Abstract
Probe electrospray ionization mass spectrometry (PESI‐MS) is an ambient ionization‐based mass spectrometry method that surpasses the original electrospray ionization technique in features such as the rapidity of analysis, simplicity of the equipment and procedure, and lower cost. This study found that the PESI‐MS system with machine learning has the potential to establish a lipid‐based diagnosis of breast cancer with higher accuracy, using a simpler approach.

Rapid mass spectrometry for breast cancer
Introduction
Breast cancer is the most common cancer in women, and detection at an early stage of the disease assures a good prognosis. Initial screening is often based on imaging techniques such as mammography, but many women have dense breasts, which makes the sensitivity of mammography unsatisfactory1. Pathological examination of a tissue biopsy is a routine approach in establishing a breast cancer diagnosis, but this technique requires multiple steps over several days. No alternative or supportive procedures are currently available in a clinical setting. More accurate and less time‐consuming diagnostic methods using small specimens are needed.
Previous studies2, 3, 4 have been directed towards characterization of tissues, including intensive research for new genetic or metabolic markers. Using metabolomics, markers in tissues and body fluids can be identified3, 4. Liquid chromatography tandem mass spectrometry (LC‐MS/MS) analysis is good at identifying breast cancer‐specific molecules, but it is a time‐consuming procedure, starting from preparation of the column to purifying specific components from tissue extracts. To mitigate this situation, probe electrospray ionization mass spectrometry (PESI‐MS) is applied to a biological system5, enabling the analysis of almost ‘raw’ samples with no specific, laborious pretreatments.
PESI‐MS is an ambient ionization‐based mass spectrometry (MS) method that surpasses the original electrospray ionization technique in features such as the rapidity of analysis, simplicity of the equipment and procedure, and the low cost per capita6. For example, PESI‐MS has been used by the present authors for the rapid analysis of biological samples to discriminate cancers7, 8. Additionally, machine learning has been combined with PESI‐MS to diagnose various diseases with reference to information such as medical images and clinical data9, 10.
Methods
Full details of the study design, methods employed, and statistical analysis can be found in Appendix S1 (supporting information).
Results and Discussion
PESI‐MS and LC‐MS/MS were used to study breast cancer as summarized in Fig. 1. To create a database, 46 sets of spectra were acquired from small samples (10 mg) of normal and cancerous tissues in both positive and negative ion modes of PESI‐MS. Breast cancer‐specific spectra in each ion mode were analysed by eMSTAT Solution software (Shimadzu Corporation, Kyoto, Japan).
Figure 1.
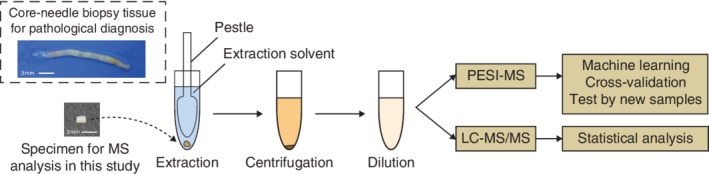
Study design A representative 10‐mg fragment of specimen is shown on the left with core‐needle biopsy tissue for comparison of sizes. The procedure of sample preparation for the probe electrospray ionization mass spectrometry (PESI‐MS) and liquid chromatography tandem mass spectrometry (LC‐MS/MS) analyses is depicted on the right.
Partial least squares discriminant analysis (PLS‐DA) plots showed clear differences between the non‐cancerous and cancerous groups with both ion modes (Fig. 2 a,b). This suggests that the instrumentation can distinguish breast cancer from normal breast tissue, even with specimens much smaller than those obtained by core‐needle biopsy.
Figure 2.
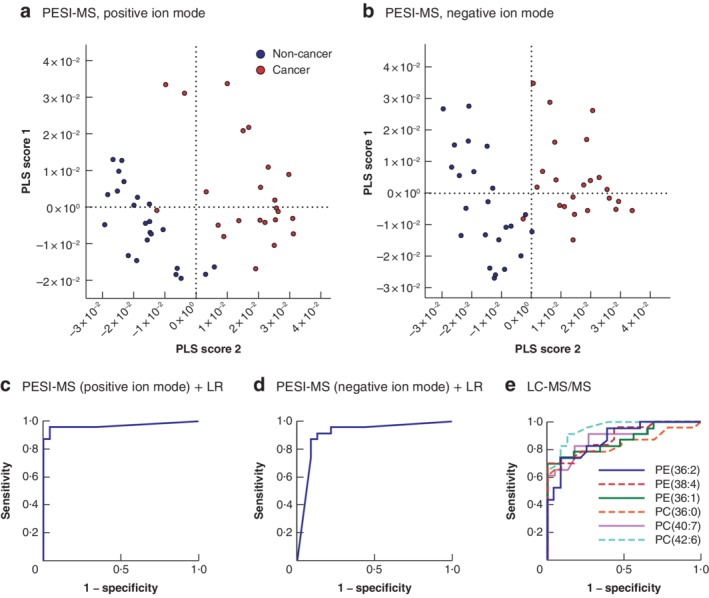
Probe electrospray ionization mass spectrometry combined with logistic regression analysis showed better prediction of breast cancer than biomarkers based on liquid chromatography tandem mass spectrometry a,b Scatter plots of partial least squares discriminant analysis scores of mass spectra from 46 specimens in a positive and b negative ion modes. c,d Depiction of receiver operating characteristic (ROC) curves for c positive and d negative ion modes using sensitivity and specificity scores. Area under the ROC curve (AUC) values indicate the evaluation of the discrimination algorithm: c AUC = 0·967; d AUC = 0·910. e ROC curves giving AUC values for phosphatidylethanolamine (PE) (36:2): AUC = 0·870; PE(38:4): AUC = 0·860; PE(36:1): AUC = 0·886; phosphatidylcholine (PC) (36:0): AUC = 0·836; PC(40:7): AUC = 0·889; PC(42:6): AUC = 0·952. PESI‐MS, probe electrospray ionization mass spectrometry; PLS, partial least squares; LR, logistic regression; LC‐MS/MS, liquid chromatography tandem mass spectrometry.
All mass spectra obtained from 23 patients with breast cancer were further analysed by logistic regression to establish an algorithm for distinguishing breast cancer from normal breast tissue. To test the predictive power of the constructed algorithm, leave‐one‐out cross‐validation was performed. Ninety‐two data sets (23 non‐cancerous and 23 cancerous samples analysed using two ion modes) were used to evaluate the potential of the system for breast cancer diagnosis. According to the relative cumulative frequency distribution plots, the thresholds of probability were 0·480–0·625 and 0·370–0·465 in the positive and negative ion modes respectively (Fig. S1, supporting information). At these points, sensitivity, specificity and accuracy in the positive ion mode were all 95·7 per cent, whereas those in the negative ion mode were 87·0, 91·3 and 89·1 per cent respectively (Table S1, supporting information). The area under the receiver operating characteristic curve (AUC) was calculated to be 0·967 and 0·910 for the positive and negative ion modes respectively (Fig. 2 c,d).
The system was further validated using specimens from additional patients and 24 data sets (6 non‐cancerous and 6 cancerous samples in both ion modes) and the generated algorithm. This testing achieved complete (100 per cent) discrimination in the positive ion mode (Table S2, supporting information). Data from the negative ion mode yielded one mismatch among the non‐cancerous samples (92 per cent accuracy).
Collectively, these results strongly indicate that this diagnostic system can distinguish breast cancer from normal breast tissue. Previous reports showed the usefulness of this system in discriminating squamous cell carcinoma of head and neck regions from normal mucosa11, and the present study of breast cancer indicates that this technology can be applied to epithelial tissues regardless of their origin. Moreover, because breast cancer is heterogeneous and comprises glands and a large proportion of stroma, this study has revealed the versatility of the instrumentation.
The present study demonstrates that this system can discriminate breast cancer from normal breast tissue without identifying the molecules. However, it is vital to focus on the specific molecules that are critical to the pathogenesis and pathophysiology of breast cancer. To address this issue, LC‐MS/MS was used to enumerate the candidate molecular markers. Some species of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) were increased significantly in breast cancer (Table S3, supporting information). AUC values were calculated for those species with significantly lower P values: PE(36:2), PE(38:4), PE(36:1), PC(36:0), PC(40:7) and PC(42:6) (Fig. 2e; Fig. S2, supporting information). Interestingly, none of the specific molecules exceeded the power of the instrumentation employing machine learning with positive ion mode. Because PC(42:6) gave the highest AUC value (0·952) among the individual molecules, this can be assumed to be a candidate molecular marker of breast cancer. Although data for PE(36:2), PE(38:4), PE(36:1), PC(40:6) and PC(30:0) have been reported in the previous studies12, 13, the finding of PC(42:6) as a potential molecular marker is novel. This result may be attributed to study population differences and/or the precision of the LC‐MS/MS used here.
The expression of oestrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor (HER) 2 in breast cancer provides valuable guidance for systemic treatments. The expression of the proliferation marker Ki‐67 is used to establish the aggressiveness of the cancer. All of these can be combined as surrogate markers for the molecular subtypes of breast cancer. To investigate whether these profiles were predicted by this instrumentation, 28 samples judged as cancer were classified by machine learning into two subgroups according to the value of each criterion. Spectra were acquired by PESI‐MS and analysed by eMSTAT Solution™ software. PLS‐DA could discriminate two groups categorized by the signature of Ki‐67, HER2, histological tumour grade or PR (Fig. S3, supporting information). In this study, a high Ki‐67 value overlapped completely with a low ER value, but pT and pN groups were less separated (Fig. S3, supporting information). These results suggest that the instrumentation has the potential to assess several breast cancer properties, such as aggressiveness. To launch a diagnostic system for breast cancer subgrouping on to the market, more mass spectra and clinical data need to be entered into the database.
This approach using PESI‐MS requires only a small amount of tissue with minimal pretreatments, and enables instantaneous judgement using machine learning. Ambient MS techniques, including PESI‐MS, have been studied in diagnostics. Rapid evaporative ionization mass spectrometry (REIMS) and desorption electrospray ionization (DESI) MS were used to study breast cancer diagnosis14, 15. REIMS has been commercialized as an intelligent knife (iKnife™; Waters Corporation, Milford, Massachusetts, USA), which analyses the mass spectra of vapours released by an electric surgical knife. The MasSpec Pen (Ms Pen Technologies Inc., Tulsa, Oklahoma, USA), combined with DESI‐MS, can analyse directly the surface of the solid specimens16. Recently, another ambient MS system, internal extractive electrospray ionization mass spectrometry (iEESI‐MS), was used for the discrimination of lung cancer by analysis of metabolites and lipids in tissue extracts17. This would also be applicable for extracts of breast cancer tissue. As PESI‐MS is available for solid and liquid samples18, 19, the combination of MS analysis with existing procedures such as core‐needle biopsy and liquid biopsy will be more useful and more accurate for breast cancer screening in the future. Before clinical implementation, rigorous validation studies with different MS methods will be mandatory.
Supporting information
Appendix S1 Methods
Table S1 Sensitivity, specificity and accuracy in positive and negative ion modes
Table S2 Further validation of sensitivity, specificity and accuracy in positive and negative ion modes
Table S3 Analysis of candidate molecular markers
Fig. S1 Establishment of a discrimination algorithm for breast cancer
Fig. S2 The six significantly changed phospholipids identified by liquid chromatography tandem mass spectrometry
Fig. S3 Discrimination of breast cancer subgroups
Acknowledgements
The authors thank M. Nagai, A. Osuga, A. Manita, T. Kozakai and M. Tanzawa for their technical assistance in this study. The study was performed in collaboration with the Shimadzu Corporation, which gave financial support.
Disclosure: S. Takeda receives financial support from Shimadzu Corporation.
References
- 1. Oiwa M, Endo T, Suda N, Morita T, Sato Y, Kawasaki T et al Can quantitative evaluation of mammographic breast density, ‘volumetric measurement’, predict the masking risk with dense breast tissue? Investigation by comparison with subjective visual estimation by Japanese radiologists. Breast Cancer 2019; 26: 349–358. [DOI] [PubMed] [Google Scholar]
- 2. Saparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF et al Adjuvant chemotherapy guided by a 21‐gene expression assay in breast cancer. N Engl J Med 2018; 379: 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silva C, Prestrelo R, Silva P, Tomás H, Câmara JS. Breast cancer metabolomics: from analytical platforms to multivariate data analysis. A review. Metabolites 2019; 9: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z, Li Z, Li H, Jiang Y. Metabolomics: a promising diagnostic and therapeutic implement for breast cancer. Onco Targets Ther 2019; 12: 6797–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takeda S, Yoshimura K, Hiraoka K. Innovations in analytical oncology – status quo of mass spectrometry‐based diagnostics for malignant tumor. J Anal Oncol 2012; 1: 74–80. [Google Scholar]
- 6. Huang MZ, Cheng SC, Cho YT, Shiea J. Ambient ionization mass spectrometry: a tutorial. Anal Chim Acta 2011; 702: 1–15. [DOI] [PubMed] [Google Scholar]
- 7. Yoshimura K, Chen LC, Mandal MK, Nakazawa T, Yu Z, Uchiyama T et al Analysis of renal cell carcinoma as a first step for developing mass spectrometry‐based diagnostics. J Am Soc Mass Spectrom 2012; 23: 1741–1749. [DOI] [PubMed] [Google Scholar]
- 8. Yoshimura K, Mandal MK, Hara M, Fujii H, Chen LC, Tanabe K et al Real‐time diagnosis of chemically induced hepatocellular carcinoma using a novel mass spectrometry‐based technique. Anal Biochem 2013; 441: 32–37. [DOI] [PubMed] [Google Scholar]
- 9. Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine learning for medical imaging. Radiographics 2017; 37: 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ko J, Baldassano SN, Loh P‐L, Kording K, Litt B, Issadore D. Machine learning to detect signatures of disease in liquid biopsies ‐ a user's guide. Lab Chip 2018; 18: 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ashizawa K, Yoshimura K, Johno H, Inoue T, Katoh R, Funayama S et al Construction of mass spectra database and diagnosis algorithm for head and neck squamous cell carcinoma. Oral Oncol 2017; 75: 111–119. [DOI] [PubMed] [Google Scholar]
- 12. Cífková E, Holčapek M, Lísa M, Vrána D, Gatĕk J, Melichar B. Determination of lipidomic differences between human breast cancer and surrounding normal tissues using HILIC‐HPLC/ESI‐MS and multivariate data analysis. Anal Bioanal Chem 2015; 407: 991–1002. [DOI] [PubMed] [Google Scholar]
- 13. Hilvo M, Denkert C, Lehtinen L, Müller B, Brockmöller S, Seppänen‐Laakso T et al Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res 2011; 71: 3236–3246. [DOI] [PubMed] [Google Scholar]
- 14. Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ, Richardson AL et al Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci U S A 2014; 111: 15 184–15 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. St John ER, Balog J, Mckenzie JS, Rossi M, Covington A, Muirhead L et al Rapid evaporative ionisation mass spectrometry of electrosurgical vapours for the identification of breast pathology: towards an intelligent knife for breast cancer surgery. Breast Cancer Res 2017; 19: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown HM, Pirro V, Cooks RG. From DESI to the MasSpec Pen: ambient ionization mass spectrometry for tissue analysis and intrasurgical cancer diagnosis. Clin Chem 2018; 64: 628–630. [DOI] [PubMed] [Google Scholar]
- 17. Lu H, Zhang H, Chingin K, Wei Y, Xu J, Ke M et al Sequential detection of lipids, metabolites, and proteins in one tissue for improved cancer differentiation accuracy. Anal Chem 2019; 91: 10 532–10 540. [DOI] [PubMed] [Google Scholar]
- 18. Yoshimura K, Yamada Y, Ninomiya S, Chung WY, Chang Y‐T, Dennison AR et al Real‐time analysis of living animals and rapid screening of human fluid samples using remote sampling electrospray ionization mass spectrometry. J Pharm Biomed Anal 2019; 172: 372–378. [DOI] [PubMed] [Google Scholar]
- 19. Takeda S, Yoshimura K, Tanihata H. Sample preparation for probe electrospray ionization mass spectrometry. J Vis Exp 2020. 10.3791/59942. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1 Methods
Table S1 Sensitivity, specificity and accuracy in positive and negative ion modes
Table S2 Further validation of sensitivity, specificity and accuracy in positive and negative ion modes
Table S3 Analysis of candidate molecular markers
Fig. S1 Establishment of a discrimination algorithm for breast cancer
Fig. S2 The six significantly changed phospholipids identified by liquid chromatography tandem mass spectrometry
Fig. S3 Discrimination of breast cancer subgroups


