Abstract
Plants retain the ability to produce new organs throughout their life cycles. Continuous aboveground organogenesis is achieved by meristems, which are mainly organized, established, and maintained in the shoot apex and leaf axils. This paper will focus on reviewing the recent progress in understanding the regulation of shoot apical meristem and axillary meristem development. We discuss the genetics of plant meristems, the role of plant hormones and environmental factors in meristem development, and the impact of epigenetic factors on meristem organization and function.
Keywords: shoot apical meristem, axillary meristem, transcription, plant hormones, epigenetics
1. Introduction
Plants are unique in their ability to continuously produce new organs throughout their life cycles. The process of continuous organogenesis depends on the activity of pluripotent cells. Some of these stem cells are located at the tips of shoot and root known as apical meristems. During embryogenesis, the shoot apical meristem (SAM) and root apical meristem (RAM) are established in the shoot apex and root apex, respectively. In monocots, such as rice and maize, the SAM is formed laterally, at the base of a single cotyledon [1]. In dicots, such as Arabidopsis, the SAM is established centrally, between two cotyledons. In Arabidopsis, during post-embryonic development, the SAM together with other meristems generates aboveground organs and the RAM contributes to underground architecture with the help of other meristems. Axillary meristems (AMs) form in the leaf axils to enable branching. During reproductive growth, the SAM transits into inflorescence meristem (IM). Then, the IM generates the floral meristem (FM), which produces floral organs [2].
SAM has dual functions of generating leaves, stems, and floral organs, as well as maintaining a pool of pluripotent cells in the center. The SAM contains three functional zones: The central zone (CZ), peripheral zone (PZ), and rib zone (RZ). The CZ is composed of slowly dividing pluripotent stem cells. The PZ, that surrounds the CZ, comprises rapidly dividing cells. Some progenies of the stem cells are displaced from the CZ into the PZ, where they give rise to lateral organs. The organizing center (OC), which is located within the rib meristem beneath the CZ, maintains the stem cell population [3]. In Arabidopsis, the SAM can also be classified into three clonally distinct layers of stem cells (L1–L3). The surface layer L1 forms the epidermis, L2 forms the subepidermal layer, and the L3 layer gives rise to the remaining inner corpus tissues of the shoot [4]. In Arabidopsis, AMs locate in leaf axils and share common features with the SAM such as generating leaves, stems, and maintaining pluripotent stem cells. The SAM and AMs together give rise to aboveground parts of the plant and determine plant architecture. In this review, we will summarize recent progress in the research on SAM and AM development and discuss how plant hormones, epigenetic factors, and environmental factors participate in the regulation of meristem development. All studies were conducted in Arabidopsis unless otherwise mentioned.
2. Establishment of the SAM
The SAM is established during embryogenesis. In Arabidopsis, the zygote divides asymmetrically to produce an apical cell and a basal cell. The smaller apical cell generates the embryo, while the larger basal cell forms the suspensor [5]. Asymmetric distribution of auxin mediated by the auxin efflux carrier PIN-FORMED (PIN) promotes the formation of an “apical-basal axis” in the embryo [6]. In the eight-cell embryo, the upper four cells give rise to the shoot meristem, whereas the lower cells generate the hypocotyl. A round of periclinal division gives rise to the 16-cell globular stage embryo. During the globular stage, the shoot meristem primordium is established. Establishment of the radial pattern of the embryo is a prerequisite for SAM formation and is regulated by two types of transcription factors (TFs): The class III homeodomain-leucine zipper (HD-ZIPIII) and KANADI (KAN) proteins [7]. The HD-ZIPIII proteins contain an HD-Zip domain, which is involved in DNA binding and protein dimerization [8]. The HD-ZIPIII transcription factor family consists of five members, namely REVOLUTA (REV), PHABULOSA (PHB), PHAVOLUTA (PHV), CORONA (CNA), and ATHB8 [9]. REV, PHB, and PHV begin to be expressed at the 16-cell globular stage, and their expression is confined to the central region of the embryo at the late globular embryo stage [7]. Consequently, the central region specifies the identity of the central cells. Meanwhile, KAN specifies peripheral identity in the developing embryo [10]. Bilateral symmetry is established at a later stage of embryo development in a process involving the interplay of auxin, the SHOOT MERISTEMLESS (STM) gene, and the CUP SHAPED COTYLEDON (CUC) genes [5]. STM, a member of the KNOTTED-LIKE HOMEOBOX1 (KNOX1) gene family, encodes a class I KNOTTED-LIKE protein [11]. It maintains stem cell identity and regulates SAM cell division. The NAC domain transcription factors CUC1, CUC2, and CUC3 are involved in initiating the SAM [12]. CUC1 and CUC2 promote STM expression, and STM can both promote and repress CUC1/2 gene expression, implying the existence of a positive and negative feedback loop between these genes [5,13,14]. Expression of the CUC genes is confined to a narrow strip, which is mediated by the auxin efflux carrier protein PIN1 and AUXIN RESPONSE FACTOR 5/MONOPTEROS (ARF5/MP) [12,15]. During the early stages of globular development, STM is expressed in the median region, which consists of a presumptive SAM region and a boundary region of cotyledon margins [16]. The cotyledons arise from the peripheral region, where AINTEGUMENTA (ANT) but not STM is expressed [17]. In summary, HD-ZIPIII genes, STM, and CUCs establish the radial and bilateral symmetry of the globular embryo, and thereafter, the SAM forms.
3. Maintenance of Stem Cells in Meristems
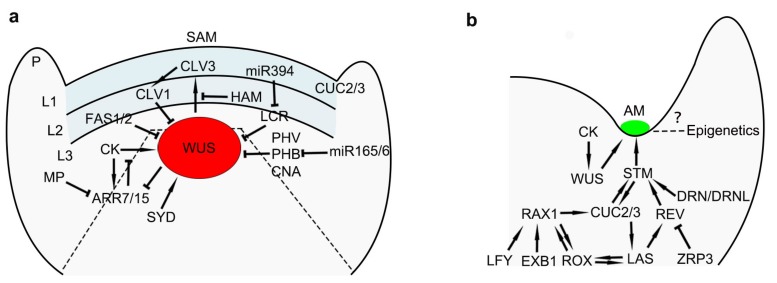
The SAM harbors pluripotent stem cells. In Arabidopsis, the CLAVATA (CLV)-WUSCHEL (WUS) negative feedback loop is the central genetic mechanism that coordinates stem cell proliferation with differentiation in the meristem (Figure 1a) [18,19,20]. WUS, a homeodomain transcription factor, is required for regulation of meristem activity and stem cell maintenance [3,21]. Mutants for WUS fail to maintain stem cells, leading to the loss of the meristems and termination of organogenesis [22]. The CLV (CLV1, CLV2, and CLV3) genes play a role in promoting the progression of meristem cells toward organ initiation, which is opposite to that of WUS. Mutants for any of these genes lead to delayed organ initiation and accumulation of meristem cells [23,24,25]. WUS is initially expressed at the 16-cell embryo stage, when the SAM is not evident. The expression of WUS is restricted to the SAM OC [3]. Then, WUS migrates from the OC into the CZ region and activates CLV3 expression exclusively in the outermost apical layer of the SAM by binding to CLV3 promoter elements [3,25,26]. WUS-mediated CLV3 activation is inhibited by the HAIRY MERISTEM (HAM) proteins [27]. CLV3 encodes a putative ligand of the CLV1 receptor kinase, which is expressed throughout most of the SAM [28]. CLV3/CLV1 restrict WUS transcription to the OC via a receptor kinase-signaling cascade and thus limit the size of the OC [18,19,20,26,29]. This negative feedback loop coordinating stem cell proliferation and differentiation, appears to be conserved in diverse plant species [30,31]. It is possible that, CLV signaling and WUS are linked through the CLV3 control of transcriptional regulators of WUS, such as STIMPY (STIP), SPLAYED (SYD), BARD1, OBERON1 (OBE1), and OBERON2 (OBE2) [1]. SYD and BARD1 interact directly with the WUS promoter [32,33]; however, they have opposite functions. SYD is a SNF2 chromatin remodeling ATPase that promotes WUS transcription. In the syd mutant, the size of the meristems and transcriptional level of WUS are reduced. The BARD1 protein appears to be a repressor of WUS. In the bard1 mutant, the meristem size is increased and WUS expression is upregulated. The Arabidopsis OBE1 and OBE2 genes are implicated in maintaining the expression of CLV3 and WUS and maintaining stem cell activity [34].
Figure 1.
Conceptual models showing regulators in shoot apical meristem (SAM) and axillary meristems (AM) development. (a) A diagram illustrates the network in SAM regulation; P: Primordium. (b) A model shows factors involved in the regulation of AM development. Arrows and inhibition symbols indicate activation and repression, respectively.
The KNOX genes, including the Arabidopsis STM, have been proven to prevent the differentiation of stem cells. STM and WUS have distinct and complementary functions in the SAM. STM maintains the meristem cell fate and WUS specifies a subset of cells at the shoot apex as stem cells. In the stm mutant, the establishment and maintenance of the SAM are abolished. Thus, STM is vital for maintenance of meristematic activity. STM is expressed throughout the SAM [11] and prevents meristem cells from differentiating by promoting cytokinin synthesis [35]. In addition to STM, there are three other KNOX genes (KNAT1, KNAT2, and KNAT6) in Arabidopsis. They are localized at the shoot apex and function redundantly with STM [36]. These KNOX genes restrict the expression of the ASYMMETRIC LEAVES1 (AS1) and ASYMMETRIC LEAVES2 (AS2) genes to organ primordia, thus preventing ectopic organ initiation. AS1 and AS2 can in turn repress KNOX gene expression [37,38]. The interactions between KNOX transcription factors, which are expressed in the meristems, and AS1/AS2, which are expressed in the organ primordia, indicates that these genes are crucial for distinguishing stem cells and organ founder cells [36].
4. Initiation of AMs
Different from the SAM, AMs initiate at the base of adaxial side of leaves and develop at post-embryo stages. The microenvironment of leaf axils and developmental stages of leaves impact AM development. In seed plants, AMs enable lateral shoot branching. The development of a lateral shoot includes two steps: Initiation and outgrowth/dormancy. During AM initiation, a bump forms in the leaf axil and develops into an axillary bud (Figure 2). Owing to its subtle shape, AM development has been less studied. Several regulatory genes involved in AM initiation have been identified (Figure 1b), some of which participate in both SAM and AM development. For example, WUS and CLV3, which are required for SAM maintenance, are involved in AM initiation, and the meristem marker genes STM and CUC also participate in AM initiation. In weak stm mutants, which can survive to reproduce, there are significantly fewer axillary buds. The initiation of AMs requires a meristematic cell population continuously expressing STM, and the absence of STM-expressing cells abolishes bud formation [39]. STM expression is dynamic and is associated with two phases of stem cell division. During the first phase, in leaf axils younger than P9 (the ninth earliest leaf primordium), STM is expressed at a low level that is sufficient for stem cell competence but not for AM initiation [39]. It was recently reported that ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1) interacts with STM. The ATH1-STM heterodimer maintains STM expression and meristematic cell fate in the early leaf axil [40]. During the second phase, in leaf axils older than P10, STM expression is upregulated by REV and other transcription factors, and this propels bump formation. A transcriptional network contributes to the dynamic regulation of STM. DORNRÖSCHEN (DRN)/ENHANCER OF SHOOT REGENERATION1 (ESR1) and its homolog DORNRÖSCHEN-LIKE (DRNL)/ESR2, which encode APETALA2 (AP2) type transcription factors, promoting AM initiation by activating STM [41]. In Arabidopsis mutants for these genes, AM initiation is compromised in the early rosette leaves [41,42]. During AM initiation, DRN/DRNL interact with REV to promote STM expression in leaves older than P10, while LITTLE ZIPPER3 (ZRP3), another REV-interacting protein interferes with the DRN/DRNL-REV complex, to prevent STM expression in leaf axils younger than P9 [41]. After the STM-induced gain of meristematic activity, de novo activation of the homeodomain transcription factor WUS by cytokinin signaling promotes AM initiation in the leaf axils around the P13 leaves [43]. WUS expression is also regulated by CLV3 in the AMs [44]. In addition, CUC genes are also expressed in both the SAM and AMs. In single and triple CUC gene mutants, AM formation is blocked to different extents due to the redundant functions of the CUC genes [45].
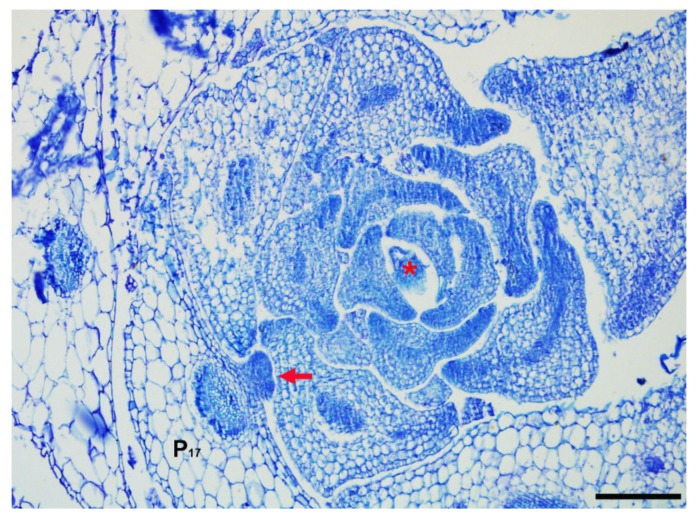
Figure 2.
Axillary meristem and shoot apical meristem in Arabidopsis. A bump corresponding to an axillary meristem initiated in a mature leaf axil of P17 (the seventeenth earliest leaf primordium) is shown. The shoot apical meristem is located in the shoot apex. Arrow indicates axillary meristem and asterisk indicates shoot apical meristem. Bar = 100 µm.
Different from the aforementioned genes, some transcription factors, including LATERAL SUPPRESSOR (LAS) and REGULATOR OF AXILLARY MERISTEMS (RAX), specifically regulate AM initiation. LAS encodes a GRAS family transcription factor, which is expressed in the leaf axil. Loss of function of LAS leads to severe defects in axillary bud formation [46]. LAS expression can be upregulated by CUC2 [42]. The downstream target of LAS is REV, which encodes an HD-ZIP transcription factor as mentioned above, and is another gene involved in both SAM and AM initiation. REV binds to the STM promoter and directly upregulates STM expression in the leaf axil [39]. The RAX family genes, which encode R2R3 MYB transcription factors, are expressed in boundary regions and are specific regulators of AM initiation. RAX1, RAX2, and RAX3 have redundant functions in promoting AM initiation. In rax1 rax2 rax3 triple mutants, most axillary buds cannot form [47]. RAX1 expression is regulated by a WRKY transcription factor EXCESSIVE BRANCHES1 (EXB1). Overexpression of EXB1 leads to a bushy phenotype with excessive AM initiation [48]. LEAFY (LFY), which is required for the vegetative-reproductive stage transition, also upregulates RAX1 expression to promote precocious AM development in rosette leaves [49]. REGULATOR OF AXILLARY MERISTEM FORMATION (ROX), which is regulated by RAX and LAS, is expressed in the leaf axils and promote AM initiation downstream of RAX and LAS [50]. As high throughput data and in-depth gene function data accumulate, the elaborate mechanisms underlying AM development will be unveiled.
5. Plant Hormones Regulate Meristem Development
Plant hormones, primarily auxin, cytokinin (CK), brassinosteroids (BRs), gibberellins (GA), and strigolactones (SLs) are signaling molecules that play important roles in a variety of physiological processes [51]. The interaction between cytokinin and auxin is required for the regulation of SAM, AM, and FM development [52,53,54,55]. In early embryogenesis, cytokinin and auxin control meristem formation. Stem cell maintenance requires high levels of cytokinin, which are achieved by repression of two negative regulators of cytokinin signaling, the ARABIDOPSIS RESPONSE REGULATORS (ARR7 and ARR15). As previously mentioned, WUS defines the stem cell niche in the SAM. WUS is also required for stem cell definition during AM formation. Cytokinin signaling activates the expression of WUS in the leaf axil of P13, which is a requisite for AM initiation [43]. The WUS and auxin response factor ARF5/MP are both crucial for repression of ARR7 and ARR15 [55].
Auxin is required for organogenesis [56]. The boundary regions between the SAM and leaf primordia, show low auxin concentrations at an early stage before AM initiation, which was demonstrated using the auxin indicators DII and DR5 [57,58,59]. Ectopic application of an auxin signaling inhibitor resulted in supernumerary axillary buds indicating that a relatively low auxin level is required for boundary formation and the subsequent AM initiation. A high level of auxin in the boundary blocks the following STM expression in the leaf axil confirming that proper auxin is essential for boundary formation [39]. However, the role of auxin in AM initiation after boundary formation is not clear. The asymmetric distribution of auxin plays a vital role in the establishment of primordium, which is mediated by PIN proteins. Cytokinin can modulate the auxin distribution through transcriptional regulation of the PIN genes. Another factor required for auxin transcriptional response is MP/ARF5 [60]. MP regulates the polar localization of PIN1, which promotes the asymmetric distribution of auxin [61]. In addition, PINOID (PID), which encodes a serine-threonine protein kinase, regulates the polar subcellular localization of the PIN auxin efflux regulators [62]. In pin1 and pid mutants, AMs are defective, again implying that auxin signals correlate with AM initiation.
Axillary buds can grow out into shoot branches or be inhibited in the leaf axil. The fate of an axillary bud is controlled by hormonal signals. Auxin, cytokinin, and strigolactones are important players during bud outgrowth. In pea, the export of auxin from axillary buds is a prerequisite for bud outgrowth, which is illustrated by the breaking of apical dominance [63,64,65]. Cytokinin plays a positive role late in AM development, and direct application of cytokinin promotes bud outgrowth in pea and Jatropha curcas [66,67,68]. Strigolactones, which act antagonistically to cytokinin in bud outgrowth, inhibit shoot branching [68,69].
6. Epigenetic Regulation of Meristem Development
Epigenetic regulation, including DNA methylation, histone modifications, chromatin remodeling, and noncoding RNAs, plays important roles in plant development. The abovementioned regulators required for SAM maintenance, such as WUS and STM, are subject to epigenetic regulation. Chromatin remodelers regulate meristem activity by regulating WUS expression. Mutations in chromatin factors result in SAM dysfunction [70]. FASCIATA1 (FAS1) and FASCIATA 2 (FAS2) are two subunits of the CHROMATIN ASSEMBLY FACTOR 1 (CAF-1) complex. The FAS1 and FAS2 genes play a role in SAM organization. Mutations in FAS1 and FAS2 lead to aberrant SAMs with abnormal expression of WUS [71], thus leading to defects in shoot development. In addition, SYD and BRAHMA (BRM) which are SWITCH2/SNF2-related ATP-dependent chromatin remodeling factors, are expressed in meristems and involved in shoot development [72,73]. In the syd mutant, SAM formation is impaired and WUS expression is decreased. SYD regulates WUS expression by directly binding to the WUS promoter, indicating that WUS is a direct target of SYD [32]. In addition, the SYD binding protein BARD1 regulates SAM organization and maintenance by limiting WUS expression to the OC [33].
Histone modifications, such as methylation, acetylation, phosphorylation, and ubiquitination, can regulate gene expression. In Arabidopsis, STM is expressed exclusively in the SAM and silenced in lateral organ primordia. This expression pattern is considered to be important for SAM maintenance. Repression of STM in primordia is accomplished by a repressive mark, Histone 3 Lysine 27 trimethylation (H3K27m3), in the STM promoter. Polycomb Repressive Complex 2 (PRC2) establishes and maintains the H3K27m3 modification at target loci. In Arabidopsis, PRC2-like complexes consist of CURLY LEAF (CLF), SWINGER (SWN), EMBRYONIC FLOWER2 (EMF2), FERTILIZATION INDEPENDENT ENDOSPERM (FIE), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) [74]. In plants with mutations in these genes, H3K27 methylation was abolished at the STM locus, and STM was ectopically expressed [75]. PRC1 is required for maintaining the repressive marks established by PRC2, and there is evidence indicating that PRC1 also regulates STM expression [76,77].
Small RNAs are categorized into two major classes, miRNAs and siRNAs, based on the mode of biogenesis in the plant. Small RNAs play pleiotropic roles in plant development. They repress the target mRNAs mainly through post-transcriptional slicing and in part by translational inhibition. The RNA-induced silencing complex (RISC) consists of small RNAs and ARGONAUTE (AGO) proteins and is a key effector for target silencing [78]. Some MIR genes have been reported to be required for meristem development. For example, miR165/166 acting together with AGO1 regulate HD-ZIPIII genes, which are critical for stem cell maintenance, by cleaving their mRNAs [9,79,80]. ZWILLE/AGO10 (ZLL/AGO10), which is another important ARGONAUTE protein closely related to AGO1, also regulates HD-ZIPIII gene expression. Several studies have shown that ZLL/AGO10 and AGO1 have antagonistic functions with AGO10 functioning as a decoy protein. ZLL/AGO10 sequesters miR165/166, and also prevents the incorporation of miR165/166 into AGO1 [81], leading to HD-ZIPIII accumulation and SAM maintenance [82]. In addition, ZLL/AGO10 accelerates the degradation of miR165/166 by SMALL RNA DEGRADING NUCLEASES (SDNs) exonucleases, thereby favoring HD-ZIPIII accumulation [83].
The abovementioned STM-WUS-CLV pathway controls SAM maintenance. miRNAs also play vital roles in SAM maintenance by regulating targets in the STM-WUS-CLV pathway [84]. The decoy protein, ZLL/AGO10, is required for potentiating CLV3 and WUS activity in a non-cell-autonomous manner [85]. WUS expression is also indirectly regulated by miR394, which is synthesized in the L1 layer of the SAM. miR394 can diffuse from the L1 layer into the OC region, where it represses the F-box protein LEAF CURLING RESPONSIVENESS (LCR), a suppressor of WUS. Repression of LCR by miR394 is important for stem cell maintenance [29]. The NAC domain transcription factors CUC1, and CUC2 regulate organ boundary formation, and SAM and AM initiation, are targeted by miR164. In the miR164 mutant, levels of CUC transcripts increased, which resulted in the formation of bud-like structures in the leaf margin [45,86]. AP2 is a second class A gene that regulates FM termination. miR172 downregulates AP2 expression through translation inhibition, which in turn represses stem cell fate and leads to the determination of FM identity [82,87,88,89]. Taken together, these studies indicate that miRNAs play an important role in SAM development; however, although ago1 and ago10 mutants display defective branches [90], much less is known about the roles of miRNAs in AM development (Figure 1b).
7. Environmental Factors Impact Meristem Development
Environmental signals, such as light, sugar, and nitrate modulate meristem development. These in vitro signals always interplay with plant hormones to regulate meristems. Light is not only a key energy source of plants but also an important player in shoot meristem development. In the dark, plants show skotomorphogenesis and SAM is dormant. In the light, plants display photomorphogenesis and meristems are activated [91]. Light activates WUS expression through CK signaling and also integrates with metabolic signals for stem cell activation in the SAM of Arabidopsis [92]. In tomato, light controls the initiation of leaves through auxin and CK [93]. The energy from light generates sugars and oxygen, which regulates meristem activity by converging on the TARGET OF RAPAMYCIN (TOR) kinase. TOR, which can respond to nutrient, energy, hormone, and environmental cues, is essential for metabolism, biogenesis, organ growth, and development transitions [94,95,96]. Auxin can substitute light to activate the TOR kinase in the presence of sucrose in Arabidopsis [97]. During SAM development, Arabidopsis plants adapt their organogenesis rate to the availability of nitrate in the soil. Cytokinin mediates this process by modulating WUS expression [98]. Oxygen is another important factor affecting meristem development. Recent studies confirmed that SAM develops in hypoxic conditions. Hypoxia protects LITTLE ZIPPER 2 (ZPR2), which is required for the activity of HD-ZIPIII transcription factors, thereby regulating shoot meristem activity [99]. Reactive oxygen species (ROS) play essentials roles in plants [100,101,102]. The mutation of an ATP-dependent mitochondrial protease, AtFTSH4, leads to the accumulation of oxidative stress in SAM at 30 °C and affects meristematic fate [103]. ROS contain the superoxide anion (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (OH). Different forms of ROS have different roles in stem cell regulation. The superoxide anion (O2−) activates WUS and maintains stem cell fate. In contrast, hydrogen peroxide (H2O2) promotes stem cell differentiation [104]. From these results, ROS are evidenced to be vital players in the maintenance of shoot stem cell niche [105].
How these environmental cues regulate AMs is still not clear. The outgrowth of lateral buds is regulated by environmental signals. Prolonged exposure to low temperatures can reactivate buds of the plants [106]. Another example is the inhibition of bud outgrowth when plants are subjected to far-red light, in which signal is mediated by the photoreceptor PHYTOCHROME B (PHYB). Consistent with this, in the sorghum and Arabidopsis phyB mutants, the plants have fewer branches [107,108]. There is evidence that nutrient deficiency also causes the inhibition of bud outgrowth [106]. In summary, because plants are immotile, environmental factors play rather important roles in regulating AM development. Although little is known about how the environment affects AM initiation, it is obvious that exogenous signals cooperate with plant hormones to influence bud growth and the whole plant architecture.
8. Future Perspectives
8.1. Cooperation of SAM and AM Development will Facilitate Systematic Design for High Yield Breeds in Agriculture
As environmental factors are unpredictable, modulation of plant development to shape fitness and achieve great yield in a variable environment is promising. Apical meristem and lateral branches are vital components of the aboveground architecture of the plant. A strong meristematic SAM and highly productive AMs are ideal traits pyramiding yield. The continuous activity of SAM was proposed to give rise to more lateral organs. A number of approaches have been adopted to screen diverse germplasms for prolonged meristem activity. However, longer periods of vegetative growth from the shoot apex are often accompanied by delayed maturity and an unintended reduction in yield [109]. Lateral shoots and inflorescence architecture determine seed number and yield, so alternative methods were used to increase the number of lateral branches and inflorescences through activation of AM initiation or outgrowth of lateral buds. However, few mutants with a larger number of AMs were obtained, potentially because of the tight regulation of AM initiation in leaf axils. In contrast, several bushy mutants showing increased bud outgrowth were identified, but grain yield was not significantly improved due to limitation of biomass in the whole plant and reduced growing density [110,111,112,113]. The relationship between the shoot apex and lateral branch is elucidated majorly by shoot apical dominance and in part by feedback from lateral organs. Different from canonical mutants with obvious phenotypes and a significant reduction in production, transformative modulation of gene expression at various levels (i.e., DNA, RNA, and protein) is prospective to provide more elite cultivars [114]. By taking advantage of recent genomic editing methods, the systematic design of innovative breeds through precise control of the SAM and AMs has become attainable [109].
8.2. Plant Hormones and Transcriptional Regulation Tailors Shoot
Plant hormones and transcription factors (TFs) play essential roles in plant development. A substantial number of genes important for meristem development, such as STM and WUS encode TFs. Transcriptional networks in SAMs centered on STM, WUS, and CLVs have been elucidated in detail [115,116]. More recently, a series of studies unveiled the transcriptional network underlying AM initiation, which features TFs specially expressed in AMs [117]. Altered expression of factors common to SAM and AM development always lead to defects in both types of meristems, with effects ranging from serious to lethal. In contrast, modulation of AM-specific genes allows specific control of AMs. Knockout mutants of AM-specific genes lead to fewer branches without defects in apical shoots [46,47]. Regarding yield, overexpression lines or knockout mutants of canonical meristem genes always display pleiotropic phenotypes along with a reduction or slight increase in yield, mostly because of the dominant function of TFs in plant development [9,118]. Inducible lines make it possible to control specific developmental stages, but these lines consume a lot of resources and application in the field seems impractical. Genomic editing enables precise control of gene expression through editing cis elements in the promoters or other noncoding regions at the nucleotide-scale. The ability to perform such precise editing has the potential to accelerate breeding for a high yield. Genome editing methods have already been applied in breeding of tomatoes and other crops [109,119,120].
8.3. Epigenetic Regulation Sculpts Shoot Architecture
Epigenetic regulation sculpts plant architecture [121]. Although the canonical epigenetic pathways have been uncovered, the function of epigenetic regulation in plant development needs to be elaborated. Genetic modification of DNA and changes in histone levels modify gene expression without altering DNA sequence. As outlined above, meristem modulators such as WUS and STM are modified at the epigenetic level to ensure appropriate expression during meristem formation. However, epigenetic modifications usually affect the whole genome. Tissue-specific modulation of epigenetic regulators is more feasible than constitutive expression. Genomic editing of DNA modification sites also provides an alternative means for construction of new transformants. miRNAs are a vital participant in plant development, and miRNA mutants display morphological phenotypes. Identifying miRNAs and analyzing their roles in AM initiation remains imperative. After being transcribed, targeted mRNAs, some of which are crucial for meristem development, are subjected to miRNA-mediated post-transcriptional gene silencing (PTGS) and translational inhibition. These regulatory pathways provide possibilities for modulating gene expression. In addition, the secondary structure of RNAs also affects PTGS and translation efficiency. As an increasing number of modifications are identified in either mRNAs or small RNAs, new methods for optimizing gene expression at the RNA level will shed light on the precise control of meristem development.
AMs located in leaf axils provide a good model for studying stem cell differentiation and meristem organization in a microenvironment. AMs are compact and are suitable for studying the short-distance movement of molecules. A further study of the movement of molecules among various cell types within AMs will aid the discovery of new mechanisms underlying plant development.
Acknowledgments
We apologize to those authors whose research could not be cited owing to space limits.
Abbreviations
| SAM | Shoot apical meristem |
| RAM | Root apical meristem |
| AMs | Axillary meristems |
| IM | Inflorescence meristem |
| FM | Floral meristem |
| CZ | Central zone |
| PZ | Peripheral zone |
| RZ | rib zone |
| OC | Organizing center |
| L1 | Layer 1 |
| L2 | Layer 2 |
| L3 | Layer 3 |
| PIN | PIN-FORMED |
| HD-ZIPIII | Class III homeodomain-leucine zipper |
| KAN | KANADI |
| REV | REVOLUTA |
| PHB | PHABULOSA |
| PHV | PHAVOLUTA |
| CNA | CORONA |
| STM | SHOOT MERISTEMLESS |
| CUC | CUP SHAPED COTYLEDON |
| KNOX1 | KNOTTED-LIKE HOMEOBOX1 |
| ARF5 | AUXIN RESPONSE FACTOR5 |
| MP | MONOPTEROS |
| ANT | AINTEGUMENTA |
| CLV | CLAVATA |
| WUS | WUSCHEL |
| HAM | HAIRY MERISTEM |
| OBE1 | OBERON1 |
| OBE2 | OBERON2 |
| SYD | SPLAYED |
| STIP | STIMPY |
| BRAD1 | BRCA1 associated RING domain 1 |
| AS1 | ASYMMETRIC LEAVES1 |
| AS2 | ASYMMETRIC LEAVES2 |
| DRN | DORNRÖSCHEN |
| ESR1 | ENHANCER OF SHOOT REGENERATION1 |
| DRNL | DORNRÖSCHEN-LIKE |
| ZPR3 | LITTLE ZIPPER3 |
| LAS | LATERAL SUPPRESSOR |
| RAX | REGULATOR OF AXILLARY MERISTEMS |
| EXB1 | EXCESSIVE BRANCHES1 |
| LFY | LEAFY |
| ROX | REGULATOR OF AXILLARY MERISTEM FORMATION |
| CK | Cytokinin |
| BRs | Brassinosteroids |
| GA | Gibberellins |
| SLs | Strigolactones |
| ARR7 | ARABIDOPSIS RESPONSE REGULATOR7 |
| PID | PINOID |
| PHYB | PHYTOCHROME B |
| FAS1 | FASCIATA1 |
| FAS2 | FASCIATA2 |
| BRM | BRAHMA |
| H3K27m3 | Histone 3 Lysine 27 trimethylation |
| PRC2 | Polycomb Repressive complex 2 |
| CLF | CURLY LEAF |
| SWN | SWINGER |
| EMF2 | EMBRYONIC FLOWER2 |
| FIE | FERTILIZATION INDEPENDENT ENDOSPERM |
| MSI1 | MULTICOPY SUPPRESSOR OF IRA1 |
| miRNAs | microRNAs |
| siRNAs | Small RNAs |
| RISC | RNA-induced silencing complex |
| AGO | ARGONAUTE |
| ZLL | ZWILLE |
| SDNs | SMALL RNA DEGRADING NUCLEASES |
| LCR | LEAF CURLING RESPONSIVENESS |
| AP2 | APETALA2 |
| TFs | Transcription factors |
| ATH1 | ARABIDOPSIS THALIANA HOMEOBOX GENE1 |
Author Contributions
Z.X., L.L., and C.Z. wrote the article. All authors have read and agree to the published version of the manuscript.
Funding
Research in the Zhang lab is supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24010106-2) to C.Z., the National Natural Science Foundation of China (31900391), and the Youth Innovation Promotion Association CAS (2020083) to Z.X.
References
- 1.Barton M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010;341:95–113. doi: 10.1016/j.ydbio.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann K., Pajoro A., Angenent G.C. Regulation of transcription in plants: Mechanisms controlling developmental switches. Nat. Rev. Genet. 2010;11:830–842. doi: 10.1038/nrg2885. [DOI] [PubMed] [Google Scholar]
- 3.Mayer K.F., Schoof H., Haecker A., Lenhard M., Jurgens G., Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/S0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 4.Satina S., Blakeslee A.F., Avery A.G. Demonstration of the three germ layers in the shoot apex of Datura by means of induced polyploidy in periclinal chimeras. Am. J. Bot. 1940;27:895–905. doi: 10.1002/j.1537-2197.1940.tb13952.x. [DOI] [Google Scholar]
- 5.Bosca S., Knauer S., Laux T. Embryonic development in Arabidopsis thaliana: From the zygote division to the shoot meristem. Front. Plant Sci. 2011;2:93. doi: 10.3389/fpls.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y.H., Liu Y.B., Zhang X.S. Auxin-cytokinin interaction regulates meristem development. Mol. Plant. 2011;4:616–625. doi: 10.1093/mp/ssr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery J.F., Floyd S.K., Alvarez J., Eshed Y., Hawker N.P., Izhaki A., Baum S.F., Bowman J.L. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 8.Sessa G., Steindler C., Morelli G., Ruberti I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 1998;38:609–622. doi: 10.1023/A:1006016319613. [DOI] [PubMed] [Google Scholar]
- 9.Prigge M.J., Otsuga D., Alonso J.M., Ecker J.R., Drews G.N., Clark S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. KANADI regulates organ polarity in Arabidopsis. Nature. 2001;411:706–709. doi: 10.1038/35079629. [DOI] [PubMed] [Google Scholar]
- 11.Long J.A., Moan E.I., Medford J.I., Barton M.K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 12.Aida M., Ishida T., Tasaka M. Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: Interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development. 1999;126:1563–1570. doi: 10.1242/dev.126.8.1563. [DOI] [PubMed] [Google Scholar]
- 13.Scofield S., Murison A., Jones A., Fozard J., Aida M., Band L.R., Bennett M., Murray J.A.H. Coordination of meristem and boundary functions by transcription factors in the SHOOT MERISTEMLESS regulatory network. Development. 2018;145:dev157081. doi: 10.1242/dev.157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spinelli S.V., Martin A.P., Viola I.L., Gonzalez D.H., Palatnik J.F. A mechanistic link between STM and CUC1 during Arabidopsis development. Plant Physiol. 2011;156:1894–1904. doi: 10.1104/pp.111.177709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aida M., Vernoux T., Furutani M., Traas J., Tasaka M. Roles of PIN-FORMED1 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development. 2002;129:3965–3974. doi: 10.1242/dev.129.17.3965. [DOI] [PubMed] [Google Scholar]
- 16.Aida M., Ishida T., Fukaki H., Fujisawa H., Tasaka M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long J.A., Barton M.K. The development of apical embryonic pattern in Arabidopsis. Development. 1998;125:3027–3035. doi: 10.1242/dev.125.16.3027. [DOI] [PubMed] [Google Scholar]
- 18.Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M., Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 19.Schoof H., Lenhard M., Haecker A., Mayer K.F., Jurgens G., Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 20.Ha C.M., Jun J.H., Fletcher J.C. Shoot apical meristem form and function. Curr. Top. Dev. Biol. 2010;91:103–140. doi: 10.1016/S0070-2153(10)91004-1. [DOI] [PubMed] [Google Scholar]
- 21.Yadav R.K., Perales M., Gruel J., Ohno C., Heisler M., Girke T., Jonsson H., Reddy G.V. Plant stem cell maintenance involves direct transcriptional repression of differentiation program. Mol. Syst. Biol. 2013;9:654. doi: 10.1038/msb.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laux T., Mayer K.F., Berger J., Jurgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 23.Clark S.E., Running M.P., Meyerowitz E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- 24.Kayes J.M., Clark S.E. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- 25.Fletcher J.C., Brand U., Running M.P., Simon R., Meyerowitz E.M. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 26.Yadav R.K., Perales M., Gruel J., Girke T., Jonsson H., Reddy G.V. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–2030. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y., Yan A., Han H., Li T., Geng Y., Liu X., Meyerowitz E.M. HAIRY MERISTEM with WUSCHEL confines CLAVATA3 expression to the outer apical meristem layers. Science. 2018;361:502–506. doi: 10.1126/science.aar8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark S.E., Williams R.W., Meyerowitz E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 29.Knauer S., Holt A.L., Rubio-Somoza I., Tucker E.J., Hinze A., Pisch M., Javelle M., Timmermans M.C., Tucker M.R., Laux T. A protodermal miR394 signal defines a region of stem cell competence in the Arabidopsis shoot meristem. Dev. Cell. 2013;24:125–132. doi: 10.1016/j.devcel.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Xu C., Liberatore K.L., MacAlister C.A., Huang Z., Chu Y.H., Jiang K., Brooks C., Ogawa-Ohnishi M., Xiong G., Pauly M., et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 31.Somssich M., Je B.I., Simon R., Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 32.Kwon C.S., Chen C., Wagner D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005;19:992–1003. doi: 10.1101/gad.1276305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han P., Li Q., Zhu Y.X. Mutation of Arabidopsis BARD1 causes meristem defects by failing to confine WUSCHEL expression to the organizing center. Plant Cell. 2008;20:1482–1493. doi: 10.1105/tpc.108.058867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiga S., Furumizu C., Yokoyama R., Kurata T., Sato S., Kato T., Tabata S., Suzuki M., Komeda Y. The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development. 2008;135:1751–1759. doi: 10.1242/dev.014993. [DOI] [PubMed] [Google Scholar]
- 35.Yanai O., Shani E., Dolezal K., Tarkowski P., Sablowski R., Sandberg G., Samach A., Ori N. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 36.Carles C.C., Fletcher J.C. Shoot apical meristem maintenance: The art of a dynamic balance. Trends Plant Sci. 2003;8:394–401. doi: 10.1016/S1360-1385(03)00164-X. [DOI] [PubMed] [Google Scholar]
- 37.Byrne M.E., Simorowski J., Martienssen R.A. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- 38.Semiarti E., Ueno Y., Tsukaya H., Iwakawa H., Machida C., Machida Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 39.Shi B., Zhang C., Tian C., Wang J., Wang Q., Xu T., Xu Y., Ohno C., Sablowski R., Heisler M.G., et al. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS Genet. 2016;12:e1006168. doi: 10.1371/journal.pgen.1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao X., Wang J., Xiong Y., Yang H., Yang M., Ye P., Bencivenga S., Sablowski R., Jiao Y. A Self-Activation Loop Maintains Meristematic Cell Fate for Branching. Curr. Biol. 2020;30:1–12. doi: 10.1016/j.cub.2020.03.031. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C., Wang J., Wenkel S., Chandler J.W., Werr W., Jiao Y. Spatiotemporal control of axillary meristem formation by interacting transcriptional regulators. Development. 2018;145:dev158352. doi: 10.1242/dev.158352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian C., Zhang X., He J., Yu H., Wang Y., Shi B., Han Y., Wang G., Feng X., Zhang C., et al. An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 2014;10:755. doi: 10.15252/msb.20145470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., Zhao Z., Wang J.W., Jiao Y. Cytokinin Signaling Activates WUSCHEL Expression during Axillary Meristem Initiation. Plant Cell. 2017;29:1373–1387. doi: 10.1105/tpc.16.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xin W., Wang Z., Liang Y., Wang Y., Hu Y. Dynamic expression reveals a two-step patterning of WUS and CLV3 during axillary shoot meristem formation in Arabidopsis. J. Plant Physiol. 2017;214:1–6. doi: 10.1016/j.jplph.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Raman S., Greb T., Peaucelle A., Blein T., Laufs P., Theres K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008;55:65–76. doi: 10.1111/j.1365-313X.2008.03483.x. [DOI] [PubMed] [Google Scholar]
- 46.Greb T., Clarenz O., Schafer E., Muller D., Herrero R., Schmitz G., Theres K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003;17:1175–1187. doi: 10.1101/gad.260703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller D., Schmitz G., Theres K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell. 2006;18:586–597. doi: 10.1105/tpc.105.038745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo D., Zhang J., Wang X., Han X., Wei B., Wang J., Li B., Yu H., Huang Q., Gu H., et al. The WRKY transcription factor WRKY71/EXB1 controls shoot branching by transcriptionally regulating RAX genes in Arabidopsis. Plant Cell. 2015;27:3112–3127. doi: 10.1105/tpc.15.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chahtane H., Vachon G., Le Masson M., Thevenon E., Perigon S., Mihajlovic N., Kalinina A., Michard R., Moyroud E., Monniaux M., et al. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013;74:678–696. doi: 10.1111/tpj.12156. [DOI] [PubMed] [Google Scholar]
- 50.Yang F., Wang Q., Schmitz G., Muller D., Theres K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012;71:61–70. doi: 10.1111/j.1365-313X.2012.04970.x. [DOI] [PubMed] [Google Scholar]
- 51.Oliva M., Farcot E., Vernoux T. Plant hormone signaling during development: Insights from computational models. Curr. Opin. Plant Biol. 2013;16:19–24. doi: 10.1016/j.pbi.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 52.Depuydt S., Hardtke C.S. Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 2011;21:R365–R373. doi: 10.1016/j.cub.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 53.Sablowski R. Plant stem cell niches: From signalling to execution. Curr. Opin. Plant Biol. 2011;14:4–9. doi: 10.1016/j.pbi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Luo L., Zeng J., Wu H., Tian Z., Zhao Z. A Molecular Framework for Auxin-Controlled Homeostasis of Shoot Stem Cells in Arabidopsis. Mol. Plant. 2018;11:899–913. doi: 10.1016/j.molp.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 55.Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., Lohmann J.U. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
- 56.Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426:255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 57.Vernoux T., Brunoud G., Farcot E., Morin V., Van den Daele H., Legrand J., Oliva M., Das P., Larrieu A., Wells D., et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulmasov T., Liu Z.B., Hagen G., Guilfoyle T.J. Composite structure of auxin response elements. Plant Cell. 1995;7:1611–1623. doi: 10.1105/tpc.7.10.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q., Kohlen W., Rossmann S., Vernoux T., Theres K. Auxin depletion from the leaf axil conditions competence for axillary meristem formation in Arabidopsis and Tomato. Plant Cell. 2014;26:2068–2079. doi: 10.1105/tpc.114.123059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lavy M., Estelle M. Mechanisms of auxin signaling. Development. 2016;143:3226–3229. doi: 10.1242/dev.131870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhatia N., Bozorg B., Larsson A., Ohno C., Jonsson H., Heisler M.G. Auxin Acts through MONOPTEROS to Regulate Plant Cell Polarity and Pattern Phyllotaxis. Curr. Biol. 2016;26:3202–3208. doi: 10.1016/j.cub.2016.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Friml J., Yang X., Michniewicz M., Weijers D., Quint A., Tietz O., Benjamins R., Ouwerkerk P.B., Ljung K., Sandberg G., et al. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science. 2004;306:862–865. doi: 10.1126/science.1100618. [DOI] [PubMed] [Google Scholar]
- 63.Balla J., Kalousek P., Reinohl V., Friml J., Prochazka S. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J. 2011;65:571–577. doi: 10.1111/j.1365-313X.2010.04443.x. [DOI] [PubMed] [Google Scholar]
- 64.Balla J., Medvedova Z., Kalousek P., Matijescukova N., Friml J., Reinohl V., Prochazka S. Auxin flow-mediated competition between axillary buds to restore apical dominance. Sci. Rep. 2016;6:35955. doi: 10.1038/srep35955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller D., Leyser O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011;107:1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimizu-Sato S., Mori H. Control of outgrowth and dormancy in axillary buds. Plant Physiol. 2001;127:1405–1413. doi: 10.1104/pp.010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ni J., Zhao M.L., Chen M.S., Pan B.Z., Tao Y.B., Xu Z.F. Comparative transcriptome analysis of axillary buds in response to the shoot branching regulators gibberellin A3 and 6-benzyladenine in Jatropha curcas. Sci. Rep. 2017;7:11417. doi: 10.1038/s41598-017-11588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dun E.A., de Saint Germain A., Rameau C., Beveridge C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012;158:487–498. doi: 10.1104/pp.111.186783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dun E.A., Brewer P.B., Beveridge C.A. Strigolactones: Discovery of the elusive shoot branching hormone. Trends Plant Sci. 2009;14:364–372. doi: 10.1016/j.tplants.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 70.Guyomarc’h S., Bertrand C., Delarue M., Zhou D.X. Regulation of meristem activity by chromatin remodelling. Trends Plant Sci. 2005;10:332–338. doi: 10.1016/j.tplants.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 71.Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/S0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 72.Wagner D., Meyerowitz E.M. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 2002;12:85–94. doi: 10.1016/S0960-9822(01)00651-0. [DOI] [PubMed] [Google Scholar]
- 73.Farrona S., Hurtado L., Bowman J.L., Reyes J.C. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131:4965–4975. doi: 10.1242/dev.01363. [DOI] [PubMed] [Google Scholar]
- 74.Pien S., Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim. Biophys. Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 75.Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 2006;25:4638–4649. doi: 10.1038/sj.emboj.7601311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu L., Shen W.H. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 2008;18:1966–1971. doi: 10.1016/j.cub.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X., Germann S., Blus B.J., Khorasanizadeh S., Gaudin V., Jacobsen S.E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 78.Chen X. Small RNAs in development-insights from plants. Curr. Opin. Genet. Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Byrne M.E. Shoot meristem function and leaf polarity: The role of class III HD-ZIP genes. PLoS Genet. 2006;2:e89. doi: 10.1371/journal.pgen.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang Z., Zhang X. Argonautes compete for miR165/166 to regulate shoot apical meristem development. Curr. Opin. Plant Biol. 2012;15:652–658. doi: 10.1016/j.pbi.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu H., Hu F., Wang R., Zhou X., Sze S.H., Liou L.W., Barefoot A., Dickman M., Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ji L., Liu X., Yan J., Wang W., Yumul R.E., Kim Y.J., Dinh T.T., Liu J., Cui X., Zheng B., et al. ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 2011;7:e1001358. doi: 10.1371/journal.pgen.1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Y., Ji L., Le B.H., Zhai J., Chen J., Luscher E., Gao L., Liu C., Cao X., Mo B., et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLoS Biol. 2017;15:e2001272. doi: 10.1371/journal.pbio.2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wheeler B.S. Small RNAs, big impact: Small RNA pathways in transposon control and their effect on the host stress response. Chromosome Res. 2013;21:587–600. doi: 10.1007/s10577-013-9394-4. [DOI] [PubMed] [Google Scholar]
- 85.Tucker M.R., Hinze A., Tucker E.J., Takada S., Jurgens G., Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- 86.Laufs P., Peaucelle A., Morin H., Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development. 2004;131:4311–4322. doi: 10.1242/dev.01320. [DOI] [PubMed] [Google Scholar]
- 87.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang C.Y., Chen Y.Q., Liu Q. Sculpting the meristem: The roles of miRNAs in plant stem cells. Biochem. Biophys. Res. Commun. 2011;409:363–366. doi: 10.1016/j.bbrc.2011.04.123. [DOI] [PubMed] [Google Scholar]
- 89.Aukerman M.J., Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lynn K., Fernandez A., Aida M., Sedbrook J., Tasaka M., Masson P., Barton M.K. The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development. 1999;126:469–481. doi: 10.1242/dev.126.3.469. [DOI] [PubMed] [Google Scholar]
- 91.Walbot V. Arabidopsis thaliana genome. A green chapter in the book of life. Nature. 2000;408:794–795. doi: 10.1038/35048685. [DOI] [PubMed] [Google Scholar]
- 92.Pfeiffer A., Janocha D., Dong Y., Medzihradszky A., Schone S., Daum G., Suzaki T., Forner J., Langenecker T., Rempel E., et al. Integration of light and metabolic signals for stem cell activation at the shoot apical meristem. eLife. 2016;5:e17023. doi: 10.7554/eLife.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshida S., Mandel T., Kuhlemeier C. Stem cell activation by light guides plant organogenesis. Genes Dev. 2011;25:1439–1450. doi: 10.1101/gad.631211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Considine M.J. Oxygen, Energy, and Light Signalling Direct Meristem Fate. Trends Plant Sci. 2018;23:1–3. doi: 10.1016/j.tplants.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 95.Pfeiffer A., Wenzl C., Lohmann J.U. Beyond flexibility: Controlling stem cells in an ever changing environment. Curr. Opin. Plant Biol. 2017;35:117–123. doi: 10.1016/j.pbi.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 96.Wu Y., Shi L., Li L., Fu L., Liu Y., Xiong Y., Sheen J. Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J. Exp. Bot. 2019;70:2227–2238. doi: 10.1093/jxb/erz028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X., Cai W., Liu Y., Li H., Fu L., Liu Z., Xu L., Liu H., Xu T., Xiong Y. Differential TOR activation and cell proliferation in Arabidopsis root and shoot apexes. Proc. Natl. Acad. Sci.USA. 2017;114:2765–2770. doi: 10.1073/pnas.1618782114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Landrein B., Formosa-Jordan P., Malivert A., Schuster C., Melnyk C.W., Yang W., Turnbull C., Meyerowitz E.M., Locke J.C.W., Jonsson H. Nitrate modulates stem cell dynamics in Arabidopsis shoot meristems through cytokinins. Proc. Natl. Acad. Sci. USA. 2018;115:1382–1387. doi: 10.1073/pnas.1718670115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Weits D.A., Kunkowska A.B., Kamps N.C.W., Portz K.M.S., Packbier N.K., Nemec Venza Z., Gaillochet C., Lohmann J.U., Pedersen O., van Dongen J.T., et al. An apical hypoxic niche sets the pace of shoot meristem activity. Nature. 2019;569:714–717. doi: 10.1038/s41586-019-1203-6. [DOI] [PubMed] [Google Scholar]
- 100.Swanson S., Gilroy S. ROS in plant development. Physiol. Plant. 2010;138:384–392. doi: 10.1111/j.1399-3054.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 101.Schippers J.H., Foyer C.H., van Dongen J.T. Redox regulation in shoot growth, SAM maintenance and flowering. Curr. Opin. Plant Biol. 2016;29:121–128. doi: 10.1016/j.pbi.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 102.Tsukagoshi H., Busch W., Benfey P.N. Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 103.Dolzblasz A., Smakowska E., Gola E.M., Sokolowska K., Kicia M., Janska H. The mitochondrial protease AtFTSH4 safeguards Arabidopsis shoot apical meristem function. Sci. Rep. 2016;6:28315. doi: 10.1038/srep28315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zeng J., Dong Z., Wu H., Tian Z., Zhao Z. Redox regulation of plant stem cell fate. EMBO J. 2017;36:2844–2855. doi: 10.15252/embj.201695955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang S., Yu Q., Zhang Y., Jia Y., Wan S., Kong X., Ding Z. ROS: The Fine-Tuner of Plant Stem Cell Fate. Trends Plant Sci. 2018;23:850–853. doi: 10.1016/j.tplants.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 106.Domagalska M.A., Leyser O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011;12:211–221. doi: 10.1038/nrm3088. [DOI] [PubMed] [Google Scholar]
- 107.Kebrom T.H., Burson B.L., Finlayson S.A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006;140:1109–1117. doi: 10.1104/pp.105.074856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Finlayson S.A., Krishnareddy S.R., Kebrom T.H., Casal J.J. Phytochrome regulation of branching in Arabidopsis. Plant Physiol. 2010;152:1914–1927. doi: 10.1104/pp.109.148833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li T., Yang X., Yu Y., Si X., Zhai X., Zhang H., Dong W., Gao C., Xu C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018;36:1160–1163. doi: 10.1038/nbt.4273. [DOI] [PubMed] [Google Scholar]
- 110.Stirnberg P., Furner I.J., Ottoline Leyser H.M. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007;50:80–94. doi: 10.1111/j.1365-313X.2007.03032.x. [DOI] [PubMed] [Google Scholar]
- 111.Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S.M., Li J. Strigolactone Signaling in Arabidopsis Regulates Shoot Development by Targeting D53-Like SMXL Repressor Proteins for Ubiquitination and Degradation. Plant Cell. 2015;27:3128–3142. doi: 10.1105/tpc.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang B., Smith S.M., Li J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018;69:437–468. doi: 10.1146/annurev-arplant-042817-040422. [DOI] [PubMed] [Google Scholar]
- 113.Crawford S., Shinohara N., Sieberer T., Williamson L., George G., Hepworth J., Muller D., Domagalska M.A., Leyser O. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development. 2010;137:2905–2913. doi: 10.1242/dev.051987. [DOI] [PubMed] [Google Scholar]
- 114.Tang G., Yan J., Gu Y., Qiao M., Fan R., Mao Y., Tang X. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58:118–125. doi: 10.1016/j.ymeth.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Han H., Liu X., Zhou Y. Transcriptional circuits in control of shoot stem cell homeostasis. Curr. Opin. Plant Biol. 2020;53:50–56. doi: 10.1016/j.pbi.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 116.Shi B., Vernoux T. Patterning at the shoot apical meristem and phyllotaxis. Curr. Top. Dev. Biol. 2019;131:81–107. doi: 10.1016/bs.ctdb.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 117.Cao X., Jiao Y. Control of cell fate during axillary meristem initiation. Cell. Mol. Life Sci. 2019 doi: 10.1007/s00018-019-03407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Talbert P.B., Adler H.T., Parks D.W., Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–2735. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- 119.Jin S., Zong Y., Gao Q., Zhu Z.X., Wang Y.P., Qin P., Liang C.Z., Wang D.W., Qiu J.L., Zhang F., et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 120.Kwon C.T., Heo J., Lemmon Z.H., Capua Y., Hutton S.F., Van Eck J., Park S.J., Lippman Z.B. Rapid customization of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 2020;38:182–188. doi: 10.1038/s41587-019-0361-2. [DOI] [PubMed] [Google Scholar]
- 121.Eisen T.J., Eichhorn S.W., Subtelny A.O., Lin K.S., McGeary S.E., Gupta S., Bartel D.P. The Dynamics of Cytoplasmic mRNA Metabolism. Mol. Cell. 2020;77:786–799.e10. doi: 10.1016/j.molcel.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]