Abstract
Fecal microbiota in seven different monogastric animal species, elephant, horse, human, marmoset, mouse, pig and, rat were compared using the same analytical protocol of 16S rRNA metagenome. Fecal microbiota in herbivores showed higher alpha diversity than omnivores except for pigs. Additionally, principal coordinate analysis based on weighted UniFrac distance demonstrated that herbivores and pigs clustered together, whereas other animal species were separately aggregated. In view of butyrate‐ and lactate‐producing bacteria, predominant genera were different depending on animal species. For example, the abundance of Faecalibacterium, a known butyrate producer, was 8.02% ± 3.22% in human while it was less than 1% in other animal species. Additionally, Bifidobacterium was a predominant lactate producer in human and marmoset, while it was rarely detected in other omnivores. The abundance of lactate‐producing bacteria in herbivores was notably lower than omnivores. On the other hand, herbivores as well as pig possess Fibrobacter, a cellulolytic bacterium. This study demonstrated that fecal microbiota in herbivorous animals is similar, sharing some common features such as higher alpha diversity and higher abundance of cellulolytic bacterium. On the other hand, omnivorous animals seem to possess unique fecal microbiota. It is of interest that pigs, although omnivore, have fecal microbiota showing some common features with herbivores.
Keywords: fecal microbiota, feeding habitat, herbivore, monogastric animal, omnivore
1. INTRODUCTION
The intestine harbors a dense, delicately balanced microbial population that mainly consists of bacteria (O'Hara & Shanahan, 2006). In general, the gut microbiota plays three major roles in the host's physiology: breaks down and utilizes dietary fiber that otherwise the host would not be able to, such as resistant polysaccharide; prevents adherence of pathogenic bacteria by competing for ecological niches and nutritional components; and synthesizes certain compounds such as vitamins, serotonin, and dopamine that helps promote the host's homeostasis.
Except for animals artificially bred germ‐free, all animal species possess to a microbiota in the gut. The microbiota is found at all locations in the intestinal tract, but its communities and their numbers vary depending on the environment of the location (e.g., oxygen concentration, pH, etc.) (Kovatcheva‐Datchary, Tremaroli, & Bäckhed, 2013). In hindgut fermenters, bacteria can be found in larger numbers in the hindgut than in the foregut, because microbial fermentation takes place mainly in the hindgut. (Stevens & Hume, 1998). Differences in the communities in the microbiota of animals are thought to be defined by different habitats and feeding preferences, as well as by putatively diverse genetic/immunological profiles (Ley et al., 2008; Wen & Duffy, 2017). While ruminants are preferentially herbivorous, monogastric animals can be categorized as herbivores, carnivores, or omnivores. Furthermore, there are clear differences between the structure of gastrointestinal tracts of monogastric herbivorous and carnivorous animals. For example, monogastric herbivores generally have a large cecum and/or colon for bacterial fermentation of fiber (Chivers & Hladik, 1980; Clauss et al., 2003).
In gastrointestinal research, metagenomic analysis of 16S rRNA gene is now widely used to obtain comprehensive and comparative information of the microbiota. Indeed, 16S rRNA gene metagenomic analysis has been used to study the gastrointestinal microbiota in a variety of animal species (Chassaing et al., 2015; Chung et al., 2016; Costa et al., 2015; Fan, Tang, Qu, Cao, & Huo, 2014; Ilmberger et al., 2014; Kim, Nguyen, Guevarra, Lee, & Unno, 2015), but only a few studies have used this technique to comprehensively compare the gut microbial profiles of different animal species. For example, O' Donnell, Harris, Ross, and O'Toole (2017) used 16S rRNA gene metagenome pyrosequencing analyses to compare the gastrointestinal microbiota of herbivorous animals, although pigs were included as an out‐group control. More recently, Nagpal et al. (2018) conducted a comparison between the gut microbiota of humans and animal models such as rats, mice, and macaques. Although these workers demonstrated that the gut microbiota was unique in each animal species, the study did not include herbivores. Separately, Ley et al. (2008) evaluated the gastrointestinal microbiota in 59 mammalian species using 16S rRNA gene cloning‐based analysis but they did not study the species at metagenomic level. In addition, Muegge et al. (2011) compared the microbiota of 33 mammalian species and demonstrated the adaptation of gut microbiota to diet. Nonetheless, the methodology was limited, because both studies used a relatively small number of samples per animal species (1–3) and in the study by Ley et al. (2008), the number of sequence reads per sample was smaller than 300, due to the methodological restrictions.
In the present study, we aimed to accumulate the knowledge of uniqueness or similarity of gut microbiota among monogastric animal species with relation to feeding habitat. The gastrointestinal microbiota of wide range of monogastric animals, namely two herbivorous and five omnivorous species, was comprehensively compared using the same protocol for 16S rRNA gene metagenomic analysis. For a statistical comparison, we collected at least five samples per animal species. Fecal samples were collected from elephants (Elephas maximus), horses (Equus caballus), humans (Homo sapiens), marmosets (Callithrix jacchus), mice (Mus musculus), pigs (Sus scrofa), and rats (Rattus norvegicus). Due to variation of the bacterial taxonomy among animal species evaluated, the abundance of more than 200 genera was significantly different. Therefore, in the present work, the abundance of butyrate‐ and lactate‐producing bacteria in each animal species is particularly discussed. This is due to their reported health‐promoting effects on humans (Louis & Flint, 2009; Masood, Qadir, Shirazi, & Khan, 2011) and pigs (Tsukahara, Hashizume, Koyama, & Ushida, 2006; Yoshida, Tsukahara, & Ushida, 2009) and due to plausibility of selection of bacterial genera that produce these organic acids. While butyrate‐ and lactate‐producing pathways are relatively conserved, in particular, bacterial genera, acetate‐, and propionate‐producing pathways are widely distributed among bacterial groups (Morrison & Preston, 2016; Reichardt et al., 2014).
2. MATERIALS AND METHODS
2.1. Sample collection
Fresh fecal samples were collected from Asian elephants reared at the Okinawa Zoo and Museum (Okinawa, Japan), Thoroughbred horses at the Hidaka Training and Research Center (Hokkaido, Japan), common marmosets at the Research Resources Division, RIKEN center for Brain Science (Saitama, Japan), C57BL/6 mice at Kyoto Prefectural University (Kyoto, Japan), LW (Landrace × Large white) pigs at two commercial farms in Japan, and Sprague‐Dawley rats at Shizuoka University (Shizuoka, Japan). All animals were adults. Except for those of humans, fecal samples were frozen immediately (−20ºC) after collection. The samples from each animal species were collected as a part of different experiments but none were used in the published articles. With regard to human samples, data from feces of healthy Japanese adults obtained in our previous work were used (Kawada, Naito, Andoh, Ozeki, & Inoue, 2019). Samples used for metagenomic analysis are listed in Table 1.
Table 1.
Animals and their diets, and 16S gene amplicon sequence reads generated in this study
| Animal | Binomial nomenclature | n | Male/Female | Diets | Housing | Sequence read |
|---|---|---|---|---|---|---|
| Elephant | Elephas maximus | 9 | 0/9 | Fruits and vegetables | Individual | 16,256 ± 8,923 |
| Horse | Equus caballus | 5 | 0/5 | Grasses and concentrated feed | Individual | 14,350 ± 4,677 |
| Human a | Homo sapiens | 5 | 3/2 | Not recorded | Individual | 20,834 ± 4,904 |
| Marmoset | Callithrix jacchus | 9 | 0/9 | Described in Shigeno et al.(2018) | Individual | 22,063 ± 4,798 |
| Mouse | Mus musculus | 6 | 0/6 | Labo MR Stock b | Single Herd | 29,271 ± 2,722 |
| Pig | Sus scrofa | 20 | 0/20 | Commercial diet for sow b | Individual | 13,801 ± 3,419 |
| Rat | Rattus norvegicus | 8 | 8/0 | AIN−93G c | Individual | 18,459 ± 9,339 |
The data obtained from freshly collected but not stored feces in the previous study were used.
Nihon Nosan Kogyo (Yokohama, Japan).
Japan CLEA (Tokyo, Japan).
2.2. Metagenomic analysis of 16S rRNA genes
DNA extraction from samples and library preparation for analysis of Illumina MiSeq Next Generation Sequencer (Illumina, San Diego, CA, USA) were carried out as described by Inoue et al.(2016).
De‐multiplexing of sequences as per dual indices was carried out using Quantitative Insights Into Microbial Ecology (QIIME) open‐source bioinformatics pipeline v1.9.0, (Caporaso et al., 2010), allowing a Phred score higher than Q21, a maximum of three errors in barcodes, and no ambiguous bases (Ns). Afterward, sequence analysis tool VSEARCH v2.4.3 (Rognes, Flouri, Nichols, Quince, & Mahé, 2016) was used for further filtering (maximum expected error = 1.0), removing replicates and chimeras, discarding clusters with less than two reads, and clustering OTU (identity 99%). This is because, clustering with 97% identity, a threshold often used, found to be too lenient for the dataset in this study, causing wrong taxonomy assignment. Taxonomy assignment of the resulting OTU was carried out using RDP classifier v2.10.2 (Wang, Garrity, Tiedje, & Cole, 2007) with the Greengenes database (published May, 2013). Default software settings were used for all analyses unless otherwise stated.
2.3. Statistical analysis
Alpha‐diversity indices Chao1 (richness) and Shannon (evenness) were calculated with the R “phyloseq” package (McMurdie & Holmes, 2013). Weighted UniFrac distance‐based principal coordinate analysis plot was generated with QIIME. Based on the Bray–Curtis distance, similarity of fecal microbial composition between samples at genus level was calculated with the R “vegan” package (https://CRAN.R-project.org/package=vegan).
Using Kruskal–Wallis and PERMANOVA (permutational multivariate analysis of variance), respectively, statistical comparison of alpha‐ and beta diversity was conducted with QIIME2 v2019.1.0 (Bolyen et al., 2019). The relative abundance of bacterial genera was statistically compared with the Kruskal–Wallis test followed by the Scheffe's post hoc test, using STAMP software (Parks, Tyson, Hugenholtz, & Beiko, 2014). Differences were considered significant when p < .05.
3. RESULTS
3.1. Alpha diversity
In total, 1,120,121 reads were obtained (average: 18,066 reads/sample; range: 6,469 to 36,123 reads). As shown in Figure 1, rarefaction curve reached an apparent plateau at 5,000 reads, suggesting that most variation present in the samples was covered.
Figure 1.
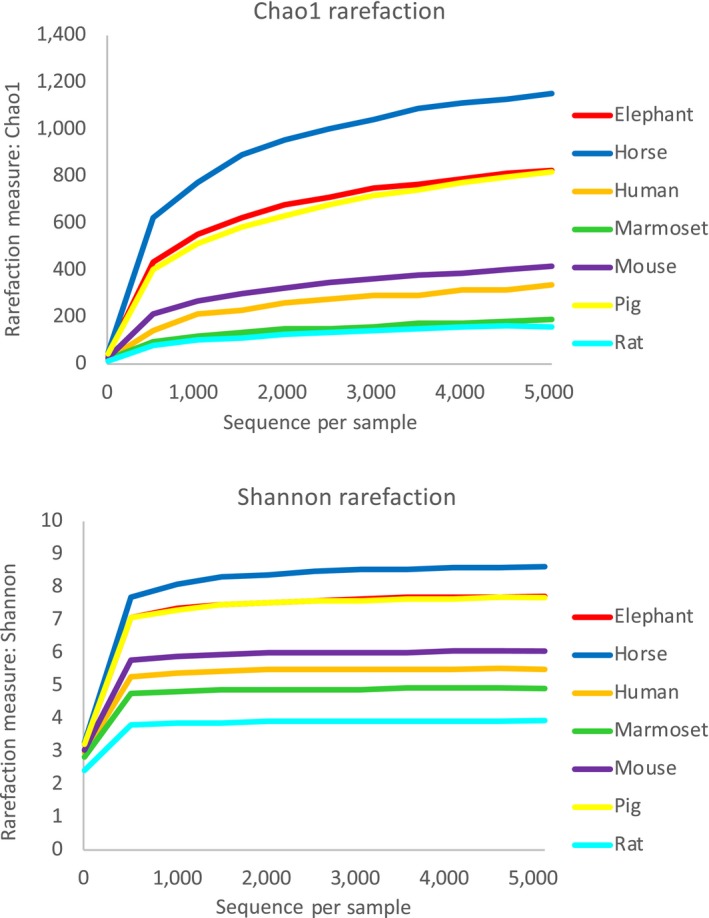
Rarefaction curves of each animal species. Rarefaction curves were constructed using Chao1 and Shannon indices
With regard to alpha diversity, both Chao1 and Shannon indices in elephant and horse samples were significantly higher than in those of humans, marmosets, mice, and rats. Moreover, horse samples showed the highest Chao1 and Shannon indices in all animal species evaluated in the present study. Interestingly, although pigs are categorized as omnivores (Olsen, Hansen, Jespersen, Marckmann, & Bladbjerg, 1999), the alpha diversity of their samples was as high as that of elephants, which are herbivores by definition (Figure 2).
Figure 2.

Chao1 and Shannon alpha diversity of each animal species. Chao1 and Shannon indices were calculated with the R package “phyloseq” and statistically analyzed with the Kruskal–Wallis test, followed by a pairwise comparison using QIIME2 software. Boxplots with different letters are significantly different (p < .05)
3.2. Beta diversity
The principal coordinate analysis (PCoA) plot based on weighted UniFrac distance showed that samples from the same animal species aggregated closely. However, samples from elephant, horse, and pig clustered all together (Figure 3). Similarity of fecal microbiota within the same animal species (intraspecies similarity) was higher than that between different animal species (inter‐species similarity) (Table 2). Interestingly, at 83.24 ± 3.08%, inter‐species similarity between elephants and horses was very high, followed by that between these herbivores, pigs, and mice at >60%. However inter‐species similarity between ohter omnivorous animals, namely marmosets, human and rats was lower than 50%. The inter‐species similarity between humans and other animal species was at most 33.32 ± 10.36% which was obtained with marmosets. Intraspecies similarity was higher in elephants (88.62 ± 3.13%), horses (92.83 ± 1.51%), mice (88.54 ± 3.11%), and pigs (80.79 ± 4.56%) than in humans (67.26 ± 5.76%), marmosets (55.89 ± 14.90%), and rats (56.22 ± 14.80%).
Figure 3.
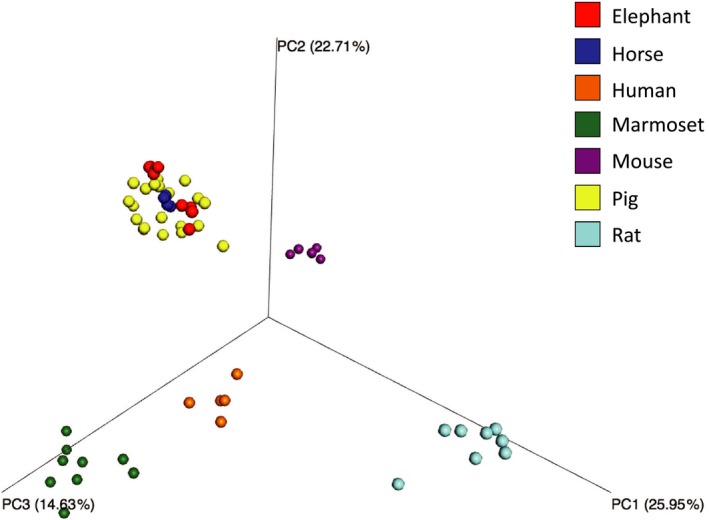
Principal coordinate analysis plot of the fecal microbiota of seven monogastric animal species. Beta diversity based on weighted UniFrac distance was calculated and the plot was generated by QIIME. The PERMANNOVA calculated by QIIME2 indicated that the fecal microbiota differed significantly between animal species (p < .01)
Table 2.
Similarity of fecal microbiota among seven monogastric animal species
| Elephant | Horse | Human | Marmoset | Mouse | Pig | Rat | |
|---|---|---|---|---|---|---|---|
| Elephant | 88.62 ± 3.13 | 83.24 ± 3.08 | 18.27 ± 5.76 | 18.50 ± 5.85 | 64.43 ± 2.75 | 62.28 ± 5.96 | 11.21 ± 4.99 |
| Horse | 92.83 ± 1.51 | 20.32 ± 4.26 | 21.50 ± 5.97 | 65.30 ± 2.43 | 67.13 ± 5.49 | 15.29 ± 5.89 | |
| Human | 67.26 ± 5.76 | 33.32 ± 10.36 | 26.32 ± 4.26 | 24.66 ± 6.63 | 21.56 ± 5.99 | ||
| Marmoset | 55.89 ± 0.14.90 | 24.59 ± 07.99 | 28.38 ± 5.10 | 12.22 ± 5.75 | |||
| Mouse | 88.54 ± 3.11 | 59.21 ± 4.83 | 17.20 ± 6.01 | ||||
| Pig | 80.79 ± 4.56 | 17.34 ± 5.63 | |||||
| Rat | 56.22 ± 14.80 |
Similarity based on Bray–Curtis distance was calculated by R package "vegan". Values (%) are expressed as mean ± SD.
3.3. Taxonomy
At phylum level, the abundance of Bacteroidetes and Firmicutes accounted for more than 80% in most animal species, except for marmosets and rats (Figure 4). In marmosets, Bacteroidetes (30.1 ± 8.0%) and Firmicutes (34.0 ± 6.9%) were most abundant as well as other animal species but considerable abundance of Actinobacteria (24.0 ± 13.6%) was detected. In rats, Firmicutes (65.8 ± 14.3%) was the most abundant followed by Verrucomicrobia (25.2 ± 15.4%).
Figure 4.
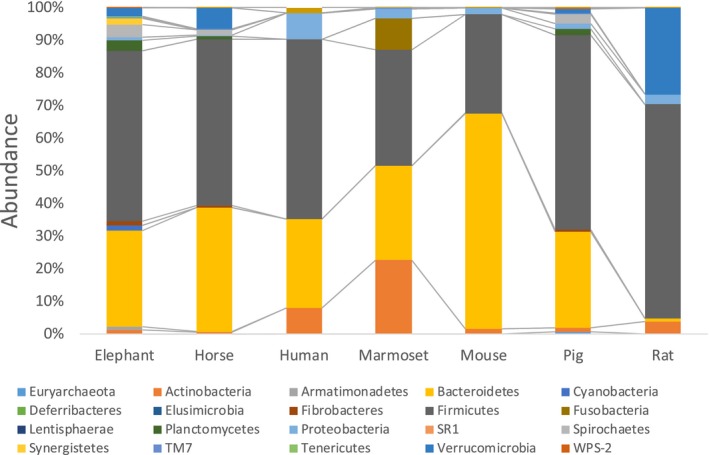
Mean abundances at the phylum level in fecal microbiota of each animal species
At genus level, the abundance of 212 bacterial genera was significantly different in the samples of the animal species studied (Table S1). When analyzing major butyrate‐producing bacterial genera (Anand, Kaur, & Mande, 2016; Counotte, Prins, Janssen, & Debie, 1981; Eeckhaut et al., 2008; Gophna, Konikoff, & Nielsen, 2017; Li, Wu, Baldwin, Li, & Li, 2012; Louis & Flint, 2009) (Table 3), human samples contained a variety of butyrate producers of which the most predominant was Faecalibacterium (8.02 ± 3.22%), followed by Coprococcus (3.69 ± 2.76%) and Roseburia (3.11 ± 1.67%). In samples of the other omnivorous animals, the most predominant butyrate producer was Megasphaera (5.15 ± 2.76%, marmosets), Oscillospira (2.78 ± 1.74% and 5.75 ± 1.32%, pigs and mice, respectively), and Coprococcus (0.61 ± 0.40%, rats). The total abundance of butyrate producers was >5% in humans, pigs, and marmosets. However, at 3.94% and 1.11%, the abundance of butyrate‐producing bacterial genera in mice and rats, respectively, was relatively small. In addition, the total abundance of butyrate‐producing bacteria in horses (3.87%) and elephants (4.09%) was similar to that of mice. Although Butyrivibrio and Pseudobutyrivibrio, major butyrate‐producing bacterial genera in the rumen, were detected in elephants and horses, their abundance was lower than that of other butyrate‐producing bacteria such as Roseburia (elephants) and Oscillospira (horses).
Table 3.
Major known butyrate‐ and lactate‐producing bacteria in fecal microbiota
| Taxonomy | p values | Elephant | Horse | Human | Marmoset | Mouse | Pig | Rat | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| family | genus | Abundance (%) | Abundance (%) | Abundance (%) | Abundance (%) | Abundance (%) | Abundance (%) | Abundance (%) | ||||||||
| Butyrate‐producing bacteria | ||||||||||||||||
| Ruminococcaceae | Butyricicoccus | <.01 | <0.01 | b | <0.01 | b | 0.72 ± 0.43 | a | 0.14 ± 0.11 | b | 0.09 ± 0.08 | b | 0.25 ± 0.25 | b | <0.01 | b |
| [Odoribacteraceae | Butyricimonas | <.01 | 0.00 | b | 0.00 | b | 0.00 | b | 0.00 | b | 0.15 ± 0.09 | a | 0.00 | b | <0.01 | b |
| Lachnospiraceae | Butyrivibrio | <.01 | 0.06 ± 0.04 | 0.00 | 0.00 | 0.01 ± 0.02 | 0.00 | 0.08 ± 0.10 | 0.00 | |||||||
| Lachnospiraceae | Coprococcus | <.01 | 1.24 ± 0.18 | b | 0.73 ± 0.16 | b | 3.69 ± 2.76 | a | <0.01 | b | 0.92 ± 0.82 | b | 1.02 ± 0.34 | b | 0.61 ± 0.40 | b |
| Ruminococcaceae | Faecalibacterium | <.01 | 0.04 ± 0.02 | b | 0.06 ± 0.01 | b | 8.02 ± 3.22 | a | 0.02 ± 0.03 | b | 0.00 ± 0.00 | b | 0.61 ± 0.45 | b | 0.15 ± 0.12 | b |
| Veillonellaceae | Megasphaera | <.01 | 0.00 | b | 0.01 ± 0.01 | b | 1.05 ± 2.01 | b | 5.15 ± 2.76 | a | <0.01 | b | 1.26 ± 0.92 | b | <0.01 | b |
| Ruminococcaceae | Oscillospira | <.01 | 0.85 ± 0.53 | bc | 2.00 ± 0.73 | bc | 1.29 ± 1.17 | bc | 0.32 ± 0.42 | c | 2.78 ± 1.74 | b | 5.75 ± 1.32 | a | 0.36 ± 0.13 | c |
| Lachnospiraceae | Roseburia | <.01 | 1.37 ± 0.32 | b | 0.95 ± 0.30 | bc | 3.11 ± 1.67 | a | <0.01 | c | <0.01 | c | 0.59 ± 0.48 | bc | <0.01 | c |
| Sum of above listed butyrate‐producing bacteria | 3.56 ± 0.74 | 3.75 ± 0.98 | 17.89 ± 4.30 | 5.64 ± 2.82 | 3.94 ± 2.76 | 9.55 ± 1.69 | 1.11 ± 0.58 | |||||||||
| Lactate‐producing bacteria | ||||||||||||||||
| Bifidobacteriaceae | Bifidobacterium | <.01 | <0.01 | b | 0.01 ± 0.01 | b | 7.07 ± 2.41 | ab | 15.36 ± 11.01 | a | <0.01 | b | 0.76 ± 0.81 | b | 2.36 ± 5.92 | b |
| Coriobacteriaceae | Enterococcus | <.01 | 0.02 ± 0.02 | a | 0.00 | b | 0.00 | b | <0.01 | b | 0.00 | b | 0.00 | b | 0.00 | b |
| Enterococcaceae | Enterococcus | <.01 | 0.01 ± 0.01 | ab | 0.00 | ab | <0.01 | ab | 0.01 ± 0.03 | ab | <0.01 | ab | <0.01 | b | 0.03 ± 0.03 | a |
| Lactobacillaceae | Lactobacillus | <.01 | 0.04 ± 0.04 | b | 0.62 ± 0.77 | b | 0.13 ± 0.05 | b | 0.02 ± 0.02 | b | 8.34 ± 4.15 | a | 3.47 ± 2.27 | b | 12.58 ± 5.58 | a |
| Streptococcaceae | Lactococcus | <.01 | 0.00 | ab | 0.00 | ab | 0.03 ± 0.05 | ab | 0.01 ± 0.02 | ab | 0.18 ± 0.10 | ab | 0.00 | b | 0.56 ± 0.91 | a |
| Streptococcaceae | Streptococcus | <.01 | 0.19 ± 0.38 | b | 0.10 ± 0.17 | ab | 1.25 ± 0.70 | ab | 0.13 ± 0.19 | b | 0.01 ± 0.01 | b | 4.07 ± 3.87 | a | 0.24 ± 0.25 | b |
| Staphylococcaceae | Staphylococcus | <.01 | <0.01 | b | 0.00 | b | <0.01 | b | <0.01 | b | 0.22 ± 0.08 | a | 0.00 | b | 0.04 ± 0.03 | b |
| Sum of above listed lactate‐producing bacteria | 0.26 ± 0.45 | 0.73 ± 0.76 | 8.47 ± 2.58 | 15.53 ± 11.64 | 8.52 ± 4.50 | 8.31 ± 5.46 | 15.76 ± 8.95 | |||||||||
Values are expressed as mean ± SD. Values with different letters are significantly different (p < .05). Zero value of the abundance means it was not detected in all samples.
Regarding major lactate‐producing bacteria (Kandler, 1983; Pessione, 2012), the predominant genera were Bifidobacterium in humans and marmosets (7.07% ± 2.41% and 15.36% ± 11.01%, respectively) and Lactobacillus in rats, mice, and pigs (12.58% ± 5.58%, 8.34% ± 4.15%, and 3.47% ± 2.27%, respectively) (Table 3). Fecal samples of pigs also contained a considerable abundance of Streptococcus (4.07% ± 3.87%). It is worth noting that while lactate‐producing bacterial genera were barely detected in samples of herbivores, especially elephants, as per our results, Lactobacillus seemed to be predominant in those of horses (0.62% ± 0.77%).
Genus Fibrobacter, a well‐known cellulose‐degrading bacterium (Béra‐Maillet, Ribot, & Forano, 2004), was readily detected in samples of elephants (1.22 Quantitative Insights Into Microbial Ecology ±1.01%) and horses (0.75 Quantitative Insights Into Microbial Ecology ±0.47%), and in lower abundance, in pigs (0.47 ± 0.39%) (Table S1).
4. DISCUSSION
While all animal species have a microbial population in the gut that plays essential roles in the host's physiology, it has been suggested that variation of the microbial communities in the gut microbiota depends on the habitats, feeding preferences, and the structure of the gastrointestinal tract of animals (Ikeda‐Ohtsubo et al., 2018). However, this notion is likely to be somewhat biased, because although in the past workers used a variety of analytical methods, the obtained information arose mainly from several studies focusing on the gut microbiota of a single animal species (Chassaing et al., 2015; Chung et al., 2016; Costa et al., 2015; Fan et al., 2014; Fernandes et al., 2014; Ilmberger et al., 2014; Kim et al., 2015; Shigeno et al., 2018).
Hence, to obtain a comprehensive insight into the similarities or dissimilarities between the fecal microbiota of different animal species, in the present work, we compared the microbial population in fecal samples of seven different monogastric animals, using exactly the same analytical protocol.
It must be underlined that in the present study, animal species were reared at different facilities and fed different diets. Thus, at first sight, a lack of normalization and control of the rearing environments including housing condition and diets of studied animals might be considered as limiting factors in the present work. Nonetheless, it should be noted that although pigs that housed individually were reared at two different farms, their samples were clustered together in the PCoA analysis plot (Figure 3). Likewise, human sample data from our previous study were also clustered together, even though diets were not controlled, meaning the diet preferences of the sample donors were completely randomized and mostly unknown (Kawada et al., 2019). In the study by O’Donnell et al. (2017), clear differences in the microbiota of different herbivores were observed even though the animals were reared at the same farm and given the same diet, which seems to demonstrate that rearing sites and diets can be hardly considered limiting factors. Therefore, we believe that it was worth comparing the fecal microbiota of the animal species studied in the present work even though rearing sites and diets were not under control.
The alpha diversity was significantly higher in elephants and horses than in omnivores except for pigs, suggesting that a complex amalgam of bacterial communities is required to efficiently degrade and utilize fiber such as cellulose, hemicellulose, and lignin. While rarely detected in omnivorous animals except for pigs, Fibrobacter (a well‐known cellulolytic bacterium) and Treponema [a bacterium reported to enhance cellulolytic activity in rumen (Stanton & Canale‐Parola, 1980)] were detected in both elephants and horses as reported previously (Fernandes et al., 2014; Ilmberger et al., 2014). Similarly, in the present study, a high alpha diversity and a high abundance of Fibrobacter and Treponema, closely resembling those detected in herbivores, were also observed in pigs. Although an effect of diet should not be ruled out, the presence of Fibrobacter and occurrence of high cellulolytic activity in pigs, especially when compared with humans, were previously shown (Sunvold, Hussein, Fahey, Merchen, & Reinhart, 1995; Varel, 1987), suggesting that the gut microbiota of pigs are better equipped to utilize fiber than other omnivorous animals. Thus, our results seem to be in total concordance with the previous work.
Beta diversity and similarity analyses showed that herbivores have similar fecal microbiota. Remarkably, pigs showed a higher inter‐species similarity with herbivorous animals than did other omnivorous animals. Conversely, in the PCoA plot, humans, marmosets, mice, and rats clustered by species and in consequence, their inter‐species similarity was notably lower. These results seem to indicate that fecal microbiota profiles of these omnivores are unique to each species. The species specificity of the gut microbiota in humans, nonhuman primates, mice, and rats was also reported by Nagpal et al. (2018). These workers also reported that the gut microbiota of nonhuman primates was more similar to that of humans than that of rodents. Although Nagpal et al. (2018) studied macaques but not marmosets, our result is in harmony with their study. Therefore, the fact that two independent studies have now demonstrated divergence between the gut microbiota profiles of rodents and humans should be taken into account when interpreting the results from gut microbiota‐related research in which rodents were used as experimental models. For example, utilization of a given substrate—nondigestible fiber—is likely to be different in humans and rodents. Similarly, the same caution should be taken when attempting to extrapolate results from research using pigs as experimental models to describe gut microbial profiles in humans.
Taxonomic analysis revealed that, at phylum level, Bacteroidetes and Firmicutes were commonly present at high abundance in fecal microbiota of all animal species. On the other hand, at genus level, all animal species showed characteristic composition of fecal microbiota. With respect to butyrate‐ and lactate‐producing bacterial genera, characteristic features for each animal species were observed. In humans, Faecalibacterium, Coprococcus, and Roseburia were found to be the major butyrate producers in the present work and elsewhere (Louis & Flint, 2009). In the other omnivores evaluated, Megasphaera and/or Oscillospira were the major butyrate producers. Herbivores have a variety of butyrate producers including Roseburia, Oscillospira, and Pseudobutyrivibrio. However, in the present study, the abundance of these bacteria was relatively lower compared with that of humans and pigs. Thus, it can be theorized that herbivores likely have other butyrate‐producing bacterial species in the gut microbiota, for example, some species in genus Clostridium and Prevotella (Esquivel‐Elizondo, Ilhan, Garcia‐Pena, & Krajmalnik‐Brown, 2017), because n‐butyrate was previously detected in the feces of elephants and horses (Breves & Stuck, 1995). In marmosets and humans, the predominant lactate producer was Bifidobacterium, but Lactobacillus was predominant in the other omnivores. It has been shown that, while Lactobacillus acts as lactate producer in the porcine large intestine, Megasphaera becomes a lactate‐utilizing butyrate producer, fittingly interacting with each other to produce butyrate (Tsukahara et al., 2006). Interestingly, our results show that butyrate production via lactate degradation may be the major metabolism pathway not only in pigs but also in marmosets (Table 3). Lastly, elephant and horse samples had an unexpected lower abundance of lactate producers, because no bacterial genera exceeded 1% in the gut microbiota. This result also agrees with previous work conducted on these animal species in separate occasions (Fernandes et al., 2014; Ilmberger et al., 2014).
To conclude, the present work demonstrated that the fecal microbiota in herbivorous animals share certain features such as a high alpha diversity and a high abundance of fiber‐degrading bacteria. In contrast, omnivorous animals, especially primates, seem to have unique fecal microbiota. For future studies, it can be recommended to add an analysis of short‐chain fatty acid concentrations and controlled environment sites and diets to obtain more precise information on the species specificity of gut microbiota. Uniqueness or similarity of gut microbiota in carnivores would also be of interest. Finally, similarities in the microbial profiles of herbivores and pigs, which may be of interest for feed producers, should be elucidated further.
Supporting information
Table S1
ACKNOWLEDGMENTS
We thank Mr. Yuki Kawada and Mr. So Morishima (Kyoto Prefectural University) for skilful technical assistance.
Kobayashi R, Nagaoka K, Nishimura N, et al. Comparison of the fecal microbiota of two monogastric herbivorous and five omnivorous mammals. Anim Sci J. 2020;91:e13366 10.1111/asj.13366
REFERENCES
- Anand, S. , Kaur, H. , & Mande, S. S. (2016). Comparative in silico analysis of butyrate production pathways in gut commensals and pathogens. Frontiers in Microbiology, 7, 1945 10.3389/fmicb.2016.01945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béra‐Maillet, C. , Ribot, Y. , & Forano, E. (2004). Fiber‐degrading systems of different strains of the genus Fibrobacter. Applied and Environmental Microbiology, 70(4), 2172–2179. 10.1128/AEM.70.4.2172-2179.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E. , Rideout, J. R. , Dillon, M. R. , Bokulich, N. A. , Abnet, C. C. , Al‐Ghalith, G. A. , … Caporaso, J. G. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology, 37(8), 852–857. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breves, G. , & Stuck, K. (1995). Short‐chain fatty acids in the hindgut In Cummings J. H., Rombeau J. L., & Sakata T. (Eds.), Physiological and clinical aspects of short‐chain fatty acids (pp. 73–85). Cambridge, UK: Cambridge University Press. [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7(5), 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing, B. , Koren, O. , Goodrich, J. K. , Poole, A. C. , Srinivasan, S. , Ley, R. E. , & Gewirtz, A. T. (2015). Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature, 519(7541), 92–96. 10.1038/nature14232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers, D. J. , & Hladik, C. M. (1980). Morphology of the gastrointestinal tract in primates: Comparisons with other mammals in relation to diet. Journal of Morphology, 166(3), 337–386. 10.1002/jmor.1051660306 [DOI] [PubMed] [Google Scholar]
- Chung, W. S. F. , Walker, A. W. , Louis, P. , Parkhill, J. , Vermeiren, J. , Bosscher, D. , … Flint, H. J. (2016). Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biology, 14, 3 10.1186/s12915-015-0224-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauss, M. , Frey, R. , Kiefer, B. , Lechner‐Doll, M. , Loehlein, W. , Polster, C. , … Streich, W. J. (2003). The maximum attainable body size of herbivorous mammals: Morphophysiological constraints on foregut, and adaptations of hindgut fermenters. Oecologia, 136(1), 14–27. 10.1007/s00442-003-1254-z [DOI] [PubMed] [Google Scholar]
- Costa, M. C. , Stämpfli, H. R. , Arroyo, L. G. , Allen‐Vercoe, E. , Gomes, R. G. , & Weese, J. S. (2015). Changes in the equine fecal microbiota associated with the use of systemic antimicrobial drugs. BMC Veterinary Research, 11, 19 10.1186/s12917-015-0335-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counotte, G. H. , Prins, R. A. , Janssen, R. H. , & Debie, M. J. (1981). Role of Megasphaera elsdenii in the fermentation of dl‐[2‐C]lactate in the Rumen of Dairy Cattle. Applied and Environmental Microbiology, 42(4), 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhaut, V. , Van Immerseel, F. , Teirlynck, E. , Pasmans, F. , Fievez, V. , Snauwaert, C. , … Vandamme, P. (2008). Butyricicoccus pullicaecorum gen. nov., sp. nov., an anaerobic, butyrate‐producing bacterium isolated from the caecal content of a broiler chicken. International Journal of Systematic and Evolutionary Microbiology, 58(Pt 12), 2799–2802. 10.1099/ijs.0.65730-0 [DOI] [PubMed] [Google Scholar]
- Esquivel‐Elizondo, S. , Ilhan, Z. E. , Garcia‐Pena, E. I. , & Krajmalnik‐Brown, R. (2017). Insights into butyrate production in a controlled fermentation system via gene predictions. mSystems, 2(4), e00051‐17 10.1128/mSystems.00051-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W. , Tang, Y. , Qu, Y. , Cao, F. , & Huo, G. (2014). Infant formula supplemented with low protein and high carbohydrate alters the intestinal microbiota in neonatal SD rats. BMC Microbiology, 14, 279 10.1186/s12866-014-0279-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, K. A. , Kittelmann, S. , Rogers, C. W. , Gee, E. K. , Bolwell, C. F. , Bermingham, E. N. , & Thomas, D. G. (2014). Faecal microbiota of forage‐fed horses in New Zealand and the population dynamics of microbial communities following dietary change. PLoS ONE, 9(11), e112846 10.1371/journal.pone.0112846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gophna, U. , Konikoff, T. , & Nielsen, H. B. (2017). Oscillospira and related bacteria – From metagenomic species to metabolic features. Environmental Microbiology, 19(3), 835–841. 10.1111/1462-2920.13658 [DOI] [PubMed] [Google Scholar]
- Ikeda‐Ohtsubo, W. , Brugman, S. , Warden, C. H. , Rebel, J. M. J. , Folkerts, G. , & Pieterse, C. M. J. (2018). How can we define "Optimal Microbiota?": A comparative review of structure and functions of microbiota of animals, fish, and plants in agriculture. Frontiers in Nutrition, 5, 90 10.3389/fnut.2018.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilmberger, N. , Güllert, S. , Dannenberg, J. , Rabausch, U. , Torres, J. , Wemheuer, B. , … Streit, W. R. (2014). A comparative metagenome survey of the fecal microbiota of a breast‐ and a plant‐fed Asian elephant reveals an unexpectedly high diversity of glycoside hydrolase family enzymes. PLoS ONE, 9(9), e106707 10.1371/journal.pone.0106707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, R. , Sakaue, Y. , Sawai, C. , Sawai, T. , Ozeki, M. , Romero‐Pérez, G. A. , & Tsukahara, T. (2016). A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Bioscience Biotechnology and Biochemistry, 80(12), 2450–2458. 10.1080/09168451.2016.1222267 [DOI] [PubMed] [Google Scholar]
- Kandler, O. (1983). Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek, 49(3), 209–224. 10.1007/BF0039949 [DOI] [PubMed] [Google Scholar]
- Kawada, Y. , Naito, Y. , Andoh, A. , Ozeki, M. , & Inoue, R. (2019). Effect of storage and DNA extraction method on 16S rRNA‐profiled fecal microbiota in Japanese adults. Journal of Clinical Biochemistry and Nutrition, 64(2), 106–111. 10.3164/jcbn.18-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. , Nguyen, S. G. , Guevarra, R. B. , Lee, I. , & Unno, T. (2015). Analysis of swine fecal microbiota at various growth stages. Archives of Microbiology, 197(6), 753–759. 10.1007/s00203-015-1108-1 [DOI] [PubMed] [Google Scholar]
- Kovatcheva‐Datchary, P. , Tremaroli, V. , & Bäckhed, F. (2013). The Gut Microbiota. In Rosenberg E., DeLong E. F., Stackebrandt E., Thompson F., & Lory S. (Eds.), The Prokaryotes (pp. 3–24). Germany: Springer‐Verlag, Berlin Heidelberg. [Google Scholar]
- Ley, R. E. , Hamady, M. , Lozupone, C. , Turnbaugh, P. J. , Ramey, R. R. , Bircher, J. S. , … Gordon, J. I. (2008). Evolution of mammals and their gut microbes. Science, 320(5883), 1647–1651. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R. W. , Wu, S. , Baldwin, R. L. , Li, W. , & Li, C. (2012). Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PLoS ONE, 7(1), e29392 10.1371/journal.pone.0029392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, P. , & Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate‐producing bacteria from the human large intestine. FEMS Microbiology Letters, 294(1), 1–8. 10.1111/j.1574-6968.2009.01514.x [DOI] [PubMed] [Google Scholar]
- Masood, M. I. , Qadir, M. I. , Shirazi, J. H. , & Khan, I. U. (2011). Beneficial effects of lactic acid bacteria on human beings. Criticla Revies in Microbiology, 37(1), 91–98. 10.3109/1040841X.2010.536522 [DOI] [PubMed] [Google Scholar]
- McMurdie, P. J. , & Holmes, S. (2013). phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE, 8(4), e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, D. J. , & Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes, 7(3), 189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge, B. D. , Kuczynski, J. , Knights, D. , Clemente, J. C. , Gonzalez, A. , Fontana, L. , … Gordon, J. I. (2011). Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science, 332(6032), 970–974. 10.1126/science.1198719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, R. , Wang, S. , Solberg Woods, L. C. , Seshie, O. , Chung, S. T. , Shively, C. A. , … Yadav, H. (2018). Comparative microbiome signatures and short‐chain fatty acids in mouse, rat, non‐human primate, and human feces. Frontiers in Microbiology, 9, 2897 10.3389/fmicb.2018.02897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’ Donnell, M. M. , Harris, H. M. B. , Ross, R. P. , & O'Toole, P. W. (2017). Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals. MicrobiologyOpen, 6(5), e00509 10.1002/mbo3.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hara, A. M. , & Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Reports, 7(7), 688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A. K. , Hansen, A. K. , Jespersen, J. , Marckmann, P. , & Bladbjerg, E. M. (1999). The pig as a model in blood coagulation and fibrinolysis research. Scandinavian Journal of Laboratory Animal Science, 26(4), 214–224. [Google Scholar]
- Parks, D. H. , Tyson, G. W. , Hugenholtz, P. , & Beiko, R. G. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics, 30(21), 3123–3124. 10.1093/bioinformatics/btu494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessione, E. (2012). Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Frontiers in Cellular and Infection Microbiology, 2, 86 10.3389/fcimb.2012.00086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt, N. , Duncan, S. H. , Young, P. , Belenguer, A. , McWilliam Leitch, C. , Scott, K. P. , … Louis, P. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. The ISMI Journal, 8(6), 1323–1335. 10.1038/ismej.2014.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognes, T. , Flouri, T. , Nichols, B. , Quince, C. , & Mahé, F. (2016). VSEARCH: A versatile open source tool for metagenomics. PeerJ, 4, e2584 10.7717/peerj.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeno, Y. , Toyama, M. , Nakamura, M. , Niimi, K. , Takahashi, E. , & Benno, Y. (2018). Comparison of gut microbiota composition between laboratory‐bred marmosets (Callithrix jacchus) with chronic diarrhea and healthy animals using terminal restriction fragment length polymorphism analysis. Microbiology and Immunology, 62(11), 702–710. 10.1111/1348-0421.12655 [DOI] [PubMed] [Google Scholar]
- Stanton, T. B. , & Canale‐Parola, E. (1980). Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Archives of Microbiology, 127(2), 145–156. [DOI] [PubMed] [Google Scholar]
- Stevens, C. E. , & Hume, I. D. (1998). Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiological Reviews, 78(2), 393–427. 10.1152/physrev.1998.78.2.393 [DOI] [PubMed] [Google Scholar]
- Sunvold, G. D. , Hussein, H. S. , Fahey, G. C. Jr , Merchen, N. R. , & Reinhart, G. A. (1995). In vitro fermentation of cellulose, beet pulp, citrus pulp, and citrus pectin using fecal inoculum from cats, dogs, horses, humans, and pigs and ruminal fluid from cattle. Journal of Animal Science, 73(12), 3639–3648. 10.2527/1995.73123639x [DOI] [PubMed] [Google Scholar]
- Tsukahara, T. , Hashizume, K. , Koyama, H. , & Ushida, K. (2006). Stimulation of butyrate production through the metabolic interaction among lactic acid bacteria, Lactobacillus acidophilus, and lactic acid‐utilizing bacteria, Megasphaera elsdenii, in porcine cecal digesta. Animal Science Journal, 77(4), 454–461. 10.1111/j.1740-0929.2006.00372.x [DOI] [Google Scholar]
- Varel, V. H. (1987). Activity of fiber‐degrading microorganisms in the pig large intestine. Journal of Animal Science, 65(2), 488–496. 10.2527/jas1987.652488x [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73(16), 5261–5267. 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, L. , & Duffy, A. (2017). Factors Influencing the Gut Microbiota, Inflammation, and Type 2 Diabetes. Journal of Nutrition, 147(7), 1468S–1475S. 10.3945/jn.116.240754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, Y. , Tsukahara, T. , & Ushida, K. (2009). Oral administration of Lactobacillus plantarum Lq80 and Megasphaera elsdenii iNP‐001 induces efficient recovery from mucosal atrophy in the small and the large intestines of weaning piglets. Animal Science Journal, 80(6), 709–715. 10.1111/j.1740-0929.2009.00692.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


