Abstract
Background
MicroRNAs (miRNAs) play an important role in the development and progression of breast cancer (BC). The purpose of the present study was to identify plasma miRNAs enabling early diagnosis of BC.
Materials and Methods
Expression levels of seven plasma miRNAs (miR‐23a‐3p, miR‐29b‐2‐5p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, miR‐152‐3p, and miR‐182‐5p) in 106 patients with newly diagnosed BC and 96 healthy participants were analyzed by qRT‐PCR. We also evaluated the relationship between the expression levels of these miRNAs and clinicopathological features of patients with BC.
Results
Compared with healthy controls, we found that miR‐23a‐3p (p = .025), miR‐130a‐5p (p = .006), miR‐144‐3p (p = .040), miR‐148a‐3p (p = .023), and miR‐152‐3p (p = .019) were downregulated in the plasma of patients with BC. MiR‐130a‐5p, miR‐144‐3p, and miR‐152‐3p were downexpressed in BC tissues as well as plasma. The expression of the miR‐23a‐3p, miR‐144‐3p, and miR‐152‐3p was related to ER positive and PR positive. Besides, miR‐23a‐3p, miR‐144‐3p, and miR‐152‐3p did show the significant difference in the staging compromised to the control, especially in stage I‐II. Moreover, we also found that miR‐144‐3p and miR‐148a‐3p were associated with lymph node invasion.
Conclusions
The expression levels of the miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p were lower in patients with BC compared to healthy controls and were associated with ex hormone receptor, clinical stage, and lymph node metastasis, indicating the diagnostic potential of these miRNAs in BC.
Keywords: biomarker, breast cancer, miRNAs, plasma
We identified significant reduction of miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p in the plasma of the patients compared with the controls. MiR‐130a‐5p, miR‐144‐3p, and miR‐152‐3p were downexpressed in BC tissues as well as plasma in silico analysis. The relative expression of these five miRNAs was closely associated with the clinicopathologic features of the BC such as the expression of ER, PR, and HER2, histological tumor grades, and lymph node metastasis.
1. INTRODUCTION
Breast cancer (BC) is the most commonly diagnosed cancer and the leading cause of cancer death among females (Bray et al., 2018). In China, there were about 268,600 newly diagnosed female BC cases, accounting for 15% of all new cancers in women, and 69,500 deaths in 2015(Chen et al., 2016). Although the mortality rates have continued to decrease over the years due to the advances made in early diagnosis and treatment, tens of thousands of women die from this disease each year. Early detection of BC is one of the main prerequisites for successful treatment and reduction in mortality from this disease (Coleman, 2017). Moreover, it also plays a critical role in the diagnosis of BC, monitoring of the disease progression and response to treatment (Nattinger & Mitchell, 2016). Despite the fact that mammography, ultrasonography, and magnetic resonance imaging diagnosis for BC are the currently used noninvasive screening tool, there are some concerns about the high rates of misdiagnosis, missed diagnoses, and overdiagnosis of this malignancy (Bleyer & Welch, 2012). Thus, there is a need for the development of novel noninvasive biomarkers to improve the early diagnosis of malignant breast lesions. Available evidence suggests that serum/plasma miRNA levels may be used as noninvasive biomarkers for early BC diagnosis (Hamam et al., 2017).
MicroRNAs (miRNAs) are a class of small, approximately 19–25 nucleotides of noncoding RNAs that regulate gene expression at post‐transcriptional level (Bartel, 2004). Evidence suggests that miRNA is associated with the development and progression in a number of human diseases, including cancer (Cai et al., 2017; Ding et al., 2017; Li et al., 2013). MiRNAs are involved in multiple biological processes of BC, such as differentiation, proliferation, apoptosis, development, and tumorigenesis (Yahya & Elsayed, 2015). MiRNA expression may be used as potential biomarkers for the diagnosis, prognosis, and therapy. Nevertheless, most of the studies describe the miRNA expression in cell lines and primary tumor tissue to date. Recently, several reports have proved that circulating miRNAs derived from epithelial tumors were deregulated in different types of diseases, including cancer (Cheng, 2015). The distinct biological properties of the serum/plasma miRNAs including the remarkable stability, availability for rapid and accurate quantification, the possibility of their repeated measurement, and a direct link with disease states make them attractive candidates for the development of novel markers (McDonald, Milosevic, Reddi, Grebe, & Algeciras‐Schimnich, 2011).
Recently, several reports have shown the dysregulated expression of miR‐23a‐3p, miR‐29b‐2‐5p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, miR‐152‐3p, and miR‐182‐5p in tumor tissue was associated with the development of BC (Chen, Cheng, Chen, Chen, & Shen, 2017; Gu et al., 2018; Kong, Zhang, Li, Shao, & Fang, 2018). However, there have been a few reports on the role of plasma miRNAs in BC. Here, we investigated and compared the expression of these seven plasma miRNAs in patients with BC and healthy controls. Moreover, potential relationships between these plasma miRNAs level and existing clinicopathological features of BC, such as hormone receptor status, stage, clinical T, and lymph node invasion, were statistically analyzed. Our data showed that miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p were significantly downregulated in plasma of patient with BC and could be served as the potential noninvasive molecular biomarkers for the early detection of BC.
2. SUBJECTS AND METHODS
2.1. Study subjects and sample collection
The study consisted 106 patients with BC and 96 healthy age‐matched volunteers from the First Affiliated Hospital of Xi'an Jiaotong University. All participants were genetically unrelated ethnic Han Chinese females. Patients with BC were newly diagnosed and histopathologically confirmed by biopsy and pathology. No patients had undergone chemotherapy or radiotherapy, surgical therapy, or systemic therapy before sample collection. The healthy controls had no history of cancer, no family history of BC, and no endocrine or reproductive system diseases by physical examination. Clinical characteristics information was collected through interviewer‐administered questionnaires and/or medical records.
Five milliliters of blood sample from each participant was collected into EDTA tube for miRNA analysis. Plasma was subsequently isolated by centrifugation at 3,000 rpm for 10 min at 4°C for the isolation of RNA. This study was approved by the institutional ethics committees of the First Affiliated Hospital of Xi'an Jiaotong University. All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration. Written informed consent was obtained from all individuals who participated in the study.
2.2. Plasma RNA extraction and miRNA qRT‐PCR
Total RNA (including miRNAs) was extracted from plasma using the miRcute Serum/plasma miRNA isolation kit (cat. no. DP501; Tiangen) following the manufacturer's instructions. After washing miRNAs were eluted in 30 μl of RNase‐free ddH2O. Quality of RNA was assessed by NanoDrop 2000C (Thermo Scientific). Chromatographic characteristics and integrity of all RNA samples were determined by RNA 6000 Nano LabChip (Agilent Technologies) and Agilent 2100 Bioanalyzer system (Agilent Technologies). Then, RNA samples were converted into cDNA immediately.
The reverse transcription (RT) reactions of U6 and miRNAs (miR‐23a‐3p, miR‐29b‐2‐5p, miR‐130a‐5p miR‐144‐3p, miR‐148a‐3p, miR‐152‐3p, and miR‐182‐5p) were performed by the miRcute Plus miRNA First‐Strand cDNA Synthesis Kit (cat. no. KR211; Tiangen) with Oligo (dT)‐Universal Tag primer. The synthesis reaction consisted of 10 μl 2 × miRNA RT reaction buffer, 2 μl miRNA RT enzyme Mix, 0.6–2 μg total RNA, and RNase‐free ddH2O up to 20 μl. The synthesis reaction conditions were 42°C for 60 min and 95°C for 3 min, and hold at 4°C. After the reaction, the product was diluted with 200 μl RNase‐free water and kept at −20°C until further use.
SYBR Green‐based qPCR profiling was performed using miRcute Plus miRNA qPCR Detection Kit (cat. no. FP411; Tiangen) in ABI 7500 Real‐Time PCR System (Applied Biosystems) according to the manufacturer's instructions. The miRNA‐specific forward primers sequences were designed based on the miRNA sequences obtained from the miRBase database (Table S1). The amplification reaction system consisted of 10 μl 2 × miRcute plus miRNA premix (with SYBR&ROX), 0.4 μl forward primer (10 μM), 0.4 μl universal reverse primer (10 μM), 2 μl cDNA, and RNase‐free ddH2O up to 20 μl. The reaction was carried out at 95°C for 15 min, followed by 40 cycles at 94°C for 20 s and 60°C for 34 s, and hold at 4°C. Triplicate wells were measured for each sample. RT reactions and SYBR Green Real‐Time PCR were carried out in a blinded manner. Appropriate negative controls (ddH2O or no template control) were used in both cDNA synthesis and RT‐qPCR reactions. The levels of miRNAs were normalized using U6 as reference RNA. The relative expression quantity (RQ) of miRNA was calculated as RQ = 2−ΔΔ Ct. ΔΔCt = mean value of the study group (Ct miRNA − Ct U6) – mean value of the control group (Ct miRNA − Ct U6).
2.3. Statistical analyses
All analyses were performed with SPSS 18.0 software (SPSS Institute). Baseline characteristics were presented as mean ± standard deviation (SD) for continuous data and as number (percentages) for categorical parameters. Independent samples t test was used to evaluate the age distribution. Plasma miRNA levels between multiple groups were compared using the independent samples t test. The Spearman correlation coefficient was used for correlation analysis of expression levels of these miRNAs. The median expression level of miRNAs was used as the cutoff point to divide patients with BC into low and high expression groups. A chi‐square test and one‐way ANOVA were used to assess the association between miRNA levels and clinicopathological characteristics between cases and controls. Diagnostic accuracy of candidate miRNAs or their combinations was assessed by receiver operating characteristic (ROC) curves analysis, and the area under the ROC curve (AUC) was also calculated. All tests were two‐sided test and p values less than .05 was considered to be statistically significant. Statistical significance was established at *p < .05 or **p < .01.
2.4. TCGA data
Due to the purpose of relatively early diagnosis in this study and plasma collected precancer therapy, tumor tissue was not available from patients recruited for this study. In addition, plasma collected from all patients enrolled in this study occurred prior to chemotherapy administration and/or tumor biopsy. Therefore, in order to compare differences in miRNA expression levels in tumor tissue versus plasma, publicly available data from the TCGA (Cancer Genome Atlas Network, 2012) were used. The miRNA expression data of the cancer tissues from patients with BC were downloaded from TCGA (https://portal.gdc.cancer.gov/) in the form of data matrices documenting patterns of miRNA expression for 870 BC tissue samples and 78 normal tissues at age 20–75 years. The tissue level expression of miRNA (miRNA‐seq), available as RPKM (reads per kilobase of transcript per million) values, was obtained using the Illumina HiSeq platform. The expression level of miRNA was normalized using Limma package of R language (Smyth, 2004).Then, we analyzed the expression trends of a given miRNA across tissue and plasma samples. In addition, the miRNA expression and prognostic significance in BC were evaluated using Kaplan‐Meier Plotter database (http://kmplot.com/analysis/index.php?p=service%26cancer=breast_mirna).
3. RESULTS
3.1. Characteristics of study subjects
A total of 202 women which included 106 patients (median age 53.19 ± 6.41 years) with BC and 96 healthy controls (median age 54.81 ± 8.38 years) were enrolled in the study. There were no significant differences in age between the patients with BC and the controls (p = .122). Characteristics of study subjects were summarized in Table 1, including age, hormone‐receptor‐positive status, HER2 status, stage, T classification, nodal status, and pathological subtypes.
Table 1.
Characteristics of patients with breast cancer and controls
| Variables | Case (n = 106) | Control (n = 96) | p | |||
|---|---|---|---|---|---|---|
| Number | % | Number | % | |||
| Age | Mean ± SD (year) | 53.19 ± 6.41 | 54.81 ± 8.38 | .122 | ||
| ER status | Positive | 66 | 62.3 | |||
| Negative | 31 | 29.2 | ||||
| Missing | 9 | 8.5 | ||||
| PR status | Positive | 57 | 53.8 | |||
| Negative | 39 | 36.8 | ||||
| Missing | 10 | 9.4 | ||||
| HER2 status | Positive | 30 | 28.3 | |||
| Negative | 49 | 46.2 | ||||
| Missing | 27 | 25.5 | ||||
| Stage | I‐II | 63 | 59.4 | |||
| III‐IV | 38 | 35.8 | ||||
| Missing | 5 | 4.7 | ||||
| Clinical T | 1–2 | 86 | 81.1 | |||
| 3–4 | 14 | 13.2 | ||||
| Missing | 6 | 5.6 | ||||
| Lymph node invasion | Yes | 55 | 51.9 | |||
| No | 46 | 43.4 | ||||
| Missing | 5 | 4.7 | ||||
| Pathological subtypes | Luminal B | 52 | 47.7 | |||
| Luminal A | 23 | 21.1 | ||||
| Her‐2 | 19 | 17.4 | ||||
| Triple negative | 10 | 9.2 | ||||
| Missing | 5 | 4.6 | ||||
Bold p values were calculated by independent samples T test.
Abbreviations: ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; RP, progesterone receptor.
3.2. Significantly differentiated plasma miRNAs between the controls and BC patients
We measured plasma miRNA levels of miR‐23a‐3p, miR‐29b‐2‐5p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, miR‐152‐3p, and miR‐182‐5p between the controls and patients with BC. We found that miR‐23a‐3p (p = .025), miR‐130a‐5p (p = .006), miR‐144‐3p (p = .040), miR‐148a‐3p (p = .023), and miR‐152‐3p (p = .019) were downregulated in the plasma of patients with BC when compared with healthy women (Table 2 and Figure 1).
Table 2.
Expression changes of plasma miRNA in BC cases and healthy controls
| miRNAs | Plasma | Tissue (TCGA) | ||||
|---|---|---|---|---|---|---|
| Controls (M ± SD) | BC patients (M ± SD) | p | Controls (M ± SD) | BC patients (M ± SD) | p | |
| miR‐23a‐3p | 65.37 ± 16.43 | 24.48 ± 7.35 | .025 * | 3,545.35 ± 127.80 | 3,476.15 ± 77.20 | .644 |
| miR‐130a‐5p | 200.62 ± 46.65 | 66.38 ± 12.32 | .006 ** | 134.48 ± 66.14 | 84.61 ± 21.77 | <.001 ** |
| miR‐144‐3p | 39.87 ± 10.36 | 16.54 ± 4.30 | .040 * | 613.89 ± 154.24 | 70.48 ± 28.67 | .001 ** |
| miR‐148a‐3p | 167.03 ± 49.49 | 50.90 ± 8.83 | .023 * | 30,384.03 ± 2,351.92 | 55,066.80 ± 1,650.86 | <.001 ** |
| miR‐152‐3p | 333.18 ± 94.81 | 100.13 ± 22.44 | .019 * | 293.70 ± 112.36 | 261.84 ± 161.79 | .023 * |
Abbreviations: BC, breast cancer; M, mean values; SD, standard deviation.
Bold indicates statistical significance of expression level with
p < .05,
p < .01.
Figure 1.
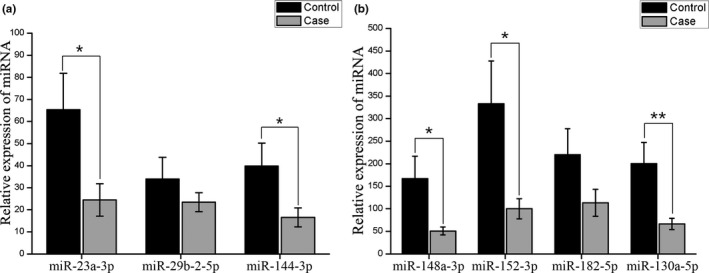
The expression of seven plasma miRNAs in breast cancer patients and healthy controls. Statistical significance of expression level with * for p < .05 and ** for p < .01
Next, tissue expression trends in the corresponding five miRNAs were measured (Table 2). MiRNA expression values in BC tissue (n = 870) and noncancerous tissue (n = 78) were obtained from publically available TCGA RNASeq data. Expression trends for some miRNAs (miR‐130a‐5p, miR‐144‐3p and miR‐152‐3p) were similar in both tissue and plasma. The expression of miR‐23a‐3p and miR‐148a‐3p in tissue and plasma levels showed a reversal of the trend. MiR‐148a‐3p was a significantly increased in BC tissue levels (TCGA) compared with nontumor tissue samples, and miR‐23a‐3p showed no significantly different.
In addition, the correlation analyses showed that these plasma miRNA expression levels had strong correlations, and their correlation coefficients were 0.315 (miR‐23a‐3p vs. miR‐144‐3p, p = .001), 0.584 (miR‐23a‐3p vs. miR‐148a‐3p, p < .001), 0.289 (miR‐23a‐3p vs. miR‐152‐3p, p = .003), 0.412 (miR‐130a‐5p vs. miR‐148a‐3p, p < .001), 0.391 (miR‐144‐3p vs. miR‐148a‐3p, p < .001), 0.671 (miR‐144‐3p vs. miR‐152‐3p, p < .001), and 0.481 (miR‐148a‐3p vs. miR‐152‐3p, p < .001), respectively (Table 3).
Table 3.
Coefficients of correlation among five miRNAs
| miRNAs | miR‐23a‐3p | miR‐130a‐5p | miR‐144‐3p | miR‐148a‐3p | miR‐152‐3p | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | |
| miR‐23a‐3p | 1.000 | — | ||||||||
| miR‐130a‐5p | 0.146 | .154 | 1.000 | — | ||||||
| miR‐144‐3p | 0.315 | .001 ** | 0.060 | .557 | 1.000 | — | ||||
| miR‐148a‐3p | 0.584 | <.001 ** | 0.412 | <.001 ** | 0.391 | <.001 ** | 1.000 | — | ||
| miR‐152‐3p | 0.289 | .003 ** | 0.185 | .068 | 0.671 | <.001 ** | 0.481 | <.001 ** | 1.000 | — |
Bold indicates statistical significance of expression level with
p < .05,
p < .01.
3.3. The diagnosis role of candidate miRNAs for BC
Receiver operating characteristics (ROC) curve was used to evaluate the ability of significantly differentiated plasma miRNAs to distinguish patients with BC from healthy controls (Figure 2). For combined plasma miRNA panel (miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p), the AUC was 0.699 with 95% CI: 0.622–0.775 and the sensitivity and specificity were 0.865 and 0.459, respectively.
Figure 2.
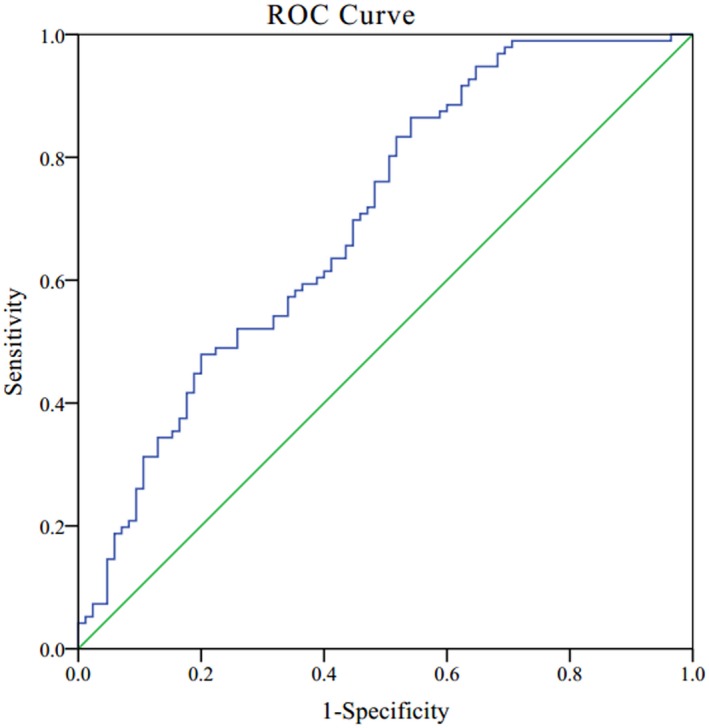
Receiver operating characteristics (ROC) curve of combined miRNA panel to differentiate patients with breast cancer from healthy controls
3.4. Correlation of plasma miRNA levels to the status of ER, PR and HER2
Moreover, we have stratified the patients to examine the associations between the plasma levels of the designated miRNAs and the status of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). We found that the miR‐23a‐3p (p = .037), miR‐130a‐5p (p = .007), miR‐144‐3p (p = .043), miR‐148a‐3p (p = .032), and miR‐152‐3p (p = .037) did show significant difference in ER‐positive patients compromised to the controls (Figure S1). MiR‐130a‐5p (p = .020), miR‐148a‐3p (p = .023), and miR‐152‐3p (p = .012) also showed significant differences between ER‐negative patients and the controls. As shown in Figure S2, the miR‐23a‐3p (p = .004), miR‐130a‐5p (p = .010), miR‐144‐3p (p = .006), miR‐148a‐3p (p = .035), and miR‐152‐3p (p = .010) were significantly downregulated in the patients with PR positive compared with the controls. Furthermore, miR‐130a‐5p (p = .011) and miR‐148a‐3p (p = .023) expressed statistically significantly different between two groups: PR‐negative patients and the controls. For HER2 status (Figure S3), we found that the miR‐23a‐3p (p = .002), miR‐130a‐5p (p = .007), miR‐144‐3p (p = .025), and miR‐152‐3p (p = .012) displayed significant differences in HER2‐negative patients with comparison to the healthy controls. The significant difference miR‐130a‐5p between HER2‐positive patients and the controls was observed (p = .044, Table 4).
Table 4.
The correlation of relative levels of miRNAs with breast cancer patients' clinic pathological parameters
| Characteristics | miR‐23a‐3p | miR‐130a‐5p | miR‐144‐3p | miR‐148a‐3p | miR‐152‐3p | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | p value | Low | High | p value | Low | High | p value | Low | High | p value | Low | High | p value | |
| Age (years) | |||||||||||||||
| ≤54 | 54 | 9 | .481 | 46 | 14 | .534 | 47 | 15 | .577 | 46 | 17 | .267 | 51 | 11 | .271 |
| >54 | 33 | 8 | 27 | 11 | 33 | 8 | 33 | 7 | 30 | 11 | |||||
| ER status | |||||||||||||||
| Positive | 51 | 13 | .377 | 48 | 13 | .150 | 49 | 15 | .734 | 50 | 14 | .393 | 47 | 17 | .291 |
| Negative | 27 | 4 | 18 | 10 | 22 | 8 | 21 | 9 | 25 | 5 | |||||
| PR status | |||||||||||||||
| Positive | 44 | 11 | .567 | 42 | 12 | .292 | 43 | 12 | .433 | 43 | 13 | .677 | 42 | 13 | .996 |
| Negative | 33 | 6 | 23 | 11 | 27 | 11 | 27 | 10 | 29 | 9 | |||||
| HER2 status | |||||||||||||||
| Positive | 22 | 7 | .127 | 15 | 11 | .044 * | 20 | 8 | .619 | 19 | 8 | .555 | 22 | 6 | .687 |
| Negative | 41 | 5 | 36 | 9 | 36 | 11 | 36 | 11 | 35 | 12 | |||||
| Stage | |||||||||||||||
| I‐II | 50 | 13 | .227 | 41 | 17 | .320 | 48 | 14 | .967 | 46 | 16 | .691 | 48 | 13 | .971 |
| III‐IV | 32 | 4 | 28 | 7 | 28 | 8 | 28 | 8 | 29 | 8 | |||||
| Clinical T | |||||||||||||||
| 1–2 | 70 | 15 | .814 | 59 | 20 | .678 | 64 | 21 | .171 | 65 | 19 | .218 | 64 | 20 | .189 |
| 3–4 | 11 | 2 | 2 | 4 | 12 | 1 | 8 | 5 | 12 | 1 | |||||
| Lymph node invasion | |||||||||||||||
| No | 43 | 10 | .631 | 36 | 13 | .866 | 37 | 16 | .041 * | 36 | 16 | .124 | 39 | 13 | .360 |
| Yes | 39 | 7 | 33 | 11 | 40 | 6 | 38 | 8 | 38 | 8 | |||||
| Pathological subtypes | |||||||||||||||
| Luminal A | 20 | 2 | .723 | 21 | 1 | .683 | 19 | 3 | .661 | 18 | 4 | .996 | 20 | 1 | .088 |
| Luminal B | 39 | 9 | 39 | 4 | 41 | 7 | 39 | 9 | 34 | 14 | |||||
| Her‐2 | 16 | 3 | 16 | 3 | 15 | 3 | 15 | 3 | 16 | 2 | |||||
| Triple negative | 9 | 1 | 9 | 1 | 7 | 3 | 8 | 2 | 8 | 2 | |||||
Bold indicates statistical significance of expression level with
p < .05.
3.5. Correlation of plasma miRNA levels to the status of pathological data
We also evaluated the correlation of these miRNAs and the pathological data, such as BC stage, T classification, nodal status, and pathological subtypes. Compared with normal controls, lower levels of miR‐23a‐3p (p = .025), miR‐130a‐5p (p = .021), miR‐144‐3p (p = .011), miR‐148a‐3p (p = .032), and miR‐152‐3p (p = .013) were also observed in patients with stage I‐II, and miR‐130a‐5p (p = .002) and miR‐148a‐3p (p = .020) in patients with stage III‐IV (Figure S4). Stratified by T classification, we observed that significant difference in the expression of miR‐130a‐5p (p = .009), miR‐144‐3p (p = .042), miR‐148a‐3p (p = .029), and miR‐152‐3p (p = .024) between patients with clinical T1‐2 and the controls, as shown in Figure S5. The expression of miR‐23a‐3p level was significantly lower in BC with clinical T3‐4 than in the health controls (p = .003).
Expression of miR‐23a‐3p (p = .002), miR‐130a‐5p (p = .011), miR‐144‐3p (p = .002), miR‐148a‐3p (p = .008), and miR‐152‐3p (p = .008) was significantly different between patients without lymph node metastasis and the controls (Figure S6). MiR‐130a‐5p (p = .008) was also significantly different between patients with lymph node metastasis and the controls. Furthermore, miR‐148a‐3p expression was higher in patients with lymph node metastasis as compared to patients without lymph node metastasis (p = .030). Our results also suggested that patients with lymph node metastasis appeared to have higher expression of miR‐144‐3p (p = .041, Table 4).
In addition, lower levels of miR‐23a‐3p (p = .005) and miR‐152‐3p (p = .004) in patients with luminal A subtype, miR‐130a‐5p (p = .019) in patients with luminal B subtype and miR‐152‐3p (p = .010) in patients with Her‐2 subtype were observed compared with normal controls (Figure S7). MiR‐130a‐5p (p = .022) was also different between patients with Her‐2 subtype and patients with triple negative. The expression of miR‐152‐3p level was significantly lower in patients with luminal A subtype than in patients with luminal B subtype (p = .010).
3.6. miRNA expression and prognosis based on TCGA database
Based on Kaplan‐Meier Plotter database (Figure 3), the expression of miR‐130a‐5p [hazard ratio (HR) = 0.78; 95% CI: 0.64–0.96; logrank p = .021], miR‐148a‐3p (HR = 0.70; 95% CI: 0.57–0.86; logrank p = .00062), and miR‐152‐3p (HR = 0.78; 95% CI: 0.63–0.96; logrank p = .021) was found to be associated with BC prognosis.
Figure 3.
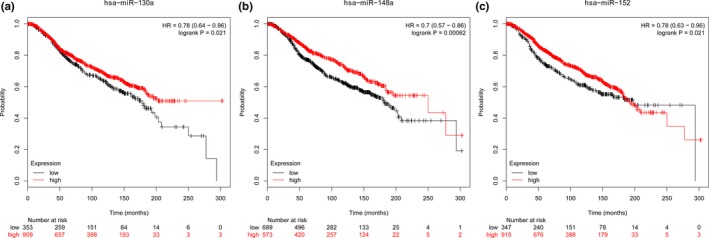
Kaplan–Meier Plotter of miRNA expression and breast cancer (BC) prognosis. The expression of miR‐130a‐5p (a), miR‐148a‐3p (b), and miR‐152‐3p (c) was associated with BC survival. The Kaplan–Meier plots were generated by Kaplan–Meier Plotter database (http://kmplot.com/analysis/index.php?p=service%26cancer=breast_mirna). p‐values (logrank test) .05 means the data is statistically significant
4. DISCUSSION
An important area in current BC research is the identification of novel biomarkers for early diagnosis. Currently, several studies have demonstrated that circulating miRNAs are differentially expressed in patients with BC and are potential biomarkers useful in BC diagnosis (Grimaldi & Incoronato, 2019; Incoronato & Grimaldi, 2019; Swellam, El Magdoub, Hassan, Hefny, & Sobeih, 2018). In this study, we have identified significant reduction of miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p in the plasma of the patients compared with the controls, indicating the diagnostic potential of these miRNAs in BC.
In BC, miR‐23a was involved in invasion, migration, lymph node metastasis, and survival (Eissa, Matboli, & Shehata, 2015; Ma, Li, et al., 2017). Moreover, miR‐23a acted as negative regulators of PR expression in ER‐positive BC (Gilam et al., 2017). Several studies have suggested miR‐130a expression was downregulated in BC tissues and cells, and associated with the patients with recurrence (Sueta, Yamamoto, Mtt, Yamamoto‐Ibusuki, & Iwase, 2017). Besides, overexpression of miR‐130a was able to inhibit cell proliferation, invasion, and migration in MCF7 and MDA‐MB‐435 cells (Pan et al., 2015). MiR‐144 suppressed BC cell proliferation, migration, and invasion and induced cell cycle arrest and cell apoptosis by repressing CEP55 (Yin, Cai, Meng, Sui, & Jiang, 2018). Previous studies showed that miR‐148a could suppress the estrogen‐induced viability and migration of BC cells via inhibition of ERα protein expression (Ma, Feng, et al., 2017). MiR‐152‐3p might serve as a tumor suppressor in human BC cells via negatively regulating PIK3CA expression to inhibit the activation of AKT and RPS6, leading to suppression of BC cell proliferation (Ge, Wang, Kong, Gao, & Sun, 2017). These studies have indicated that miR‐23a‐3p, miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p played important roles in human BC tumorigenesis.
Plasma levels of miRNA are not necessarily a reflection of tissue levels (Cookson et al., 2012). We also compared the differences in miRNA expression levels in tumor tissue versus plasma. Notably, miR‐130a‐5p, miR‐144‐3p, and miR‐152‐3p were downexpressed in BC tissues as well as plasma; therefore, these miRNAs might directly reflect BC tumor biology. We also found that the relative expression of these five miRNAs was closely associated with the clinicopathologic features of the BC such as the expression of sex hormone receptor (ER, PR, and HER2), histological tumor grades, and lymph node metastasis. In clinic, the expression level of hormone receptors including ER, PR, and HER2 and tumor grade is often used for classification and target therapy indicators of BCs (Schettini et al., 2016). Our results showed that the expression of miR‐23a‐3p, miR‐144‐3p, and miR‐152‐3p was related to ER positive and PR positive, indicating that these plasma miRNA levels might separate the ER‐positive and PR‐positive subtypes of BC. Stratified by BC stage, miR‐23a‐3p, miR‐144‐3p, and miR‐152‐3p did show significant differences in the staging compromised to the control, especially in stage I‐II. Besides, the expression of the miR‐130a‐5p, miR‐144‐3p, miR‐148a‐3p, and miR‐152‐3p in the plasma showed a significant difference between patients with T1‐2 and the controls. Moreover, we also found that miR‐144‐3p and miR‐148a‐3p were associated with lymph node invasion. Consistent with our results, one study indicated that miR‐144‐3p served as early detection markers of metastasis in BC (Madhavan et al., 2016). The findings suggest that the deregulation of the miRNAs might affect critical molecular events involved in tumor progression and could be used to identify the different nature of breast tissues. Thus, measurements of miRNAs as the biochemical markers can help to diagnose the different stages of BC prior to clinical investigations on the samples. However, further studies in a large population should be needed to validate the feasibility of these miRNAs as novel noninvasive biomarkers.
Our study presents several limitations. First, the size of patients with BC and normal controls were still relatively small, limiting the evaluation on these miRNAs as predictive biomarkers in early detection of BC. Second, the follow‐up results, such as disease recurrence and disease‐free survival were lacked, which prevented us from conducting prognosis analyses. Further validations in large, independent prospective cohorts are recommended.
5. CONCLUSION
In conclusion, we identified five miRNAs as being downregulated in the plasma of patients with BC compared to healthy control individuals. Strikingly, expression trends for miR‐130a‐5p, miR‐144‐3p, and miR‐152‐3p were similar in both tissue and plasma. Besides, they were also significantly associated with sex hormone receptor, clinical stage, and lymph node metastasis. All of plasma miRNAs them exhibit great potential in serving as promising noninvasive candidates for BC detection. Future prospective studies using a large cohort will be needed to confirm our findings.
CONFLICT OF INTEREST
The authors declare that there are no competing interests regarding the publication of this paper.
AUTHORS CONTRIBUTION
The work presented here was carried out in collaboration between all authors. Xu Li and Wenjing Zou carried out the molecular genetic studies and drafted the manuscript. Yuzhen Wang and Zijun Liao performed the statistical analyses and interpreted the results. Lina Li, Yang Zhai, and Lingxiao Zhang collected clinical information about patients and performed the extraction of plasma RNA and miRNA qRT‐PCR experiments. Shanzhi Gu and Xinhan Zhao conceived the study, worked on associated data collection and their interpretation, participated in the design and coordination of the study, and funded the study. All authors read and approved the final manuscript.
Supporting information
ACKNOWLEDGEMENT
We are grateful to the individuals who participated in this study. We also thank the clinicians and hospital staff who contributed to the sample and data collection for this study.
Li X, Zou W, Wang Y, et al. Plasma‐based microRNA signatures in early diagnosis of breast cancer. Mol Genet Genomic Med. 2020;8:e1092 10.1002/mgg3.1092
Xu Li and Wenjing Zou are co‐first authors.
Contributor Information
Shanzhi Gu, Email: gushanzhi@mail.xjtu.edu.cn.
Xinhan Zhao, Email: zhaoxinhan@mail.xjtu.edu.cn.
REFERENCES
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bleyer, A. , & Welch, H. G. (2012). Effect of three decades of screening mammography on breast‐cancer incidence. New England Journal of Medicine, 367(21), 1998–2005. 10.1056/NEJMoa1206809 [DOI] [PubMed] [Google Scholar]
- Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. , Torre, L. A. , & Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 68(6), 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Cai, J. , Zhang, Y. , Huang, S. , Yan, M. , Li, J. , Jin, T. , & Bao, S. (2017). MiR‐100‐5p, miR‐199a‐3p and miR‐199b‐5p induce autophagic death of endometrial carcinoma cell through targeting mTOR. International Journal of Clinical and Experimental Pathology, 10(9), 9262–9272. [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network . (2012). Comprehensive molecular portraits of human breast tumours. Nature, 490(7418), 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. J. , Cheng, Y. M. , Chen, C. C. , Chen, Y. C. , & Shen, C. J. (2017). MiR‐148a and miR‐152 reduce tamoxifen resistance in ER+ breast cancer via downregulating ALCAM. Biochemical and Biophysical Research Communications, 483(2), 840–846. 10.1016/j.bbrc.2017.01.012 [DOI] [PubMed] [Google Scholar]
- Chen, W. , Zheng, R. , Baade, P. D. , Zhang, S. , Zeng, H. , Bray, F. , … He, J. (2016). Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians, 66(2), 115–132. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- Cheng, G. (2015). Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Advanced Drug Delivery Reviews, 81, 75–93. 10.1016/j.addr.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Coleman, C. (2017). Early detection and screening for breast cancer. Seminars in Oncology Nursing, 33(2), 141–155. 10.1016/j.soncn.2017.02.009 [DOI] [PubMed] [Google Scholar]
- Cookson, V. J. , Bentley, M. A. , Hogan, B. V. , Horgan, K. , Hayward, B. E. , Hazelwood, L. D. , & Hughes, T. A. (2012). Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cellular Oncology, 35(4), 301–308. 10.1007/s13402-012-0089-1 [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Tian, Z. , Yang, H. , Yao, H. , He, P. , Ouyang, Y. , & Jin, T. (2017). MicroRNA expression profiles of whole blood in chronic obstructive pulmonary disease. International Journal of Clinical and Experimental Pathology, 10, 4860–4865. [Google Scholar]
- Eissa, S. , Matboli, M. , & Shehata, H. H. (2015). Breast tissue‐based microRNA panel highlights microRNA‐23a and selected target genes as putative biomarkers for breast cancer. Translational Research, 165(3), 417–427. 10.1016/j.trsl.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Ge, S. , Wang, D. , Kong, Q. , Gao, W. , & Sun, J. (2017). Function of miR‐152 as a tumor suppressor in human breast cancer by targeting PIK3CA. Oncology Research, 25(8), 1363–1371. 10.3727/096504017x14878536973557 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gilam, A. , Shai, A. , Ashkenazi, I. , Sarid, L. A. , Drobot, A. , Bickel, A. , & Shomron, N. (2017). MicroRNA regulation of progesterone receptor in breast cancer. Oncotarget, 8(16), 25963–25976. 10.18632/oncotarget.15657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi, A. M. , & Incoronato, M. (2019). Clinical translatability of "identified" circulating miRNAs for diagnosing breast cancer: Overview and update. Cancers, 11(7), 10.3390/cancers11070901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, J. , Wang, Y. , Wang, X. , Zhou, D. , Wang, X. , Zhou, M. , & He, Z. (2018). Effect of the LncRNA GAS5‐MiR‐23a‐ATG3 axis in regulating autophagy in patients with breast cancer. Cellular Physiology and Biochemistry, 48(1), 194–207. 10.1159/000491718 [DOI] [PubMed] [Google Scholar]
- Hamam, R. , Hamam, D. , Alsaleh, K. A. , Kassem, M. , Zaher, W. , Alfayez, M. , … Alajez, N. M. (2017). Circulating microRNAs in breast cancer: Novel diagnostic and prognostic biomarkers. Cell Death & Disease, 8(9), e3045 10.1038/cddis.2017.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incoronato, M. , Grimaldi, A. M. , Mirabelli, P. , Cavaliere, C. , Parente, C. A. , Franzese, M. , … Salvatore, M. (2019). Circulating miRNAs in untreated breast cancer: An exploratory multimodality morpho‐functional study. Cancers, 11(6), 10.3390/cancers11060876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, X. , Zhang, J. , Li, J. , Shao, J. , & Fang, L. (2018). MiR‐130a‐3p inhibits migration and invasion by regulating RAB5B in human breast cancer stem cell‐like cells. Biochemical and Biophysical Research Communications, 501(2), 486–493. 10.1016/j.bbrc.2018.05.018 [DOI] [PubMed] [Google Scholar]
- Li, G. , Zhang, Z. , Tu, Y. , Jin, T. , Liang, H. , Cui, G. , … Gao, G. (2013). Correlation of microRNA‐372 upregulation with poor prognosis in human glioma. Diagnostic Pathology, 8, 1 10.1186/1746-1596-8-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, F. , Feng, Y. , Li, W. , Li, Z. , Liu, T. , & Li, L. (2017). miR‐148a Suppresses estrogen‐induced viability and migration of breast cancer cells via inhibition of estrogen receptor alpha expression. Experimental and Therapeutic Medicine, 13(5), 2515–2522. 10.3892/etm.2017.4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, F. , Li, W. , Liu, C. , Li, W. , Yu, H. , Lei, B. , … Qian, C. (2017). MiR‐23a promotes TGF‐β1‐induced EMT and tumor metastasis in breast cancer cells by directly targeting CDH1 and activating Wnt/β‐catenin signaling. Oncotarget, 8(41), 69538–69550. 10.18632/oncotarget.18422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan, D. , Peng, C. , Wallwiener, M. , Zucknick, M. , Nees, J. , Schott, S. , … Burwinkel, B. (2016). Circulating miRNAs with prognostic value in metastatic breast cancer and for early detection of metastasis. Carcinogenesis, 37(5), 461–470. 10.1093/carcin/bgw008 [DOI] [PubMed] [Google Scholar]
- McDonald, J. S. , Milosevic, D. , Reddi, H. V. , Grebe, S. K. , & Algeciras‐Schimnich, A. (2011). Analysis of circulating microRNA: Preanalytical and analytical challenges. Clinical Chemistry, 57(6), 833–840. 10.1373/clinchem.2010.157198 [DOI] [PubMed] [Google Scholar]
- Nattinger, A. B. , & Mitchell, J. L. (2016). Breast cancer screening and prevention. Annals of Internal Medicine, 164(11), http://Itc81-itc96.10.7326/aitc201606070 [DOI] [PubMed] [Google Scholar]
- Pan, Y. , Wang, R. , Zhang, F. , Chen, Y. , Lv, Q. , Long, G. , & Yang, K. (2015). MicroRNA‐130a inhibits cell proliferation, invasion and migration in human breast cancer by targeting the RAB5A. International Journal of Clinical and Experimental Pathology, 8(1), 384–393. [PMC free article] [PubMed] [Google Scholar]
- Schettini, F. , Buono, G. , Cardalesi, C. , Desideri, I. , De Placido, S. , & Del Mastro, L. (2016). Hormone receptor/human epidermal growth factor receptor 2‐positive breast cancer: Where we are now and where we are going. Cancer Treatment Reviews, 46, 20–26. 10.1016/j.ctrv.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Smyth, G. K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology, 3, 1–25. 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- Sueta, A. , Yamamoto, Y. , Tomiguchi, M. , Takeshita, T. , Yamamoto‐Ibusuki, M. , & Iwase, H. (2017). Differential expression of exosomal miRNAs between breast cancer patients with and without recurrence. Oncotarget, 8(41), 69934–69944. 10.18632/oncotarget.19482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swellam, M. , El Magdoub, H. M. , Hassan, N. M. , Hefny, M. M. , & Sobeih, M. E. (2018). Potential diagnostic role of circulating MiRNAs in breast cancer: Implications on clinicopathological characters. Clinical Biochemistry, 56, 47–54. 10.1016/j.clinbiochem.2018.04.013 [DOI] [PubMed] [Google Scholar]
- Yahya, S. M. , & Elsayed, G. H. (2015). A summary for molecular regulations of miRNAs in breast cancer. Clinical Biochemistry, 48(6), 388–396. 10.1016/j.clinbiochem.2014.12.013 [DOI] [PubMed] [Google Scholar]
- Yin, Y. , Cai, J. , Meng, F. , Sui, C. , & Jiang, Y. (2018). MiR‐144 suppresses proliferation, invasion, and migration of breast cancer cells through inhibiting CEP55. Cancer Biology & Therapy, 19(4), 306–315. 10.1080/15384047.2017.1416934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


