Abstract
Recent advances in high-speed atomic force microscopy (HS-AFM) have made it possible to study the conformational dynamics of single unlabeled transmembrane channels and transporters. Improving environmental control with the integration of a non-disturbing buffer exchange system, which in turn allows the gradual change of conditions during HS-AFM operation, has provided a breakthrough towards the performance of structural titration experiments. Further advancements in temporal resolution with the use of line scanning and height spectroscopy techniques show how high-speed atomic force microscopy can measure millisecond to microsecond dynamics, pushing this method beyond current spatial and temporal limits offered by less direct techniques.
Introduction
Since the developments that have allowed a revolution in cryo-EM [1,2], solving membrane protein structures is becoming more and more common [3]. Whilst static structures provide vast amounts of information they are often only half of a complex story concerning how a protein behaves in reality. How these structures change and react dynamically to and with their environment is now key to developing a full biological understanding. Therefore, focus must turn to functional and mechanistic studies, specifically addressing conformational dynamics and state transitions. In this effort, techniques like double electron-electron resonance (DEER), electron paramagnetic resonance (EPR) and Förster resonance energy transfer (FRET) have major impact [4–6]; however advancements in HS-AFM are now allowing comparable and even faster dynamics to not only be detected but to simultaneously provide real-space submolecular structural information [7,8•,9,10•]. Video rate HS-AFM imaging provides 3D information under physiological conditions at ~50–100ms per image, whilst reducing the dimensionality to single line (2D) and single point (1D) measurements can drastically increase the temporal resolution to ~1ms and 10μs respectively [11••]. These features, unique to HS-AFM, provide a toolbox of methods that can surpass the time resolution of many techniques, whilst measuring membrane proteins in native-like conditions, i.e. in membranes, immersed in physiological buffer solution, at controllable temperature, without labeling, fixing or staining the proteins. Additionally, the integration of non-disturbing buffer exchange, pulsed UV-laser [10•] and accurate temperature control systems [12•] allow the gradual and rapid changes of the environmental conditions during HS-AFM operation.
The advent of HS-AFM to reach unprecedented scan rates has been driven by a combination of various technical developments, including improved cantilevers, sample-stage scanners, cantilever deflection detection and feedback systems [13]. As with conventional AFM, HS-AFM has been extended to high-speed force spectroscopy (HS-FS) [14,15], and active high frequency microrheology (HF-MR) [16] applications, reaching previously inaccessible dynamic ranges. Indeed HS-FS has reached pulling velocities directly comparable to molecular dynamics simulations [14,15]. Finally, HS-AFM imaging has successfully been applied to various biological systems such as molecular motors [8•], membrane associated proteins [17•], large macromolecular systems [18] and protein-DNA complexes [19]. However, in this short review, we focus on recent and ongoing advancements in HS-AFM imaging studies of transmembrane proteins, i.e. channels and transporters, that have provided a breakthrough towards analyzing membrane protein structural-dynamics under controlled exposure to environmental stimuli. We further discuss a most recent development, HS-AFM height spectroscopy (HS-AFM-HS), that will have immediate impact in this field as it allows analysis of structural dynamics at unprecedented rate. While in this review we discuss membrane proteins in large membrane patches, the analysis of membrane proteins in lipid nanodiscs is showing promise as a powerful platform to study dynamics from different orientations [20].
Channel Dynamics
When opened, ion channels create an electrical response by allowing the passage of ions across membranes down electrochemical gradients. The opening can be controlled by varying stimuli such as temperature, pH, small signaling molecules, voltage and mechanical stress. Given that this family of proteins is often involved in sensing and signal transduction processes, their sensitivity to and interactions with drugs have great physiological and clinical significance. The open/close activity of channels can be directly detected by ion conductance measurements, however the structural dynamics involved in gating have often to be inferred between structures locked in specific states, provided they are available. Performing ensemble averaging may mask the full dynamic range of the channel gating mechanism and potentially overlook intermediates, fluctuations and unpredictable behaviors.
The pentameric Cys-loop receptor ligand-gated ion channel (pLGIC) superfamily contains a number of channels fundamental to synaptic transmission [21,22]. X-ray studies of a proton-gated prokaryotic homolog of pLGIC, Gloeobacter violaceus ligand-gated ion channel (GLIC), have provided great insight into the open and closed structures of these channels [23,24]. However, EPR spectroscopy has suggested larger conformational changes to occur than visible in the crystallized states [25]. By combining HS-AFM imaging with a buffer exchange system to titrate a pH change, the reversible conformational changes and supramolecular rearrangements of GLIC channels have now been characterized [26••]. At pH3.4 images of the GLIC channels reconstituted into membranes show the pentameric extracellular domain protruding out of the membrane with a central cavity (Fig 1a). These structural features are consistent with the high-resolution structure in the active/desensitized state. Significant conformational changes could then be observed by slowly titrating pH7.4 buffer into the fluid chamber whilst HS-AFM imaging (Fig 1e), with the channels shifting conformation to a state with a tightened extracellular domain (Fig 1b). Additionally, the channels underwent a 2D packing reorganization from a seemingly disordered arrangement with specific protein-protein interactions between pentamers and five neighboring channels, to a hexagonal, less dense packing without specific interaction between channels (mismatch between oligomeric state and number of nearest neighbors) (Fig 1d). This rearrangement was interpreted to potentially be representative of inter-molecular gating cooperativity. Returning the buffer to pH3.4 showed that for the same set of ~70 molecules the process is fully reversible and that tip sample interactions for over 3,000 images did not damage the proteins (Fig 1c). Adapting single particle cryo-EM imaging processing methods to classify HS-AFM movies of GLIC molecules with the reference-free classification algorithm RELION allowed structural conformations to be unbiasedly searched for over time and pH. Additionally, classification provided evidence for the short-term existence of an asymmetric pentamer at early stages of activation during the HS-AFM structural titration experiment (Fig 1f). Two aspects in these experiments deserve special mention. First, the titration rate is very slow, typically 0.16 μL s−1 to exchange the fluid cell volume of 150μL, and thus on the time scale of molecular reactions the conditions can be considered equilibrated at any time point during the experiment. Second, while the scan rate is of the order of 1 s−1 and the entire titration experiment takes almost 1 hour, each molecule is imaged over ~15 scan lines corresponding to ~30 ms. Thus, the experiment reveals conformations adopted by single molecules during short instants over extended periods of time under well-defined conditions.
Figure 1. HS-AFM structural titration experiments of GLIC channels.
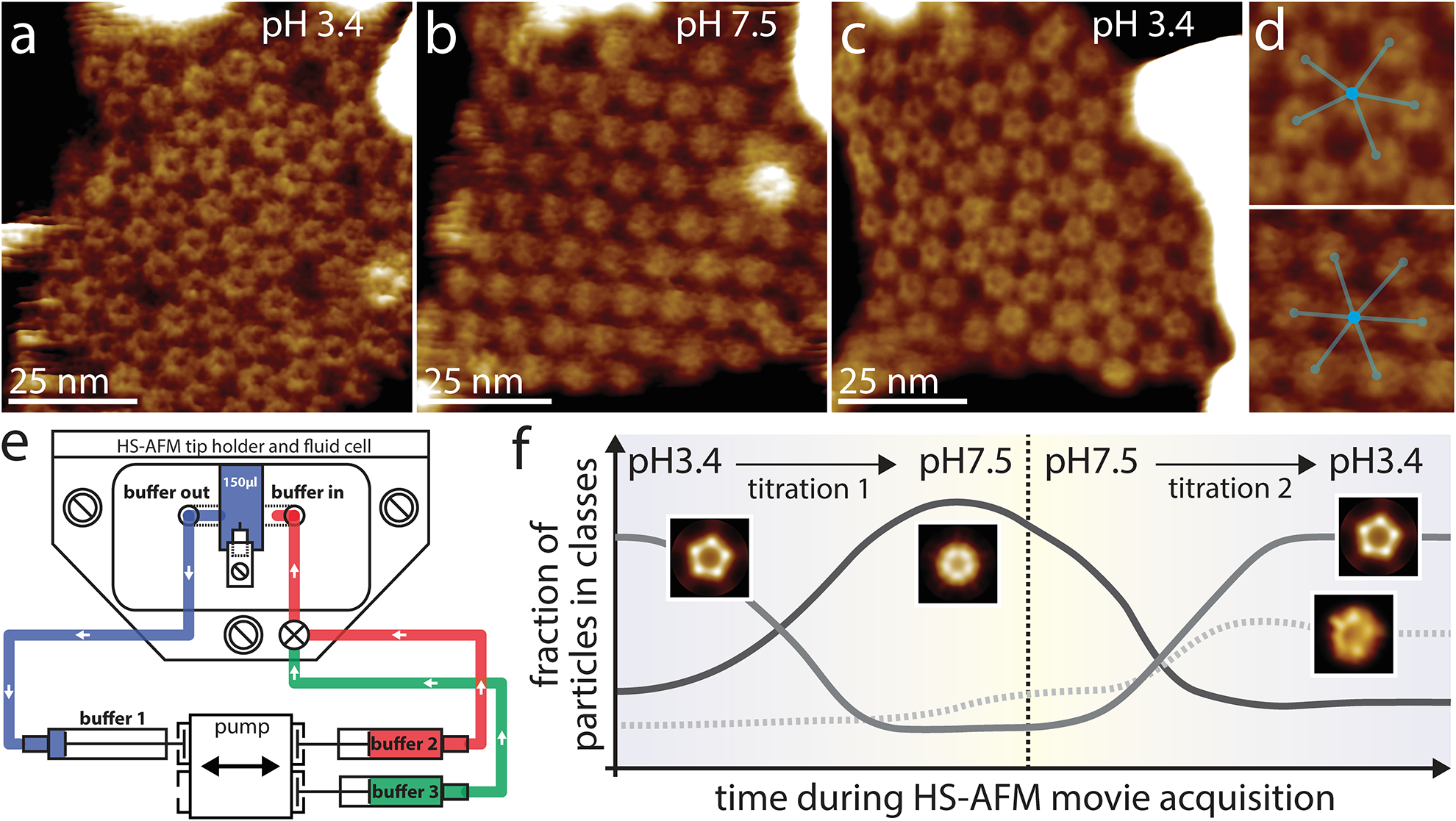
During HS-AFM operation, the pH in the fluid chamber was gradually exchanged through a buffer exchange microfluidic device from pH 3.4 (a) to pH 7.5 (b) and back to pH 3.4 (c). Analysis of the supramolecular assemblies show reversible rearrangements from seemingly disordered arrangements, where channels assemble in trimers of pentamers and most channels have 5 neighbors (yellow outlines), to hexagonal packing and back again (the white dashed outlines and the inset in (c) highlight higher-order structures in the different conditions). (d) Zoom-in of the molecular arrangements at pH3.4 (top) and pH7.5 (bottom) with ball/stick overlays to highlight how 5 neighbors are preferred at low pH over 6 at higher pH. (e) Schematic representation of the coupling of the buffer exchange system to the HS-AFM fluid cell. The ‘buffer in’ and ‘buffer out’ channels are placed in proximity to the HS-AFM cantilever (central element inside the fluid cell). The fluid cell contains initially 150ul of buffer 1 (blue) that is connected to the receiving syringe. Buffer 2 (red) is the first buffer to be injected into the fluid cell. Note the slight gap between buffer 2 and the fluid cell, representing a tiny bubble in the tubing to avoid involuntary mixing of buffer 2 before activation of the pumping system. By the use of a switch in the tubing system, buffer 3 (green) can later be injected into the fluid cell. Buffer 3 can be identical to buffer 1, allowing for a reversibility experiment. (f) Abundance of GLIC channel conformations as a function of time as determined by reference-free 2D classification. At the beginning and end of the movie, at pH 3.4, the active/desensitized-state, a class where the ECDs form a flower-shaped pentamer surrounding a central cavity are most abundant. In the middle of the movie, at pH 7.5, the closed-state conditions, a class with narrowed ECDs dominates. An asymmetric class exists throughout the experiment, but peaks upon re-exposure to low activating pH.
HS-AFM has recently provided new insight into the molecular mechanisms of another family of channels: cyclic nucleotide-gated (CNG) channels. CNG channels are non-selective cation channels that play a central role in signal transduction, specifically involved in neuronal excitability in the brain and pace making in heart cells as well as various sensory pathways [27,28]. They are regulated by the binding of cyclic nucleotides (cAMP or cGMP) to a specialized intracellular cyclic nucleotide-binding domain (CNBD) which results in conformational changes to open the pore. This family of channels has a tetrameric arrangement of subunits surrounding a central pore with each subunit consisting of six transmembrane helices. The first four helices form a voltage sensor (S1-S4) and the last two form the pore domain (S5-S6). The CNBD is then C-terminally connected to the final pore domain helix (S6) via a C-linker [29]. However, despite the recent advances in structural determination of CNG channels, the conformational changes associated with cyclic nucleotide gating remain unknown. The prokaryotic homolog from Spirochaeta thermophila potassium (K+) channel, SthK, is a good functional and structural model of CNG channels. Three SthK structures have been solved, in apo, cGMP- bound and cAMP-bound states [30]. However, all these structures are in the resting state, and the absence of an open state highlights the difficulty in capturing infrequent states with cryo-EM. HS-AFM imaging of SthK 2D-crystals in membranes has shown the conformational changes associated with ligand-binding [31••]. When flowing cGMP through the fluid chamber large changes in surface topography and 2D-crystal packing were observed with the initially cAMP-bound channels (Fig 2 a,b,c), interpreted based on electrophysiology experiments to represent the conformational changes associated with gating from an activated to an inactivated state. This process was reversible when cAMP was titrated back into the fluid chamber. The same effect, although less sharp, was also observed when cAMP was removed from the solution. The four CNBDs in the tetramer moved vertically ~0.6nm towards the membrane and spread ~0.4nm, when transitioning from cGMP (closed) to cAMP (active/open). Most importantly, the CNBDs underwent a ~25° clockwise rotation (viewed from the cytosolic side) with respect to the pore domain (Fig 2d). Structural analysis suggests that such a rotation by the CNBDs upon cAMP activation should results in a ~1.3nm displacement on the periphery of the C-linker. Translated by the C-linker to the pore domain this movement causes pulling on the bundle-crossing helices S6, that form a right-handed iris diaphragm, with a clockwise torque, thus opening the pore. These global and coordinated structural transitions, indicating long-range interactions between distant and adjacent protein domains, can likely be extended to other cyclic nucleotide-modulated channels and possibly to other channels controlled by binding of small intracellular ligands to carboxyl terminal regulatory domains.
Figure 2. Dynamics of ligand-induced conformational changes in SthK by real-time HS-AFM imaging.
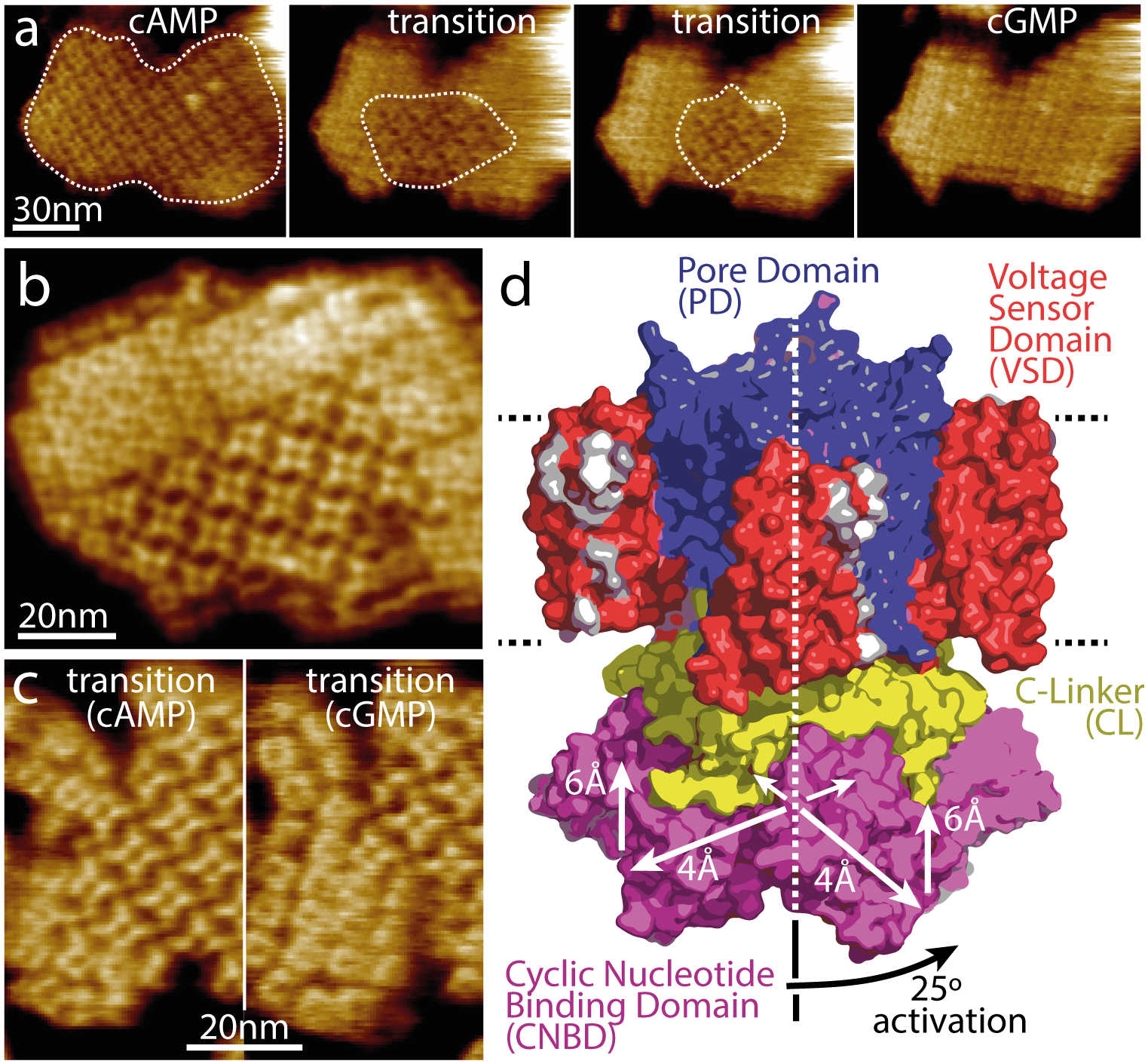
(a) HS-AFM time-lapse high-resolution image sequence of a SthK 2D-crystal initially in 0.1mM cAMP and exposing CNBDs. Upon addition of 7mM cGMP, SthK channels undergo a conformational change progressively from the borders to the center of the membrane patch (dotted outline). (b) High-resolution topography of a membrane containing well-ordered channels in both conformations. (c) High-resolution topographs during a cAMP to cGMP transition where the majority of the molecules are in the cAMP (left) and in the cGMP (right) conformation, respectively. (d) Model of SthK in the activated state: Upon activation, the CNBDs rotate by ~25° clockwise (when viewed from the intracellular side) and move by ~6 Å towards the membrane and by ~4 Å outwards from the four-fold axis (note, this activated state illustration is a cartoon using domains of the SthK structure repositioned according to the displacements found by HS-AFM).
Transporter and Pump Conformational Dynamics
Whilst channels passively allow solutes to pass (or not) across membranes, transporters physically assist solutes. Generally, via the input of energy (ATP) or a chemical potential, transporters act against concentration gradients with multiple steps/conformational changes and therefore offer a more complex set of dynamics to decipher.
Glutamate transporters are vital for the regulation of glutamate concentration in the synaptic cleft and thus their dysfunction is associated with numerous neurological diseases [32]. The transporter cycle of glutamate transporters has been inferred from X-ray structures of a homolog from the archaebacterium Pyrococcus horikoshii, sodium/aspartate symporter GltPh, in outward and inward facing conformations [33,34]. HS-AFM imaging has recently enabled the first direct dynamics study of the transport mechanism using purified GltPh, reconstituted into lipid bilayers [35••]. GltPh forms a bowl-shaped homotrimer in which three peripheral transport domains are centrally connected by a trimerization domain [33]. In the absence of aspartate and Na+ (apo conditions) HS-AFM images clearly map the peripheral transport domains of the GltPh trimers which protrude ~2nm above the membrane (Fig 3a, left). Imaging over time at 1 s−1 revealed how each transport domain moved ~2nm down from this protruding outward facing state to an inward facing state where there is almost no protrusion above the membrane (Fig 3a, right). These conformational changes were consistent with an elevator mechanism proposed by the static crystal structures in each state as well as suggested by donor-acceptor distance dynamics in single molecule FRET studies [36,37]. By monitoring the dwell-time of each protomer in each state, the transition energetics and state probabilities could be calculated for a range of conditions including transport (Na+ + Asp), apo (no substrate), saturating sodium (Na+) and in the presence of a selective, non-transportable inhibitor (TBOA). In addition to showing varying transport dynamics depending on the substrates available, these results showed that the outward facing state is preferably adopted by GltPh, suggested to be important for exposing the substrate binding sites for a new transport cycle. As well as measuring single protomer dynamics, HS-AFM imaging of the transporter allowed assessment of transport domain interplay within the trimer. Analyzing all possible correlations between conformations of protomers revealed that each protomer acted independently and without order with respect to their location within the trimer.
Figure 3. Active domain motions in the glutamate transporter GltPh and the light-driven proton pump bR.
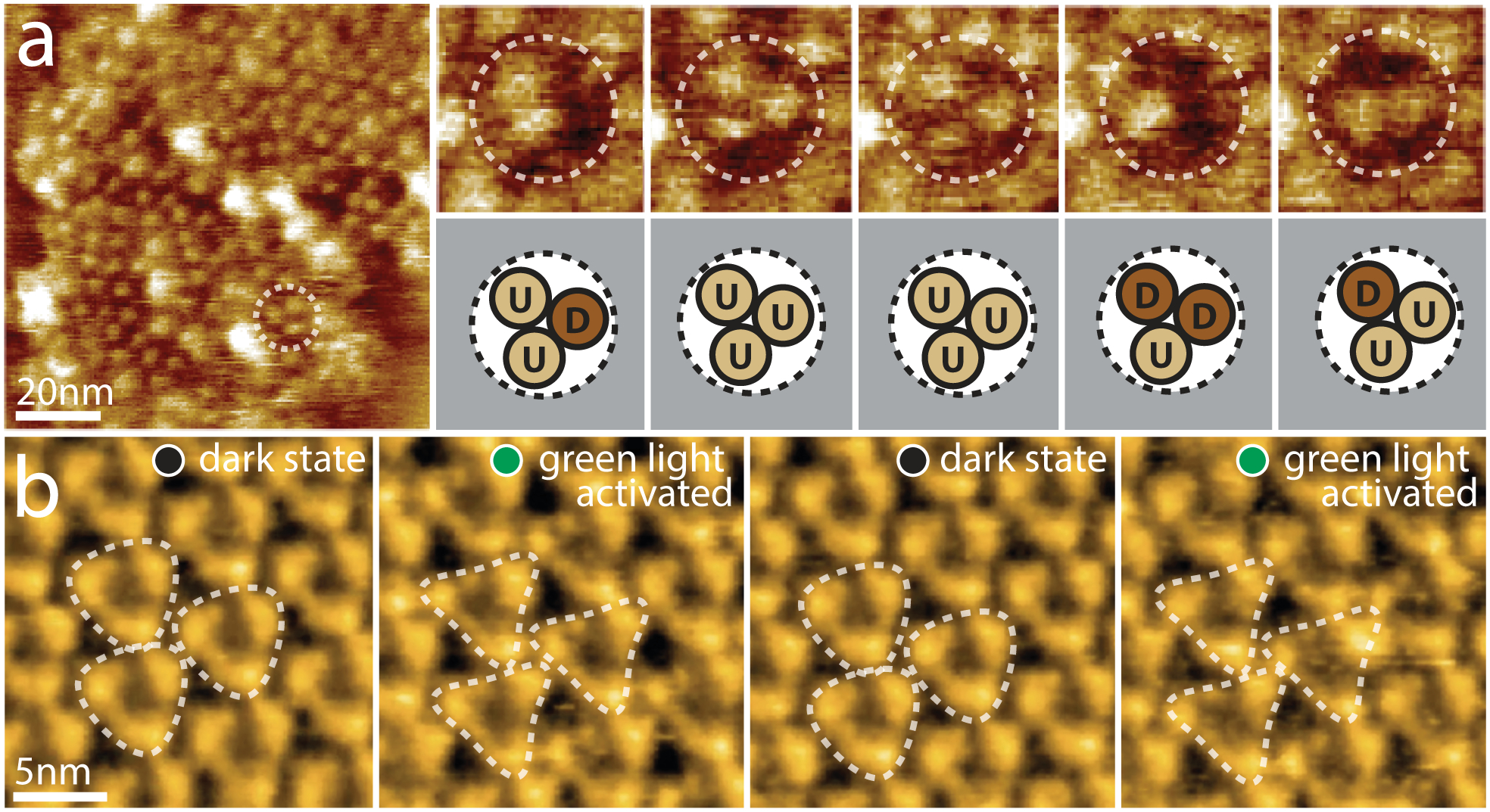
(a) Direct visualization of GltPh transport domain elevator movements by HS-AFM. Left: A typical HS-AFM image of a GltPh reconstituted membrane displays densely packed GltPh trimers (dashed outline). Right: Conformational dynamics of a representative GltPh trimer under substrate-free conditions (imaging rate: 1 s−1, frame size: 20nm). Each GltPh protomer in the trimer (top) shows reversible conformational alternation between outward facing (up, U) and inward facing (down, D) states (bottom). (b) HS-AFM movie frames of D96N bacteriorhodopsin (bR) exposed to repeated dark and green light illumination cycles (imaging rate: 1 s−1). bR trimers are highlighted by the white dashed triangles. Under illumination, conformational changes result in significant changes in the topography, notably a movement of the E-F loop outwards from the 3-fold axis (E-F loops of neighboring activated trimers interact closely in the activated state).
Whilst the vertical elevator motion of GltPh is clearly detected by the Ångstrom sensitivity of HS-AFM, the lateral motions of individual loops have also been detected in bacteriorhodopsin (bR), a light-driven proton pump. Despite numerous previous structural and biophysical studies, the structural amplitude and intermolecular cross-talk remained elusive. By applying HS-AFM imaging to a slow photocycle bR mutant (D96N), the conformational changes of the cytoplasmic domains have been observed [38••]. Upon light illumination, HS-AFM revealed a ~0.8nm lateral outward displacement of the E–F loops of each bR (Fig 3b) (loop assignment based on resting state crystallography structures of bR [39,40]). Allocating AFM topography to high resolution EM and X-ray crystallography structural data in this way shows how the combination of these techniques can be extremely powerful to assign the structural dynamics. Illumination with different intensities showed how the probability of conformational changes depended on the inter-trimer bR–bR contacts which produce both positive and negative cooperative effects. More recent HS-AFM studies of a number of bR mutants have provided further insights into the importance of the trimer arrangement for bR function [41].
These studies demonstrate the unique capability of HS-AFM to study dynamic processes at the single unlabeled membrane protein level for the characterization of conformational changes that are difficult to capture using low signal to noise techniques which rely on ensemble averaging.
Millisecond and Microsecond Protein Dynamics
Whilst, as shown in this review, the conformational changes of GLIC and Sthk channels could be clearly observed over seconds and minutes depending on their ligand-induced gating state, GltPh and bR have dynamics that may occur outside the current temporal resolution of HS-AFM. Additionally, many membrane proteins have dynamics in the millisecond-microsecond range and thus are too fast to be resolved directly in HS-AFM imaging mode (Fig. 4a, 50–100ms temporal resolution). Progress in developing faster HS-AFM is ongoing, but it is unlikely to reach sub-millisecond imaging resolution in the near future [42,43]. Therefore, to bypass current imaging speed limitations and gain orders of magnitude in temporal resolution one can reduce scanning to a single line [11••,42–44] (HS-AFM line scanning, HS-AFM-LS) or a single point (HS-AFM height spectroscopy, HS-AFM-HS), giving millisecond and microsecond temporal resolution, respectively, without losing z-accuracy. The capabilities of these two approaches have been recently demonstrated [11••] through the study of annexin self-assembly at membranes, a process important for cell membrane repair [47]. By halting the y-piezo, aka HS-AFM line scanning, the movements of annexin trimers within a 2D-lattice were monitored at millisecond rates. The rotational displacement between two preferred orientations could be observed by the movement of the apex of the trimer which rotated in 60° intervals as it interacted with the surrounding six annexin trimers (Fig. 4b). Analysis of the line scanning kymographs showed the trimer spent an equal amount of time in each orientation with average dwell-times of 35ms consistent with symmetric interactions with the neighboring lattice. Additionally, the 2.4ms time resolution allowed the rotational velocity in both clockwise and counterclockwise directions to be captured mid rotation.
Figure 4. HS-AFM line scanning (HS-AFM-LS) and HS-AFM height spectroscopy (HS-AFM-HS): Increasing the temporal resolution by reducing the dimensionality of data acquisition.
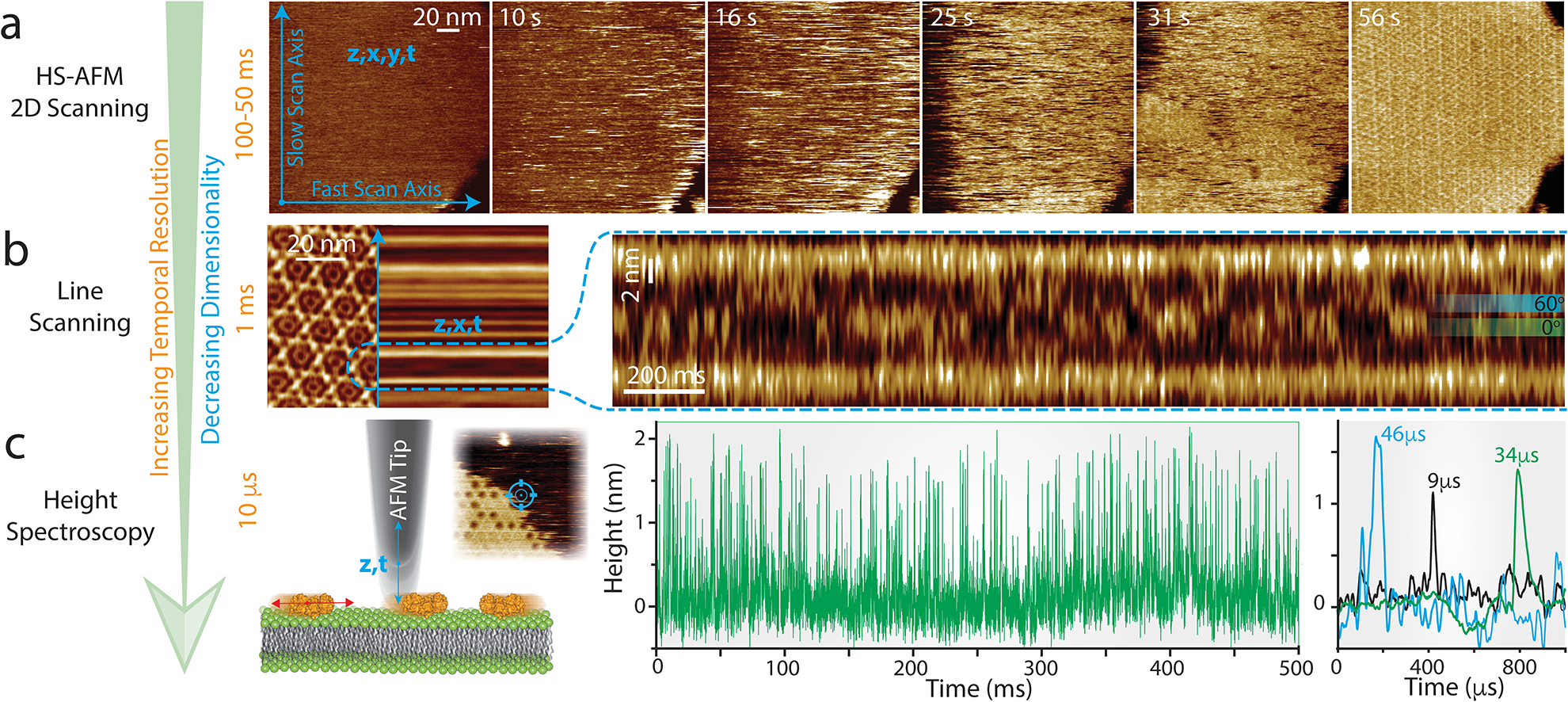
(a) HS-AFM 2D-scanning movie of A5 membrane-binding, self-assembly and formation of p6 2D-crystals upon UV-illumination-induced Ca2+-release. Blue arrows illustrate the slow (vertical) and the fast (horizontal) scan axes. Images can be captured at up to 10–20 images per second. (b) Left: averaged HS-AFM image of an A5 p6-lattice overlaid with the subsequent line scanning kymograph, obtained by scanning repeatedly the central x-direction line as illustrated by the blue arrow with a maximum rate of 1000–2000 lines per second. Right: line scanning kymograph across one protomer of the non-p6 trimer, outlined by the semicircular dashed line in the 2D image (left), at a rate of 417 lines per second (2.4 ms/line). Overlaid on the kymography are the positions of the 0° and 60° states that the trimer alternately adopts. (c) Left: schematic showing the principle of HS-AFM height spectroscopy (HS-AFM-HS) allowing 10 μs temporal resolution. The AFM tip is oscillated in z at a fixed x,y-position, detecting single molecule dynamics such as diffusion under the tip (inset: HS-AFM image of an A5 p6-lattice partially covering the membrane surface during self-assembly, HS-AFM-HS is performed at a fixed position at the center of the image as illustrated by the target). Right: Height/time traces obtained by HS-AFM-HS allowing determination of the local A5 concentration and diffusion rates (the colored traces on the very right are a zoomed overlay of three traces displaying diffusion events under the tip in the low microsecond time range).
Halting the x-piezo as well as the y-piezo allowed a further 100-fold gain in time resolution whilst maintaining Ångstrom accuracy height data (z, t). With ~10μs temporal resolution, this method termed high-speed AFM height spectroscopy (HS-AFM-HS), was used in an approach inspired by fluorescence spectroscopy to measure the mobility of rapidly diffusing membrane-bound molecules as they diffused under the tip (Fig. 4c). Due to the tip size being much smaller than the diffusing molecules of interest, not only could diffusion coefficients be measured but also the oligomeric state and surface concentrations of the molecules. Using HS-AFM-HS to measure mobile membrane-bound annexin trimers showed sharp peaks in the height signal as the trimers diffused under the tip. Increasing the surface concentration of trimers by either increasing the calcium or annexin concentrations in solution resulted in more frequent and stepwise longer-lived dwell-times up until a complete lattice was formed. Analysis of the surface concentration dependent dwell-times was consistent with two processes: oligomer formation and diffusion reduction due to crowding. This data gave access to biochemical and biophysical parameters including affinities and association/dissociation kinetics describing entirely and quantitatively the Annexin-V membrane-association process. Furthermore, HS-AFM-HS was used to also measure the movements of trimers free to rotate in the lattice, providing a direct comparison to line-scanning measurements and demonstrating the ability to use this technique to make microsecond measurements at specific positions on proteins. This demonstration indicates direct applicability of HS-AFM-HS for the analysis of fast membrane protein dynamics such as ligand binding and unbinding events in channels, transport cycles of mammalian transporters or ATPase-driven processes.
Conclusions
Since its development the most exciting and challenging applications of HS-AFM have been the combined real-space and real-time measurements of proteins in action. Until recently, successful observations of membrane protein dynamics have been limited despite their importance in biology (Table 1). Developments in HS-AFM environmental control and stability are now showing the power of this technique to measure single membrane protein conformational changes in response to a range of stimuli (Table 1, bold). The advent of HS-AFM-LS and HS-AFM-HS methods which work in series with HS-AFM imaging pushes the temporal range to timescales not accessible to other techniques, all without the use of labels. This leap in temporal resolution opens up the range of systems that can be studied and gives access to fast dynamic processes that were so far inaccessible, such as ligand-induced oligomerization of receptors and transporters, the conformational dynamics of transporters, receptors and channels during transport, signaling and gating.
Table 1).
List of transmembrane proteins studied by HS-AFM and the related dynamic parameters that were investigated.
| Membrane Protein | Function | Species | Characterized Dynamics | Reference: |
|---|---|---|---|---|
| Bacteriorhodopsin (bR) | Light driven proton pump | Halobacterium salinarum | Photoactivated conformation changes | [38••,39,46,47] |
| PoXeR and various other microbial rhodopsins | Light driven pumps | Parvularcula oceani and serval other eubacteria/archaea | Oligomerisation | [50,51] |
| ATP-synthase |
ATP synthesis | Halobacterium salinarum | Membrane mediated protein-protein interactions | [52] |
| Aquaporin-0 (AQP0) Connexin (Cx) |
Water and solute transport Cell-cell adhesion |
Sheep lens fiber cells | Protein microdomain formation, dynamics and adhesion | [53,54] |
| SecYEG and GlpF | Protein translocation channel and aquaglyceroporin | Escherichia coli | Protein docking and conformational dynamics | [55] |
| P2X | ATP-gated ion channel | Human astrocytoma cells | ATP induced conformational changes | [56] |
| GltPh | Na+/L-Asp symport | Pyrococcus horikoshii | Transport dynamics | [35••] |
| SERCA | ATP-driven Ca2+-pump | Rabbit | Transport dynamics | [57] |
| OmpF | Large non-specific channel | Escherichia coli | Diffusion and Interaction | [9,58] |
| KcsA | Potassium channel | Streptomyces lividans | Conformational dynamics and toxin binding | [59,60] |
| TWIK1 | Potassium channel | Mouse | Structural properties | [61] |
| SthK | Potassium channel | Spirochaeta thermophila | Ligand induced conformational dynamics | [31••] |
| MloK1 | Potassium channel | Mesorhizobium loti | Ligand induced conformational dynamics | [62] |
| GLIC | Ligand gated ion channel | Gloeobacter violaceus | Ligand induced conformational dynamics | [26••] |
| Listeriolysin O | Pore forming toxin | Listeria monocytogenes | Assembly kinetics and membrane disruption | [63] |
| Lysenin | Pore forming toxin | Eisenia fetida | Diffusion, Interaction and membrane insertion | [64,65] |
| OmpLA | Phospholipid hydrolysis | Escherichia coli | Ca2+-dependent membrane degradation | [66] |
Highlights.
HS-AFM detects conformational changes of unlabeled membrane proteins
In situ responses to pH, ligands, temperature and light can be visualized
Developments in techniques now allow microsecond temporal resolution
Footnotes
Conflict of interest statement
Nothing declared.
References:
• of special interest
•• of outstanding interest
- 1.Scheres SHW: RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 2012, 180:519530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao M, Cao E, Julius D, Cheng Y: Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013, 504:107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Y: Membrane protein structural biology in the era of single particle cryo-EM. Curr Opin Struct Biol 2018, 52:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drew D, Boudker O: Shared Molecular Mechanisms of Membrane Transporters. Annu Rev Biochem 2016, 85:543–572. [DOI] [PubMed] [Google Scholar]
- 5.Bavi N, Martinac AD, Cortes DM, Bavi O, Ridone P, Nomura T, Hill AP, Martinac B, Perozo E: Structural Dynamics of the MscL C-terminal Domain. Sci Rep 2017, 7:17229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basak S, Schmandt N, Gicheru Y, Chakrapani S: Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchihashi T, Iino R, Ando T, Noji H: High-Speed Atomic Force Microscopy Reveals Rotary Catalysis of Rotorless F1-ATPase. Science (80-) 2011, 333:755 LP–758. [DOI] [PubMed] [Google Scholar]
- 8.Kodera N, Yamamoto D, Ishikawa R, Ando T: Video imaging of walking myosin V by high-speed atomic force microscopy. Nature 2010, 468:72–76. [DOI] [PubMed] [Google Scholar]; • Milestone paper illustrating the power of HS-AFM to take films of biomolecules, myosin-V walking on actin, in action. The paper also nicely illustrates the low invasiveness of the technique.
- 9.Casuso I, Khao J, Chami M, Paul-Gilloteaux P, Husain M, Duneau J-P, Stahlberg H, Sturgis JN, Scheuring S: Characterization of the motion of membrane proteins using high-speed atomic force microscopy. Nat Nanotech 2012, 7:525–529. [DOI] [PubMed] [Google Scholar]
- 10.Miyagi A, Chipot C, Rangl M, Scheuring S: High-speed atomic force microscopy showsthat annexin V stabilizes membranes on thesecond timescale. Nat Nanotechnol 2016, 11:783–790. [DOI] [PubMed] [Google Scholar]; • Introducing slow and fast changes of environmental conditions during HS-AFM imaging through coupling of a constant pressure and constant flow buffer exchange system and UV-laser assisted uncaging of caged compounds, respectively.
- 11.Heath GR, Scheuring S: High-speed AFM height spectroscopy reveals μs-dynamics of unlabeled biomolecules. Nat Commun 2018, 9:4983. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Milestone paper introducing HS-AFM height spectroscopy measurements allowing height data to be captured with 10 μs temporal resoltuion. This technique will allow direct detection of membrane protein dynamics at unprecedented timescales.
- 12.Takahashi H, Miyagi A, Redondo-Morata L, Scheuring S: Temperature-Controlled High-Speed AFM: Real-Time Observation of Ripple Phase Transitions. Small 2016, 12:6106–6113. [DOI] [PubMed] [Google Scholar]; • Technical development showcasing how HS-AFM movies can be acquired during temperature-sweep cycles. Temperature controlled HS-AFM will allow the direct dynamic study of temperature sensitive channels, temperature dependent enzymatic action, and of use for the study of mammalian systems.
- 13.Ando T: High-speed atomic force microscopy coming of age. Nanotechnology 2012, 23:62001. [DOI] [PubMed] [Google Scholar]
- 14.Rico F, Gonzalez L, Casuso I, Puig-Vidal M, Scheuring S: High-Speed Force Spectroscopy Unfolds Titin at the Velocity of Molecular Dynamics Simulations. Science (80-) 2013, 342:741–743. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi H, Rico F, Chipot C, Scheuring S: α-Helix Unwinding as Force Buffer in Spectrins. ACS Nano 2018, 12:2719–2727. [DOI] [PubMed] [Google Scholar]
- 16.Rigato A, Miyagi A, Scheuring S, Rico F: High-frequency microrheology reveals cytoskeleton dynamics in living cells. Nat Phys 2017, 13:771–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A: Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 2015, 163:866–79. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Milestone paper in which HS-AFM describes the assembly and conformational dynamics of the ESCRT-III filament polymer spirals, which are highly variable in size and structure. The data is suggestive of a functional model.
- 18.Sakiyama Y, Mazur A, Kapinos LE, Lim RYH: Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol 2016, 11:719–723. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z, Hashemi M, Warren G, Bianco PR, Lyubchenko YL: Dynamics of the Interaction of RecG Protein with Stalled Replication Forks. Biochemistry 2018, 57:1967–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haruyama T, Sugano Y, Kodera N, Tanaka Y, Konno H, Correspondence TT, Uchihashi T, Ando T, Tsukazaki T: Single-Unit Imaging of Membrane Protein-Embedded Nanodiscs from Two Oriented Sides by High-Speed Atomic Force Microscopy. Structure 2019, 27:152–160. [DOI] [PubMed] [Google Scholar]
- 21.Du J, Lü W, Wu S, Cheng Y, Gouaux E: Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature 2015, 526:224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sine SM, Engel AG: Recent advances in Cys-loop receptor structure and function. Nature 2006, 440:448–455. [DOI] [PubMed] [Google Scholar]
- 23.Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux J-P, Delarue M, Corringer P-J: X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 2009, 457:111–114. [DOI] [PubMed] [Google Scholar]
- 24.Sauguet L, Shahsavar A, Poitevin F, Huon C, Menny A, Nemecz À, Haouz A, Changeux J-P, Corringer P-J, Delarue M: Crystal structures of a pentameric ligand-gated ion channel provide a mechanism for activation. Proc Natl Acad Sci U S A 2014, 111:966–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velisetty P, Chalamalasetti SV, Chakrapani S: Structural basis for allosteric coupling at the membrane-protein interface in Gloeobacter violaceus ligand-gated ion channel (GLIC). J Biol Chem 2014, 289:3013–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan Y, Kao K, Lefebvre S, Marchesi A, Corringer P-J, Hite RK, Scheuring S: Structural titration of receptor ion channel GLIC gating by HS-AFM. Proc Natl Acad Sci 2018, 115:10333–10338. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Direct structural titration experiment observing the same pH-gated GLIC channels over extended period of time while being exposed to a pH gradient. The data reports structural changes on the single molecule level as well as on the level of the supramolecular arrangement of the channels.
- 27.DiFrancesco JC, DiFrancesco D: Dysfunctional HCN ion channels in neurological diseases. Front Cell Neurosci 2015, 6:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pifferi S, Boccaccio A, Menini A: Cyclic nucleotide-gated ion channels in sensory transduction. FEBS Lett 2006, 580:2853–2859. [DOI] [PubMed] [Google Scholar]
- 29.Paoletti P, Young EC, Siegelbaum SA: C-Linker of cyclic nucleotide-gated channels controls coupling of ligand binding to channel gating. J Gen Physiol 1999, 113:17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rheinberger J, Gao X, Schmidpeter PA, Nimigean CM: Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchesi A, Gao X, Adaixo R, Rheinberger J, Stahlberg H, Nimigean C, Scheuring S: An iris diaphragm mechanism to gate a cyclic nucleotide-gated ion channel. Nat Commun 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• HS-AFM movies show the structural changes of cyclic nucleotide gated SthK channels binding cAMP or cGMP. The cyclic nucleotide binding domains undergo conformational changes that are accompanied with a concerted rotation of the extramembranous domains proposed to induce channel gating in an iris-like manner.
- 32.Takahashi K, Foster JB, Lin C-LG: Glutamate transporter EAAT2: regulation, function, and potential as a therapeutic target for neurological and psychiatric disease. Cell Mol Life Sci 2015, 72:3489–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yernool D, Boudker O, Jin Y, Gouaux E: Structure of a glutamate transporter homologue from Pyrococcus horikoshii. Nature 2004, 431:811–818. [DOI] [PubMed] [Google Scholar]
- 34.Reyes N, Ginter C, Boudker O: Transport mechanism of a bacterial homologue of glutamate transporters. Nature 2009, 462:880–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruan Y, Miyagi A, Wang X, Chami M, Boudker O, Scheuring S: Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc Natl Acad Sci U S A 2017, 114:1584–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• First direct HS-AFM visualization of a transmembrane transporter’s transport dynamics and how it depends on substrate availability. The work provided a conclusive confirmation of the proposed ‘elevator’ mechanism of transport domain motion and showed that each protomer in the trimer acted fully independently.
- 36.Akyuz N, Altman RB, Blanchard SC, Boudker O: Transport dynamics in a glutamate transporter homologue. Nature 2013, 502:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akyuz N, Georgieva ER, Zhou Z, Stolzenberg S, Cuendet MA, Khelashvili G, Altman RB, Terry DS, Freed JH, Weinstein H, et al. : Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature 2015, 518:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata M, Yamashita H, Uchihashi T, Kandori H, Ando T: High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat Nanotechnol 2010, 5:208–212. [DOI] [PubMed] [Google Scholar]; •• First use of light stimulation into the HS-AFM fluid cell to induce structural changes in bacteriorhodopsin during HS-AFM imaging. The HS-AFM movies reveal rather large conformational changes in the bR loops that have eluded analysis by other techniques and illustrated the functional importance of the lattice of trimers.
- 39.Kimura Y, Vassylyev DG, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y: Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature 1997, 389:206–211. [DOI] [PubMed] [Google Scholar]
- 40.Luecke H, Schobert B, Richter H-T, Cartailler J-P, Lanyi JK: Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol 1999, 291:899–911. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita H, Inoue K, Shibata M, Uchihashi T, Sasaki J, Kandori H, Ando T: Role of trimer–trimer interaction of bacteriorhodopsin studied by optical spectroscopy and high-speed atomic force microscopy. J Struct Biol 2013, 184:2–11. [DOI] [PubMed] [Google Scholar]
- 42.Ando T: High-speed atomic force microscopy and its future prospects. Biophys Rev 2018, 10:285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyagi A, Scheuring S: A novel phase-shift-based amplitude detector for a high-speed atomic force microscope. Rev Sci Instrum 2018, 89:083704. [DOI] [PubMed] [Google Scholar]
- 44.Yokokawa M, Wada C, Ando T, Sakai N, Yagi A, Yoshimura SH, Takeyasu K: Fast-scanning atomic force microscopy reveals the ATP/ADP-dependent conformational changes of GroEL. EMBO J 2006, 25:4567–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Viani MB, Pietrasanta LI, Thompson JB, Chand A, Gebeshuber IC, Kindt JH, Richter M, Hansma HG, Hansma PK: Probing protein-protein interactions in real time. Nat Struct Biol 2000, 7:644–647. [DOI] [PubMed] [Google Scholar]
- 46.Yokokawa M, Takeyasu K: Motion of the Ca2+-pump captured. FEBS J 2011, 278:3025–3031. [DOI] [PubMed] [Google Scholar]
- 47.Bouter A, Gounou C, Bérat R, Tan S, Gallois B, Granier T, d’Estaintot BL, Pöschl E, Brachvogel B, Brisson AR: Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat Commun 2011, 2:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita H, Voïtchovsky K, Uchihashi T, Contera SA, Ryan JF, Ando T: Dynamics of bacteriorhodopsin 2D crystal observed by high-speed atomic force microscopy. J Struct Biol 2009, 167:153–158. [DOI] [PubMed] [Google Scholar]
- 49.Shibata M, Uchihashi T, Yamashita H, Kandori H, Ando T: Structural Changes in Bacteriorhodopsin in Response to Alternate Illumination Observed by High-Speed Atomic Force Microscopy. Angew Chemie 2011, 123:4502–4505. [DOI] [PubMed] [Google Scholar]
- 50.Inoue K, Ito S, Kato Y, Nomura Y, Shibata M, Uchihashi T, Tsunoda SP, Kandori H: A natural light-driven inward proton pump. Nat Commun 2016, 7:13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shibata M, Inoue K, Ikeda K, Konno M, Singh M, Kataoka C, Abe-Yoshizumi R, Kandori H, Uchihashi T: Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci Rep 2018, 8:8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casuso I, Sens P, Rico F, Scheuring S: Experimental evidence for membrane-mediated protein-protein interaction. Biophys J 2010, 99:L47–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colom A, Casuso I, Boudier T, Scheuring S: High-Speed Atomic Force Microscopy: Cooperative Adhesion and Dynamic Equilibrium of Junctional Microdomain Membrane Proteins. J Mol Biol 2012, 423:249–256. [DOI] [PubMed] [Google Scholar]
- 54.Colom A, Casuso I, Rico F, Scheuring S: A hybrid high-speed atomic force–optical microscope for visualizing single membrane proteins on eukaryotic cells. Nat Commun 2013, 4:2155. [DOI] [PubMed] [Google Scholar]
- 55.Karner A, Nimmervoll B, Plochberger B, Klotzsch E, Horner A, Knyazev DG, Kuttner R, Winkler K, Winter L, Siligan C, et al. : Tuning membrane protein mobility by confinement into nanodomains. Nat Nanotechnol 2017, 12:260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shinozaki Y, Sumitomo K, Tsuda M, Koizumi S, Inoue K, Torimitsu K: Direct Observation of ATP-Induced Conformational Changes in Single P2X4 Receptors. PLoS Biol 2009, 7:e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yokokawa M, Takeyasu K: Motion of the Ca2+-pump captured. FEBS J 2011, 278:3025–3031. [DOI] [PubMed] [Google Scholar]
- 58.Yamashita H, Taoka A, Uchihashi T, Asano T, Ando T, Fukumori Y: Single-Molecule Imaging on Living Bacterial Cell Surface by High-Speed AFM. J Mol Biol 2012, 422:300–309. [DOI] [PubMed] [Google Scholar]
- 59.Sumino A, Yamamoto D, Sumikama T, Iwamoto M, Dewa T, Oiki S: Structure and Dynamics of Membrane-embedded KcsA Potassium Channel Revealed by Atomic Force Microscopy. Seibutsu Butsuri 2015, 55:005–010. [Google Scholar]
- 60.Sumino A, Uchihashi T, Oiki S: Oriented Reconstitution of the Full-Length KcsA Potassium Channel in a Lipid Bilayer for AFM Imaging. J Phys Chem Lett 2017, 8:785–793. [DOI] [PubMed] [Google Scholar]
- 61.Tsukamoto H, Higashi M, Motoki H, Watanabe H, Ganser C, Nakajo K, Kubo Y, Uchihashi T, Furutani Y: Structural properties determining low K+ affinity of the selectivity filter in the TWIK1 K+ channel. J Biol Chem 2018, 293:6969–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rangl M, Miyagi A, Kowal J, Stahlberg H, Nimigean CM, Scheuring S: Real-time visualization of conformational changes within single MloK1 cyclic nucleotide-modulated channels. Nat Commun 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruan Y, Rezelj S, Bedina Zavec A, Anderluh G, Scheuring S: Listeriolysin O Membrane Damaging Activity Involves Arc Formation and Lineaction -- Implication for Listeria monocytogenes Escape from Phagocytic Vacuole. PLOS Pathog 2016, 12:e1005597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Munguira I, Casuso I, Takahashi H, Rico F, Miyagi A, Chami M, Scheuring S: Glasslike Membrane Protein Diffusion in a Crowded Membrane. ACS Nano 2016, 10:2584–2590. [DOI] [PubMed] [Google Scholar]
- 65.Munguira ILB, Takahashi H, Casuso I, Scheuring S: Lysenin Toxin Membrane Insertion Is pH-Dependent but Independent of Neighboring Lysenins. Biophys J 2017, 113:2029–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rangl M, Rima L, Klement J, Miyagi A, Keller S, Scheuring S: Real-time Visualization of Phospholipid Degradation by Outer Membrane Phospholipase A using High-Speed Atomic Force Microscopy. J Mol Biol 2017, 429:977–986. [DOI] [PubMed] [Google Scholar]


