Abstract
The process of drug discovery and drug development consumes billions of dollars to bring a new drug to the market. Drug development is time consuming and sometimes, the failure rates are high. Thus, the pharmaceutical industry is looking for a better option for new drug discovery. Drug repositioning is a good alternative technology that has demonstrated many advantages over de novo drug development, the most important one being shorter drug development timelines. In the last two decades, drug repositioning has made tremendous impact on drug development technologies. In this review, we focus on the recent advances in drug repositioning technologies and discuss the repositioned drugs used for inflammatory diseases such as sepsis, asthma, and atopic dermatitis.
Keywords: Drug repositioning, Inflammatory disease, Sepsis, Asthma, Atopic dermatitis
INTRODUCTION
Since the earlyrug repositioning has evolved and generated considerable interest, especially during the last 10 years. Bringing new drugs to the market using conventional drug discovery technologies is a time-consuming and expensive process, due to which many drug companies have turned towards drug repositioning (Sekhon, 2013). Another major factor that has led to an increased interest in drug repositioning is the high failure rate of new drugs. Drug repositioning provides a promising avenue to identify better and safer treatment options without the full cost or time required for de novo drug development (Ashburn and Thor, 2004; Chong and Sullivan, 2007; Tobinick, 2009; Sleigh and Barton, 2010; Phelps, 2011; Sardana et al., 2011). Discovery of novel drug targets is another big challenge in the drug development process. In the treatment of diseases more than 400 proteins are used as drug targets and many known drug targets are involved in multiple pathologies (Sekhon, 2013). Historically, researchers came to know about drug repositioning due to some unexpected discoveries during late stage clinical trials or post-approval steps. One such classic example is sildenafil that was unsuccessful in its development as a new drug for common hypertension but became immensely successful as a drug for male erectile impotence and was later established as a drug to treat pulmonary arterial hypertension (Sekhon, 2013). Research is ongoing to find alternate uses of existing approved or generic drugs to treat many diseases (Fig. 1).
Fig. 1.
Diagram showing the drug repositioning steps (Fig. modified from Sekhon, 2013).
Drug repositioning can be defined as finding new uses of existing drugs that are outside the scope of the original indication (Jang et al., 2016). Since the last decade, drug repositioning has gained increased importance due to the enormous rise in the cost of new drug discovery (O’Connor and Roth, 2005; Chong and Sullivan, 2007). Many researchers are applying repositioning strategies to discover novel therapeutic applications of known drugs, since 90% of the new drug candidates fail in the early stages of safety and efficacy tests during de novo drug discovery. For the development of drugs, various target protein and drug characteristics, based on chemical structures and the properties of ligands and receptors, have been used to identify new targets for existing drugs (Jang et al., 2016). According to the Keiser hypothesis, structurally similar chemicals have comparable properties, including biological activities (Keiser et al., 2009).
The docking method is a common strategy used in drug repositioning. This method focuses on the physical interactions between drugs and targets (Zahler et al., 2007; Kinnings et al., 2009; Chang et al., 2010). This method mainly deals the chemical and physical binding to the targets of protein of interest. This type of interaction has been reported to play an important role in the development or treatment of the disease. Systematic approaches, such as the use of chemical structures, amino acid sequences of target proteins, and chemical-protein interaction network, can be utilized to find new molecular targets for the existing drugs (Yamanishi et al., 2008, 2010; Mei et al., 2013). Three-dimensional structural libraries of both chemical compounds and target proteins are required for the physical binding stimulations between drugs and their targets (Jang et al., 2016).
Currently, studies are ongoing to identify novel uses of existing drugs by analyzing the patterns of both drug-associated and disease-associated gene sets. Results from these studies suggest there are some problems in computational approaches used to discover new drug indications, with respect to the experimental outcomes of molecular activity (Lamb et al., 2006; Iorio et al., 2010; Sirota et al., 2011). Recently, some researchers have focused on electronic clinical data, collected when medical services are provided to the patients in medical institutions, and includes information encompassing a variety of medical history such as prescriptions, diagnoses, and laboratory test results (Holmes et al., 2011; Roque et al., 2011; Jung et al., 2013). This clinical information can be used to investigate the hidden connections between diseases and clinical variables (Jensen et al., 2012). The clinical notes have explicit information about disease-drug relationships which are useful in drug repositioning (Jung et al., 2013, 2014).
DATA SOURCES
The drug bank maintains a drug database that includes 6,811 drug entities, 1,578 approved drugs, and their 22,143 synonyms (Wishart et al., 2008). National Health and Nutrition Examination survey (NHANES) is a data source for electronic clinical information (available at http://www.cdc.gov/nchs/nhanes.htm) that has data related to the demographics, dietary habits, health related questions, and results of laboratory tests.
DRUG REPOSITIONING PROCESS
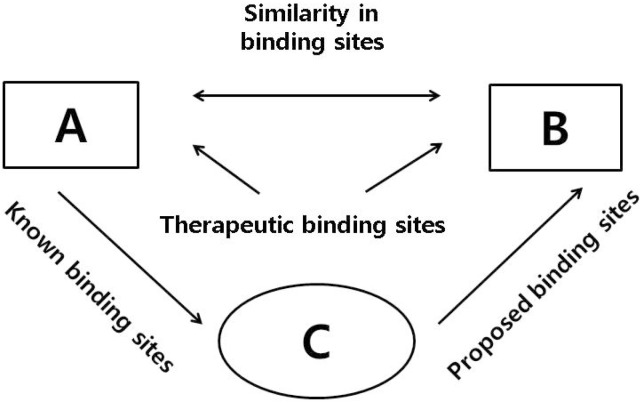
Drug discovery and design are based on drug-target interaction (Jang et al., 2016). Important and necessary steps for reducing the burden of disease and finding new uses for drugs can be found by computational methods. These methods use two main strategies as follows: 1. drug/target-based (based on the chemical or pharmaceutical perspective), 2. disease-based (based on the clinical perspective of a disease or its pathology and symptomatology). The drug/target-based methods include chemical similarity, molecular activity similarity, and molecular docking, while disease-based methods include side-effect similarity, shared molecular pathology, and associative indicative transfer (Dudley et al., 2011a). Currently, in silico methods such as data mining, bioinformatics, and novel screening platforms are being used in drug repositioning projects for the identification and screening of potential repositioning candidates (Li et al., 2006; Yildirim et al., 2007; Campillos et al., 2008; Yamanishi et al., 2008; Kieser et al., 2009; Morrow et al., 2010; Chen and Zei, 2011; Dudley et al., 2011b; Ekins et al., 2011; Haupt et al., 2011; Sanseau and Koehler, 2011; Lounkine et al., 2012; Phatak and Zhang, 2013). Some researchers have also developed computational methods to represent and align biding sites (Fig. 2) (Haupt and Shoroeder, 2011).
Fig. 2.
Exploitation of binding sites similarities between A and B for binding of a compound (Fig. modified from Sekhon, 2013).
According to pharmacological study, drug molecules often interact with multiple targets. Prediction of polypharmacology of known drugs and new molecules against multiple targets is more useful for drug repositioning (Liu et al., 2013).
ADVANTAGES OF DRUG REPOSITIONING
Adoption or integration of drug repositioning strategy offers some valuable advantages. These include easy availability of the active ingredients, increased success rates, decreased time to launch, reduced drug development costs, availability of numerous druggable compounds in the drug libraries that have the potential to be repurposed, integration of standard process of resource utilization, and de-risking and acceleration of drug development process using repurposing technology. Since the existing drugs have already undergone and cleared significant number of toxicology and safety assessments, the chances of failure of the repositioned drugs are greatly reduced. Drug repositioning candidates include known compounds with new targets or those with known mechanism for new indications (Oprea et al., 2011).
DRUG REPOSITIONING BASED ON PATHWAY
Few researchers have focused on pathway-based approach for drug repositioning. Several studies have reported success of drug repositioning through manual analysis of casual associations in drug-involved pathways (Sivachenko et al., 2006; Kotelnikova et al., 2010; Cramer et al., 2012; Strittmatter, 2012). For example, bexarotene, an anti-cancer drug has been used for the treatment of Alzheimer’s disease (AD) (Cramer et al., 2012). The nuclear receptor PPAR (peroxisome proliferator-activated receptor) is activated by bexarotene and LXR (liver X receptor), in coordination with RXR (retinoid X receptor), and then up-regulates the expression of the ApoE (apolipoprotein E) gene (Li and Lu, 2013). This entire process plays a key role in the clearance of amyloid beta (Aβ) from the brain, thus resulting in the alleviation of AD. One drug and one disease chain causality was examined and took the following advantage in the bexarotene-related pathways knowledge: 1. drug-target (bexarotene is an RXR agonist); 2. target involved pathway (LXR: RXR activation pathway); 3. transcriptional responses in a given pathway (increased ApoE gene expression in the LXR: RXR activation pathway); 4. genetic mechanism of disease (ApoE is associated with AD). Pathway based on this computational method has been used to build a network of casual chains between drugs and diseases. This method was built by a multi-layer casual network, consisting of chains from drug to target, target to pathway, pathway to downstream gene, and gene to disease (Li and Lu, 2013).
THE USE OF ZEBRAFISH TO SCREENREPOSITIONING DRUGS FOR INFLAMMATORYDISEASES
The host is usually protected from infection and injury to maintain homeostasis by an inflammatory response, which is a complex reflective process (Medzhitov, 2008). Usually, illness and death are majorly caused by inflammatory disorders such as auto-immune diseases, allergies, asthma, and sepsis. Drug repositioning is a faster and cheaper approach than traditional drug discovery (Hall et al., 2014).
Zebrafish is a valuable drug discovery platform. To define and characterize drug activity in a high-content manner, whole live bioassay approach is carried out using embryos and larvae of zebrafish. Furthermore, zebrafish is a well-established model for studying leukocyte behavior. Zebrafish embryos populated with neutrophil and macrophage lineages two days post-fertilization function similar to human neutrophil and macrophage lineages (Hall et al., 2014). Zebrafish is suited for in vivo phenotypic drug screening and larval zebrafish has revealed several new drugs that interfere with multiple complex behaviors used by high-throughput behavioral profiling (Rihel et al., 2010). The suppression of neutrophil recruitment is an initial selection criterion of anti-inflammatory activity in zebrafish tail fin wounds. A total of 251 drugs, with significant anti-inflammatory effect, (p<0.05) have been identified from zebrafish screening. This represents 22.4% of the drugs in the library. About 43.9% of non-steroidal anti-inflammatory drugs and 51.9% of corticosteroids, present in the library, were identified as having conserved anti-inflammatory activity in the zebrafish screen (Hall et al., 2014). Table 1 shows the 12 most significant hits from the zebrafish anti-inflammatory screen.
Table 1.
Drug names, formulas, and mechanism of action of the 12 most significant hits from the zebrafish anti-inflammatory screen
| Drug name | Generic names | Therapeutic group | Mechanism of action |
|---|---|---|---|
| Acetohexamide | Dymelor | Antidiabetic (type-II noninsulin dependent) | Blocks ATP-sensitive K plus channels/stimulates insulin release |
| Alfuzosin hydrochloride | Uroxatral, Rilif, Tevax, Unibenestan, Urion, UroXatral OD, Weiping, Xatger, Zofu | Antihypertensive | Alpha 1-adrenergic antagonist |
| Amodiaquine | Amadaquin, Amobin, Amochin, Basoquin, Trimalact, Camoquin, Creaquine, Larimal | Antimalarial | Heme polymerase inhibitor |
| Chlorophenesin carbamate | Cloricool, Kalsont, Maolate, Muslax Rinlaxer, Skenesin, Steacol | Muscle relaxant | Blocks nerve impulses |
| Clonidine hydrochloride | Clonidine, Mylan, Clonidural, Cloniprex, Clonistada, Clonnirit, Clophelin | Antihypertensive | Alpha 2 agonist/imidazoline agonist |
| Etodolac | Etodol, Etodolac, Apotex, ETOFACT, EtoGesic | Anti-inflammatory (NSAID) | Cox inhibitor |
| Fludrocortisone acetate | Flrinef, Astonin-H, Cortineff, Florineff Acetaat, Astonin, Merk, Lonikan | Mineralocorticoid | Causes kidneys to retain sodium |
| Mafenide hydrochloride | Sulfamylon, Abamide, Emilene, Homonal, Malfamin, Marfanil | Antibacterial | Inhibitor of folic acid biosynthesis |
| Methyldopa | Aldomet, Aldochlor, Aldopren, Aldotensin, Alfametildopa, Datleal, Dopagrand | Antihypertensive | L-aromatic amino acid decarboxylase inhibitor |
| Nefopam hydrochloride | Acupainlex, Acupan, Acuten, Anton, Benoton, Glosic, Ketopen, Licopam, Neforex, Paton, Sezen, Tonfupin, Xripa | Analgesic | Unknown |
| Niflumic acid | Donalgin, Flogovital, Forenal, Niflactol, Niflam, Landdruma | Anti-inflammatory (NSAID) | Cox inhibitor |
| Pinacidil | Pindac | Vasodilator | K+ channel Ca2+ activator |
NSAID, nonsteroidal anti-inflammatory drugs.
REPOSITIONING DRUGS OF SEPSIS
A systematic inflammatory response syndrome caused by infection is called sepsis. In well-developed and high-income countries, severe sepsis (sepsis accompanied by acute organ dysfunction) is the leading cause of death and a common cause of death among critically ill patients in intensive care units (Russell, 2006). Since 2001, recombinant activated protein C (APC), approved by FDA was the only drug available for the treatment of sepsis and septic shock. However, in October 2011, APC was withdrawn from the market due to side effects and lack of efficacy (Bernard et al., 2001). Researchers and physicians have continued to search for new therapeutic approaches and targets against sepsis. Therefore, drug repositioning can be used in these situations where the currently used drugs are not efficient (Kwak et al., 2015).
Methylthiouracil (MTU)
The systematic name of MTU is 6-methyl-2-thiouracil, as per International Union of Pure and Applied Chemistry (IUPAC). Molecular formula and weight of MTU are C5H6N2OS and 142.18 DA, respectively. It is slightly soluble in benzene, diethyl ether, ethanol, and methanol and insoluble in water. In the mid-1940s, MTU was introduced as a thionamide, an antithyroid drug for the treatment of hypothyroidism (Glock, 1946; Reddy and Kaul, 1979; Kwak et al., 2015). MTU has been repositioned to treat sepsis that includes multiple organ failure by cecum ligation and puncture (CLP) such as liver injury, renal injury, and overall tissue injury (Kwak et al., 2015). MTU has demonstrated anti-sepsis effects through inhibition of the release of high mobility group box 1 protein (HMGB1) and HMGB1-mediated inflammatory responses. MTU inhibited the expression HMGB1 receptors such as toll-like receptor 2 (TLR2) and TLR4. MTU also inhibited HMGB1 mediated hyper-permeability via suppression of the activation of p38. The translocation of NF-kB from cytosol to nucleus is also inhibited by MTU (Kwak et al., 2015).
Simvastatin
Simvastatin belongs to the statin class of drugs and is widely used to control hypercholesterolemia and hypertriglyceridemia. It reduces cholesterol synthesis by inhibiting 3-hydroxy-3-methylglutaryl-CoA reductase and is widely used in hyperlipidemia to reduce the risk of atherosclerotic complications (Robinson, 2007). Simvastatin has been repositioned as an anti-sepsis drug and has demonstrated improved survival rates in septic patients with multiple organ dysfunction syndrome (Schmidt et al., 2006).
Simvastatin has shown to prevent the loss of integrity of the blood brain barrier (BBB), induced by polymicrobial sepsis (Yang et al., 2015). In addition, it also possesses anti-inflammatory and anti-oxidative properties and it is possible that its preservative properties may act by mitigating the detrimental effects of systemic inflammation on the BBB integrity (Yang et al., 2015).
Mangiferin
Mangiferin, a xanthone glucoside, is an active phytochemical present in various plants, including Mangifera indica L (Matkowski et al., 2013). Mangiferin has been reported to possess antioxidant, antitumor, antiviral, and immunomodulatory activities (Guha et al., 1996). In addition, the aqueous extract of M. indica leaves was shown to possess hypoglycemic activity (Aderibigbe et al., 1999). Mangiferin has been repurposed to treat sepsis-induced acute kidney injury (AKI). Sepsis-induced AKI is a multifactorial disease that involves inflammatory reactions by systemic cytokine storm or local cytokine production (Zarjou and Agarwal, 2011) and tubular dysfunction, induced by oxidative stress. The activation of NOD-like receptor family pyrin domain containing 3 (NLRP3) inflammasome contributes in the pathogenesis of lipopolysaccharide-induced rat kidney proximal tubular cell (RPTC) apoptosis and CLP-induced sepsis (He et al., 2014). Mangiferin ameliorates CLP-induced oxidative and inflammation-mediated renal damage via enhancement of Nrf2 expression and prevention of NLRP3 activation. Sepsis-induced AKI is attenuated via NLRP3 inflammasome inhibition, reduction in the secretion of IL-1β and IL-18 secretion, and upregulation of Nrf2 expression by mangiferin (He et al, 2014).
REPOSITIONING DRUGS OF ASTHMA
Asthma has a complex and multifactorial pathogenesis, affecting over 300 million people worldwide (Papi et al., 2018). Inflammation in asthma is primarily mediated by Th2 lymphocytes and is characterized by pulmonary eosinophilia, production of Th2 cytokines and allergen-specific immunoglobulin E (IgE), mucus hypersecretion, expression of inflammatory factors such as inducible nitric oxide synthase (iNOS), and airway hyperresponsiveness (Barnes, 2015, 2017). Airway inflammation, airway hyperreactivity (AHR), goblet cell metaplasia, and increased levels of Th2 cytokines and IgE play key roles in allergic asthma (Bateman et al., 2008; Holgate, 2008; Hamid and Tulic, 2009). The inflammatory cell influx in asthma is triggered by the endothelial cell adhesion molecules, such as vascular cell adhesion molecule 1 (VCAM-1) and intracellular adhesion molecule (ICAM-1) (Woodside and Vanderslice, 2008; Hasegawa et al., 2010). Table 2 shows repositioned drugs targeting various inflammatory mediators.
Table 2.
Repositioned drugs for targeting inflammatory mediators
| Drug | Inflammatory mediators for target | Disease |
|---|---|---|
| Artemisia apiacea Hance | IκBα, NF-κB p65, p38, RANTES, IL-8, IL-6, and TARC | Atopic dermatitis |
| Heparin | iNOS, Il-4, ARG1, and ARG2 | Asthma |
| Mangiferin | Nrf2 expression, IL-1β & IL-18, and NLRP3 | Sepsis |
| Methylthiouracil | TLR2, TLR4, RAGE, and p38, NF-κB | Sepsis |
| Rapamycin | mTOR, IL-13, and IgE | Asthma |
| Rifampicin | Histamine, β-HEX, PGD2, proinflammatory cytokines, TNF-α, and COX-2 | Atopic dermatitis |
| Simvastatin | IL-1, IL-6, IL-8, IL-12, CD4 T–cell, Th2, ICAM-1, and VCAM-1 | Sepsis and Asthma |
TLR2, toll-like receptor 2; TLR4, toll-like receptor 4; RAGE, receptor for advanced glycation end products; p38, protein kinase 38; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; Nrf2, nuclear factor erythroid 2 (NFE2)-related factor 2; IL, interleukin; ICAM-1, intercellular adhesion molecule 1; VCAM-1, vascular cell adhesion protein 1; mTOR, mammalian target of rapamycin; IgE, immunoglobulin E; iNOS, inducible nitric oxide synthase; ARG1, arginase 1; ARG2, arginase 2; β-HEX, β-N-acetylhexosaminidase. Hexosaminidase A; PGD2, prostaglandin D2; TNF-α, tumor necrosis factor alpha.
Rapamycin
Rapamycin, also known as sirolimus, is used to coat coronary stents, prevent organ transplant rejection, and treat a rare lung disease called lymphangioleiomyomatosis (Vezina et al., 1975). This compound has immunosuppressant functions and antiproliferative properties due to mTOR inhibition (Waldner et al., 2016).
Glucocorticoids and bronchodilators are the first-line therapy for asthma. However, these therapies are not effective in all the patients. Rapamycin has been repositioned as a new treatment option for asthma. Rapamycin attenuates allergic asthma through inhibition of mTOR and suppression of various key mediators. Allergen-induced IL-13 and leukotriene levels have been reported to be suppressed by rapamycin. Additionally, AHR, IL-13, and IgE were completely decreased by rapamycin (Mushaben et al., 2011).
Heparin
Heparin is commonly known as unfractionated heparin (UFH) and is used as an anticoagulant. It is mainly used to treat and prevent deep vein thrombosis and arterial thromboembolism. In addition, it is also used in the treatment of heart attacks and unstable angina. Various low molecular weight derivatives of heparin (LMWH) have shown to possess anti-inflammatory properties and were found to be therapeutically active against various inflammatory diseases, including asthma (Mousavi et al., 2015; Oduah et al., 2016; Alaez-verson et al., 2017).
LMWH and sulphate non-coagulant heparin (S-NACH) are highly effective in blocking different asthma traits in mice (Ghonim et al., 2018). Th2 inflammation is blocked via impeding IL-4 mediated signal transduction by S-NACH. S-NACH treatment blocked the expression of IL-4 induced expression of iNOS and reduced the basal levels of ARG1 and ARG2 (Ghonim et al., 2018). S-NACH has also demonstrated protective effects against lung dysfunction by reducing inflammation.
Simvastatin
Statins also have shown anti-inflammatory effects and are proposed as novel drugs for treating inflammatory diseases, including asthma (Samson et al., 2006; Kim et al., 2007; Menzies et al., 2007; Ostroukhova et al., 2009; Maneechotesuwan et al., 2010; Huang et al., 2011; Lokhandwala et al., 2012). Statins suppress inflammation via reduction in the levels of secretory cytokines and chemokines, such as IL-1, IL-6, IL-8, IL-12, and tumor necrosis factor-α (TNF-α) in asthma. Additionally, airway inflammation, remodeling, and AHR are attenuated by simvastatin in a dose-dependent manner through inhibition of the RhoA pathway. Simvastatin significantly reduces the CD4+ T cells numbers and the CD4/CD8 ratio along with attenuation of AHR, airway inflammation, and Th2 cytokine levels. After simvastatin treatment, the expression of ICAM-1 and VCAM-1 decreased significantly (Liu et al., 2014). Furthermore, treatment with simvastatin resulted in a decrease in the expression of cell adhesion molecules, inhibition of the migration of inflammatory cells from the blood into the airways and sites of inflammation, and reduction in augmentation of IL-4 and IL-13 and eosinophils. Simvastatin attenuated airway responsiveness and allergic inflammation, via reduction of T-cell influx, thereby resulting in a decrease in the expression of cell adhesion molecules (Liu et al., 2014).
Repositioning drugs of atopic dermatitis
Atopic dermatitis (AD) is an inflammatory skin disorder associated with epidermal hyper-reactivity to allergens (Leung et al., 2004). AD is associated with elevated serum levels of IgE due to increased inflammatory infiltration (Bergmann et al., 1998; Chang and Shiung, 2006). Mast cells release histamine, which is a major mediator of hypersensitivity and a potent vasoactive agent observed near allergic lesions (Petersen et al., 1996). Mast cells induce allergic inflammation through the release of pro-inflammatory cytokines, such as TNF-α, interleukin-4 (IL-4), IL-8, and IL-13 (Kalesnikoff and Galli, 2008).
Rifampicin
Rifampicin, also known as rifampin, is an antibiotic used to treat several types of bacterial infections such as tuberculosis, leprosy, and Legionnaires disease (Eule et al., 1974). It is a polyketide belonging to the chemical class of compounds known as ansamycins. Bacterial RNA synthesis is inhibited by rifampicin via inhibition of bacterial DNA-dependent RNA polymerase (Wehrli et al., 1968). Rifampicin suppresses inflammatory effects and plays a major role in relieving neuropathic pain and in immune modulation. In addition, it also exhibits therapeutic effects against psoriasis in clinical practice (Bellahsene and Forsgren, 1980; Tsankov and Angelova, 2003). Rifampicin showed anti-AD activity and effectively decreased the elevated serum levels of IgE and IL-4, which are characteristic features of AD (Kim et al., 2017). Thus, rifampicin can be considered as an effective drug for the treatment of AD based on the reduction in the severity of symptoms and IgE production. Mast cells and basophils store β-hexosaminidase (β-HEX) in secretory granules. When mast cells are activated, β-HEX is released along with histamine, which in turn acts as a biomarker for allergic reactions. Rifampicin decreases both β-HEX and histamine secretion. Rifampicin is used as a new anti-allergic agent owing to its suppressive effect of degranulation of mast cell by controlling intracellular calcium concentrations (Kim et al., 2017).
Artemisia apiacea Hance
Artemisia apiacea Hance is a herb commonly found on the river beaches of eastern Asia. It is used as a traditional medicine in various east Asian countries including China, Korea, and Japan to treat fever, eczema, and jaundice. It is a common oriental medicine used for treating malaria, jaundice, and dyspepsia (Ryu et al., 2013). Artemisinin was isolated and identified in the early 1970s as an active antimalarial ingredient. Thereafter, various studies showed that it possesses anti-inflammatory, antipyretic, antibacterial, antiparasitic, and immunosuppressive effects. Recently, A. apiacea was repositioned as anti-inflammatory and anti-atopic dermatitis drug (Ryu et al., 2013). Ethanolic extracts of A. apiacea Hance (EAH) inhibited the production of chemokines and pro-inflammatory cytokines, such as RANTES, IL-8, IL-6, and TARC, and inhibited the activation of p38, ERK without JNK inhibition. The expression of proinflammatory cytokines and chemokines was also found to be regulated by EAH via the p38/NF-κB pathway in allergic inflammation. EAH inhibited the degradation of IκBα and the translocation of NF-κB p65 (Yang et al., 2018). Furthermore, skin lesions were improved and thickness of dorsal skin, ear skin thickness was reduced by EAH in 2,4-dinitrochlorobenzene-induced AD-like model. These results suggested that EAH possessed therapeutic properties to reduce AD-like symptoms in mice (Yang et al., 2018).
CLOSING REMARK
Drug repositioning has become a significant drug development strategy since its advent in the 1990s. Several repositioned drugs are already being marketed as drug candidates for disease that have failed drug indications. Drug repositioning offers promising success rate in drug development and safer treatments without the full cost or time required for de novo drug development. It allows alternative therapeutic use of approved drugs for the treatment of many diseases.
Inflammation is a protective response involving immune cells, blood vessels, and molecular mediators. The main aims of inflammation are to eliminate the initial cause of cell injury, clear out necrotic cells and tissue damaged from original insult and the inflammatory process, and initiate tissue repair. Additionally, inflammation is driven by various inflammatory mediators, such as TLR-2, TLR-4, iNOS, and interleukins.
In this review, we mainly focused on repositioned drugs used for the treatment of inflammatory diseases such as sepsis, asthma, and atopic dermatitis. First, we summarized the advances in drug repositioning methods till date. Then, we discussed three repositioned drugs used to treat sepsis, namely, methylthiouracil, mangiferin, and simvastatin. These drugs mitigate sepsis via reduction in inflammatory cytokines, inflammatory receptors, and inflammasomes. We also discussed three repositioned asthma drugs, namely, rapamycin, heparin, and simvastatin. These drugs inhibit the inflammatory cytokines and IgE to ameliorate asthma. Rifampicin, a repurposed drug used for the treatment of atopic dermatitis, decreases the levels of histamine, intracellular calcium, and pro-inflammatory cytokines and reduces COX-2 expression, thereby alleviating atopic dermatitis.
ACKNOWLEDGMENTS
This paper was supported by the Fund of the Sahmyook University in 2017.
REFERENCES
- Aderibigbe A. O., Emudianughe T. S., Lowal B. A. Antihyperglycaemic effect of Mangifera indica in rat. Phytother. Res. 1999;13:504–507. doi: 10.1002/(SICI)1099-1573(199909)13:6<504::AID-PTR533>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Alaez-Verson C. R., Lantero E., Fernandez-Busquets X. Heparin: new life for an old drug. Nanomedicine (Lond.) 2017;12:1727–1744. doi: 10.2217/nnm-2017-0127. [DOI] [PubMed] [Google Scholar]
- Ashburn T. T., Thor K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- Barnes P. J. Cellular and molecular mechanisms of asthma and COPD. Clin. Sci. (Lond.) 2017;131:1541–1558. doi: 10.1042/CS20160487. [DOI] [PubMed] [Google Scholar]
- Barnes P. J. Therapeutic approaches to asthma-chronic obstructive pulmonary disease overlap syndromes. J. Allergy Clin. Immunol. 2015;136:531–545. doi: 10.1016/j.jaci.2015.05.052. [DOI] [PubMed] [Google Scholar]
- Bateman E. D., Hurd S. S., Barnes P. J., Bousquet J., Drazen J. M., FitzGerald J. M., Gibson P., Ohta K., O'Byrne P., Pedersen S. E., Pizzichini E., Sullivan S. D., Wenzel S. E., Zar H. J. Global strategy for asthma management and prevention: GINA executive summary. Eur. Respir. J. 2008;31:143–178. doi: 10.1183/09031936.00138707. [DOI] [PubMed] [Google Scholar]
- Bellahsene A., Forsgren A. Effect of rifampin on the immune response in mice. Infect. Immun. 1980;27:15–20. doi: 10.1128/IAI.27.1.15-20.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann R. L., Edenharter G., Bergmann K. E., Forster J., Bauer C. P., Wahn V., Zepp F., Wahn U. Atopic dermatitis in early infancy predicts allergic airway disease at 5 years. Clin. Exp.Allergy. 1998;28:965–970. doi: 10.1046/j.1365-2222.1998.00371.x. [DOI] [PubMed] [Google Scholar]
- Bernard G. R., Vincent J. L, Laterre P. F., LaRosa S. P., Dhainau J. F., Lopez-Rodriguez A., Steingrub J. S., Garber G. E., Helterbrand J. D., Ely E. W., Fisher C. J., Jr. Recombinant human protein C Worldwide Evaluation in Severe Sepsis (PROWESS) study group. Efficacy and safety of recombinant human activated protein C for severe sepsis. N. Engl. J. Med. 2001;344:699–709. doi: 10.1056/NEJM200103083441001. [DOI] [PubMed] [Google Scholar]
- Campillos M., Kuhn M., Gavin A. C., Jensen L. J., Bork P. Drug target identification using side-effect similarity. Science. 2008;321:263–266. doi: 10.1126/science.1158140. [DOI] [PubMed] [Google Scholar]
- Chang M. W., Ayeni C., Breuer S., Torbett B. E. Virtual screening for HIV protease inhibitors: a comparison of AutoDock 4 and Vina. PLoS ONE. 2010;5:e11955. doi: 10.1371/journal.pone.0011955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang , Shiung Y. Y. Anti-IgE as a mast cell-stabilizing therapeutic agent. J. Allergy Clin. Immunol. 2006;117:1203–1212. doi: 10.1016/j.jaci.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Chen Y. Z., Zhi D. G. Ligand-protein inverse docking and its potential use in the computer search of protein targets of a small molecule. Proteins. 2011;43:217–226. doi: 10.1002/1097-0134(20010501)43:2<217::AID-PROT1032>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Chong C. R., Sullivan D. J. New uses for old drugs. Nature. 2007;448:645–646. doi: 10.1038/448645a. [DOI] [PubMed] [Google Scholar]
- Cramer P. E., Cirrito J. R., Wesson D. W., Lee C. Y., Karlo J. C., Zinn A. E., Casali B. T., Restivo J. L., Goebel W. D., James M. J., Brunden K. R., Wilson D. A., Landreth G. E. ApoE-directed therapeutics rapidly clear beta-amyloid and reverse deficits in AD mouse models. Science. 2012;335:1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. T., Deshpande T., Butte A. J. Exploiting drug-disease relationships for computational drug repositioning. Brief. Bioinformatics. 2011a;12:303–311. doi: 10.1093/bib/bbr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J. T., Sirota M., Shenoy M., Pai R. K., Roedder S., Chiang A. P., Morgan A. A., Sarwal M. M., Pasricha P. J., Butte A. J. Computational repositioning of the anticonvulsant topiramate for inflammatory bowel disease. Sci. Transl. Med. 2011b;3:96ra76. doi: 10.1126/scitranslmed.3002648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekins S., Williams A. J., Krasowski M. D., Joel S., Freundlich J. S. In silico repositioning of approved drugs for rare and neglected diseases. Drug Discov. Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Eule H., Werner E., Winsel K., Iwainsky H. Intermittent chemotherapy of pulmonary tuberculosis using rifampicin and isoniazid for primary treatment: the influence of various factors on the frequency of side-effects. Tubercle. 1974;55:81–89. doi: 10.1016/0041-3879(74)90069-5. [DOI] [PubMed] [Google Scholar]
- Ghonim M. A., Wang J., Ibba S. V., Luu H. H., Pyakurel K., Benslimane I., Mousa S., Boulares A. H. Sulfated non-anticoagulant heparin blocks Th2-induced asthma by modulating the IL-4/signal transducer and activator of transcription 6/Janus kinase 1 pathway. J. Transl. Med. 2018;16:243. doi: 10.1186/s12967-018-1621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glock G. E. Methyl-thiouracil and thiouracil as antithyroid drugs. Br. J. Pharmacol. Chemother. 1946;1:127–134. doi: 10.1111/j.1476-5381.1946.tb00031.x. [DOI] [PubMed] [Google Scholar]
- Guha S., Ghosal S., Chattopadhyay U. Antitumor, immunomodulatory and anti-HIV effect of mangiferin, a naturally occurring glucosylxanthone. Chemotherapy. 1996;42:443–451. doi: 10.1159/000239478. [DOI] [PubMed] [Google Scholar]
- Hall C. J., Wicker S. M., Chien A. T., Tromp A., Lawrence L. M., Sun X., Krissansen G. W., Crosier K. E., Crosier P. S. Repositioning drugs for inflammatory disease - fishing for new anti-inflammatory agents. Dis. Model. Mech. 2014;7:1069–1081. doi: 10.1242/dmm.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid Q., Tulic M. Immunobiology of asthma. Annu. Rev. Physiol. 2009;71:489–507. doi: 10.1146/annurev.physiol.010908.163200. [DOI] [PubMed] [Google Scholar]
- Hasegawa A., Hayashi K., Kishimoto H., Yang M., Tofukuji S., Suzuki K., Nakajima H., Hoffman R. M., Shirai M., Nakayama T. Color-coded real-time cellular imaging of lung T-lymphocyte accumulation and focus formation in a mouse asthma model. J. Allergy Clin. Immunol. 2010;125:461–468. doi: 10.1016/j.jaci.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Haupt V. J., Schroeder M. Old friends in new guise: repositioning of known drugs with structural bioinformatics. Brief. Bioinformatics. 2011;12:312–326. doi: 10.1093/bib/bbr011. [DOI] [PubMed] [Google Scholar]
- He L., Peng X., Zhu J., Chen X., Liu H., Tang C., Dong Z., Liu F., Peng Y. Mangiferin attenuate sepsis-induced acute kidney injury via antioxidant and anti-inflammatory effects. Am. J. Nephrol. 2014;40:441–450. doi: 10.1159/000369220. [DOI] [PubMed] [Google Scholar]
- Holgate S. T. Pathogenesis of asthma. Clin. Exp. Allergy. 2008;38:872–897. doi: 10.1111/j.1365-2222.2008.02971.x. [DOI] [PubMed] [Google Scholar]
- Holmes A. B., Hawson A., Liu F., Friedman C., Khiabanian H., Rabadan R. Discovering disease associations by integrating electronic clinical data and medical literature. PLoS ONE. 2011;6:e21132. doi: 10.1371/journal.pone.0021132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C. C., Chan W. L., Chen Y. C., Chen T. J., Chou K. T., Lin S. J., Chen J. W., Leu H. B. Statin use in patients with asthma: a nationwide population-based study. Eur. J. Clin. Invest. 2011;41:507–512. doi: 10.1111/j.1365-2362.2010.02434.x. [DOI] [PubMed] [Google Scholar]
- Iorio F., Bosotti R., Scacheri E., Belcastro V., Mithbaokar P., Ferriero R., Murino L., Tagliaferri R., Brunetti-Pierri N., Isacchi A., di Bernardo D. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. U.S.A. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang D., Lee S., Lee J., Kim K., Lee D. Inferring new drug indications using the complementarity between clinical disease signatures and drug effects. J. Biomed. Inform. 2016;59:248–257. doi: 10.1016/j.jbi.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Jensen P. B., Jensen L. J., Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 2012;13:395–405. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- Jung K., LePendu P., Chen W. S., Iyer S. V., Readhead B., Dudley J. T., Shah N. H. Automated detection of off-label drug use. PLoS ONE. 2014;9:e89324. doi: 10.1371/journal.pone.0089324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K., LePendu P., Shah Automated detection of systematic off-label drug use in free text of electronic medical records. AMIA Jt. Summits Transl. Sci. Proc. 2013;2013:94–98. [PMC free article] [PubMed] [Google Scholar]
- Kalesnikoff J., Galli S. J. New developments in mast cell biology. Nat. Immunol. 2008;9:1215–1223. doi: 10.1038/ni.f.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser M. J., Setola V., Irwin J. J., Laggner C., Abbas A. I., Hufeisen S. J., Jensen N. H., Kuijer M. B., Matos R. C., Tran T. B., Whaley R., Glennon R. A., Hert J., Thomas K. L., Edwards D. D., Shoichet B. K., Roth B. L. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y., Ryu S. Y., Lim J. E., Lee Y. S., Ro J. Y. Anti-inflammatory mechanism of simvastatin in mouse allergic asthma model. Eur. J. Pharmacol. 2007;557:76–86. doi: 10.1016/j.ejphar.2006.11.027. [DOI] [PubMed] [Google Scholar]
- Kim S. H., Lee K. M., Lee G. S., Seong J. W., Kang T. J. Rifampicin alleviates atopic dermatitis-like response in vivo and in vitro. Biomol. Ther. (Seoul) 2017;25:634–640. doi: 10.4062/biomolther.2017.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnings S. L., Liu N., Buchmeier N., Tonge P. J., Xie L., Bourne P. E. Drug discovery using chemical systems biology: repositioning the safe medicine Comtan to treat multi-drug and extensively drug resistant tuberculosis. PLoS Comput. Biol. 2009;5:e1000423. doi: 10.1371/journal.pcbi.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotelnikova E., Yuryev A., Mazo I., Daraselia N. Computational approaches for drug repositioning and combination therapy design. J. Bioinform. Comput. Biol. 2010;8:593–606. doi: 10.1142/S0219720010004732. [DOI] [PubMed] [Google Scholar]
- Kwak S., Ku S. K., Kang H., Baek M. C., Bae J. S. Methylthiouracil, a new treatment option for sepsis. Vascul. Pharmacol. 2015;88:1–10. doi: 10.1016/j.vph.2015.07.013. [DOI] [PubMed] [Google Scholar]
- Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Leung D. Y., Boguniewicz M., Howell M. D., Nomura I., Hamid Q. A. New insights into atopic dermatitis. J. Clin. Invest. 2004;113:651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu Z. Pathway-based drug repositioning using causal inference. BMC Bioinformatics. 2013;14(Suppl 16):S3. doi: 10.1186/1471-2105-14-S16-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Y., An J., Jones S. J. A large-scale computational approach to drug repositioning. Genome Inform. 2006;17:239–247. [PubMed] [Google Scholar]
- Liu J., Suh D., Yang E., Lee S., Park H., Shin Y. Attenuation of airway inflammation by simvastatin and the implications for asthma treatment: is the jury still out? Exp. Mol. Med. 2014;46:e113. doi: 10.1038/emm.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhu F., Ma X. H., Shi Z., Yang S. Y., Wei Y. Q., Chen Y. Z. Predicting targeted polypharmacology for drug repositioning and multi- target drug discovery. Curr. Med. Chem. 2013;20:1646–1661. doi: 10.2174/0929867311320130005. [DOI] [PubMed] [Google Scholar]
- Lokhandwala T., West-Strum D., Banahan B. F., Bentley J. P., Yang Y. Do statins improve outcomes in patients with asthma on inhaled corticosteroid therapy? A retrospective cohort analysis. BMJ Open. 2012;2:e001279. doi: 10.1136/bmjopen-2012-001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounkine E., Keiser M. J., Whitebread S., Mikhailov D., Hamon J., Jenkins J. L., Lavan P., Weber E., Doak A. K., Cote S., Shoichet B. K., Urban L. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–367. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maneechotesuwan K., Ekjiratrakul W., Kasetsinsombat K., Wongkajornsilp A., Barnes P. J. Statins enhance the anti-inflammatory effects of inhaled corticosteroids in asthmatic patients through increased induction of indoleamine 2 , 3-dioxygenase. J. Allergy Clin. Immunol. 2010;126:754–762.e1. doi: 10.1016/j.jaci.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Matkowski A., Kuś P., Góralska E., Woźniak D. Mangiferin - a bioactive xanthonoid, not only from mango and not just antioxidant. Mini Rev. Med. Chem. 2013;13:439–455. doi: 10.2174/138955713804999838. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- Mei J. P., Kwoh C. K., Yang P., Li X. L., Zheng J. Drug-target interaction prediction by learning from local information and neighbors. Bioinformatics. 2013;29:238–245. doi: 10.1093/bioinformatics/bts670. [DOI] [PubMed] [Google Scholar]
- Menzies D., Nair A., Meldrum K. T., Fleming D., Barnes M., Lipworth B. J. Simvastatin does not exhibit therapeutic anti-inflammatory effects in asthma. J. Allergy Clin. Immunol. 2007;119:328–335. doi: 10.1016/j.jaci.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Morrow J. K., Tian L., Zhang S. Molecular networks in drug discovery. Crit. Rev. Biomed. Eng. 2010;38:143–156. doi: 10.1615/CritRevBiomedEng.v38.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousavi S., Moradi M., Khorshidahmad T., Motamedi M. Anti-inflammatory effects of heparin and its derivatives: a systematic review. Adv. Pharmacol. Sci. 2015;2015:507151. doi: 10.1155/2015/507151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushaben E. M., Kramer E. L., Brandt E. B., Khurana Hershey G. K., Le Cras T. D. Rapamycin attenuates airway hyperreactivity, goblet cells, and IgE in experimental allergic asthma. J. Immunol. 2011;187:5756–5763. doi: 10.4049/jimmunol.1102133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor K. A., Roth B. L. Finding new tricks for old drugs: an efficient route for public-sector drug discovery. Nat. Rev. Drug Discov. 2005;4:1005–1014. doi: 10.1038/nrd1900. [DOI] [PubMed] [Google Scholar]
- Oduah E. I., Linhardt R. J., Sharfstein S. T. Heparin: past, present, and future. Pharmaceuticals (Basel) 2016;9:e38. doi: 10.3390/ph9030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprea T. I., Nielsen S. K., Ursu O., Yang J. J., Taboureau O., Mathias S. L., Kouskoumvekaki L., Sklar L. A., Bologa C. G. Associating drugs, targets and clinical outcomes into an integrated network affords a new platform for computer-aided drug repurposing. Mol. Inform. 2011;30:100–111. doi: 10.1002/minf.201100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostroukhova M., Kouides R. W., Friedman E. The effect of statin therapy on allergic patients with asthma. Ann. Allergy Asthma Immunol. 2009;103:463–468. doi: 10.1016/S1081-1206(10)60261-X. [DOI] [PubMed] [Google Scholar]
- Papi A., Brightling C., Pedersen S. E., Reddel H. K. Asthma. Lancet. 2018;391:783–800. doi: 10.1016/S0140-6736(17)33311-1. [DOI] [PubMed] [Google Scholar]
- Petersen L. J., Mosbech H., Skov P. S. Allergen-induced histamine release in intact human skin in vivo assessed by skin microdialysis technique: characterization of factors influencing histamine releasability. J. Allergy Clin. Immunol. 1996;97:672–679. doi: 10.1016/S0091-6749(96)70313-5. [DOI] [PubMed] [Google Scholar]
- Phatak S. S., Zhang S. A novel multi-modal drug repurposing approach for identification of potent ACK1 inhibitors. Pac. Symp. Biocomput. 2013;2013:29–40. doi: 10.1142/9789814447973_0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps K. Repositioning drugs to enhance a product's lifecycle. Drug Discov. Today Ther. Strateg. 2011;8:97–101. doi: 10.1016/j.ddstr.2011.09.006. [DOI] [Google Scholar]
- Reddy A. R., Kaul A. Effect of methyl thiouracil on radioiodine thyroidal retention in rats. Radiat. Environ. Biophys. 1979;16:347–354. doi: 10.1007/BF01340572. [DOI] [PubMed] [Google Scholar]
- Rihel J., Prober D. A., Arvanites A., Lam K., Zimmerman S., Jang S., Haggarty S. J., Kokel D., Rubin L. L., Peterson R. T., Schier A. F. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. G. Simvastatin: present and future perspectives. Expert Opin. Pharmacother. 2007;8:2159–2127. doi: 10.1517/14656566.8.13.2159. [DOI] [PubMed] [Google Scholar]
- Roque F. S., Jensen P. B., Schmock H., Dalgaard M., Andreatta M., Hansen T., Søeby K., Bredkjær S., Juul A., Werge T., Jensen L. J., Brunak S. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLoS Comput. Biol. 2011;7:e1002141. doi: 10.1371/journal.pcbi.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. A. Management of sepsis. N. Engl. J. Med. 2006;355:1699–1713. doi: 10.1056/NEJMra043632. [DOI] [PubMed] [Google Scholar]
- Ryu J. C., Park S. M., Hwangbo M., Byun S. H., Ku S. K., Kim Y. W., Kim S. C., Jee S. Y., Cho I. J. Methanol extract of Artemisia apiacea Hance attenuates the expression of inflammatory mediators via NF-kappaB inactivation. Evid. Based Complement. Altern. Med. 2013;2013:494681. doi: 10.1155/2013/494681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson K. T., Minoguchi K., Tanaka A., Oda N., Yokoe T., Yamamoto Y., Yamamoto M., Ohta S., Adachi M. Inhibitory effects of fluvastatin on cytokine and chemokine production by peripheral blood mononuclear cells in patients with allergic asthma. Clin. Exp. Allergy. 2006;36:475–482. doi: 10.1111/j.1365-2222.2006.02470.x. [DOI] [PubMed] [Google Scholar]
- Sanseau P., Koehler J. Editorial: computational methods for drug repurposing. Brief. Bioinformatics. 2011;12:301–302. doi: 10.1093/bib/bbr047. [DOI] [PubMed] [Google Scholar]
- Sardana D., Zhu C., Zhang M., Gudivada R. C., Yang L., Jegga A. G. Drug repositioning for orphan diseases. Brief. Bioinformatics. 2011;12:346–356. doi: 10.1093/bib/bbr021. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Hennen R., Keller A., Russ M., Müller-Werdan U., Werdan K., Buerke M. Association of statin therapy and increased survival in patients with multiple organ dysfunction syndrome. Intensive Care Med. 2006;32:1248–1251. doi: 10.1007/s00134-006-0246-y. [DOI] [PubMed] [Google Scholar]
- Sekhon B. S. Repositioning drugs and biologics: retargeting old/existing drugs for potential new therapeutic applications. J. Pharm. Educ. Res. 2013;4:1–15. [Google Scholar]
- Sirota M., Dudley J. T., Kim J., Chiang A. P., Morgan A. A., Sweet-Cordero A., Sage J., Butte A. J. Discovery and preclinical validation of drug indications using compendia of public gene expression data. Sci. Transl. Med. 2011;3:96ra77. doi: 10.1126/scitranslmed.3001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivachenko A., Kalinin A., Yuryev A. Pathway analysis for design of promiscuous drugs and selective drug mixtures. Curr. Drug Discov. Technol. 2006;3:269–277. doi: 10.2174/157016306780368117. [DOI] [PubMed] [Google Scholar]
- Sleigh S. H., Barton C. L. Repurposing strategies for therapeutics. Pharm. Med. 2010;24:151–159. doi: 10.1007/BF03256811. [DOI] [Google Scholar]
- Strittmatter W. J. Medicine. Old drug, new hope for Alzheimer's disease. Science. 2012;35:1447–1448. doi: 10.1126/science.1220725. [DOI] [PubMed] [Google Scholar]
- Tobinick E. L. The value of drug repositioning in the current pharmaceutical market. Drug News Perspect. 2009;22:119–125. doi: 10.1358/dnp.2009.22.2.1303818. [DOI] [PubMed] [Google Scholar]
- Tsankov N., Angelova I. Rifampin in dermatology. Clin. Dermatol. 2003;21:50–55. doi: 10.1016/S0738-081X(02)00328-0. [DOI] [PubMed] [Google Scholar]
- Vézina C., Kudelski A., Sehgal S. N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- Waldner M., Fantus D., Solari M., Thomson A. W. New perspectives on mTOR inhibitors (rapamycin, rapalogs and TORKinibs) in transplantation. Br. J. Clin. Pharmacol. 2016;82:1158–1170. doi: 10.1111/bcp.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrli W., Knusel F., Schmid K., Staehelin M. Interaction of rifamycin with bacterial RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1968;61:667–673. doi: 10.1073/pnas.61.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S., Knox C., Guo A. C., Cheng D., Shrivastava S., Tzur D., Gautam B., Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodside D. G., Vanderslice P. Cell adhesion antagonists: therapeutic potential in asthma and chronic obstructive pulmonary disease. BioDrugs. 2008;22:85–100. doi: 10.2165/00063030-200822020-00002. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y., Araki M., Gutteridge A., Honda W., Kanehisa M. Prediction of drug-target interaction networks from the integration of chemical and genomic spaces. Bioinformatics. 2008;24:i232–i240. doi: 10.1093/bioinformatics/btn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y., Kotera M., Kanehisa M., Goto S. Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework. Bioinformatics. 2010;26:i246–i254. doi: 10.1093/bioinformatics/btq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. H., Kao M. C., Shih P. C., Li K. Y., Tsai P. S., Huang C. J. Simvastatin attenuates sepsis-induced blood-brain barrier integrity loss. J. Surg. Res. 2015;194:591–598. doi: 10.1016/j.jss.2014.11.030. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Lee E., Lee B., Cho W. K., Ma J. Y., Park K. I. Ethanolic extracts of Artemisia apiacea Hance improved atopic dermatitis-like skin lesions in vivo and suppressed TNF-alpha/IFN-gamma-induced proinflammatory chemokine production in vitro. Nutrients. 2018;10:806. doi: 10.3390/nu10070806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M. A., Goh K. I., Cusick M. E., Barabási A. L., Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Zahler S., Tietze S., Totzke F., Kubbutat M., Meijer L., Vollmar A. M., Apostolakis J. Inverse in silico screening for identification of kinase inhibitor targets. Chem. Biol. 2007;14:1207–1214. doi: 10.1016/j.chembiol.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Zarjou A., Agarwal A. Sepsis and acute kidney injury. J. Am. Soc. Nephrol. 2011;22:999–1006. doi: 10.1681/ASN.2010050484. [DOI] [PubMed] [Google Scholar]