Abstract
Ion pairing between the major phospholipids of the Staphylococcus aureus plasma membrane (phosphatidylglycerol – PG and lysyl‐phosphatidylglycerol – LPG) confers resistance to antimicrobial peptides and other antibiotics. We developed 3adLPG, a stable synthetic analogue which can substitute for the highy‐labile native LPG, in biophysical experiments examining the membrane‐protecting role of lipid ion pairing, in S. aureus and other important bacteria. Here we examine the surface charge and lipid packing characteristics of synthetic biomimetic mixtures of DPPG and DP3adLPG in Langmuir monolayers, using a combination of complementary surface‐probing techniques such as infrared reflection‐absorption spectroscopy and grazing‐incidence x‐ray diffraction. The resultant phase diagram for the ion paired lipids sheds light on the mixing behavior of lipids in monolayer models of resistant phenotype bacterial membranes, and provides a platform for future biophysical studies.
Keywords: aminoacyl lipids, antimicrobial resistance, catanionic monolayers, membrane biophysics, Staphylococcus aureus
Lipid ion pairing in antibiotic‐resistant bacteria: A biomimetic binary mixed monolayer is used to model the effect of ion pairing between synthetic Staphylococcus aureus phospholipids on lipid packing and interfacial charge. The resultant phase diagram aids our understanding of how membrane composition plasticity may tune the bilayer physicochemical properties in response to antibiotic stress.
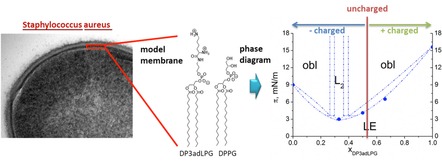
The aminoacyl lipids produced by a wide range of bacteria are becoming increasingly recognized as clinically relevant virulence factors, due to the role they play in phenotypic adaptations to the physical and biochemical stressors which confer intrinsic defence against infection.1, 2 The most widely studied of these lipids is lysyl‐phosphatidylglycerol (LPG), for which the genomic regulation and biosynthetic pathways in Staphylococcus aureus, have recently been elucidated in some detail.3, 4 Data from microbiological assays, has shown that an increased proportion of LPG in S. aureus membranes correlates with resistance to both host defensive peptides5 and membrane‐active therapeutic antibiotics.6 The mechanisms facilitating such resistances are assumed to involve the tuning of target membrane physical properties, notably those of interfacial charge and lipid ordering, which are influenced by the LPG content in the bacteria membranes.7 Biophysical investigations into these phenomena have been hampered by the labile nature of native LPG, which is readily hydrolysed under mild conditions, therefore exposing any such study to the risk of artefact.8, 9 To this end, stable LPG analogues have been synthesized. One such analogue, lysyl‐phosphatidylethanolamine (LPE) exhibited an inhibitory effect on antimicrobial peptide activity when incorporated into vesicles containing phosphatidylglycerol (PG).10 A second analogue, 3‐aza‐dehydroxy lysyl‐phosphatidylglycerol (3adLPG), facilitated enhanced membrane ordering and antimicrobial peptide resistance when mixed with PG under mildly acidic conditions,11 which are known to promote PG/LPG ion pair formation in model bacterial membranes.12 In order to gain a better understanding of the influence of lipid ion pairing on membrane structure and interfacial properties, we conducted a high‐resolution study of biomimetic mixtures of dipalmitoyl‐3adLPG and DPPG in Langmuir monolayers.
The natural phospholipid composition (Figure 1A) observed in different S. aureus strains is dominated by PG (≈40–70 %) and LPG (≈30–55 %) with saturated and iso‐branched fatty acids. Cardiolipin (CL) is present to 4–9 %.7 To simplify the lipid composition for the purposes of our biophysical investigations, only binary mixtures of DPPG and DP3adLPG (Figure 1B) were studied. Since CL is fully deprotonated at physiological pH13 and is known to promote negative curvature,14 its exclusion from the monolayer model removed the possibility that it might cause distortions in the chain packing. The use of lipids possessing only palmitoyl chains allowed us to focus on the charge interaction between the head groups, using total reflection x‐ray fluorescence (TXRF), and to determine a phase diagram for condensed monolayers using grazing incidence x‐ray diffraction (GIXD).
Figure 1.
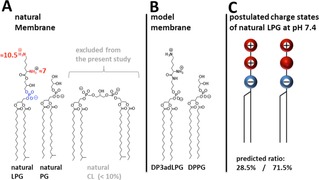
The natural lipid composition of the bacterial cell membrane of S. aureus. The major components are phosphatidylglycerol (PG), lysyl‐phosphatidylglycerol (LPG), and cardiolipin (CL).7 The numbers represent the pKa values according to literature for LPG.9 B) The components of the model system for biophysical investigations: 1,2‐dipalmitoyl‐sn‐phosphatidylglycerol (DPPG) and 1,2‐dipalmitoyl‐sn‐3‐aza‐dehydroxy lysyl‐phosphatidylglycerol (DP3adLPG). C) Predicted charge states of LPG at pH 7.4 according to the pKa value of the α‐amine given in A (calculated using the Henderson‐Hasselbalch equation).
In all of our experiments, premixed lipid solutions were spread as Langmuir monolayers at the gas/liquid interface on a pH 7.4 solution with the addition of 1 mM CsBr for the TRXF experiments. In earlier work,15, 16 we encountered the problem that the ultrapure water used in the subphase contained traces of calcium. To avoid the competition of divalent calcium ions with the monovalent cesium ions for interaction with the negatively charged lipid phosphate groups,17 50 μM EDTA was added. At pH 7.4, DPPG is assumed to be fully ionized18, 19 whereas DP3adLPG can be either zwitterionic or positively charged, in the same way as the native LPG (Figure 1C).9
To make comparisons between the charge states of the different DPPG/DP3adLPG mixed monolayers, TRXF measurements17 were performed at a surface pressure of 30 mN ⋅ m−1 (Figure 2), the physiological lateral pressure in membranes,20 when all of the different compositions were in the condensed phase (Figure 3A). Figure 2A and B show the Lα and Lβ bands of the subphase Cs+ and the Kα signal of Br−, both of which exhibited high intensities, allowing quantitative evaluation of the integrated values (Figure 2C). The pure DPPG monolayer has a high negative charge which attracts of Cs+ ions to the interface. The addition of DP3adLPG at a 0.33 mole fraction reduces the charge and therefore the amount of attracted Cs+. At xDP3adLPG=0.5 a charge neutral monolayer would expected if both amino groups of DP3adLPG were protonated and full ion pairing occurs.12 However, the residual negative charge of the monolayer proves that this is not the case. Assuming that DPPG is fully deprotonated at pH 7.4 (Figure 1B),15, 16 at xDP3adLPG=0.5 half of the total lipid molecules would be expected to carry a net negative charge. The residual negative charge in the system could only result from the presence of a mixture of positively charged DP3adLPG (phosphate deprotonated and both amines protonated) and the zwitterionic species (phosphate deprotonated, ϵ amine protonated and α amine uncharged) (Figure 1C). The small irregular shapes of the condensed phase domains observed in the 1 : 1 DPPG/DP3adLPG monolayer (Supporting Information Figure S2) are also indicative of a charge imbalance in the lipid mixture. The extrapolated point of zero charge is at xDP3adLPG=0.561 (Supporting Information Figure S1A). Examining the evolution of the bromide signal as a function of xDP3adLPG supports the same interpretation (Figure 2C), where monolayers with an xDP3adLPG >0.5 attract Br− to the interface in proportion with DP3adLPG concentration. At xDP3adLPG=0.5 and lower, the signal is negative because of the repulsive forces between the negatively charged monolayers and bromide which reduces the bromide concentration at the interface below the buffer value. Extrapolating the decrease in Br− intensity (Supporting Information Figure S1B) suggests that the interface would be neutralised at xDP3adLPG=0.524, a value slightly lower than that estimated from the Cs+ signal. Averaging both values implies that approximately 15 % of the DP3adLPG was zwitterionic, with the remaining 85 % carrying a net positive charge. Using this ratio of the different protonation states, the Henderson‐Hasselbalch equation gives a predicted DP3adLPG α‐amine pKa of 8.15. Due to the sensitivity of the TXRF measurements, it can be readily seen (Figure 2) that when one charged lipid is in the minority, its corresponding counter ion is repelled from the interface. This clearly indicates the formation of a neutral ion paired compound21 between the PG and 3adLPG, when the latter carries a net positive charge (Figure 1C). In this respect, 3adLPG is both structurally and functionally analogous to the native bacterial lipid.9, 12
Figure 2.
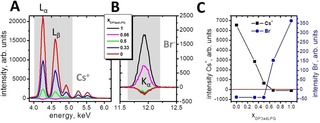
Sections of the buffer corrected TRXF spectra of the caesium region (A) and the bromide region (B) of Langmuir monolayers composed of DP3adLPG, DPPG and different mixtures on a subphase adjusted to pH 7.4 containing 1 mM CsBr and 0.05 mM EDTA. C) Intensities of the Lα and Lβ bands of the Cs signal (grey shaded area in A: Lα=4.286 keV, Lβ1=4.619 keV, and Lβ2=4.935 keV) and the Kα signal of Br (grey shaded area in B: Kα=11.924 keV) as a function of the mole fraction of DP3adLPG.
Figure 3.
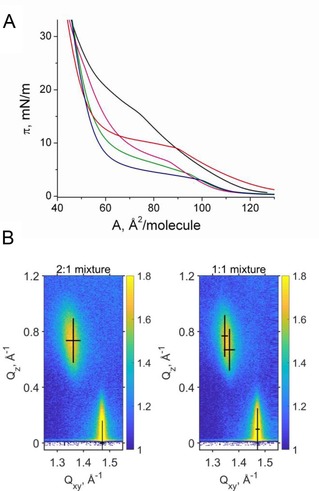
A) Pressure‐area isotherms of DPPG (red), DP3adLPG (black) and the DPPG/DP3adLPG mixtures 2 : 1 (blue), 1 : 1 (green) and 1 : 2 (magenta) on a pH 7.4 subphase containing 1 mM CsBr at 20 °C. B) GIXD data (scattered intensity vs. the vertical and horizontal components of the scattering vector) of the DPPG/DP3adLPG 2 : 1 and 1 : 1 mixtures at 30 mN ⋅ m−1 and the fitted Bragg peak and rod positions (black crosses). The width and the length of the crosses represent the FWHM of the peak and the corresponding rod.
The extent to which ion pairing between DPPG and DP3adLPG influenced their packing behaviour was probed using a number of monolayer techniques in addition to GIXD. Langmuir isotherms show that both the pure lipids and their various mixtures exhibit a first‐order phase transition from the liquid expanded (LE) to a liquid condensed (LC) phase (Figure 3A). The transition is characterized by a pronounced LE/LC coexistence region (see fluorescence microscopy images in Supporting Information Figure S2 and infrared reflection‐absorption spectroscopy experiments in Figures S3 and S4). The transition pressures of the mixtures are clearly lower compared with those of the pure lipids (Figure S5), with the DPPG/DP3adLPG 2 : 1 mixture exhibiting the lowest value. This suggests that the 2 : 1 mixture forms condensed phases more readily than the other mixtures, a phenomenon which has hitherto not been observed for native LPG, as a previous study on PG/LPG mixed monolayers used lipids which did not undergo first‐order transitions.12
The in‐plane structure of the condensed phases was determined at the Angstrom level by GIXD. The monolayer of DPPG is characterized by three diffraction peaks above the horizon (Qz>0) in the wide‐angle region (at high Qxy) at all the lateral pressures investigated. Three Bragg peaks are typical for an oblique lattice structure with tilted chains (Figure 3B). The Bragg peak positions, their full‐widths at half‐maximum (FWHM) and all lattice parameters obtained for DPPG, DP3adLPG and the different mixtures at 20 °C and different surface pressures are listed in Supporting Information Tables S1–S5. The cross‐sectional chain area of A0=19.7 Å2 indicates reduced rotational freedom, but it is slightly larger compared with the reported values of DPPG on water containing 1 mM CsBr.18 Plotting 1/cos(t) vs. the lateral pressure and extrapolating to zero tilt angle allows the determination of the tilting transition pressure.22 The extrapolated value (69.6 mN ⋅ m−1) is too high to be experimentally determined. Interestingly, this value is much larger compared to the one determined on a water subphase (50.5 mN ⋅ m−1).18 This shows clearly that on water (pH ∼5.8) with 1 mM CsBr, DPPG is only partially ionized. However, the ionization degree depends not only on the subphase pH but also on the charge state of a mixing partner. In mixtures with DHDAB (positively charged molecule), the ionization degree of DPPG is interestingly higher compared with the pure monolayer.18 For this reason, we chose to conduct our experiments at pH 7.4, in order to avoid the presence of protonated DPPG.
DP3adLPG exhibits a higher LE/LC transition pressure compared to DPPG (Figure 3A). The head group is quite large leading to a larger molecular in‐plane area of 48.0 Å2 (at 30 mN ⋅ m−1) compared to 45.2 Å2 for DPPG. The lattice structure is the same (three diffraction peaks of an oblique lattice structure) but the cross‐sectional area of the chains is with 20.0 Å2 larger and typical for a rotator phase, as a result of accommodating the bulky head group.
Two of the DPPG/DP3adLPG mixtures, 1 : 1 and 1 : 2, exhibit the same oblique in‐plane lattice structure. However, the mixture DPPG/DP3adLPG 2 : 1 displays only two diffraction peaks characteristic of a rectangular in‐plane lattice. The chains are tilted in the direction of the nearest neighbour (NN).
Based on isotherm and GIXD data, a putative phase diagram has been constructed (Figure 4A). The exact dimensions of the two‐phase co‐existence regions are unknown and would require many more mixtures to be studied what was clearly not the aim of this work. But it indicates the formation of a congruent melting compound in the binary system. The two lipids DPPG and DP3adLPG are completely miscible in the liquid‐like LE phase. However, the miscibility is not ideal as the molecular area vs. mole fraction (Figure S6) demonstrates. The molecular area in the mixtures is clearly much smaller compared to the expected one of ideal mixtures or completely de‐mixed systems indicating preferred interactions between the two unlike compounds. This suggests that even in the fluid state, the lipids are associated through head group‐driven ion pairing, which may explain the ordering effect these associations confer on fluid phase bilayers.11 In the condensed phase of the monolayer, the congruent melting compound DPPG/DP3adLPG 2 : 1 forms an L2 phase separated from the oblique phases on both sides of the phase diagram by small miscibility gaps. This indicates that in addition to the proportion of DP3adLPG regulating the surface charge through ion pairing with DPPG, it also alters chain packing and monolayer condensation, but in a somewhat unexpected way. Although the near surface neutrality of the 1 : 1 mixture demonstrates a higher degree of ion pairing and thus presumably a more condensed and stressor resistant combination,11, 12 it is the 2 : 1 mixture which, although more anionic, appears to have the greater propensity to form a stable condensed phase. How this proportion of the lipids might affect bacterial physiology in nature requires further study, however, it should be noted that for a number of S. aureus strains, when grown at physiological pH, the proportion of anionic to cationic lipids in their membranes is approximately 2 : 1.7, 23
Figure 4.
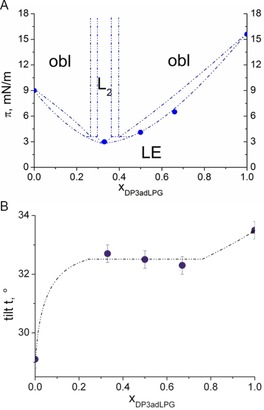
A) Phase diagram (lateral pressure π versus the mole‐fraction of DP3adLPG) of the DPPG/DP3adLPG system. The phases observed by GIXD are obl (oblique), L2 (orthorhombic with NN tilt). The first‐order transition pressures (•) from the disordered LE to an ordered LC phase are determined by pressure‐area isotherms. B) Tilt angle t at 30 mN ⋅ m−1 (the lateral pressure in biological membranes20) versus the mole fraction xDP3adLPG.
Despite the differences in lattice geometries between the 2 : 1 and the other DPPG/DP3adLPG mixtures, in all cases the tilt angle of the chains decreases with increasing lateral pressure (Figure S7). Comparing the tilt angles at 30 mN ⋅ m−1 displays an interesting behaviour of the mixtures (Figure 4B). The tilt angle is quite constant over a wide composition range showing that the thickness of the membranes does not depend on the composition. Therefore, despite alteration in membrane charge and condensation potential, altering DP3adLPG content should not affect the thickness of bilayers in which it is combined with DPPG. It should be noted that due to the differences in the calculated α‐amine pKa of native LPG (pKa 7) and DP3adLPG (pKa 8.15) the pH conditions modelled in this study would be comparable to the native lipid at pH 6.25 (according to the Henderson‐Hasselbalch equation). In S. aureus, the proton gradient across the plasma membrane ensures that the outer leaflet has a pH of 6.25–6.65,24 which means that our monolayer model provides an accurate mimetic for the environment‐facing half of the lipid bilayer. Thus, 3adLPG is not only useful in biomimetic model membranes used to assess antimicrobial peptide mechanisms against S. aureus,11 but should also fulfil the need for more biorelevant lipid environments for the biophysical study of bacterial membrane proteins.25
It is increasingly recognised that in order to improve our knowledge of the preventative and therapeutic interventions which can be made in host/pathogen interactions, a multi‐disciplinary approach is required. Bacterial membrane biophysics can play a very important role in this field, by contributing to research elucidating the mechanisms of susceptibility and resistance to novel therapeutics such as antimicrobial peptides,26 and facilitating structure and functional studies on membrane‐associated bacterial virulence factors. The validity of such biophysical research relies on the biological relevance of the model systems employed, which becomes more important as they necessarily increase in complexity, to improve their biomimetic proximity. One way to ensure this would be to use lipids extracted from bacteria.25 However, for studies which require membranes of defined composition whose physical properties may be tuned to suit specific purposes, especially those related to S. aureus and other aminoacyl lipid containing pathogens, 3adLPG may prove to be a valuable tool.
Experimental Section
Experimental details are given in the Supporting Information.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities on the PETRA III ring, and R. Kirchhof for the technical support on the setup at P08.
C. Wölk, H. Youssef, T. Guttenberg, H. Marbach, G. Vizcay-Barrena, C. Shen, G. Brezesinski, R. D. Harvey, ChemPhysChem 2020, 21, 702.
Contributor Information
Dr. Christian Wölk, Email: christian.woelk@medizin.uni-leipzig.de
Dr. Richard D. Harvey, Email: richard.harvey@pharmazie.uni-halle.de.
References
- 1. Roy H., IUBMB Life 2009, 61, 940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hornef M. W., Normark S., Henriques-Normark B., Rhen M., Chem. Immunol. Allergy 2005, 86, 72–98. [DOI] [PubMed] [Google Scholar]
- 3. Slavetinsky C., Kuhn S., Peschel A., Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1310–1318. [DOI] [PubMed] [Google Scholar]
- 4. Ernst C. M., Slavetinsky C. J., Kuhn S., Hauser J. N., Nega M., Mishra N. N., Gekeler C., Bayer A. S., Peschel A., mBio 2018, 9, e01659–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mukhopadhyay K., Whitmire W., Xiong Y. Q., Molden J., Jones T., Peschel A., Staubitz P., Adler-Moore J., McNamara P. J., Proctor R. A., Yeaman M. R., Bayer A. S., Microbiology-Sgm 2007, 153, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 6. Ernst C. M., Peschel A., Int. J. Med. Microbiol. 2019, 309, 359–363. [DOI] [PubMed] [Google Scholar]
- 7. Rehal R. P., Marbach H., Hubbard A. T. M., Sacranie A. A., Sebastiani F., Fragneto G., Harvey R. D., Chem. Phys. Lipids 2017, 206, 60–70. [DOI] [PubMed] [Google Scholar]
- 8. Danner S., Pabst G., Lohner K., Hickel A., Biophys. J. 2008, 94, 2150–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tocanne J. F., Ververgaert P. H. J. T., Verkleij A. J., van Deenen L. L. M., Chem. Phys. Lipids 1974, 12, 201–219. [DOI] [PubMed] [Google Scholar]
- 10. Cox E., Michalak A., Pagentine S., Seaton P., Pokorny A., Biochim. Biophys. Acta Biomembr. 2014, 1838, 2198–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rehal R., Gaffney P. R. J., Hubbard A. T. M., Barker R. D., Harvey R. D., Eur. J. Pharm. Sci. 2019, 128, 43–53. [DOI] [PubMed] [Google Scholar]
- 12. Tocanne J. F., Ververgaert P. H. J., Verkleij A. J., Van Deenen L. L. M., Chem. Phys. Lipids 1974, 12, 220–231. [DOI] [PubMed] [Google Scholar]
- 13. Sathappa M., Alder N. N., Biochim. Biophys. Acta Biomembr. 2016, 1858, 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzaki K., Sugishita K., Ishibe N., Ueha M., Nakata S., Miyajima K., Epand R. M., Biochemistry 1998, 37, 11856–11863. [DOI] [PubMed] [Google Scholar]
- 15. Sacré M. M., Tocanne J. F., Chem. Phys. Lipids 1977, 18, 334–354. [DOI] [PubMed] [Google Scholar]
- 16. Grigoriev D., Krustev R., Miller R., Pison U., J. Phys. Chem. B 1999, 103, 1013–1018. [Google Scholar]
- 17. Sturm M., Gutowski O., Brezesinski G., ChemPhysChem 2019, 20, cphc.201900126. [DOI] [PubMed] [Google Scholar]
- 18. Wölk C., Hause G., Gutowski O., Harvey R. D., Brezesinski G., Chem. Phys. Lipids 2019, 225, 104827. [DOI] [PubMed] [Google Scholar]
- 19. Brezesinski G., Schneck E., Langmuir 2019, 35, 8531–8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh D., Biochim. Biophys. Acta Rev. Biomembr. 1996, 1286, 183–223. [DOI] [PubMed] [Google Scholar]
- 21. Vaknin D., Bu W., J. Phys. Chem. Lett. 2010, 1, 1936–1940. [Google Scholar]
- 22. Bringezu F., Dobner B., Brezesinski G., Chem. Eur. J. 2002, 8, 3203–3210. [DOI] [PubMed] [Google Scholar]
- 23. Gould R. M., Lennarz W. J., J. Bacteriol. 1970, 104, 1135–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins S., Hamilton W., J. Bacteriol. 1976, 126, 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nickels J. D., Chatterjee S., Mostofian B., Stanley C. B., Ohl M., Zolnierczuk P., Schulz R., Myles D. A. A., Standaert R. F., Elkins J. G., Cheng X., Katsaras J., J. Phys. Chem. Lett. 2017, 8, 4214–4217. [DOI] [PubMed] [Google Scholar]
- 26. Arendt W., Hebecker S., Jager S., Nimtz M., Moser J., J. Bacteriol. 2012, 194, 1401–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


