Abstract
Reddish purple Chinese cabbage (RPCC) is a popular variety of Brassica rapa (AA = 20). It is rich in anthocyanins, which have many health benefits. We detected novel anthocyanins including cyanidin 3-(feruloyl) diglucoside-5-(malonoyl) glucoside and pelargonidin 3-(caffeoyl) diglucoside-5-(malonoyl) glucoside in RPCC. Analyses of transcriptome data revealed 32,395 genes including 3345 differentially expressed genes (DEGs) between 3-week-old RPCC and green Chinese cabbage (GCC). The DEGs included 218 transcription factor (TF) genes and some functionally uncharacterized genes. Sixty DEGs identified from the transcriptome data were analyzed in 3-, 6- and 9-week old seedlings by RT-qPCR, and 35 of them had higher transcript levels in RPCC than in GCC. We detected cis-regulatory motifs of MYB, bHLH, WRKY, bZIP and AP2/ERF TFs in anthocyanin biosynthetic gene promoters. A network analysis revealed that MYB75, MYB90, and MYBL2 strongly interact with anthocyanin biosynthetic genes. Our results show that the late biosynthesis genes BrDFR, BrLDOX, BrUF3GT, BrUGT75c1-1, Br5MAT, BrAT-1, BrAT-2, BrTT19-1, and BrTT19-2 and the regulatory MYB genes BrMYB90, BrMYB75, and BrMYBL2-1 are highly expressed in RPCC, indicative of their important roles in anthocyanin biosynthesis, modification, and accumulation. Finally, we propose a model anthocyanin biosynthesis pathway that includes the unique anthocyanin pigments and genes specific to RPCC.
Keywords: anthocyanins, anthocyanin biosynthetic genes, cis-regulatory motifs, DEGs, network analysis, qRT-PCR, reddish purple Chinese cabbage, transcriptome, transcription factors
1. Introduction
Introgression breeding is an important traditional breeding technique for transferring key agronomic traits between two distinct species [1]. Using this technique, improvements have been made to many Brassica traits, such as disease resistance, male sterility, seed color, oil quality traits, and other morphological traits [1,2]. Some purple Brassicaceae lines have also been generated [3,4]. Red Chinese cabbage (Brassica rapa ssp. pekinensis L.) is reddish purple in color and rich in anthocyanins. This vibrantly colored variety is a popular addition to salads and it has important antioxidant properties [4].
Anthocyanins are a class of secondary metabolites that are synthesized through the phenylpropanoid pathway [5]. These water-soluble compounds with red, purple, or blue colors are synthesized in the cytosol and stored in the vacuole [6]. Anthocyanins play crucial roles in reducing damage from, and in defense responses against, abiotic stresses such as ultraviolet exposure, wounding, high light, chilling, pollution, osmotic stress, and nutrient deficiency, as well as biotic stresses such as pathogen infection [7].
The important structural genes in the anthocyanin biosynthetic pathway are those encoding phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumaroyl CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), UDP-flavonoid glucosyl transferase (UFGT) and glutathione S-transferase (GST) [8]. The late anthocyanin biosynthesis genes are regulated by a complex of transcription factors (TFs), MYB-bHLH-WD40, known as the MBW complex. In vivo assays in Antirrhinum revealed that a flower-specific MYB protein activates the transcription of genes involved in phenylpropanoid biosynthesis [9,10]. Four MYB TFs, encoded by PAP1/MYB75, PAP2/MYB90, MYB113, and MYB114, have been identified to control anthocyanin biosynthesis in vegetative tissues of Arabidopsis [11,12]. Previous biochemical and genetic studies have shown that TTG1 (WD40), GL3/EGL3/TT8 (bHLH) and PAP1/PAP2/MYB113/MYB114 (MYB) are components of potential WBM complexes that activate anthocyanin biosynthesis [13,14]. Interestingly, recent studies on proanthocyanidin and anthocyanin biosynthesis pathways in Arabidopsis and Petunia suggest that WRKY TFs regulate color accumulation along with the MBW complex [10,15]. Similarly, studies on bZIP-TFs revealed the mechanisms by which they regulate anthocyanin accumulation in apples [16,17].
Secondary metabolites are abundant in Chinese cabbage. Previous studies have shown that some secondary metabolites are specific to particular species, cultivars, or varieties. Such metabolites are usually detected by high performance liquid chromatography (HPLC) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) [4,18,19,20]. Recent metabolic profiling studies have demonstrated that cyanidin derivatives are highly accumulated in Brassica species [18,21,22,23]. It is difficult to identify all the genes related to specific traits in a plant system, but transcriptome sequencing allows for the prediction of functional genes in the plant genome. Several RNA-seq studies have been conducted for Brassica species, and their results have led to the identification of sets of genes that are expressed under various conditions and/or in certain genotypes. This information is very useful for the functional characterization of target genes [24,25,26,27].
Reddish purple Chinese cabbage (RPCC) is a variety of B. rapa. It is an economically important leafy vegetable that is widely cultivated and consumed in East Asian countries, especially Korea, due to its health promoting properties [28]. To date, the underlying molecular mechanisms and the genes regulating the anthocyanin pigments responsible for its vivid red color are unexplored, and may differ from those reported previously for other Brassica lines. In this study, we used a next generation sequencing (NGS) based RNA-sequencing approach to identify a novel set of genes involved in anthocyanin biosynthesis, and the regulation of this pathway, in a red Chinese cabbage variety. A comprehensive analysis of this plant including computational, leaf chemotype, and expressional abundance analyses shows the significance of this variety.
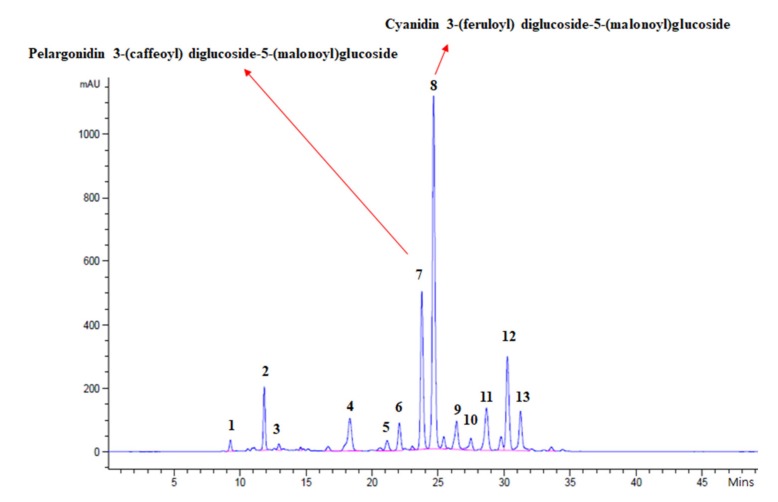
2. Results
2.1. Estimation of Anthocyanin Content in RPCC and GCC Leaf Samples
The red color is a distinguishing feature of some Brassica species, and is due to the accumulation of anthocyanins [29]. To determine the types of anthocyanins that confer color in RPCC, we analyzed anthocyanins in the innermost and outermost leaves of 9-week-old GCC and RPCC plants (Table 1 and Table S1) by HPLC-MS/MS. We detected 13 anthocyanin pigments, 12 of which were derivatives of cyanidin. The other one was a pelargonidin derivative. The most abundant pigment in the innermost leaves of RPCC was cyanidin 3-(feruloyl) diglucoside-5-(malonoyl) glucoside (peak 8) with a concentration of 12.63 ± 1.37 mg/g dry weight, followed by pelargonidin 3-(caffeoyl) diglucoside-5-(malonoyl) glucoside (peak 7) with a concentration of 5.66 ± 0.60 mg/g dry weight (Figure 1; Table 1 and Table S1). In the outermost leaves of RPCC, the most abundant pigments were cyanidin 3-(feruloyl) diglucoside-5-(malonoyl) glucoside and cyanidin 3-O-(sinapoyl)(feruloyl) diglucoside-5-O-(malonyl) glucoside (Figure 1; Table 1 and Table S1). The total amount of anthocyanins was 32.31 ± 2.84 mg/g dry weight in the innermost leaf and 10.17 ± 1.69 mg/g dry weight in the outermost leaf of RPCC. No anthocyanins were detected in the GCC leaves (Table 1). Interestingly, the RPCC pigment peaks 8 and 7 and a few other pigments identified in this study (Table S1) had not been reported previously. This result indicates that cyanidin 3-(feruloyl) diglucoside-5-(malonoyl) glucoside and pelargonidin 3-(caffeoyl) diglucoside-5-(malonoyl) glucoside are specific to this variety of RPCC, and contribute to its color.
Table 1.
Total anthocyanin pigments identified in outer and inner leaf tissues of reddish purple and green Chinese cabbage.
| No. of Pigments | Trivial Names | RPCC_IL | RPCC_OL | GCC_IL | GCC_OL |
|---|---|---|---|---|---|
| 1 | Cyanidin 3-diglucoside-5-glucoside | 0.26 ± 0.00 | 0.04 ± 0.05 | ND | ND |
| 2 | Cyanidin 3-diglucoside-5-(malonyl)glucoside | 1.74 ± 0.23 | 0.29 ± 0.23 | ND | ND |
| 3 | Cyanidin 3-(feruloyl)diglucoside-5-glucoside | 0.19 ± 0.01 | 0.00 ± 0.00 | ND | ND |
| 4 | Cyanidin 3-(caffeoyl)diglucoside-5-(malonyl)glucoside | 1.93 ± 0.23 | 0.26 ± 0.19 | ND | ND |
| 5 | Cyanidin 3-(p-coumaroyl)diglucoside-5-glucoside | 0.34 ± 0.03 | 0.13 ± 0.09 | ND | ND |
| 6 | Cyanidin 3-(feruloyl)diglucoside-5-glucoside | 0.76 ± 0.09 | 0.08 ± 0.06 | ND | ND |
| 7 | Pelargonidin 3-(caffeoyl)diglucoside-5-(malonoyl)glucoside | 5.66 ± 0.60 | 0.81 ± 0.58 | ND | ND |
| 8 | Cyanidin 3-(feruloyl)diglucoside-5-(malonoyl)glucoside | 12.63 ± 1.37 | 2.19 ± 1.60 | ND | ND |
| 9 | Cyanidin 3-(feruloyl)(feruloyl)diglucoside-5-glucoside | 1.25 ± 0.09 | 0.19 ± 0.14 | ND | ND |
| 10 | Cyanidin 3-O-(sinapoyl)(feruloyl)diglucoside-5-O-glucoside | 0.41 ± 0.06 | 0.16 ± 0.12 | ND | ND |
| 11 | Cyanidin 3-O-(p-coumaroyl)(sinapoyl)diglucoside-5-O-(malonyl)glucoside | 1.92 ± 0.12 | 0.61 ± 0.45 | ND | ND |
| 12 | Cyanidin 3-O-(sinapoyl)(feruloyl)diglucoside-5-O-(malonyl)glucoside | 3.55 ± 0.26 | 1.24 ± 0.92 | ND | ND |
| 13 | Cyanidin 3-O-(p-coumaroyl)(sinapoyl)diglucoside-5-O-(malonyl)glucoside | 1.68 ± 0.12 | 0.77 ± 0.61 | ND | ND |
| Total | 32.31 ± 2.84 | 10.17 ± 1.69 | ND | ND |
RPCC-reddish purple Chinese cabbage; GCC- green Chinese cabbage; IL-inner leaf; OL- outer leaf; anthocyanin content-mg/g dry weight.
Figure 1.
HPLC chromatogram of anthocyanin pigments detected in leaf extract of reddish purple Chinese cabbage at 520 nm. Horizontal axis shows retention time (min); vertical axis indicates strength of the peak (mAU).
2.2. RNA-Sequencing of RPCC and GCC Samples
To identify differences in gene transcription between RPCC and GCC, we obtained and sequenced 53,127,646 (GCC) and 48,179,516 (RPCC) total reads, making up 5.37 Gb (giga bases) and 4.87 Gb, respectively (Table 2) with a GC content of 47.6% and 47.2%, respectively. After filtering, 38,611,432 and 34,924,846 clean reads were obtained for the GCC and RPCC samples, respectively, with Q30 values of 94.4% and 94.1%, respectively. A total 32,395 non-redundant transcripts were identified with varying lengths, most in the range of 501 to 1000 nucleotide bases, followed by 200–500 and 1001–1500 nucleotide bases (Figure S1a). Among the identified transcripts, 90.2% were annotated to the following public databases: NR-Viridiplantae (88.61%)¸ Phytozome (88.05%), UniProtKB–Viridiplantae (85.31%), KOG (84.06%), GO (83.72%), InterProscan (68.43%), and KEGG (18.87%) (Figure S1b).
Table 2.
Summary of RNA sequence data.
| Reddish Purple Chinese Cabbage (RPCC) | Green Chinese Cabbage (GCC) | |
|---|---|---|
| Total Reads | 48,179,516 | 53,127,646 |
| Total Bases | 4866,131,116 | 5365,892,246 |
| Total Bases(Gb) | 4.87 Gb | 5.37 Gb |
| GC_ Count | 2320,659,939 | 2532,951,037 |
| N_ Zero Reads | 48,028,490 | 52,961,958 |
| N5_ Less Reads | 48,091,492 | 53,032,280 |
| N_ Rate | 0.03% | 0.03% |
| Q20_More Bases | 4699,696,370 | 5195,093,244 |
| Q30_ More Bases | 4579,319,019 | 5069,583,507 |
| Clean reads | 34,924,846 | 38,611,432 |
| Clean bases | 3073,838,581 | 3400,979,929 |
2.3. Identification of DEGs
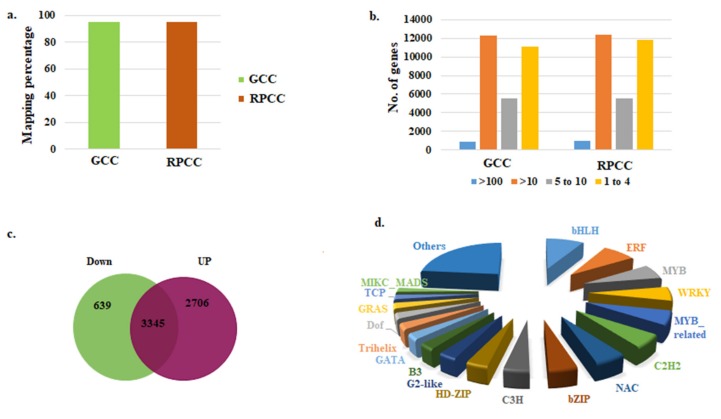
The cDNA libraries of RPCC and GCC were mapped to the B. rapa reference genome with coverage of 95.2% and 94.66%, respectively (Figure 2a). Among the mapped and annotated DEGs, the highest proportion had RPKM (reads per kilobase per million mapped reads) values of >10, followed by RPKM values of 1 to 4 (Figure 2b). Among the predicted 3345 DEGs, 2706 were up-regulated and 639 were down-regulated in RPCC vs. GCC (Figure 2c; Table S2 and Table S3). Of them, 643 of the up-regulated DEGs and 354 of the down-regulated DEGs between RPCC and GCC samples were not functionally characterized (Table S2 and Table S3). The DEGs included many ABGs. A Bland–Altman (MA) plot was constructed to show the differentiation of gene expression between the two samples by plotting the values onto M (log ratio) and A (mean average) scales. The differences in gene expression are shown on the MA plot, where genes with ≥1-fold expression values are shown in red and those with negative log2 fold values are shown in green (Figure S2).
Figure 2.
RNA-seq data for reddish purple Chinese cabbage. (a) Percentage of transcripts mapped to reference genome; (b) Gene expression values (RPKM); (c) Differentially expressed genes (DEGs) between two genotypes [green (GCC) and reddish purple (RPCC)]; (d) Transcription factor families identified in the transcriptome.
2.4. Identification of Transcription Factor Genes
Transcription factors play crucial roles in regulating gene expression, and many TFs control ABGs [30]. Hence, we searched for TFs that regulate anthocyanin biosynthesis in B. rapa. We detected 1625 TF genes in 54 TF families from our transcriptome data (Figure 2d; Table S4 and Table S5). The proportions of TF genes in different TF families were as follows: 8.7% in the basic helix loop helix (bHLH) family, 7.6% in the ethylene response factor (ERF) family, 6.05% in the myeloblastosis (MYB) family, 5.77% in the WRKY family, and 5.72% in the MYB-related family (Figure 2d). Additionally, 218 TF genes with log2fold change expression ≥1 and 37 TFs with log2fold change expression ≤ 1 were identified (Table S4 and Table S5). A few TF genes had very high log2fold change values (5–11.3), including MYB90 (Bra004162), MYB75 (Bra039763), three RRTF1s (Bra017656, Bra011529, Bra034624), CBF4 (Bra028290), MYBL2 (Bra016164), TTG2 (Bra023112), DDF1 (Bra019777) and ERF-13 (Bra037630) (Table 3). These DEGs may be involved in various functions related to pigment accumulation in RPCC.
Table 3.
Summary of important regulatory transcriptional factor family genes identified from transcriptome data.
| BRAD ID | E-Value | Identity | TAIR Description | Log2 Fold Change | |||
|---|---|---|---|---|---|---|---|
| A. thaliana Id | TF Family | Gene Annotation | Gene Description | RPCC vs. GCC | |||
| AP2/ERF Transcription Factors | |||||||
| Bra011782 | 0 | 100 | AT4G37750 | AP2 | ANT, CKC, CKC1, DRG | Integrase-type DNA-binding superfamily protein | 2.21 |
| Bra017656 | 0 | 99.88 | AT4G34410 | ERF | RRTF1 | Redox responsive transcription factor 1 | 6.90 |
| Bra011529 | 0 | 99.75 | AT4G34410 | ERF | RRTF1 | Redox responsive transcription factor 1 | 6.72 |
| Bra019087 | 0 | 100 | AT2G20350 | ERF | − | Integrase-type DNA-binding superfamily protein | 6.50 |
| Bra034624 | 0 | 99.8 | AT4G34410 | ERF | RRTF1 | Redox responsive transcription factor 1 | 6.16 |
| Bra028290 | 0 | 100 | AT5G51990 | ERF | CBF4, DREB1D | C-repeat-binding factor 4 | 5.94 |
| Bra019777 | 0 | 98.04 | AT1G12610 | ERF | DDF1 | Integrase-type DNA-binding superfamily protein | 5.06 |
| Bra037630 | 0 | 96.93 | AT2G44840 | ERF | ATERF13, EREBP, ERF13 | Ethylene-responsive element binding factor 13 | 5.00 |
| Bra015882 | 0 | 94.68 | AT1G74930 | ERF | ORA47 | Integrase-type DNA-binding superfamily protein | 4.96 |
| Bra027612 | 0 | 100 | AT1G63030 | ERF | DDF2 | Integrase-type DNA-binding superfamily protein | 4.88 |
| Bra016763 | 0 | 100 | AT1G12610 | ERF | DDF1 | Integrase-type DNA-binding superfamily protein | 4.84 |
| Bra024539 | 0 | 99.48 | AT1G22810 | ERF | − | Integrase-type DNA-binding superfamily protein | 4.79 |
| Bra026963 | 0 | 98.87 | AT1G12610 | ERF | DDF1 | Integrase-type DNA-binding superfamily protein | 4.72 |
| Bra031069 | 0 | 100 | AT1G19210 | ERF | − | Integrase-type DNA-binding superfamily protein | 4.64 |
| Bra028759 | 0 | 94.66 | AT5G05410 | ERF | DREB2,DREB2A | DRE-binding protein 2A | 4.58 |
| Bra015660 | 0 | 100 | AT1G77640 | ERF | − | Integrase-type DNA-binding superfamily protein | 4.04 |
| Bra028291 | 0 | 100 | AT5G52020 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.98 |
| Bra016400 | 0 | 100 | AT1G21910 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.95 |
| Bra032901 | 0 | 100 | AT1G28370 | ERF | ATERF11,ERF11 | ERF domain protein 11 | 3.66 |
| Bra014925 | 0 | 100 | AT3G23230 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.64 |
| Bra027614 | 0 | 85 | AT1G12630 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.62 |
| Bra002377 | 0 | 100 | AT5G21960 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.62 |
| Bra010881 | 0 | 99.46 | AT1G28360 | ERF | ATERF12,ERF12 | ERF domain protein 12 | 3.57 |
| Bra016136 | 0 | 100 | AT1G71450 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.36 |
| Bra003780 | 0 | 100 | AT1G74930 | ERF | ORA47 | Integrase-type DNA-binding superfamily protein | 3.29 |
| Bra036022 | 0 | 99.44 | AT1G21910 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.28 |
| Bra016518 | 3.00E−162 | 95.32 | AT1G19210 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.23 |
| Bra029147 | 0 | 100 | AT5G52020 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.21 |
| Bra024953 | 0 | 100 | AT5G47230 | ERF | ATERF-5, ATERF5, ERF5 | Ethylene responsive element binding factor 5 | 3.14 |
| Bra040309 | 0 | 99.15 | AT1G44830 | ERF | − | Integrase-type DNA-binding superfamily protein | 3.14 |
| Bra032665 | 0 | 100 | AT2G44840 | ERF | ATERF13, EREBP, ERF13 | Ethylene-responsive element binding factor 13 | 3.00 |
| Bra030957 | 0 | 99.3 | AT1G53170 | ERF | ATERF-8, ATERF8, ERF8 | Ethylene response factor 8 | 2.99 |
| Bra010880 | 0 | 98.27 | AT1G28370 | ERF | ATERF11, ERF11 | ERF domain protein 11 | 2.91 |
| Bra011383 | 0 | 98.7 | AT4G32800 | ERF | − | Integrase-type DNA-binding superfamily protein | 2.91 |
| Bra015478 | 1.00E−10 | 74.84 | AT2G44840 | ERF | ATERF13, EREBP, ERF13 | Ethylene-responsive element binding factor 13 | 2.86 |
| Bra021048 | 0 | 97.12 | AT4G17500 | ERF | ATERF-1, ERF-1 | Ethylene responsive element binding factor 1 | 2.63 |
| Bra024954 | 0 | 99.54 | AT5G47220 | ERF | ATERF-2, ATERF2, ERF2 | Ethylene responsive element binding factor 2 | 2.61 |
| Bra008952 | 0 | 98.76 | AT5G11590 | ERF | TINY2 | Integrase-type DNA-binding superfamily protein | 2.53 |
| Bra035732 | 0 | 97.33 | AT5G51190 | ERF | − | Integrase-type DNA-binding superfamily protein | 2.51 |
| Bra040158 | 0 | 100 | AT4G17490 | ERF | ATERF6, ERF-6-6, ERF6 | Ethylene responsive element binding factor 6 | 2.48 |
| Bra017493 | 0 | 98.76 | AT5G47230 | ERF | ATERF-5, ATERF5, ERF5 | Ethylene responsive element binding factor 5 | 2.33 |
| Bra040159 | 0 | 100 | AT4G17500 | ERF | ATERF-1,ERF-1 | Ethylene responsive element binding factor 1 | 2.22 |
| Bra034535 | 0 | 100 | AT4G32800 | ERF | − | Integrase-type DNA-binding superfamily protein | 2.11 |
| Bra027736 | 0 | 97.6 | AT1G64380 | ERF | − | Integrase-type DNA-binding superfamily protein | 2.00 |
| bHLH transcription factors | |||||||
| Bra033690 | 0 | 100 | AT5G43650 | bHLH | BHLH92 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 4.69 |
| Bra027501 | 0 | 100 | AT5G43650 | bHLH | BHLH92 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 2.68 |
| Bra035639 | 0 | 97.42 | AT5G56960 | bHLH | − | Basic helix-loop-helix (bHLH) DNA-binding family protein | 2.21 |
| Bra036640 | 0 | 100 | AT1G62975 | bHLH | − | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein | 2.20 |
| bZIP transcription factors | |||||||
| Bra010035 | 0 | 94.65 | AT5G49450 | bZIP | AtbZIP1,bZIP1 | Basic leucine-zipper 1 | 3.10 |
| C2H2 transcription factors | |||||||
| Bra006692 | 1.00E−160 | 88.04 | AT5G59820 | C2H2 | RHL41, ZAT12 | C2H2-type zinc finger family protein | 4.46 |
| Bra002528 | 0 | 98.96 | AT5G59820 | C2H2 | RHL41, ZAT12 | C2H2-type zinc finger family protein | 3.43 |
| Bra022436 | 0 | 97.64 | AT3G19580 | C2H2 | AZF2, ZF2 | Zinc-finger protein 2 | 2.99 |
| Bra010922 | 0 | 99.57 | AT1G27730 | C2H2 | STZ, ZAT10 | Salt tolerance zinc finger | 2.96 |
| Bra001752 | 0 | 99.87 | AT3G19580 | C2H2 | AZF2, ZF2 | Zinc-finger protein 2 | 2.96 |
| Bra032845 | 0 | 100 | AT1G27730 | C2H2 | STZ, ZAT10 | Salt tolerance zinc finger | 2.81 |
| Bra038219 | 0 | 98.11 | AT3G19580 | C2H2 | AZF2, ZF2 | Zinc-finger protein 2 | 2.50 |
| C3H transcription factors | |||||||
| Bra000170 | 1.00E−156 | 100 | AT2G40140 | C3H | ATSZF2, CZF1, SZF2, ZFAR1 | Zinc finger (CCCH-type) family protein | 2.77 |
| Bra007205 | 0 | 98.79 | AT3G55980 | C3H | ATSZF1, SZF1 | Salt-inducible zinc finger 1 | 2.41 |
| Bra004982 | 4.00E−135 | 81.1 | AT2G40140 | C3H | ATSZF2, CZF1, SZF2, ZFAR1 | Zinc finger (CCCH-type) family protein | 2.16 |
| Dof transcription factors | |||||||
| Bra014297 | 0 | 98.01 | AT1G51700 | Dof | ADOF1, DOF1 | DOF zinc finger protein 1 | 2.18 |
| GRAS transcription factors | |||||||
| Bra021063 | 0 | 99.75 | AT4G17230 | GRAS | SCL13 | SCARECROW-like 13 | 2.06 |
| Bra033813 | 0 | 91.33 | AT3G46600 | GRAS | − | GRAS family transcription factor | 2.02 |
| HD-ZIP transcription factors | |||||||
| Bra005259 | 0 | 100 | AT2G36610 | HD-ZIP | ATHB22, HB22 | Homeobox protein 22 | 3.85 |
| Bra016300 | 0 | 100 | AT1G26960 | HD-ZIP | AtHB23, HB23 | Homeobox protein 23 | 2.02 |
| LBD transcription factors | |||||||
| Bra021433 | 5.00E-124 | 83.53 | AT3G02550 | LBD | LBD41 | LOB domain-containing protein 41 | 2.90 |
| MADS transcription factors | |||||||
| Bra017376 | 0 | 99.84 | AT2G03710 | MIKC_ MADS | AGL3,SEP4 | K-box region and MADS-box transcription factor family protein | 2.33 |
| Bra024521 | 0 | 99.59 | AT1G22590 | M-type_ MADS | AGL87 | AGAMOUS-like 87 | 2.10 |
| Bra005166 | 0 | 100 | AT2G28700 | M-type_ MADS | AGL46 | AGAMOUS-like 46 | 2.05 |
| MYB transcription factors | |||||||
| Bra004162 | 2.00E−111 | 84.01 | AT1G66390 | MYB | ATMYB90, MYB90, PAP2 | MYB domain protein 90 | 11.38 |
| Bra039763 | 0 | 92.86 | AT1G56650 | MYB | ATMYB75, MYB75, PAP1, SIAA1 | Production of anthocyanin pigment 1 | 6.87 |
| Bra029990 | 0 | 100 | AT3G50060 | MYB | MYB77 | MYB domain protein 77 | 3.89 |
| Bra012910 | 0 | 100 | AT3G50060 | MYB | MYB77 | MYB domain protein 77 | 2.82 |
| Bra013000 | 9.00E−118 | 100 | AT5G60890 | MYB | ATMYB34, ATR1, MYB34 | MYB domain protein 34 | 2.51 |
| Bra016164 | 0 | 100 | AT1G71030 | MYB_ related | ATMYBL2, MYBL2 | MYB-like 2 | 5.55 |
| Bra007957 | 0 | 97.7 | AT1G71030 | MYB_ related | ATMYBL2, MYBL2 | MYB-like 2 | 4.06 |
| Bra022637 | 5.00E−79 | 89.91 | AT5G53200 | MYB_ related | TRY | Homeodomain-like superfamily protein | 2.34 |
| NAC transcription factors | |||||||
| Bra008553 | 0 | 94.59 | AT4G01550 | NAC | anac069, NAC069 | NAC domain containing protein 69 | 4.31 |
| Bra020188 | 0 | 99.8 | AT5G22380 | NAC | anac090, NAC090 | NAC domain containing protein 90 | 3.14 |
| Bra006624 | 0 | 99.58 | AT5G22380 | NAC | anac090, NAC090 | NAC domain containing protein 90 | 2.86 |
| Bra027238 | 0 | 100 | AT3G15500 | NAC | ANAC055,ATNAC3,NAC055, NAC3 | NAC domain containing protein 3 | 2.62 |
| Bra037283 | 0 | 99.52 | AT2G17040 | NAC | anac036, NAC036 | NAC domain containing protein 36 | 2.17 |
| Bra013034 | 0 | 98.94 | AT2G17040 | NAC | anac036, NAC036 | NAC domain containing protein 36 | 2.08 |
| WRKY transcription factors | |||||||
| Bra023112 | 0 | 99.53 | AT2G37260 | WRKY | ATWRKY44, DSL1,TTG2, WRKY44 | WRKY family transcription factor family protein | 5.13 |
| Bra014426 | 0 | 99.88 | AT2G46400 | WRKY | ATWRKY46, WRKY46 | WRKY DNA-binding protein 46 | 3.87 |
| Bra003588 | 0 | 99.5 | AT1G80840 | WRKY | ATWRKY40, WRKY40 | WRKY DNA-binding protein 40 | 3.81 |
| Bra035148 | 0 | 100 | AT1G80840 | WRKY | ATWRKY40, WRKY40 | WRKY DNA-binding protein 40 | 3.69 |
| Bra005210 | 0 | 100 | AT2G37260 | WRKY | ATWRKY44, DSL1,TTG2, WRKY44 | WRKY family transcription factor family protein | 3.54 |
| Bra035147 | 0 | 99.52 | AT1G80850 | WRKY | − | DNA glycosylase superfamily protein | 3.53 |
| Bra033158 | 0 | 98.49 | AT4G11070 | WRKY | AtWRKY41, WRKY41 | WRKY family transcription factor | 2.95 |
| Bra020196 | 0 | 90.34 | AT5G22570 | WRKY | ATWRKY38, WRKY38 | WRKY DNA-binding protein 38 | 2.87 |
| Bra010032 | 0 | 99.58 | AT5G49520 | WRKY | ATWRKY48, WRKY48 | WRKY DNA-binding protein 48 | 2.86 |
| Bra019265 | 0 | 100 | AT4G23810 | WRKY | ATWRKY53, WRKY53 | WRKY family transcription factor | 2.65 |
| Bra013731 | 0 | 99.57 | AT4G23800 | WRKY | − | HMG (high mobility group) box protein | 2.41 |
| Bra023998 | 0 | 100 | AT4G31550 | WRKY | WRKY11 | WRKY DNA-binding protein 11 | 2.27 |
| Bra031900 | 0 | 98.34 | AT5G64810 | WRKY | WRKY51 | WRKY DNA-binding protein 51 | 2.13 |
2.5. Functional Characteristics of DEGs and KEGG Pathway Enrichment Analysis
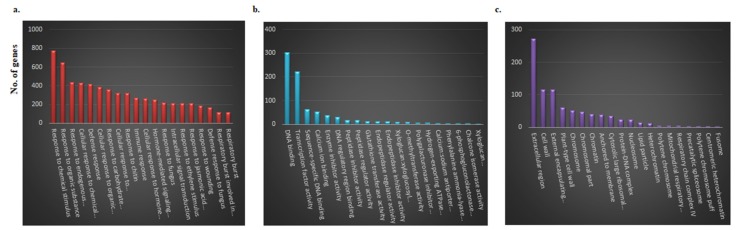
We performed GO analyses to annotate the DEGs. The top 20 enriched terms in each GO category of DEGs were selected based on significance (p < 0.05) and are summarized in Figure 3. In the BP category, the majority of DEGs were involved in “response to chemical stimulus” followed by “response to organic substance”, “response to endogenous stimulus”, and “cellular response to chemical stimulus” processes. Similarly, in the MF category, the majority of genes were involved in “DNA binding”, followed by “transcription factor activity”, “sequence specific DNA binding” and “calcium ion binding”. In the CC category, many of the DEGs were related to “extracellular region”, “cell wall”, “external encapsulating structure” and “plant type cell wall” (Figure 3). We further categorized the up-regulated and down-regulated genes. Among the up-regulated DEGs, a total of 385 GO terms were identified, comprising 328 in the BP category, 35 in the MF category, and 22 in the CC category. Among the down-regulated DEGs, a total of 210 GO terms were identified, comprising 133 terms in the BP category, 50 in the MF category, and 22 in the CC category (Table S6.) In the BP category, a few genes were involved in “biosynthetic and metabolic processes of anthocyanins, ethylene signaling, flavonoids and phenylpropanoids”, “catabolic and metabolic processes of L-phenylalanine”, “cinnamic acid biosynthesis”, “response to absence of light”, and “response to temperature stimulus”. In the MF category, some genes were involved in “phenylalanine ammonia-lyase activity” and “O-methyltransferase activity” (Table S6.). Some of the down-regulated genes were involved in “biosynthesis process”, “negative regulation of cellular metabolic process”, and “response to UV-C” (Table S6).
Figure 3.
Gene ontology (GO) analysis of differentially expressed genes (DEGs) between red and green Chinese cabbage. DEGs were grouped into three categories: (a) Biological process; (b) molecular function; and (c) cellular component. X-axis shows gene annotation term; y-axis shows number of genes.
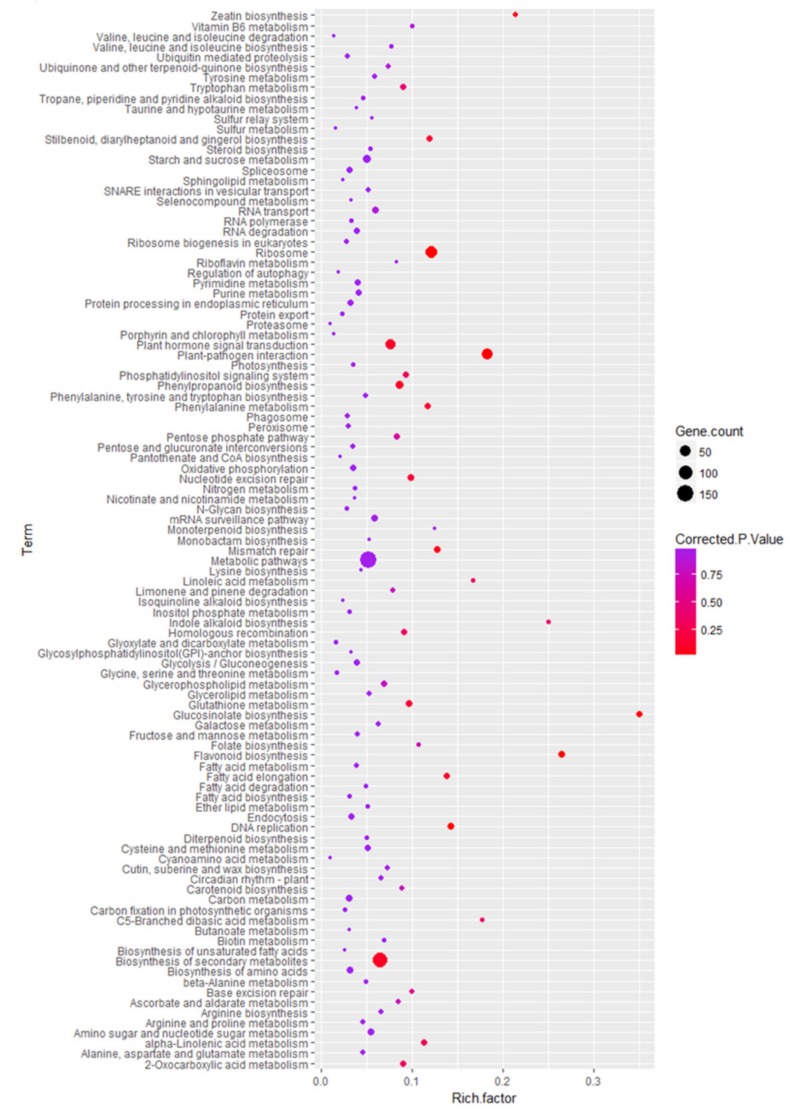
Next, we conducted a KEGG enrichment analysis to identify pathways significantly enriched with DEGs. The pathways enriched with up-regulated DEGs were “biosynthesis of secondary metabolites”, “metabolic pathways”, “biosynthesis of flavonoids and phenylpropanoids”, “metabolism of fructose, mannose, starch, and sucrose”, and several others (Figure 4 and Table S7). The down-regulated DEGs were involved in 46 types of functions related to organ development, and other functions related to plant growth and development (Table S7).
Figure 4.
KEGG enrichment analysis of differentially expressed anthocyanin biosynthetic genes. X-axis shows KEGG terms and y-axis shows enrichment factor. Gene count and corrected p-values are shown on right.
2.6. Genes Related to Anthocyanin Biosynthesis Identified from Transcriptome Data
From the transcriptome data, we identified 255 ABGs (Table S8) comprising 58 phenylpropanoid biosynthetic genes (PBGs), 56 early biosynthetic genes (EBGs), 67 late biosynthetic genes (LBGs), 19 anthocyanin transporter genes (ATGs), 29 other anthocyanin biosynthesis regulatory genes (OABRGs) and 26 regulatory TF genes (Table S8). The main PBGs were those encoding cinnamyl alcohol dehydrogenase (CAD), caffeoyl CoA O-methyltransferase (CCoAMT), cinnamoyl-CoA reductase (CCR), cinnamate 4-hydroxylase (C4H), 4-coumarate: coenzyme A ligase (4CL), O-methyltransferase (OMT), and phenylalanine ammonia lyase (PAL). These genes are involved in different stages of the phenylpropanoid biosynthesis pathway and showed large differences in transcript levels (−0.7 to 3.2-fold change) between RPCC and GCC (Table S8). Similarly, EBGs encoding chalcone isomerase (CHI), chalcone synthase (CHS), flavanone-3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), and flavonol synthase (FLS) showed large differences in their transcript levels between RPCC and GCC (−1.1 to 4.9-fold change) (Table S8). The LBGs showing large differences in transcript levels (−1.9 to 9.3-fold change) between RPCC and GCC encoded beta glucosidase (BGLU), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), 5-O-glucoside-6-O-malonyltransferase (5MAT), UDP-glucose: flavonoid 3-o-glucosyltransferase (UF3GT), and UDP-glucosyltransferases (UGT). Genes for anthocyanin transporters and regulatory TFs included those encoding glutathione S-transferase 26/TRANSPARENT TESTA 19 (GST26/TT19), multidrug and toxic compound extrusion (MATE), basic helix-loop-helix 32 (BHLH32), ENHANCER OF GLABRA 3 (EGL3), GLABRA 3 (GL3), myeloblastosis protein 75 (MYB75), myeloblastosis protein 90 (MYB90), TRANSPARENT TESTA 8 (TT8) and TRANSPARENT TESTA GLABRA 1 (TTG1), and TRANSPARENT TESTA GLABRA (TTG2). These genes showed differences in expression between RPCC and GCC ranging from a −0.5 to 11.3-fold change. Interestingly, we identified a few OABRGs with high log2fold expression values (≥ 2) (Table S8). These results indicate that ABGs have vital roles in anthocyanin biosynthesis, transportation, and accumulation in the leaf tissues of RPCC at the seedling stage (3 weeks).
2.7. Expression Analysis of Anthocyanin Biosynthetic Genes by qRT-PCR
In general, gene expression studies can demonstrate the biological activity of genes in plants. We confirmed the reproducibility and accuracy of DEGs identified in our transcriptome data through qRT-PCR analyses. Although transcriptome sequencing was performed using samples from 3-week-old plants (seedlings), we checked the expression profiles of genes in samples from 6-week-old (rosette stage) and 9-week-old (heading stage) plants of GCC and RPCC. This analysis of gene expression at three developmental stages sheds light on the expression profile of ABGs and their effect on anthocyanin biosynthesis and accumulation throughout plant development. In general, the anthocyanin biosynthesis pathway can be classified into three phases; 1. The phenylpropanoid pathway; 2. Early steps of the flavonoid pathway; and 3. The anthocyanin pathway [31]. For the validation of gene expression, we selected 60 genes identified from the transcriptome data and from a previous study [26] that are involved in various stages of anthocyanin biosynthesis (Table 4; Table S9).
Table 4.
Details of 60 genes selected for validation by qRT_PCR analysis.
| Given I.D | BRAD Id | Gene Position V 1.5 | A. thaliana Id | Gene Annotation | |||||
|---|---|---|---|---|---|---|---|---|---|
| B. rapa Id | Identity | E-Value | Chromosome | Start | End | Strand | |||
| Phenylpropanoid pathway genes | |||||||||
| BrPAL1 | Bra017210 | 98.94 | 0 | A04 | 16132008 | 16134574 | - | AT2G37040 | PAL1 |
| BrPAL2 | Bra003126 | 98.68 | 0 | A07 | 14754085 | 14756786 | - | AT3G53260 | PAL2 |
| BrPAL4 | Bra029831 | 100 | 0 | A05 | 22819640 | 22826303 | - | AT3G10340 | PAL4 |
| BrC4H | Bra021637 | 97.23 | 0 | A04 | 13688684 | 13690602 | - | AT2G30490 | C4H |
| Br4CL2 | Bra031266 | 100 | 0 | A05 | 17255035 | 17257878 | + | AT3G21240 | 4CL2,AT4CL2 |
| Early anthocyanin biosynthesis genes | |||||||||
| BrCHS | Bra006224 | 99.5 | 0 | A03 | 2596137 | 2597594 | + | AT5G13930 | CHS,TT4 |
| BrCHI | Bra007142 | 99.47 | 0 | A09 | 29055564 | 29057157 | - | AT3G55120 | CHI,TT5 |
| BrCHI1 | Bra009101 | 100 | 0 | A10 | 15229294 | 15230280 | - | AT5G05270 | CHI1 |
| BrF3H | Bra036828 | 99.91 | 0 | A09 | 27095567 | 27097080 | + | AT3G51240 | F3H,TT6 |
| BrF3′H-1 | Bra009312 | 100 | 0 | A10 | 14356094 | 14358845 | - | AT5G07990 | F3′H,TT7 |
| BrF3′H-2 | Bra020459 | 90.02 | 0 | A02 | 5846392 | 5848174 | - | AT5G57220 | F3′H,TT7 |
| BrF3′H-3 | Bra019366 | 99.68 | 0 | A03 | 24701345 | 24702922 | + | AT4G22690 | F3′H,TT7 |
| BrF3′H-4 | Bra030246 | 100 | 0 | A04 | 10022729 | 10024935 | + | AT2G22330 | F3′H,TT7 |
| BrF3′H-5 | Bra011280 | 94.06 | 0 | A01 | 2958371 | 2960458 | - | AT1G13080 | F3′H,TT7 |
| Late anthocyanin biosynthesis genes | |||||||||
| BrDFR | Bra027457 | 100 | 0 | A09 | 10926334 | 10927890 | - | AT5G42800 | DFR,M318,TT3 |
| BrLDOX | Bra013652 | 98.48 | 0 | A01 | 6885692 | 6887113 | - | AT4G22880 | LDOX |
| BrUF3GT | Bra003021 | 99.81 | 0 | A10 | 6063162 | 6064651 | - | AT5G54060 | UF3GT |
| BrUGT75C1-1 | Bra038445 | 100 | 0 | A08 | 8755970 | 8757334 | - | AT4G14090 | UGT75C1 |
| BrUGT75C1-2 | Bra039545 | 100 | 0 | A01 | 11553894 | 11555345 | + | AT4G15490 | UGT75C1 |
| BrUGT73B2 | Bra034610 | 100 | 0 | A08 | 11760868 | 11762581 | + | AT4G34138 | UGT73B2 |
| BrUGT78D2 | Bra023594 | - | - | A02 | 3087246 | 3088800 | - | AT5G17050 | UGT78D2 |
| Br5MAT | Bra036208 | 98.82 | 0 | A09 | 1925788 | 1927140 | - | AT3G29590 | 5MAT |
| BrAT-1 | Bra030550 | 98.52 | 0 | A08 | 20600205 | 20602987 | + | AT1G03940 | Acyl-transferase family protein-1 |
| BrAT-2 | Bra034255 | 100 | 0 | A04 | 11806054 | 11806788 | - | AT3G29680 | Acyl-transferase family protein |
| Anthocyanin transporter genes | |||||||||
| BrMATE2-1 | Bra031776 | 88.51 | 0 | A09 | 36544120 | 36546929 | + | AT1G11670 | TT12 |
| BrMATE-2 | Bra027073 | 99.8 | 0 | A09 | 8642073 | 8645111 | + | AT1G61890 | TT12 |
| BrTT19-1 | Bra008570 | 100 | 0 | A10 | 11677671 | 11678470 | - | AT5G17220 | GST26,TT19 |
| BrTT19-2 | Bra023602 | 99.69 | 0 | A02 | 3117740 | 3118547 | + | AT5G17220 | TT19 |
| Other anthocyanin biosynthesis genes | |||||||||
| BrOMT1-2 | Bra011292 | 99.13 | 0 | A01 | 2896202 | 2897686 | - | AT1G77520 | OMT1 |
| BrCCR2 | Bra008438 | 100 | 0 | A02 | 14649009 | 14650738 | - | AT1G80820 | CCR2 |
| BrRNS1-1 | Bra026570 | 99.86 | 0 | A02 | 20391492 | 20392500 | + | AT2G02990 | RNS1 |
| BrCCoAMT | Bra033968 | 100 | 0 | A02 | 9548647 | 9549923 | + | AT1G67980 | CCoAMT |
| BrFLS3 | Bra029212 | 99.68 | 0 | A02 | 25974752 | 25976735 | - | AT5G63590 | ATFLS3,FLS3 |
| BrLAC17 | Bra006683 | 90.74 | 0 | A03 | 4611941 | 4613968 | - | AT5G60020 | LAC17 |
| BrOxygenase protein | Bra012691 | 100 | 0 | A03 | 22738914 | 22741827 | - | AT4G16770 | oxygenase superfamily protein |
| BrSCPL10-2 | Bra025601 | 98.52 | 0 | A04 | 7905792 | 7908695 | - | AT2G22980 | SCPL10 |
| BrBGLU10 | Bra037647 | 100 | 0 | A04 | 18448229 | 18455545 | + | AT3G60120 | BGLU10 |
| BrFLS1 | Bra022378 | 100 | 0 | A05 | 18803675 | 18805312 | + | AT3G19010 | FLS1 |
| BrIRX12 | Bra005140 | 99.73 | 0 | A05 | 3679277 | 3682262 | - | AT2G38080 | IRX12 |
| BrBGLU46-1 | Bra018969 | 99.24 | 0 | A06 | 976174 | 979392 | - | AT1G52400 | BGLU46 |
| BrOST2 | Bra024452 | 96.24 | 9.00E−83 | A06 | 16477147 | 16483736 | - | AT2G18960 | AHA1,HA1,OST2,PMA |
| BrSCPL10-1 | Bra012153 | 88.53 | 0 | A07 | 11924527 | 11938309 | + | AT2G23000 | SCPL10 |
| BrOMT1-1 | Bra012269 | 98.11 | 0 | A07 | 11175574 | 11176865 | + | AT1G21100 | OMT1 |
| BrOMT | Bra003707 | 100 | 3.00E−43 | A07 | 17900548 | 17902240 | + | AT1G76790 | O-methyltransferase family protein |
| BrCAD1-2 | Bra010819 | 99.59 | 0 | A08 | 15846126 | 15848906 | + | AT1G29690 | CAD1 |
| BrCAD1-3 | Bra010879 | 99.78 | 0 | A08 | 16130363 | 16132914 | - | AT1G28380 | CAD1 |
| BrCCoAOMT1 | Bra034600 | 100 | 0 | A08 | 11803933 | 11805048 | - | AT4G34050 | CCoAOMT1 |
| BrCAD1-1 | Bra026804 | 100 | 0 | A09 | 35535138 | 35538610 | - | AT1G14780 | CAD1 |
| BrRNS1-2 | Bra026846 | 100 | 0 | A09 | 35723513 | 35724548 | + | AT1G14220 | RNS1 |
| BrCCR5 | Bra008743 | 100 | 0 | A10 | 12472468 | 12475087 | + | AT5G14700 | CCR5 |
| BrBGLU46-2 | Bra002978 | 98.56 | 0 | A10 | 6383388 | 6385871 | - | AT5G54570 | BGLU46 |
| BrOMT1-3 | Bra003009 | 100 | 0 | A10 | 6154442 | 6160001 | + | AT5G54160 | ATOMT1,OMT1 |
| Regulatory transcription factors | |||||||||
| BrMYBR1 | Bra012149 | 100 | 0 | A07 | 11952841 | 11953731 | - | AT5G67300 | MYBR1 |
| BrMYBL2-1 | Bra016164 | 100 | 0 | A07 | 22386380 | 22387236 | - | AT1G71030 | MYB-like 2 |
| BrMYB15 | Bra001907 | 99.77 | 0 | A03 | 19315200 | 19316380 | + | AT3G23250 | MYB15 |
| BrMYB51 | Bra025666 | 97.36 | 0 | A06 | 6841688 | 6842966 | + | AT1G18570 | MYB51 |
| BrMYB51 | Bra016553 | 100 | 0 | A08 | 18248352 | 18249890 | - | AT1G18570 | MYB51 |
| BrMYB75 | Bra039763 | 92.86 | 0 | A02 | 8839008 | 8840737 | + | AT1G56650 | PAP1,MYB75 |
| BrMYB77 | Bra012910 | 100 | 0 | A03 | 21598456 | 21599337 | - | AT3G50060 | MYB77 |
| BrMYB90 | Bra004162 | 84.01 | 2.00E−111 | A07 | 20426416 | 20431671 | + | AT1G66390 | MYB90 |
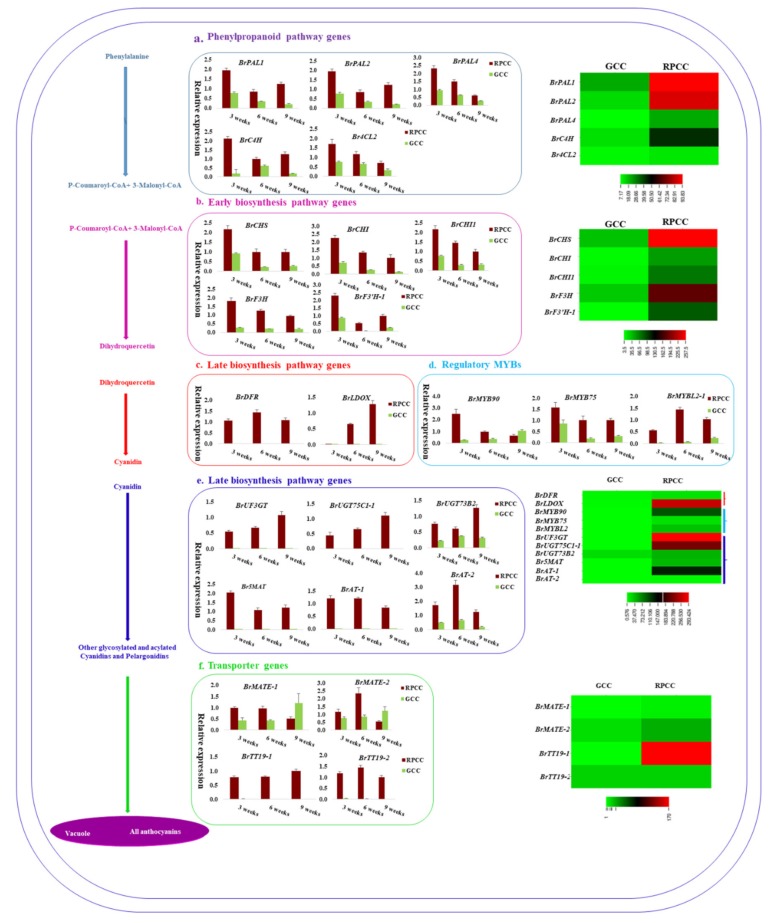
In the phenylpropanoid pathway, three PAL genes (BrPAL1, BrPAL2 and BrPAL4), BrC4H, and the 4CL homolog Br4CL2 were detected in GCC and RPCC at three developmental stages. The transcriptome data from 3-week-old plants (heat map, Figure 5a) and the qRT-PCR analyses showed that the transcript levels of these genes were much higher in RPCC than in GCC. Gene expression was also compared among stages (seedling, rosette, and heading stages) (Figure 5a). Similar to PBGs, EBGs such as BrCHS, BrCHI and BrCHI1, BrF3H, and BrF3′H-1 showed similar expression patterns in both the transcriptome analysis (heat map, Figure 5b) and qRT-PCR analyses (Figure 5b). Unlike PBGs and EBGs, the LBG BrDFR was expressed only in the RPCC at all stages, while the LBG BrLDOX was expressed at lower levels at the early stage. The BrLDOX transcript levels gradually increased from the rosette to the heading stage in RPCC (Figure 5c). The transcript levels of MYBs including BrMYB90, BrMYB75, and BrMYBL2-1 varied among different stages. BrMYB90 and BrMYB75 showed maximum transcript levels in RPCC at the seedling stage, while BrMYBL2-1 had higher transcript levels at the rosette and heading stages than at the seedling stage in RPCC (Figure 5d).
Figure 5.
Validation of anthocyanin biosynthetic genes (ABGs) detected in transcriptome data by qRT-PCR analyses of reddish purple (RPCC) and green (GCC) leaf tissue samples. (a) Phenylpropanoid pathway genes; (b) early biosynthesis pathway genes; (c) and (e) late biosynthesis pathway genes; (d) regulatory MYB genes; and (f) transporter genes. Gene expression levels were normalized against that of Actin. Error bars are based on mean of three technical replicates. Schematic representation of anthocyanin biosynthetic pathway is shown in left corner. Heatmaps in middle and right corner indicate transcript abundance of ABGs.
The downstream LBGs are involved in the acylation, glycosylation, and methylation of anthocyanins and include genes encoding acyltransferase (AT), glycosyltransferase (GT) and O-methyltransferase (OMT) [32,33]. The downstream LBGs selected from the transcriptome data for qRT validation included BrUF3GT, BrUGT75C1-1, BrUGT73B2, Br5MAT, BrAT-1, and BrAT-2. BrUF3GT, BrUGT75C1-1, and BrUGT73B2 showed increased expression while Br5MAT and BrAT-1 showed decreased expression from the seedling to heading stages in RPCC. The highest transcript level of BrAT-2 was at the rosette stage in RPCC (Figure 5e). Interestingly, all the LBGs including regulatory MYB (RM) genes showed low or no expression in GCC compared with RPCC in the transcriptome data (heat map, Figure 5e) and in the qRT-PCR analyses (Figure 5e).
Among many anthocyanin transporter genes, four B. rapa transporter genes including BrMATE-1, BrMATE-2, and BrTT19-1 showed differential expression between GCC and RPCC (heat map, Figure 5f). At the seedling and rosette stages, both BrMATE-1 and BrMATE-2 were expressed at higher levels in RPCC than in GCC. BrTT19-1 and BrTT19-2 transcript levels were high at all stages in RPCC, but at negligible or undetectable levels in GCC at all stages (Figure 5f). The remaining ABGs, showed diverse expression patterns in the qRT-PCR analyses (Figure S3). Most of the analyzed genes had higher transcript levels in RPCC than in GCC (Figure S3).
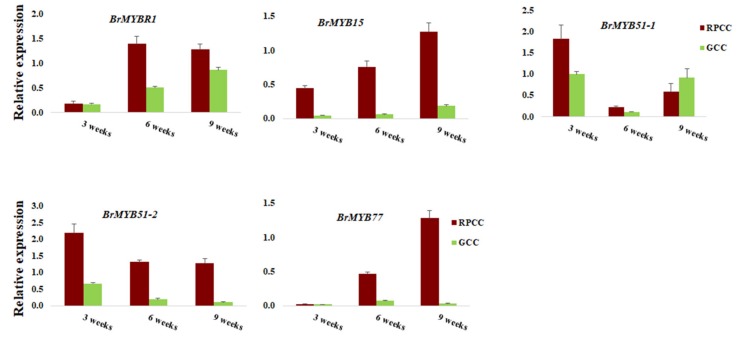
We also conducted qRT-PCR analyses for some MYB TF genes showing differences in expression levels between RPCC and GCC in transcriptome data (Table S4). Among the five selected MYB genes, BrMYB15 and BrMYB51-2 showed >1-fold expression in RPCC than in GCC at all stages, and BrMYB77 transcript levels increased from the rosette to the heading stage in RPCC but not in GCC (Figure 6).
Figure 6.
qRT-PCR validation of MYBs with high transcript abundance in transcriptome data. qRT-PCR expression values were normalized against that of Actin. Error bars are based on mean of three technical replicates.
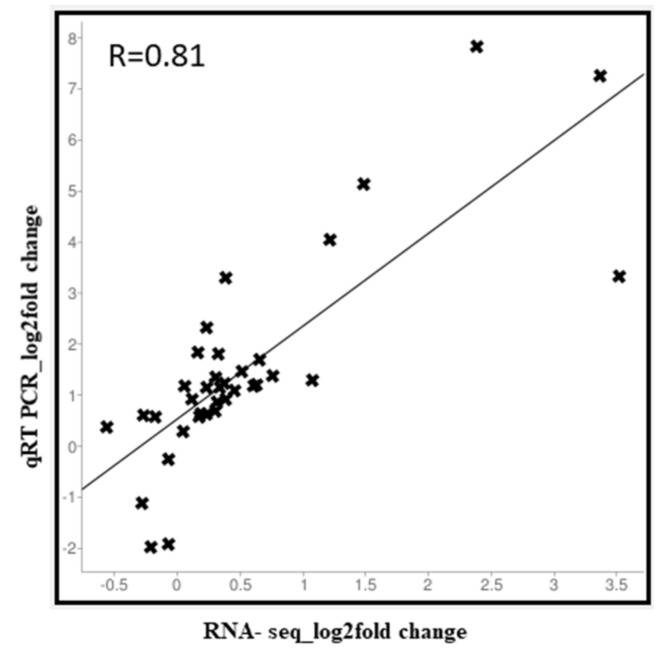
A correlation analysis revealed a strong correlation between the transcriptome data and qRT-PCR data (R = 0.81) (Figure 7). Overall, similar trends in gene expression were detected from transcriptome data and qRT-PCR analyses. The expression patterns of PBGs, EBGs, LBGs, TGs, and RMs implied that LBGs, TGs and RMs play crucial roles in anthocyanin biosynthesis during different developmental stages of RPCC.
Figure 7.
Correlation analysis between RNA-seq and qRT-PCR methods. Log2fold values of RNA-seq data (x-axis) are plotted against log2fold values of qRT-PCR (y-axis) data.
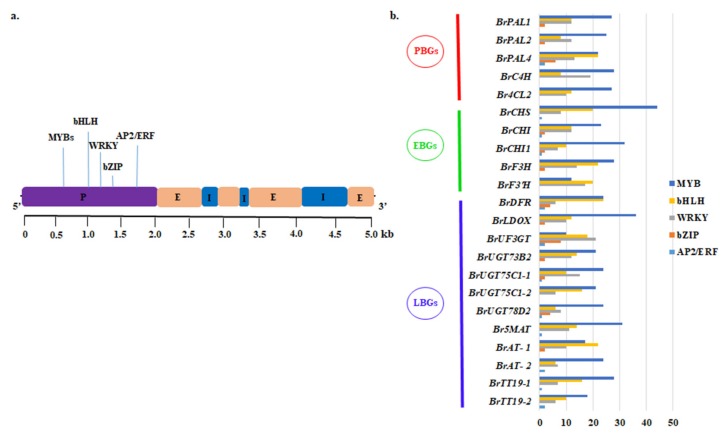
2.8. Promoter Analysis of Anthocyanin Biosynthetic Genes
Cis-regulatory elements (CREs) are binding sites for TFs in the promoters of target genes. To identify CREs, we analyzed the promoter regions of 22 important ABGs. The 2-kb region upstream of the transcription start site (TSS) was extracted and analyzed by the New PLACE program to find CRE motifs (Figure 8a). Among the predicted CREs, most were binding elements for MYB, bHLH, WRKY, bZIP and Ap2/ERF TFs (Figure 8b and Table S10). Those binding to MYB TFs were the most abundant, followed by those binding to bHLH, WRKY, bZIP and AP2/ERF TFs (Figure 8b). MYB CREs have been found in the promoters of genes related to secondary metabolism, flavonoid biosynthesis, anthocyanin biosynthesis, and plant defense (Table S10). The bHLH and bZIP CREs are known to be involved in the light response, tissue specific activation of phenylpropanoid biosynthetic genes, sugar repression, seed development, and the biosynthesis of phenylpropanoids, lignin, and flavonoids. AP2/ERF CREs are involved in functions related to the ethylene response, the jasmonate response, and secondary metabolism (Table S10). Therefore, the results of our study and other studies [34,35] indicate that MYB, and bHLH CREs regulate the expression of genes at all stages in the anthocyanin biosynthesis pathway.
Figure 8.
Cis-regulatory elements predicted in upstream promoter regions of anthocyanin biosynthetic genes (ABGs). (a) Example of plant gene organization and important cis-elements in promoter. (b) Number of each type of cis-element identified in ABGs. P, promoter; E, exon; I, intron.
2.9. Regulatory Network Analysis of Anthocyanin Biosynthesis Genes
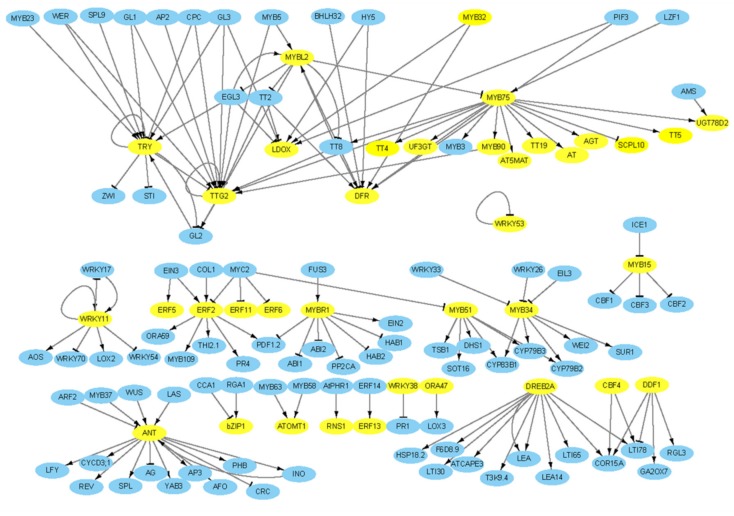
To identify the interactions among anthocyanin biosynthetic genes and the transcription factor genes including MYB, bHLH, WRKY, bZIP, and AP2/ERF (with log2fold change > 2) (Table 3 and Table 4), a putative interactive network was constructed (Figure 9). Among them, 37 B. rapa genes (yellow circles) showed 147 interactions, which could be classified into two types: activation (↓/↑) and repression (┴) (Figure 9). The gene network results showed that MYB75 interacts with a gene encoding an acyl transferase family protein, as well as MYB90, 5MAT, TT5, AGT, TT4, UGT78D2, TT19, DFR, and UF3GT, and activates them to promote anthocyanin biosynthesis. Two LBGs (DFR and LDOX) are positively regulated by TFs such as PIF3, MYB32, HY5, and TT2 (Figure 9) and DFR is also positively regulated by the TT8 TF. Besides gene–gene interactions, this network analysis revealed many other interactions among TFs and structural genes involved in various functions. Among the repressors, the MYBL2 TF interacts with MYB75, DFR, TT2, TT8, GL2, and EGL3; MYB75 represses SCPL10; both EGL3 and GL3 repress LDOX; and bHLH32 represses DFR (Figure 9). These results indicate that interactions between TFs and their target genes play a vital role in the regulation of anthocyanin biosynthesis and other metabolic functions related to the growth and development of RPCC.
Figure 9.
Gene regulatory network of anthocyanin biosynthetic genes and important transcription factors. DEGs detected from our transcriptome data are shown in yellow: other interactive genes involved in various functions including anthocyanin biosynthesis are shown in blue. Up tack (┴) indicates repressors and arrow (↓/↑) indicates activators.
3. Discussion
The red color in Chinese cabbage has been introduced through different techniques of introgression breeding. Among the introduced varieties, Xie et al. [3] introgressed the red color phenotype by crossing a Chinese cabbage variety with red color Brassica juncea through the embryo rescue technique. While in our study, RPCC and red Chinese cabbage (RCC) reported by Lee et al. [4], have been developed through interspecific-crossing between Chinese cabbage and red cabbage.
3.1. Novel Anthocyanin Pigments Responsible for the Color of RPCC
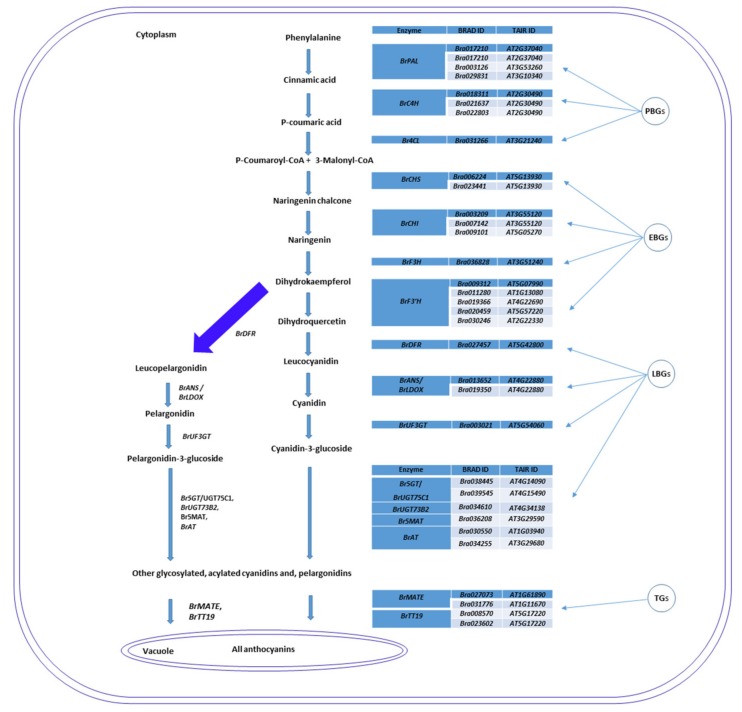
Red Chinese cabbage is an economically important variety that is rich in various secondary metabolites including anthocyanins [4]. This variety accumulates red pigments at an early stage of plant growth (seedling stage), so it is an ideal system to study the genes and regulatory TFs that are involved in regulating color (anthocyanin) accumulation at the early stages of plant development (Figure 10). Fruits and vegetables contain five main types of anthocyanins, with different frequencies of occurrence: cyanidin (50%), delphinidin (12%), pelargonidin (12%), peonidin (12%), malvidin (7%), and petunidin (7%) [36]. We detected 11 anthocyanin variants in the RPCC samples, approximately 85% of which were cyanidin isoforms. Thus, our results and those of other studies show that cyanidin derivatives are the most abundant type of anthocyanins in red Chinese cabbage [4]. In addition, the anthocyanin pigments accumulated in RPCC and in previous studies [3,4] are entirely different, indicating that the color accumulation and regulation are due to different sets of anthocyanin pigments. In accordance with the results of the HPLC analysis, we propose a model of the anthocyanin biosynthesis pathway that generates cyanidin 3-(feruloyl) diglucoside-5-(malonoyl) glucoside, and pelargonidin 3-(caffeoyl) diglucoside-5-(malonoyl) glucoside in RPCC (Figure 11). Most of the anthocyanin components identified in this study have been detected in radish or other Brassica crops but not in red Chinese cabbage [20,21,22,23,37,38,39,40,41,42,43]. Hence, 11 of the 13 pigments identified in our study were detected for the first time in RPCC (Table S1). Joo et al [28] have proved that the anthocyanin rich extract has the ability to lower the risk of vascular inflammatory diseases.
Figure 10.
Chinese cabbage at young and mature stages. (a). Green (GCC) and (b). reddish purple (RPCC).
Figure 11.
Schematic representation of anthocyanin biosynthetic pathway based on anthocyanin pigments and important anthocyanin biosynthetic genes (ABGs) identified from transcriptome data.
3.2. Anthocyanin Biosynthesis Genes Are Differentially Regulated in RPCC
Transcriptome sequencing is an advanced NGS technique that can be used to predict novel genes, gene function, and genome evolution. Comparative transcriptome sequencing between two different phenotypes, GCC and RPCC, revealed differences in the expression levels of genes involved in anthocyanin biosynthesis and the regulation of this pathway [26,44]. The transcriptome sequencing of RPCC and GCC at the seedling stage revealed 3345 DEGs, which included unique genes with unknown functions and TF genes. About 255 DEGs were involved in various functions related to phenylpropanoids, lignins, flavonols, and anthocyanins. Further qRT-PCR analyses showed that PBGs and EBGs were expressed at levels 0.5- to 1.0-fold higher at the seedling stage than at the other two stages (rosette and heading) in RPCC. Many of these genes encoded proteins involved in the early phases of anthocyanin biosynthesis [7,45,46]. Our results indicate that BrPAL, BrPAL2, BrPAL4, BrC4H, Br4CL2, BrCHS, BrCHI, BrCHI1, BrF3H, and BrF3′H-1 may be involved in the early phase of anthocyanin biosynthesis (i.e., the conversion of phenylalanine to dihydroquercetin) in RPCC. As shown in Figure 5e, the important LBGs BrDFR and BrLDOX, whose encoded products catalyze the conversion of dihydroquercetin to cyanidin at the late stage of anthocyanin biosynthesis, were expressed in RPCC at all stages, but not in GCC at any stage. Previous studies have shown that DFR plays crucial roles in anthocyanin accumulation in many plant species under different abiotic stress conditions [47,48]. Similarly, in Arabidopsis, sucrose and jasmonic acid have been shown to induce LBGs such as DFR, LDOX, and UF3GT, leading to anthocyanin accumulation [45,49]. Interestingly, we detected high transcript levels of the LBGs BrDFR, BrLDOX, and BrUF3GT in RPCC but not in GCC, indicating that anthocyanin biosynthesis occurs in RPCC under normal growth conditions without induction by external factors such as sugars or hormones.
Some downstream LBGs are involved in p-coumaroylation (At3AT1: At1g03940), glucosylation (UGT75C1:At4g14090) and malonylation (At5MAT: At3g29590) [33,50,51,52]; the orthologs of these genes (BrAT-1, BrUGT75C1-1, and Br5MAT) were expressed only in RPCC. Accordingly, the HPLC analyses detected p-coumaroyl (At3AT1: At1g03940) diglucoside (UGT75C1: At4g14090), indicating that anthocyanins have been modified as reported previously [33]. Our qRT-PCR analyses showed that the transcript levels of BrUGT73B2 and BrAT-2, whose encoded proteins catalyze p-coumaroylation and glucosylation, respectively, were much higher in RPCC than in GCC at the rosette and heading stages. TT19 encodes a transporter involved in the movement and accumulation of anthocyanins in the Brassicaceae [53,54]. In this study, two TT19 paralogs, BrTT19-1 and BrTT19-2, which have a common ortholog in A. thaliana (AtTT19: AT5G17220), showed very high transcript levels in RPCC, indicating that transport of anthocyanins from the cytosol to the vacuole is an important process in RPCC.
3.3. Differential Expression of MYBs Regulates Reddish Purple Color Accumulation
In general, ABGs are controlled by various TFs, including the MYB, bHLH, and WD40 TFs that make up the most well-known complex in plants, the MBW complex [55,56]. A study using the transcriptome approach in red Chinese cabbage identified that the anthocyanin pathway regulating genes are TT8 (Bra037887) and PAP1 (c3563g1i2) [3]. Most TFs that are known to regulate anthocyanins are MYB TFs. A study on grapes reported that the MYBA transcription factor regulates the anthocyanin biosynthesis pathway through controlling the expression of UFGT [57]. Because TFs, especially MYB, are known to be important for controlling expression of ABGs, we searched our transcription data for TF sequences. As a result, we identified 25 TF genes with log2fold change > 3: 12 TF genes with log2fold change > 4, five TF genes with log2fold change > 5 and > 6, and one TF gene with log2fold change > 10 (Table 3). They included two important MYB genes: MYB90 (log2fold change, 11.3) and MYB75 (log2fold change, 6.8), which are homologs of PRODUCTION OF ANTHOCYANIN PIGMENT 2 (PAP2) and PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1). These genes showed similar transcript levels in qRT-PCR and transcriptome analyses, and their high expression levels in RPCC suggested that they might be involved in the positive regulation of LBGs during anthocyanin biosynthesis [13,58]. We identified duplicate copies of MYBL2, Bra016164 (BrMYBL2-1) and Bra007957 (BrMYBL2-2), in B. rapa, which are orthologs of A. thaliana AtMYBL2 (AT1G71030). The transcript levels of both BrMYBL2-1 and BrMYBL2-2 were very high, as revealed by transcriptome analysis (log2fold change of 5.5 and 4.06, respectively) and qRT-PCR analysis. A previous study detected a similar expression pattern of MYBL2 in B. rapa, indicative of its role as a positive regulator [59]. However, other studies on the Brassicaceae have demonstrated that MYBL2 can function as a negative regulator of anthocyanin biosynthesis [60,61]. Functional characterization of BrMYBL2-1 and Bra007957 (BrMYBL2-2) will clarify the molecular mechanisms of these genes in RPCC. Our results also showed that MYB binding motifs are highly conserved not only in LBGs but in most of the analyzed ABGs.
4. Materials and Methods
4.1. Plant Material and Sample Collection
The RPCC was developed (through introgression hybridization) and registered by the Kwonnong Seed Company (Cheongju, S. Korea) as described by Lee et al. [4]. The lines used in this study, green Chinese cabbage (GCC) and RPCC, belong to B. rapa L. ssp. pekinensis and were selected on the basis of their distinct color phenotypes (Figure 1). Seeds of GCC and RPCC were germinated in a growth chamber under a 16-h light/8-h dark photoperiod at 24 °C. Leaf samples (innermost and outermost leaves) were collected from three biological replicates at three growth stages: the seedling, rosette, and heading stages (at 3-, 6-, and 9-weeks-old, respectively). The leaf samples were stored at −70 °C until further analysis.
4.2. Anthocyanin Extraction and HPLC Analysis
The total anthocyanin content of freeze-dried outer and inner leaf tissues of 9-week-old GCC and RPCC was determined by HPLC and LC-MS/MS, as described previously [37]. Each 100-mg lyophilized leaf sample was mixed with 2 mL water:formic acid (95:5 v/v) followed by 5 min vortexing and 20 min sonication. The sample was centrifuged at 9200× g for 15 min at 4 °C and the supernatant was filtered through a 0.45-µm PTFE hydrophilic syringe filter. From this filtrate, 10 μL was used for estimating anthocyanin content. The sample was injected into an Agilent 1200 series HPLC connected to a 4000 Q-Trap LC-ECI-MS/MS system. A Synergy 4µL Polar-RP 80A column (250 × 4.6 mm i.d., particle size 4 µm; Phenomenex, Torrance, CA, USA) with a Security Guard Cartridge (AQ C18, 4 × 3.0 mm KJO-4282; Phenomenex, Torrance, CA, USA) were used. Anthocyanin pigments were detected at 520 nm. The oven temperature was set to 40 °C. The composition of the mobile phase was as follows: solvent A: water:formic acid (95:5 v/v), and solvent B (acetonitrile:formic acid, 95:5 v/v). The gradient conditions were as follows: 0–8 min, 8–13 min, 13% solvent B; 13–20 min, 20–23 min, 17% solvent B; 23–30 min, 30–40 min, 20% solvent B; 40–40.1 min, 5% solvent B; and 40.1–50 min, 5% solvent B. The anthocyanin concentration in each sample was measured by comparison of the area of each peak with that of the external standard (cyanidin-3-O-glucoside) on the HPLC chromatogram. Mean ± SD values were calculated from the three replicates of each sample.
4.3. Sequence Pre-Processing and Assembly
For transcriptome analysis, total RNA was extracted from leaf tissues of 3-week-old plants using an RNeasy Mini kit (Qiagen, Valencia, CA, USA) and sequenced with the Illumina Hi-seq2000 platform by SEEDERS Inc. (Korea). The sequence data was submitted to the NCBI database and are available under the accession number PRJNA612946. The raw reads were trimmed using the Dynamic-Trim and Length-Sort programs of the Solexa QA [62] package. Based on the Dynamic-Trim (phred score ≥ 20) and Length-Sort (short read length ≥ 25bp) parameters, clean reads were obtained. The clean reads were assembled according to the protocols of Velvet (version 1.2.08) and Oases (version 0.2.08) software [63]. The optimal k-mer was selected based on the max length, average length, and N50 according to the total length of the assembled sequence.
4.4. Mapping and Annotation of Transcripts
To identify gene function, the transcripts were used as queries in BlastX searches against the amino acid sequences in the BRAD [64] and KEGG databases with the following parameters: filter criterion: e-value ≤ 1e−10, best hits. Mapping was performed using Bowtie2 (v2.1.0) software with the following limitation: mismatch ≤ 2 bp, computed by the penalty method) [65]. Transcript levels were normalized using the R package of DESeq [66], and this software was also used to calculate the gene expression values for each sample with data deviation.
4.5. Identification of Transcription Factors
To identify TFs, sequences of all B. rapa 4127 TFs were downloaded from the Plant Transcription Factor Database [67] (http://planttfdb.cbi.pku.edu.cn/). The total assembled transcripts were compared and analyzed using BlastX software with the parameters e-value ≤ 1e−50 and identity ≥ 50, and annotated.
4.6. Prediction of Differentially Expressed Genes
Differentially expressed genes (DEGs) were defined as those with at least a log2fold difference in transcript levels between the RPCC and GCC samples. Up-regulated genes were those with log2 fold-change greater than 1, and down-regulated genes were those with log2 fold-change less than −1 [22].
4.7. Expression Analyses
Leaf samples collected from 3-, 6-, and 9-week-old plants were collected and immediately frozen in liquid nitrogen. Total RNA was extracted using an RNeasy Mini kit (Qiagen). cDNA was synthesized using a Reverse Transcription System kit (Promega, Madison, WI, USA). The resulting cDNA was used as a template for qRT-PCR analyses, which were conducted with the CFX96 Real-Time system (Bio-Rad, Hercules, CA, USA). qRT-PCR analyses were conducted for 60 anthocyanin biosynthetic genes (ABGs) with three biological replicates and three technical replicates using the following conditions: 95 °C for 3 min; 39 cycles of 95 °C for 15 s and 58 °C for 20 s. The relative expression levels were determined by normalizing the data with the comparative Ct method 2−[ΔΔCt] [68] using the Actin gene as a reference.
4.8. Gene Ontology Annotation and KEGG Enrichment Analyses
Gene ontology (GO) [69] alignments were performed using total transcripts and ‘GO DB’ with the threshold of ‘counts ≥ 1’ and ‘GO depth’ set to 3. Genes were classified into three functional categories, BP (Biological Process), CC (Cellular Component), and MF (Molecular Function). Then, gene annotation (filter criterion: e-value ≤ 1e−10, best hits) was performed through comparisons with amino acid sequences in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using BLASTX.
To predict the functional characteristics of up- and down-regulated genes, we first computed p-values based on Fisher’s exact test; these values were taken as indicators of significance of the gene in the respective function. Further KEGG enrichment analysis was performed using the KOBAS online tool (http://kobas.cbi.pku.edu.cn/kobas3) [70]. These analyses provided pathway annotations for the transcripts.
4.9. Identification of Cis-Regulatory Elements
The 2-kb region upstream from the transcription start site of selected genes was screened to identify cis-regulatory motifs using the New PLACE web tool [71].
4.10. Network Analysis
We carried out network analysis to construct an active gene-to-gene regulatory network of genes positively correlated with the anthocyanin biosynthetic pathway. The STRING 10.0 database (http://string-db.org/) was used to obtain an interaction network of these genes in B. rapa [72], according to orthologous genes in A. thaliana. Every link has a score from 0 to 1, where 1 is considered as the highest confidence link for reconstruction [73].
5. Conclusions
In conclusion, 11 of the 13 pigments detected in RPCC are reported for the first time for this variety. Analyses of the transcriptome data from two varieties at the seedling stage revealed many unique transcripts including DEGs and TF genes that are involved in a multitude of functions in growth and development. Our results show that many DEGs between the red and green varieties are involved in the biosynthesis of secondary metabolites such as phenylpropanoids, lignins, flavonoids, and anthocyanins, and in the regulation of these biosynthetic pathways. Further qRT-PCR expression analyses confirmed that ABGs and many TFs play essential roles in anthocyanin biosynthesis. The gene-to-gene interaction network illustrates the possible regulatory mechanism of MYBs with ABGs during anthocyanin biosynthesis in RPCC. Overall, our study describes the pigments in RPCC, identifies the important anthocyanin biosynthetic genes and TF genes that control the anthocyanin biosynthesis pathway, and proposes a model for the possible interaction mechanism between ABGs and TFs.
Acknowledgments
We thank Jennifer Smith, from Liwen Bianji, Edanz Group China (www.liwen bianji.cn/ac), for editing the English text of a draft of this manuscript.
Abbreviations
| ABGs | Anthocyanin biosynthetic genes |
| ABP | Anthocyanin biosynthetic pathway |
| AP2/ERF | Apetala 2/ethylene response factor |
| AT | Acyltransferases |
| bHLH | Basic helix-loop-helix |
| DEGs | Differentially expressed genes |
| DFR | Dihydroflavonol-4-reductase |
| EBGs | Early biosynthetic genes |
| GCC | Green Chinese cabbage |
| HPLC | High performance liquid chromatography |
| LBGs | Late biosynthetic genes |
| LDOX | Leucoanthocyanidin dioxygenases |
| MATE | Multidrug and toxic compound extrusion |
| MYB | Myeloblastosis |
| 5MAT | 5-O-glucoside-6-O-malonyltransferase |
| PBGs | Phenylpropanoid biosynthetic genes |
| qRT-PCR | Quantitative real-time polymerase chain reaction |
| RPCC | Reddish purple Chinese cabbage |
| RPKM | Reads per kilo base per million mapped reads |
| TT19 | Transparent Testa 19 |
| UF3GT | UDP- glucose: flavonoid 3-O-glucosyltransferase |
| UGT75c1 | UDP-glucosyltransferase 75c1 |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/8/2901/s1. References [74,75,76,77,78,79,80,81,82,83,84,85] are cited in Supplementary Materials. Figure S1. Statistics of non-redundant transcripts predicted in transcriptome data. (a) Length distribution of transcripts. (b) Transcripts annotated in public databases. Figure S2 MA plot of DEGs between green (G) vs. reddish purple (RP) Chinese cabbage identified from transcriptome data. X-axis shows log2fold change and y-axis shows differentially expressed genes. Red and green dots indicate up-regulated and down-regulated genes, respectively. Figure S3. Additional ABGs showing differential expression in transcriptome data validated by qRT-PCR. Expression values were normalized against that of Actin. Error bars are based on mean of three technical replicates. Table S1. Total anthocyanin pigments identified in reddish purple Chinese cabbage leaf samples. Table S2. Total up-regulated DEG transcripts between RPCC and GCC with annotations from BRAD and TAIR databases. Table S3. Total down-regulated DEG transcripts between RPCC and GCC with annotations from BRAD and TAIR databases. Table S4. Transcription factor genes up-regulated between RPCC and GCC samples, as identified from transcriptome data. Table S5. Transcription factor genes down-regulated between RPCC and GCC, as identified from transcriptome data. Table S6. Gene ontology (GO) annotation of DEG transcripts predicted from transcriptome data. GO annotations are shown for up-regulated genes (left) and down-regulated genes (right). Table S7. KEGG pathway annotation of DEG transcripts predicted in transcriptome data. KEGG annotations are shown for up-regulated genes (left) and down-regulated genes (right). Table S8. Genes identified from transcriptome data that are involved in phenylpropanoid and anthocyanin pathways with expression values and GO and KEGG annotations. Table S9. List of primer sequences used for qRT-PCR of anthocyanin biosynthetic genes. Table S10. Cis-regulatory elements (CREs) identified in upstream promoter regions of anthocyanin biosynthetic genes.
Author Contributions
Conceptualization, J.J.R., S.R.C., S.S.C., S.Y.Y., M.-H.O., S.U.P., S.-J.K., J.L., O.H.K. and Y.P.L.; Data curation, J.J.R., S.H.O.; Formal analysis, J.J.R., S.S.C., S.Y.Y., S.H.O., H.K., C.Y.L., S.U.P., K.S.-J.; Funding acquisition, S.R.C. and YPL; Investigation, J.J.R., S.Y.Y., H.K., C.Y.L., J.L. and O.H.K.; Methodology, J.J.R., S.S.C., S.H.O., H.K., C.Y.L., J.L. and O.H.K.; Project administration, S.R.C. and Y.P.L.; Resources, S.R.C., J.L., O.H.K. and Y.P.L.; Software, J.J.R. and M.-S.K.; Supervision, S.R.C. and Y.P.L.; Validation, J.J.R. and S.S.; Writing–original draft, J.J.R.; Writing–review & editing, J.J.R., S.R.C., S.S.C., S.Y.Y., M.-S.K.,S.S., M.-H.O., S.U.P., S.-J. K., J.L., O.H.K. and Y.P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, and Forestry (IPET) through Golden Seed Project (213006-05-4-SBD30, 213006-05-4-SB110), funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA), Ministry of Oceans and Fisheries (MOF), Rural Development Administration (RDA) and Korea Forest Services (KFS).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Katche E., Quezada-Martinez D., Katche E.I., Vasquez-Teuber P., Mason A.S. Interspecific hybridization for Brassica crop improvement Crop Breeding. Genet. Genom. 2019;1:e190007 [Google Scholar]
- 2.Wang G.-X., Lv J., Zhang J., Han S., Zong M., Guo N., Zeng X.-Y., Zhang Y.-Y., Wang Y.-P., Liu F. Genetic and epigenetic alterations of Brassica nigra introgression lines from somatic hybridization: A resource for cauliflower improvement. Front. Plant Sci. 2016;7:1258. doi: 10.3389/fpls.2016.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie L., Li F., Zhang S., Zhang H., Qian W., Li P., Zhang S., Sun R. Mining for candidate genes in an introgression line by using RNA sequencing: The anthocyanin over accumulation phenotype in Brassica. Front. Plant Sci. 2016;7:1245. doi: 10.3389/fpls.2016.01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H., Oh I.-N., Kim J., Jung D., Cuong N.P., Kim Y., Lee J.C., Kwon O., Park S.U., Lim Y.P., et al. Phenolic compound profiles and their seasonal variations in new red-phenotype head-forming Chinese cabbages. Lwt-Food Sci. Tech. 2018;90:433–439. doi: 10.1016/j.lwt.2017.12.056. [DOI] [Google Scholar]
- 5.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka Y., Sasaki N., Ohmiya A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Tikunov Y., Schouten R.E., Marcelis L.F.M., Visser R.G.F., Bovy A. anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018;6:52. doi: 10.3389/fchem.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petroni K., Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Sablowski R.W.M., Moyano E., Cullanez-Macia F.A., Schuch W., Martin C., Bevan M. A flower specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J. 1994;13:128–137. doi: 10.1002/j.1460-2075.1994.tb06242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd A., Brockman A., Aguirre L., Campbell A., Bean A., Cantero A., Gonzalez A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017;58:1431–1441. doi: 10.1093/pcp/pcx075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 12.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez A., Zhao M., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 14.Xie X., Xie X.B., Li S., Zhang R.F., Zhao J., Chen Y.C., Zhao Q., Yao Y.X., You C.X., Zhang X.S., et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Env. 2012;35:1884–1897. doi: 10.1111/j.1365-3040.2012.02523.x. [DOI] [PubMed] [Google Scholar]
- 15.Verweij W., Spelt C.E., Bliek M., de Vries M., Wit N., Faraco M. Functionally similar WRKY proteins regulate vacuolar acidification in Petunia and hair development in Arabidopsis. Plant Cell. 2016;28:786–803. doi: 10.1105/tpc.15.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An J., Qu F., Yao J., Wang X.N., You C.X., Wang X.F., Hao Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017;4:1702. doi: 10.1038/hortres.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.An J.P., Yao J.F., Xu R.R., You C.X., Wang X.F., Hao Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Env. 2018;41:2678–2692. doi: 10.1111/pce.13393. [DOI] [PubMed] [Google Scholar]
- 18.Guo N., Wu J., Zheng S., Cheng F., Liu B., Liang J., Cui Y., Wang X. Anthocyanin profile characterization and quantitative trait locus mapping in zicaitai (Brassica rapa L. ssp.chinensis var. purpurea) Mol. Breed. 2015;35:113. doi: 10.1007/s11032-015-0237-1. [DOI] [Google Scholar]
- 19.He Q., Zhang Z., Zhang L. Anthocyanin accumulation, antioxidant ability and stability, and a transcriptional analysis of anthocyanin biosynthesis in purple heading Chinese cabbage (Brassica rapa L. ssp. pekinensis) J. Agricul. Food Chem. 2016;64:132–145. doi: 10.1021/acs.jafc.5b04674. [DOI] [PubMed] [Google Scholar]
- 20.Oh Y.S., Lee J.H., Yoon S.H., Oh C.H., Choi D.S., Choe E., Jung M.Y. Characterization and quantification of anthocyanins in grape juices obtained from the grapes cultivated in Korea by HPLC/DAD, HPLC/MS, and HPLC/MS/MS. J. Food Sci. 2008;73:C378–C389. doi: 10.1111/j.1750-3841.2008.00756.x. [DOI] [PubMed] [Google Scholar]
- 21.Moreno D.A., Perez-Balibrea S., Ferreres F., Gil-Izquierdo A., Garcia-Viguera C. Acylated anthocyanins in Broccoli sprouts. Food Chem. 2010;123:358–363. doi: 10.1016/j.foodchem.2010.04.044. [DOI] [Google Scholar]
- 22.Jeon J., Kim J.K., Kim H., Kim Y.J., Park Y.J., Kim S.J., Kim C., Park S.U. Transcriptome analysis and metabolic profiling of green and red kale (Brassica oleracea var. acephala) seedlings. Food Chem. 2018;241:7–13. doi: 10.1016/j.foodchem.2017.08.067. [DOI] [PubMed] [Google Scholar]
- 23.Jeon J., Lim C.J., Kim J.K., Park S.U. Comparative metabolic profiling of green and purple pakchoi (Brassica rapa Subsp. Chinensis) Molecules. 2018;23:1613. doi: 10.3390/molecules23071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong C., Wang X., Yu J., Wu J., Li W., Huang J., Dong C., Hua W., Liu S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013;14:689. doi: 10.1186/1471-2164-14-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S., Zhou G., Huang X., Hu J., Wang B., Lin C., Li X., Jia Y., Wang A. Transcriptome analysis of non-heading Chinese cabbage under heat stress by RNA-seq and marker identification. Euphytica. 2017;213:109. doi: 10.1007/s10681-017-1891-7. [DOI] [Google Scholar]
- 26.Zhang L., Xu B., Wu T., Yang Y., Fan L., Wen M., Sui J. Transcriptomic profiling of two pak choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in anthocyanin biosynthetic pathway. BMC Genom. 2017;18:288. doi: 10.1186/s12864-017-3677-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eom S.H., Baek S.-A., Kim J.K., Hyun T.K. Transcriptome analysis in Chinese cabbage (Brassica rapa ssp pekinensis) provides the role of glucosinolate metabolism in response to drought stress. Molecules. 2018;23:1186. doi: 10.3390/molecules23051186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joo H.K., Choi S., Lee Y.R., Lee E.O., Park M.S., Park K.B., Kim C.S., Lim Y.P., Park J.T., Jeon B.H. Anthocyanin-rich extract from red Chinese cabbage alleviates vascular inflammation in endothelial cells and Apo E–/–mice. Int. J. Mol. Sci. 2018;19:816. doi: 10.3390/ijms19030816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren J., Liu Z., Niu R., Feng H. Mapping of Re, a gene conferring the red leaf trait in ornamental kale (Brassica oleracea L. var. acephala) Plant Breed. 2015;134:494–500. doi: 10.1111/pbr.12286. [DOI] [Google Scholar]
- 30.Outchkourov N., Karlova R., Hoelscher M., Schrama X., Blilou I., Jongedijk E., Simon C.D., van Dijk A.D.J., Bosch D., Hall R.D., et al. Transcription factor-mediated control of anthocyanin biosynthesis in vegetative tissues. Plant Physiol. 2018;176:1862–1878. doi: 10.1104/pp.17.01662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bu C., Zhang Q., Zeng J., Cao X., Hao Z., Qiao D., Cao Y., Xu H. Identification of a novel anthocyanin synthesis pathway in the fungus Aspergillus sydowii H-1. BMC Genom. 2020;21:29. doi: 10.1186/s12864-019-6442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng J., Wei G., Zhou H., Gu C., Vimolmangkang S., Liao L., Han Y. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014;166:1044–1058. doi: 10.1104/pp.114.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki N., Nishizaki Y., Ozeki Y., Miyahara T. The role of acyl-glucose in anthocyanin modifications. Molecules. 2014;19:18747–18766. doi: 10.3390/molecules191118747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartmann U., Sagasser M., Mehrtens F., Stracke R., Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, bZIP, and bHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 2005;57:155–171. doi: 10.1007/s11103-004-6910-0. [DOI] [PubMed] [Google Scholar]
- 35.Yu M., Man Y., Wang Y. Light- and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int. J. Mol. Sci. 2019;20:5228. doi: 10.3390/ijms20205228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castañeda-Ovando A., Pacheco-Hernández M.L., Páez-Hernández E., Rodríguez J.A., Galán-Vidal C.A. Chemical studies of anthocyanins: A review. Food Chem. 2009;113:859–871. doi: 10.1016/j.foodchem.2008.09.001. [DOI] [Google Scholar]
- 37.Park C.H., Yeo H.J., Kim N.S., Eun P.Y., Kim S.-J., Arasu M.V., Al-Dhabi N.A., Park S.Y., Kim J.K., Park S.U. Metabolic profiling of pale green and purple kohlrabi (Brassica oleracea var. gongylodes) Appl. Biol. Chem. 2017;60:249–257. doi: 10.1007/s13765-017-0274-z. [DOI] [Google Scholar]
- 38.Wu X., Prior R.L. Identification and characterization of anthocyanins by High-performance liquid chromatography–electrospray ionization–tandem mass spectrometry in common foods in the United States: Vegetables, nuts, and grains. J. Agric. Food Chem. 2005;53:3101–3113. doi: 10.1021/jf0478861. [DOI] [PubMed] [Google Scholar]
- 39.Charron C.S., Clevidence B.A., Britz S.J., Novotny J.A. Effect of dose size on bioavailability of acylated and non-acylated anthocyanins from red cabbage (Brassica oleracea L. var. Capitata) J. Agric. Food Chem. 2007;55:5354–5362. doi: 10.1021/jf0710736. [DOI] [PubMed] [Google Scholar]
- 40.Park N.I., Xu H., Li X., Jang I.H., Park S., Ahn G.H., Lim Y.P., Kim S.J., Park S.U. Anthocyanin accumulation and expression of anthocyanin biosynthetic genes in radish (Raphanus sativus) J. Agric. Food Chem. 2011;59:6034–6039. doi: 10.1021/jf200824c. [DOI] [PubMed] [Google Scholar]
- 41.Sun J., Xiao Z., Lin L.-Z., Lester G.E., Wang Q., Harnly J.M., Chen P. Profiling Polyphenols in Five Brassica Species Microgreens by UHPLC-PDA-ESI/HRMSn. J. Agric. Food Chem. 2013;61:10960–10970. doi: 10.1021/jf401802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu J., Liu W., Yuan L., Guan W.Q., Brennan C.S., Zhang Y.Y., Zhang J., Wang Z.D. The influence of postharvest UV-C treatment on anthocyanin biosynthesis in fresh-cut red cabbage. Sci. Rep. 2017;7:5232. doi: 10.1038/s41598-017-04778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami G., Nath U.K., Park J.-I., Hossain M.R., Biswas M.K., Kim H.T., Kim H.R., Nou I.S. Transcriptional regulation of anthocyanin biosynthesis in a high-anthocyanin resynthesized Brassica napus cultivar. J. Biol. Res. 2018;25:19. doi: 10.1186/s40709-018-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuan Y., Chiu L.-W., Li L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta. 2009;230:1141–1153. doi: 10.1007/s00425-009-1013-4. [DOI] [PubMed] [Google Scholar]
- 45.Solfanelli C., Poggi A., Loreti E., Alpi A., Perata P. Sucrose-specific induction of the anthocyanin biosynthetic path-way in Arabidopsis. Plant Physiol. 2006;140:637–646. doi: 10.1104/pp.105.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo N., Han S., Zong M., Wang G., Zheng S., Liu F. Identification and differential expression analysis of anthocyanin biosynthetic genes in leaf color variants of ornamental kale. BMC Genom. 2019;20:564. doi: 10.1186/s12864-019-5910-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H., Fan W., Li H., Yang J., Huang J., Zhang P. Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PLoS ONE. 2013;8:e78484. doi: 10.1371/journal.pone.0078484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed N.U., Park J.I., Jung H.J., Yang T.J., Hur Y., Nou I.S. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica Rapa. Gene. 2014;550:46–55. doi: 10.1016/j.gene.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Li T., Jia K.P., Lian H.L., Yang X., Li L., Yang H.Q. Jasmonic acid enhancement of anthocyanin accumulation is de-pendent on phytochrome A signaling pathway under far-red light in Arabidopsis. Biochem. Biophys. Res. Commun. 2014;454:78–83. doi: 10.1016/j.bbrc.2014.10.059. [DOI] [PubMed] [Google Scholar]
- 50.Tohge T., Nishiyama Y., Hirai M.Y., Yano M., Nakajima J., Awazuhara M., Inoue E., Takahashi H., Goodenowe D.B., Kitayama M., et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- 51.Luo J., Nishiyama Y., Fuell C., Taguchi G., Elliott K., Hill L., Tanaka Y., Kitayama M., Yamazaki M., Bailey P., et al. Convergent evolution in the BAHD family of acyl transferases: Identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana. Plant J. 2007;50:678–695. doi: 10.1111/j.1365-313X.2007.03079.x. [DOI] [PubMed] [Google Scholar]
- 52.D’Auria J.C., Reichelt M., Luck K., Svatos A., Gershenzon J. Identification and characterization of the BAHD acyltransferase malonyl CoA: Anthocyanidin 5-O-glucoside-6’’-O-malonyltransferase (At5MAT) in Arabidopsis. Thaliana. FEBS Lett. 2007;581:872–878. doi: 10.1016/j.febslet.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y., Li H., Huang J.-R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant. 2012;5:387–400. doi: 10.1093/mp/ssr110. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y., Tang Y., Zhang M., Cai F., Qin J., Wang Q., Liu C., Wang G., Xu L., Yang L., et al. Molecular cloning and functional characterization of a glutathione S-transferase involved in both anthocyanin and proanthocyanidin accumulation in Camelina sativa (Brassicaceae) Genet. Mol. Res. 2012;11:4711–4719. doi: 10.4238/2012.September.25.4. [DOI] [PubMed] [Google Scholar]
- 55.Ramsay N.A., Glover B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Zhang B., Chopra D., Schrader A., Hülskamp M. Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J. Exp. Bot. 2019;70:3197–3209. doi: 10.1093/jxb/erz155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kobayashi S., Ishimaru M., Hiraoka K., Honda C. Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta. 2002;215:924–933. doi: 10.1007/s00425-002-0830-5. [DOI] [PubMed] [Google Scholar]
- 58.Borevitz J.O., Xia Y.J., Blount J., Dixon R.A., Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mushtaq M.A., Pan Q., Chen D., Zhang Q., Ge X., Li Z. Comparative leaves transcriptome analysis emphasizing on accumulation of anthocyanins in Brassica: Molecular regulation and potential interaction with photosynthesis. Front. Plant Sci. 2016;7:311. doi: 10.3389/fpls.2016.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsui K., Umemura Y., Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008;55:954–967. doi: 10.1111/j.1365-313X.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- 61.Zhang X., Zhang K., Wu J., Guo N., Liang J., Wang X., Cheng F. QTL-Seq and sequence assembly rapidly mapped the gene BrMYBL21 for the purple trait in Brassica rapa. Sci. Rep. 2020;10:2328. doi: 10.1038/s41598-020-58916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox M.P., Peterson D.A., Biggs P.J. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schulz M.H., Zerbino D.R., Vingron M., Birney E. Oases: Robust denovo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics. 2012;28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun J.H., Bancroft I., Cheng F. The genome of the mesopolyploid crop species Brassica Rapa. Nat. Genet. 2011;28:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 65.Langmead B., Salizberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin J.P., Tian F., Yang D.C., Meng Y.Q., Kong L., Luo J.C., Gao G. Plant TFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S., Kong L., Gao G., Li C.Y., Wei L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim M.-S., Hong S., Devaraj S.P., Im S., Kim J.-R., Lim Y.P. Identification and characterization of the leaf specific networks of inner and rosette leaves in Brassica rapa. Biochem. Biophys. Res. Commun. 2017;490:821–826. doi: 10.1016/j.bbrc.2017.06.123. [DOI] [PubMed] [Google Scholar]
- 74.Wong D.C.J., Gutierrez R.L., Gambetta G.A., Castellarin S.D. Genome-wide analysis of cis-regulatory element structure and discovery of motif-driven gene co-expression networks in grapevine. DNA Res. 2017;24:311–326. doi: 10.1093/dnares/dsw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chakravarthy S., Tuori R.P., Dascenzo M.D., Fobert P.R., Despres C., Martin G.B. The tomato transcription factor Pti4 regulates defence-related gene expression via GCC box and non-GCC box cis-elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abe H., Urao T., Ito T., Seki M., Shinozaki K., Shinozaki K.-Y. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abe H., Shinozaki K.-Y., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–1868. doi: 10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Planchais S., Perennes C., Glab N., Mironov V., Inze D., Bergounioux C. Characterization of cis-acting element involved in cell cycle phase-independent activation of Arath; CycB1;1 transcription and identification of putative regulatory proteins. Plant Mol. Biol. 2002;50:111–127. doi: 10.1023/A:1016018711532. [DOI] [PubMed] [Google Scholar]
- 79.Gubler F., Kalla R., Roberts J.K., Jacobsen J.V. Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pl alpha-amylase gene promoter. Plant Cell. 1995;7:1879–1891. doi: 10.2307/3870195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tamagnone L., Merida A., Parr A., Mackay S., Culianez-Macia F.A., Roberts K., Martin C. (1998). The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell. 1998;10:135–154. doi: 10.1105/tpc.10.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Baranowskij N., Frohberg C., Prat S., Willmitzer L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. Embo J. 1994;13:5383–5392. doi: 10.1002/j.1460-2075.1994.tb06873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yamamoto S., Nakano T., Suzuki K., Shinshi H. Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochem. Biophys. Acta. 2004;1679:279–287. doi: 10.1016/j.bbaexp.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 83.Toyofuku K., Umemura T., Yamaguchi J. Promoter elements required for sugar-repression of the RAmy3D gene for alpha-amylase in rice. FEBS Lett. 1998;428:275–280. doi: 10.1016/S0014-5793(98)00518-3. [DOI] [PubMed] [Google Scholar]
- 84.Izawa T., Foster R., Nakajima M., Shimamoto K., Chua N.-H. The rice bZIP transcriptional activator RITA-1 is highly expressed during seed development. Plant Cell. 1994;6:1277–1287. doi: 10.1105/tpc.6.9.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown R.L., Kazan K., McGrath K.C., Maclean D.J., Manners J.M. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.