Abstract
Ryanodine receptor ion channels (RyR1s) release Ca2+ ions from the sarcoplasmic reticulum to regulate skeletal muscle contraction. By whole-exome sequencing, we identified the heterozygous RYR1 variant c.14767_14772del resulting in the in-frame deletion p.(Phe4923_Phe4924del) in two brothers with a lethal form of the fetal akinesia deformation syndrome (FADS). The two deleted phenylalanines (RyR1-Δ4923FF4924)are located in theS6 pore-lining helix of RyR1. Clinical features in one of the two siblings included severe hypotonia, thin ribs, swallowing inability, and respiratory insufficiency that caused early death. Functional consequences of the RyR1-Δ4923FF4924 variant were determined using recombinant 2,200-kDa homotetrameric and heterotetrameric RyR1 channel complexes that were expressed in HEK293 cells and characterized by cellular, electrophysiological, and computational methods. Cellular Ca2+ release in response to caffeine indicated that the homotetrameric variant formed caffeine-sensitive Ca2+ conducting channels in HEK293 cells. In contrast, the homotetrameric channel complex was not activated by Ca2+ and did not conduct Ca2+ based on single-channel measurements. The computational analysis suggested decreased protein stability and loss of salt bridge interactions between RyR1-R4944 and RyR1-D4938, increasing the electrostatic interaction energy of Ca2+ in a region 20 Å from the mutant site. Co-expression of wild-type and mutant RyR1s resulted in Ca2+-dependent channel activities that displayed intermediate Ca2+ conductances and suggested maintenance of a reduced Ca2+ release in the two patients. Our findings reveal that the RYR1 pore variant p.(Phe4923_Phe4924del) attenuates the flow of Ca2+through heterotetrameric channels, but alone was not sufficient to cause FADS, indicating additional genetic factors to be involved.
Keywords: Fetal akinesia, ryanodine receptor, sarcoplasmic reticulum, single-channel recordings, molecular dynamics simulations
1. Introduction
Skeletal muscle type 1 ryanodine receptor ion channels (RyR1s) rapidly release Ca2+ from the sarcoplasmic reticulum (SR) into the myoplasm to initiate muscle contraction. The 2,200 kDa RyR1s are comprised of four RyR1 subunits of ~5000 residues and four FK506 binding proteins (FKBP) of ~110 residues [1–3]. RyR1 is controlled in skeletal muscle by Cav1.1 voltage-gated channel and Ca2+ by a not well-understood mechanism. Studies with membrane isolates and purified preparations show that micromolar Ca2+ activates and millimolar Ca2+ inhibits RyR1. Exogenous ligands include the plant alkaloid ryanodine and caffeine. Ryanodine modifies thegating and ion conductance properties by binding with nanomolar affinity and high specificity to the open RyRs. Caffeine activates the RyRs at millimolar concentrations without altering their ion conductance properties.
A large number of RYR1 variants have been linked to autosomal dominantly and recessively inherited skeletal muscle myopathies, including central core disease (CCD), multi minicore disease, core-rod myopathy, and congenital neuromuscular disease [4–6]. Core diseases result in weakening of skeletal muscle function and manifest as cores that lack mitochondria and oxidative enzymes [7]. Dominantly acting CCD-associated RYR1 variants tend to be located in the pore region [8–11], whereas recessive variants widely spread throughout the RYR1 gene [4–6]. The spectrum of ryanodinopathies was broadened by the identification of biallelic RYR1 variants associated with fetal akinesia deformation sequence (FADS)/arthrogryposis multiplex congenita and lethal multiple pterygium syndrome (LMPS) [7, 12–18]. These recessive RYR1 variants likely cause complete protein loss or loss-of-function of the encoded protein, which can lead to lethal outcomes.
We performed whole-exome sequencing and identified a heterozygous deletion of 6 bp (c.14767_14772del), resulting in the in-frame deletion of two phenylalanines [p.(Phe4923_Phe4924del)] in the carboxyl-terminal pore region of RyR1 in two brothers with FADS. The index patient, patient 2, died at the age of 10 weeks, while the first affected child, patient 1, died on the first day of life. The two affected siblings inherited the heterozygous RYR1 variant from their healthy father, who is a somatic mosaic. No second variant on the maternal RYR1 allele was identified that could explain the severe, autosomal recessive FADS in the two affected boys. The deletion of the two phenylalanines was arbitrarily assigned to the most 3’ position, according to the sequence variant nomenclature [19], and deletes the third and fourth phenylalanine (RYR1-Δ4923FF4924)in a stretch of four successive phenylalanines located in thehumanS6 pore-lining helix of the RyR1 C-terminus.
To determine the functional effects of the pore deletion mutant, HEK293 cells were transfected with rabbit wild-type RyR1 (RyR1-WT) and RyR1-Δ4922FF4923 (which is analogous to Δ4923FF4924 in human RYR1) expression vectors. Homotetrameric and heterotetrameric channel complexes were characterized by cellular Ca2+ release measurements, single-channel recordings, and computational methods. The results indicated that homotetrameric RyR1-Δ4922FF4923 channels were not activated by Ca2+ and did not conduct Ca2+ in single-channel measurements, whereas heterotetrameric channels composed of wild-type and mutant subunits maintained a Ca2+-dependent channel activity but exhibited reduced Ca2+ conductances compared to wild type. The findings suggest that the RYR1 pore variant p.(Phe4923_Phe4924del) contributed to but alone was not sufficient to cause the severe FADS in the two brothers.
2. Materials and Methods
2.1. Materials
[3H]Ryanodine was obtained from Perkin Elmer Life Sciences, protease inhibitors from Sigma-Aldrich, and phospholipids from Avanti Polar Lipids.
2.2. Patients
All investigations were part of an ethically approved protocol (Hamburg Medical Chamber; PV3802) and were undertaken with prior informed consent.
2.3. Exome sequencing and sequence data analysis
Targeted enrichment and massively parallel sequencing were performed on genomic DNA extracted from the leukocytes of patient 2 and his parents. Enrichment of the whole exome was performed according to the manufacturer’s protocols using the Nextera Enrichment Kit (62 Mb) (Illumina). Captured libraries were then loaded and sequenced onto the HiSeq2500 platform (Illumina). Trimmomatic was employed to remove adapters, low quality (phred quality score < 5) bases from the 3’ ends of sequence reads [20]. Reads shorter than 36 bp were subsequently removed. Further processing was performed following the Genome Analysis Toolkit’s (GATK) best practice recommendations. Briefly, trimmed reads were aligned to the human reference genome (UCSC GRCh37/hg19) using the Burrows-Wheeler Aligner (BWA mem v0.7.12). Duplicate reads were marked with Picard tools (v1.141). GATK (v3.4) was employed for indel realignment, base quality score recalibration, calling variants using the HaplotypeCaller, joint genotyping, and variant quality score recalibration. AnnoVar (v2015–03-22) was used to functionally annotate and filter alterations against public databases (dbSNP138, 1000 Genomes Project, and ExAC and gnomAD Browsers). Only private (absent in public database) and rare (with a minor allele frequency <0.5% and not present in the homozygous state in public databases or the parents) exonic and intronic variants at exon-intron boundaries ranging from −40 to +40 were retained.
2.4. Variant validation
For patient 1, DNA was extracted from blood plasma (Nucleo Spin Plasma XS, Macherey-Nagel). Sequence validation and segregation analysis for all candidate variants in the family were performed by Sanger sequencing. The sequence of the primer pairs designed to amplify exon 102 of the RYR1 gene and exon-intron boundaries (NM_000540.3) are as follows: forward primer 5’-cctgaccatttctggctgtt-3’ and reverse primer 5’-cccactcccagcactgac-3’. Amplicons were directly sequenced using the ABI BigDye Terminator Sequencing Kit (Applied Biosystems) and an automated capillary sequencer (ABI 3500; Applied Biosystems). Sequence electropherograms were analysed using the Sequence Pilot software (JSI medical systems).
2.5. RNA isolation and cDNA synthesis
Total RNA was extracted from a frozen muscle biopsy of patient 2 (RNeasy Fibrous Tissue Mini Kit, Qiagen). 250 ng total RNA was reverse transcribed (Superscript III RT, ThermoFisher) using random hexamers, and 1 μl of the reverse transcription reaction was utilized to amplify a 245-bp RYR1 cDNA fragment encompassing the c.14767_14772del variant (forward primer 5′-caacaagagcgaggatgagg-3′, reverse primer 5′-ctcggagctcaccaaaagc −3′). The PCR product was directly sequenced.
2.6. PCR product cloning and colony PCR
Exon 102 of RYR1 and adjacent intronic sequences were amplified from leukocyte-derived DNA of the father of patients 1 and 2. The PCR product was cloned into the pCR2.1 TOPO TA Cloning Vector (ThermoFisher). Individual E. coli clones were subjected to colony PCR followed by Sanger sequencing to haplotype determination.
2.7. Preparation of wild-type and mutant channels
pCMV5-RyR1-ΔFF was prepared by a gene synthesis method using full-length rabbit cDNA [21] and a proprietary protocol (Genewiz Inc, South Plainfield, NJ). Wild-type and mutant expression vectors were transiently expressed in HEK293 cells using jetPRIME reagent (Polyplus) according to the manufacturer’s instructions. Cells were maintained at 37°C and 5% CO2 in Dulbecco’s Modified Eagle Medium containing 10% fetal bovine serum and replated the day before transfection. RyR1 cDNAs were transiently expressed in HEK293 cells in 10-cm tissue culture dishes using 10 μg cDNA/dish. Cells were grown at 35°C for 72 h, washed twice with PBS containing cOmplete protease inhibitor cocktail (Roche), harvested in the same solution by scrapping, collected by centrifugation and stored in a liquid N2 freezer. To prepare membrane fractions, cells were resuspended in 0.15 M KCl, 0.3 M sucrose, 20 mM imidazole, pH 7 solution containing 1 mM EGTA, 1 mM glutathione disulfide and cOmplete protease inhibitor cocktail and homogenized using a Tekmar Tissumizer for 5 s, setting 35,000 rpm. Cell homogenates were centrifuged, pellets were washed and resuspended in 0.15 M KCl, 0.3 M sucrose, 20 mM imidazole, pH 7 solution containing cOmplete protease inhibitor cocktail and stored in a liquid N2 freezer [22].
2.8. Cellular Ca2+ release
Stored Ca2+ release was determinedas described [22]. HEK293 cells were grown on glass coverslips and incubated with 5 μM Fluo 4-AM. After washing away excess Fluo-4 AM, cellular Ca2+ release was induced by the addition of 5– 8 mM caffeine and measured in individual cells using EasyRatioPro (Photon Technology International, Lawrenceville, NJ) or Sola SEII Light Engine and NIS Elements software (Nikon Instruments, Melville NY). On the coverslips, 30–60 cells were analyzed.
2.9. SDS-PAGE and immunoblot analysis
Proteins in crude membrane fractions of HEK293 cells (20 μg protein/lane) were separated using 3 – 12% acrylamide gradient SDS-PAGE, transferred to nitrocellulose membranes and probed using primary rabbit anti-RyR1 polyclonal antibody 6425 prepared by ψProSci (Poway, CA) against RyR1 FIKGLDSFSGKPRGSG sequence peptide [23]. Immunoblots were developed using horse-radish peroxidase-linked secondary anti-rabbit IgG antibody (Cell Signaling, Danvers, MA). RyR1s were quantified using Bio-Rad ChemiDoc Imaging System and Image Quant TL software.
2.10. [3H]Ryanodine binding
Ryanodine binds with high specificity and nanomolar affinity to RyR1 and is widely used to probe for RyR1 activity and expression [24]. RyR1-WT and RyR1-ΔFF protein levels were incubated for 4–5 h at 24°C in 20 mM imidazole, pH 7.0, 0.6 M KCl, 150 μM Ca2+, protease inhibitors, and near-saturating concentration of 20 nM [3H]ryanodine. Nonspecific binding was determined using 2.5 μM unlabeled ryanodine. Bound [3H]ryanodine was determined using a filter assay [22].
2.11. Single-channel recordings
In single-channel measurements, membrane fractions isolated from HEK293 cells were fused with Mueller-Rudin type planar lipid bilayers containing a 5:3:2 mixture of bovine brain phosphatidylethanolamine, phosphatidylserine, and phosphatidylcholine (25 mg of total phospholipid/ml n-decane) [22]. A small aliquot of membranes was added to the cis cytosolic bilayer chamber and fused in the presence of an osmotic gradient containing 0.25 M cis cytosolic KCl and 0.02 M trans SR luminal KCl in 2 μM free Ca2+ (0.23 mM EGTA and 0.21 mM Ca2+) and 20 mM KHEPES, pH 7.4). After the appearance of channel activity, the trans SR luminal KCl concentration was increased to 0.25 M KCl. Bilayer potential was set to 0 mV using symmetric cis and trans 0.25 M KCl, 2 μM Ca2+, 20 mM HEPES, pH 7.4 solutions. 2 μM Ca in the trans bath was not reduced to nominally zero, as it did not noticeably affect channel conductances and activities. The trans side of the bilayer was defined as ground. Electrical signals were filtered at 2 kHz, digitized at 10 kHz,and analyzed at 50% threshold setting [25, 26]. Data acquisition and analysis of 2 min recordings were performed using commercially available software (pClamp, Axon Instruments, CA). Channel activities were also recorded after increasing the Ca2+ concentration in the trans bilayer chamber to 10 mM using 87mM Ca2+ solution. The reversal potential was measured to determine Ca2+/K+ permeability ratios using a modified Goldman- Hodgkin-Katz equation [25, 26]. Channel open probabilities (Po) in multichannel recordings were calculated using the equation Po =ƩiPo,i/N, where N is the total number of channels and Po,i is the channel open probability of the ith channel. The number of current levels was determined with 2 μM cis cytosolic Ca2+.
2.12. Computational methods
The transmembrane domain residues 4559–5037 of the ATP/caffeine (CFF)/Ca2+ bound-open RyR1 cryo-electron micrograph (EM) structure (PDB ID: 5TAL) [27] were used for in silico mutagenesis and molecular dynamics simulations. Missing segment residues 4588–4625 were modeled using Modeller-9v19 [28] and Gromacs-2019 [29]. We used homology modeling tool Modeller-9v19 [28] to remove deleted residues and assign geometrical constraints between the two flanking residues to ensure that planar peptide bond constraints were not violated. For the secondary structures adjacent to the inserted/deleted residues, Modeller allowed full conformational flexibility. The side chains were optimized and re-packed to confirm that the backbone dihedral angles are within the allowed regions of the Ramachandran plot. From 40 structures of RyR1-ΔFF, the structure with the least molecular objective function was selected as best model for molecular dynamics simulations using Gromacs-2019 [29]. The RyR1-WT structure was simulated for comparative analysis of the mutant structures. Simulated systems were prepared by inserting RyR1-WT and RyR1-ΔFF structures into POPC-lipid bilayers using the CHARMM-GUI tool [30, 31]. Protein atoms were initially restrained to a starting conformation with a harmonic potential of 2 kcal/nm2/mole. Restraints were slowly released by gradually reducing the potential in several equilibrated short molecular dynamics (MD) simulations. The equilibrated membrane-inserted RyR1 structure was placed in a box with dimensions 203.8 Å x 203.8 Å x 145.9 Å and explicitly solvated using the TIP3P water model. Under isobaric conditions, solvent density was maintained at 1 bar and 310 K using Parrinello-Rahman barostat [32] with a coupling constant of 0.1 ps. The systems were equilibrated for 10 ns with a simulation time step of 2 fs. The final production run was carried out for 100 ns using a time step of 2 fs with all bonds constrained using the LINCS algorithm [33]. Long-range electrostatic interactions were evaluated using Particle mesh Ewald with 12 Å cutoff [34]. All MD simulations were performed using the CHARMM36 force field [35, 36]. Five structures sampled evenly over the last 50 ns of the simulation trajectories of RyR1-WT and RyR1-ΔFF were subjected to pore profile calculations using HOLE [37]. To assess the electrostatics of calcium and potassium ion passage through the pore, the Poisson-Boltzmann (PB) equation was used with APBSmem [38, 39]. Two focusing layers with a grid length of 90 Å and 97 grid points in the x, y, and z dimensions were determined. Ca2+ and K+ concentrations (as Cl− salts) were 0.1 M, the temperature was 298 K, and non-linear Poisson-Boltzmann method was utilized in Adaptive Poisson-Boltzmann Solver. The upper and lower exclusions were set at 16 Å to exclude the lipid bilayer from the pore region. Membrane thickness and head group thickness were set at 42.5 Å and 7 Å respectively according to previous calculations [40, 41]. The dielectric constant for lipid bilayer acyl chains and protein were set at 2.0, whereas the dielectric constant for solvent and lipid head groups was set at 80.0. The step size for ion movement through the pore was 1.0 Å along the channel axis and the ionic radii used for Ca2+ and K+ were 1.03 and 1.41 Å, respectively. All the structural figures in the current study were rendered using the PyMOL Molecular Graphics System [42].
2.13. Biochemical assays and data analysis
All free Ca2+ concentrations were established with the use of a Ca2+ selective electrode. Differences between samples were analyzed using the Student’s t-test. p<0.05 was considered significant.
3. Results
3.1. Identification of the heterozygous RYR1 variant
c. 14767_14772del/p.(Phe4923_Phe4924del) in two siblings with severe FADS
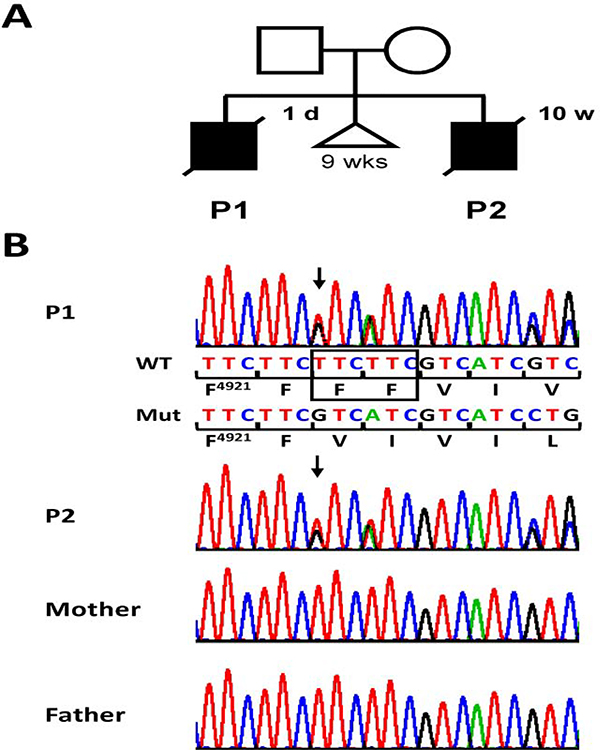
Parents of the index patient (patient 2) are non-consanguineous. The first pregnancy was complicated by contractions. The prematurely delivered male fetus (patient 1) died at day one due to prematurity and intractable cardio-respiratory insufficiency (Fig. 1A). Deformity of his thorax and scoliosis were noticed. Radiograph of the thorax showed thin ribs and confirmed scoliosis. A subsequent pregnancy ended spontaneously of unknown cause at 9 weeks of gestation (Fig. 1A). Patient 2 is the product of his mother’s third pregnancy, complicated by transient increased nuchal translucency and polyhydramnios. Delivery occurred spontaneously at 30 + 6 weeks of gestation. Weight was 1550 g (mean), length 40 cm (mean), and occipital frontal circumference 31 cm (+0.5 SDS). His face was mildly dysmorphic with a large appearing skull, retrognathia, and low set ears as well as medial incisors of the maxilla. Scoliosis, flexion contractures of knee joints, and ulnar deviation of hands were noted. Poor respiratory movements necessitated immediate intubation and artificial respiration. Muscle tone was markedly low, and the infant showed no reaction to touch and pain stimuli. Only sporadic movements of hands or feet were noticed. Radiology disclosed lung hypoplasia, thin ribs (one fractured after resuscitation), osteopenic bones with fractures of both humeri, left ulna, and occiput. The clinical course was characterized by persistence of marked muscular hypotonia, severe respiratory insufficiency, and inability to swallow necessitating constant tube feeding. With agreement of the parents and under guidance of an ethical committee, therapeutic measures were terminated resulting in the infant’s death at age 10 weeks. Clinical presentation of patients 1 and 2 was suggestive of FADS.
Figure 1. Heterozygous RYR1 variant c.14767_14772del in two siblings with a lethal form of FADS.
(A) Pedigree of the family with two affected boys, patient 1 (P1) and 2 (P2). The triangle indicates a spontaneous abortion of unknown cause at 9 weeks (wks) of gestation. Age at death is indicated at top right of the symbol. d, days; w, weeks. (B) Partial sequence electropherograms showing the RYR1 c.14767_14772del/p.(Phe4923_Phe4924del) variant in the heterozygous state in blood plasma-derived DNA of patient 1 and leukocyte-derived DNA of patient 2. The variant was not visible in the sequence from leukocyte-derived DNA of both the healthy mother and father. Wild-type (WT) and mutant (Mut) sequence and encoded amino acid residues in the one-letter code [beginning with phenylalanine (F) at position 4921] are depicted below the top sequence electropherogram. Deleted bases are marked by a black rectangle in the wild-type sequence. Arrows point to the start of the heterozygous in-frame deletion.
To identify the molecular basis of FADS in the two brothers, we performed trio whole-exome sequencing in patient 2 and his healthy parents. The heterozygous 6-bp deletion c.14767_14772del in exon 102 of the RYR1 gene, predicting the in-frame deletion of two phenylalanines [p.(Phe4923_Phe4924del)], was identified as the top candidate. This variant was absent in both parents and in population databases (1000 Genomes Project, EVS, ExAC, and gnomAD browser). Segregation analysis of the c.14767_14772del variant confirmed the presence of the heterozygous 6-bp deletion in patient 2 and also patient 1, the first child of the couple (Fig. 1B). The variant was not detectable in leukocyte-derived DNA of both parents by Sanger sequencing (Fig. 1B). As FADS is caused by rare recessive (biallelic) RYR1 variants [7, 12–18], we analysed the trio exome data for a second, heterozygous RYR1 variant in patient 2 that could underlie, together with the heterozygous c.14767_14772del variant, the FADS phenotype, but could not detect any other, possibly pathogenic RYR1 variant. The presence of a shared heterozygous RYR1 variant in two affected brothers and absence of the variant in leukocyte-derived DNA of both parents suggested germline and/or somatic mosaicism of the variant in mother or father. By cloning the RYR1-exon 102 amplicon followed by Sanger-sequencing of 127 individual colony PCR products, we confirmed the father to be a mosaic carrier (4% of his leukocytes were heterozygous for the RYR1 variant) (Supplemental Fig. S1). Low mosaicism of the RYR1 variant c.14767_14772del in white blood cells of the father suggests that he also carried the RYR1 variant in some of his sperm cells (germline mosaicism), explaining inheritance of the RYR1 variant by his two sons.
The RYR1 gene has been shown to undergo polymorphic and developmentally regulated allele silencing. For example, RYR1 was monoallelically expressed in skeletal and smooth muscles in 6 of 11 patients with recessive core myopathies [43]. We next studied RYR1 transcripts in skeletal muscle-derived mRNA of patient 2 to investigate the possible monoallelic expression of the c.14767_14772del-bearing RYR1 allele that may underlie the FADS phenotype. RYR1 transcription analysis indicated biallelic expression with a 1:1 ratio of mutant and wild-type mRNAs in the skeletal muscle of patient 2 (Supplemental Fig. S2). The data suggest that both the paternal and maternal RYR1 gene copy were equally expressed in patient 2’s skeletal muscle.
In summary, we identified the heterozygous RYR1 variant
c.14767_14772del/p.(Phe4923_Phe4924del) in two siblings with a lethal form of FADS. We were unable to detect a second, pathogenic RYR1 event in the exome data set of patient 2 that could be responsible for an autosomal recessively inherited FADS form. However, the absence of the RYR1 variant c.14767_14772del in the general population and localization of the deleted phenylalanines at positions 4923 and 4924 in the carboxyl-terminal pore region of RyR1 suggested a possible contribution of the RYR1 variant to the severe clinical course of the two affected sibs. We, therefore, characterized the functional consequences of the RYR1 variant by cellular, electrophysiological, and computational methods.
3.2. Cellular and electrophysiological characterization of rabbit RyR1-Δ4922FF4923
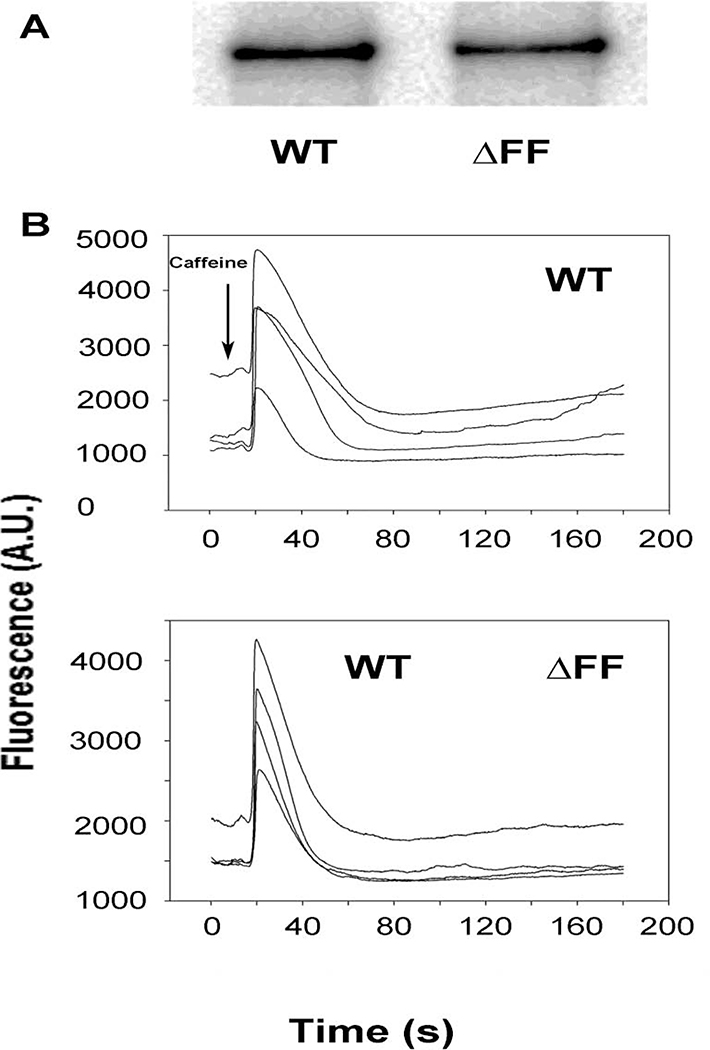
Immunoblot analysis indicated that the RyR1-ΔFF protein level in HEK293 cells was ~65 % of WT (Fig. 2A, Table 1). Using the Ca2+ releasing drug caffeine [44], a Ca2+ release response was retained in ~85% of HEK293 cells transfected with RyR1-ΔFF compared to WT (Fig. 2B, Table 1). This suggested that the homotetrameric RyR1-ΔFF channel complex expressed caffeine-sensitive Ca2+-conducting channels in HEK293 cells. RyR1-ΔFF function was further evaluated using HEK293 membrane isolates and a [3H]ryanodine binding assay that showed RyR1-ΔFF had a Bmax of [3H]ryanodine binding that was ~5% compared to WT (Table 1). The result suggested, compared to the Ca2+ response in HEK293 cells, a loss of function during membrane isolation or the binding assay.
Figure 2. Immunoblot and caffeine-induced Ca2+ release measured in HEK293 cells expressing RyR1-WT or RyR1-ΔFF.
(A) Immunoblots of HEK293 cells transfected with pCMV5-RyR1-WT or pCMV5-RyR1-ΔFF show 565 kDa WT and ΔFF bands. Both bands are absent in cells transfected with pCMV5 vector (not shown). (B) Ca2+ transients in HEK293 cells expressing WT-RyR1 or RyR1-ΔFF as changes of Fluo-4 fluorescence before and following the addition of 8 mM caffeine (arrow) to the bath solution. A.U., arbitrary units.
Table 1.
Properties of homotetrameric RyR1-WT and RyR1-ΔFF channels
| RyR1-WT | RyR1-ΔFF | |
|---|---|---|
| Immunoblots (%WT) | 100 | 66.9±12.7(4)* |
| Caffeine Response (% WT) | 100 | 86.5±13.5(4) |
| Bmax of [3H]Ry binding (%WT) | 100 | 4.6±2.7(4)* |
| Po at 2 μM Ca2+ | 0.11±0.03(13)a | 0.45±0.19(6) |
| Po at 0.1 μM Ca2+ | 0.01±0.01(9)a | 0.58±0.16(6)* |
| γK+ (pS) | 766±11(12)a | 354±76(6)* |
| ICa (+10 mM Ca trans) (pA) | −2.3±0.1(9)a | 0.1±0.1(6)* |
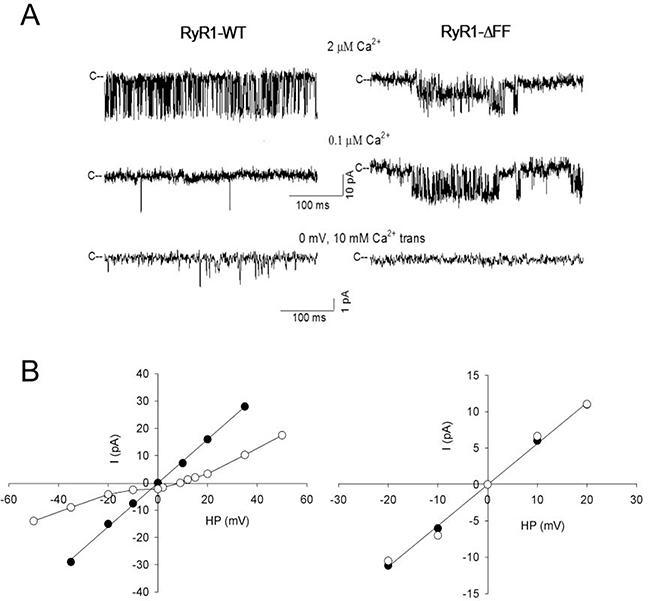
Membrane fractions containing RyR1-WT or RyR1-ΔFF were fused with planar lipid bilayers to determine their gating and ion permeation properties. Single channels were recorded with 0.25 M KCl on both sides of the bilayer, taking advantage of the impermeability of RyR1 to Cl− and the high conduction of K+ relative to Ca2+[3]. Channel open probability (Po) of RyR1-WT in presence of 2 μM cytosolic Ca2+ was 0.11 ± 0.03 (n=13) and K+ conductance was 766 ± 11 pS (n=12) (Fig. 3, Table 1). Reduction of cytosolic Ca2+ to 0.1 μM decreased RyR1-WT Po to near zero. In the presence of 10 mM SR luminal Ca2+, a Ca2+ current of −2.3±0.1 pA (n=9)was obtained at 0 mV. In contrast, RyR1-ΔFF exhibited reduced variable K+ conductance, while maintaining a similar Po at 2 μM and 0.1 μM cytosolic Ca2+ and failing to conduct Ca2+. The results suggest that the RyR1-ΔFF variant had a major impact on the function of the channel resulting in loss of Ca2+-dependent channel activity and Ca2+ conductance in lipid bilayers. We previously reported that purified loss-of-function channels exhibited similar variable K+ conductances (e.g. RyR1-G4898E: 410 ± 50 pS; RyR1-G4898R: 352 ± 61 pS; RyR1-Δ4926/I4927: 621 ± 66 pS, Ref. 10). However, we could not rule out the possibility of trace contaminate channels in the bilayers, as variable K+ conductances were occasionally observed in recordings using crude WT membrane preparations.
Figure 3. Single-channel measurements of homotetrameric RyR1-WT and RyR1-ΔFF channel complexes.
(A) Representative single-channel currents at −20 mV (upper and middle traces) or 0 mV (bottom traces) shown as downward deflections from the closed states (c−) in symmetrical 0.25 M KCl with 2 μM Ca2+ in the cis chamber (upper traces) and after the subsequent addition of EGTA to yield free Ca2+ of 0.1 μM (middle traces) or after the addition of 10 mM Ca2+ to the trans chamber (bottom traces). (B) Representative current-voltage relationships in 0.25 M symmetrical KCl (●) and after the addition of 10 mM trans Ca2+ (○). Current and time scales for single-channel traces are shown. Averaged Po values and ion permeation properties are summarized in Table 1.
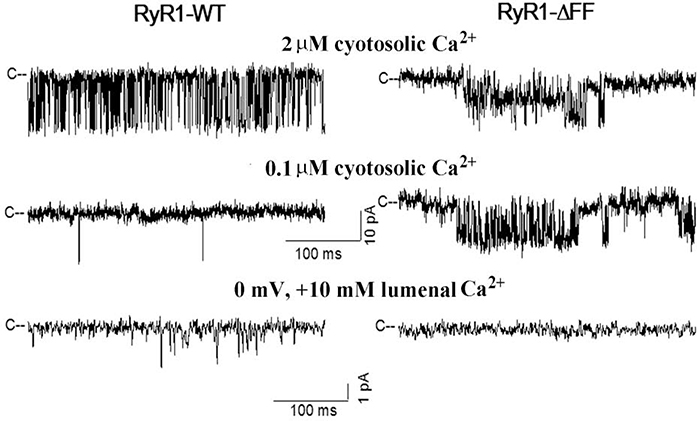
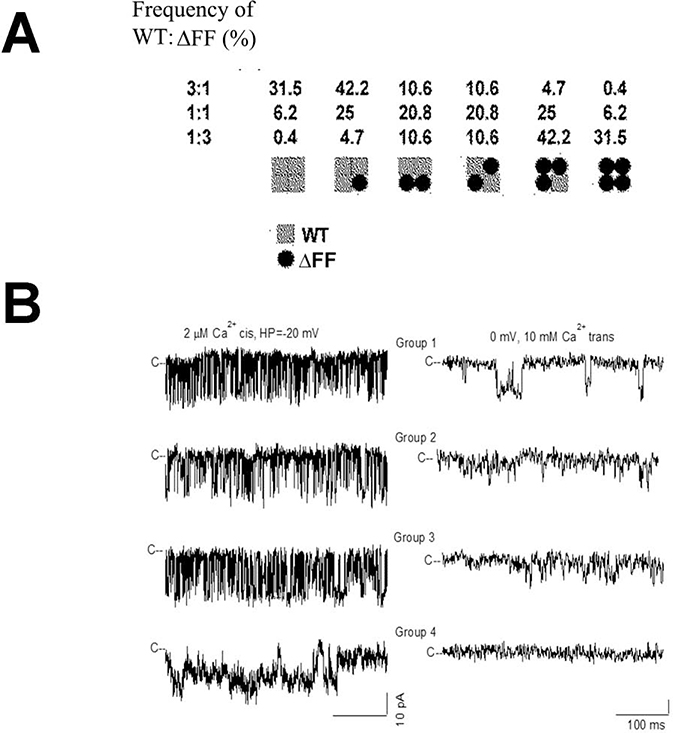
The four 565-kDa subunit composition of RyR1 suggests that in patients heterozygous for WT and ΔFF subunits heterotetrameric channels are formed that differ in their gating and ion permeation properties. To address this, HEK293 cells were co-transfected with RyR1-WT and RyR1-ΔFF expression vectors in ratios of 3:1, 1:1 and 1:3. Assuming uniform distribution of WT and mutant subunits, six channel groups were predicted (Fig. 4A). We observed 4 groups of channels that differed in their gating and ion permeation characteristics in 29 single-channel recordings (Fig. 4B, Table 2). Group 1 exhibited single-channel properties that suggested homotetrameric channels corresponding to RyR1-WT. Groups 2 and 3 had channel open probabilities (Po) and K+ conductances similar to WT, but a reduced Ca2+ current (−1.8 pA and −1.4 pA vs −2.3 pA for WT) and Ca2+ over K+ selectivity (3.6 and 1. 6 vs 6.5 for WT). Like homotetrameric RyR1-ΔFF channels, Group 4 channels had elevated Po,s at 2 μM and 0.1 μM cytosolic Ca2+ and failed to conduct Ca2+, while maintaining K+ conductance close to WT. Additionally, we observed one single channel that had reduced K+ conductance (494 pS vs 772 for WT). Time analysis indicated that Group 1 (WT) channel (Fig. 4B left panel) had mean closed and open times that were similar to Group 2 and 3 channels (Supplemental Fig. S3). By comparison, the loss-of-function Group 4 channel had an increased mean closed time, while maintaining mean open time similar to Group 1–3 channels. The PCa/PK values of the four groups of channels differed significantly as determined by Anova (1 way) followed by Tukey Test (Supplemental Figure S4).
Figure 4. Single-channel measurements of homotetrameric and heterotetrameric RyR1-WT and RyR1-ΔFF channel complexes.
(A) Subunit distribution and frequency of channel complexes in cells expressing WT or ΔFF subunits at the indicated ratios, assuming uniform distribution of subunits. (B) Representative single-channel measurements were performed and analyzed as in Fig. 3. Current and time scales for single-channel traces are as shown. Scale bars apply to all four left and right traces, respectively. Averaged Po values and ion permeation properties are summarized in Table 2.
Table 2.
Single-channel recordings of membrane isolates from HEK293 cells co-transfected with pCMV5-RyR1-WT and pCMV5-RyR1-ΔFF.
| Po | γK+ | ICa (+10 mM Ca trans) | PCa/PK | ||
|---|---|---|---|---|---|
| 2 μM Ca2+ | 0.1 μM Ca2+ | (pS) | (pA) | ||
| Group 1 (WT) | 0.17±0.05(6) | 0.01±0.01(6) | 772±16(6) | −2.3±0.1(6) | 6.5±0.1(6) |
| Group 2 | 0.21±0.02(10) | 0.03±0.01(10) | 769±11(10) | −1.8±0.1*(10) | 3.6±0.2*(10) |
| Group 3 | 0.21±0.03(5) | 0.01±0.01(5) | 798±20(5) | −1.4±0.1*(5) | 1.6±0.2*(5) |
| Group 4 | 0.65±0.13(7) | 0.70±0.12(7) | 733±14(7) | 0.1±0.1*(7) | ND |
Data denote means ± S.E.M
p<0.05 compared to WT
ND, not determined
A regression coefficient of 0.97 (n = 9) indicated that the number of experimentally determined channel groups in Table 2 was in good agreement with calculated channel groups assuming uniform distribution of wild-type and mutant subunits expressed in HEK293 cells (in parenthesis, Table 3). The results suggest that heterotetrameric channels composed of WT and mutant subunits conduct Ca2+ and respond to cytosolic Ca2+.
Table 3.
Comparison of the number of channel types determined from single channel measurements and calculation
| RyR expression vector ratio | RyR1 channel types | ||
|---|---|---|---|
| WT:ΔFF | WT | Heterotetrameric | ΔFF |
| 3:1 | 5 (4.1) | 7 (8.8) | 1 (0.1) |
| 1:1 | 1 (0.8) | 12 (11.4) | 0 (0.8) |
| 1:3 | 0 (0.1) | 3 (2.0) | 0 (0.9) |
Number of experimentally determined homotetrameric WT (Group 1, Table 2) heterotetrameric (Groups 2–4) and homotetrameric ΔFF (Group 5) channels at the indicated expression vector ratios using 7 μg total DNA/dish. In parentheses are the number of predicted channel types assuming uniform distribution of WT and ΔFF subunits in channel complexes.
3.3. Molecular dynamics simulations
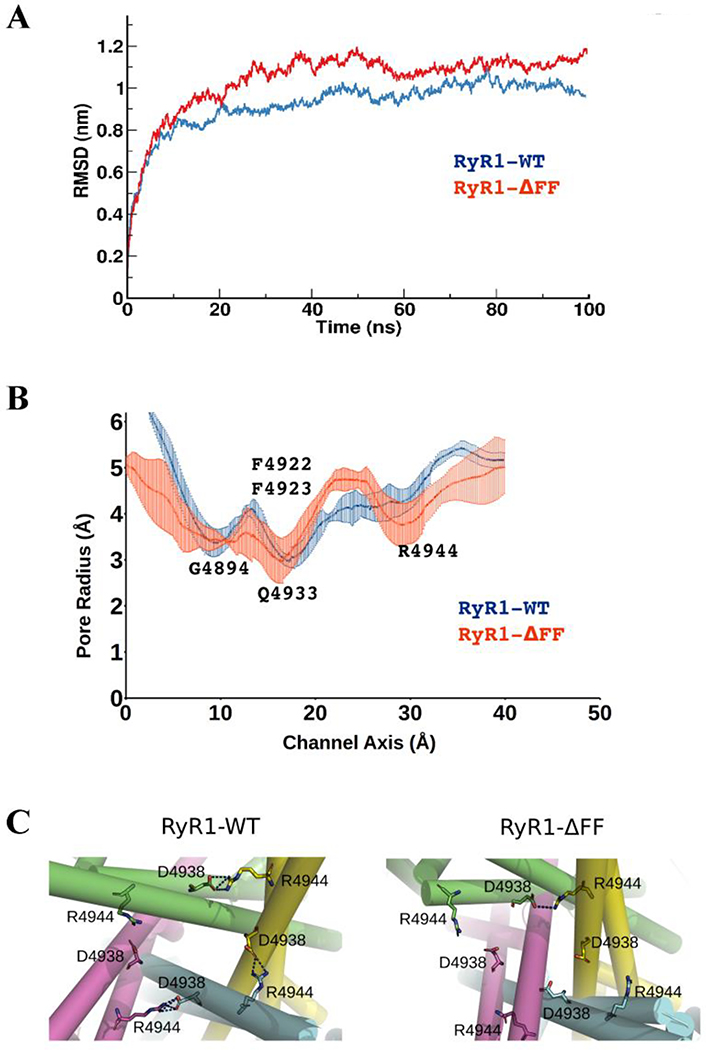
The effects of the ΔFF deletion on the RyR1 pore structure and stability were modeled. We linked the residues flanking the deletion without major changes taking place in the helical registry and performed molecular dynamics simulations. To characterize the effects of the ΔFF deletion on the stability of the pore structure, the root mean square deviations (RMSD) of Cα atoms of WT and ΔFF were plotted with respect to the simulation time (Fig. 5A). The RMSD plots show that during the last 60 ns of the trajectory, RyR1-WT was structural stable throughout the simulation (RMSD ~ 0.95 nm). A higher RMSD (RMSD ~ 1.2 nm) with increased fluctuations for RyR1-ΔFF suggested that the deletion of the two phenylalanine residues in transmembrane S6 helix disturbed favorable interactions in the pore vestibule or formed energetically unfavorable contacts.
Figure 5. Pore properties of homotetrameric RyR1-WT and RyR1-ΔFF channel complexes.
(A) Structural stability of RyR1-WT (blue) and RyR1-ΔFF (red) quantified through backbone RMSD measurement. (B) Pore profiles of RyR1-WT and RyR1-ΔFF mutant channels estimated using the HOLE program. RyR1-ΔFF significantly increased channel fluctuations at Q4933 and R4944 and reduced the pore radius from 4.5 Å to 3.5 Å at R4944, which is approximately 20 Å from the deletion site. (C) Salt-bridges in (a) RyR1-WT and (b) RyR1-ΔFF. Three out of four chains in RyR1-WT show inter-subunit salt bridges (blue dotted lines) between R4944 and D4938, whereas, one out four chains in RyR1-ΔFF show R4944−D4938 salt bridge due to ΔFF deletion.
The computational data predict that the flow of ions in the RyR1-ΔFF channel might have been affected at two sites. The data in Fig. 5B suggest that, while deletion of ΔFF maintained a similar pore profile near the selectivity filter site (G4894), pore stability decreased at the pore Q4933 restriction site of the open channel [45, 46], as indicated by increased fluctuations. At R4944, protein stability and pore radius decreased. Computational analysis of cryo-EM densities predicted that RyR1-R4944 formed inter-subunit salt bridges (blue dotted lines) with RyR1-D4938 in RyR1-WT, and the ΔFF deletion attenuated the number of salt-bridge interactions between R4944 and D4938 from 3–4 chains in WT to one chain in the deletion mutant (Fig. 5C). Thus, loss of salt bridge interactions between R4944 and D4938 in RyR1-ΔFF distorted the stability and structure of the RyR1 pore at a region 20Å from the variant site.
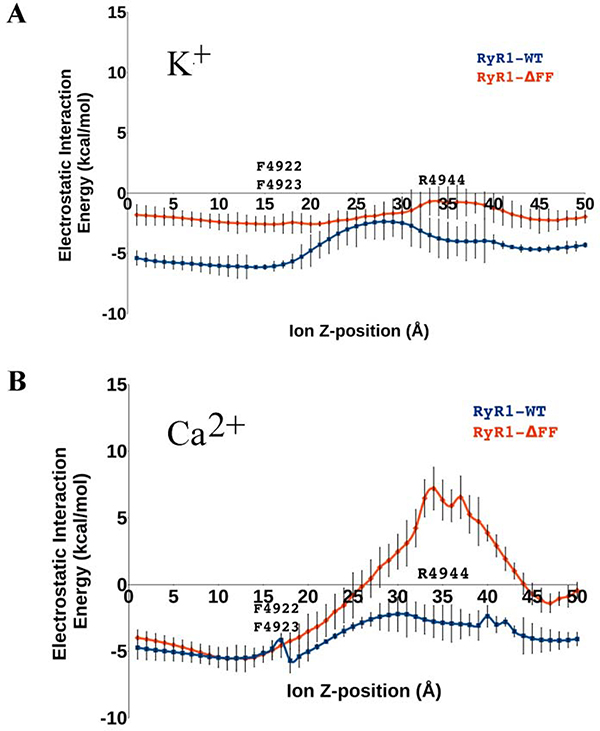
To estimate the ion conductance property of RyR1-WT and RyR1-ΔFF, the electrostatic interaction energies of Ca2+ and K+ were determined relative to the RyR1 pore-lining residues as the two ions passed through the pore. Figs. 6A and 6B show that the electrostatic interaction energies of RyR1-WT with Ca2+ and K+ ions fluctuated between −6 and −2 kcal/mol, respectively, which signified favorable interactions with pore-lining residues. RyR1-ΔFF exhibited divergent interaction energy profiles for Ca2+ and K+ ions, the mutant having a greater effect on Ca2+ carrying 2+ charges and K+ carrying 1+ charge. Specifically, RyR1-ΔFF raised the electrostatic interaction energy of the Ca2+ ion to +7 kcal/mol at a region 20Å from the variant site (Fig. 6B). The ion conductance data corroborated well with the experimental K+ and Ca2+ conductances of RyR1-ΔFF, showing a reduced K+ conductance and loss of Ca2+ conductance.
Figure 6. Electrostatic interaction energies of K+ (A) and Ca2+ (B) ions in homotetrameric RyR1-WT and RyR1-ΔFF channel complexes.
The plots summarize alterations in the electrostatics of Ca2+ and K+ ion passage through the pore along the channel axis in RyR1-WT (blue) and RyR1-ΔFF (red). Error bars represent the difference between the means of electrostatic interaction energy with 95% confidence interval.
4. Discussion
Here we report that the homotetrameric RyR1-ΔFF channel expresses caffeine-sensitive and Ca2+-conducting channels in HEK293 cells. In contrast, negligible [3H]ryanodine binding and loss of Ca2+ conductance and regulation by Ca2+ was observed for homotetrameric RyR1-ΔFF channels in single-channel measurements. Computational data predicted that the deletion of two phenylalanines affected channel stability of the RyR1 pore and resulted in an electrostatic energy barrier for the Ca2+ ions at a region 20Å from the variant site. In contrast, heterotetrameric RyR1s composed of WT and mutant subunits formed channels that conducted Ca2+ and responded to a change in cytosolic Ca2+. The findings suggest that, while the homotetrameric RyR1-ΔFF was unstable and lost function on removal from HEK293 cells, heterotetrameric RyR1s composed of different ratios of WT and ΔFF subunits formed channels that partially restored the WT phenotype in single-channel experiments.
Previous studies have indicated that co-expression of RyR1-WT and mutant subunits in HEK293 cells resulted in the formation of heterotetrameric channels [8, 10]. Four different arrangements of WT and ΔFF subunits are predicted for heterotetrameric WT:ΔFF channel complexes (Fig. 4A). We detected 3 groups of channel recordings (Groups 2–4) that differed from WT and ΔFF. Group 2 and 3 channels responded to cytosolic Ca2+ and had a K+ conductance comparable to WT, but had a reduced Ca2+ current and Ca2+/K+ selectivity compared to WT. This suggests that shortening of the S6 helices and potentially disrupting the interaction with neighboring amino acid residues only modestly impacted the pore structure of RyR1:ΔFF complexes comprised of 1 or 2 mutant subunits. Group 4 single-channel recordings showed a loss of Ca2+ permeation and regulation by Ca2+. This suggested that the presence of a larger number, possibly three ΔFF subunits in the RyR1:ΔFF complex, resulted in channels unable to conduct Ca2+ and to respond to Ca2+ in single-channel measurements. We conclude that in the two patients the expression of RyR1-WT and RyR1-ΔFF subunits resulted in the formation of heterotetrameric channel complexes that differed, depending on their WT:mutant subunit composition, in Ca2+ conductance and Ca2+ regulation.
The computational analysis corroborates the experimental data by predicting that the homotetrameric ΔFF variant conducts K+ but not Ca2+. The initial structure considered for molecular dynamics simulations had undisturbed helical registry after deletion of two residues and by linking the residues flanking next to the deletion. However, the deletion within the helix may have resulted in significant rearrangement of the structure, such as complete unwinding of the helix, or a shift of the entire helix relative to other regions. We consider such a scenario to be unlikely as we would not have observed a functional ΔFF variant in HEK cells and partial restoration of WT channel activity by inserting WT subunits in the mutant channel.
Results with RyR1-ΔFF are reminiscent of the results with RyR1-ΔV4926/I4927, a double S6 pore deletion variant [10]. RyR1-ΔV4926/I4927 was present as a heterozygous 6-bp in-frame deletion in a patient with CCD [47]. Transfection of HEK293 cells with RyR1-ΔV4926/I4927 expression vector resulted in channels unable to conduct Ca2+, whereas co-expression of WT and ΔV4926/I4927 resulted in two groups of channels that maintained a Ca2+-dependent channel activity comparable to WT but exhibited reduced Ca2+ conductance compared to WT. The results suggested that, as in the present study, at least two WT subunits are required in heterotetrameric channel complexes to maintain conducting Ca2+.
Several studies with RYR1 variants in individuals with FADS and related lethal recessive disorders have been reported [7, 12–18]. Some of the RYR1 variants identified in fetuses with FADS/LMPS phenotype, such as p.(Gly4782Arg) and p.(Leu4976Pro), are located in the RyR1 transmembrane domain [13], and may have potentially affected the Ca2+ conductance. The heterozygous missense variant p.(Gly4899Glu) in the RyR1 S6 pore-lining segment has been reported in a newborn girl with severe hypotonia, thin ribs, swallowing difficulty, respiratory distress, and cyanosis. The RYR1 variant was inherited from her mother who had a classic form of CCD [7]. However, the severe disorder in the female newborn suggested the presence of a second RYR1 mutation in trans that was not confirmed. A 9-year-old boy with congenital myopathy was reported to carry the 3-bp deletion c.14770_14772del in exon 102 that caused loss of the single phenylalanine at position 4924 [p.(Phe4924del)] [48] instead of the two neighboring phenylalanines 4923 and 4924 in the two siblings reported here. No information on zygosity of the RYR1 variant was given, and his healthy parents were not tested [48]. Together, the data demonstrate that variants in the transmembrane pore region of RyR1 are associated with congenital myopathies of various severity. These and our data further suggest that the p.(Phe4923_Phe4924del) variant in RYR1 possibly was one genetic factor that contributed to fetal akinesia in the two brothers reported here, while the second RYR1 variant was not identified in both patients. Alternatively, mutation in another gene cannot be excluded as the molecular basis of FADS in the two siblings.
4. Conclusion
In conclusion, the RyR1-ΔFFvariant located in the pore-lining, transmembrane-spanning segment of the S6 helix was shown to form functional channels in HEK293 cells but to be associated with decreased pore stability, altered pore structure and loss of function when isolated from HEK293 cells. Co-expression of RyR1-WT and ΔFF subunits resulted in Ca2+-dependent channel activities that displayed intermediate Ca2+ conductances. We conclude that the heterozygous RYR1 variant p.(Phe4923_Phe4924del) alone was insufficient to cause fetal akinesia, and additional genetic factors were involved.
Supplementary Material
Highlights -FF.
De novo RYR1 c.14767_14772del p.(Phe4923_Phe4924del) pore variant in FADS
Homotetrameric channel has an unstable pore structure
Heterotetramercic channel complexes composed of WT and mutant subunits conduct Ca2+
RYR1 p.(Phe4923_Phe4924del) contributed to but was not sufficient to cause FADS
Acknowledgements
The studies were supported by grants from the Deutsche Forschungsgemeinschaft (KU 1240/10-1 to K.K.; KO 4576/1-2 to F.K. and K.K.) and National Institute of Health Grant AR018687 (G.M. and N.V.D.).
ABBREVIATIONS
- EM
electron microscopy
- FADS
fetal akinesia deformation syndrome
- HEK
human embryonic kidney
- RyR1
type-1 ryanodine receptor
- SR
sarcoplasmic reticulum
- WT
wild type
Footnotes
Conflicts of interests
No conflicts of interests are declared by the authors
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Franzini-Armstrong C, Protasi F, Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions, Physiol Rev, 77 (1997) 699–729. [DOI] [PubMed] [Google Scholar]
- [2].Lanner JT, Georgiou DK, Joshi AD, Hamilton SL, Ryanodine receptors: structure, expression, molecular details, and function in calcium release, Cold Spring Harb Perspect Biol, 2 (2010) a003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Meissner G, The structural basis of ryanodine receptor ion channel function, J Gen Physiol, 149 (2017) 1065–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Klein A, Lillis S, Munteanu I, Scoto M, Zhou H, Quinlivan R, Straub V, Manzur AY, Roper H, Jeannet PY, Rakowicz W, Jones DH, Jensen UB, Wraige E, Trump N, Schara U, Lochmuller H, Sarkozy A, Kingston H, Norwood F, Damian M, Kirschner J, Longman C, Roberts M, Auer-Grumbach M, Hughes I, Bushby K, Sewry C, Robb S, Abbs S, Jungbluth H, Muntoni F, Clinical and genetic findings in a large cohort of patients with ryanodine receptor 1 gene-associated myopathies, Hum Mutat, 33 (2012) 981–988. [DOI] [PubMed] [Google Scholar]
- [5].Lawal TA, Todd JJ, Meilleur KG, Ryanodine Receptor 1-Related Myopathies: Diagnostic and Therapeutic Approaches, Neurotherapeutics, 15 (2018) 885–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jungbluth H, Treves S, Zorzato F, Sarkozy A, Ochala J, Sewry C, Phadke R, Gautel M, Muntoni F, Congenital myopathies: disorders of excitation-contraction coupling and muscle contraction, Nat Rev Neurol, 14 (2018) 151–167. [DOI] [PubMed] [Google Scholar]
- [7].Romero NB, Monnier N, Viollet L, Cortey A, Chevallay M, Leroy JP, Lunardi J, Fardeau M, Dominant and recessive central core disease associated with RYR1 mutations and fetal akinesia, Brain, 126 (2003) 2341–2349. [DOI] [PubMed] [Google Scholar]
- [8].Loy RE, Orynbayev M, Xu L, Andronache Z, Apostol S, Zvaritch E, MacLennan DH, Meissner G, Melzer W, Dirksen RT, Muscle weakness in Ryr1I4895T/WT knock-in mice as a result of reduced ryanodine receptor Ca2+ ion permeation and release from the sarcoplasmic reticulum, J Gen Physiol, 137 (2011) 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lyfenko AD, Ducreux S, Wang Y, Xu L, Zorzato F, Ferreiro A, Meissner G, Treves S, Dirksen RT, Two central core disease (CCD) deletions in the C-terminal region of RYR1 alter muscle excitation-contraction (EC) coupling by distinct mechanisms, Hum Mutat, 28 (2007) 61–68. [DOI] [PubMed] [Google Scholar]
- [10].Xu L, Wang Y, Yamaguchi N, Pasek DA, Meissner G, Single channel properties of heterotetrameric mutant RyR1 ion channels linked to core myopathies, J Biol Chem, 283 (2008) 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou H, Yamaguchi N, Xu L, Wang Y, Sewry C, Jungbluth H, Zorzato F, Bertini E, Muntoni F, Meissner G, Treves S, Characterization of recessive RYR1 mutations in core myopathies, Hum Mol Genet, 15 (2006) 2791–2803. [DOI] [PubMed] [Google Scholar]
- [12].Rajcan-Separovic E, Next generation sequencing in recurrent pregnancy loss-approaches and outcomes, Eur J Med Genet, (2019)Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [13].Alkhunaizi E, Shuster S, Shannon P, Siu VM, Darilek S, Mohila CA, Boissel S, Ellezam B, Fallet-Bianco C, Laberge AM, Zandberg J, Injeyan M, Hazrati LN, Hamdan F, Chitayat D, Homozygous/compound heterozygote RYR1 gene variants: Expanding the clinical spectrum, Am J Med Genet A, 179 (2019) 386–396. [DOI] [PubMed] [Google Scholar]
- [14].Suzumori N, Inagaki H, Ohtani A, Kumagai K, Takeda E, Yoshihara H, Sawada Y, Inuzuka S, Iwagaki S, Takahashi Y, Kurahashi H, Sugiura-Ogasawara M, Compound heterozygous RYR1 mutations by whole exome sequencing in a family with three repeated affected fetuses with fetal akinesia, Eur J Obstet Gynecol Reprod Biol, 230 (2018) 200–202. [DOI] [PubMed] [Google Scholar]
- [15].Casey J, Flood K, Ennis S, Doyle E, Farrell M, Lynch SA, Intra-familial variability associated with recessive RYR1 mutation diagnosed prenatally by exome sequencing, Prenat Diagn, 36 (2016) 1020–1026. [DOI] [PubMed] [Google Scholar]
- [16].Kariminejad A, Ghaderi-Sohi S, Hossein-Nejad Nedai H, Varasteh V, Moslemi AR, Tajsharghi H, Lethal multiple pterygium syndrome, the extreme end of the RYR1 spectrum, BMC Musculoskelet Disord, 17 (2016) 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McKie AB, Alsaedi A, Vogt J, Stuurman KE, Weiss MM, Shakeel H, Tee L, Morgan NV, Nikkels PG, van Haaften G, Park SM, van der Smagt JJ, Bugiani M, Maher ER, Germline mutations in RYR1 are associated with foetal akinesia deformation sequence/lethal multiple pterygium syndrome, Acta Neuropathol Commun, 2 (2014) 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ellard S, Kivuva E, Turnpenny P, Stals K, Johnson M, Xie W, Caswell R, Lango Allen H, An exome sequencing strategy to diagnose lethal autosomal recessive disorders, Eur J Hum Genet, 23 (2015) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, Roux AF, Smith T, Antonarakis SE, Taschner PE, HGVS Recommendations for the Description of Sequence Variants: 2016 Update, Hum Mutat, 37 (2016) 564–569. [DOI] [PubMed] [Google Scholar]
- [20].Bolger AM, Lohse M, Usadel B, Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30 (2014) 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gao L, Balshaw D, Xu L, Tripathy A, Xin C, Meissner G, Evidence for a role of the lumenal M3-M4 loop in skeletal muscle Ca2+ release channel (ryanodine receptor) activity and conductance, Biophys J, 79 (2000) 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang Y, Xu L, Pasek DA, Gillespie D, Meissner G, Probing the role of negatively charged amino acid residues in ion permeation of skeletal muscle ryanodine receptor, Biophys J, 89 (2005) 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chirasani VR, Xu L, Addis HG, Pasek DA, Dokholyan NV, Meissner G, Yamaguchi N, A central core disease mutation in the Ca2+ binding site of skeletal muscle ryanodine receptor impairs single channel regulation, Am J Physiol Cell Physiol, 317 (2019) C358–C365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sutko JL, Airey JA, Welch W, Ruest L, The pharmacology of ryanodine and related compounds, Pharmacol Rev, 49 (1997) 53–98. [PubMed] [Google Scholar]
- [25].Xu L, Wang Y, Gillespie D, Meissner G, Two rings of negative charges in the cytosolic vestibule of type-1 ryanodine receptor modulate ion fluxes, Biophys J, 90 (2006) 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mei Y, Xu L, Mowrey DD, Mendez Giraldez R, Wang Y, Pasek DA, Dokholyan NV, Meissner G, Channel Gating Dependence on Pore Lining Helix Glycine Residues in Skeletal Muscle Ryanodine Receptor, J Biol Chem, 290 (2015) 17535–17545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].des Georges A, Clarke OB, Zalk R, Yuan Q, Condon KJ, Grassucci RA, Hendrickson WA, Marks AR, Frank J, Structural Basis for Gating and Activation of RyR1, Cell, 167 (2016) 145–157.e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Webb B, Sali A, Comparative Protein Structure Modeling Using MODELLER, Curr Protoc Bioinformatics, 54 (2016) 5.6.1–5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hess B, Kutzner C, van der Spoel D, Lindahl E, GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation, J Chem Theory Comput, 4 (2008) 435–447. [DOI] [PubMed] [Google Scholar]
- [30].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: a web-based graphical user interface for CHARMM, J Comput Chem, 29 (2008) 1859–1865. [DOI] [PubMed] [Google Scholar]
- [31].Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL 3rd, MacKerell AD Jr., Klauda JB, Im W, CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field, J Chem Theory Comput, 12 (2016) 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Parrinello M, Rahman A, Polymorphic transitions in single crystals: A new molecular dynamics method, J. Appl. Physics, 52 (1981) 7182–7190. [Google Scholar]
- [33].Hess B, Bekker H, Berendsen HJC, Fraaije JGEM, LINCS: A linear constraint solver for molecular simulations., J. Comput. Chem, 18 (1997) 1463–1472. [Google Scholar]
- [34].Essmann U, Perera L, Berkowitz M, T. Darden L, Lee H, Pedersen LG, A smooth particle mesh Ewald method, J. Chem. Phys, 103 (1995) 8577–8593. [Google Scholar]
- [35].Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD, Optimization of the Additive CHARMM All-Atom Protein Force Field Targeting Improved Sampling of Backbone and Side-Chain and Dihedral Angles, J.Chem.Theory Comput, 8 (2012) 3257–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Klauda JB, Monje V, Kim T, Im W, Improving the CHARMM force field for polyunsaturated fatty acid chains, J Phys Chem B, 116 (2012) 9424–9431. [DOI] [PubMed] [Google Scholar]
- [37].Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS, HOLE: a program for the analysis of the pore dimensions of ion channel structural models, J Mol Graph, 14 (1996) 354–360, 376. [DOI] [PubMed] [Google Scholar]
- [38].Callenberg KM, Choudhary OP, de Forest GL, Gohara DW, Baker NA, Grabe M, APBSmem: a graphical interface for electrostatic calculations at the membrane, PLoS One,5 (2010) e12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA, Electrostatics of nanosystems: application to microtubules and the ribosome, Proc Natl Acad Sci U S A, 98 (2001) 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Xu L, Mowrey DD, Chirasani VR, Wang Y, Pasek DA, Dokholyan NV, Meissner G, G4941K substitution in the pore-lining S6 helix of the skeletal muscle ryanodine receptor increases RyR1 sensitivity to cytosolic and luminal Ca2+, J Biol Chem, 293 (2018) 2015–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Marcoline FV, Bethel N, Guerriero CJ, Brodsky JL, Grabe M, Membrane Protein Properties Revealed through Data-Rich Electrostatics Calculations, Structure, 23 (2015) 1526–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].The Pymol Molecular Grapics System, Versin 2.0 Schrodinger, LLC. [Google Scholar]
- [43].Zhou H, Brockington M, Jungbluth H, Monk D, Stanier P, Sewry CA, Moore GE, Muntoni F, Epigenetic allele silencing unveils recessive RYR1 mutations in core myopathies, Am J Hum Genet, 79 (2006) 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rousseau E, Meissner G, Single cardiac sarcoplasmic reticulum Ca2+-release channel: activation by caffeine, Am J Physiol, 256 (1989) H328–H333. [DOI] [PubMed] [Google Scholar]
- [45].Peng W, Shen H, Wu J, Guo W, Pan X, Wang R, Chen SR, Yan N, Structural basis for the gating mechanism of the type 2 ryanodine receptor RyR2, Science, 354 (2016) aah5324. [DOI] [PubMed] [Google Scholar]
- [46].Mowrey DD, Xu L, Mei Y, Pasek DA, Meissner G, Dokholyan NV, Ion-pulling simulations provide insights into the mechanisms of channel opening of the skeletal muscle ryanodine receptor, J Biol Chem, 292 (2017) 12947–12958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Davis MR, Haan E, Jungbluth H, Sewry C, North K, Muntoni F, Kuntzer T, Lamont P, Bankier A, Tomlinson P, Sanchez A, Walsh P, Nagarajan L, Oley C, Colley A, Gedeon A, Quinlivan R, Dixon J, James D, Muller CR, Laing NG, Principal mutation hotspot for central core disease and related myopathies in the C-terminal transmembrane region of the RYR1 gene, Neuromuscul Disord, 13 (2003) 151–157. [DOI] [PubMed] [Google Scholar]
- [48].Snoeck M, van Engelen BG, Kusters B, Lammens M, Meijer R, Molenaar JP, Raaphorst J, Verschuuren-Bemelmans CC, Straathof CS, Sie LT, de Coo IF, van der Pol WL, de Visser M, Scheffer H, Treves S, Jungbluth H, Voermans NC, Kamsteeg EJ, RYR1-related myopathies: a wide spectrum of phenotypes throughout life, Eur J Neurol, 22 (2015) 1094–1112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.