Abstract
Zika virus (ZIKV) nonstructural protein 5 (NS5) plays a critical role in viral RNA replication and mediates key virus-host cell interactions. As with other flavivirus NS5 proteins, ZIKV NS5 is primarily found in the nucleus. We previously reported that the NS5 protein of dengue virus, another flavivirus, localized to centrosomes during cell division. Here we show that ZIKV NS5 also relocalizes from the nucleus to centrosomes during mitosis. In infected cells with supernumerary centrosomes, NS5 was present at all centrosomes. Transient expression of NS5 in uninfected cells confirmed that centrosomal localization was independent of other viral proteins. Live-cell imaging demonstrated that NS5-GFP accumulated at centrosomes shortly after break down of nuclear membrane and remained there through mitosis. Cells expressing NS5-GFP took longer to complete mitosis than control cells. Finally, an analysis of ZIKV NS5 binding partners revealed several centrosomal proteins, providing potential direct links between NS5 and centrosomes.
Keywords: Zika virus, flavivirus, nonstructural protein NS5, RNA-dependent RNA polymerase, centrosome
1. Introduction
ZIKV was first isolated in 1947 from a sentinel rhesus macaque found in the Zika forest of Uganda (Dick et al., 1952). ZIKV is a mosquito-borne, positive-sense, single-stranded RNA virus belonging to the family Flaviviridae, which includes dengue virus (DENV), West Nile virus and Japanese encephalitis virus. Clinical manifestations of ZIKV infection are usually mild and include fever, rashes, arthralgia, myalgia and conjunctivitis (Petersen et al., 2016). In 2007, the first major ZIKV outbreak was reported in the Yap islands, but no significant pathologies were detected (Baud et al., 2017; Duffy et al., 2009). However, during subsequent outbreaks in French Polynesia and Brazil, ZIKV became a global health concern when an increased incidence of microcephaly was reported in children born to mothers who were infected during pregnancy (Baud et al., 2017).
The ZIKV genome encodes ten proteins that are translated as a single polyprotein. Processing by viral and cellular proteases releases the structural proteins C, prM and E, and the nonstructural (NS) proteins 1, 2A, 2B, 3, 4A, 4B and 5. NS5 is the largest and most highly conserved of these. All flavivirus NS5 proteins contain a methyltransferase domain at the N-terminus and an RNA-dependent RNA polymerase (RdRp) at the C-terminus. The structures of flavivirus methyltransferase and RdRp domains are very similar, with some differences in the relative orientation of the two domains (Coloma et al., 2016; Coutard et al., 2017; Duan et al., 2017; Godoy et al., 2017; Stephen et al., 2016; Upadhyay et al., 2017; Wang et al., 2017; Zhang et al., 2017; Zhao et al., 2017; Zhou et al., 2017). ZIKV NS5 can also form higher order oligomers that appear as extended fibrils in vitro or spherical structures in infected cells (Ng et al., 2019; Zhao et al., 2017). In addition to its direct role in viral RNA synthesis and capping, NS5 mediates important virus-host cell interactions. ZIKV NS5 interferes with both the expression of interferon and with the downstream interferon signaling pathway (Grant et al., 2016; Lin et al., 2019; Xia et al., 2018); stimulates an inflammatory response by binding to and activating the NLRP3 inflammasome (He et al., 2018; Wang et al., 2018); and interacts with multiple cellular splicing factors, leading to reduction in pre-mRNA processing (Coyaud et al., 2018; De Maio et al., 2016). Despite the fact that flavivirus RNA replication occurs in the cytoplasm, many NS5 proteins, including ZIKV NS5, are found primarily in the nucleus due to nuclear localization signals in the linker between the methyltransferase and RdRp domains or at the C-terminus (Grant et al., 2016; Ng et al., 2019; Pryor et al., 2007; Tay et al., 2016).
In a previous study, we reported that DENV NS5 interacted with centrosomal proteins (Khadka et al., 2011). Although DENV NS5 from serotypes 2-4 is predominantly in the nucleus (Tay et al., 2016), DENV2 NS5 localized at centrosomes in mitotic cells (Khadka et al., 2011). Centrosomes are the primary microtubule organizing centers (MTOC) of vertebrate cells, consisting of pair of centrioles embedded in pericentriolar material (PCM), a dynamic matrix of proteins (Andersen et al., 2003; Conduit et al., 2015). Centrosomes increase the efficiency of bipolar spindle assembly and contribute to error-free segregation of chromosomes (Arquint et al., 2014; Conduit et al., 2015; Sir et al., 2013). Recently, supernumerary and structurally defective centrosomes were observed in ZIKV-infected cells (Gabriel et al., 2017; Souza et al., 2016; Wolf et al., 2017).
Since NS5 is one of the most highly conserved flavivirus proteins, we predicted that ZIKV NS5 also relocalizes to centrosomes during mitosis. We report here that this is indeed the case. In the absence of other ZIKV proteins, NS5 localized at the centrosomes in transiently transfected cells and stable cell lines. We also demonstrated that NS5 remained at centrosomes throughout mitosis and that stable cells expressing NS5-GFP took longer to complete mitosis than control cells. Finally, we identified centrosomal and spindle pole proteins that interact with ZIKV NS5, providing a potential link between NS5 and centrosomes.
2. Materials and methods
2.1. Plasmids
Plasmids pcDNA5/FRT/TO-NS5, pcDNA5/FRT/TO-NS5-GFP, pcDNA5/FRT/TO-nls-GFP, pZKVRLuc, pZKVRLuc (ΔDD) and pCAG-FLpO (a gift from Massimo Scanziani; Addgene plasmid # 60662; http://n2t.net/addgene:60662; RRID:Addgene_60662; (Matsuda and Cepko, 2007)) were used in this study. pcDNA5/FRT/TO-NS5 was cloned by amplifying ZIKV (strain FSS13025) NS5 from pFLZIKV (Shan et al., 2016) and inserting the resulting PCR product into the pcDNA5/FRT/TO vector backbone at the NotI and XhoI sites of pCDNA5-FRT-TO-SBP-TNRC6A (a gift from Kumiko Ui-Tei, Addgene plasmid # 42044; http://n2t.net/addgene:42044; RRID:Addgene_42044; (Nishi et al., 2013)). Similarly, pcDNA5/FRT/TO-NS5-GFP was constructed by sequentially inserting GFP and NS5 into the pcDNA5/FRT/TO vector backbone. To construct pcDNA5/FRT/TO-nls-GFP, an NLS was added to the 5’ end of GFP through two successive rounds of PCR with forward primers (ATGCCCAAGAAAAAGCGGAAAGTGGGCAGCGGCGTGAGCAAGGGCGAGGAG) and (AGCGTTTCCACCGCGGTGGCGGCCGCCACCATGCCCAAGAAAAAGCGGAA), and reverse primer (TTCCGCCGCTACCTCCGCCGGATCCGCCTCCCTTGTACAGCTCGTCCATGC, used in both rounds), followed by insertion into pcDNA5/FRT/TO. The ZIKV replicon plasmid pZKVRLuc was generated using the infectious pFLZIKV (strain FSS13025) cDNA clone as the backbone (Shan et al., 2016). A region encompassing RLuc-FMDV2A was amplified by overlap extension PCR to generate a T7 promoter-5’UTR-Nter21-RLuc-FMDV2A-E23-ZIKV-NS1 fusion with unique AgeI and SphI restriction sites at the ends. This fragment was then inserted in pFLZIKV digested with the same restriction enzymes. A replication defective replicon pZKVRluc (ΔDD) was generated by deleting the conserved GDD motif (corresponding to residues Gly664, Asp665, and Asp666) within the RdRp domain of NS5 by site directed mutagenesis. All constructs were verified by sequencing.
2.2. Antibodies
The following antibodies were used in this study: mouse anti-ZIKV NS5 (BioFront Technologies, BF-6A), chicken anti-ZIKV NS5 (generously provided by Dr. Sonja Best, National Institute of Allergy and Infectious Diseases) (Grant et al., 2016), rabbit anti-GFP (Invitrogen, A-6455), anti-actin (Sigma Aldrich, A1978), rabbit anti-pericentrin (Abcam, ab4448), Alexa-594 anti-mouse IgG (Invitrogen, A11032), Alexa-488 anti-chicken IgG (Invitrogen, A11039), Alexa-488 anti-rabbit IgG (Invitogen, A11034), Alexa-594 anti-rabbit IgG (Invitogen, A11012), IRDye800 anti-rabbit IgG (LI-COR, 926-32211), IRDye800 anti-chicken IgG (LI-COR, 92532218), and IRDye 680 anti-mouse IgG (LI-COR, 925-68070).
2.3. Cells:
Human embryonic kidney-293 (HEK-293), HEK-293T and Huh7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco™) + 10% FBS (Atlanta Biologicals) at 37° C in 5% CO2. Flp-In™ T-REx™ 293 cells (Invitrogen) were maintained in DMEM + 10% FBS + Zeocin (Gibco™) 100 μg/mL. Stable Flp-In™ T-REx™ 293 cells expressing transgenes were maintained in DMEM + 10% FBS supplemented with 200 μg/mL hygromycin (Gibco™). Vero cells were grown at 37° C in DMEM supplemented with 10% FBS in the presence of 5% CO2.
2.4. Transfections
HEK-293 were washed with phosphate-buffered saline (PBS) and replaced with fresh media. Circular plasmids were incubated with Lipofectamine2000 (Life Technologies) and DMEM for 5 minutes at room temperature. The transfection mix was added to the cells and media were replaced after 4 h.
2.5. ZIKV infection
ZIKV was propagated in Vero cells at 37° C in DMEM supplemented with 2% FBS in the presence of 5% CO2. The culture supernatant was clarified by centrifugation and filtered using 0.25μM filter. Huh7 cells were infected with ZIKV (strain H/PF/2013) at a multiplicity of infection (MOI) of 0.5.
2.6. Immunofluorescence assays
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Following two washes with PBS, cells were blocked in PBS with 1% bovine serum albumin (BSA). Cells were then incubated with primary antibodies overnight at 4° C, washed twice with PBS and incubated with secondary antibodies for 2 h at room temperature. After washing twice, cells were incubated with Hoechst stain.
2.7. Replicon rescue assay
pZKVRLuc and pZKVRLuc (ΔDD) were linearized using ClaI, purified on GFX columns (GE Healthcare), and incubated with T7 RNA polymerase (New England Biolabs) at 37° C for 1.5 h for in vitro transcription of RNA. HEK-293T cells were transfected with 5 μg of pZKVRLuc or pZKVRLuc (ΔDD) RNA using Lipofectamine 3000 (Thermo Fisher Scientific). Culture medium was replaced 4-6 h after transfection. At 12 h after transfection, pcDNA5/FRT/TO-NS5 was transfected into pZKVRLuc (ΔDD)-transfected HEK-293T cells using Lipofectamine 3000. Cells were washed once with PBS and lysed using lysis buffer (Promega) 60 h after transfection. Luciferase activity was measured using a luminescence microplate reader (LMAXII 384, Molecular Devices) per the manufacturer’s protocol.
2.8. Generation and induction of cell lines stably expressing NS5-GFP or nls-GFP
Tetracycline-inducible Flp-In™ T-REx™ cells stable cells expressing NS5-GFP and nls-GFP were generated as per the manufacturer’s protocol (Invitrogen). Flp-In™ T-REx™ 293 cells were grown overnight in DMEM + 10% FBS. Cells were cotransfected with pCAG-FLpO (1 μg) and either circular pcDNA5/FRT/TO-NS5-GFP or circular pcDNA5/FRT/TO-nls-GFP (100 ng) using Lipofectamine 2000 (Thermo Fisher Scientific). On the following day, cells were transferred to a 10 cm plate. At 48 h after transfection, 200 μg/mL hygromycin was added. After approximately 2 weeks, surviving cells were pooled and passaged. Expression of transgenes was induced by adding 2 ug/mL tetracycline (Sigma).
2.9. Imaging
Imaging of immunofluorescence assays was performed using a Nikon A1 confocal system on a Nikon Eclipse Ti microscope. To confirm expression of NS5-GFP and nls-GFP, cells were visualized using Zeiss Axio Observer 7 microscope. Time-lapse live-cell imaging was performed using an IncuCyte® S3 Live-Cell Analysis System with a 20X objective. Cells were placed in the instrument 20 - 24 h after transfection or induction with tetracycline.
2.10. Mitotic time measurement
The time required to complete mitosis was measured by analyzing images acquired at 10-minute intervals using an IncuCyte® S3 Live-Cell Analysis System. Rounding of cells was used as the indicator for onset of mitotic phase. The time point just prior to mitotic cell rounding was taken as the start of mitosis and the time at which rounding was no longer observed was taken as the end. The cells arrested in mitosis for more than 150 min and or that underwent mitotic catastrophe were excluded.
2.11. Yeast two-hybrid screen
NS5 cDNA was amplified from genomic ZIKV (strain H/PF/2013) RNA by RT-PCR. PCR products corresponding to full-length ZIKV NS5, the methyltransferase domain, and the RdRp domain were cloned into the yeast two-hybrid DNA-binding domain (DBD) plasmid pOBD2 via in vivo homologous recombination in the yeast strain R2HMet (MATa ura3–52 ade2–101 trp1– 901 leu2– 3,112 his3–200 met2Δ::hisG gal4Δ gal80Δ) (LaCount et al., 2005). The propensity of the DBD constructs to self-activate expression of auxotrophic reporter genes was assessed by growth on yeast two-hybrid selective media. The lowest concentration of 3-amino-1,2,4-triazole (3-AT) that suppressed growth was used for the yeast two-hybrid screens. The pOBD2 constructs were screened against cDNA libraries derived from human liver, macrophages, and interferon-treated macrophages. Yeast expressing interacting proteins were selected on synthetic deficient medium lacking tryptophan, leucine, uracil, histidine and adenine (SD-TLUHA) and 1 mM 3-AT. Human gene inserts from colonies that grew on SD-TLUHA were PCR-amplified, sequenced, and identified by searching the NCBI RefSeq mRNA database using BLASTN (Altschul et al., 1990).
To confirm yeast two-hybrid interactions, the human gene inserts were PCR-amplified and cloned into the activation domain plasmid pOAD102 by in vivo homologous recombination in the yeast strain BK100 (MATa ura3-52 ade2-101 trp1-901 leu2-3,112 his3-200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ) (LaCount et al., 2005). The resulting prey constructs were retested against all ZIKV NS5 bait constructs by assessing yeast growth on SD-TLUH and 1 mM 3-AT. Interactions were considered positive if at least two out of three independent assays demonstrated growth greater than negatives control strains.
3. Results
3.1. ZIKV NS5 localizes to the centrosomes in infected cells during mitosis
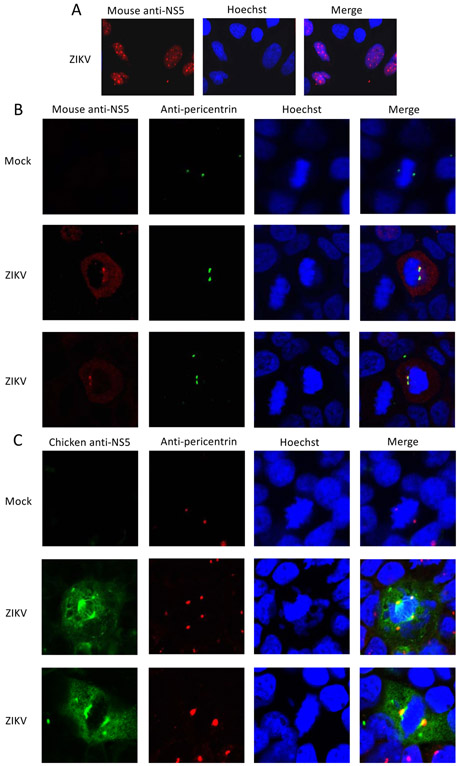
To test the hypothesis that ZIKV NS5 localizes to centrosomes during mitosis, immunofluorescence assays were performed on ZIKV-infected cells. As previously reported, NS5 was found in the nucleus in cells during interphase at punctate structures that are most likely Cajal bodies (Fig. 1A) (Coyaud et al., 2018; Grant et al., 2016). However, in cells undergoing mitosis, NS5 accumulated at centrosomes based on co-localization with the centrosomal protein pericentrin (Fig. 1B and C, Supplemental Fig. 1). NS5 localization was independently assessed with two different anti-ZIKV antibodies and each detected the presence of NS5 at centrosomes in mitotic cells. Some ZIKV-infected cells had supernumerary centrosomes and NS5 was detected at each centrosome in these cells as well (Fig. 1B and C, Supplemental Fig. 1).
Fig. 1. NS5 localizes at centrosomes in ZIKV-infected cells during mitosis.
ZIKV-infected Huh7 cells were fixed, stained with mouse anti-ZIKV NS5 (A and B, red), chicken anti-ZIKV NS5 (C, red), anti-pericentrin antibodies (B and C, green), and Hoechst (blue), and imaged by confocal microscopy. Panel A shows cells in interphase, whereas panels B and C show cells undergoing mitosis. A single ZIKV-infected cell is imaged in two focal planes to highlight the supernumerary centrosomes (B).
3.2. ZIKV NS5 localizes to centrosomes during mitosis independent of other viral proteins
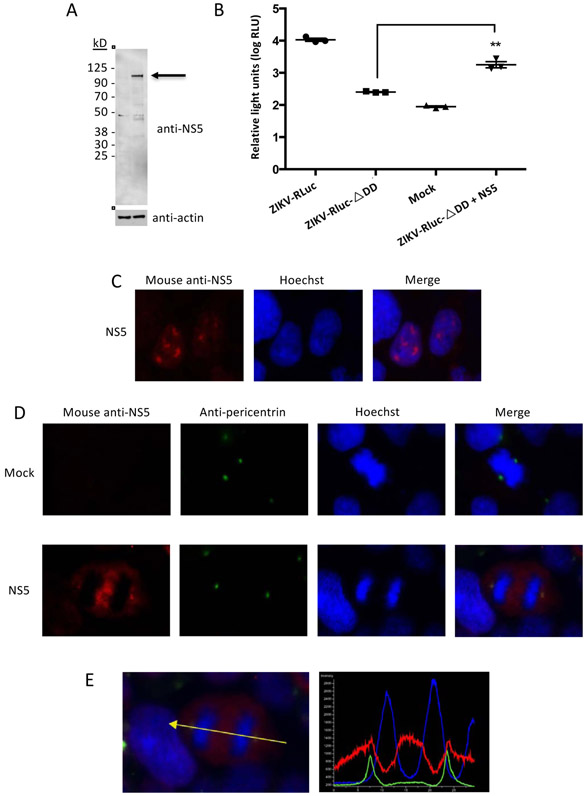
Flavivirus NS3 was previously shown to interact with centrosomal proteins (Coyaud et al., 2018; Khadka et al., 2011). Earlier studies also demonstrated that DENV NS5 interacts with NS3 (Kapoor et al., 1995). Thus, ZIKV NS5 could be recruited to centrosomes via its association with NS3. To investigate this possibility, HEK-293 cells were transfected with an NS5 expression plasmid and immunofluorescence assays were performed. Expression of full-length NS5 was verified by immunoblot (Fig. 2A). Transiently expressed ZIKV NS5 restored luciferase expression from a ZIKV replicon containing a mutation in the RdRp active site, indicating that the NS5 was functional (Fig. 2B). Consistent with our results from infected cells, NS5 was primarily found in the nucleus during interphase, but co-localized with pericentrin at centrosomes in cells undergoing mitosis (Fig. 2C-E). Thus, NS5 can associate with centrosomes independently of NS3 or any other viral protein.
Fig. 2. Transiently expressed NS5 localizes to centrosomes.
A. Immunoblot analysis of ZIKV NS5 expression in transiently transfected cells. Molecular weight markers are indicated at left. Arrow indicates full-length NS5 (predicted size 103 kDa).
B. Transiently expressed ZIKV NS5 rescues the activity of a replication-deficient ZIKV replicon. HEK-293T cells were sequentially transfected with a replication-defective ZIKV replicon bearing a mutation in the RdRp active site and a ZIKV NS5 expression plasmid, and assayed for luciferase activity. The graph depicts the average relative luminescence units (RLU) (±SD). The experiment was repeated twice with three technical replicates each. **Unpaired t-test p < 0.05.
C and D. Transiently expressed ZIKV NS5 localized to the nucleus during interphase (C) and to centrosomes in dividing cells (D). HEK-293 cells were transfected with a ZIKV NS5 expression plasmid, fixed, permeabilized, and stained with mouse anti-NS5 (red), anti-pericentrin antibodies (green) and Hoechst (blue).
E. Pixel intensities of NS5 (red), pericentrin (green) and chromosomal DNA (blue) along the line connecting the centrosomes were plotted using NIS-Elements AR software.
3.3. ZIKV NS5 association with centrosomes is maintained during mitosis
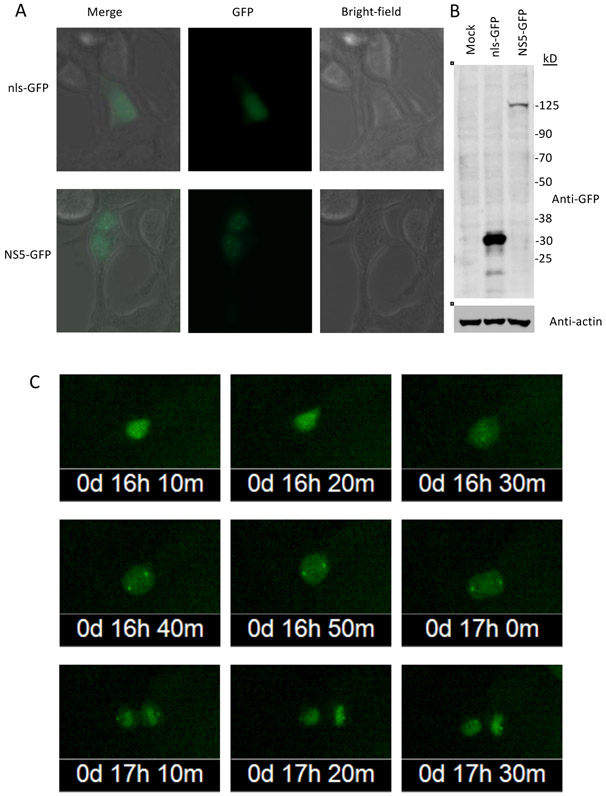
To further study the association of NS5 with centrosomes during mitosis, cells were transfected with plasmids expressing NS5-GFP or nls-GFP and visualized with time-lapse live cell imaging. Expression of NS5-GFP in HEK-293 cells was confirmed by immunoblot assay and fluorescent microscopy, the latter of which showed punctate nuclear distribution of NS5-GFP in non-dividing cells (Fig. 3A). Upon break down of the nuclear envelope during mitosis, nuclear-localized ZIKV NS5-GFP dispersed into the cytoplasm and soon accumulated at centrosomes of the cells (Fig. 3B). The localization at the centrosomes was maintained through the mitosis, even after the nuclear envelope was reassembled in telophase and most NS5-GFP relocated to the nucleus. In control cells expressing nls-GFP, no such accumulation of GFP was seen at centrosomes (Supplemental Fig. 2).
Fig. 3. Association of NS5-GFP with the centrosomes is maintained through mitosis.
HEK-293 cells were transiently transfected with NS5-GFP and nls-GFP expression plasmids.
A. NS5-GFP and nls-GFP expression were monitored by fluorescent microscopy (40X magnification).
B. Immunoblot analysis of NS5-GFP and nls-GFP expression. Blots were probed with anti-GFP antibodies. Molecular weight markers are shown at right (sizes in kDa). Predicted sizes of NS5-GFP and nls-GFP are 131 and 28 kDa, respectively.
C. Time-lapse images of an HEK-293 cell transiently expressing NS5-GFP undergoing mitosis.
3.4. Cells stably expressing ZIKV NS5 take longer to complete mitosis
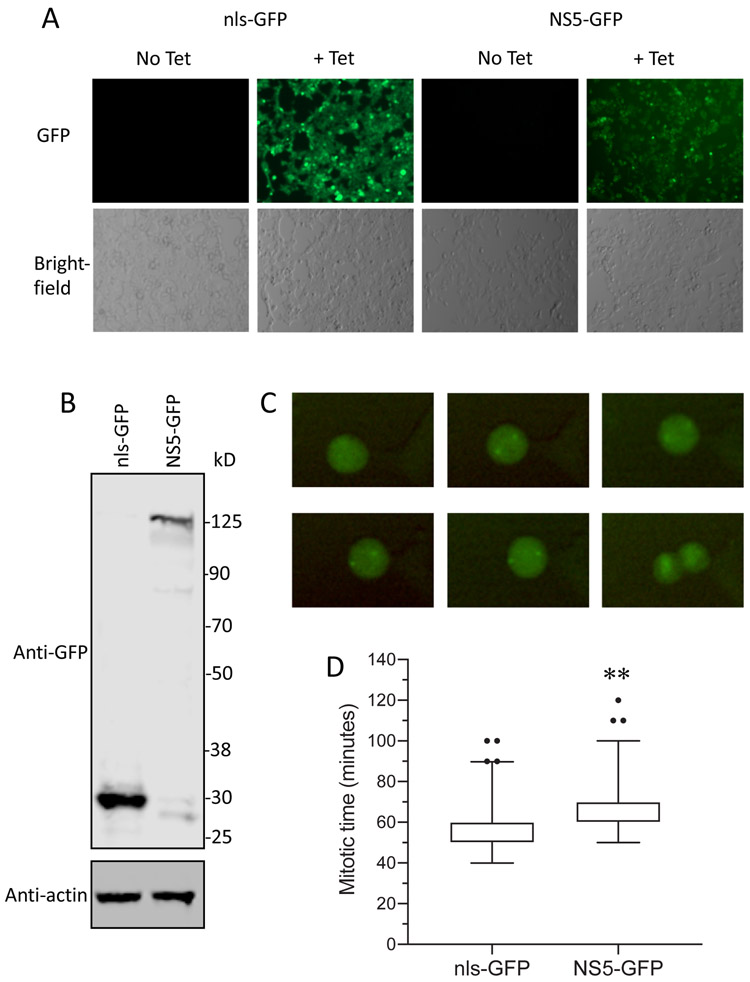
The association of NS5 with centrosomes raised the possibility that centrosome function might be affected by NS5. Since centrosomes play a key role in the separation of chromosomes during mitosis, we asked if expression of NS5-GFP affected the kinetics of mitosis in tetracycline-inducible stable cell lines expressing NS5-GFP or nls-GFP. Expression of full-length NS5-GFP and nls-GFP was verified by fluorescent microscopy (Fig. 4A) and immunoblotting (Fig. 4B). Accumulation of NS5-GFP at centrosomes during mitosis was confirmed by time-lapse live cell imaging (Fig. 4C, and Supplemental videos 1 and 2). NS5-GFP persisted at this extranuclear location even after most of the NS5-GFP reentered the nucleus upon reassembly of the nuclear membrane (Supplemental videos 1 and 2). To assess the kinetics of cell division, time-lapse images from 160 cells were analyzed. Cells expressing NS5-GFP cell took significantly longer to complete mitosis than cells expressing nls-GFP (unpaired T-test, p < 0.001) (Fig. 4D).
Fig. 4. NS5-GFP expression increases the time required to complete mitosis.
Expression of NS5-GFP and nls-GFP was induced by adding 2ug/mL tetracycline to the culture media.
A. Fluorescent and bright-field microscopic images of NS5-GFP and nls-GFP cell lines in the absence and presence of tetracycline.
B. Immunoblot analysis of lysates from NS5-GFP and nls-GFP cell lines in the presence of tetracycline. The blot was probed with anti-GFP and anti-actin antibodies. Molecular weight markers are shown at right (sizes in kDa).
C. Time-lapse images of a Flp-In™ T-REx™ 293 cell stably expressing NS5-GFP undergoing mitosis.
D. Flp-In™ T-REx™ 293 cells stably expressing ZIKV NS5-GFP or nls-GFP were analyzed for the time required to complete mitosis. A total of 160 cells from three independent experiments were analyzed for each cell line and the data were pooled. ** Unpaired t-test, p < 0.001.
3.5. ZIKV NS5 interacts with centrosomal proteins
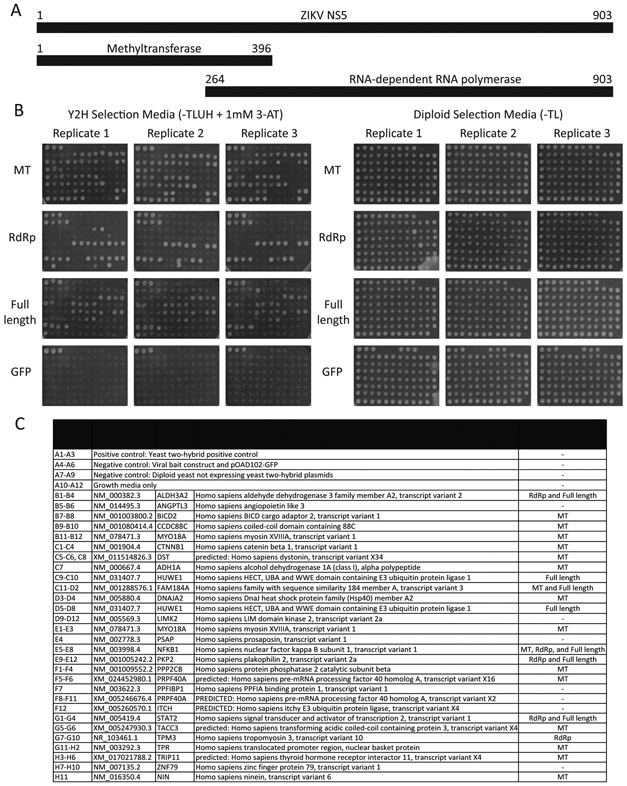
To begin to address the mechanism by which NS5 is targeted to centrosomes, we performed yeast two-hybrid screens to identify cellular proteins that interacted with NS5. Gal4 DNA binding domain constructs expressing full-length ZIKV NS5, the methyltransferase domain or the RdRp domain were screened against three human cDNA libraries. Overall, 168 pairs of interactions were identified, representing 29 human genes. Of these, 20 interactions were confirmed in the yeast two-hybrid assay after retesting the interactions in freshly transformed cells (Fig. 5, and Supplemental Table 1). The interacting human proteins included eight that are stable components of the centrosome or that are recruited to the centrosome or the spindle pole at some point during the cell cycle (Table 1). Thus, as we previously reported for DENV NS5, ZIKV NS5 has the potential to make multiple connections to the centrosome (Khadka et al., 2011).
Fig. 5. ZIKV NS5-host cell protein interactions.
A. ZIKV NS5 constructs used for yeast two-hybrid screens. ZIKV NS5 gene fragments were fused to the 3’ end of the Gal4 DNA-binding domain. Numbers indicate amino acids.
B. Human genes identified in the yeast two-hybrid screens were cloned into the yeast two-hybrid activation domain vector and retested against all three ZIKV NS5 constructs by assessing yeast growth on SD-TLUH plus 1 mM 3-AT. Interactions were considered positive if at least two out of three independent assays demonstrated growth greater than negative control strains.
C. Plate map of constructs tested for interaction with ZIKV NS5.
Table 1.
ZIKV NS5 interactions with centrosomal proteins and proteins that affect centrosome function.
| Gene | Protein | Localization* | Centrosomal Function |
|---|---|---|---|
| BICD2 | BICD cargo adaptor 2 | Centrosome, cytoplasm, Golgi apparatus, nuclear envelope, nuclear pore complex, plasma membrane | Positioning of centrosome at nuclear envelope and dynein recruitment prior to mitosis; triggers apical migration of nucleus necessary for cell division of neural stem cells (Baffet et al., 2015; Splinter et al., 2010) |
| CCDC88C | Daple | Centrosome, cytoplasm, pericentriolar recycling endosomes (Aznar et al., 2017) | |
| CTNNB1 | Catenin β-1/ β-catetnin | Centrosome, spindle pole, cytoskeleton, cilium basal body, plasma membrane, nucleus, cell junction, cytoplasm | Centrosome separation; bipolar spindle pole assembly; microtubule reorganization at centrosomes (Bahmanyar et al., 2008; Huang et al., 2007; Kaplan et al., 2004; Kobayashi et al., 2015). |
| NIN | Ninein | Centrosome, centriole, spindle pole, nucleus, plasma membrane | Anchoring of microtubules at the centrosomes; nucleation by docking gamma tubulin; centrosomal assembly following mitosis (Delgehyr et al., 2005; Ou et al., 2002). |
| TACC3 | Transforming acidic coiled-coil containing protein 3 | Centrosome, spindle fibers, spindle pole, cytoplasm | spindle pole assembly and stabilization; microtubule growth at centrosomes (Burgess et al., 2015; Thakur et al., 2014). |
| TPR | Nucleoprotein TPR/Translocated promoter region protein | Centrosome (Kobayashi et al., 2015); mitotic spindle, kinetochore, nucleus, nuclear membrane, nuclear pore complex, cytoplasm | Spindle pole organization; bipolar spindle pole assembly (Kobayashi et al., 2015) |
| TRIP11 | Thyroid hormone receptor interactor 11/ Golgi-associated microtubule-binding protein 210 (GMAP-210) | Golgi apparatus, nucleus, cytoskeleton, negative end of centrosome nucleated microtubules. | Positioning and linking Golgi to centrosome (Infante et al., 1999; Rios et al., 2004). |
Source: Uniprot subcellular location and/ or GO cellular component except (Aznar et al., 2017; Kobayashi et al., 2015).
4. Discussion
Here we report that ZIKV NS5 accumulated at centrosomes during mitosis in infected cells. In cells with supernumerary centrosomes, ZIKV NS5 was present at all centrosomes. Although flavivirus NS3 and NS5 bind to each other (Kapoor et al., 1995), and ZIKV NS3 localizes to centrosomes (Cortese et al., 2017; Coyaud et al., 2015), NS5 transiently expressed in cells in the absence of other viral proteins localized to the centrosome independently of other viral proteins. Localization of NS5-GFP at the centrosomes occurred shortly after breakdown of the nuclear membrane and was maintained throughout mitosis. NS5-GFP was detected at centrosomes even after most of the NS5-GFP relocated to the nucleus, though the length of time NS5-GFP was detected at centrosomes after the cell completed mitosis varied. Expression of ZIKV NS5-GFP increased the length of time cells required to complete mitosis, as has been observed in cells with reduced levels or function of centrosomal proteins (Fong et al., 2016; Lambrus et al., 2016; Lambrus et al., 2015; Meitinger et al., 2016; Wong et al., 2015).
Based on our yeast two-hybrid analyses, ZIKV NS5 has the potential to target multiple centrosomal proteins and proteins involved in centrosomal function (Table 1). Although other studies have identified cellular proteins that interact with ZIKV proteins (Coyaud et al., 2018; Kovanich et al., 2019; Shah et al., 2018), this is the first to report interactions with centrosomal proteins. Such interactions may have been missed because NS5 primarily associates with centrosomes during mitosis and such cells are a minority of the total cell population at any given time.
Of the centrosomal proteins that interacted with NS5, two localize to the minus end of microtubules (NIN and TRIP11) (Bouckson-Castaing et al., 1996; Infante et al., 1999; Mogensen et al., 2000; Ou et al., 2002). NIN organizes microtubules at both centrosomal and non-centrosomal MTOCs, and associates with the subdistal appendages of centrosomes, which are damaged during ZIKV infection (Gabriel et al., 2017; Mogensen et al., 2000). TRIP11 connects the minus ends of centrosomal microtubules to the cis-Golgi network (Infante et al., 1999). Other ZIKV NS5-interactors are dynein adaptors or dynein binding partners (for example, NIN, BICD2, CTNNB1 and TPR (Hoogenraad et al., 2003; Ligon et al., 2001; Nakano et al., 2010; Redwine et al., 2017; Splinter et al., 2010)). Dynein-dynactin complexes are exploited by other viruses to target viral proteins to centrosomes (Lakadamyali et al., 2003; Liu et al., 2014; Petit et al., 2003). Thus, these interactions may promote translocation of NS5 to the centrosome following nuclear membrane breakdown.
Interest in potential links between ZIKV and centrosomes has been stimulated by the correlation of maternal ZIKV infections with an increase in the incidence of congenital microcephaly (Baud et al., 2017) and by the fact that more than half the genes implicated in genetically-linked autosomal recessive primary microcephaly (MCPH) encode proteins involved in centrosome function (Jayaraman et al., 2018). Recent studies have reported extra centrosomes, either clustered in foci during interphase or at multiple poles during mitosis, in ZIKV-infected cells (Souza et al., 2016; Wolf et al., 2017). The levels of centrosomal proteins CEP152, CEP164, CPAP, PCNT, and PLK were reduced following infection and TBK1 relocalized from centrosomes to the mitochondrion (Gabriel et al., 2017; Li et al., 2016; McDougall et al., 2019; Onorati et al., 2016). Defects in chromosome separation, spindle orientation and the spindle plane were also noted (Gabriel et al., 2017; McDougall et al., 2019; Wolf et al., 2017). Finally, centrosomes from infected cells had structural defects in the appendages located at the minus end of the microtubule bundle (Gabriel et al., 2017). At least some of these effects appear to be mediated by interferon rather than the direct action of ZIKV proteins (McDougall et al., 2019; Onorati et al., 2016).
Although multiple links between ZIKV and centrosomes have been discovered, flaviviruses not associated with microcephaly also interact with centrosomes. We previously reported DENV, a virus closely related to ZIKV that is not associated with microcephaly, interacted with multiple centrosomal proteins; we further showed that DENV NS5 accumulated at centrosomes in mitotic cells (Khadka et al., 2011). Although both DENV and ZIKV NS5 bind to centrosomal proteins, different centrosomal proteins were implicated (Khadka et al., 2011). This suggests that interactions between NS5 and centrosomes may be a general feature of flaviviruses, but the specific interactions may vary. Interactions between DENV NS3 and centrosomal proteins were also identified, as has been observed with ZIKV NS3 (Coyaud et al., 2018; Khadka et al., 2011). Since DENV infection has not been linked to increased risks of microcephaly, the importance of the association of flavivirus proteins with centrosomes is unclear. The interactions of flavivirus proteins with centrosomes may not be causally related to the development of microcephaly. Alternatively, the effect of viral proteins on centrosome function may be different for DENV and ZIKV, or may depend on the specific cell types infected. ZIKV is able to cross the placental barrier and infect cell types, such as neural progenitor cells, that are not infected by DENV (Merfeld et al., 2017).
In conclusion, we show that ZIKV NS5 associates with the centrosomes during mitosis. This association occurs independently of other viral proteins and is maintained through mitosis. Interactions between nonstructural proteins and centrosomes appears to be a general feature of flaviviruses. The molecular mechanism of these associations and their effect on centrosomal function and viral replication will require further investigation.
Supplementary Material
Supplemental Fig. 1. NS5 localizes at centrosomes in dividing ZIKV-infected cells.
Confocal microscopic images of fixed ZIKV-infected Huh7 cells were stained with anti-ZIKV NS5 and anti-pericentrin antibodies; DNA was labelled with Hoechst (left panels). Pixel intensities of NS5, centrosomes and DNA along the line connecting the centrosomes in ZIKV-infected cells were plotted using NIS-Elements AR software (right panels).
A. NS5, red (stained with mouse anti-NS5); pericentrin, green; Hoechst, blue.
B. NS5, green (stained with chicken anti-NS5); pericentrin, red; Hoechst, blue.
Supplemental Fig. 2. Live cell imaging of HEK-293 cells expressing nls-GFP undergoing mitosis.
A and B. Time-lapse images of HEK-293 cells transiently transfected with an nls-GFP expression plasmid. Images were acquired using an IncuCyte® S3 Live-Cell Analysis System at 20X magnification.
Supplemental Videos 1 and 2. Time lapse imaging of cells expressing ZIKV NS5-GFP.
Acknowledgements
Funding: This work was supported by the National Institutes of Health [grant numbers AI114814 and AI120943] and the Purdue Institute of Inflammation, Immunology and Infectious Disease (PI4D) Team Science Program (grant number 209263).
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, 1990. Basic local alignment search tool. J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M, 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574. [DOI] [PubMed] [Google Scholar]
- Arquint C, Gabryjonczyk AM, Nigg EA, 2014. Centrosomes as signalling centres. Philos Trans R Soc Lond B Biol Sci 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar N, Sun N, Dunkel Y, Ear J, Buschman MD, Ghosh P, 2017. A Daple-Akt feed-forward loop enhances noncanonical Wnt signals by compartmentalizing beta-catenin. Mol Biol Cell 28, 3709–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baffet AD, Hu DJ, Vallee RB, 2015. Cdk1 Activates Pre-mitotic Nuclear Envelope Dynein Recruitment and Apical Nuclear Migration in Neural Stem Cells. Dev Cell 33, 703–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH Jr., O'Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI, 2008. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev 22, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D, 2017. An update on Zika virus infection. Lancet 390, 2099–2109. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR, 1996. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci 109 ( Pt 1), 179–190. [DOI] [PubMed] [Google Scholar]
- Burgess SG, Peset I, Joseph N, Cavazza T, Vernos I, Pfuhl M, Gergely F, Bayliss R, 2015. Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly. PLoS Genet 11, e1005345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coloma J, Jain R, Rajashankar KR, Garcia-Sastre A, Aggarwal AK, 2016. Structures of NS5 Methyltransferase from Zika Virus. Cell reports 16, 3097–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit PT, Wainman A, Raff JW, 2015. Centrosome function and assembly in animal cells. Nat Rev Mol Cell Biol 16, 611–624. [DOI] [PubMed] [Google Scholar]
- Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, Schorb M, Pruunsild P, Schwab Y, Chatel-Chaix L, Ruggieri A, Bartenschlager R, 2017. Ultrastructural Characterization of Zika Virus Replication Factories. Cell reports 18, 2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Barral K, Lichiere J, Selisko B, Martin B, Aouadi W, Lombardia MO, Debart F, Vasseur JJ, Guillemot JC, Canard B, Decroly E, 2017. Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E, Mis M, Laurent EM, Dunham WH, Couzens AL, Robitaille M, Gingras AC, Angers S, Raught B, 2015. BioID-based identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics 14, 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E, Ranadheera C, Cheng D, Goncalves J, Dyakov BJA, Laurent EMN, St-Germain J, Pelletier L, Gingras AC, Brumell JH, Kim PK, Safronetz D, Raught B , 2018. Global interactomics uncovers extensive organellar targeting by Zika virus. Mol Cell Proteomics 17, 2242–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Maio FA, Risso G, Iglesias NG, Shah P, Pozzi B, Gebhard LG, Mammi P, Mancini E, Yanovsky MJ, Andino R, Krogan N, Srebrow A, Gamarnik AV, 2016. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog 12, e1005841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Sillibourne J, Bornens M, 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci 118, 1565–1575. [DOI] [PubMed] [Google Scholar]
- Dick GW, Kitchen SF, Haddow AJ, 1952. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 46, 509–520. [DOI] [PubMed] [Google Scholar]
- Duan W, Song H, Wang H, Chai Y, Su C, Qi J, Shi Y, Gao GF, 2017. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J 36, 919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB, 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360, 2536–2543. [DOI] [PubMed] [Google Scholar]
- Fong CS, Mazo G, Das T, Goodman J, Kim M, O'Rourke BP, Izquierdo D, Tsou MF, 2016. 53BP1 and USP28 mediate p53-dependent cell cycle arrest in response to centrosome loss and prolonged mitosis. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, Gopalakrishnan J, 2017. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20, 397–406 e395. [DOI] [PubMed] [Google Scholar]
- Godoy AS, Lima GM, Oliveira KI, Torres NU, Maluf FV, Guido RV, Oliva G, 2017. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat Commun 8, 14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Ponia SS, Tripathi S, Balasubramaniam V, Miorin L, Sourisseau M, Schwarz MC, Sanchez-Seco MP, Evans MJ, Best SM, Garcia-Sastre A, 2016. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Chen J, Zhu X, An S, Dong X, Yu J, Zhang S, Wu Y, Li G, Zhang Y, Wu J, Li M, 2018. NLRP3 Inflammasome Activation Mediates Zika Virus-Associated Inflammation. J Infect Dis 217, 1942–1951. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, de Zeeuw CI, Akhmanova A, 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J 22, 6004–6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Senga T, Hamaguchi M, 2007. A novel role of phospho-beta-catenin in microtubule regrowth at centrosome. Oncogene 26, 4357–4371. [DOI] [PubMed] [Google Scholar]
- Infante C, Ramos-Morales F, Fedriani C, Bornens M, Rios RM, 1999. GMAP-210, A cis-Golgi network-associated protein, is a minus end microtubule-binding protein. J Cell Biol 145, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman D, Bae BI, Walsh CA, 2018. The Genetics of Primary Microcephaly. Annu Rev Genomics Hum Genet 19, 177–200. [DOI] [PubMed] [Google Scholar]
- Kaplan DD, Meigs TE, Kelly P, Casey PJ, 2004. Identification of a role for beta-catenin in the establishment of a bipolar mitotic spindle. J Biol Chem 279, 10829–10832. [DOI] [PubMed] [Google Scholar]
- Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner KE, Padmanabhan R, 1995. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem 270, 19100–19106. [DOI] [PubMed] [Google Scholar]
- Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, Sengupta R, Sahasrabudhe S, Randall G, Gribskov M, Kuhn RJ, Perera R, Lacount DJ, 2011. A physical interaction network of dengue virus and human proteins. Mol. Cell. Proteom 10, M111.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Hashizume C, Dowaki T, Wong RW, 2015. Therapeutic potential of mitotic interaction between the nucleoporin Tpr and aurora kinase A. Cell Cycle 14, 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovanich D, Saisawang C, Sittipaisankul P, Ramphan S, Kalpongnukul N, Somparn P, Pisitkun T, Smith DR, 2019. Analysis of the Zika and Japanese Encephalitis Virus NS5 Interactomes. J Proteome Res 18, 3203–3218. [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, Schoenfeld LW, Ota I, Sahasrabudhe S, Kurschner C, Fields S, Hughes RE, 2005. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107. [DOI] [PubMed] [Google Scholar]
- Lakadamyali M, Rust MJ, Babcock HP, Zhuang X, 2003. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci U S A 100, 9280–9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Daggubati V, Uetake Y, Scott PM, Clutario KM, Sluder G, Holland AJ, 2016. A USP28-53BP1-p53-p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol 214, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, Sluder G, Holland AJ, 2015. p53 protects against genome instability following centriole duplication failure. J Cell Biol 210, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z, 2016. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell 19, 672. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Karki S, Tokito M, Holzbaur EL, 2001. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat Cell Biol 3, 913–917. [DOI] [PubMed] [Google Scholar]
- Lin S, Yang S, He J, Guest JD, Ma Z, Yang L, Pierce BG, Tang Q, Zhang YJ, 2019. Zika virus NS5 protein antagonizes type I interferon production via blocking TBK1 activation. Virology 527, 180–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Zhang LJ, Wang ZG, Zhang ZL, Wu QM, Sun EZ, Shi YB, Pang DW, 2014. Globally visualizing the microtubule-dependent transport behaviors of influenza virus in live cells. Anal Chem 86, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL, 2007. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A 104, 1027–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall WM, Perreira JM, Hung HF, Vertii A, Xiaofei E, Zimmerman W, Kowalik TF, Doxsey S, Brass AL, 2019. Viral infection or ifn-alpha alters mitotic spindle orientation by modulating pericentrin levels. iScience 12, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Anzola JV, Kaulich M, Richardson A, Stender JD, Benner C, Glass CK, Dowdy SF, Desai A, Shiau AK, Oegema K, 2016. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J Cell Biol 214, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merfeld E, Ben-Avi L, Kennon M, Cerveny KL, 2017. Potential mechanisms of Zika-linked microcephaly. Wiley Interdiscip Rev Dev Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M, 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci 113 ( Pt 17), 3013–3023. [DOI] [PubMed] [Google Scholar]
- Nakano H, Funasaka T, Hashizume C, Wong RW, 2010. Nucleoporin translocated promoter region (Tpr) associates with dynein complex, preventing chromosome lagging formation during mitosis. J Biol Chem 285, 10841–10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng IHW, Chan KW, Tan MJA, Gwee CP, Smith KM, Jeffress SJ, Saw WG, Swarbrick CMD, Watanabe S, Jans DA, Gruber G, Forwood JK, Vasudevan SG, 2019. Zika virus NS5 forms supramolecular nuclear bodies that sequester importin-alpha and modulate the host immune and pro-inflammatory response in neuronal cells. ACS Infect Dis 5, 932–948. [DOI] [PubMed] [Google Scholar]
- Nishi K, Nishi A, Nagasawa T, Ui-Tei K, 2013. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 19, 17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AMM, Nakagawa N, Li M, Dell'Anno MT, Gulden FO, Pochareddy S, Tebbenkamp ATN, Han W, Pletikos M, Gao T, Zhu Y, Bichsel C, Varela L, Szigeti-Buck K, Lisgo S, Zhang Y, Testen A, Gao XB, Mlakar J, Popovic M, Flamand M, Strittmatter SM, Kaczmarek LK, Anton ES, Horvath TL, Lindenbach BD, Sestan N, 2016. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell reports 16, 2576–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou YY, Mack GJ, Zhang M, Rattner JB, 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J Cell Sci 115, 1825–1835. [DOI] [PubMed] [Google Scholar]
- Petersen LR, Jamieson DJ, Honein MA, 2016. Zika Virus. N Engl J Med 375, 294–295. [DOI] [PubMed] [Google Scholar]
- Petit C, Giron ML, Tobaly-Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H, Saib A, 2003. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J Cell Sci 116, 3433–3442. [DOI] [PubMed] [Google Scholar]
- Pryor MJ, Rawlinson SM, Butcher RE, Barton CL, Waterhouse TA, Vasudevan SG, Bardin PG, Wright PJ, Jans DA, Davidson AD, 2007. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic 8, 795–807. [DOI] [PubMed] [Google Scholar]
- Redwine WB, DeSantis ME, Hollyer I, Htet ZM, Tran PT, Swanson SK, Florens L, Washburn MP, Reck-Peterson SL, 2017. The human cytoplasmic dynein interactome reveals novel activators of motility. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios RM, Sanchis A, Tassin AM, Fedriani C, Bornens M, 2004. GMAP-210 recruits gamma-tubulin complexes to cis-Golgi membranes and is required for Golgi ribbon formation. Cell 118, 323–335. [DOI] [PubMed] [Google Scholar]
- Shah PS, Link N, Jang GM, Sharp PP, Zhu T, Swaney DL, Johnson JR, Von Dollen J, Ramage HR, Satkamp L, Newton B, Huttenhain R, Petit MJ, Baum T, Everitt A, Laufman O, Tassetto M, Shales M, Stevenson E, Iglesias GN, Shokat L, Tripathi S, Balasubramaniam V, Webb LG, Aguirre S, Willsey AJ, Garcia-Sastre A, Pollard KS, Cherry S, Gamarnik AV, Marazzi I, Taunton J, Fernandez-Sesma A, Bellen HJ, Andino R, Krogan NJ, 2018. Comparative flavivirus-host protein interaction mapping reveals mechanisms of dengue and Zika virus pathogenesis. Cell 175, 1931–1945 e1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR, Yang Y, Tesh RB, Bourne N, Barrett AD, Vasilakis N, Weaver SC, Shi PY, 2016. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe 19, 891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sir JH, Putz M, Daly O, Morrison CG, Dunning M, Kilmartin JV, Gergely F, 2013. Loss of centrioles causes chromosomal instability in vertebrate somatic cells. J Cell Biol 203, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza BS, Sampaio GL, Pereira CS, Campos GS, Sardi SI, Freitas LA, Figueira CP, Paredes BD, Nonaka CK, Azevedo CM, Rocha VP, Bandeira AC, Mendez-Otero R, Dos Santos RR, Soares MB, 2016. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Scientific reports 6, 39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter D, Tanenbaum ME, Lindqvist A, Jaarsma D, Flotho A, Yu KL, Grigoriev I, Engelsma D, Haasdijk ED, Keijzer N, Demmers J, Fornerod M, Melchior F, Hoogenraad CC, Medema RH, Akhmanova A, 2010. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol 8, e1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen P, Baz M, Boivin G, Lin SX, 2016. Structural Insight into NS5 of Zika Virus Leading to the Discovery of MTase Inhibitors. J Am Chem Soc 138, 16212–16215. [DOI] [PubMed] [Google Scholar]
- Tay MY, Smith K, Ng IH, Chan KW, Zhao Y, Ooi EE, Lescar J, Luo D, Jans DA, Forwood JK, Vasudevan SG, 2016. The C-terminal 18 amino acid region of dengue virus NS5 regulates its subcellular localization and contains a conserved arginine residue essential for infectious virus production. PLoS Pathog 12, e1005886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur HC, Singh M, Nagel-Steger L, Kremer J, Prumbaum D, Fansa EK, Ezzahoini H, Nouri K, Gremer L, Abts A, Schmitt L, Raunser S, Ahmadian MR, Piekorz RP, 2014. The centrosomal adaptor TACC3 and the microtubule polymerase chTOG interact via defined C-terminal subdomains in an Aurora-A kinase-independent manner. J Biol Chem 289, 74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay AK, Cyr M, Longenecker K, Tripathi R, Sun C, Kempf DJ, 2017. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta crystallographica. Section F, Structural biology communications 73, 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Tan XF, Thurmond S, Zhang ZM, Lin A, Hai R, Song J, 2017. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat Commun 8, 14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li G, De W, Luo Z, Pan P, Tian M, Wang Y, Xiao F, Li A, Wu K, Liu X, Rao L, Liu F, Liu Y, Wu J, 2018. Zika virus infection induces host inflammatory responses by facilitating NLRP3 inflammasome assembly and interleukin-1beta secretion. Nat Commun 9, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf B, Diop F, Ferraris P, Wichit S, Busso C, Misse D, Gonczy P, 2017. Zika virus causes supernumerary foci with centriolar proteins and impaired spindle positioning. Open Biol 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, Mitchell BJ, Desai A, Gahman TC, Shiau AK, Oegema K, 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Luo H, Shan C, Muruato AE, Nunes BTD, Medeiros DBA, Zou J, Xie X, Giraldo MI, Vasconcelos PFC, Weaver SC, Wang T, Rajsbaum R, Shi PY, 2018. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat Commun 9, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Feng T, Cheng J, Li Y, Yin X, Zeng W, Jin X, Li Y, Guo F, Jin T, 2017. Structure of the NS5 methyltransferase from Zika virus and implications in inhibitor design. Biochem Biophys Res Commun 492, 624–630. [DOI] [PubMed] [Google Scholar]
- Zhao B, Yi G, Du F, Chuang YC, Vaughan RC, Sankaran B, Kao CC, Li P, 2017. Structure and function of the Zika virus full-length NS5 protein. Nat Commun 8, 14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wang F, Wang H, Chen C, Zhang T, Han X, Wang D, Chen C, Wu C, Xie W, Wang Z, Zhang L, Wang L, Yang H, 2017. The conformational changes of Zika virus methyltransferase upon converting SAM to SAH. Oncotarget 8, 14830–14834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. NS5 localizes at centrosomes in dividing ZIKV-infected cells.
Confocal microscopic images of fixed ZIKV-infected Huh7 cells were stained with anti-ZIKV NS5 and anti-pericentrin antibodies; DNA was labelled with Hoechst (left panels). Pixel intensities of NS5, centrosomes and DNA along the line connecting the centrosomes in ZIKV-infected cells were plotted using NIS-Elements AR software (right panels).
A. NS5, red (stained with mouse anti-NS5); pericentrin, green; Hoechst, blue.
B. NS5, green (stained with chicken anti-NS5); pericentrin, red; Hoechst, blue.
Supplemental Fig. 2. Live cell imaging of HEK-293 cells expressing nls-GFP undergoing mitosis.
A and B. Time-lapse images of HEK-293 cells transiently transfected with an nls-GFP expression plasmid. Images were acquired using an IncuCyte® S3 Live-Cell Analysis System at 20X magnification.
Supplemental Videos 1 and 2. Time lapse imaging of cells expressing ZIKV NS5-GFP.