This study investigated immune profiles of ulcerative colitis (UC) patients and irritable bowel syndrome (IBS) patients and healthy individuals and showed that inflammatory mechanisms leading to IBS-like symptoms in UC may be different from the mechanisms leading to IBS.
Keywords: ulcerative colitis, irritable bowel syndrome, inflammatory markers
Abstract
Background
Inflammatory mechanisms of ulcerative colitis (UC) and irritable bowel syndrome (IBS) may overlap or are part of different spectrums. However, potential links between inflammation and IBS-like symptoms in these patient groups are still unclear. The aim of this study was to determine if the systemic inflammatory protein (SIP) profiles differ between UC patients, with presence of inflammation or in remission with or without IBS-like symptoms, and IBS patients.
Methods
Serum from patients with active UC (UCA), UC patients in remission with or without IBS-like symptoms (UCR + IBS, UCR-IBS), IBS patients (IBS), and healthy subjects (HS) was analyzed using the ProSeek Multiplex Inflammation kit, which detects 92 proteins.
Results
The exploratory cohort consisted of 166 subjects (UCA, n = 40; UCR-IBS, n = 45; UCR + IBS, n = 20; IBS, n = 40; HS, n = 21). Systemic inflammatory protein profiles separated UC from non-UC (HS and IBS) patients in multivariate analysis, revealing caspase 8, axin 1, sulfotransferase 1A1, and tumor necrosis factor superfamily member 14 as the variables most important to clustering. Although minor differences were detected between UCR + IBS and UCR-IBS, SIP profiles discriminated UCA from UCR, and interleukin (IL) 17C, IL17A, chemokine ligand 9, and transforming growth factor–α characterized active inflammation. SIP profiles weakly discriminated HS from IBS, although fibroblast growth factor 21 and IL6 serum levels were higher in IBS. Results were confirmed in a validation cohort (UCA, n = 15; UCR + IBS, n = 9; IBS, n = 14).
Conclusions
SIP profiles distinguish UC patients from IBS patients, irrespective of inflammation or IBS-like symptoms, suggesting that inflammatory mechanisms of the diseases are part of different spectrums.
INTRODUCTION
Patients with ulcerative colitis (UC), a chronic inflammatory bowel disease (IBD) involving the colonic mucosa, may suffer from irritable bowel syndrome (IBS)–like symptoms during periods of remission. It has been suggested that UC patients experiencing IBS-like symptoms during remission suffer from inadequately treated inflammation.1 Likewise, there are reports of systemic and intestinal low-grade immune activation in IBS patients.2 Indeed, IBS and UC present some similarities that range from high incidence of clinical depression and/or anxiety to imbalanced microbiome and persistent immune activation.3 However, it is still unclear whether the inflammatory mechanisms of UC and IBS overlap or are part of different spectrums.3
It may sometimes be difficult to differentiate between IBS-like symptoms and an upcoming flare in IBD patients, resulting in mistakes in choice of treatment, increased morbidity, and impaired quality of life. The similarity could also suggest similar underlying immunopathology, ranging from low-grade immune activity to full-scale inflammation. We and others have demonstrated that the serum cytokine profiles of IBS patients, or at least subgroups thereof, are characterized by increased levels of proinflammatory cytokines as compared with healthy subjects.4–7 Moreover, we recently reported that UC patients in deep remission with IBS-like symptoms had higher levels of certain serum cytokines than patients without these symptoms.8
Lately, studies have focused on identifying inflammatory protein profiles, including cytokines and chemokines, in the serum of IBD patients. Interestingly, a study suggests that Crohn’s disease (CD) and UC involve different inflammatory pathways, irrespective of disease activity, based on serum protein levels.9 The same study also identified serum proteins differentiating UC and CD, respectively, from healthy subjects (HS). Increased serum levels of inflammatory proteins such as eotaxin, growth-related oncogene, and tumor necrosis factor α (TNF-α) in UC patients compared with HS has also been reported, although no proteins were found to differ between UC and CD patients.10
We propose that characterization of disease-specific serum protein profiles will improve our understanding of the potential link between inflammation and IBS symptoms in patients with gastrointestinal diseases. Therefore, in this study, we aimed to determine systemic inflammatory protein (SIP) profiles, analyzing a broad panel of inflammation-related proteins, to establish if the profiles differ between UC patients, with presence of inflammation or in remission with or without IBS-like symptoms, and IBS patients.
METHODS
Sample Collection and Inclusion of Study Subjects
Serum samples were obtained from UC and IBS patients at 4 outpatient clinics in the southwest region of Sweden, Sahlgrenska University Hospital, Södra Älvsborg Hospital, Norra Älvsborg Hospital, and Kungälv Hospital, and from healthy subjects (HS) without gastrointestinal (GI) symptoms. Samples were aliquoted and stored at –80ºC until further use. The study was approved by the Regional Ethical Review Board in Gothenburg (Dnr: 403-12, approved on August 23, 2012; Dnr 266-16, approved on May 4, 2016; Dnr 988-14, approved on February 11, 2015) and follows the ethical guidelines of the Declaration of Helsinki (1975) and its later amendments. All subjects gave verbal and written consent before participation.
Exploratory Study Population
Adult (age ≥18 years) UC patients with a histologically verified diagnosis of UC, IBS patients meeting the Rome III criteria,11 and healthy subjects were recruited. Active UC (UCA) was defined as an endoscopic Mayo score ≥1 with a total Mayo score ≥3 points.12 UCA subjects were further subdivided into mild UCA (UCAm; Mayo score 3–5) and moderate to severe UCA (UCAs; Mayo score ≥6). UC in remission (UCR) was defined by an endoscopic Mayo score of 0 and a total Mayo score ≤2 with no flares during the 3 months preceding sample collection. UCR patients fulfilling the Rome III criteria for IBS were classified as UCR with IBS-like symptoms (UCR + IBS), whereas the remaining patients were classified as UCR without IBS-like symptoms (UCR-IBS).
The diagnosis of IBS patients was based on clinical presentation and investigations excluding organic diseases when deemed necessary. A vast majority of the patients were enrolled after referral from primary care and underwent sufficient investigation to rule out other diagnoses. Patients were assessed for symptom severity by the IBS–Severity Scoring System (IBS-SSS)13 and classified into IBS subtypes according to the evaluation of a 2-week stool diary using the Bristol Stool Form scale.14 For this study, patients subclassified as having predominantly diarrhea (IBS-D) or mixed bowel habits (IBS-M) with moderate to severe IBS symptoms (IBS-SSS score >175 points) were included.
The exclusion criteria for the study were malignancy during the 5 years before enrollment, difficulties in understanding Swedish, abuse of alcohol or drugs, previous intestinal resection, celiac disease by control of negative transglutaminase antibodies, severe cardiac, pulmonary, renal, neurological, rheumatological, or psychiatric disease(s), or other significant diseases that could affect the patient’s ability to comply with the study protocol.
Healthy subjects had no current or prior history of gastrointestinal or other chronic disorders, nor had they taken any immunosuppressive agents, antibiotics, or any other medication during the 3 months before sample collection, and they reported no current GI symptoms.
Validation Cohort
For validation of the results, a separate cohort was recruited including patients with UCA, UCR + IBS, and IBS following the same inclusion and exclusion criteria as the exploratory cohort described above. Serum sample collection was executed, and disease activity for all patient groups was defined as described above.
Fecal Calprotectin
Fecal calprotectin was analyzed by a sandwich enzyme-linked immunosorbent assay (ELISA; Calprotectin ELISA; BÜHLMANN Laboratories AG, Basel, Switzerland), according to the manufacturer’s instructions. The interassay coefficient of variation was <15%, and the assay sensitivity was <10 μg/g. In the general population, the fecal calprotectin level is <50 μg/g.
Proximity Extension Immunoassay
Serum levels of 92 inflammation-related proteins were measured by proximity extension immunoassay (PEA; Olink Proteomics, Uppsala, Sweden) using the ProSeek Multiplex Inflammation Kit. Briefly, oligonucleotide-labeled antibodies are allowed to pairwise-bind to the target protein, giving rise to a reporter sequence by DNA polymerization that is further quantified by real-time polymerase chain reaction (BioMark HD platform, Fludigm, San Francisco, CA, USA).15 Data are then normalized to log2 values corresponding to protein levels in the sample (termed normalized protein expression [NPX]), which show relative quantification, meaning that no comparison of absolute levels between different proteins can be made. Samples that deviate more than 0.3 NPX from the median value of an internal control do not pass quality assessment. From the exploratory study population, 7 samples did not pass quality control (HS, n = 2; IBS, n = 3; UCR-IBS, n = 1; and UCR + IBS, n = 1), whereas all samples in the validation cohort passed. Measured proteins that did not meet the minimum level of detection were reported as missing data. From the 92 target proteins, 91 were successfully quantified, and 13 had a missing data frequency of >40% and were excluded from further analyses (Supplementary Table 1).
Multivariate Analysis
Principal component analysis (PCA) was conducted on serum protein levels using the prcomp-algorithm and visualized using the pca3d-package in R (version 3.3.2; R Core Team).16 Heat map analysis was performed in R using the heatmap function and library:Rcolorbrewer for color customization. Orthogonal Projections to Latent Structures Discriminant Analyses (OPLS-DA) were implemented to correlate selected y-variables (patient groups) to x-variables (serum proteins) in linear multivariate models using SIMCA software (version 15; MKS Data Analytics Solutions). The R2Y parameter represents the goodness of fit of the model (best possible fit: R2Y = 1). The Q2 parameter represents an estimate of the predictive ability calculated by cross-validation (best possible predictive ability: Q2 = 1). For heterogeneous biological variables, a model is considered to have a good fit with an R2Y ≥0.5 and a good predictive ability with a Q2 >0.4.17, 18 The variable influence on projection (VIP) cutoff was defined based on discriminatory power; different VIP cutoffs were tested, and the cutoff most influential for explaining the y observations, based on the R2Y and Q2 values, was selected. Outliers were removed if they were both above the Hotelling’s T2 Range Line of 95% and DModX DCrit (0.05). In the OPLS-DA loading scatter and loading column plots, each x-variable is shown in relation to y. The x-variables positioned furthest to the left or right are most closely related to the respective y-variable and thus contribute most to the model. In the loading column plot, the importance of each x-variable is represented by column bars. The larger the bar and smaller the 95% confidence interval (shown by the whiskers), the stronger and more reliable the contribution to the model.
Validation of models was performed with SIMCA software using the predict tool, where a model work set was created based on OPLS-DAs from the exploratory cohort and further complemented by the data from the validation cohort. The generated misclassification table defines classification of tested members according to the model work set and uses Fisher’s exact probability test. P values <0.05 were considered significant.
Statistical Analyses
Differences between the 2 groups were assessed using the Mann-Whitney U test and between 3 or more groups using the Kruskal-Wallis test followed by Dunn’s multiple comparison test. To correct for false-positive results in multiple comparisons, the classical 1-stage method was employed, and data were then presented as q values. Statistical analyses were performed using IBM SPSS Statistics 25; P values or q values <0.05 were considered statistically significant. Data are shown as median with interquartile range. Power analysis to estimate the size of patient cohorts was not included in the experimental design.
RESULTS
Clinical Characteristics of Study Subjects
The exploratory cohort consisted of 166 study subjects, of whom 105 patients had UC, 40 patients had IBS, and 21 were HS (Table 1). Patients with UC were grouped according to disease activity and presence or absence of IBS-like symptoms, resulting in 40 patients with active disease (UCA), 45 patients in remission without IBS-like symptoms (UCR-IBS), and 20 patients in remission with IBS-like symptoms (UCR + IBS). Further clinical characterization of UC patients revealed disease severity heterogeneity within the UCA group, resulting in 18 patients with mild disease (UCAm) and 22 patients with moderate/severe disease (UCAs). Among UC patients with IBS-like symptoms, 7 patients had predominant diarrhea (35%), 12 patients had mixed bowel habits (60%), and 1 patient had predominant constipation (5%). For IBS patients, 17 patients had predominant diarrhea (43%) and 23 patients had mixed bowel habits (67%). Patient demographics are shown in Table 1 and the study groups are summarized in Supplementary Figure 1. Age distribution and sex proportion differed between groups (P < 0.0001 and P = 0.04, respectively). Fecal calprotectin levels were significantly different between groups (P < 0.0001).
TABLE 1.
Demographic Description of Study Subjects in the Exploratory Cohort
| HS (n = 21) | IBS (n = 40) | UCR-IBS (n = 45) | UCR + IBS (n = 20) | UCA (n = 40) | |
|---|---|---|---|---|---|
| Male/female | 9/12 | 14/26 | 30/15 | 10/10 | 17/23 |
| Age, median (range), y | 25 (21–54) | 31 (20–62) | 46 (18–67) | 43 (19–66) | 39 (21–69) |
| Mayo score, low/intermediate/higha | NA | NA | 45/0/0 | 20/0/0 | 0/18/22 |
| IBS subgroup IBS-D/IBS-C/IBS-Mb | N.A | 17/0/23 | NA | 7/1/12 | NA |
| Calprotectin, median (range), μg/g | <15 (<15–31) | <15 (<15–85) | <15 (<15–50) | 28 (<15–160) | 700 (51–10,000) |
| IBD-related treatmentc | |||||
| None | 21 | 40 | 11 | 0 | 8 |
| 5-aminosalicylic acid | 0 | 0 | 34 | 17 | 28 |
| Thiopurine | 0 | 0 | 0 | 3 | 4 |
| Anti-TNF | 0 | 0 | 0 | 1 | 0 |
| Systemic steroids | 0 | 0 | 0 | 0 | 0 |
| Disease extent | |||||
| Proctitis | NA | NA | 11 | 1 | 11 |
| Left-sided colitis | NA | NA | 9 | 10 | 11 |
| Extensive colitis | NA | NA | 25 | 9 | 18 |
| IBS-SSS, moderate/severed | NA | 21/19 | NA | *e | NA |
aMayo score, disease activity index for UC, low ≤2 pts, intermediate 3–5 pts, high ≥6 pts.
bSubclassification of IBS, predominant diarrhea (IBS-D), predominant constipation (IBS-C), predominant mixed bowel habits (IBS-M).
cOngoing treatment before the study.
dIBS-SSS (IBS–severity score system), moderate >175 pts, severe >300 pts.
eNonavailable patient information.
The Systemic Inflammatory Protein Profile Discriminates Patients With UC From Patients With IBS and Healthy Subjects
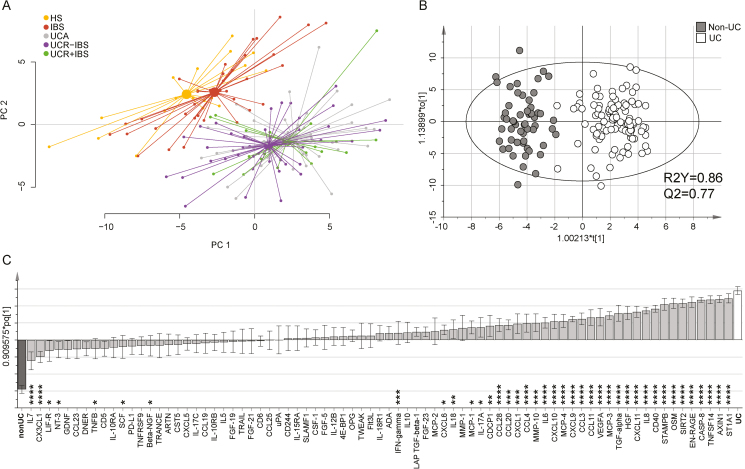
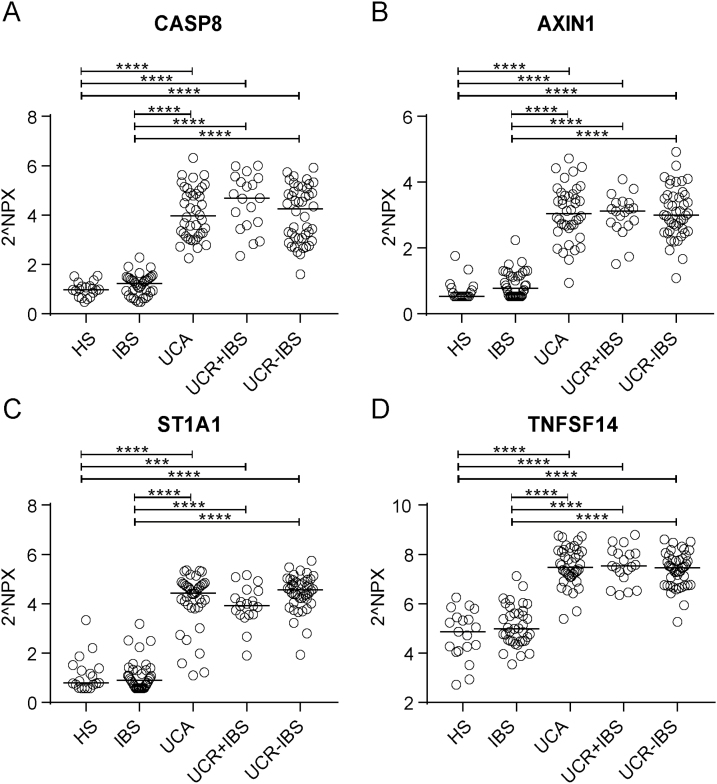
A PCA on the systemic inflammatory protein profile of all study subjects revealed 2 clusters: 1 cluster composed of IBS patients and HS and 1 cluster composed of all UC patients irrespective of disease activity or presence of IBS-like symptoms (Fig. 1A). To define the variables most important for discrimination between the 2 clusters, an OPLS-DA was performed, comparing non-UC (including HS and IBS) and UC (including all UC patient groups) (Fig. 1B). The variables most important for the clustering were caspase-8 (CASP8; q = 2 × 10–23), axin 1 (AXIN1; q = 5 × 10–23), sulfotransferase 1A1 (ST1A1; q = 9 × 10–23), and tumor necrosis factor superfamily member 14 (TNFSF14; q = 2 × 10–22), which all were found in higher levels in UC as compared with non-UC subjects (Fig. 1C). The discrimination between UC and non-UC subjects was confirmed in a heat map analysis (Supplementary Fig. 2). The aforementioned identified proteins were analyzed separately and divided for the different study groups, and almost total discrimination between HS and IBS vs the UC subgroups was revealed (Fig. 2A–D).
FIGURE 1.
Systemic inflammatory protein profile in healthy subjects, patients with IBS, and patients with UC. Serum proteins related to inflammation were analyzed by proximity extension assay and are presented in arbitrary unit NPX. A, Principle component analysis including all 77 proteins analyzed for HS (n = 19, yellow), IBS (n = 37, red), UC with active disease (n = 40, gray), UC in remission without IBS-like symptoms (n = 44, green), and UC in remission with IBS-like symptoms (n = 19, purple). Lines connect each group to a centroid that shows the combined mean of the group. B, Score scatter plot and (C) loading column plot from an OPLS-DA based on all 77 proteins analyzed in relation to non-UC (HS and IBS, n = 56, gray) and UC (all UC patients irrespective of disease activity, n = 103, white) as y-variables. R2Y defines the goodness of fit, and Q2 the goodness of prediction. The Mann-Whitney U test and false discovery rate analysis were used to compare the data in (C): *q < 0.05; **q < 0.01; ***q < 0.001; and ****q < 0.0001.
FIGURE 2.
Serum levels of CASP8, AXIN1, ST1A1, and TNFSF14 in healthy subjects, IBS patients, and UC patients. Protein expression in serum was analyzed by proximity extension assay and is presented in arbitrary unit NPX. Serum protein levels of CASP8 (A), AXIN1 (B), ST1A1 (C), and TNFSF14 (D) in HS (n = 19), IBS (n = 37), UC with active disease (n = 40), UC in remission with IBS-like symptoms (n = 19), and UC in remission without IBS-like symptoms (n = 44). Statistical difference between groups was assessed by Kruskal-Wallis test followed by Dunn’s multiple comparisons test; only significant differences are shown: ***P < 0.001; ****P < 0.0001. Each symbol represents 1 individual, and horizontal lines indicate the median of the group.
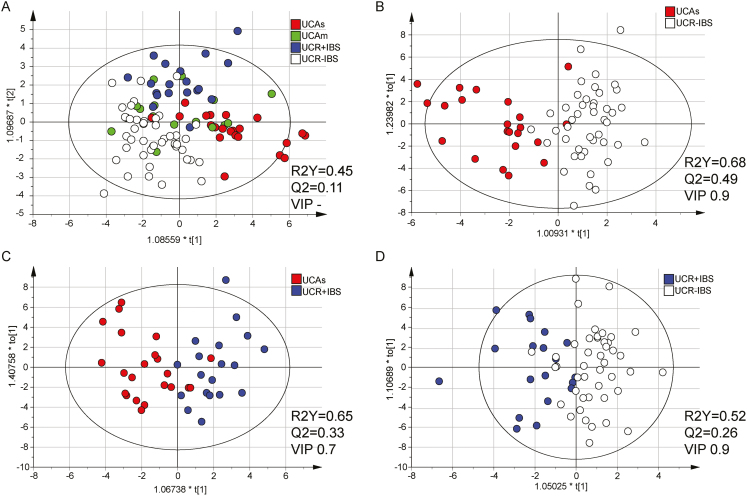
Systemic Inflammatory Protein Profiles Vary Between UC Patients With Different Disease Activities
For UC patients, we evaluated whether SIP profiles characterized inflammatory disease activity and/or presence/absence of IBS-like symptoms. As the SIP profile may differ depending on the severity of inflammation, the groups UCAs, UCAm, UCR + IBS, and UCR-IBS were compared in an OPLS-DA. Results showed weak separation between the groups, with the UCAm group scattered over the 3 other groups (Fig. 3A; Supplementary Fig. 3A). To reduce possible confounding effects from the UCAm group in discovering proteins important for inflammation and IBS-like symptoms, respectively, UCAm patients were excluded from further analyses.
FIGURE 3.
Systemic inflammatory protein profiles in patients with UC. Serum proteins related to inflammation were analyzed by proximity extension assay and are presented in arbitrary unit NPX. OPLS-DA analyses with serum proteins as x-variables and patient groups as y-variables are shown. Variable influence on projection cutoffs were used, as indicated in the figure, to select the number of x-variables resulting in the best model based on the R2Y and Q2 values. A, Score scatter plot including patients with UC with moderate/severe disease (Mayo score ≥6, n = 22, red), mild disease (Mayo score 3–5, n = 17, green), remission with IBS-like symptoms (n = 19, blue), and remission without IBS-like symptoms (n = 44, white) as y-variables. The analysis includes all 77 proteins, as no VIP was used. B, Score scatter plot of 36 proteins (VIP > 0.9) including patients with UC having severe active disease (Mayo score ≥6, n = 22, red) and UC in remission without IBS-like symptoms (n = 44, white) as y-variables. C, Score scatter plot of 53 proteins (VIP > 0.7) including patients with UC having severe active disease (Mayo score ≥6, n = 22) and UC in complete remission with IBS symptoms (n = 19, blue) as y-variables. D, Score scatter plot of 45 proteins (VIP > 0.9) including patients with UC in complete remission with IBS symptoms (n = 19, blue) and without IBS symptoms (n = 44, white) as y-variables. R2Y defines the goodness of fit, and Q2 the goodness of prediction. Based on Hotelling’s T2 Range Line and DModX DCrit, 1 outlier was excluded from the UCAm group.
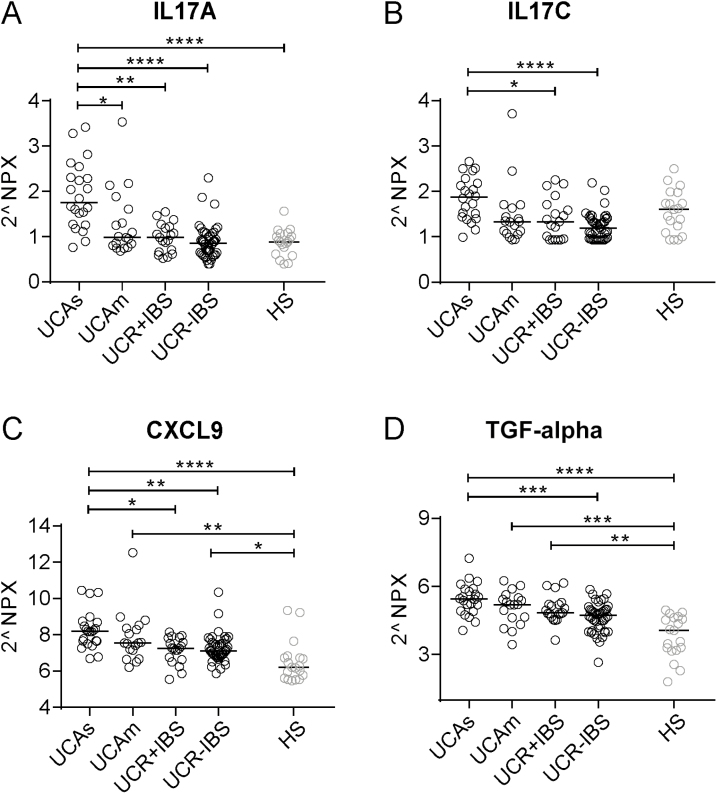
Next, the 3 groups, UCAs, UCR + IBS, and UCR-IBS, were analyzed pairwise. SIP profiles showed good separation between UCAs vs UCR-IBS (Fig. 3B) and UCAs vs UCR + IBS (Fig. 3C), whereas only weak separation was detected between UCR + IBS vs UCR-IBS (Fig. 3D). The loading scatter plots defined IL17A, IL17C, CXCL9, and TGF-α as the best proteins to characterize active inflammation, whereas no specific proteins characterized UCR + IBS or UCR-IBS (Supplementary Fig. 3B–D). The proteins IL17A, IL17C, CXCL9, and TGF-α were then analyzed separately and divided for all 4 UC study groups together with HS as the reference; the results revealed that IL17A and IL17C increased with increasing inflammatory activity (Fig. 4A, B). CXCL9 was higher in UCAs vs UCR + IBS and UCR-IBS (Fig. 4C), whereas TGF-α was higher in UCAs vs UCR-IBS (Fig. 4D). The serum levels of both CXCL9 and TGF-α were higher among most UC patients as compared with HS (Fig. 4C, D). In summary, the SIP profiles of UC patients differentiated active inflammatory disease from remission, whereas only minor differences were detected between UCR + IBS and UCR-IBS.
FIGURE 4.
Serum levels of IL17A, IL17C, CXCL9, and TGF-α patients with UC. Protein expression in serum was analyzed by proximity extension assay and is presented in arbitrary unit NPX. Serum protein levels of IL17A (A), IL17C (B), CXCL9 (C), and TGF-α (D) in patients with UC with moderate/severe disease (Mayo score ≥6, n = 22), mild disease (Mayo score 3–5, n = 18), complete remission with IBS-like symptoms (n = 19), complete remission without IBS-like symptoms (n = 44), and healthy subjects (n = 19). Statistical difference between groups was assessed by the Kruskal-Wallis test followed by Dunn’s multiple comparisons test; only significant differences are shown: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Each symbol represents 1 individual, and horizontal lines indicate the median of the group.
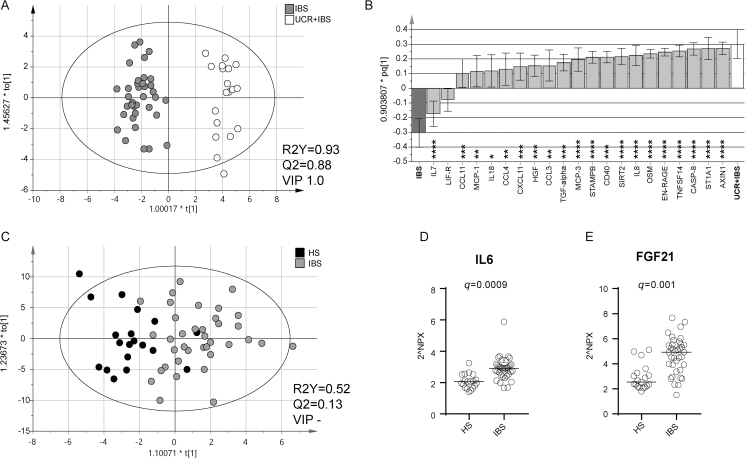
IBS Patients Deviate From UCR + IBS and HS by Their Systemic Inflammatory Protein Profiles
As already shown in the PCA (Fig. 1A), SIP profiles from non-UC subjects, comprising IBS and HS, deviated from patients with UC irrespective of disease activity. Next, an OPLS-DA analysis was performed to assess SIP profile similarities or discrepancies between IBS and UCR + IBS patients. The analysis resulted in a robust and highly predictive model, again identifying CASP8, AXIN1, ST1A1, and TNFSF14 as the proteins most important for separation between the groups (Fig. 5A, B). When comparing IBS with the other UC subgroups, similar models were obtained, IBS vs UCA (R2Y, 0.84; Q2, 0.80; VIP 1.0) and IBS vs UCR-IBS (R2Y, 0.93; Q2, 0.88; VIP 1.0), and CASP8, AXIN1, ST1A1, and TNFSF14 were identified as the most important variables. When evaluating differences between IBS and HS, the SIP profiles generated a weak model, with only moderate differences between the groups (Fig. 5C; Supplementary Fig. 4). Nevertheless, when analyzed separately, (FGF)-21 and IL6 were higher in IBS as compared with HS (Fig. 5D, E).
FIGURE 5.
Systemic inflammatory protein profiles in healthy subjects, patients with IBS, and patients with UC in remission with IBS-like symptoms. Protein expression in serum was analyzed by proximity extension assay and is presented in arbitrary unit NPX. A, Score scatter plot and (B) loading column plot from an OPLS-DA based on 21 serum proteins (VIP > 0.9) analyzed in relation to patients with IBS (n = 37, gray) and UCR + IBS (n = 19, white) as y-variables. C, Score scatter plot from an OPLS-DA based on all 77 serum proteins analyzed in relation to patients with IBS (n = 37, gray) and HS (n = 19, white) as y-variables. R2Y defines the goodness of fit, and Q2 the goodness of prediction. Serum protein levels of IL6 (D) and FGF21 (E) in IBS (n = 37) and HS (n = 19). Statistical difference between groups was assessed by the Mann-Whitney U test, followed by false discovery rate analysis including all the 77 proteins, and is shown as q values. Each symbol represents 1 individual, and horizontal lines indicate the median of the group.
Validation of Systemic Inflammatory Protein Profiles in Discrimination Between UC and IBS
Finally, the strong and highly predictive multivariate models generated between IBS and UC in the exploratory cohort described above were tested using a validation cohort consisting of IBS patients (n = 14), UCA patients (n = 15), and UCR + IBS patients (n = 9) (Table 2). To diminish the number of x-variables used, models based on the 4 variables shown to be most important for the separation between the groups (CASP8, AXIN1, ST1A1, and TNFSF14) were generated for IBS vs UCAs (R2Y, 0.85; Q2, 0.84) and IBS vs UCR + IBS (R2Y, 0.85; Q2, 0.83). Patients from the validation cohort were then tested into these models. The models correctly classified 97% of the IBS and UCA patients (Table 3A) and 100% of the IBS and UCR + IBS patients (Table 3B) in the respective models. Finally, UCA and UCR + IBS patients from the validation cohort were tested in the best model derived between UCAs vs UCR + IBS (R2Y, 0.65; Q2, 0.33; VIP 0.7) (model shown in Fig. 3C). Here, 71% of the patients were correctly classified (Table 3C).
TABLE 2.
Demographic Description of Study Subjects in the Validation Cohort
| IBS (n = 14) | UCR + IBS (n = 9) | UCA (n = 15) | |
|---|---|---|---|
| Male/female | 5/9 | 7/2 | 9/6 |
| Age, median (range), y | 28.5 (20–52) | 41 (32–69) | 48 (31–67) |
| Mayo score, low/intermediate/higha | NA | 7/2/0 | 0/6/9 |
| IBS subgroup IBS-D/IBS-C/IBS-Mb | 6/0/8 | 4/0/5 | NA |
| Calprotectin, median (range), μg/g | 18 (<15–120) | *d | 551 (34–3519) |
| Current treatment | |||
| None | 14 | 1 | 5 |
| 5-aminosalicylic acid | 0 | 7 | 10 |
| Thiopurine | 0 | 1 | 0 |
| Anti-TNF | 0 | 0 | 0 |
| Systemic steroids | 0 | 0 | 0 |
| Disease extent | |||
| Proctitis | NA | 1 | 2 |
| Left-sided colitis | NA | 3 | 5 |
| Extensive colitis | NA | 5 | 8 |
| IBS-SSS, moderate/severec | 10/4 | *d | NA |
aMayo score, disease activity index for UC, low ≤2 pts, intermediate 3–5 pts, high ≥6 pts.
bSubclassification of IBS, predominant diarrhea (IBS-D), predominant constipation (IBS-C), predominant mixed symptoms (IBS-M).
cIBS-SSS (IBS–severity score system), moderate >175 pts, severe >300 pts.
dNonavailable patient information.
TABLE 3.
Misclassification Table for Patients in the Validation Cohort Tested in Multivariate Models Generated From the Exploratory Cohort
| Model | Patients Tested | Members, No. | Correctly Classified, % | Classification Details | |
|---|---|---|---|---|---|
| a: IBS vs UCAs | IBS | UCA | |||
| R2Y: 0.85 | IBS | 14 | 92.86 | 13 | 1 |
| Q2: 0.84 | UCA | 15 | 100 | 0 | 15 |
| x-variables: 4 | Total | 29 | 96.55 | 13 | 16 |
| Fisher’s proba: 2.1 × 10–7 | |||||
| b: IBS vs UCR + IBS | IBS | UCR + IBS | |||
| R2Y: 0.85 | IBS | 14 | 100 | 14 | 0 |
| Q2: 0.83 | UCR + IBS | 9 | 100 | 0 | 9 |
| x-variables: 4 | Total | 23 | 100 | 14 | 9 |
| Fisher’s prob: 1.2 × 10–6 | |||||
| c: UCR + IBS vs UCAs | UCA | UCR + IBS | |||
| R2Y: 0.60 | UCR + IBS | 9 | 44.44 | 5 | 4 |
| Q2: 0.27 | UCA | 15 | 93.33 | 14 | 1 |
| x-variables: 36 | Total | 24 | 75 | 19 | 5 |
| Fisher’s prob: 0.047 |
aFisher’s exact probability test.
DISCUSSION
Characterizing disease-specific immune profiles of UC patients, with and without inflammation and IBS-like symptoms, and IBS patients may help to disentangle the underlying mechanisms possibly linking inflammation and IBS symptoms in these patient groups. In this exploratory study, we demonstrate that SIP profiles discriminate healthy subjects and patients with IBS from patients with UC. Furthermore, we show that SIP profiles vary between UC patients with different inflammatory activities. Additionally, IBS patients deviate from UC patients with IBS-like symptoms and healthy subjects by their SIP profiles. Thus, our data do not support shared inflammatory mechanisms of UC and IBS, or that symptoms of patients with UCR + IBS and IBS, respectively, are driven by similar mechanisms.
When comparing UC patients, with and without active inflammation, with non-UC subjects, an almost perfect separation between the 2 groups was achieved based on the overall SIP profiles, suggesting that different inflammatory mechanisms are driving UC and IBS. The proteins most important for the separation between the 2 groups, CASP8, AXIN1, ST1A1, and TNFSF14 also correctly classified at least 97% of the patients when examining UC and IBS patients in the validation cohort. Thus, only 4 serum proteins may be used to identify and separate UC patients, irrespective of disease activity, from IBS patients and healthy subjects. Among the identified proteins, TNFSF14, CASP8, and AXIN1 have previously been directly or indirectly linked to IBD. Increased expression of TNFSF14 mRNA has been reported in the inflamed intestinal mucosa of IBD patients,19 inducing barrier dysfunction,20 and CASP8 was suggested to be involved in mucosal inflammation by regulating necroptosis of Paneth cells and intestinal epithelial cells of patients with CD.21 The cytoplasmic protein AXIN1 has been described to facilitate phosphorylation and transcriptional activity of Smad3 in the transforming growth factor β (TGFβ) signaling pathway.22 In turn, Smad3 signaling is involved in gene regulation and has been demonstrated to be important for induction of Th9 and Th17 cells, which are associated with inflammation in IBD patients.23–25 In contrast, there are no previous reports of the enzyme ST1A1 being linked to the pathology of IBD. Nevertheless, ST1A1, regulating the activities of endogenous metabolites and neurotransmitters, has been shown to positively correlate with disease activity in other inflammatory conditions.26 Thus, irrespective of inflammatory activity, the SIP profiles including the abovementioned proteins are shared among UC patients, suggesting that these proteins reflect underlying disease mechanisms unrelated to inflammatory status. When examining UC patients in more detail, the SIP profiles distinguished between UC patients with different inflammatory activities and between patients with and without IBS-like symptoms. Ulcerative colitis patients with mild disease (UCAm) were the most difficult to differentiate from the other groups and shared some features regarding SIP profiles with UCAs and UCR, which was not surprising. Patients with active disease were characterized by increased levels of IL17A, IL17C, and CXCL9, all proteins well known to be involved in inflammatory processes, and TGFα, which is a growth factor important for cell proliferation and differentiation.27 These proteins, together with other proteins in the SIP profile, correctly classified 75% of patients when comparing UCA and UCR + IBS patients in the validation cohort. The good separation between the patient groups based on SIP profiles and the marked lower levels of the abovementioned proteins in UCR + IBS imply that IBS-like symptoms of UC patients in remission are not driven by the same inflammatory events as those of UC patients with active disease.
The SIP profiles characterizing UC patients in remission with and without IBS-like symptoms differed, although the model was rather weak, and no unique proteins in this panel were found to differ significantly between the patient groups. Thus, there was no evidence of any major differences in the SIP immune profiles of the groups, suggesting that low-grade inflammatory events may not be the main driving factor of the presence of IBS-like symptoms among UC patients in remission. Nevertheless, increased visceral hypersensitivity,28 along with intestinal permeability and low-grade inflammation,29 has been reported in IBD patients in remission with IBS-like symptoms as compared with IBD patients in remission without these symptoms.28 To determine if local events such as low-grade inflammation, impaired barrier integrity, or neurobiological processes drive symptoms among the UCR + IBS group, broad analysis panels exploring these parameters could be of use.
Although the differentiation between UC patients in remission with and without IBS-like symptoms was unclear, the SIP profiles of UCR + IBS and IBS patients differed substantially, and, again, the model was driven by the proteins CASP8, AXIN1, ST1A1, and TNFSF14. Thus, these proteins are present at higher concentrations in the circulation of UCR + IBS patients than in IBS patients, suggesting that the 2 patient groups are part of different mechanistic spectrums, although they present with similar symptoms.
Comparing IBS patients and healthy controls, the SIP profiles generated a weak model with only moderate differences in their SIP profiles, although higher FGF-21 and IL6 concentrations were demonstrated in IBS patients. The pro-inflammatory cytokine IL6 has repeatedly been reported to be increased in the serum of IBS patients,30, 31 especially in diarrhea-predominant IBS. However, increased serum levels of FGF-21, important for endocrine metabolic regulation and lipid homeostasis,32 have not been demonstrated previously. Recently, it was shown that dextran sulfate sodium–mediated experimental colitis induces white adipose tissue lipolysis via an IL6/FGF21-mediated signaling pathway.33 Thus, the increased levels of circulating FGF-21 and IL6 in IBS patients may reflect an altered lipolysis due to a low-grade inflammation.
A weakness of our study is that the method used to measure serum proteins, proximity extension immunoassay, is based on relative quantification, and therefore no comparison of absolute levels can be made between different proteins or with other studies where traditional ELISAs have been used. Unfortunately, despite being a highly sensitive assay, several cytokines such as IL4, IL13, and TNF were not detectable in >40% of the study population, with no difference in detection rate between patient groups, and were therefore not included in further analyses. Another possible limitation is that a substantial number of the UC patients were on treatment potentially regulating immune activity, which may have affected the serum levels of inflammatory proteins. It was also not possible to measure fecal calprotectin in the UCR + IBS group in the validation cohort, which may have affected patient selection, even though colonoscopy was performed to confirm remission. The IBS patient group consisted of only IBS-M and IBS-D patients, which reflects our attempt to compare these IBS subgroups with UC, as the patient groups may overlap regarding symptom profile in a clinical setting. Also, only the systemic and not the intestinal immune protein profiles were determined, and as systemic profiles are not a complete mirror of the mucosal events, this still needs to be investigated. Finally, there were age and sex differences between the groups that may have influenced the data. Despite these limitations, this was an exploratory study investigating possible mechanistic differences for a better understanding of these conditions, and no previous study has analyzed such a wide range of systemic inflammatory proteins characterizing UC and IBS patients. Additionally, the confirmation of our findings in a validation cohort is an important strength of our study.
In summary, this study demonstrates that SIP profiles differentiate between UC patients, irrespective of the presence of inflammation or IBS-like symptoms, and IBS patients. Thus, our data suggest that the SIP profiles reflect different underlying inflammatory mechanisms of UC and IBS. Moreover, SIP profiles differentiate UC patients with active disease and UC patients in remission. Potentially, the SIP profiles found to characterize the different patient groups in our study may enable better understanding of the mechanisms driving the diseases and be a source for development of future diagnostic biomarkers.
Supplementary Material
Supported by: The Swedish Medical Research Council (VR-M), the Healthcare Committee Region Västra Götaland, the Agreement Concerning Research and Education of Doctors (ALF) in Region Västra Götaland, the Ruth and Richard Julin Foundation, the Swedish Society of Medicine, the Wilhelm and Martina Lundgren Fund, the Adlerbertska Foundations, the Consul Thure Carlsson Memorial Foundation, and the O.E. and Edla Johansson Scientific Foundations provided funds in the form of grants for this study.
Disclaimer: This was an independent study; the views expressed in the submitted article are our own and not an official position of the institution or funder.
Conflicts of interest: The authors declare no conflicts of interest.
Author contributions: Guarantor of the article: Lena Öhman. Specific author contributions: L.M.H. contributed to study concept and design, sample preparation, acquisition, analysis and interpretation of data, statistical analysis, and drafting and writing the manuscript; M.M. contributed to study concept and design, acquisition, analysis, and interpretation of data, statistical analysis, study supervision, and drafting and writing the manuscript; G.M., B.J., H.T., J.S., M.S., and H.S. contributed to data and sample collection and critical revision of the manuscript; A.P. contributed to technical support, statistical analysis of the data, and critical revision of the manuscript; L.Ö. contributed to obtaining funding, study concept and design, acquisition, analysis, and interpretation of data, study supervision, and drafting and writing the manuscript. All authors have agreed to be accountable for all aspects of the work and approved the final version of the manuscript.
REFERENCES
- 1. Keohane J, O’Mahony C, O’Mahony L, et al. Irritable bowel syndrome-type symptoms in patients with inflammatory bowel disease: a real association or reflection of occult inflammation? Am J Gastroenterol. 2010;105:1788, 1789–1794; quiz 1795. [DOI] [PubMed] [Google Scholar]
- 2. Bennet SMP, Sundin J, Magnusson MK, et al. Altered intestinal antibacterial gene expression response profile in irritable bowel syndrome is linked to bacterial composition and immune activation. Neurogastroenterol Motil. 2018;30:e13468. [DOI] [PubMed] [Google Scholar]
- 3. Spiller R, Major G. IBS and IBD - separate entities or on a spectrum? Nat Rev Gastroenterol Hepatol. 2016;13:613–621. [DOI] [PubMed] [Google Scholar]
- 4. Bennet SM, Polster A, Törnblom H, et al. Global cytokine profiles and association with clinical characteristics in patients with irritable bowel syndrome. Am J Gastroenterol. 2016;111:1165–1176. [DOI] [PubMed] [Google Scholar]
- 5. Bennet SMP, Palsson O, Whitehead WE, et al. Systemic cytokines are elevated in a subset of patients with irritable bowel syndrome but largely unrelated to symptom characteristics. Neurogastroenterol Motil. 2018;30:e13378. [DOI] [PubMed] [Google Scholar]
- 6. Seyedmirzaee S, Hayatbakhsh MM, Ahmadi B, et al. Serum immune biomarkers in irritable bowel syndrome. Clin Res Hepatol Gastroenterol. 2016;40:631–637. [DOI] [PubMed] [Google Scholar]
- 7. Scully P, McKernan DP, Keohane J, et al. Plasma cytokine profiles in females with irritable bowel syndrome and extra-intestinal co-morbidity. Am J Gastroenterol. 2010;105:2235–2243. [DOI] [PubMed] [Google Scholar]
- 8. Jonefjäll B, Öhman L, Simrén M, et al. IBS-like symptoms in patients with ulcerative colitis in deep remission are associated with increased levels of serum cytokines and poor psychological well-being. Inflamm Bowel Dis. 2016;22:2630–2640. [DOI] [PubMed] [Google Scholar]
- 9. Andersson E, Bergemalm D, Kruse R, et al. Subphenotypes of inflammatory bowel disease are characterized by specific serum protein profiles. PLoS One. 2017;12:e0186142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Korolkova OY, Myers JN, Pellom ST, et al. Characterization of serum cytokine profile in predominantly colonic inflammatory bowel disease to delineate ulcerative and Crohn’s colitides. Clin Med Insights Gastroenterol. 2015;8:29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology. 2006;130:1480–1491. [DOI] [PubMed] [Google Scholar]
- 12. Schroeder KW, Tremaine WJ, Ilstrup DM. Coated oral 5-aminosalicylic acid therapy for mildly to moderately active ulcerative colitis. A randomized study. N Engl J Med. 1987;317:1625–1629. [DOI] [PubMed] [Google Scholar]
- 13. Francis CY, Morris J, Whorwell PJ. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment Pharmacol Ther. 1997;11:395–402. [DOI] [PubMed] [Google Scholar]
- 14. Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Gut. 1997;41:A122–A123. [DOI] [PubMed] [Google Scholar]
- 15. Assarsson E, Lundberg M, Holmquist G, et al. Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9:e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Team RDC. R: A Language and Environment for Statistical Computing. Vol. 2018 Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 17. Utmetrics. SIMCA-P and Multivariate Analysis FAQ. Vol. 2019 Umeå, Sweden: Utmetrics Inc.; 2006. [Google Scholar]
- 18. Eriksson L, Kettaneh-Wold N, Trygg J, et al. Multi‐and Megavariate Data Analysis: Part I: Basic Principles and Applications. Umeå, Sweden: Utmetrics Inc.; 2006. [Google Scholar]
- 19. Cohavy O, Zhou J, Ware CF, Targan SR. LIGHT is constitutively expressed on T and NK cells in the human gut and can be induced by CD2-mediated signaling. J Immunol. 2005;174:646–653. [DOI] [PubMed] [Google Scholar]
- 20. Clayburgh DR, Musch MW, Leitges M, et al. Coordinated epithelial NHE3 inhibition and barrier dysfunction are required for TNF-mediated diarrhea in vivo. J Clin Invest. 2006;116:2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Becker C, Watson AJ, Neurath MF. Complex roles of caspases in the pathogenesis of inflammatory bowel disease. Gastroenterology. 2013;144:283–293. [DOI] [PubMed] [Google Scholar]
- 22. Salahshor S, Woodgett JR. The links between axin and carcinogenesis. J Clin Pathol. 2005;58:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elyaman W, Bassil R, Bradshaw EM, et al. Notch receptors and Smad3 signaling cooperate in the induction of interleukin-9-producing T cells. Immunity. 2012;36:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shohan M, Sabzevary-Ghahfarokhi M, Bagheri N, et al. Intensified Th9 response is associated with the immunopathogenesis of active ulcerative colitis. Immunol Invest. 2018;47:700–711. [DOI] [PubMed] [Google Scholar]
- 25. Martinez GJ, Zhang Z, Chung Y, et al. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J Biol Chem. 2009;284:35283–35286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guillot F, Garcia A, Salou M, et al. Transcript analysis of laser capture microdissected white matter astrocytes and higher phenol sulfotransferase 1A1 expression during autoimmune neuroinflammation. J Neuroinflammation. 2015;12:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kumar V, Bustin SA, McKay IA. Transforming growth factor alpha. Cell Biol Int. 1995;19:373–388. [DOI] [PubMed] [Google Scholar]
- 28. Akbar A, Yiangou Y, Facer P, et al. Expression of the TRPV1 receptor differs in quiescent inflammatory bowel disease with or without abdominal pain. Gut. 2010;59:767–774. [DOI] [PubMed] [Google Scholar]
- 29. Vivinus-Nebot M, Frin G, Bzioueche H, et al. Functional bowel symptoms in quiescent inflammatory bowel diseases: role of epithelial barrier disruption and low grade inflammation. Gastroenterology. 2013;144:S538–S538. [DOI] [PubMed] [Google Scholar]
- 30. Bennet SMP, Palsson O, Whitehead WE, et al. Systemic cytokines are elevated in a subset of patients with irritable bowel syndrome but largely unrelated to symptom characteristics. Neurogastroenterol Motil. 2018;30:e13378. [DOI] [PubMed] [Google Scholar]
- 31. Bashashati M, Moradi M, Sarosiek I. Interleukin-6 in irritable bowel syndrome: a systematic review and meta-analysis of IL-6 (-G174C) and circulating IL-6 levels. Cytokine. 2017;99:132–138. [DOI] [PubMed] [Google Scholar]
- 32. Woo YC, Xu A, Wang Y, et al. Fibroblast growth factor 21 as an emerging metabolic regulator: clinical perspectives. Clin Endocrinol (Oxf). 2013;78:489–496. [DOI] [PubMed] [Google Scholar]
- 33. Liu L, Zhao C, Yang Y, et al. Fibroblast growth factor 21 deficiency attenuates experimental colitis-induced adipose tissue lipolysis. Gastroenterol Res Pract. 2017;2017:3089378. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.