Abstract
Purpose
Single-stranded DNA (ssDNA) library has shown to enrich shorter and more degraded DNA fragments than double-stranded DNA (dsDNA) library. In this study, we evaluated whether ssDNA libraries capture more circulating tumor DNAs (ctDNAs) in plasma cell-free DNA (cfDNA).
Materials and Methods
We prepared dsDNA, ssDNA and pure-ssDNA (capture the pre-existing ssDNA) libraries using ten plasma cfDNA samples. After low-pass whole genome sequencing, we calculated duplicate rate to estimate library complexity and compared the library insert sizes between different library methods. Finally, we estimated ctDNA content and plasma genomic abnormality (PGA) score, an indicator of ctDNA burden.
Results
27 libraries were prepared and sequenced from the ten cfDNA samples. Duplicate rate in the ssDNA and pure-ssDNA libraries was significantly lower than dsDNA libraries (p< 0.001 and p< 0.01, respectively). ctDNA content and PGA scores were consistently higher in ssDNA and pure-ssDNA libraries than in matched dsDNA libraries (p< 0.005). The higher ctDNA content in ssDNA libraries was associated with smaller library insert size.
Conclusions
ssDNA libraries preserve more diversity and capture more ctDNA than dsDNA libraries. The ssDNA library method is preferred when performing genomic analysis of ctDNA.
1. Introduction
Liquid biopsy, using cells, nucleic acids and proteins in body fluid, is a minimally invasive and real-time accessible approach for disease detection and monitoring (1, 2). Cell-free DNAs (cfDNAs) are short and degraded DNA fragments in circulation that are detectable by many genetic technologies including next-generation sequencing (NGS) (3). In cancer patients, circulating cell-free tumor DNAs (ctDNAs) shed by tumor cells, often harboring tumor-specific aberrations, are believed to have a great potential for cancer diagnosis and prognosis (4, 5). Although ctDNA analysis has become an important approach for liquid biopsy, many challenges still remain (6). Among those are low ctDNA content in high background of cfDNAs and high degradation of ctDNAs (6). Sensitive methods to enrich ctDNAs will have significant impact on clinical applications of the liquid biopsy.
Currently, double-stranded DNA (dsDNA) libraries have been widely used for DNA sequencing. However, dsDNA library preparation methods are insensitive to short, degraded, single-stranded DNA (ssDNA) as well as DNAs with single-strand breaks and modifications (7). In contrast, ssDNA library method showed an advantage of recovering shorter, degraded and fragmented DNAs. Successful applications of ssDNA libraries have been reported in ancient DNA (7), ultra-short-cfDNA in plasma (8) and the formalin-fixed and paraffin-embedded (FFPE) cancer tissue DNA (9). Studies on the fragment size of ctDNAs have revealed that ctDNAs are more fragmented (10), shorter in the principal fragment length (11, 12) and that the shorter fragments preferentially carry tumor-associated copy number aberrations (12). Size selection for fragments between 90 to 150 bp significantly enriched ctDNAs and improved detection of mutations (13). These results lead us to hypothesize that ssDNA library preparation may have some advantages to enrich ctDNAs among total cfDNAs and thus increase chance of success for liquid biopsy-based cancer genomic and genetic study.
The present study is to determine whether the ssDNA libraries could enrich more ctDNAs from the plasma cfDNAs in cancer patients when compared to traditional dsDNA method. To evaluate the pre-existing ssDNA in plasma for its potential to capture more ctDNA, we prepared the “pure” ssDNA (pure-ssDNA) library by skipping the denaturation step. The results from this study will further clarify if ssDNA library method shows any advantage in ctDNA enrichment over dsDNA method.
2. Materials and Methods
2.1. Sample Collection and cfDNA extraction
We used ten plasma samples (PS1 to PS10) with relatively high tumor DNA content from eight advanced and metastatic cancer patients based on previously published results (14, 15). Each patient had one plasma sample except patients 1 and 2, who had two plasma samples at different time points during chemotherapy. Clinical characteristics of these patients were listed in Supplementary Table S1. All plasma samples were separated within two hours after blood draw and frozen immediately at −80°C without any freeze-thaw cycle before use. These plasma samples were prepared using platelet-poor protocol (double spins, each at 2000 rpm for 10 min). After thaw on ice, we applied an additional centrifugation at 3000 rpm for 10 min before DNA extraction. We extracted cfDNAs from 800 μl of plasma using QIAamp DNA Blood Mini Kit (QIAGEN, Valencia, CA, USA) and quantified final DNA eluents using Qubit 2.0 Fluorometer (Life Technology, Carlsbad, CA, USA) with Qubit dsDNA HS Assay Kit (Invitrogen). Ten additional plasma cfDNAs (Samples 1 to 10 in Supplementary Table S2) were extracted from ten late stage cancer patients using the same protocol. dsDNA and ssDNA were quantified by combinational results from Qubit dsDNA HS Assay and ssDNA Assay kit (Invitrogen). The purified cfDNAs were stored at −80°C until use. Informed consent was obtained from all patients. This study was approved by Institutional Review Boards at Medical College of Wisconsin.
2.2. Library preparation and DNA sequencing
For each sample, 2 ng cfDNA was used to prepare dsDNA (Rubicon Genomics, Ann Arbor, Michigan) and ssDNA (Swift Bioscience, Ann Arbor, Michigan) libraries. The dsDNA library preparation method included end-repair, addition of adapters and 10 cycles of amplification. We purified these dsDNA libraries with beads ratio of 1.0 (1:1, beads to sample ratio) (Agencourt AMPure XP Beads, Beckman Coulter, Indianapolis, IN, USA). The ssDNA library method included denaturation, adaptase, extension, adaptor ligation and 10–14 cycles of PCR amplification. The ssDNA preparation process also included post-extension cleanup, post-ligation cleanup and post-PCR cleanup, with beads ratios of 2.0, 1.8 and 1.2 respectively. For seven of the ten cfDNA samples (PS1, PS2, PS6, PS7, PS8, PS9 and PS10), we also used 2 ng cfDNA to prepare pure-ssDNA libraries by skipping the DNA denaturation step to capture the pre-existing ssDNA. Because of limited availability of cfDNA, the pure-ssDNA libraries were prepared for seven of ten samples only. Processes of pure-ssDNA libraries were basically the same as ssDNA libraries except that the beads ratio of post-PCR cleanup was 1.2 for two samples (PS1 and PS2) and 1.0 for other five samples (PS6, PS7, PS8, PS9 and PS10). We diluted the library DNAs to a concentration of 10 nM and then pooled them for pair-end sequencing using Illumina HiSeq2500 (Illumina, Inc., San Diego, CA, USA).
2.3. Duplicate rate calculation
Library complexity is a key determinant of sequencing library quality. Less PCR artifacts and more diverse DNA sequences will lead to lower duplicate rate in the sequencing results. To evaluate complexity of the DNA libraries, we first mapped the sequencing data (fastq files) to human genome (hg38) and limited a total of mapped read counts to 1,000,000. We then retrieved the duplicate read counts from each mapping report (DNASTAR, Madison, WI). The duplicate rate was defined as number of duplicate reads per 100 mapped reads. To ensure compatibility, we compared duplicate rates in DNA libraries with same PCR amplification cycles (10 cycles).
2.4. ctDNA content calculation and plasma genome abnormality score
To calculate ctDNA content, we first mapped fastq files to human genome (hg38) and then binned the mapped reads into 1 Mb genomic windows (total 3113 genomic bins). The read count was adjusted to the global mean count for each sample. The read count ratio in each bin was calculated by dividing adjusted read count in patient to two unrelated healthy controls. The controls were prepared using the same library preparation protocols (either dsDNA or ssDNA method). The resulting ratios were transformed with log2 and adjusted for GC content (16). The fully normalized log2 ratios were subjected to segmentation using the copy number analysis method (Golden Helix, Bozeman, Montana). After the segmentation, we selected the most significantly deleted genomic region with segment size >20 Mb in each library pair and calculated ctDNA content using formula: 100 × (1 – 2log2 ratio), where log2 ratio is the mean log2 ratio value in the selected genomic region.
To best estimate ctDNA burden in peripheral blood as described before (14, 17), we applied a composite score algorithm, plasma genomic abnormality score (PGA score) to reflect overall ctDNA burden in peripheral blood. For each library, we summed the absolute values of the log2-ratios from ten most significant deletions/amplifications (segment size >10Mb and selected segments were patient-dependent) as PGA-Top10. A higher PGA score indicates higher ctDNA content in the plasma sample and hence, higher tumor burden in the patient.
2.5. Statistical analysis
Differences of ctDNA content and PGA scores between libraries were analyzed by student’s paired t-test. The correlation analysis was carried out by simple linear regression using IBM SPSS statistics (version 22). p value < 0.05 was considered as statistically significant.
3. Results
3.1. Mapping statistics of ssDNA and dsDNA libraries
We prepared and sequenced a total of 27 libraries including ten dsDNA libraries, ten ssDNA libraries and seven pure-ssDNA libraries, using ten cfDNAs from patient plasma. In total, we received an average of 24,264,998 raw reads and 19,495,014 mappable reads per sequencing library. Specifically, for the dsDNA libraries, the average number of mappable read was 12,847,819, ranging from 7,868,823 to 16,583,641. For the ssDNA library, average mappable read was 30,491,953 (ranging from 13,322,967 to 39,845,857) while for the pure-ssDNA library the average mappable read was 13,281,097 counts (ranging from 9,666,846 to 23,387,516). Overall, ssDNA libraries and pure-ssDNA libraries have lower mapping rate when compared to dsDNA libraries. This result is consistent with a previous study showing that ssDNA library recovers more non-human cfDNA possibly because of greater sensitivity of ssDNA library preparation to short fragment cfDNA (8). The detailed mapping statistics of each library, including total reads, mappable reads (mapped reads) and mapping rate (ratio of mapped reads to total reads), were summarized in Supplementary Table S3.
3.2. Lower duplicate rate in ssDNA libraries
Since library complexity is a key feature to estimate the quality of sequencing library preparation methods, we calculated and compared duplicate rates in these libraries with the same amplification cycles (Supplementary Table S4). Among 27 libraries, 18 libraries with ten PCR amplification cycles were selected and compared. These 18 libraries included ten dsDNA libraries, six ssDNA libraries and two pure-ssDNA libraries. We found that duplicate rates in the ssDNA libraries (mean= 0.057%, range= 0.046–0.083%) and pure-ssDNA libraries (mean= 0.06%, range= 0.054–0.065%) were significantly lower than in dsDNA libraries (mean= 0.2%, range= 0.14–0.249%) (p< 0.001 and p< 0.01, respectively) (Fig. 1).
Fig. 1.
Duplicate rates in dsDNA, ssDNA and pure-ssDNA libraries. The duplicate rates in ssDNA libraries and pure-ssDNA libraries were significantly lower than in dsDNA libraries in all libraries with same PCR cycles (p<0.01)
3.3. High proportion of pre-existing ssDNA in plasma cfDNA
To determine ssDNA proportion in cfDNA, additional plasma cfDNAs were retrieved from ten late stage cancer patients and quantified for dsDNA and ssDNA (see in Materials and Methods). The dsDNA kit is highly selective for dsDNA while the ssDNA kit is for ssDNA as well as dsDNA and RNA. Since we extracted DNA from plasma and treated the products with RNase, we could exclude the RNA contamination in the final elute. The quantification results showed a significant amount of ssDNA (1.4–2.8 folds to dsDNA) in the extracted cfDNAs (Supplementary Table S2), suggesting that the ssDNA in plasma cfDNAs may provide abundant resource for the pure-ssDNA library preparation.
3.4. Smaller inserts in ssDNA libraries
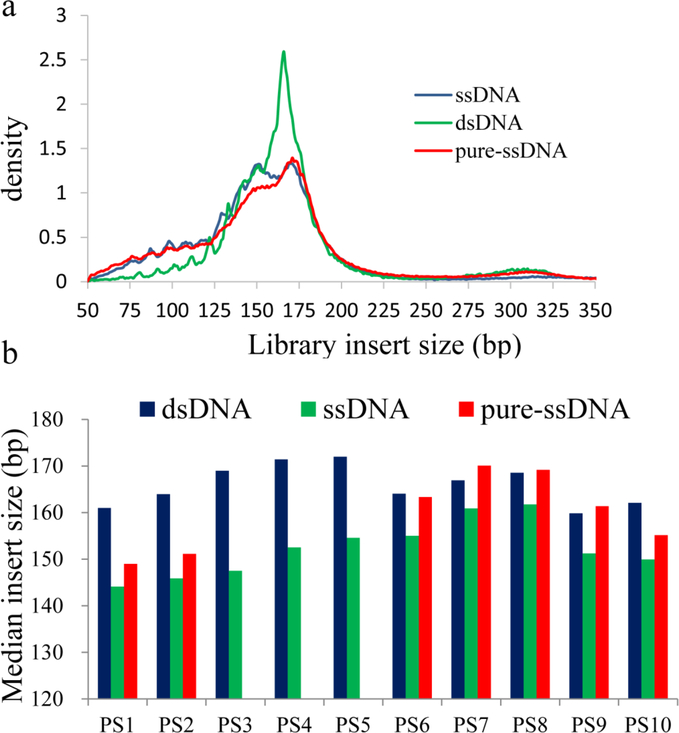
To compare fragment sizes of these libraries, we extracted median insert sizes and also generated the density plots of size distribution from the pair-end sequencing data. A single peak at 166bp could be seen in dsDNA libraries while two or more peaks from 130 to 170 bp were observed in ssDNA and pure-ssDNA libraries. 10 nt subpeaks were also clearly revealed by both methods. Besides, sub-nucleosomal cfDNA (fragment lengths below 100 bp) was significantly higher in ssDNA libraries (p<0.001) and pure-ssDNA libraries (p<0.005) when compared to dsDNA libraries. The highly fragmented cfDNA (<150bp) also showed a similar trend in both ssDNA libraries and pure-ssDNA libraries. A representative figure of density plot of the ds/ss/pure-ss library was shown in Figure 2a. More specifically, the median insert size of ssDNA libraries was on average 14 bp shorter (range from 6 bp in PS7 to 21 bp in PS3) than their matched dsDNA libraries (Fig. 2b). The median sizes of pure-ssDNA libraries were 12–13 bp shorter than their matched dsDNA libraries in two samples (PS1 and PS2). However, there was no obvious median size difference between pure-ssDNA and dsDNA libraries in other five samples (PS6 to PS10) when beads ratio was lowered in the pure-ssDNA library (see in Materials and Methods) (Fig. 2b). Possibly, the size difference may be attributable to library preparation methods (dsDNA/ssDNA) as well as the bead purification ratio.
Fig. 2.
Fragment sizes of dsDNA, ssDNA and pure-ssDNA libraries. (a). Representative size distribution of different library preparation methods. ssDNA and pure-ssDNA methods showed significant enrichment of sub-100bp fragments. (b). ssDNA libraries showed 6–21 bp smaller fragment size than their matched dsDNA libraries. The size of pure-ssDNA libraries was 12–13 bp smaller in PS1 and PS2 than dsDNA libraries but similar to dsDNA libraries in PS6 to PS10
3.5. Higher ctDNA content and PGA score in ssDNA libraries
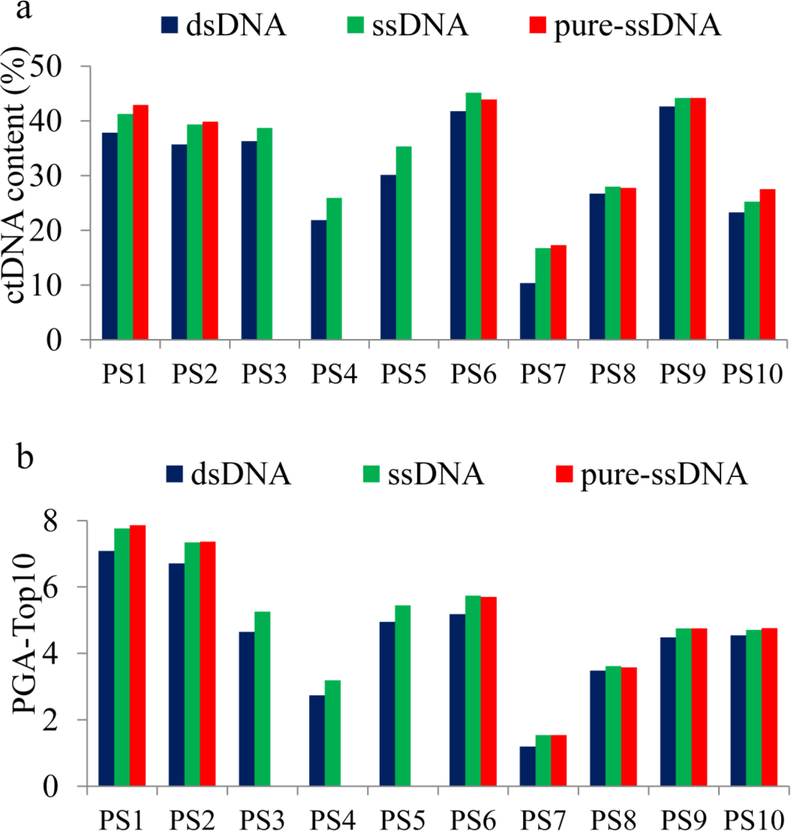
To compare the ctDNA enrichment efficiency, we performed copy number analysis and selected the most significant deletion to calculate the ctDNA content. We first binned the mapped reads (~19.5 million reads per sample) into 1 Mb genomic windows and normalized the read counts using unrelated healthy controls. We then performed segmentation analysis to estimate copy number changes and selected the most significantly deleted segment in each library pair for ctDNA content calculation (Supplementary Table S5). This analysis showed that ctDNA content was consistently higher in ssDNA libraries than in dsDNA libraries (p<0.0005). Comparing ssDNA libraries (mean ctDNA content= 34%) to their matched dsDNA libraries (mean ctDNA content= 30.7%), the increased ctDNA content ranged from 1.3% to 6.4% (mean= 3.3%). ctDNA content was also significantly higher in pure-ssDNA libraries than in their matched dsDNA libraries (p<0.005). The increased ctDNA content ranged from 1.0% to 6.9% (mean=3.6%) (Fig. 3a). Therefore, when compared to dsDNA method, both ssDNA and pure-ssDNA methods showed consistent but limited increase for ctDNA content.
Fig. 3.
ctDNA content differences between dsDNA, ssDNA and pure-ssDNA libraries. (a) ctDNA content was consistently higher in ssDNA libraries and pure-ssDNA libraries than matched dsDNA libraries; (b) Plasma ctDNA burden (PGA-Top10) was consistently higher in ssDNA libraries and pure-ssDNA libraries than matched dsDNA libraries
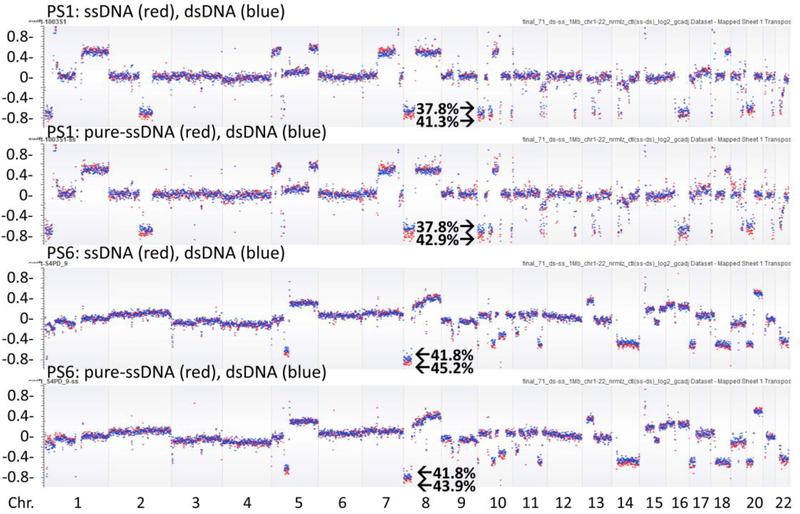
Previously, we reported a composite score algorithm (PGA score) to represent overall ctDNA burden in peripheral blood (see in Materials and Methods). To calculate the PGA scores in present study, we summed the absolute values of the log2-ratios from top ten deleted/amplified segments (designated as PGA-Top10). The chromosome coordinates of the ten segments selected (including the most significant deletion for ctDNA content calculation), the log2-ratios of the segments in each library, the calculated ctDNA content and PGA scores were listed in Supplementary Table S5. We found that, when compared to the matched dsDNA method, the PGA-Top10 was always higher in the ssDNA libraries (p<0.0001) or pure-ssDNA libraries (p<0.005) (Fig. 3b).To further demonstrate the ctDNA content difference, we plotted the log2 values of copy number along chromosomes 1–22 and observed a clear separation of copy number changes between the two library preparation methods, proving higher ctDNA in the ssDNA or the pure-ssDNA library method than in the dsDNA library method (Fig. 4).
Fig. 4.
Copy number change across the genome in different libraries. Copy number changes (log2 values) were clearly separated between ssDNA libraries or pure-ssDNA libraries (red dots) and their matched dsDNA libraries (blue dots). ctDNA content was higher in ssDNA libraries or pure-ssDNA libraries than dsDNA libraries. Segment deletions in chromosome 10p (in PS1) and 8p (in PS6) were used for ctDNA content calculation (ctDNA percentage indicated). The x axis indicated the chromosomes and the y axis indicated the log2 copy number values
3.6. Impact of library insert size on ctDNA content
To evaluate effect of fragment size on ctDNA content, we first calculated insert fragment size differences and ctDNA content differences between ten ssDNA and their paired dsDNA libraries. We then performed correlation analysis and observed a significant positive correlation (R= 0.673, p= 0.017) (Supplementary Figure S1). This data support that enrichment of smaller cfDNA fragments may partially contribute to the higher ctDNA content in these ssDNA libraries. Meanwhile, for the five pure-ssDNA libraries, from plasma sample PS6 to PS10, the median insert size of pure-ssDNA libraries were similar to dsDNA libraries, possibly due to lowered beads ratio in these pure-ssDNA libraries. However, the PGA-Top10 values of these pure-ssDNA libraries were still consistently higher than in matched dsDNA libraries. These results indicated that library insert size as well as cfDNA characteristics may contribute to ctDNA enrichment efficiency.
4. Discussion
cfDNA analysis has shown great utilities for detecting and monitoring pregnancy, hereditary diseases, cancers and therapy response. However, the quantity of target DNAs, such as ctDNAs in circulation, is often extremely low in relatively high background of the normal cfDNAs. Methods increasing ctDNA content and improving the sensitivity of detection are highly demanded. Previous studies have demonstrated that ssDNA method recovered more shorter DNA fragments than dsDNA method (7, 8). Since ctDNAs are usually shorter and degraded when compared to normal cfDNAs, it is reasonable to hypothesize that ssDNA library may enrich more ctDNAs than dsDNA libraries. To test this hypothesis, we constructed and compared both dsDNA and ssDNA libraries using cfDNA samples from advanced stage cancer patients. The ctDNA content is calculated using log2 value of a single most significant genomic deletion, which represents percentage of ctDNA in overall cfDNA background. Meanwhile, plasma genomic abnormality score (PGA score) is calculated using log2 values of top 10 most significant copy number changes, which represents a composite score for ctDNA content (14, 17). Although not equal, both ctDNA content and PGA-Top10 showed similar tendency and can be used as an indicator of overall tumor burden. Furthermore, our results showed that ssDNA may be more represented than dsDNA in plasma cfDNAs, which is in line with a previous publication in circulating exosome DNA (18). Additionally, by skipping the denaturation step to ideally capture the pre-existing ssDNA, the pure-ssDNA library also showed consistently higher ctDNA content. However, we should also note that ssDNA-specific quantification method is currently not available and the exact abundance of ssDNA in plasma needs further determination. The pure-ssDNA library work here is also preliminary and needs more verification.
Higher ctDNA content in ssDNA libraries may be attributable to following reasons: First, the ability of ssDNA library to ligate shorter and damaged cfDNAs. Consistent with Sanchez’s previous results (19), our data showed that, highly fragmented cfDNA was more enriched by ssDNA library. The range of shortening of length varied in different studies which may be related to the sampling time, cancer types and stages, as well as details of library preparation methods (8, 9, 20, 21). Besides, fragment size difference is positively correlated with the PGA score difference between the ssDNA and dsDNA libraries, suggesting that smaller insert size in ssDNA library may partially contribute to enrichment of ctDNAs. Consistently, by selecting shorter fragments in plasma samples from cancer patients, Mouliere et al. observed significant enrichment of ctDNA (13). However, library size may not be the only contributing factor. When lowering beads ratio in five pure-ssDNA libraries, we found that all these libraries still had higher ctDNA content than dsDNA libraries although median insert size was not significantly smaller than matched dsDNA libraries. Clearly, multiple factors contribute to the higher ctDNA content in ssDNA-based library methods.
Second, ssDNA library may ligate a wider variety of DNA molecules including nicked DNA, preexisting ssDNA and one strand end modified DNA. In this study, ssDNA library had the DNA denaturation step and captured both the dsDNA and preexisting ssDNA while pure-ssDNA library skipped the denaturation step and captured the preexisting ssDNA. Both the ssDNA library and pure-ssDNA library consistently showed higher ctDNA enrichment, suggesting that preexisting ssDNA and nicked DNA are abundant in plasma. It is reasonable to hypothesize that ctDNAs may be more degraded, single stranded or have more single-stranded breaks, and hence, have higher enrichment efficiency by ssDNA method. Additionally, end modifications on one strand may inhibit the capture of entire DNA fragment by dsDNA library method while availability of opposite strand provides additional chance for ssDNA library method. Thus, ssDNA library methods may have more chances to capture the differently degenerated DNA or modified DNA. Consistently, in this study, we did observe that ssDNA libraries have much lower duplicate rates, suggesting that ssDNA library method may better preserve the diversity and complexity of ctDNAs.
Third, ctDNAs may preserve specific molecular property that facilitates adaptor ligation by the ssDNA library method (22). Vong and his colleges reported that, although fetal cfDNA in maternal plasma is shorter than maternal cfDNA, ssDNA library preferentially enriches short maternally derived cfDNA but not shorter fetal DNA (21). They hypothesized that, instead of physical size selection process, there may be biological differences between fetal and maternal cfDNAs which make ssDNA library preferentially enrich maternal cfDNAs (21). In contrast to our study, Moser’s previous publication showed that ssDNA library does not preferentially enrich ctDNA although ssDNA library has shorter insert size than dsDNA libraries (20). The conflict result may be attributable to many factors including specific ligation reaction used in the ssDNA library kit (Adaptase from Swift Bioscience).
The current study also has some limitations. First, number of the samples used was relatively small. Especially, for pure-ssDNA method, only seven of ten samples had sufficient cfDNA for library preparation. More tests are needed to confirm our observation. Second, although omitting denaturation step, the pure-ssDNA method may still capture dsDNA because of the nicks and overhangs. The method specific for ssDNA is needed to inclusively evaluate ssDNAs for their characteristics. Third, when evaluating the library diversity, the PCR amplification cycles may increase amplification bias. By increasing the cfDNA input and subsequently decreasing the amplification cycles, duplicate rates of libraries can be significantly reduced. Besides, although ssDNA method consistently enriches ctDNA content, extent of this increase was very limited. It is possible to further improve the ctDNA enrichment by combination of ssDNA method with other methods such as size selection.
Together, our results showed that ssDNA library method enriched more ctDNAs when compared to dsDNA method. This study provides additional evidence that favors ssDNA over dsDNA library method for ctDNA enrichment. This study also provides preliminary evidence showing the abundance of pre-existing ssDNA in plasma. Further understanding of the causes of ctDNA enrichment among different methods may allow more sensitive liquid biopsy assay to be developed for clinical applications.
Supplementary Material
Supplementary Figure S1 Correlation between insert size difference and ctDNA burden difference. For ssDNA and their matched dsDNA libraries, a positive correlation (r= 0.673, p= 0.017) was found between insert size difference and ctDNA burden difference.
Key Points.
ssDNA sequencing libraries captured shorter and more diverse cfDNAs than dsDNA libraries.
ssDNA sequencing libraries enriched more ctDNA than dsDNA libraries.
ssDNA library method is a preferred option for genomic analysis of ctDNA.
Acknowledgments
We thank Molecular Pathology Core in Medical College of Wisconsin for timely sequencing service. We thank the scholarship from the China Scholarship Council (CSC) (201508230031 to JZ).
Funding This research was supported by the National Institute of Health (R01CA212097) to LW; the University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2017056), Natural Science Foundation of Heilongjiang Province (YQ2019H002) and the Fund Program of Heilongjiang Province for Selected Returned Overseas Professionals to JZ.
Footnotes
Conflicts of Interest The authors, JZ, JH, PZ, QL, MK, CH, and LW, declare that they have no conflict of interest.
References
- 1.Thierry AR, El Messaoudi S, Gahan PB, Anker P, Stroun M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016;35(3):347–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–48. [DOI] [PubMed] [Google Scholar]
- 3.Ignatiadis M, Dawson SJ. Circulating tumor cells and circulating tumor DNA for precision medicine: dream or reality? Ann Oncol. 2014;25(12):2304–13. [DOI] [PubMed] [Google Scholar]
- 4.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23. [DOI] [PubMed] [Google Scholar]
- 5.Otandault A, Anker P, Al Amir Dache Z, Guillaumon V, Meddeb R, Pastor B, et al. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 2019;30(3):374–84. [DOI] [PubMed] [Google Scholar]
- 6.Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18(8):1174–96. [DOI] [PubMed] [Google Scholar]
- 7.Gansauge MT, Meyer M. Single-stranded DNA library preparation for the sequencing of ancient or damaged DNA. Nat Protoc. 2013;8(4):737–48. [DOI] [PubMed] [Google Scholar]
- 8.Burnham P, Kim MS, Agbor-Enoh S, Luikart H, Valantine HA, Khush KK, et al. Single-stranded DNA library preparation uncovers the origin and diversity of ultrashort cell-free DNA in plasma. Sci Rep. 2016;6:27859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stiller M, Sucker A, Griewank K, Aust D, Baretton GB, Schadendorf D, et al. Single-strand DNA library preparation improves sequencing of formalin-fixed and paraffin-embedded (FFPE) cancer DNA. Oncotarget. 2016;7(37):59115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouliere F, Robert B, Arnau Peyrotte E, Del Rio M, Ychou M, Molina F, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6(9):e23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, et al. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016;12(7):e1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P, Chan CW, Chan KC, Cheng SH, Wong J, Wong VW, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A. 2015;112(11):E1317–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10(466). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia S, Kohli M, Du M, Dittmar RL, Lee A, Nandy D, et al. Plasma genetic and genomic abnormalities predict treatment response and clinical outcome in advanced prostate cancer. Oncotarget. 2015;6(18):16411–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Dittmar RL, Xia S, Zhang H, Du M, Huang CC, et al. Cell-free DNA copy number variations in plasma from colorectal cancer patients. Mol Oncol. 2017;11(8):1099–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36(19):e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia S, Huang CC, Le M, Dittmar R, Du M, Yuan T, et al. Genomic variations in plasma cell free DNA differentiate early stage lung cancers from normal controls. Lung Cancer. 2015;90(1):78–84. [DOI] [PubMed] [Google Scholar]
- 18.Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, et al. Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J Extracell Vesicles. 2018;7(1):1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanchez C, Snyder MW, Tanos R, Shendure J, Thierry AR. New insights into structural features and optimal detection of circulating tumor DNA determined by single-strand DNA analysis. NPJ Genom Med. 2018;3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser T, Ulz P, Zhou Q, Perakis S, Geigl JB, Speicher MR, et al. Single-Stranded DNA Library Preparation Does Not Preferentially Enrich Circulating Tumor DNA. Clin Chem. 2017;63(10):1656–9. [DOI] [PubMed] [Google Scholar]
- 21.Vong JSL, Tsang JCH, Jiang P, Lee WS, Leung TY, Chan KCA, et al. Single-Stranded DNA Library Preparation Preferentially Enriches Short Maternal DNA in Maternal Plasma. Clin Chem. 2017;63(5):1031–7. [DOI] [PubMed] [Google Scholar]
- 22.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016;164(1–2):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1 Correlation between insert size difference and ctDNA burden difference. For ssDNA and their matched dsDNA libraries, a positive correlation (r= 0.673, p= 0.017) was found between insert size difference and ctDNA burden difference.