Abstract
Background
Accurate identification Candida is important for successful therapy and epidemiology study. The aim of research is to study API 20C yeast identification system identification rate by using molecular identification as gold standard and tested the antifungal susceptibility of Candida from patients with vulvovaginal candidiasis (VVC).
Methods
In total, 3574 yeast isolates were obtained from patients with VVC. API 20C yeast identification, molecular identification and in vitro antifungal susceptibility were performed.
Results
C. albicans was the predominant Candida species [2748 isolates, 76.9%] in VVC. The isolates from vaginal samples represented 22 species based on molecular identification. The API 20C system identifies only 11 of the species encountered during the study period. Based on the API 20C system, 3273 (91.58%) isolates were correctly identified to the species level. The correct identification rate of the API 20C system for rare yeast was 15.29% (26/170 isolates). Antifungal susceptibility was tested in a total of 1844 isolates of Candida from patients with VVC. C. albicans was susceptible to most of the tested antifungals. The MICs of azoles for C. glabrata were higher than those for C. albicans. The MICs of echinocandins for C. parapsilosis were higher than those for C. albicans.
Conclusions
The API 20C yeast identification system can be used to reliably identify the most common Candida species while molecular methods are necessary for the identification of closely related, emerging, and rare yeast species. The results from this study suggest that much of the previous studies on the epidemiology of VVC should be re-thought. C. albicans was susceptible to most of the tested antifungals.
Keywords: Candidiasis, Vulvovaginal, Candida, Identification, Antifungal susceptibility
Background
An estimated 75% of women will have at least one episode of vulvovaginal candidiasis (VVC) and 40–45% will have two or more episodes [1]. The estimated probability of recurrent VVC (RVVC),which was defined as four or more episodes of symptomatic VVC within 1 year, after VVC will be 14–28% [2]. C. albicans, which is responsible for 85–95% of Candida vaginal infections, is the major aetiological agent involved in cases of VVC, followed by C. glabrata and C. parapsilosis [3–6]. Accurate identification at the species level is paramount for successful therapy and appropriate patient care. However, commonly used identification method, the API yeast identification system, has shown a rather inconsistent ability to identify clinical isolates with an overall identification rate ranging from 80 to 96% [7]. In addition, with the discovery of new, closely related Candida species and novel species, the correct identification of the isolates has become more difficult by using the common methods [8]. PCR and sequencing of relevant genes provide a rapid and accurate Candida speciation, genotyping of individual species, and finally, antifungal drug sensitivity [9]. Treatment options for refractory symptoms caused by antifungal resistant Candida are extremely limited. New therapeutic study, options and strategies are urgently needed to meet the challenge of drug resistance [10–15]. RVVC affects about 138 million women annually, with a global annual prevalence of 3871 per 100,000 women; 372 million women are affected by RVVC over their lifetime [11]. We reported the distribution of yeast isolates based on molecular identification from patients with VVC in southern China from 2003 to 2018 and compare the identification rate of molecular methods with API 20 C system. We also tested the common used and several potential clinical using antifungals.
Methods
Patients and case definition
A prospective study of 3574 consecutive patients with VVC and RVVC was conducted at the Department of Gynecology, Peking University Shenzhen Hospital from April 2003 to September 2018. The research protocol was approved by the ethics committee of the hospital, and all subjects gave their informed consent to participate. The cases of VVC in pregnancy and VVC caused by multiple Candida species were excluded. A case of VVC was defined as a patient with vulvar itching, vaginal discharge and a positive Candida culture. Confirmation was obtained by demonstration of blastoconidia and pseudohyphae on 10% potassium hydroxide preparation. Among the 3574 patients, 588 isolates (16.5%) which first found from per patient with RVVC were selected. The mean ages of patients with RVVC and VVC were 31.01 [SD 6.04] and 29.67 [SD 6.64] years, respectively (P < 0.05).
Vaginal samples and API 20C identification
A sample from the lateral vaginal wall was obtained with a sterile cotton-tipped swab. The swab was placed in a tube filled with saline prior to direct microscopic examination on a wet slide, to which a drop of 10% potassium hydroxide had been added. Culture was performed on samples obtained from all cases that had positive on 10% potassium hydroxide preparation. All specimens were plated on a Sabouraud agar plate for 24–48 h at 37 °C. Isolates were identified using a standard system, API 20C [Biomerieux, France], and stored in medium containing 2% glucose, 2% peptone and 20% glycerol at − 70 °C.
Molecular identification
Isolates were removed from the − 70 °C freezer and revived on a Sabouraud agar plate for 24–48 h at 37 °C. One single yeast colony from the isolates was suspended in a microcentrifuge tube containing 50 μL of lysis buffer for direct polymerase chain reaction (PCR) to identify fungus (Takara Biotechnology Co., Ltd., Dalian, China). The composition of the PCR mixture, and the PCR conditions were in accordance with the methods previously described [16–19]. At first, we use PCR primers of C. albicans complexes, C. glabrata complexes, and C. parapsilosis complexes to identify the three complexes, respectively. All other yeasts were identified by using PCR and sequencing. The primers used in this study are shown in Table 1.
Table 1.
The primers used in this study
| Candida species | Primer name | Forward(5′-3′) | Reverse(5′-3′) | Amplified fragment size (bp) | References |
|---|---|---|---|---|---|
| C. albicans complexes | HWP1 | GCTACCACTTCAGAATCATCATC | GCACCTTCAGTCGTAGAGACG |
C. albicans:839 and 941 C. africana:700 C. dubliniensis:569 |
Shan,2014 |
| C. glabrata complexes |
GLA NIV BRA |
CGGTTGGTGGGTGTTCTGC AGGGAGGAGTTTGTATCTTTCAAC GGGACGGTAAGTCTCCCG |
ACCAGAGGGCGCAATGTG |
C. glabrata:397 C. bracarensis: 223 C. nivariensis:293 |
Li,2014 |
| C. parapsilosis complexes |
mCPF mCOF mCMF |
TTTGCTTTGGTAGGCCTTCTA TAAGTCAACTGATTAACTAAT AACTGCAATCCTTTTCTTTCTA |
AATATCTGCAATTCATATTACT |
C. parapsilosis:171 C. orthopsilosis:109 C. metapsilosis:217 |
Asadzadeh,2015 |
| Rare yeast | NL1,NL4 | GCATATCAATAAGCGGAGGAAAAG-3’ | GGTCCGTGTTTCAAGACGG | 500–600 | Leaw,2006 |
1Shan Y, Fan S, Liu X, et al. Prevalence of Candida albicans-closely related yeasts, Candida africana and Candida dubliniensis, in vulvovaginal candidiasis. Med Mycol, 2014, 52 (6): 636–40.
2Li J, Shan Y, Fan S, et al. Prevalence of Candida nivariensis and Candida bracarensis in vulvovaginal Candidiasis. Mycopathologia, 2014, 178 (3, 4): 279–83.
3Asadzadeh M, Ahmad S, Hagen F, et al. Simple, Low-Cost Detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS One, 2015, 10 (11): e0142880.
4Leaw SN, Chang HC, Sun HF, et al. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microb, 2006, 44 (3): 693–9.
Antifungal susceptibility testing
The in vitro susceptibility tests by using the CLSI reference broth microdilution method were performed for all species isolates number less than 100 strains. C. albicans and C. glabrata were randomly selected for the test. Those include 1272 C. albicans strains (including 998 isolates from VVC and 274 from RVVC) and 267 C. glabrata strains(including 197 isolates from VVC and 70 from RVVC). The MIC of Candida for all agents was read following 24–48 h incubation. The antifungals used were amphotericin B (Sigma, USA), Anidulafungin(Selleckchem, USA), Butoconazole(Sigma, USA), Caspofungin(Sigma, USA), Clotrimazole(Sigma, USA), Fluconazole(Sigma, USA), Flucytosine(Sigma, USA), Itraconazole(Sigma, USA), Micafungin(Selleckchem),Miconazole(Sigma, USA), Nystatin (Amresco, USA), Terbinafine(Santa Cruz, USA),Terconazole(Sigma),and Voriconazole(Fluka, USA). Quality control was performed as recommended in CLSI documents M27-A3 and M60 by using ATCC 90028 which is a reference strain of C. albicans and all results of the control were within established ranges [20, 21].
Statistical analysis
All values given in tables and text are expressed as the means unless otherwise indicated. Each variable was tested for differences between groups by Student’s t test or chi-square analysis where appropriate. Statistical significance was set at P < 0.05. Statistical analysis of the data was performed using SPSS 10.0 software (SPSS Inc.; Chicago, Illinois, United States).
Results
Strain distribution and yeast identification
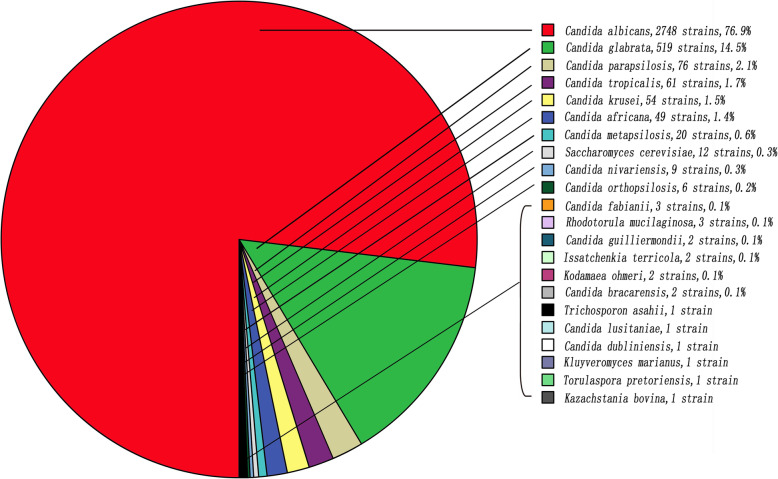
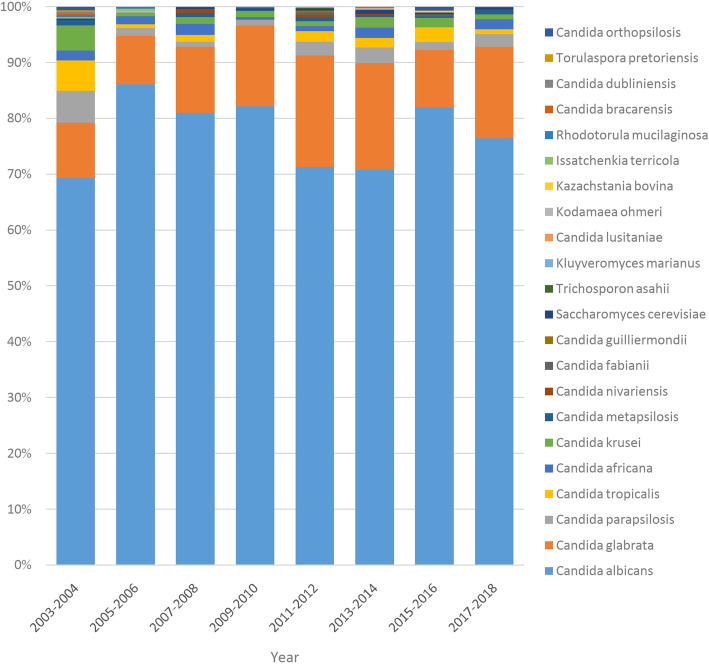
The 3574 isolates from the vaginal samples represented 22 species based on molecular identification. C. albicans were the predominant Candida species (2748 isolates, 76.9%) in VVC, followed by C. glabrata (519 isolates, 14.5%), C. parapsilosis (76 isolates, 2.1%), and C. tropicalis (61 isolates, 1.7%). Fig. 1 shows the distribution of the yeast species from all VVC based on molecular identification. Fig. 2 shows the distribution of the yeast species from all VVC by years.
Fig. 1.
The distribution of the yeast from VVC
Fig. 2.
The distribution of the yeast from VVC by years
The API 20C system identified only 11 of the species encountered during the study period (Table 2). Among the isolates analysed by the API 20C system, 3273 (91.58%) isolates were correctly identified to the species level. The correct identification rates of C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis were 98.51% (2707/2748 isolates), 84.59% (439/519 isolates), 80.26% (61/76 isolates), and 65.57% (40/61 isolates), respectively. The correct identification rate of the API 20C system for rare yeasts, including C. krusei, Saccharomyces cerevisiae, Candida africana, C. dubliniensis, C. orthopsilosis, C. metapsilosis, C. lusitaniae, C. fabianii, Trichosporon asahii, Rhodotorula, Kodamaea ohmeri, C. nivariensis, C. bracarensis, C. guilliermondii, Torulaspora pretoriensis, Kazachstania bovina, Kluyveromyces marianus, and Issatchenkia terricola, was 15.29% (26/170 isolates) (Table 2).
Table 2.
Identification of 3574 isolates of yeast from vulvovaginal candidasis based on molecular methods and API 20C
| Candida identified by molecular methods (abbreviation) | Candida identified by API 20C (isolates) | Correct identification/total,% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
CA (2866) |
CG(489) | CP (85) |
CT (57) |
CK (27) |
SC (24) |
C. famata (13) |
CL (4) |
RH (4) |
C. inconspicua (3) | C. humicola (2) | ||
| C. albicans (CA) | 2707 | 24 | 0 | 7 | 4 | 1 | 1 | 0 | 3 | 0 | 1 | 2707/2748,98.51 |
| C. africana | 48 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/49,0 |
| C. dubliniensis | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/1,0 |
| C. glabrata (CG) | 52 | 439 | 7 | 7 | 2 | 7 | 4 | 0 | 1 | 0 | 0 | 439/519,84.59 |
| C. nivariensis | 0 | 7 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0/9,0 |
| C. bracarensis | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/2,0 |
| C. parapsilosis (CP) | 7 | 4 | 61 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 61/76,80.26 |
| C. orthopsilosis | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/6,0 |
| C. metapsilosis | 3 | 5 | 8 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0/20,0 |
| C. tropicalis (CT) | 10 | 2 | 2 | 40 | 0 | 4 | 3 | 0 | 0 | 0 | 0 | 40/61,65.57 |
| C. krusei (CK) | 28 | 2 | 1 | 0 | 18 | 1 | 0 | 1 | 0 | 3 | 0 | 18/54,33.33 |
| Saccharomyces cerevisiae (SC) | 2 | 1 | 1 | 0 | 1 | 7 | 0 | 0 | 0 | 0 | 0 | 7/12,58.33 |
| C. lusitaniae (CL) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1/1,100 |
| C. fabianii | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0/3,0 |
| Trichosporon asahii () | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0/1,0 |
| Rhodotorula (RH) | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/3,0 |
| C. guilliermondii | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0/2,0 |
| Kodamaea ohmeri | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0/2,0 |
| Issatchenkia terricola | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0/2,0 |
| Torulaspora pretoriensis | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0/1,0 |
| Kazachstania bovina | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/1,0 |
| Kluyveromyces marianus | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/1,0 |
| Total | 2866 | 489 | 85 | 57 | 27 | 24 | 13 | 4 | 4 | 3 | 2 | 3273/3574,91.58 |
Antifungal susceptibility
Antifungal susceptibility was tested in a total of 1844 isolates of Candida from patients with VVC. C. albicans was susceptible to most of the tested antifungals including azole, polyenes and echinocandins. The MICs of azoles for C. glabrata were higher than those for C. albicans. The MICs of echinocandins for C. parapsilosis were higher than those for C. albicans. Some drug-resistant isolates mainly to azoles were found. C.albicans showed 7.7, 10.2 and 6.2% resistance to the fluconazole (MIC ≥8 μg/mL), itraconazole (MIC ≥1 μg/mL) and voriconazole (MIC ≥1 μg/mL), respectively. On the other hand, C.glabrate showed 3.4 and 29.1% resistance to the fluconazole (MIC ≥64 μg/mL) and itraconazole (MIC ≥1 μg/mL). In addition, a small number of C. parapsilosis were resistant to echinococcins. The resistance rates of C. parapsilosis to Anifungin (MIC ≥8 μg/mL), carpofungin (MIC ≥8 μg/mL) and micafungin (MIC ≥8 μg/mL) were 5.2, 5.2 and 1.3%, respectively. The MIC GM value of C. albicans for itraconazole, terconazole, and terbinafine in RVVC is higher than those in VVC. The MIC GM value of C. glabrata for miconazole, amphotericin B, nystatin, caspofungin, and terbinafine in RVVC is higher than those in VVC (Tables 3 and 4).
Table 3.
In vitro antifungal susceptibility of 1844 clinical isolates of Candida species as determined by the CLSI method
| Antifungal agents | ||||||||
|---|---|---|---|---|---|---|---|---|
| Candida species(n) | BUC | CLO | FLC | ITC | MIC | TEC | VRC | |
| C. albicans, n = 1272 | Range | 0.015–32 | 0.015–32 | 0.06–128 | 0.015–32 | 0.015–32 | 0.015–32 | 0.015–8 |
| GM | 0.11 | 0.05 | 0.79 | 0.09 | 0.30 | 0.15 | 0.07 | |
| MIC90 | 1 | 0.25 | 4 | 1 | 4 | 2 | 0.5 | |
| R | 7.7% | 10.2% | 6.2% | |||||
| C. africana, n = 49 | Range | 0.015–2. | 0.015–4 | 0.125–1 | 0.015–1 | 0.015-0.5 | 0.03–16 | 0.03–0.5 |
| GM | 0.04 | 0.04 | 0.25 | 0.04 | 0.04 | 0.07 | 0.04 | |
| MIC90 | 0.06 | 0.06 | 0.5 | 0.06 | 0.125 | 0.5 | 0.06 | |
| C. dubliniensis n = 1 | Range | 0.015 | 0.015 | 4 | 0.125 | 0.03 | 0.015 | 0.015 |
| C. glabrata, n = 267 | Range | 0.015–16 | 0.015–16 | 0.125–128 | 0.015–32 | 0.015–32 | 0.015–32 | 0.015–32 |
| GM | 0.21 | 0.20 | 1.48 | 0.38 | 0.25 | 0.16 | 0.09 | |
| MIC90 | 1 | 1 | 8 | 4 | 2 | 1 | 0.25 | |
| R | 3.4% | 29.1% | – | |||||
| C. nivariensis, n = 9 | Range | 0.03–1 | 0.03–0.5 | 0.125–32 | 0.03–0.5 | 0.03–16 | 0.03–1 | 0.03–4 |
| MIC50 | 0.03 | 0.06 | 2 | 0.5 | 0.03 | 0.03 | 0.06 | |
| MIC90 | 0.125 | 0.125 | 4 | 0.5 | 2 | 1 | 0.5 | |
| C. bracarensis, n = 2 | Range | 0.03–1 | 0.03–0.5 | 0.25–2 | 0.125–2 | 0.03–4 | 0.03–1 | 0.03–0.125 |
| MIC50 | 0.03 | 0.03 | 0.25 | 0.125 | 0.03 | 0.03 | 0.03 | |
| MIC90 | 1 | 0.5 | 2 | 2 | 4 | 1 | 0.125 | |
| C. parapsilosis, n = 76 | Range | 0.03–4 | 0.015–1 | 0.125–4 | 0.03–4 | 0.03–4 | 0.015–32 | 0.015–2 |
| GM | 0.18 | 0.04 | 0.47 | 0.07 | 0.54 | 0.03 | 0.04 | |
| MIC90 | 1 | 0.06 | 1 | 0.25 | 2 | 0.03 | 0.06 | |
| R | 0 | 3% | 1% | |||||
| C. metapsilosis, n = 20 | Range | 0.03–1 | 0.03–0.25 | 0.25–4 | 0.03–1 | 0.03–8 | 0.015–0.05 | 0.015–0.06 |
| GM | 0.16 | 0.04 | 0.79 | 0.08 | 0.48 | 0.04 | 0.03 | |
| MIC90 | 1 | 0.06 | 2 | 1 | 4 | 0.25 | 0.06 | |
| C. orthopsilosis, n = 6 | Range | 0.03–8 | 0.03–1 | 0.125–4 | 0.03–1 | 0.25–8 | 0.03–0.25 | 0.03–0.25 |
| MIC50 | 0.5 | 0.06 | 1.0 | 0.25 | 1.0 | 0.03 | 0.06 | |
| MIC90 | 8 | 1 | 4 | 1 | 8 | 0.25 | 0.25 | |
| C. tropicalis, n = 61 | Range | 0.03–2 | 0.015–0.5 | 0.125–128 | 0.015–32 | 0.03–32 | 0.03–2 | 0.03–4 |
| GM | 0.15 | 0.05 | 0.66 | 0.07 | 0.70 | 0.08 | 0.05 | |
| MIC90 | 1 | 0.25 | 8 | 0.125 | 8 | 0.25 | 0.25 | |
| R | 10.9% | 5.6% | 4.3% | |||||
| C. krusei, n = 54 | Range | 0.06–16 | 0.015–0.5 | 0.25–64 | 0.015–4 | 0.03–16 | 0.03–4 | 0.03–1 |
| GM | 0.85 | 0.08 | 15.69 | 0.27 | 0.72 | 0.14 | 0.21 | |
| MIC90 | 4 | 0.5 | 64 | 1 | 8 | 1 | 0.5 | |
| R | – | 12.5% | 0 | |||||
| Saccharomyces cerevisiae, n = 12 | Range | 0.015–2 | 0.015–1 | 0.125–32 | 0.015–2 | 0.06–8 | 0.015–0.5 | 0.03–1 |
| GM | 0.1 | 0.09 | 1.2 | 0.20 | 0.54 | 0.11 | 0.09 | |
| MIC90 | 0.5 | 0.5 | 8 | 2 | 4 | 0.5 | 0.25 | |
| C. guilliermondii, n = 2 | Range | 0.06–4 | 0.03–4 | 0.5–16 | 0.125–16 | 1–16 | 0.03–0.25 | 0.06–1 |
| MIC50 | 0.06 | 0.03 | 0.5 | 0.125 | 1 | 0.03 | 0.06 | |
| MIC90 | 4 | 4 | 16 | 16 | 16 | 0.25 | 1 | |
| R | 0 | 50% | 0 | |||||
| C. lusitaniae, n = 1 | Range | 0.03 | 0.03 | 0.125 | 0.03 | 0.03 | 0.03 | 0.03 |
| C.Fabianii, n = 3 | Range | 0.03–0.25 | 0.03–0.06 | 0.5–1 | 0.03–0.25 | 0.125–1 | 0.03 | 0.03 |
| MIC50 | 0.125 | 0.03 | 0.5 | 0.06 | 0.125 | 0.03 | 0.03 | |
| MIC90 | 0.25 | 0.06 | 1 | 0.25 | 1 | 0.03 | 0.03 | |
| Trichosporon asahii,n = 1 | Range | 0.03 | 0.03 | 0.25 | 0.03 | 0.06 | 0.03 | 0.03 |
| Rhodotorula, n = 3 | Range | 0.03–0.5 | 0.03–1 | 4–128 | 0.03–8 | 0.25–8 | 0.03–0.5 | 0.03–1 |
| MIC50 | 0.06 | 0.06 | 64 | 2 | 1 | 0.06 | 0.03 | |
| MIC90 | 0.5 | 1 | 128 | 8 | 8 | 0.5 | 1 | |
| Kodamaea ohmeri, n = 2 | Range | 0.125–0.5 | 0.03 | 0.25–2 | 0.125–0.25 | 0.25–0.5 | 0.03 | 0.03 |
| MIC50 | 0.125 | 0.03 | 0.25 | 0.125 | 0.25 | 0.03 | 0.03 | |
| MIC90 | 0.5 | 0.03 | 2 | 0.25 | 0.5 | 0.03 | 0.03 | |
| Issatchenkia terricola,n = 2 | Range | 1–4 | 0.06–0.125 | 32–64 | 0.25–0.5 | 0.5 | 0.25 | 0.25 |
| MIC50 | 1 | 0.06 | 32 | 0.25 | 0.5 | 0.25 | 0.25 | |
| MIC90 | 4 | 0.125 | 64 | 0.5 | 0.5 | 0.25 | 0.25 | |
| Torulaspora pretoriensis,n = 1 | Range | 0.25 | 0.03 | 8 | 0.5 | 0.5 | 0.125 | 0.125 |
| ATCC90028a | Range | 0.015–0.5 | 0.015–0.5 | 0.125–2 | 0.015–4 | 0.008–0.015 | 0.015–32 | 0.015–8 |
| GM | 0.04 | 0.03 | 0.21 | 0.08 | 0.06 | 0.03 | 0.03 | |
| MIC90 | 0.125 | 0.03 | 0.5 | 0.25 | 0.015 | 0.03 | 0.03 | |
| Candida species(n) | Antifungal agents | |||||||
| AmB | FLU | NYS | TEB | AFG | CFG | MFG | ||
| C. albicans n = 1272 | Range | 0.015–32 | 0.03–128 | 0.03–32 | 0.03–256 | 0.008–0.5 | 0.008–0.5 | 0.008–0.5 |
| GM | 0.22 | 0.70 | 1.60 | 45.11 | 0.015 | 0.1 | 0.03 | |
| MIC90 | 0.5 | 4 | 8 | 256 | 0.03 | 0.25 | 0.25 | |
| R | 3.3% | 0 | 0 | 0 | ||||
| C. africana n = 49 | Range | 0.03–32 | 0.06–8 | 0.125–4 | 0.25–256 | 0.008–0.03 | 0.015–0.5 | 0.008–0.5 |
| GM | 0.08 | 0.68 | 0.5 | 17.31 | 0.01 | 0.06 | 0.02 | |
| MIC90 | 1 | 2 | 4 | 128 | 0.015 | 0.25 | 0.06 | |
| C. dubliniensis n = 1 | Range | 0.06 | 0.06 | 0.25 | 16 | 0.008 | 0.015 | 0.008 |
| C. glabrata n = 267 | Range | 0.03–2 | 0.06–16 | 0.03–32 | 0.25–256 | 0.008–0.5 | 0.008–0.5 | 0.008–0.5 |
| GM | 0.29 | 0.18 | 3.39 | 26.62 | 0.03 | 0.11 | 0.05 | |
| MIC90 | 1 | 1 | 8 | 256 | 0.06 | 0.25 | 0.25 | |
| R | 0 | 0 | 0 | 0 | ||||
| C. nivariensis n = 9 | Range | 0.06–2 | 0.125–4 | 0.5–4 | 1–256 | 0.015–0.06 | 0.08–0.5 | 0.015–0.5 |
| MIC50 | 0.06 | 0.5 | 1 | 128 | 0.06 | 0.25 | 0.015 | |
| MIC90 | 2 | 2 | 4 | 256 | 0.06 | 0.5 | 0.5 | |
| C. bracarensis n = 2 | Range | 0.06–1 | 0.125–2 | 0.25–8 | 8–256 | 0.015–0.03 | 0.125–0.5 | 0.015–0.5 |
| MIC50 | 0.06 | 0.125 | 0.25 | 8 | 0.015 | 0.125 | 0.015 | |
| MIC90 | 1 | 2 | 8 | 256 | 0.03 | 0.5 | 0.5 | |
| C. parapsilosis n = 76 | Range | 0.03–2 | 0.125–8 | 0.03–32 | 0.25–256 | 0.008–1 | 0.008–1 | 0.008–1 |
| GM | 0.19 | 0.14 | 0.59 | 0.62 | 0.69 | 0.60 | 0.54 | |
| MIC90 | 1 | 0.125 | 4 | 32 | 0.5 | 0.5 | 0.5 | |
| R | 0 | 5.2% | 5.2% | 1.3% | ||||
| C. metapsilosis n = 20 | Range | 0.015–0.5 | 0.125–4 | 0.06–4 | 0.25–256 | 0.015–0.5 | 0.008–0.5 | 0.015–1 |
| GM | 0.10 | 0.177 | 0.46 | 2.17 | 0.17 | 0.17 | 0.39 | |
| MIC90 | 0.5 | 1 | 4 | 256 | 0.25 | 0.25 | 0.5 | |
| C. orthopsilosis n = 6 | Range | 0.06–0.25 | 0.125–2 | 0.06–8 | 0.25–128 | 0.008–1 | 0.015–1 | 0.008–0.5 |
| MIC50 | 0.125 | 1 | 0.5 | 64 | 0.008 | 0.03 | 0.25 | |
| MIC90 | 0.25 | 2 | 8 | 128 | 1 | 1 | 0.5 | |
| C. tropicalis n = 61 | Range | 0.03–1 | 0.125–32 | 0.03–8 | 0.25–256 | 0.015–0.125 | 0.008–0.5 | 0.008–0.5 |
| GM | 0.19 | 0.23 | 0.54 | 60.02 | 0.03 | 0.24 | 0.04 | |
| MIC90 | 0.5 | 1 | 4 | 256 | 0.06 | 0.5 | 0.5 | |
| R | 1.8% | 0 | 0 | 0 | ||||
| C. krusei n = 54 | Range | 0.03–1 | 0.125–32 | 0.03–4 | 16–256 | 0.015–0.5 | 0.008–1 | 0.008–0.5 |
| GM | 0.43 | 4.2 | 0.32 | 75.66 | 0.08 | 0.08 | 0.15 | |
| MIC90 | 1 | 16 | 1 | 256 | 0.125 | 0.5 | 0.25 | |
| R | 2.9% | 0 | 1.85% | 0 | ||||
| Saccharomyces cerevisiae n = 12 | Range | 0.03–4 | 0.06–8 | 0.125–32 | 0.25–256 | 0.015–0.5 | 0.008–0.5 | 0.015–0.5 |
| GM | 0.18 | 0.21 | 0.78 | 53.20 | 0.14 | 0.10 | 0.18 | |
| MIC90 | 1 | 1 | 8 | 256 | 0.5 | 0.25 | 0.25 | |
| C. guilliermondii n = 2 | Range | 0.06–0.5 | 0.125–0.25 | 0.25–0.5 | 64–128 | 0.015–0.25 | 0.25–0.5 | 0.015–0.25 |
| MIC50 | 0.06 | 0.125 | 0.25 | 64 | 0.015 | 0.25 | 0.015 | |
| MIC90 | 0.5 | 0.25 | 0.5 | 128 | 0.25 | 0.5 | 0.25 | |
| R | 0 | 0 | 0 | 0 | ||||
| C. lusitaniae n = 1 | Range | 0.125 | 0.125 | 0.25 | 0.25 | 0.03 | 0.25 | 0.015 |
| C. fabianii n = 3 | Range | 0.06–0.25 | 0.125 | 0.25 | 128 | 0.015–0.03 | 0.015–0.25 | 0.015–0.06 |
| MIC50 | 0.06 | 0.125 | 0.25 | 128 | 0.015 | 0.015 | 0.03 | |
| MIC90 | 0.25 | 0.125 | 0.25 | 128 | 0.03 | 0.25 | 0.06 | |
| Trichosporon asahii n = 1 | Range | 0.125 | 1 | 0.25 | 128 | 0.015 | 0.5 | 0.015 |
| Rhodotorula n = 3 | Range | 0.03–0.5 | 0.125 | 0.06–0.125 | 8–256 | 0.06–0.5 | 0.25–0.5 | 0.25–0.5 |
| MIC50 | 0.03 | 0.125 | 0.06 | 8 | 0.06 | 0.25 | 0.25 | |
| MIC90 | 0.5 | 0.125 | 0.125 | 256 | 0.5 | 0.5 | 0.5 | |
| Kodamaea ohmeri n = 2 | Range | 0.125–1 | 0.125 | 0.5 | 128 | 0.015 | 0.125–0.25 | 0.015–0.03 |
| MIC50 | 0.125 | 0.125 | 0.5 | 128 | 0.015 | 0.125 | 0.015 | |
| MIC90 | 1 | 0.125 | 0.5 | 128 | 0.015 | 0.25 | 0.03 | |
| Issatchenkia terricola n = 2 | Range | 0.125–0.5 | 8 | 0.5 | 128–256 | 0.06 | 0.06 | 0.008 |
| MIC50 | 0.125 | 8 | 0.5 | 128 | 0.06 | 0.06 | 0.008 | |
| MIC90 | 0.5 | 8 | 0.5 | 256 | 0.06 | 0.06 | 0.008 | |
| Torulaspora pretoriensis n = 1 | Range | 0.125 | 0.125 | 0.25 | 16 | 0.015 | 0.125 | 0.015 |
| ATCC90028a | Range | 0.03–2 | 0.125–8 | 0.25–16 | 1–256 | 0.008–0.015 | 0.015–0.5 | 0.008–0.015 |
| GM | 0.22 | 0.64 | 1.31 | 88.22 | 0.01 | 0.09 | 0.01 | |
| MIC90 | 1 | 2 | 8 | 256 | 0.015 | 0.5 | 0.015 | |
Note: GM geometry mean, BUC butoconazole, CLO Clotrimazole, FLC Fluconazole, ITC Itraconazole, VRC Voriconazole, MIC Miconazole, TEC Terconazole, AmB Amphotericin B, FLU Flucytosine, NYS Nystatin, TEB Terbinafine, AFG Anidulafungin, CFG Caspofungin, MFG Micafungin
a ATCC90028 was tested 57 times
Table 4.
In vitro antifungal susceptibility of 1539 clinical C. albicans and C. glabrata isolates from VVC and RVVC as determined by the CLSI method
| AmB | FLU | NYS | TEB | AFG | CFG | MFG | BUC | CLO | FLC | ITC | MIC | TEC | VRC | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C. albicans | |||||||||||||||
| VVC | Range | 0.15–32 | 0.030–128 | 0.03–32 | 0.03–256 | 0.008–0.5 | 0.008–0.5 | 0.008–0.5 | 0.015–32 | 0.015–32 | 0.06–128 | 0.015–32 | 0.015–32 | 0.015–32 | 0.015–8 |
| GM | 0.21 | 0.65 | 1.55 | 39.23 | 0.02 | 0.1 | 0.03 | 0.11 | 0.05 | 0.74 | 0.09 | 0.27 | 0.12 | 0.06 | |
| MIC90 | 0.5 | 4 | 8 | 256 | 0.03 | 0.25 | 0.25 | 1 | 0.25 | 4 | 1 | 4 | 2 | 0.5 | |
| RVVC | Range | 0.015–32 | 0.06–128 | 0.03–32 | 0.25–256 | 0.008–0.25 | 0.008–0.5 | 0.008–0.5 | 0.015–8 | 0.015–16 | 0.06–64 | 0.015–32 | 0.015–32 | 0.015–32 | 0.015–8 |
| GM | 0.25 | 0.94 | 1.82 | 74.50 | 0.01 | 0.1 | 0.03 | 0.13 | 0.05 | 1.03 | 0.10 | 0.46 | 0.33 | 0.09 | |
| MIC90 | 0.6 | 4 | 8 | 256 | 0.015 | 0.25 | 0.06 | 1 | 0.5 | 4 | 0.5 | 8 | 8 | 0.5 | |
| P | 0.813 | 0.435 | 0.770 | 0 | 0.242 | 0.7 | 0.566 | 0.736 | 0.341 | 0.14 | 0.041 | 0.053 | 0 | 0.34 | |
| C. glabrata | |||||||||||||||
| VVC | Range | 0.03–2 | 0.06–16 | 0.03–32 | 0.25–256 | 0.015–0.5 | 0.008–0.5 | 0.008–0.5 | 0.015–16 | 0.015–16 | 0.125–128 | 0.015–32 | 0.015–16 | 0.015–32 | 0.015–32 |
| GM | 0.33 | 0.18 | 3.88 | 40.63 | 0.03 | 0.06 | 0.05 | 0.24 | 0.23 | 1.68 | 0.42 | 0.25 | 0.15 | 0.09 | |
| MIC90 | 1 | 1 | 8 | 256 | 0.06 | 0.25 | 0.25 | 1 | 1 | 8 | 4 | 2 | 1 | 0.25 | |
| RVVC | Range | 0.03–2 | 0.06–2 | 0.03–16 | 0.25–256 | 0.008–0.06 | 0.008–0.5 | 0.008–0.5 | 0.015–8 | 0.03–4 | 0.125–128 | 0.03–32 | 0.03–32 | 0.015–32 | 0.015–32 |
| GM | 0.20 | 0.17 | 2.31 | 8.86 | 0.02 | 0.06 | 0.04 | 0.16 | 0.134 | 1.03 | 0.29 | 0.27 | 0.17 | 0.11 | |
| MIC90 | 0.5 | 1 | 8 | 256 | 0.03 | 0.25 | 0.25 | 1 | 1 | 16 | 2 | 4 | 2 | 0.5 | |
| P | 0.017 | 0.366 | 0.035 | 0.015 | 0.058 | 0.68 | 0.78 | 0.87 | 0.25 | 0.1 | 1 | 0.043 | 0.683 | 0.293 | |
Note: GM geometry mean, BUC butoconazole, CLO Clotrimazole, FLC Fluconazole, ITC Itraconazole, VRC Voriconazole, MIC Miconazole, TEC Terconazole, AmB Amphotericin B, FLU Flucytosine, NYS Nystatin, TEB Terbinafine, AFG Anidulafungin, CFG Caspofungin, MFG Micafungin. The MIC GM value of C. albicans for Itraconazole, Terconazole, and Terbinafine in RVVC is higher than those in VVC. The MIC GM value of C. glabrata for Miconazole, Amphotericin B, Nystatin, Caspofungin, and Terbinafine in RVVC is higher than those in VVC
Discussion
Strain identification and distribution
Borman reported 1781 yeast isolates submitted to the United Kingdom Mycology Reference Laboratory and found that 100 isolates (9.7%) were incorrectly identified, with error rates ranging from 5.2 to 18.2% [22]. The conventional methods such as the API ID 32 C system could not identify the rare or new recovered Candida [23]. The identification ratios (IR) at the species level of yeast were 0.89 for the API ID 32C system, 0.89 for the AuxaColor system, and 0.93 for the Vitek 2 system. Subanalysis of data showed that the Vitek 2 system was more accurate (IR: 0.94) than the API ID32C system (IR: 0.84) and the AuxaColor system (IR: 0.76) [7]. Gündeş reported the performance of API 20C Aux was with 87% (101 of 116 isolates) [24]. Two hundred and fifty-one isolates (83.7%) were correctly identified, 49 (16.2%) isolates were misidentified, and there was no species without identification using API 20C AUX. The majority of misidentified yeast isolates were among rare species (n = 45), and the majority (4/5) of Pichia kudriavzevii strains were misidentified [25]. The closely related Candida complex was identified from vaginal samples by using molecular methods [8, 16–19, 26]. Based on conventional and molecular methods, C. albicans, C. glabrata, C. parapsilosis, and C. tropicalis are the four most common Candida species from VVC. Most of previous studies were non-molecular identification or small samples based molecular identification [3–6, 27, 28]. In the current study, by using molecular identification, we found that C. albicans was still the most common Candida species in VVC, followed by C. glabrata, C. parapsilosis and C. tropicalis. The yeast species from VVC was stable in the past 16 years.
The API 20C system has a lower correct identification rate for non-albicans (33.33–84.59%) than that for C. albicans (98.51%). The system also could not identify new closely related Candida species and novel species. Compared with conventional methods by which 5–10 Candida species were identified, molecular methods identified more than 20 Candida species from vaginal samples, suggesting the necessity of molecular identification in research [3–5, 22].
Antifungal susceptibility
Most non-albicans Candida species have a higher azole MICs, and the VVC they cause are often difficult to treat [28–35]. Fluconazole-resistant C. albicans have been found in VVC [34, 35].
The antifungal prescription affects the relative distribution and susceptibility of Candida [36, 37].
An increasing number of isolates with elevated MICs were observed following fluconazole introduction rather than prior to that [37]. In our current study, C. albicans was susceptible to most of the tested antifungals. The MICs of azoles for C. glabrata were higher than those for C. albicans and the MICs of echinocandins for C. parapsilosis were higher than those for C. albicans, which were similar to a previous study [32]. The MICs of nystatin for C. albicans and C. glabrata were higher than that from the findings of other reports and those of our previous study on the use of different antifungal susceptibility tests [3, 38]. In current study, terbinafine was the less active drug against most of the tested isolates, which was similar to a previous study and may not be used for treating VVC [24, 39]. CD101, a new echinocandin antifungal agent, has been studied specifically as a possible treatment for VVC in rat and human [12–15]. The current study has shown that echinocandins including anidulafungin, caspofungin and micafungin have a low MIC for C. glabrata, which may provide an opportunity for treating azole-resistant VVC.
Conclusions
It was concluted that API 20C yeast identification system can be used to reliably identify the most common Candida species. Molecular methods are necessary for the identification of closely related, emerging, and rare yeast species, which are quite important in research. C. albicans was the predominant Candida species isolated from this sample of patients with VVC. The results from this study suggest that much of the previous studies of epidemiology for VVC should be re-thought. Resistance of vaginal C. albicans isolates to antifungal agents was infrequent.
Acknowledgements
Not Applicable.
Abbreviations
- CLSI
Clinical & Laboratory Standards Institute
- GM
Geometric mean
- MIC
Minimal inhibitory concentration
- PCR
Polymerase chain reaction
- VVC
Vulvovaginal candidiasis
- RVVC
Recurrent VVC
- SPSS
Statistical product and service solutions
Authors’ contributions
YS and YXZ designed the study and finished the study; SRF and XPL designed the study and wrote the paper; YHL and YYS joined the samples collection. The author(s) read and approved the final manuscript.
Funding
This research was supported by Shenzhen Science and Technology Grant JCYJ JCYJ20160428175005906; SZXJ2017008; Guangdong Science and Technology Grant 2014A020212037. The funding body did not have input into the design of the study and collection, analysis and interpretation of data in writing the manuscript.
Availability of data and materials
The datasets used during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The verbal informed consents were obtained from all individual participants included in the study. The study and the informed consents procedure were approved by Peking University Shenzhen Hospital Medical Ethics Committee (20140406).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
The original version of this article was revised: errors in Table 3 were corrected.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yu Shi and Yuxia Zhu contributed equally to this work.
Change history
1/24/2022
A Correction to this paper has been published: 10.1186/s12879-021-06601-x
References
- 1.Workowski KA, Bolan GA. Centers for Disease Control and Prevention. Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep. 2015;64(RR-03):1–137. [PMC free article] [PubMed] [Google Scholar]
- 2.Blostein F, Levin-Sparenberg E, Wagner J, Foxman B. Recurrent vulvovaginal candidiasis. Ann Epidemiol. 2017;27(9):575–582. doi: 10.1016/j.annepidem.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Liu XP, Fan SR, Peng YT, Zhang HP. Species distribution and susceptibility of Candida isolates from patient with vulvovaginal candidiasis in southern China from 2003 to 2012. J Mycol Med. 2014;24(2):106–111. doi: 10.1016/j.mycmed.2014.01.060. [DOI] [PubMed] [Google Scholar]
- 4.Wang FJ, Zhang D, Liu ZH, Wu WX, Bai HH, Dong HY. Species distribution and in vitro antifungal susceptibility of vulvovaginal Candida isolates in China. Chin Med J. 2016;129(10):1161–1165. doi: 10.4103/0366-6999.181964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng KP, Kuan CS, Kaur H, Na SL, Atiya N, Velayuthan RD. Candida species epidemiology 2000-2013: a laboratory-based report. Tropical Med Int Health. 2015;20(11):1447–1453. doi: 10.1111/tmi.12577. [DOI] [PubMed] [Google Scholar]
- 6.Gonçalves B, Ferreira C, Alves CT, Henriques M, Azeredo J, Silva S. Vulvovaginal candidiasis: epidemiology, microbiology and risk factors. Crit Rev Microbiol. 2016;42(6):905–927. doi: 10.3109/1040841X.2015.1091805. [DOI] [PubMed] [Google Scholar]
- 7.Posteraro B, Efremov L, Leoncini E, Amore R, Posteraro P, Ricciardi W, Sanguinetti M. Are the conventional commercial yeast identification methods still helpful in the era of new clinical microbiology diagnostics? A meta-analysis of their accuracy. J Clin Microbiol. 2015;53(8):2439–2450. doi: 10.1128/JCM.00802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Criseo G, Scordino F, Romeo O. Current methods for identifying clinically important cryptic Candida species. J Microbiol Methods. 2015;111:50–56. doi: 10.1016/j.mimet.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 9.Sobel JDand Akins RA. The role of PCR in the diagnosis of Candida vulvovaginitis-a new gold standard? Curr Infect Dis Rep. 2015;17(6):488. doi: 10.1007/s11908-015-0488-3. [DOI] [PubMed] [Google Scholar]
- 10.Sobel JDand Sobel R. Current treatment options for vulvovaginal candidiasis caused by azole-resistant Candida species. Expert Opin Pharmacother. 2018;19(9):971–977. doi: 10.1080/14656566.2018.1476490. [DOI] [PubMed] [Google Scholar]
- 11.Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R. Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis. 2018;18(11):e339–e347. doi: 10.1016/S1473-3099(18)30103-8. [DOI] [PubMed] [Google Scholar]
- 12.Brand SR, Degenhardt TP, Person K, Sobel JD, Nyirjesy P, Schotzinger RJ, Tavakkol A. A phase 2, randomized, double-blind, placebo-controlled, dose-ranging study to evaluate the efficacy and safety of orally administered VT-1161 in the treatment of recurrent vulvovaginal candidiasis. Am J Obstet Gynecol. 2018;218(6):624.e1–624.e9. doi: 10.1016/j.ajog.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Garvey EP, Hoekstra WJ, Schotzinger RJ, Sobel JD, Lilly EA, Fidel PL., Jr Efficacy of the clinical agent VT-1161 against fluconazole-sensitive and -resistant Candida albicans in a murine model of vaginal candidiasis. Antimicrob Agents Chemother. 2015;59(9):5567–5573. doi: 10.1128/AAC.00185-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boikov DA, Locke JB, James KD, Bartizal K, Sobel JD. In vitro activity of the novel echinocandin CD101 at pH 7 and 4 against Candida spp. isolates from patients with vulvovaginal candidiasis. J Antimicrob Chemother. 2017;72(5):1355–1358. doi: 10.1093/jac/dkx008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyirjesy P, Alessio C, Jandourek A, Lee JD, Sandison T. Sobel JD.CD101 topical compared with oral fluconazole for acute vulvovaginal candidiasis: a randomized controlled trial. J Low Genit Tract Dis. 2019;23(3):226–229. doi: 10.1097/LGT.0000000000000473. [DOI] [PubMed] [Google Scholar]
- 16.Shan Y, Fan S, Liu X, Li J. Prevalence of Candida albicans-closely related yeasts, Candida africana and Candida dubliniensis, in vulvovaginal candidiasis. Med Mycol. 2014;52(6):636–640. doi: 10.1093/mmy/myu003. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Shan Y, Fan S, Liu X. Prevalence of Candida nivariensis and Candida bracarensis in vulvovaginal candidiasis. Mycopathologia. 2014;178(3–4):279–283. doi: 10.1007/s11046-014-9800-2. [DOI] [PubMed] [Google Scholar]
- 18.Asadzadeh M, Ahmad S, Hagen F, Meis JF, Al-Sweih N, Khan Z. Simple, low-cost detection of Candida parapsilosis complex isolates and molecular fingerprinting of Candida orthopsilosis strains in Kuwait by ITS region sequencing and amplified fragment length polymorphism analysis. PLoS One. 2015;10(11):e0142880. doi: 10.1371/journal.pone.0142880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leaw SN, Chang HC, Sun HF, Barton R, Bouchara JP, Chang TC. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J Clin Microb. 2006;44(3):693–699. doi: 10.1128/JCM.44.3.693-699.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.CLSI . Reference method for broth dilution antifungal susceptibility testing of yeasts; third edition; CLSI document M27-A3. Wayne: Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 21.CLSI . Reference method for broth dilution antifungal susceptibility testing of yeasts; 1st ed, M60. Wayne: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 22.Borman AM, Szekely A, Palmer MD, Johnson EM. Assessment of accuracy of identification of pathogenic yeasts in microbiology laboratories in the United Kingdom. J Clin Microbiol. 2012;50(8):2639–2644. doi: 10.1128/JCM.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durán-Valle MT, Sanz-Rodríguez N, Muñoz-Paraíso C, Almagro-Moltó M, Gómez-Garcés JL. Identification of clinical yeasts by Vitek MS system compared with API ID 32 C. Med Mycol. 2014;52(4):342–349. doi: 10.1093/mmy/myt036. [DOI] [PubMed] [Google Scholar]
- 24.Gündeş SG, Gulenc S, Bingol R. Comparative performance of Fungichrom I, Candifast and API 20C aux systems in the identification of clinically significant yeasts. J Med Microbiol. 2002;50(12):1105–1110. doi: 10.1099/0022-1317-50-12-1105. [DOI] [PubMed] [Google Scholar]
- 25.Arastehfar A, Daneshnia F, Kord M, Roudbary M, Zarrinfar H, Fang W, Najafzadeh MJ, Khodavaisy S, Pan W, Liao W, Badali H, Rezaie S, Zomorodian K, Hagen F, Boekhout T. Comparison of 21-plex PCR and API 20C AUX, MALDI-TOF MS, and rDNA sequencing for a wide range of clinically isolated yeast species: improved identification by combining 21-plex PCR and API 20C AUX as an alternative strategy for developing countries. Front Cell Infect Microbiol. 2019;9:21. [DOI] [PMC free article] [PubMed]
- 26.Zhu Y, Shan Y, Fan S, Li J, Liu X. Candida parapsilosis sensu stricto and the closely related species Candida orthopsilosis and Candida metapsilosis in vulvovaginal candidiasis. Mycopathologia. 2015;179(1–2):111–118. doi: 10.1007/s11046-014-9821-x. [DOI] [PubMed] [Google Scholar]
- 27.Kalkanci A, Güzel AB, Khalil II, Aydin M, Ilkit M, Kuştimur S. Yeast vaginitis during pregnancy: susceptibility testing of 13 antifungal drugs and boric acid and the detection of four virulence factors. Med Mycol. 2012;50(6):585–593. doi: 10.3109/13693786.2012.662597. [DOI] [PubMed] [Google Scholar]
- 28.Makanjuola O, Bongomin F, Fayemiwo SA. An update on the roles of non-albicans candida species in vulvovaginitis. J Fungi (Basel) 2018;4(4):121. doi: 10.3390/jof4040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao M, Fan X, Chen SC, Wang H, Sun ZY, Liao K, Chen SL, Yan Y, Kang M, Hu ZD, Chu YZ, Hu TS, Ni YX, Zou GL, Kong F, Xu YC. Antifungal susceptibilities of Candida glabrata species complex, Candida krusei, Candida parapsilosis species complex and Candida tropicalis causing invasive candidiasis in China: 3 year national surveillance. J Antimicrob Chemother. 2015;70(3):802–810. doi: 10.1093/jac/dku460. [DOI] [PubMed] [Google Scholar]
- 30.Zhang L, Zhou S, Pan A, Li J, Liu B. Surveillance of antifungal susceptibilities in clinical isolates of Candida species at 36 hospitals in China from 2009 to 2013. Int J Infect Dis. 2015;33:1–4. doi: 10.1016/j.ijid.2014.12.033. [DOI] [PubMed] [Google Scholar]
- 31.Delarze Eand Sanglard D. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug Resist Updat. 2015;23:12–19. doi: 10.1016/j.drup.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Castanheira M, Deshpande LM, Davis AP, Rhomberg PR, Pfaller MA. Monitoring antifungal resistance in a global collection of invasive yeasts and molds: application of CLSI epidemiological cutoff values and whole-genome sequencing analysis for detection of azole resistance in Candida albicans. Antimicrob Agents Chemother. 2017;61(10):e00906–17. [DOI] [PMC free article] [PubMed]
- 33.Nagashima M, Yamagishi Y, Mikamo H. Antifungal susceptibilities of Candida species isolated from the patients with vaginal candidiasis. J Infect Chemother. 2016;22(2):124–126. doi: 10.1016/j.jiac.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Marchaim D, Lemanek L, Bheemreddy S, Kaye KS, Sobel JD. Fluconazole-resistant Candida albicans vulvovaginitis. Obstet Gynecol. 2012;120(6):1407–1414. doi: 10.1097/AOG.0b013e31827307b2. [DOI] [PubMed] [Google Scholar]
- 35.Bailly S, Maubon D, Fournier P, Pelloux H, Schwebel C, Chapuis C, Foroni L, Cornet M, Timsit JF. Impact of antifungal prescription on relative distribution and susceptibility of Candida spp. - trends over 10 years. J Inf Secur. 2016;72(1):103–111. doi: 10.1016/j.jinf.2015.09.041. [DOI] [PubMed] [Google Scholar]
- 36.Bulik CC, Sobel JD, Nailor MD. Susceptibility profile of vaginal isolates of Candida albicans prior to and following fluconazole introduction - impact of two decades. Mycoses. 2011;54(1):34–38. doi: 10.1111/j.1439-0507.2009.01752.x. [DOI] [PubMed] [Google Scholar]
- 37.Fornari G, Vicente VA, Gomes RR, Muro MD, Pinheiro RL, Ferrari C, Herkert PF, Takimura M, Carvalho NS, Queiroz-Telles F. Susceptibility and molecular characterization of Candida species from patients with vulvovaginitis. Braz J Microbiol. 2016;47(2):373–380. doi: 10.1016/j.bjm.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi XY, Yang YP, Zhang Y, Li W, Wang JD, Huang WM, Fan YM. Molecular identification and antifungal susceptibility of 186 Candida isolates from vulvovaginalcandidiasis in southern China. J Med Microbiol. 2015;64(Pt 4):390–393. doi: 10.1099/jmm.0.000024. [DOI] [PubMed] [Google Scholar]
- 39.Ferahbas A, Koc AN, Uksal U, Aygen E, Mistik S, Yildiz S. Terbinafine versus itraconazole and fluconazole in the treatment of vulvovaginal candidiasis. Am J Ther. 2006;13(4):332–336. doi: 10.1097/00045391-200607000-00010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the current study are available from the corresponding author on reasonable request.