Abstract
Background
Mental disorders, including Attention‐Deficit/Hyperactivity Disorder (ADHD), have a complex etiology, and identification of underlying genetic risk factors is challenging. This study used a multistep approach to identify and validate a novel risk gene for ADHD and psychiatric comorbidity.
Methods
In a single family, severely affected by ADHD and cooccurring disorders, we applied single nucleotide polymorphism (SNP)‐array analysis to detect copy‐number variations (CNVs) linked to disease. Genes present in the identified CNV were subsequently tested for their association with ADHD in the largest data set currently available (n = 55,374); this gene‐set and gene‐based association analyses were based on common genetic variants. Significant findings were taken forward for functional validation using Drosophila melanogaster as biological model system, altering gene expression using the GAL4‐UAS system and a pan‐neuronal driver, and subsequently characterizing locomotor activity and sleep as functional readouts.
Results
We identified a copy number gain in 8p23.3, which segregated with psychiatric phenotypes in the family and was confirmed by quantitative RT‐PCR. Common genetic variants in this locus were associated with ADHD, especially those in FBXO25 and TDRP. Overexpression of the FBXO25 orthologue in two Drosophila models consistently led to increased locomotor activity and reduced sleep compared with the genetic background control.
Conclusions
We combine ADHD risk gene identification in an individual family with genetic association testing in a large case–control data set and functional validation in a model system, together providing an important illustration of an integrative approach suggesting that FBXO25 contributes to key features of ADHD and comorbid neuropsychiatric disorders.
Keywords: ADHD, psychiatric comorbidities, FBXO25, TDRP, Drosophila melanogaster
Introduction
Attention‐deficit/hyperactivity disorder (ADHD) is a neuropsychiatric disorder with a high prevalence, affecting 5%–6% of children and 2.5%–4.9% of adults (Faraone et al., 2015). Individuals with this disorder are at high risk of developing comorbid psychiatric disorders across the entire life span (Franke et al., 2018). Twin and adoption studies have shown that ADHD is highly heritable, with heritability estimates of approximately 80% in both children and adults (Faraone et al., 2005; Larsson et al., 2014). ADHD is a clinically and etiologically heterogeneous disorder with a complex, multifactorial background. The identification of genes affecting the disorder has been challenging (Demontis et al., 2019; Franke et al., 2012; Gizer et al., 2009). In the last decade, genome‐wide association studies (GWAS) have been performed with increasing sample sizes. The first genome‐wide significant risk variants for ADHD from a GWAS meta‐analysis have recently been reported, all of which have small individual effect sizes (Demontis et al., 2019). In addition to studies of common genetic variants contributing to ADHD, other studies have focused on rare variants, which potentially have larger effect sizes. First exome sequencing studies are coming available (Corominas et al., 2018; Demontis et al., 2016; Hawi et al., 2016; Kim et al., 2017), but so far, most studies investigated copy‐number variations (CNVs) and identified rare CNVs in patients with ADHD (Akutagava‐Martins et al., 2014; Elia et al., 2010; Elia et al., 2011; Jarick et al., 2014; Lesch et al., 2011; Lesch et al., 2008; Martin et al., 2014; Ramos‐Quiroga et al., 2014; Williams et al., 2012; Williams et al., 2010; Yang et al., 2013). While most studies used case–control designs, CNVs have been identified in studies using a family‐based approach (Lesch et al., 2008). Now that first risk factors for ADHD have been identified from common and rare variant approaches, it has become evident that at least part of ADHD’s comorbidity with disorders like major depression and psychotic disorders, such as bipolar disorder and schizophrenia, is based on genetic factors (Demontis et al., 2019; van Hulzen et al., 2017; Lee et al., 2013).
If a variant is only seen once in a case–control study of rare variants or in a single family, it is challenging to demonstrate the association with a psychiatric phenotype based on statistics. Similarly, in studies of common genetic risk factors for psychiatric phenotypes, it is often difficult to go beyond association and find evidence for the causal involvement of a specific gene, as variants tested through GWAS are just anonymous markers. An effective way forward in this is the use of suitable animal models, where one can specifically manipulate the expression of individual genes of interest. Around 75% of human genes have a clear orthologue in the fruit fly Drosophila melanogaster (van der Voet et al., 2014). The Drosophila model combines a vast genetic toolbox based on decades of research with a large range of quantifiable behaviors (van Alphen & van Swinderen, 2013) and is inexpensive in its use. We have recently shown that Drosophila is an appropriate model for ADHD; flies exhibited increased locomotor activity and decreased sleep upon dysregulation of validated ADHD candidate genes (van der Voet et al., 2016).
In this study, we used a multistep approach to identify and validate a novel risk gene for ADHD and comorbid psychiatric conditions. In a single family, severely affected by ADHD and comorbid disorders (especially psychosis), we applied genome‐wide single nucleotide polymorphism (SNP)‐based array analysis to detect rare copy number variants (CNVs) cosegregating with disease. Genes present in the CNV were tested – as a set and individually – for association with ADHD and other relevant psychiatric disorders based on common genetic variants, using the largest internationally available data sets. The significantly associated gene FBXO25 was taken forward for functional validation in Drosophila. According to the nature of the CNV (copy number gain), we characterized the effects of CG11658 (dFBXO25) overexpression using locomotor activity and sleep behavior as functional readouts.
Methods and materials
Study participants and cohorts
Dutch family
The male index patient had diagnoses of ADHD, anxiety disorder, and multiple complex developmental disorder (MCDD) at the age of four years; he also showed aggressive behavior and suicidal tendencies, had experienced a psychotic episode, and reported somatic abnormalities including gastric reflux and clinodactyly (Appendix S1). His parents (mother diagnosed with ADHD, father without diagnosis), the mother’s brother (psychotic episode, aggressive behavior, alcohol addiction, childhood ADHD symptoms, and currently homeless), the mother’s sister (learning difficulties), and two of the mother’s cousins (diagnosed with ADHD) were also ascertained for genetic testing (see Figure 1 for the pedigree and Table S1 and Appendix S1 for more detailed description of psychiatric diagnoses and relevant symptoms). For these members of the family, saliva (using Oragene containers; DNA Genotek, Ottawa, ON, Canada) or blood was collected. All human material was collected after approval by the local ethic committees, and participants in this study gave written informed consent.
Figure 1.
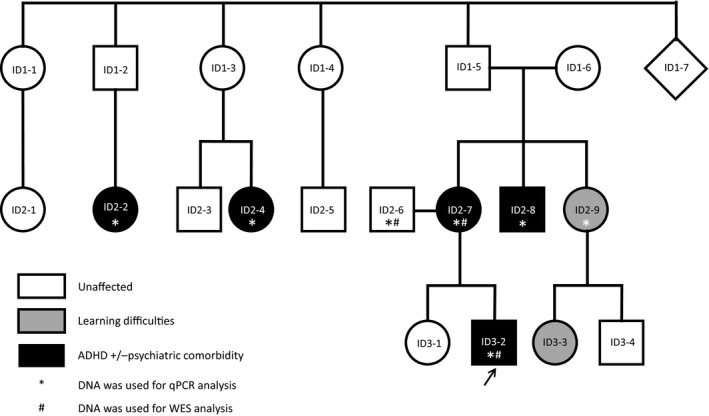
Pedigree of the family investigated in this study. Patients with ADHD with or without psychiatric comorbidity are depicted in black (see Appendix S1 and Table S1 for an exhaustive description of their phenotypes), unaffected family members are shown by white symbols, and grey symbols represent family members with learning difficulties. The index patient is marked by a diagonal arrow. An asterisk beneath an individual’s code indicates that DNA was used for qRT‐PCR analysis. The # indicates that DNA was used for WES. ID = individual
GWAS meta‐analyses data sets for ADHD and schizophrenia
We obtained genome‐wide association study meta‐analysis (GWAS‐MA) results in the form of summary statistics (p‐values and odds ratios) from the largest currently available data sets on ADHD and schizophrenia (SCZ). GWAS‐MA data on ADHD were obtained from the ADHD Working Group of the Psychiatric Genomics Consortium (PGC) and the ADHD iPSYCH‐SSI‐Broad collaboration. Data were downloaded from (https://www.med.unc.edu/pgc/results-and-downloads; n = 55,374; Demontis et al., 2019). GWAS‐MA data on schizophrenia were obtained from the SCZ working group of the PGC (n = 82,315; https://www.med.unc.edu/pgc/results-and-downloads; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Only variants with an imputation quality score INFO >0.8 and minor allele frequency (MAF) >0.01 were taken forward for further analyses. Details of inclusion criteria, genotyping, and phenotype characteristics are described in the original publications (Demontis et al., 2019; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) and the Appendix S1.
Copy number analysis in the Dutch family
Genome‐wide single nucleotide polymorphism (SNP)‐based array analysis was performed on DNA from peripheral blood of the index patient and his parents using the Affymetrix CytoScan HD array platform (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s protocols. The probe values from 2.6 million markers, consisting of 750,000 SNPs and 1.9 million nonpolymorphic probes, were analyzed using the Affymetrix software package ChAS (Chromosome Analysis Suite) at an average resolution of approximately 20 kb and based on human genome build 37 (hg19). Detection and reporting criteria were used as previously described (Kooper et al., 2014). Follow‐up testing by fluorescence in situ hybridization (FISH) was performed with a subtelomeric 8pter probe (TelVysion 8p (D8S504); Abbott Molecular, Des Plaines, IL).
The observed CNV was subsequently confirmed by quantitative real‐time PCR (qRT‐PCR) in the index patient and his mother, and three additional family members were analyzed using this method. Genomic DNA was either isolated from saliva or from EDTA blood samples according to manufacturer’s protocol at the department of Human Genetics of the Radboud University Medical Center, Nijmegen, The Netherlands. PrimeTime® Mini qRT‐PCR assay (Integrated DNA Technologies [IDT], Coralville, IA) was used to determine the copy number of FBXO25 (for details see Appendix S1). Primer sequences are displayed in Table S2. Results were analyzed with the 7500 Software v2.0.6 (Life Technologies) using an automatic threshold (Appendix S1). Data were visualized using GraphPad prism (version 5.03), and the mean and a 95% confidence interval are shown.
Gene‐set and gene‐based analyses in the PGC + iPSYCH ADHD and PGC schizophrenia data sets
The cumulative effect of common variants in the set of protein‐coding genes altered in the family (as a set and individually) was tested using summary statistics of the ADHD and the SCZ GWAS‐MA (Appendix S1; Demontis et al., 2019; Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Statistical analyses were performed using the Multi‐marker Analysis of GenoMic Annotation (MAGMA) software package (version 1.05; http://ctglab.nl/software/magma; de Leeuw et al., 2015) using the SNP‐wide mean model for gene‐based association analysis (for detailed description see Appendix S1). A correction for linkage disequilibrium (LD) was applied, based on the estimates from the 1,000 Genomes Phase 1 European ancestry samples (Genomes Project et al., 2010). Protein‐coding genes were considered gene‐wide significant, if they reached the Bonferroni‐corrected threshold adjusted for the number of genes tested (three tests; p < .0167).
Brain gene expression
We determined the messenger‐RNA (mRNA) expression of genes located within the identified CNV. Using the publicly available data set provided by the Human Brain Transcriptome Project (Kang et al., 2011) at http://hbatlas.org, we assessed mRNA expression trajectories in six regions of the developing and adult human brain. Spanning periods from embryonic development to late adulthood, this data set provides genome‐wide exon‐level transcriptome data generated using the Affymetrix GeneChip Human Exon 1.0 SS Arrays from over 1,340 tissue samples from both hemispheres of postmortem human brains (n = 57)(GTEx Consortium, 2013).
Functional characterization of Fbxo25 in Drosophila
Drosophila stocks and maintenance
Conditional overexpression of CG11658 (two‐to‐one orthologue of FBXO25 and FBXO32) was achieved by using the GAL4‐UAS system and the pan‐neuronal driver UAS‐Dcr‐2 hs(X); nSyb‐GAL4. UAS overexpression lines were obtained from the stock centers in Kyoto and Bloomington (Fbxo25 overexpression‐1: Kyoto stock 203566 and FBXO25 overexpression‐2: Bloomington stock 17663). The genetic background of the overexpression lines was isogenized for eight generations using a wild‐type Iso31 background (gift from A. Sehgal (Kumar et al., 2012)). Flies were raised on standard medium (cornmeal, sugar, and yeast) and maintained on a 12 hours light dark cycle. Crosses with the pan‐neuronal driver UAS‐Dcr‐2 hs(X); nSyb‐Gal4 and the UAS overexpression lines were raised at 28°C. For the control condition, the same pan‐neuronal driver was crossed to the isogenic wild‐type background.
Locomotor activity and sleep measurement
Locomotor activity and sleep were assessed with the Drosophila Activity Monitor system (DAM), which measures locomotor activity by infrared photoelectric barriers (Trikinetics, Waltham, MA). Single adult flies were transferred to monitor tubes with standard food. Activity counts were collected in 30‐s bins and analyzed in 1‐min bins. Activity of individual 3–5 day‐old male flies was recorded over 4 days in a 12 hr light:dark cycle and subsequently 2 days in constant darkness. According to the standard in the field, five minutes of inactivity of a fly was defined as sleep. The analysis was performed with the pySolo software package (https://www.pysolo.net/), and activity and sleep in the light‐off period (relative night) (between 12 and 24 hr) were analyzed. Activity counts represent the amount of infrared detection beam passes of the fly. Statistical analysis was performed with the GraphPad Prism software package 5.03 for Windows (GraphPad Software, San Diego, CA, http://www.graphpad.com). T‐tests were performed, adding a Welch correction, when variances of groups were significantly different.
Quantification of relative gene expression
Flies carrying the inducible UAS element allowing for overexpression of CG11658 (isogenized strains of Bloomington 17663 and Kyoto 203566) and their isogenic controls were crossed to flies carrying the yw; UAS‐Dcr‐2 hs(X); nSyb‐GAL4 driver. Crosses were raised at 28°C. Per condition, three biological replicates of 10 heads of 3‐day‐old male flies were collected and snap frozen. RNA was extracted using the ARCTURUS PicoPure RNA Isolation Kit (Applied Biosystems, Waltham, MA), and total RNA concentration was measured with the Nanodrop (Thermo Fisher Scientific, Waltham, MA). Complementary cDNA was synthesized using iScript cDNA Synthesis Kit (Bio‐Rad, Hercules, CA). Relative gene expression per sample (three biological replicates) was quantified in technical triplicates using a powerSYBR Green PCR master mix (Applied Biosystems) on a 7900HT Real‐Time PCR Systems (Applied Biosystems). Crossing‐point (CP) values of the CG11658 (dFBXO25) gene were normalized to the reference genes αTub84B and elF‐1A. For primer sequences see Table S2.
Whole‐exome sequencing (WES) in Dutch family
Genomic DNA of the index patient and both parents isolated from blood was used for clinical whole‐exome sequencing (WES) to search for de novo mutations using protocols described previously (Neveling et al., 2013). The WES was performed in a diagnostic setting as part of the routine clinical assessment of the child. In short, exome capture was performed with the Agilent SureSelect Human All Exon enrichment kit v5 (Agilent Technologies, Santa Clara, CA). WES was performed on the Illumina HiSeq platform (BGI, Copenhagen, Denmark), and median target coverage was 100.2. Data were analyzed with Burrows–Wheeler Alignment (BWA) read alignment (Li & Durbin, 2009) and Genome Analysis Toolkit (GATK) variant calling (McKenna et al., 2010) software packages. Variants were annotated using an in‐house developed pipeline (Neveling et al., 2013). Prioritization of variants was done by an in‐house designed ‘variant interface’ and manual curation in line with international standards and guidelines for the interpretation of sequence variants (Richards et al., 2015). Only variants with a minimum coverage of 10 reads were considered for further analysis.
For an overview of the study flow, see Figure S1.
Results
8p23.3 microduplication identified in a family with ADHD‐affected members
In a family with several members affected with ADHD in the presence or absence of psychiatric comorbidity (Figure 1), SNP‐array analysis revealed that the index patient, his mother, and mother’s brother carried a copy number gain of approximately 540 kb at the distal end of the short arm of chromosome 8 (arr[hg19] 8p23.3(158,049–541,637)x3), which spanned 590 probes (Figure 2). Follow‐up testing by FISH revealed that the gain in copy number was the result of a duplication rather than an unbalanced translocation. The microduplication encompassed three protein‐coding genes, ZNF596, FBXO25, and TDRP, and the pseudo‐gene RPL23AP53. The copy number gain was confirmed by qRT‐PCR through analysis of FBXO25 (Figure S2). The qRT‐PCR analysis showed that duplication of FBXO25 was absent in one ADHD‐affected family member, but was present in the mother’s sister, who had learning difficulties (Figure S2).
Figure 2.
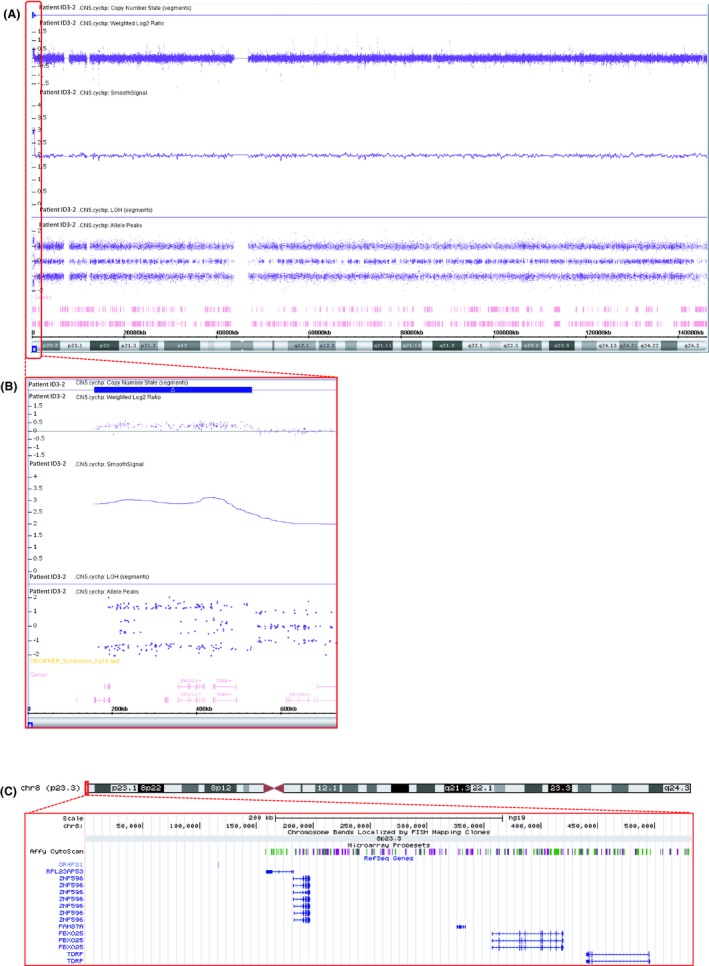
Microduplication of 8p23.3 in the index patient ID3‐2. (A) Array CGH plot of chromosome 8 of patient ID3‐2 with a terminal duplication of 535 kb in 8p23.3 spanning 590 probes (arr[hg19] 8p23.3(158,049–541,637)x3). Zoom‐in of this region is shown in panel (B). (C) Schematic representation of chromosome 8 with the 8p23.3 region enlarged in the lower part of the figure (screenshot of the UCSC Genome Browser Build 37/hg19; http://genome.ucsc.edu/); the small vertical bars show the probe coverage of the CytoScan HD array platform, the horizontal bars represent the genes in the duplicated 8p23.3 region (RPL23AP53, ZNF596, FBXO25, and TDRP)
Common variants in the 8p23.3 locus are associated with ADHD
Focusing on protein‐coding genes within the 8p23.3 duplicated region (ZNF596, FBXO25, and TDRP), we performed a gene‐set analysis to provide additional evidence for a role of genetic variation in this region in ADHD and/or psychotic disorders, as several family members also showed psychotic symptoms. This was done using summary statistics of ADHD GWAS‐MA data of 20,183 patients with ADHD and 35,191 controls (Demontis et al., 2019) and SCZ GWAS‐MA summary statistics from the analysis of 34,241 cases and 45,604 controls (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). Joint analysis of the three genes showed significant association of the gene‐set with ADHD in both self‐contained and competitive tests (p self‐contained = .000184 and p competitive = .039151, see also Figure S3a). No significance was observed for association with SCZ (p self‐contained = 0.7123 and p competitive = 0.90596, see also Figure S3b).
To identify the most likely candidate gene for ADHD among the three protein‐coding genes in the microduplication, the genes were subsequently tested individually. Gene‐based analysis revealed that the two more proximal genes, FBXO25 and TDRP, were significantly associated with ADHD (pFBXO25 = .010756 and pTDRP = .000285; Table S3).
Brain gene expression
We determined the distribution of mRNA expression for FBXO25 and TDRP in the developing and adult human brain using data on six brain regions available from the Human Brain Transcriptome Project. As the mRNA expression figures in Figure S4 show, both genes appear to be widely expressed, both in the developing and the adult human brain.
Overexpression of the FBXO25 orthologue in Drosophila induces increased activity and reduced sleep
To further establish the relevance of the associated genes as candidates for ADHD, we set out to study the effect of overexpression on ADHD‐relevant behavioral phenotypes in Drosophila melanogaster. Only FBXO25 has a direct, as yet uncharacterized orthologue (CG11658) in the fruit fly, from now on referred as Fbxo25. As the duplication also includes the promoter region of FBXO25, we induced overexpression of Fbxo25 using two unrelated Drosophila lines. Quantitative RT‐PCR confirmed the overexpression of this gene: in overexpression‐line 1, expression of Fbxo25 was 450% compared with the control line; in overexpression‐line 2, it was 650%. Using earlier established tests of locomotor activity and sleep as ADHD‐relevant readouts (van der Voet et al., 2016), we found adult flies from either line to exhibit significantly elevated activity and reduced sleep in the relative night period (12 hr dark) compared with an appropriate genetic background control (Figure 3). In line with the qRT‐PCR results, overexpression‐line 2 showed the more severe behavioral phenotypes (Figure 3).
Figure 3.
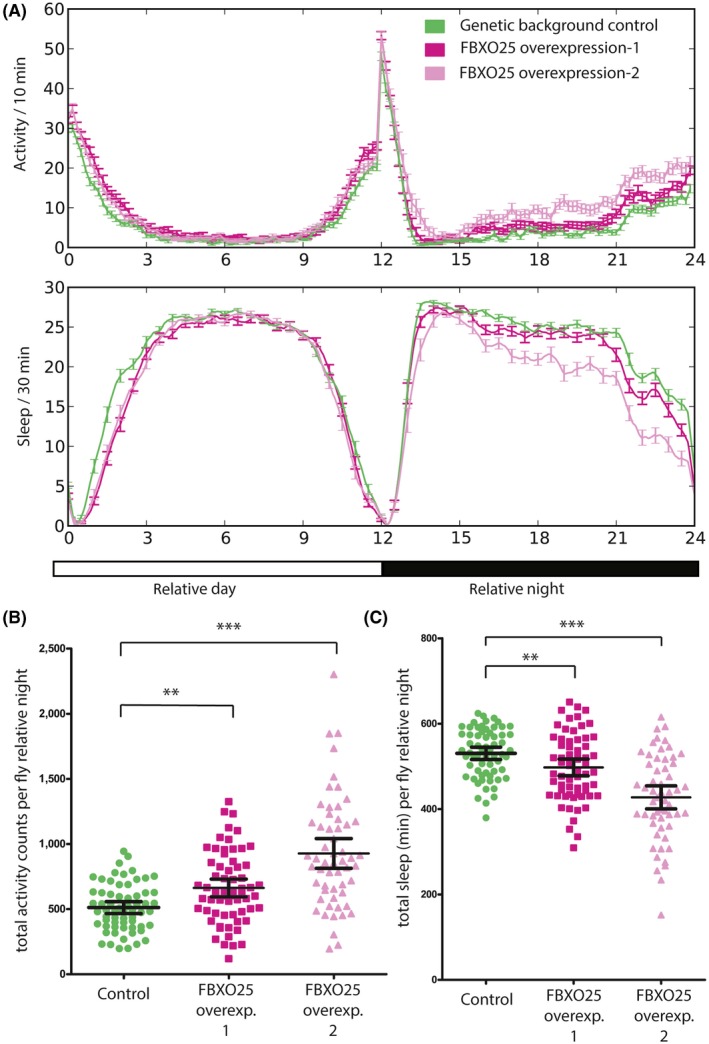
Activity monitoring upon pan‐neuronal overexpression of the FBXO25 orthologue in Drosophila melanogaster. (A) Activity and sleep profiles reveal increased activity (top graph) and decreased sleep (bottom graph) in the relative night (12–24 hr). (B) Quantification of the activity counts (amount of infrared detection beam passes) of individual flies in FBXO25 overexpression–1 (n = 64) and overexpression–2 (n = 56) lines shows that the activity in the relative night is significantly and consistently higher in both compared with control (n = 63) flies (FBXO25 overexpression–1: p = .0013; FBXO25 overexpression‐2: p < .0001). (C) Quantification of sleep in minutes of single flies shows that both FBXO25 overexpression lines sleep significantly less in the relative night compared with control flies (FBXO25 overexpression–1: p = .0076; FBXO25 overexpression‐2: p < .0001). **p < .001, ***p < .0001
Whole‐exome sequencing
Given the additional somatic features and early onset of severe psychiatric problems in the index patient, we performed clinical WES to identify additional rare variants that may contribute to the more severe clinical phenotype. Upon trio whole‐exome data analysis of the index patient and his parents, one de novo candidate variant was found in the index patient. This was a heterozygous insertion variant in DUSP16 (c.813_814insGC [p.Arg272fs] [GenBank: NM_030640.2]) leading to a frameshift, located at the end of exon 5, it possibly affects the nearest splice site. This frameshift variant is not present in gnomAD (r.2.0.2, https://gnomad.broadinstitute.org/). Exploratory gene‐wide testing of common variants in DUSP16 in the ADHD and SCZ GWAS‐MA data did not reveal evidence for association with ADHD (p = .39186), but the gene showed nominal association with schizophrenia (p = .0175).
Discussion
In this study, we identified and functionally validated a novel candidate gene for ADHD and comorbid psychiatric phenotypes. We report a microduplication in band p23.3 of chromosome 8, which segregated with a broad spectrum of psychiatric problems, including ADHD, in a family with several affected individuals in two generations. Common genetic variants in this locus were associated with ADHD risk; in particular, we observed significant gene‐based associations for the genes in the proximal part of the microduplication, FBXO25, and TDRP. One of the two genes, FBXO25, has an orthologue in Drosophila melanogaster, and we were able to confirm that its overexpression, as expected to arise from a copy number gain, leads to a night‐specific increase in locomotor activity and decrease in sleep. Since we have previously identified the same behavioral signature in several Drosophila models of ADHD (van der Voet et al., 2016) and similar phenotypes have also been suggested as readout for mania and circadian dysregulation (Guo et al., 2018; Padiath et al., 2004; Ries et al., 2017), these findings support FBXO25 as a novel risk gene for ADHD and comorbid mental disorders.
The identified microduplication in 8p23.3 has not previously been reported in conjunction with ADHD or other neuropsychiatric disorders. A recent study combined data from eleven CNV studies that had identified rare CNVs occurring in patients with ADHD (Harich et al., 2019). We queried these existing data sources and found an additional ADHD patient, among 6,176 reported ones, who carried a duplication encompassing 77% of the FBXO25 gene and including the promoter region. This duplication was not found in 25,026 controls (Elia et al., 2010; Harich et al., 2019).
The microduplication cosegregated with different psychiatric phenotypes, ranging from mild learning difficulties to ADHD, psychosis, substance use, and aggressive behavior. Of the two cousins of the index patient’s mother, each with an ADHD diagnosis, one had a normal copy number for FBXO25, and for the second cousin no results were obtained due to poor quality DNA. Since ADHD is a rather common disorder in the population (Faraone et al., 2015), we can only speculate that there may be additional variants contributing to psychiatric risk (e.g. coming from common variants (Demontis et al., 2019) and/or from environmental risk factors) present in this family, which contribute to the ADHD phenotype in addition to the structural variant. In that way, ADHD may not behave as a strictly monogenic disorder in any of the affected individuals in this pedigree, which is in line with findings from previous rare variant studies of the disorder (Corominas et al., 2018), and reminiscent of findings in previous linkage studies of ADHD (Lesch et al., 2008; Zhou et al., 2008) and other neurodevelopmental disorders (e.g. autism (Chapman et al., 2015)). Indeed, WES revealed an additional de novo frameshift variant in the DUSP16 gene in the index patient, who also showed additional somatic features, anxiety disorder, and psychiatric episodes at very young age. A recent study reported that Dusp16 plays a critical role in neurogenesis by balancing neural progenitor cell proliferation and neural differentiation (Zega et al., 2017). Moreover, mouse mutants lacking a functional Dusp16 gene developed fully‐penetrant congenital obstructive hydrocephalus together with brain overgrowth (Zega et al., 2017). When focusing on common genetic variants, we also found nominally significant association of DUSP16 with schizophrenia. Therefore, it may be suggested that this de novo de novo variant could have a modifying role in the early development of the patient’s severe psychiatric phenotypes.
After association testing of the microduplication region in the data from the world‐wide largest genome‐wide association data set on ADHD risk, we identified two genes, the F‐box protein 25 (FBXO25) and the testis development‐related protein (TDRP) to be associated with ADHD risk. While both genes are expressed in the developing and adult brain, a function in the nervous system has not been described for either, thus far. FBXO25 codes for an E3 ligase, which provides the substrate specificity for ubiquitin‐dependent destruction or inhibition of transcription factors (Jang et al., 2011). FBXO25 plays a role in cardiomyocyte development, and reduced expression of FBXO25 appears to be involved in cell proliferation and migration in nonsmall‐cell lung cancer (Jeong et al., 2015; Jiang et al., 2016). Knockdown of the gene in a cellular system has been linked to decreased phosphorylation of ERK1, whereas overexpression has an opposite effect on the MAPK signaling pathway (Teixeira et al., 2017). MAPK signaling plays a role in different psychiatric disorders, including ADHD and autism (Poelmans et al., 2013; Vithayathil et al., 2018). Moreover, the brain relies on ubiquitination to fine‐tune protein content in order to react to new stimuli (Hegde & van Leeuwen, 2017). Another E3 ligase, FBXO33, has been the top‐finding of the first case–control genome‐wide association study of persistent ADHD (Sanchez‐Mora et al., 2015). Further F‐box genes have been found associated with neurodevelopmental disorders, too, as for example, reported in a patient with intellectual disability and seizures carrying a deletion of FBXO28 (Au et al., 2014). Another study reports de novo variants in FBXO11 in 20 patients, of which 17 show behavioral problems, such as autistic‐like features and ADHD‐related traits (Gregor et al., 2018). TDRP, the second gene in the microduplication, is known to be involved in sperm motility (Mao et al., 2016). A rare missense variant in TDRP has been reported in monozygotic twins with gender dysphoria (Morimoto et al., 2017). Otherwise, little is known about this gene up to now.
Many different complex behaviors can be studied in Drosophila, and it is a model for a wide range of neuropsychiatric phenotypes (van Alphen & van Swinderen, 2013). Among the behaviors that can be robustly assessed in Drosophila are locomotor activity and sleep, which are known to undergo a well‐regulated day/night rhythm. These phenotypes have earlier been linked to genes implicated in mania and circadian dysregulation disorders (Guo et al., 2018; Padiath et al., 2004; Ries et al., 2017). In a previous study, we also reported increased activity and decreased sleep upon knockdown of genes that have been highly implicated in ADHD, encoding the dopamine transporter, latrophilin, and neurofibromin 1 (van der Voet et al., 2016). Further experiments strongly suggested this phenotype to result from abnormal activity of the dopaminergic system. Upon overexpression of the FBXO25 orthologue described here, we recapitulated this phenotype. This demonstrates that overexpression of FBXO25 in the nervous system is sufficient to trigger increased locomotor activity, suggesting that it can contribute to psychiatric symptoms in our family. This is also the first report providing evidence for FBXO25 having a role in the nervous system.
The findings described here need to be interpreted in light of several strengths and limitations of our study. The main strength of this study is its interdisciplinary setting and comprehensive approach across multiple levels. For this, we integrated complementary evidence from genetic, genomic, and animal model experiments to examine the potential causes of complex psychiatric phenotypes including ADHD segregating across generations in a single family. We combined rare variants approaches (CNV analysis and WES) with an analysis of common variants in the identified CNV using the largest meta‐analysis data sets currently available. For our functional validation of the identified candidate gene FBXO25, we used an unconventional, well‐validated animal model. A clear limitation of our study is that another identified candidate gene, TDRP, is not conserved in Drosophila. We were therefore not able to collect data that would argue for or against a contribution of TDRP overexpression to the complex psychiatric phenotypes seen in the family, in addition to FBXO25. Similarly, it would be of interest to investigate the role of DUSP16 as a possible modifier of the phenotype of the index patient. In general, it would be of interest to further assess the specificity of FBXO25 for ADHD. Given the fact that in this family many other psychiatric traits, cooccurring with ADHD, are present, future studies may want to investigate such cross‐disorder characteristics of FBXO25. Also, it has to be tested whether this gene or locus is specifically causal for the common syndrome of ADHD and it may well be informative regarding physiological processes contributing to other neuropsychiatric syndromes, which does merit further in depth exploration to confirm and extend this novel observation.
In conclusion, this study provides an important illustration of the potential of such an across‐species approach and by integrating genetic and genomics studies with biological validation, we identified FBXO25 as a novel risk gene for ADHD and comorbid psychiatric disorders. This convergence generates novel hypotheses for future studies to discover the underlying molecular mechanisms and identify proteins regulated by FBXO25 in order to better understand the etiology of the aberrant behavior caused by variation in this gene.
Supporting information
Appendix S1. Supplementary methods.
Table S1 . Overview of clinical diagnoses and relevant psychiatric symptoms in the family.
Table S2 . Primer sequences for quantitative RT‐PCR.
Table S3 . Gene‐based association results for the three protein‐coding genes within the 8p23.3 locus using data from the ADHD GWAS‐MA (Demontis et al., 2019).
Figure S1 . Design and flow of the study.
Figure S2 . Validating gain of copy number of FBXO25 gene by PrimeTime® Mini qPCR assay.
Figure S3 . Regional association plot for 8p23.3 locus (±100 kb) using (A) the ADHD GWAS‐MA data (Demontis et al., 2019) and (B) the schizophrenia GWAS‐MA data (Schizophrenia Working Group of the Psychiatric Genomics, 2014).
Figure S4 . Expression trajectories of FBXO25 and TDRP (C8orf42) in the developing and adult human brain.
Acknowledgements
This work was partly carried out on the Dutch national e‐infrastructure with the support of SURF Foundation. B.H. was funded by a Radboud University Medical Center PhD grant. M.vdV. was supported by the Netherlands Organization for Scientific Research (NWO) Veni grant (91.614.084). B.F. and M.K. are supported by funding from a personal Vici NWO grant (016‐130‐669, to B.F.) and from funding for the Dutch National Science Agenda NeurolabNL project (grant 400‐17‐602). This work was also supported by a NWO TOP grant (912‐12‐109, to A.S.), the European Community’s Seventh Framework Programme (FP7/2007 – 2013) under grant agreement no. 602805 (Aggressotype), and the European Community’s Horizon 2020 Programme (H2020/2014 – 2020) under grant agreements no. 667302 (CoCA) and no. 643051 (MiND). The authors wish to thank all participants who took part in the study. B.F. received educational speaking fees from Shire and Medice. The remaining authors have declared that they have no competing or potential conflicts of interest.
Key points.
Integration of data across multiple levels can help in gene identification for psychiatric disorders.
Microduplication cosegregated with different psychiatric phenotypes, ranging from learning difficulties to ADHD, psychosis, substance use, and aggressive behavior.
First report providing evidence for FBXO25, a gene involved in ubiquitination, having a role in the nervous system.
De novo variant in DUSP16 may have modifying role in early development of the patient’s severe psychiatric phenotypes. Information about common variants may be used to support decisions about (clinical) relevance of rare genetic variants.
Conflict of interest statement: See Acknowledgements for full disclosures.
Contributor Information
Annette Schenck, Email: Annette.Schenck@radboudumc.nl.
Barbara Franke, Email: Barbara.Franke@radboudumc.nl.
References
- Akutagava‐Martins, G.C. , Salatino‐Oliveira, A. , Genro, J.P. , Contini, V. , Polanczyk, G. , Zeni, C. , … & Hutz, M.H. (2014). Glutamatergic copy number variants and their role in attention‐deficit/hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 165b, 502–509. [DOI] [PubMed] [Google Scholar]
- Au, P.Y. , Argiropoulos, B. , Parboosingh, J.S. , & Micheil Innes, A. (2014). Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. American Journal of Medical Genetics. Part A, 164a, 441–448. [DOI] [PubMed] [Google Scholar]
- Chapman, N.H. , Nato, A.Q., Jr , Bernier, R. , Ankenman, K. , Sohi, H. , Munson, J. , … & Wijsman, E.M. (2015). Whole exome sequencing in extended families with autism spectrum disorder implicates four candidate genes. Human Genetics, 134, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corominas, J. , Klein, M. , Zayats, T. , Rivero, O. , Ziegler, G.C. , Pauper, M. , … & Lesch, K.‐P. (2018). Identification of ADHD risk genes in extended pedigrees by combining linkage analysis and whole‐exome sequencing. Molecular Psychiatry. 10.1038/s41380-018-0210-6. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw, C.A. , Mooij, J.M. , Heskes, T. , & Posthuma, D. (2015). MAGMA: generalized gene‐set analysis of GWAS data. PLoS Computational Biology, 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis, D. , Lescai, F. , Borglum, A. , Glerup, S. , Ostergaard, S.D. , Mors, O. , … & Franke, B. (2016). Whole‐exome sequencing reveals increased burden of rare functional and disruptive variants in candidate risk genes in individuals with persistent attention‐deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry, 55, 521–523. [DOI] [PubMed] [Google Scholar]
- Demontis, D. , Walters, R.K. , Martin, J. , Mattheisen, M. , Als, T.D. , Agerbo, E. , … & Neale, B.M. (2019). Discovery of the first genome‐wide significant risk loci for attention deficit/hyperactivity disorder. Nature Genetics, 51, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, J. , Gai, X. , Xie, H.M. , Perin, J.C. , Geiger, E. , Glessner, J.T. , … & White, P.S. (2010). Rare structural variants found in attention‐deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry, 15, 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, J. , Glessner, J.T. , Wang, K. , Takahashi, N. , Shtir, C.J. , Hadley, D. , … & Hakonarson, H. (2011). Genome‐wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nature Genetics, 44, 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraone, S.V. , Asherson, P. , Banaschewski, T. , Biederman, J. , Buitelaar, J.K. , Ramos‐Quiroga, J.A. , … & Franke, B. (2015). Attention‐deficit/hyperactivity disorder. Nature Reviews Disease Primers, 1, 15020. [DOI] [PubMed] [Google Scholar]
- Faraone, S.V. , Perlis, R.H. , Doyle, A.E. , Smoller, J.W. , Goralnick, J.J. , Holmgren, M.A. , & Sklar, P. (2005). Molecular genetics of attention‐deficit/hyperactivity disorder. Biological Psychiatry, 57, 1313–1323. [DOI] [PubMed] [Google Scholar]
- Franke, B. , Faraone, S.V. , Asherson, P. , Buitelaar, J. , Bau, C.H. , Ramos‐Quiroga, J.A. , … & Reif, A. (2012). The genetics of attention deficit/hyperactivity disorder in adults, a review. Molecular Psychiatry, 17, 960–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, B. , Michelini, G. , Asherson, P. , Banaschewski, T. , Bilbow, A. , Buitelaar, J.K. , … & Reif, A. (2018). Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. European Neuropsychopharmacology, 28, 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium , Abecasis, G.R. , Altshuler, D. , Auton, A. , Brooks, L.D. , Durbin, R.M. , … & McVean, G.A. (2010). A map of human genome variation from population‐scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizer, I.R. , Ficks, C. , & Waldman, I.D. (2009). Candidate gene studies of ADHD: A meta‐analytic review. Human Genetics, 126, 51–90. [DOI] [PubMed] [Google Scholar]
- Gregor, A. , Sadleir, L.G. , Asadollahi, R. , Azzarello‐Burri, S. , Battaglia, A. , … & Zweier, C. (2018). De Novo Variants in the F‐Box Protein FBXO11 in 20 Individuals with a Variable Neurodevelopmental Disorder. American Journal of Human Genetics, 103, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2013). The Genotype‐Tissue Expression (GTEx) project. Nature Genetics, 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, F. , Holla, M. , Diaz, M.M. , & Rosbash, M. (2018). A circadian output circuit controls sleep‐wake arousal in drosophila. Neuron, 100, 624–635. e4. [DOI] [PubMed] [Google Scholar]
- Harich, B. , van der Voet, M. , Klein, M. , Fenckova, M. , Cizek, P. , Franke, B. , & Schenck, A. (2019). From rare copy number variations to biological processes in ADHD. bioRxiv, 762419 10.1101/762419 [DOI] [PubMed] [Google Scholar]
- Hawi, Z. , Cummins, T.D. , Tong, J. , Arcos‐Burgos, M. , Zhao, Q. , Matthews, N ., … & Bellgrove, M.A. (2016). Rare DNA variants in the brain‐derived neurotrophic factor gene increase risk for attention‐deficit hyperactivity disorder: A next‐generation sequencing study. Molecular Psychiatry, 22, 580–584. [DOI] [PubMed] [Google Scholar]
- Hegde, A.N. , & van Leeuwen, F.W. (2017). Editorial: ubiquitin and the brain: Roles of proteolysis in the normal and abnormal nervous system. Frontiers in Molecular Neuroscience, 10, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, J.W. , Lee, W.Y. , Lee, J.H. , Moon, S.H. , Kim, C.H. , & Chung, H.M. (2011). A novel Fbxo25 acts as an E3 ligase for destructing cardiac specific transcription factors. Biochemical and Biophysical Research Communications, 410, 183–188. [DOI] [PubMed] [Google Scholar]
- Jarick, I. , Volckmar, A.L. , Putter, C. , Pechlivanis, S. , Nguyen, T.T. , Dauvermann, M.R. , … & Hinney, A. (2014). Genome‐wide analysis of rare copy number variations reveals PARK2 as a candidate gene for attention‐deficit/hyperactivity disorder. Molecular Psychiatry, 19, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, H.S. , Jung, E.S. , Sim, Y.J. , Kim, S.J. , Jang, J.W. , Hong, K.S. , … & Kim, K.‐S. (2015). Fbxo25 controls Tbx5 and Nkx2‐5 transcriptional activity to regulate cardiomyocyte development. Biochimica et Biophysica Acta (BBA) ‐ Gene Regulatory Mechanisms, 1849, 709–721. [DOI] [PubMed] [Google Scholar]
- Jiang, G.Y. , Zhang, X.P. , Wang, L. , Lin, X.Y. , Yu, J.H. , Wang, E.H. , & Zhang, Y. (2016). FBXO25 promotes cell proliferation, invasion, and migration of NSCLC. Tumour Biology, 37, 14311–14319. [DOI] [PubMed] [Google Scholar]
- Kang, H.J. , Kawasawa, Y.I. , Cheng, F. , Zhu, Y. , Xu, X. , Li, M. , … & Šestan, N. (2011). Spatio‐temporal transcriptome of the human brain. Nature, 478, 483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.S. , Burt, A.A. , Ranchalis, J.E. , Wilmot, B. , Smith, J.D. , Patterson, K.E. , … & Jarvik, G.P. (2017). Sequencing of sporadic Attention‐Deficit Hyperactivity Disorder (ADHD) identifies novel and potentially pathogenic de novo variants and excludes overlap with genes associated with autism spectrum disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 174, 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooper, A.J. , Faas, B.H. , Feenstra, I. , de Leeuw, N. , & Smeets, D.F. (2014). Best diagnostic approach for the genetic evaluation of fetuses after intrauterine death in first, second or third trimester: QF‐PCR, karyotyping and/or genome wide SNP array analysis. Molecular Cytogenetics, 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Chen, D. , & Sehgal, A. (2012). Dopamine acts through Cryptochrome to promote acute arousal in Drosophila. Genes and Development, 26, 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson, H. , Chang, Z. , D'Onofrio, B.M. , & Lichtenstein, P. (2014). The heritability of clinically diagnosed attention deficit hyperactivity disorder across the lifespan. Psychological Medicine, 44, 2223–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.H. , Ripke, S. , Neale, B.M. , Faraone, S.V. , Purcell, S.M. , Perlis, R.H. , … & International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) . (2013). Genetic relationship between five psychiatric disorders estimated from genome‐wide SNPs. Nature Genetics, 45, 984–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch, K.P. , Selch, S. , Renner, T.J. , Jacob, C. , Nguyen, T.T. , Hahn, T. , … & Ullmann, R. (2011). Genome‐wide copy number variation analysis in attention‐deficit/hyperactivity disorder: association with neuropeptide Y gene dosage in an extended pedigree. Molecular Psychiatry, 16, 491–503. [DOI] [PubMed] [Google Scholar]
- Lesch, K.P. , Timmesfeld, N. , Renner, T.J. , Halperin, R. , Roser, C. , Nguyen, T.T. , … & Jacob, C. (2008). Molecular genetics of adult ADHD: converging evidence from genome‐wide association and extended pedigree linkage studies. Journal of Neural Transmission, 115, 1573–1585. [DOI] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, S. , Wu, F. , Cao, X. , He, M. , Liu, N. , Wu, H. , Yang, Z. , Ding, Q. , & Wang, X. (2016). TDRP deficiency contributes to low sperm motility and is a potential risk factor for male infertility. American Journal of Translational Research, 8, 177–187. [PMC free article] [PubMed] [Google Scholar]
- Martin, J. , Cooper, M. , Hamshere, M.L. , Pocklington, A. , Scherer, S.W. , Kent, L. , … & Holmans, P. (2014). Biological overlap of attention‐deficit/hyperactivity disorder and autism spectrum disorder: evidence from copy number variants. Journal of the American Academy of Child and Adolescent Psychiatry, 53, 761–770.e726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , … & DePristo, M.A. (2010). The Genome Analysis Toolkit: a MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Research, 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, Y. , Ono, S. , Imamura, A. , Okazaki, Y. , Kinoshita, A. , Mishima, H. , … & Kurotaki, N. (2017). Deep sequencing reveals variations in somatic cell mosaic mutations between monozygotic twins with discordant psychiatric disease. Human Genome Variation, 4, 17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling, K. , Feenstra, I. , Gilissen, C. , Hoefsloot, L.H. , Kamsteeg, E.J. , Mensenkamp, A.R. , … & Nelen, M.R. (2013). A post‐hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Human Mutation, 34, 1721–1726. [DOI] [PubMed] [Google Scholar]
- Padiath, Q.S. , Paranjpe, D. , Jain, S. , & Sharma, V.K. (2004). Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiology International, 21, 43–55. [DOI] [PubMed] [Google Scholar]
- Poelmans, G. , Franke, B. , Pauls, D.L. , Glennon, J.C. , & Buitelaar, J.K. (2013). AKAPs integrate genetic findings for autism spectrum disorders. Translational Psychiatry, 3, e270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos‐Quiroga, J.A. , Sanchez‐Mora, C. , Casas, M. , Garcia‐Martinez, I. , Bosch, R. , Nogueira, M. , … & Ribasés, M. (2014). Genome‐wide copy number variation analysis in adult attention‐deficit and hyperactivity disorder. Journal of Psychiatric Research, 49, 60–67. [DOI] [PubMed] [Google Scholar]
- Richards, S. , Aziz, N. , Bale, S. , Bick, D. , Das, S. , Gastier‐Foster, J. , … & Rehm, H.L. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries, A.S. , Hermanns, T. , Poeck, B. , & Strauss, R. (2017). Serotonin modulates a depression‐like state in Drosophila responsive to lithium treatment. Nature Communications, 8, 15738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Mora, C. , Ramos‐Quiroga, J.A. , Bosch, R. , Corrales, M. , Garcia‐Martinez, I. , Nogueira, M. , … & Ribasés, M. (2015). Case‐control genome‐wide association study of persistent attention‐deficit hyperactivity disorder identifies FBXO33 as a novel susceptibility gene for the disorder. Neuropsychopharmacology, 40, 915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia‐associated genetic loci. Nature, 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, F.R. , Manfiolli, A.O. , Vieira, N.A. , Medeiros, A.C. , Coelho, P.O. , Santiago Guimaraes, D. , … & Gomes, M.D. (2017). FBXO25 regulates MAPK signaling pathway through inhibition of ERK1/2 phosphorylation. Archives of Biochemistry and Biophysics, 621, 38–45. [DOI] [PubMed] [Google Scholar]
- van Alphen, B. , & van Swinderen, B. (2013). Drosophila strategies to study psychiatric disorders. Brain Research Bulletin, 92, 1–11. [DOI] [PubMed] [Google Scholar]
- van der Voet, M. , Harich, B. , Franke, B. , & Schenck, A. (2016). ADHD‐associated dopamine transporter, latrophilin and neurofibromin share a dopamine‐related locomotor signature in Drosophila. Molecular Psychiatry, 21, 565–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Voet, M. , Nijhof, B. , Oortveld, M.A. , & Schenck, A. (2014). Drosophila models of early onset cognitive disorders and their clinical applications. Neuroscience and Biobehavioral Reviews, 46(Pt 2), 326–342. [DOI] [PubMed] [Google Scholar]
- van Hulzen, K.J.E. , Scholz, C.J. , Franke, B. , Ripke, S. , Klein, M. , McQuillin, A. , … & Reif, A. (2017). Genetic overlap between attention‐deficit/hyperactivity disorder and bipolar disorder: evidence from genome‐wide association study meta‐analysis. Biological Psychiatry, 82, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithayathil, J. , Pucilowska, J. , & Landreth, G.E. (2018). ERK/MAPK signaling and autism spectrum disorders. Progress in Brain Research, 241, 63–112. [DOI] [PubMed] [Google Scholar]
- Williams, N.M. , Franke, B. , Mick, E. , Anney, R.J. , Freitag, C.M. , Gill, M. , … & Faraone, S.V. (2012). Genome‐wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. American Journal of Psychiatry, 169, 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, N.M. , Zaharieva, I. , Martin, A. , Langley, K. , Mantripragada, K. , Fossdal, R. , … & Thapar, A. (2010). Rare chromosomal deletions and duplications in attention‐deficit hyperactivity disorder: a genome‐wide analysis. Lancet, 376, 1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L. , Neale, B.M. , Liu, L. , Lee, S.H. , Wray, N.R. , Ji, N. , … & Wang, Y. (2013). Polygenic transmission and complex neuro developmental network for attention deficit hyperactivity disorder: genome‐wide association study of both common and rare variants. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 162b, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zega, K. , Jovanovic, V.M. , Vitic, Z. , Niedzielska, M. , Knaapi, L. , Jukic, M.M. , … & Brodski, C. (2017). Dusp16 deficiency causes congenital obstructive hydrocephalus and brain overgrowth by expansion of the neural progenitor pool. Frontiers in Molecular Neuroscience, 10, 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, K. , Dempfle, A. , Arcos‐Burgos, M. , Bakker, S.C. , Banaschewski, T. , Biederman, J. , … & Asherson, P. (2008). Meta‐analysis of genome‐wide linkage scans of attention deficit hyperactivity disorder. American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics: The Official Publication of the International Society of Psychiatric Genetics, 147B, 1392–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supplementary methods.
Table S1 . Overview of clinical diagnoses and relevant psychiatric symptoms in the family.
Table S2 . Primer sequences for quantitative RT‐PCR.
Table S3 . Gene‐based association results for the three protein‐coding genes within the 8p23.3 locus using data from the ADHD GWAS‐MA (Demontis et al., 2019).
Figure S1 . Design and flow of the study.
Figure S2 . Validating gain of copy number of FBXO25 gene by PrimeTime® Mini qPCR assay.
Figure S3 . Regional association plot for 8p23.3 locus (±100 kb) using (A) the ADHD GWAS‐MA data (Demontis et al., 2019) and (B) the schizophrenia GWAS‐MA data (Schizophrenia Working Group of the Psychiatric Genomics, 2014).
Figure S4 . Expression trajectories of FBXO25 and TDRP (C8orf42) in the developing and adult human brain.


