose transcription does not depend on p38 MAPK pmk-1.
Venn diagrams showing the overlap of genes that are induced at each larval stage compared with genes that are induced in olrn-1(ums9) mutants are shown Figure S5A. See also Figure S5.
The bZIP transcription factor ZIP-2 and the G protein-coupled receptor FSHR-1 each function in parallel to the p38 MAPK PMK-1 pathway to promote host defense during an intestinal infection with P. aeruginosa (Estes et al., 2010; Powell et al., 2009; Reddy et al., 2016). Loss-of-function mutants of zip-2 and fshr-1 are hypersusceptible to killing by P. aeruginosa and are unable to upregulate a suite of PMK-1-independent immune effectors (Estes et al., 2010; Powell et al., 2009; Reddy et al., 2016). We examined the genes that are transcriptionally upregulated in the olrn-1(ums9) mutants to determine whether olrn-1 also suppresses the ZIP-2 or the FSHR-1 pathway. However, the overlap of zip-2-regulated genes (Figure S2A) or fshr-1-regulated genes (Figure S2B) with the genes that were upregulated in the olrn-1(ums9) mutant was not significant. In addition, we found that targets of the FOXO transcription factor DAF-16 were not significantly over-represented among olrn-1-regulated genes (Figure S2C).
Altogether, these data demonstrate that OLRN-1 targets the p38 MAPK PMK-1 immune pathway to suppress immune effector expression in the intestine and modulate host susceptibility to bacterial infection.
Promotion of Intestinal Immune Homeostasis by olrn-1 Is Required to Ensure Reproduction and Development
We previously observed that aberrant activation of immune defenses in the intestine by a gain-of-function mutation in the p38 MAPKKK nsy-1 (nsy-1(ums8)) or by exogenous treatment with an immunostimulatory small molecule slows nematode development (Cheesman et al., 2016). Like nsy-1(ums8) mutants, olrn-1(ums9) mutants take a longer time to reach adulthood than wild-type animals (Figures 4A and 4B). Reintroduction of olrn-1 under the control of its own promoter restored wild-type developmental rates to the olrn-1(ums9) mutant (Figures 4A and 4B).
Figure 4. Promotion of Intestinal Immune Homeostasis by olrn-1 Is Required to Ensure Reproduction and Development.
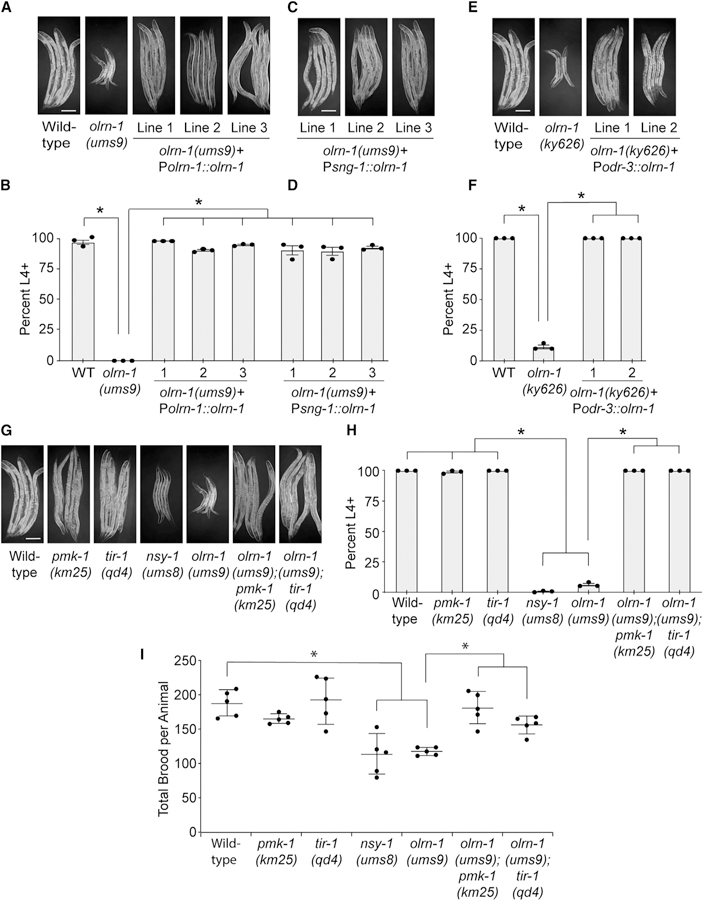
(A–H) Development assays were performed with the indicated genotypes. The stage of the animals was recorded at the same time point, approximately 72 h after eggs from C. elegans of the indicated genotypes were laid. In (B), (D), (F), and (H), data are presented as the average number of animals for each genotype that were at L4 or older (percent L4+) from three independent replicates, with error bars representing SEM. *p < 0.05 by one-way ANOVA. The stage that was recorded for each animal in these assays and the sample size for all replicates are presented in Table S2. The animals in (A), (C), (E), and (G) were from the same trial and were photographed together. The same photographs of the control genotypes [wild-type and olrn-1(ums9)] are shown in (A), (E), and (G) to make interpretation of the figure easier.
(I) Brood sizes from animals of the indicated genotypes were quantified. Each data point is the average brood size from two animals. *p < 0.05 by one-way ANOVA for the indicated comparison. The data for each replicate are presented in Table S2.
Scale bars in (A), (C), (E), and (G) are 100 μm. See also Figure S3.
To determine whether the developmental delay in the olrn-1(ums9) mutant is a consequence of de-repression of the p38 MAPK PMK-1 pathway, we compared the developmental rates of the pmk-1(km25), tir-1(qd4), atf-7(qd22 qd130), and olrn-1(ums9) single mutants to the olrn-1(ums9);pmk-1(km25), the olrn-1(ums9);tir-1(qd4), and the olrn-1(ums9);atf-7(qd22 qd130) double mutants (Figures 4G, 4H, and S3). ATF-7 is the transcription factor that functions downstream of p38 MAPK PMK-1 to control the basal expression of immune effectors (Shivers et al., 2010). The pmk-1(km25), tir-1(qd4), and atf-7(qd22 qd130) null mutants each fully suppressed the developmental delay of the olrn-1(ums9) mutant (Figures 4G, 4H, and S3). In addition, the brood sizes of both nsy-1(ums8) gain-of-function mutants and olrn-1(ums9) mutants are significantly smaller than those of wild-type animals (Figure 4I). Consistent with our observations in the C. elegans development experiments, both pmk-1(km25) and tir-1(qd4) mutations fully suppressed the small brood sizes of olrn-1(ums9) mutants (Figure 4I). Thus, neuronal olrn-1 prevents the deleterious effects of aberrant immune activation on nematode development and reproductive fitness.
Expression of olrn-1 in Chemosensory Neurons Is Sufficient to Regulate Innate Immunity in the Intestinal Epithelium
OLRN-1 is expressed in AWC chemosensory neurons, where it acts cell autonomously to promote olfactory receptor expression during nematode development (Bauer Huang et al., 2007). However, OLRN-1 is not expressed in the intestinal epithelium (Bauer Huang et al., 2007), where the p38 MAPK PMK-1 pathway coordinates the tissue autonomous expression of immune effectors and resistance to pathogen infection (Shivers et al., 2009). Therefore, we hypothesized that OLRN-1 acts in sensory neurons to control p38 MAPK PMK-1 transcriptional responses in the intestine.
We introduced an extrachromosomal array containing olrn-1 under the control of a pan-neuronal promoter (Psng-1::olrn-1) into the olrn-1(ums9) mutant. Neuronal expression of olrn-1 in three independent lines rescued the constitutive expression of Pirg-4::GFP in the intestine of olrn-1(ums9) mutants (Figure 5A). We confirmed that neuronal expression of olrn-1 was sufficient to suppress the constitutive activation of endogenous irg-4 (Figure 5B) and irg-5 (Figure 5C) in the olrn-1(ums9) mutant. Consistent with these gene expression data, neuronal expression of olrn-1 in three independent lines suppressed the pathogen-resistance phenotype of the olrn-1(ums9) mutant (Figure 5D).
Figure 5. Expression of olrn-1 in Chemosensory Neurons Is Sufficient to Regulate Innate Immunity in the Intestinal Epithelium.
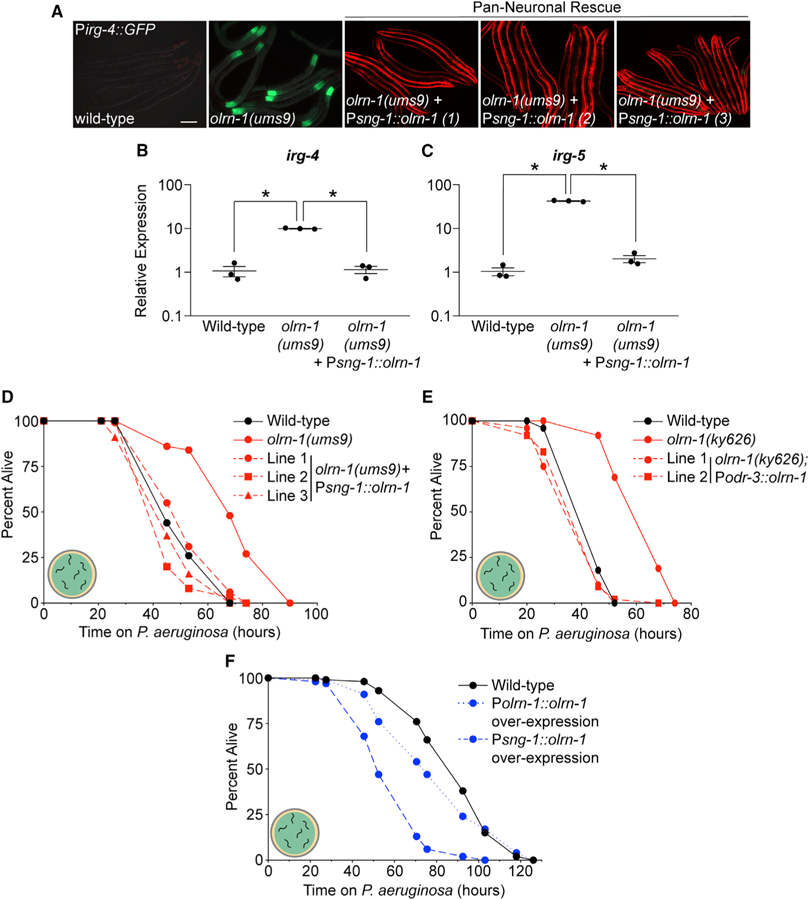
(A) Three independent lines of olrn-1 under the control of a pan-neuronal promoter (sng-1) in the olrn-1(ums9) mutant, along with the olrn-1(ums9) mutant, are shown. Pmyo-3::mCherry expression indicates the presence of an extrachromosomal array that contains the Psng-1::olrn-1 construct. Scale bar is 100 μm.
(B and C) qRT-PCR data show irg-4 (B) and irg-5 (C) expression in animals of the indicated genotypes. Data are the average of three independent replicates, each normalized to a control gene, with error bars representing SEM. Data are presented as the value relative to the average expression from all replicates of the indicated gene in wild-type animals. *p < 0.05 by one-way ANOVA for the indicated comparison.
(D–F) C. elegans pathogenesis assay conducted with a large lawn of P. aeruginosa and C. elegans of indicated genotypes at L4 is shown. Data are representative of three trials.
Sample sizes, mean lifespan, and p values for all trials are shown in Table S2. See also Figure S4.
We also expressed olrn-1 in the olrn-1(ums9) mutant under the odr-3 promoter (Podr-3::olrn-1), which drives gene expression in a specific subset of chemosensory neurons (Roayaie et al., 1998). A Podr-3::GFP transcription reporter is expressed strongly in AWC neurons and weakly or inconsistently in AWB, AWA, ASH, and ADF neurons (Roayaie et al., 1998). Historically, the odr-3 promoter has been a useful tool to characterize AWC-dependent mechanisms (Bauer Huang et al., 2007). As observed in our experiments with the Psng-1::olrn-1 constructs (Figure 5D), expression of olrn-1 under the odr-3 promoter fully suppressed the pathogen-resistance phenotype of the olrn-1(ky626) mutant (Figure 5E). Importantly, this experiment, which uses a second heterologous promoter to direct olrn-1 expression in neurons, confirms that olrn-1 activity in neurons is necessary to modulate resistance to a bacterial pathogen. Multi-copy expression of olrn-1 in a wild-type background, both under its own promoter and specifically in neurons, rendered C. elegans more susceptible to killing by P. aeruginosa (Figure 5F).
In addition, expression of olrn-1 only in neurons (under the sng-1 promoter) (Figures 4C and 4D) or in chemosensory neurons (using the odr-3 promoter) (Figures 4E and 4F) was sufficient to rescue the developmental delay of olrn-1 mutants, as assessed in multiple independent lines carrying these rescue constructs.
In summary, neuronal olrn-1 is necessary and sufficient to control pathogen resistance and promote intestinal immune homeostasis. In addition, we show that expression of olrn-1 in chemosensory neurons is sufficient to regulate innate immunity in the C. elegans intestinal epithelium.
Immune Effectors Regulated by Neuronal olrn-1 Are Dynamically Expressed during Nematode Development
During C. elegans development, olrn-1 suppresses the TIR-1/ NSY-1/SEK-1 cassette in AWC neurons, which uses either PMK-1 or PMK-2 p38 MAPK to promote left-right asymmetry of the odorant receptors str-2 and srsx-3 in AWC neurons (Bauer Huang et al., 2007; Chuang and Bargmann, 2005; Pagano et al., 2015; Torayama et al., 2007; Troemel et al., 1999). Differentiation of odorant receptors in AWC neurons is a required developmental step for C. elegans to sense and move toward diverse attractive stimuli (Wes and Bargmann, 2001). Adult C. elegans have one AWC neuron that expresses str-2, called AWCON by convention, and one AWC neuron that expresses the srsx-3 chemoreceptor instead (AWCOFF). Forward genetic screens for mutants with two AWCON or two AWCOFF neurons defined the genetic pathway that controls olfactory receptor development in AWC neurons. This work revealed that low olrn-1 activity causes activation (de-repression) of the TIR-1/NSY-1/SEK-1/(PMK-1 or PMK-2) signaling cassette in AWCOFF neurons (Bauer Huang et al., 2007; Troemel et al., 1997, 1999).
Given that the TIR-1/NSY-1/SEK-1/PMK-1 pathway is required both in neurons for the development of odorant receptors and in the intestine for innate immunity, we determined the tissues in which this signaling cassette is sufficient for the promotion of immune homeostasis by neuronal olrn-1. A Pges-1::sek-1::GFP construct, which directs sek-1 expression in intestinal epithelial cells (Shivers et al., 2009), was introduced into the olrn-1(ums9);sek-1(km4) double mutant. Reconstitution of the p38 MAPKK sek-1 in the intestine restored Pirg-4::GFP expression in the olrn-1(ums9);sek-1(km4) double mutant (Figure 6A). To confirm our results with the Pges-1::sek-1::GFP construct using a different heterologous promoter, we introduced a Pvha-6::pmk-1 construct (Bolz et al., 2010) into the olrn-1(ums9);pmk-1(km25) double mutant. Like Pges-1, Pvha-6 directs gene expression specifically in intestinal epithelial cells (McGhee et al., 1990; Oka et al., 2001). Consistent with our observations with the Pges-1::sek-1 construct (Figure 6A), intestinal expression of pmk-1 was sufficient to restore the pathogen resistance phenotype of the olrn-1(ums9) mutant to the olrn-1(ums9);pmk-1(km25) double mutant (Figure 6B). However, expression of sek-1 under the control of a neuronal-specific promoter (Punc-119::sek-1) (Shivers et al., 2009) in the olrn-1(ums9);sek-1(km4) double mutant did not restore constitutive activation of Pirg-4::GFP in the olrn-1(ums9) mutant background (Figure 6A). Thus, the p38 MAPK PMK-1 pathway in intestinal epithelial cells, but not in neurons, is sufficient for the modulation of immune effector expression and resistance to P. aeruginosa infection by neuronal olrn-1.
The function of the innexin gene nsy-5 and the claudin/calcium channel γ subunit gene nsy-4 in promoting AWC neuron differentiation, as defined in genetic studies, is similar to that of olrn-1 (Bauer Huang et al., 2007; Chuang et al., 2007; Vanhoven et al., 2006). Like olrn-1 mutants, both nsy-4(ky616) and nsy-5(ky634) loss-of-function mutants have two AWCOFF neurons, one of which never differentiates into an AWCON neuron (Bauer Huang et al., 2007). Interestingly, the immune effectors irg-4 (Figure S4A) and irg-5 (Figure S4B) are not constitutively induced in nsy-4(ky616) and nsy-5(ky634) loss-of-function mutants as they are in the olrn-1 mutants. These data indicate that olrn-1 has distinct roles in regulating immune effector expression in the intestine and promoting olfactory receptor development in AWC neurons.
Considering the role of olrn-1 in controlling olfactory receptor development in AWC neurons, we asked whether olrn-1-dependent immune effectors are differentially expressed during larval development. We compared the RNA-seq transcriptome profiles of wild-type C. elegans at each larval stage and identified 707, 580, 717, 602, and 100 genes that were expressed at significantly higher levels in wild-type animals at the first larval stage (L1), the second larval stage (L2), the third larval stage (L3), the fourth larval stage (L4), and the young adult stage (YA) compared with adult animals, respectively (p < 0.05, greater than 2-fold change). Genes that are expressed at higher levels in olrn-1(ums9) mutants are significantly enriched among the genes that are developmentally regulated in wild-type animals, including a significant number of the genes identified in Figure 2D that are targets of the olrn-1-p38 MAPK pmk-1 signaling axis (Figures 6C and S5A). In particular, innate immune genes were enriched among the genes expressed higher in L3 animals compared with adults (p value for GO term enrichment 4.8 × 10−5), as were genes induced during P. aeruginosa infection (hypergeometric p value 7.9e–10). In addition, we identified 100 genes that were expressed higher in young adult compared with adult animals, but neither olrn-1-regulated genes nor immune effectors in general were enriched in this dataset (Figure S5A). Using qRT-PCR, we confirmed that the olrn-1-dependent immune effectors irg-4 (Figure S5B) and irg-5 (Figure S5C) were expressed at higher levels in animals at L2/L3 compared with young adult animals. Low activity of OLRN-1 leads to de-repression of the p38 MAPK pathway in AWC neurons (Bauer Huang et al., 2007). Altogether, the data in this manuscript suggest that low levels of olrn-1, as recapitulated in multiple loss-of-function mutant alleles, de-repress the p38 MAPK PMK-1 pathway in intestine to promote pathogen resistance during C. elegans development.
DISCUSSION
This study demonstrates that innate immunity in C. elegans intestinal epithelial cells and the development of AWC neurons are linked by a single neuronal protein. Neuronal olrn-1 functions cell autonomously in AWC neurons to ensure olfactory receptor expression and cell non-autonomously to suppress the p38 MAPK PMK-1 immune pathway in the intestine. We propose that low activity of neuronal OLRN-1 de-represses the p38 MAPK PMK-1 pathway to prime the immune response in the intestine to handle challenges from bacterial pathogens encountered during larval development.
The decision of a nematode to move toward nutritious bacteria and away from potential pathogens involves the integration of multiple inputs from different sensory neurons, including the chemosensation of pathogen-derived metabolites, recognizing attractive odors from food sources, monitoring oxygen availability, and sensing mechanical stimuli (Chang et al., 2011; Ha et al., 2010; Meisel et al., 2014; Pradel et al., 2007; Reddy et al., 2009; Zhang et al., 2005). AWC neurons detect attractive odorants and induce chemotaxis toward the stimuli (Bargmann et al., 1993; Troemel et al., 1997), but they do not control aversion responses to bacterial pathogens (Chang et al., 2011; Meisel et al., 2014; Zhang et al., 2005). As such, we did not observe defects in the ability of olrn-1 mutants to avoid P. aeruginosa. It is intriguing that immune control in the intestine is linked to the development of neurons that promote chemotaxis. These data may speak to the importance of detecting attractive odorants from food sources for the ability of nematodes to develop in microbe-rich habitats.
We found that olrn-1 is required to suppress the p38 MAPK PMK-1 pathway to prevent the deleterious effects of immune hyperactivation on C. elegans development and fecundity. These data support previous studies, which found that activation of the p38 MAPK PMK-1 pathway constitutes a source of physiological stress for C. elegans. For example, induction of immune effector expression during pathogen infection or by treatment with an immunostimulatory small molecule causes endoplasmic reticulum stress, which requires compensatory activation of the unfolded protein response (UPR) (Peterson et al., 2019; Richardson et al., 2010). In addition, we previously found that hyperactivation of immune defenses in the intestine by a gain-of-function mutation in the p38 MAPKKK nsy-1 (nsy-1(ums8)) or by exogenous treatment with an immunostimulatory small molecule dramatically slowed nematode development in a manner dependent on the p38 MAPK PMK-1 pathway (Cheesman et al., 2016). Third, control of the p38 MAPK PMK-1 innate immune pathway by the C/EBP transcription factor CEBP-1 and the Tribbles kinase NIPI-3 is essential for development (Kim et al., 2016). Finally, other mechanisms that globally suppress p38 MAPK PMK-1 activity promote reproduction (Amrit et al., 2019). Altogether, these studies emphasize the physiological importance of mechanisms that control the activity of the p38 MAPK PMK-1 pathway and demonstrate that immune homeostasis is required for evolutionary fitness.
Two other neuronal mechanisms have been shown to suppress intestinal epithelial immune responses in C. elegans. Inhibition of dopamine signaling between CEP and ASG neurons suppresses immune activation in the nematode intestine (Cao and Aballay, 2016). In addition, the GPCR (G protein-coupled receptor) OCTR-1 functions in ASH and ASI neurons to negatively regulate a noncanonical branch of the UPR (Sun et al., 2011); downregulate protein synthesis (Liu et al., 2016); modulate the activity of multiple parallel stress response pathways, including pmk-1, daf-16, and ced-1 (Cao et al., 2017; Sellegounder et al., 2018; Sun et al., 2011); and affect pathogen-avoidance behavior (Cao et al., 2017). A mutation in the neuropeptide gene nlp-20 suppressed the immune effector induction and pathogen resistance phenotypes of octr-1 loss-of-function mutants, suggesting that this neuropeptide may signal to the intestine to suppress innate immunity (Cao et al., 2017). We do not know how OLRN-1 and AWC neurons communicate with the intestine to regulate the p38 MAPK PMK-1 innate immune pathway. Because sensory neurons do not directly synapse with intestinal epithelial cells in C. elegans, we hypothesize that neurons communicate with the intestine via a relayed signal, such as a neuropeptide or another mechanism. In any case, immune control by CEP, ASG, ASH, and ASI neurons in C. elegans is not known to be integrated with the development of olfactory receptors in neurons, as we found is the case for the activity of OLRN-1 in AWC neurons.
Of note, OLRN-1 contains two putative transmembrane domains and is related to Drosophila Raw. In flies, Raw inhibits JNK MAPK signaling during development to promote dorsal closure of the fly embryo (Bauer Huang et al., 2007; Byars et al., 1999). The inhibitory effect of OLRN-1 and a related Drosophila protein on MAPK signaling suggests that a common mechanism may be employed by these proteins to suppress signal transduction.
The human intestine is densely innervated, and it is becoming increasingly appreciated that the enteric nervous system plays a key role in the maintenance of immune homeostasis (Tracey, 2010, 2014). However, disentangling the contribution of individual sensory neurons in the regulation of intestinal inflammation has been challenging. In this regard, C. elegans has emerged as a useful system to define the wiring of neural-immune connections and to characterize the mechanisms that integrate immune control in the intestine with the physiology of the organism as a whole (Labed et al., 2018; Reddy et al., 2009; Styer et al., 2008). Here, we show that a single neuronal gene links regulation of innate immunity with the development of olfactory neuron receptors in C. elegans. Our findings suggest that integration of neuronal development programs with control of host immune defenses allows survival of animals that develop in microbe-rich environments.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All requests for reagents and further information regarding the protocols in this manuscript may be directed to the Lead Contact, Read Pukkila-Worley (Read.Pukkila-Worley@umassmed.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
C. elegans strains were maintained on standard nematode growth media (NGM) plates with E. coli OP50 as a food source, as described (Brenner, 1974). Mutant C. elegans and transgenic strains used in this study are presented in the Key Resources Table. Pseudomonas aeruginosa strain PA14 was used for all studies (Rahme et al., 1995).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Total PMK-1 Antibody | Peterson et al., 2019 | N/A |
| Anti-p38 MAPK, phospho (Thr180 / Tyr182) Antibody, Unconjugated | Cell Signaling Technology | Cat# 9211 |
| RRID:AB_331641 | ||
| Lot# 25 | ||
| Monoclonal Anti-alpha-Tubulin antibody produced in mouse | Sigma-Aldrich | Cat# T5168 |
| RRID:AB_477579 | ||
| Lot# 038M4813V | ||
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat# 7074 |
| RRID:AB_2099233 | ||
| Lot# 28 | ||
| Goat Anti-Mouse IgG - H&L Polyclonal Antibody, HRP Conjugated | Abcam | Cat# ab6789 |
| RRID:AB_955439 | ||
| Lot# GR3216437–2 | ||
|
Bacterial and Virus Strains | ||
| Escherichia coli OP50 | Brenner, 1974 | RRID:WB-STRAIN:OP50 |
| Pseudomonas aeruginosa (UCBPP-PA14) | Rahme et al., 1995 | RRID:WB-STRAIN:PA14 |
|
Chemicals, Peptides, and Recombinant Proteins | ||
| 5-Fluoro-2-deoxyuridine | Sigma-Aldrich | Cat# CAF0503 |
| (—) Tetramisole hydrochloride | Sigma-Aldrich | Cat# L9756–10G |
| EcoRI Restriction Enzyme | New England BioLabs | Cat# R0101 |
| Q5 High-Fidelity DNA Polymerase | New England BioLabs | Cat# M0491S |
| Isopropyl-β-D-thiogalactoside | Research Products International | CAT# L56000 |
| Trizol | Thermo Fisher Scientific | Cat# 15596018 |
| Proteinase K | New England BioLabs | Cat# P8107S |
| Phenol:chloroform:isoamyl alcohol | Thermo Fisher | Cat# 15593031 |
|
Critical Commercial Assays | ||
| Gibson Assembly Cloning Kit | New England BioLabs | Cat# E5510S |
| iScript gDNA Clear cDNA Synthesis Kit | Bio-Rad | Cat# 172–5034 |
| iTaq Universal SYBR Green Supermix | Bio-Rad | Cat# 1725120 |
|
Deposited Data | ||
| Raw and analyzed mRNA-Seq data | This study | GEO:GSE130959 |
|
Experimental Models: Organisms/Strains | ||
| C. elegans: Strain: N2 (Bristol) | Caenorhabditis Genetics Center | WB Cat# WBStrain00000001; RRID:WB-STRAIN:WBStrain00000001 |
| C. elegans: Strain: KU25 pmk-1(km25) | Kim et al., 2002 | WB Cat# WBStrain00024040; RRID:WB-STRAIN:WBStrain00024040 |
|
C. elegans: Strain: AU306 agIs44 [Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
Pukkila-Worley et al., 2014 | N/A |
|
C. elegans: Strain: AY101 acIs101 [PDB09.1(Pirg-5::GFP); pRF4(rol-6(su1006))] |
Bolz et al., 2010 | WB Cat# WBStrain00000322; RRID:WB-STRAIN:WBStrain00000322 |
|
C. elegans: Strain: RPW40 olrn-1(ums9); agIs44 [Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW31 olrn-1(ums11); agIs44 [Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW222 olrn-1(ky626); agIs44 [Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW211 olrn-1(ums9); acIs101 [PDB09.1(Pirg-5::GFP); pRF4(rol-6(su1006))] |
This study | N/A |
| C. elegans: Strain: ZD101 tir-1(qd4) | Shivers et al., 2009 | WB Cat# WBStrain00040806; RRID:WB-STRAIN:WBStrain00040806 |
|
C. elegans: Strain: RPW43 nsy-1(ums8); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
Cheesman et al., 2016 | WormBase ID: WBVar02145386 |
| C. elegans: Strain: RPW 129 olrn-1(ums9);pmk-1(km25) | This study | N/A |
|
C. elegans: Strain: RPW189 olrn-1(ums9);tir-1(qd4); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW131 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry]; umsEx4 [Porln-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW133 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry];umsEx6 [Porln-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW134 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry];umsEx7 [Porln-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW159 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry];umsEx20 [Psng-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW160 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry];umsEx21 [Psng-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: RPW162 orln-1(ums9); agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry];umsEx23 [Psng-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry] |
This study | N/A |
|
C. elegans: Strain: CX7517 olrn-1(ky626); kyEx914 [Podr-3::olrn-1;Podr-1::mCherry;coel::GFP] |
Bauer Huang et al., 2007 | Laboratory of Cornelia Bargmann: Strain ID CX7517 |
|
C. elegans: Strain: CX7521 olrn-1(ky626); kyEx918 [Podr-3::olrn-1;Podr-1::mCherry;coel::GFP] |
Bauer Huang et al., 2007 | Laboratory of Cornelia Bargmann: Strain ID CX7521 |
| C. elegans: Strain: olrn-1(ky626) | Bauer Huang et al., 2007 | WormBase ID: WBVar00088455 |
|
C. elegans: Strain: RPW207 agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry]; umsEx4 (Porln-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry) |
This study | N/A |
|
C. elegans: Strain: RPW209 agIs44[Pirg-4::GFP::unc-54–3′UTR; Pmyo-2::mCherry]; umsEx21 (Psng-1::orln-1;3′UTRorln-1; Pmyo-3::mCherry) |
This study | N/A |
|
C. elegans: Strain: RPW216 atf-7(qd22 qd130); agIs219[T24B8.5::GFP]; olrn-1(ums9) |
This study | N/A |
|
C. elegans: Strain: RPW218 orln-1 (ums9); pmk-1(km25); acEx102 [vha-6p::pmk-1::GFP + rol-6(su1006)] |
This study | N/A |
|
C. elegans: Strain: AY102 pmk-1(km25); acEx102 [vha-6p::pmk-1::GFP + rol-6(su1006)] |
Bolz et al., 2010 | WB Cat# WBStrain00000323; RRID:WB-STRAIN:WBStrain000003233 |
|
C. elegans: Strain: ZD193 sek-1(km4);qdEx4 [Pges-1::sek-1(cDNA)::GFP::unc-54-3′ UTR + myo-2p::mStrawberry::unc-54-3′ UTR] |
Shivers et al., 2009 | WB Cat# WBStrain00040807; RRID:WB-STRAIN:WBStrain00040807 |
|
C. elegans: Strain: ZD202 sek-1(km4);qdEx8 [Punc-119::sek-1(cDNA)::GFP::unc-54-3′ UTR + myo-2p::mStrawberry::unc-54-3′ UTR] |
Shivers et al., 2009 | WB Cat# WBStrain00040808; RRID:WB-STRAIN:WBStrain00040808 |
| C. elegans: Strain: RPW220 olrn-1(ums9); sek-1(km4); agIs44 | This study | N/A |
| C. elegans: Strain: RPW227 olrn-1(ums9); sek-1(km4); agIs44; qdEx4 | This study | N/A |
| C. elegans: Strain: RPW224 olrn-1(ums9); sek-1(km4); agIs44; qdEx8 | This study | N/A |
| C. elegans: Strain: CX5757 kyIs140 I; nsy-4(ky616) IV. |
Caenorhabditis Genetics Center |
WB Cat# WBStrain00005274; RRID:WB-STRAIN:WBStrain00005274 |
| C. elegans: Strain: (CX6161) inx-19(ky634) I. |
Caenorhabditis Genetics Center |
WB Cat# WBStrain00005278; RRID:WB-STRAIN:WBStrain00005278 |
|
Oligonucleotides | ||
| olrn-1 Rescue PCR FWD primer: CAGAACCAGATTCTCGGAATGA | IDT | N/A |
| olrn-1 Rescue PCR REV primer: AGAGGAAGAGAGACAGGATGAA | IDT | N/A |
| Gibson Assembly Primer for Psng-1::olrn-1b construct: CCCCCCCTCGAGGTCGACGGTATCGATAAGCTTGATATCGttgagcagcgactaacaaaa |
IDT | N/A |
| Gibson Assembly Primer for Psng-1::olrn-1b construct: ACCTGACACTAATTTCTCTTGGCGCTGAACATCTAGTCATgctaaaataaaagaaatata |
IDT | N/A |
| Gibson Assembly Primer for Psng-1::olrn-1b construct: atgactagatgttcagcgcc |
IDT | N/A |
| Gibson Assembly Primer for Psng-1::olrn-1b construct: GGCGGCCGCTCTAGAACTAGTGGATCCCCCGGGCTGCAGGtttcatatatcttatgccgt |
IDT | N/A |
|
Recombinant DNA | ||
| Plasmid: pBluescript SK(−) | Addgene | N/A |
|
Software and Algorithms | ||
| GraphPad Prism 8 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| OASIS 2 | Han et al., 2016 | https://sbi.postech.ac.kr/oasis2/ |
| Adobe Creative Cloud | Adobe | https://www.adobe.com/creativecloud.html |
| ApE: A plasmid Editor | M. Wayne Davis | http://jorgensen.biology.utah.edu/wayned/ape/ |
| Microsoft Excel | Microsoft | https://office.live.com/start/Excel.aspx |
| FastQC (Version 0.11.8) | Wingett and Andrews, 2018 | https://www.bioinformatics.babraham.ac.uk/projects/download.html |
| Multiqc (Version 1.7) | Ewels et al., 2016 | https://github.com/ewels/MultiQC |
| Trimmomatic (Version 0.36) | Bolger et al., 2014 | http://www.usadellab.org/cms/?page=trimmomatic |
| HISAT2 (Version 2.1.0) | Kim et al., 2015; Pertea et al., 2016 | https://ccb.jhu.edu/software/hisat2/index.shtml |
| Samtools (Version 1.3.1) | Li, 2011; Li et al., 2009 | https://sourceforge.net/projects/samtools/files/samtools/1.3.1/ |
| Stringtie (Version 1.3.4) | Pertea et al., 2015, 2016 | https://ccb.jhu.edu/software/stringtie/ |
| Ballgown (Version 3.8) | Pertea et al., 2016 | https://www.bioconductor.org/packages/release/bioc/html/ballgown.html |
| R Console (Version 3.5) | The R Foundation | https://www.r-project.org/ |
METHOD DETAILS
Forward genetic screen
Ethyl methanesulfonate (EMS) mutagenesis was performed on strain agIs44 as previously described (Cheesman et al., 2016). Synchronized F2 progeny representing approximately 40,000 haploid genomes were screened for animals that constitutively express agIs44 GFP fluorescence. Three alleles were identified (ums9, ums10 and ums11). To identify the causative mutations, pooled genomes from 52 recombinants for ums9 and 3 recombinants for ums11 following a 2X outcross to N2 were sequenced along with the agIs44 parent strain. All recombinants constitutively expressed agIs44 GFP. Homozygous variants from WS220 (ce10) C. elegans reference genome that were present in the 2X outcrossed samples, but not in agIs44, were identified using Cloud Map (Minevich et al., 2012). We were unable to identify the causative mutation in ums10.
C. elegans Bacterial Infection and Other Assays
‘‘Slow killing’’ P. aeruginosa infection experiments were performed as previously described (Foster et al., 2020; Tan et al., 1999). The wild-type control for these assays is agIs44. In brief, a single colony of P. aeruginosa PA14 was inoculated into 5 mL of Luria-Bertani (LB) medium, and allowed to incubate at 37° for 15 hr. 10 μL of this culture was spread onto 35-mm tissue culture plates containing 4 mL of slow kill agar (0.35% peptone, 0.3% sodium chloride, 1.7% agar, 5 mg/ml cholesterol, 25 mM potassium phosphate, 1 mM magnesium sulfate, 1 mM calcium chloride). Plates were incubated for 24 hours at 37°C, and approximately 24 hours at 25°C. Approximately one hour before the start of the assay, 0.1 mg/ml 5-fluorodeoxyuridine (FUDR) was added to the medium to prevent progeny from hatching. For all pathogenesis assays that studied C. elegans with extrachromosomal arrays, control genotypes, which did not express the array, were obtained from siblings isolated from the same plates as nematodes that contained the array. C. elegans lifespan assays were conducted with animals grown on nematode growth media agar at 20°C in the presence of 40 μg/mL 5-fluoro-2′-deoxyuridine. All pathogenesis and lifespan assays were conducted with nematodes at the L4 larval stage. To obtain stage-matched animals at the L4 larval stage for the pathogenesis and lifespan assays, olrn-1 mutant animals at the L1 larval stage were added to growth plates (NGM with E. coli OP50) approximately 24 hours before wild-type L1 larval stage animals were added to growth plates. Three trials of each pathogenesis assay were performed. Sample sizes, mean lifespan, and p values for all trials are shown in Table S2.
The propensity of C. elegans to avoid a small lawn (10 μL) of P. aeruginosa was determined by counting the number of C. elegans on or off the lawn at 4, 8, 16, 24 and 30 hours after synchronized L4 were placed on the bacteria (Foster et al., 2020). C. elegans development assays were performed as previously described (Cheesman et al., 2016; Foster et al., 2020). Brood sizes were quantified from five independent plates, each with two animals per plate. Animals were transferred to new plates each day to facilitate scoring of the progeny. Data for all replicates of the development and brood size assays are shown in Table S2.
Colony forming units of P. aeruginosa were quantified in the intestine of C. elegans as previously described with some modifications (Foster et al., 2020; Singh and Aballay, 2019). Briefly, C. elegans animals were exposed to P. aeruginosa for 24 hours. Animals were then picked to NGM plates lacking bacteria and incubated for 10 minutes to remove external P. aeruginosa. Animals were then transferred to a second NGM plate after which 10–11 animals per replicate were collected, washed with M9 buffer containing 25 mM levamisole and 0.01% Triton X-100, and ground with 1.0 mm silicon carbide beads. CFUs were quantified from serial dilutions of the lysate.
Generation of transgenic C. elegans strains
To generate olrn-1 rescue lines, primers 5′-CAG AAC CAG ATT CTC GGA ATG A-3′ and 5′-AGA GGA AGA GAG ACA GGA TGA A-3′ were used to amplify the entire olrn-1 locus. The resulting PCR product (30 ng/μl), the Pmyo-3::mCherry co-injection marker (15 ng/μl) and pBluescript SK (−) vector (155 ng/μl) were microinjected into olrn-1(ums9);agIs44 animals to generate the arrays umsEx4, umsEx6 and umsEx7.
The olrn-1 neuron-specific rescue construct was generated using Gibson assembly to fuse 2kb of the sng-1 promoter (amplified using primers 5′-CCC CCC CTC GAG GTC GAC GGT ATC GAT AAG CTT GAT ATC GTT GAG CAG CGA CTA ACA AAA-3′ and 5′-ACC TGA CAC TAA TTT CTC TTG GCG CTG AAC ATC TAG TCA TGC TAA AAT AAA AGA AAT ATA-3′) with 2960 bp of olrn-1b coding region + 667bp 3′ UTR (amplified using primers 5′-ATGACTAGATGTTCAGCGCC-3′ and 5′-GGC GGC CGC TCT AGA ACT AGT GGA TCC CCC GGG CTG CAG GTT TCA TAT ATC TTA TGC CGT —3′) in pBluescript vector linearized with EcoR1 (Gibson et al., 2009). The plasmid (30 ng/μl), the Pmyo-3::mCherry co-injection marker (15 ng/μl) and pBluescript SK (−) vector (155 ng/μl) were microinjected into olrn-1(ums9);agIs44 animals to generate the arrays umsEx20, umsEx21 and umsEx23.
Gene expression analyses and bioinformatics
For the RNA-seq experiment of olrn-1(ums9) and wild-type animals, synchronized L1 stage C. elegans were grown to the L4/young adult stage. RNA was isolated using TriReagent (Sigma-Aldrich), purified on a column (QIAGEN), and analyzed by mRNA-seq using the BGISEQ-500 platform (BGI Americas Corp). The quality of raw sequencing data was evaluated by FastQC (version 0.11.8) and Multiqc (version 1.7) (Ewels et al., 2016; Wingett and Andrews, 2018). Low-quality reads were trimmed using Trimmomatic (version 0.36) (Bolger et al., 2014). The trimmed reads were mapped to the C. elegans reference genome, WS220/ce11 [University of California Santa Cruz (UCSC) genome browser] using HISAT2 (version 2.1.0) (Kim et al., 2015; Pertea et al., 2016). The sequence alignment map (SAM) files were then converted to sorted BAM files using Samtools (Version 1.3.1) (Li, 2011; Li et al., 2009). The general transfer format (GTF) annotation file (WS220/ce11) was downloaded from the UCSC genome website, and the assembled GTF file was generated for each sample using Stringtie (version 1.3.4) (Pertea et al., 2015, 2016). Stringtie was then used to compare each sample against the merged assembly, estimate transcript abundance, and to generate a count table for Ballgown analysis (Pertea et al., 2016). The Ballgown package from the Bioconductor software suite (version 3.8) was used to run a custom R script in R console (R Version 3.5) to analyze the differential gene expression, visualize the data, and perform statistical tests for differential expression with multiple test correction. A gene was considered to be differentially regulated if its fold change versus wild-type was greater than two, the adjusted p value was less than 0.05, and its RPKM was greater than one.
To examine genes that are differentially expressed during C. elegans development, raw base-called fastq files were downloaded from the European Nucleotide Archive (accession number PRJEB31791). For each sample, reads were aligned to the WS220/ce11 on UCSC genome website using minimap2 (version 2.14-r883) (Li, 2018). Genomic alignments were run with the following parameters: -ax splice -k14 -uf -secondary = no -G 25000 -t 24. The resulting sam files were converted to bam format using samtools view with parameters: -b –F 2048 (Li et al., 2009). Read filtering and splice isoform identification were analyzed as described (Roach et al., 2019). The GTF (WS220/ce11) annotation file was downloaded from UCSC genome website and the assembled GTF file was generated for each sample using Stringtie (version 1.3.4). Stringtie was used to compare each sample against the merged assembly, to estimate transcript abundance, and to generate a count table for Ballgown analysis. The Ballgown package from the Bioconductor software suite (version 3.8) was used to run a custom R script in R console (R Version is 3.5) to analyze differential gene expression, visualize the data and perform statistical tests for differential expression with multiple test correction. Differential gene expression was defined as a fold change (FC) versus wild-type greater than 2, adjusted P value less than 0.05 and RPKM greater than one.
For the qRT-PCR studies, RNA was reverse transcribed to cDNA using the RETROscript Kit (Life Technologies) and analyzed using a CFX1000 machine (Bio-Rad) using previously published primers (Cheesman et al., 2016; Peterson et al., 2019; Troemel et al., 2006). All values were normalized against the control gene snb-1. Fold change was calculated using the Pfaffl method (Pfaffl, 2001). The analysis of irg-4 and irg-5 expression using nanoString was performed as previously described (Anderson et al., 2019; Cheesman et al., 2016; Pukkila-Worley et al., 2014). Counts from each gene in wild-type and olrn-1(ums9) animals were normalized to three control genes: snb-1, ama-1 and act-1.
Immunoblot Analyses
Protein lysates from stage-matched C. elegans grown to the young L4 larval stage on E. coli OP50 on NGM agar were prepared as previously described (Cheesman et al., 2016; Peterson et al., 2019). Harvested animals were washed twice with M9 buffer, incubated in a roller at room temperature for 15 minutes to allow the nematode intestine to clear of bacteria, washed an additional time and flash frozen in RIPA Buffer (Cell Signaling Technology, Inc.) using an ethanol and dry ice bath. Samples were lysed by sonication and centrifuged. Protein was quantified from the supernatant of each sample using Bradford Reagent (Bio-Rad Laboratories, Inc.). Laemmli buffer (Bio-Rad Laboratories, Inc.) was added to a concentration of 1X and the total protein from each sample was resolved on Nu-Page 4%–12% gels (Life Technologies), transferred to nitrocellulose membranes (Life Technologies), blocked with 5% BSA in TBST and probed with a 1:1000 dilution of an antibody that recognizes the doubly-phosphorylated TGY motif of PMK-1 (Cell Signaling Technology), a previously characterized total PMK-1 antibody (Peterson et al., 2019) or a monoclonal anti-tubulin antibody (Sigma-Aldrich, Clone B-5–1-2). Horseradish peroxidase (HRP)-conjugated anti-rabbit (Cell Signaling Technology) and anti-mouse IgG secondary antibodies (Abcam) were used at a dilution of 1:10,000 to detect the primary antibodies following the addition of ECL reagents (Thermo Fisher Scientific), which were visualized using a ChemiDoc MP Imaging System (BioRad). The band intensities were quantified using Image Lab software version 6.0.1 (BioRad), and the ratio of active phosphorylated PMK-1 to total PMK-1 was calculated with all samples normalized to the ratio of wild-type control animals.
Microscopy
Nematodes were mounted onto agar pads, paralyzed with 10 mM levamisole (Sigma) and photographed using a Zeiss AXIO Imager Z2 microscope with a Zeiss Axiocam 506 mono camera and Zen 2.3 (Zeiss) software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differences in survival of C. elegans in the P. aeruginosa pathogenesis assays were determined with the log-rank test after survival curves were estimated for each group with the Kaplan-Meier method. OASIS 2 was used for these statistical analyses (Han et al., 2016). qRT-PCR studies, lawn occupancy studies, intestinal CFU quantification, western blot band intensity quantification, and developmental assays are presented as the mean ± standard error. p values were calculated using one-way ANOVA in Prism 8 (GraphPad Software), unless otherwise indicated in the figure legend. Sample sizes, mean lifespan, and p values for all trials are shown in Table S2.
DATA AND CODE AVAILABILITY
The accession number for the mRNA-seq dataset reported in this paper is GEO:GSE130959. All other data are available in the manuscript.
Supplementary Material
Highlights.
The olfactory neuron gene olrn-1 regulates intestinal immunity in C. elegans
Neuronal olrn-1 suppresses the p38 MAPK PMK-1 immune pathway in the intestine
Promotion of immune homeostasis by olrn-1 ensures evolutionary fitness
olrn-1-dependent immune effectors are dynamically regulated during development
ACKNOWLEDGMENTS
The authors thank Cornelia Bargmann for providing nematode strains and acknowledge Cole Haynes, Robert Dowen, and Melanie Trombly for helpful discussions and critical reading of the manuscript. This research was supported by R01 AI130289 (to R.P.W.), an Innovator Award from the Kenneth Rainin Foundation (to R.P.W.), a Child Health Research Award from the Charles H. Hood Foundation (to R.P.W.), T32 AI095213 (to K.J.F.), and T32 AI132152 (to N.D.P.). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.03.042.
DECLARATION OF INTERESTS
R.P.W. has submitted a patent application related to this work (U.S. patent application, 16/069,399, filed July 11, 2018). The authors declare no other potential competing interests.
REFERENCES
- Amrit FRG, Naim N, Ratnappan R, Loose J, Mason C, Steenberge L, McClendon BT, Wang G, Driscoll M, Yanowitz JL, and Ghazi A (2019). The longevity-promoting factor, TCER-1, widely represses stress resistance and innate immunity. Nat. Commun 10, 3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SM, Cheesman HK, Peterson ND, Salisbury JE, Soukas AA, and Pukkila-Worley R (2019). The fatty acid oleate is required for innate immune activation and pathogen defense in Caenorhabditis elegans. PLoS Pathog 15, e1007893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, and Horvitz HR (1993). Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74, 515–527. [DOI] [PubMed] [Google Scholar]
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, Katsura I, van der Linden A, Sengupta P, and Bargmann CI (2007). Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev 2, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz DD, Tenor JL, and Aballay A (2010). A conserved PMK-1/p38 MAPK is required in Caenorhabditis elegans tissue-specific immune response to Yersinia pestis infection. J. Biol. Chem 285, 10832–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MR, Ghosh SK, Wang P, and Hanover JA (2014). Conserved nutrient sensor O-GlcNAc transferase is integral to C. elegans pathogen-specific immunity. PLoS ONE 9, e113231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byars CL, Bates KL, and Letsou A (1999). The dorsal-open group gene raw is required for restricted DJNK signaling during closure. Development 126, 4913–4923. [DOI] [PubMed] [Google Scholar]
- Cao X, and Aballay A (2016). Neural Inhibition of Dopaminergic Signaling Enhances Immunity in a Cell-Non-autonomous Manner. Curr. Biol 26, 2329–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Kajino-Sakamoto R, Doss A, and Aballay A (2017). Distinct Roles of Sensory Neurons in Mediating Pathogen Avoidance and Neuropeptide-Dependent Immune Regulation. Cell Rep 21, 1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Paek J, and Kim DH (2011). Natural polymorphisms in C. elegans HECW-1 E3 ligase affect pathogen avoidance behaviour. Nature 480, 525–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman HK, Feinbaum RL, Thekkiniath J, Dowen RH, Conery AL, and Pukkila-Worley R (2016). Aberrant Activation of p38 MAP Kinase-Dependent Innate Immune Responses Is Toxic to Caenorhabditis elegans. G3 (Bethesda) 6, 541–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, and Bargmann CI (2005). A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev 19, 270–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, and Bargmann CI (2007). An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell 129, 787–799. [DOI] [PubMed] [Google Scholar]
- Estes KA, Dunbar TL, Powell JR, Ausubel FM, and Troemel ER (2010). bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Käller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KJ, McEwan DL, and Pukkila-Worley R (2020). Measurements of Innate Immune Function in C. elegans In Aging: Methods and Protocols, Curran SP, ed. (Springer Nature; (in press)). [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Ha HI, Hendricks M, Shen Y, Gabel CV, Fang-Yen C, Qin Y, Colón-Ramos D, Shen K, Samuel AD, and Zhang Y (2010). Functional organization of a neural network for aversive olfactory learning in Caenorhabditis elegans. Neuron 68, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni S, Ji Z, Hoque M, Rust N, Sharpe H, Eberhard R, Browne C, Hengartner MO, Mellor J, Tian B, and Furger A (2012). Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 30-end-seq. Nucleic Acids Res 40, 6304–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Lee D, Lee H, Kim D, Son HG, Yang JS, Lee SV, and Kim S (2016). OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7, 56147–56152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, and Ewbank JJ (2018). Signaling in the innate immune response. WormBook 2018, 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan MW, and Ausubel FM (2002). A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KW, Thakur N, Piggott CA, Omi S, Polanowska J, Jin Y, and Pujol N (2016). Coordinated inhibition of C/EBP by Tribbles in multiple tissues is essential for Caenorhabditis elegans development. BMC Biol 14, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labed SA, Wani KA, Jagadeesan S, Hakkim A, Najibi M, and Irazoqui JE (2018). Intestinal Epithelial Wnt Signaling Mediates Acetylcholine-Triggered Host Defense against Infection. Immunity 48, 963–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H (2018). Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Sub-group (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, and Ausubel FM (2004). Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. USA 101, 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Sellegounder D, and Sun J (2016). Neuronal GPCR OCTR-1 regulates innate immunity by controlling protein synthesis in Caenorhabditis elegans. Sci. Rep 6, 36832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee JD, Birchall JC, Chung MA, Cottrell DA, Edgar LG, Svendsen PC, and Ferrari DC (1990). Production of null mutants in the major intestinal esterase gene (ges-1) of the nematode Caenorhabditis elegans. Genetics 125, 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, and Kim DH (2014). Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EV, Grandi LN, Giannini JA, Robinson JD, and Powell JR (2015). The Conserved G-Protein Coupled Receptor FSHR-1 Regulates Protective Host Responses to Infection and Oxidative Stress. PLoS ONE 10, e0137403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minevich G, Park DS, Blankenberg D, Poole RJ, and Hobert O (2012). CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics 192, 1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar M, and Tan MW (2008). Gamma-linolenic and stearidonic acids are required for basal immunity in Caenorhabditis elegans through their effects on p38 MAP kinase activity. PLoS Genet 4, e1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka T, Toyomura T, Honjo K, Wada Y, and Futai M (2001). Four subunit a isoforms of Caenorhabditis elegans vacuolar H+-ATPase. Cell-specific expression during development. J. Biol. Chem 276, 33079–33085. [DOI] [PubMed] [Google Scholar]
- Pagano DJ, Kingston ER, and Kim DH (2015). Tissue expression pattern of PMK-2 p38 MAPK is established by the miR-58 family in C. elegans. PLoS Genet 11, e1004997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, and Salzberg SL (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc 11, 1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ND, Cheesman HK, Liu P, Anderson SM, Foster KJ, Chhaya R, Perrat P, Thekkiniath J, Yang Q, Haynes CM, and Pukkila-Worley R (2019). The nuclear hormone receptor NHR-86 controls anti-pathogen responses in C. elegans. PLoS Genet 15, e1007935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Kim DH, and Ausubel FM (2009). The G protein-coupled receptor FSHR-1 is required for the Caenorhabditis elegans innate immune response. Proc. Natl. Acad. Sci. USA 106, 2782–2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradel E, Zhang Y, Pujol N, Matsuyama T, Bargmann CI, and Ewbank JJ (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R (2016). Surveillance Immunity: An Emerging Paradigm of Innate Defense Activation in Caenorhabditis elegans. PLoS Pathog 12, e1005795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Feinbaum R, Kirienko NV, Larkins-Ford J, Conery AL, and Ausubel FM (2012). Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet 8, e1002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila-Worley R, Feinbaum RL, McEwan DL, Conery AL, and Ausubel FM (2014). The evolutionarily conserved mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog 10, e1004143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, and Ausubel FM (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. [DOI] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, and Kim DH (2009). A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323, 382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Dunbar TL, Nargund AM, Haynes CM, and Troemel ER (2016). The C. elegans CCAAT-Enhancer-Binding Protein Gamma Is Required for Surveillance Immunity. Cell Rep 14, 1581–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CE, Kooistra T, and Kim DH (2010). An essential role for XBP-1 in host protection against immune activation in C. elegans. Nature 463, 1092–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach NP, Sadowski N, Alessi AF, Timp W, Taylor J, and Kim JK (2019). The full-length transcriptome of C. elegans using direct RNA sequencing. bioRxiv 10.1101/598763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, and Bargmann CI (1998). The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20, 55–67. [DOI] [PubMed] [Google Scholar]
- Sellegounder D, Yuan CH, Wibisono P, Liu Y, and Sun J (2018). Octopaminergic Signaling Mediates Neural Regulation of Innate Immunity in Caenorhabditis elegans. MBio 9, e01645–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, and Tan MW (2006). A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103, 14086–14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers RP, Kooistra T, Chu SW, Pagano DJ, and Kim DH (2009). Tissue-specific activities of an immune signaling module regulate physiological responses to pathogenic and nutritional bacteria in C. elegans. Cell Host Microbe 6, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, Whitney JK, Kamanzi O, Matsumoto K, Hisamoto N, and Kim DH (2010). Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 6, e1000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, and Aballay A (2019). Microbial Colonization Activates an Immune Fight-and-Flight Response via Neuroendocrine Signaling. Dev. Cell 49, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, and Aballay A (2008). Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Singh V, Kajino-Sakamoto R, and Aballay A (2011). Neuronal GPCR controls innate immunity by regulating noncanonical unfolded protein response genes. Science 332, 729–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MW, Mahajan-Miklos S, and Ausubel FM (1999). Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torayama I, Ishihara T, and Katsura I (2007). Caenorhabditis elegans integrates the signals of butanone and food to enhance chemotaxis to butanone. J. Neurosci 27, 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey KJ (2010). Understanding immunity requires more than immunology. Nat. Immunol 11, 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey KJ (2014). Approaching the next revolution? Evolutionary integration of neural and immune pathogen sensing and response. Cold Spring Harb. Perspect. Biol 7, a016360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, and Bargmann CI (1997). Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91, 161–169. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, and Bargmann CI (1999). Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99, 387–398. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, and Kim DH (2006). p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2, e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhoven MK, Bauer Huang SL, Albin SD, and Bargmann CI (2006). The claudin superfamily protein nsy-4 biases lateral signaling to generate left-right asymmetry in C. elegans olfactory neurons. Neuron 51, 291–302. [DOI] [PubMed] [Google Scholar]
- Wes PD, and Bargmann CI (2001). C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410, 698–701. [DOI] [PubMed] [Google Scholar]
- Wingett SW, and Andrews S (2018). FastQ Screen: A tool for multi-genome mapping and quality control. F1000Res 7, 1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lu H, and Bargmann CI (2005). Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the mRNA-seq dataset reported in this paper is GEO:GSE130959. All other data are available in the manuscript.


