Abstract
In biliary tract cancer (BTC), tissue biopsies to guide treatment are rarely feasible, thus implementing liquid biopsy approaches to improve patient management represents a priority. So far, studies on circulating tumor cells (CTCs) in BTC are insufficient to promote their use in patient clinical management and are limited to EpCAM‐enriched CTCs evaluated with the CellSearch. We applied a single‐cell protocol allowing identification not only of epithelial CTCs (eCTCs), but also of nonconventional CTCs (ncCTCs) lacking epithelial and leukocyte markers, but presenting aberrant genomes as confirmed by copy number alterations and therefore representing a distinct subpopulation of bona fide CTCs. In 41 blood samples longitudinally collected from 21 patients with advanced‐stage BTC, addition of ncCTC to classic eCTC led to a CTC‐positivity increase from 19% to 83%. Patients presenting with at least 1 eCTC/10 ml of blood at baseline prior to treatment start had a significantly shorter median disease‐specific survival (DSS) compared to those lacking eCTCs (9 months vs. 19 months, p = 0.03 by log‐rank test). No differences in DSS were observed according to ncCTC‐positivity, conversely, variations in ncCTC counts during, and at the end of treatment, were associated with the RECIST response supporting their role in treatment monitoring. Moreover, in 88 ncCTCs collected at different times during treatment, unsupervised clustering evidenced segregation of cells by patient's best response, allowing identification of genomic regions possibly involved in resistance mechanisms. The presence of ncCTCs beside eCTCs opens the way to exploiting liquid biopsy for optimizing clinical management in BTC.
Keywords: biliary tract cancer, liquid biopsy, single cell, WGS, copy number alterations
Short abstract
What's new?
Late diagnosis of advanced biliary tract cancer (BTC) limits tissue biopsy for molecular analyses, resulting in missed opportunities for personalized therapy. Meanwhile, circulating tumor cells (CTCs) are promising tissue surrogates, but current CTC‐based methods detect only a fraction of BTC patients. Here, using unbiased CTC‐enrichment, coupled with identification and recovery of single cells, the authors identify a novel CTC subpopulation detectable in all BTC patient samples prior to treatment. The presence of even a single epithelial CTC was associated with reduced disease‐specific survival. This novel approach to CTC detection could be useful for treatment‐response monitoring and molecular characterization in BTC.
Abbreviations
- BL
baseline
- BTC
biliary tract cancer
- CNAs
copy number alterations
- CTC
circulating tumor cells
- DSS
disease‐specific survival
- DT
during treatment
- eCTC
epithelial CTCs
- EOT
end of treatment
- FU
follow‐up
- lpWGS
low‐pass whole‐genome sequencing
- LST
large‐scale state transition
- mCTC
mesenchymal CTCs
- ncCTC
nonconventional CTCs
- PD
progressive disease
- PR
partial response
- RT
room temperature
- SD
stable disease
- WBC
white blood cells
Introduction
Biliary tract cancer (BTC) is an extremely aggressive and fatal epithelial cancer that arises from the biliary tree1, 2 with an incidence of about 1.67 per 100,000 people in the US3 and an estimated 5‐year survival rate less than 20%.4 Because of the lack of specific symptoms and of effective preventive regimens, more than 65% of BTC patients are diagnosed at a late stage of the disease.5
Standard first‐line chemotherapy (Cisplatin plus Gemcitabine) yields an overall survival benefit of less than 12 months6 and the mean progression‐free survival, response rate and disease control rate of second‐line therapy are 3.2 months, 7.7 and 49.5%, respectively.7
Currently, based upon molecular features of their primary tumor or subsequent tissue biopsies, only a small percentage of patients are selected for entering protocols with targeted treatments, and mostly only after progression under standard therapies, thus highlighting the unmet need for new treatment approaches. Although recent biological descriptions of BTC do definitely support a molecular stratification with direct therapeutic implications,1, 8, 9, 10, 11, 12 the limited access to tissue in advanced cases restrains the use of personalized treatment approaches.
The detection in the blood of patients with solid tumors of circulating tumor cells (CTCs), which are derived from primary and metastatic lesions, is potentially informative on disease progression and on treatment resistance. Indeed, in the clinical management of patients with various solid tumors, CTCs have been used to predict prognosis, monitor treatment efficacy and more recently also to predict treatment resistance. Due to their direct involvement in the metastatic cascade such cells represent a peculiar biomarker that can reveal much on cancer evolution and on treatment sensitivity/resistance.13
Scanty literature data limited to EpCAM‐enriched CTCs evaluated with the CellSearch, are available for patients with advanced BTC.14, 15, 16, 17 Such studies report low numbers of epithelial CTCs (eCTCs) with positivity rates ranging from 17% to 46%, depending on the case series and on the chosen positivity cutoff. A statistically significant association between baseline eCTC status and overall survival was independently suggested by the study of Valle and colleagues including 95 patients with advanced BTC who entered the ABC‐03 trial and using the 1‐CTC positivity cutoff, and by that of Yang and colleagues considering 88 prospectively collected patients with advanced disease stratified by the 2‐CTC positivity cutoff. Whereas the prognostic role of baseline eCTC determinations demonstrated for various solid tumors seems to hold also in BTC patients, eCTC did not prove to be treatment‐predictive in the ABC‐03 study.17 With BTC patients in particular, due to the lack of effective treatments, we are in a context where gathering the full message contained in by CTCs is a priority. Thus, we need to overcome the limits of studies considering the only enumeration of eCTCs by implementing biomarker‐independent approaches that identify and isolate all CTC subpopulations. In fact, only coupling phenotypic heterogeneity of CTCs with their molecular analysis would permit to really evaluate their validity in the clinics. In keeping with these issues and with the conclusions of other papers,17 we aimed at isolating and characterizing the entire CTC population consisting of eCTC and in nonconventional CTCs (ncCTCs) that have lost epithelial features.18 In fact, treatment‐associated changes in CTCs and loss of epithelial features have been reported in the literature also in other tumors19, 20 thus highlighting the importance of isolating and characterizing the entire CTC population for a proper treatment response monitoring and for exploiting the informative content on treatment resistance/sensitivity mechanisms in the bulk CTC population.
Furthermore, in light of what is emerging about targetable genomic alterations in BTC,8, 9, 10, 11, 12, 21, 22 the possibility to relay on single tumor cells collected during the entire treatment trajectory would allow addressing hurdles dictated by tumor heterogeneity and clonal evolution under treatment selective pressure.
Under this perspective, we present here preliminary data (i) showing the possibility to detect CTCs in almost 100% of patients with advanced BTC (thus overcoming constraints in obtaining tissue biopsies), (ii) confirming the already known prognostic role of eCTC collected at baseline, (iii) supporting the role of ncCTCs in treatment monitoring and (iv) suggesting that single‐cell CTC analysis is not only possible but also informative.
Materials and Methods
Patient information
This was a prospective, monocentric, observational study consecutively recruiting patients with a confirmed diagnosis of metastatic/unresectable cholangiocarcinoma between January 2015 and March 2017. Due to the explorative nature of the study, no statistical hypothesis was postulated. The number of enrolled patients was however consistent with the entropy‐based approach to sample size in translational clinical trials as proposed by Piantadosi.23
Patients have been treated and followed up as per clinical practice, with frequent clinical evaluations and tumor assessment with chest/abdomen CT scans and/or MRI performed every 2–3 months. The treatment efficacy was assessed as per RECIST v1.1 criteria. Clinical information was collected from medical records and included demographic data, anatomic localization, tumor extension and treatment history. The patient's vital status was updated to the end of June 2018.
All CTC evaluations were carried out without the knowledge of the patient's clinical status.
Blood samples were longitudinally collected at times corresponding to baseline (BL), that is, before initiation of a new treatment line, during treatment (DT) close to clinical and imaging evaluations, at the end of treatment (EOT) and at subsequent follow‐up (FU) or new treatment lines. All subjects have signed a consensus form accepting participation in our study, which was approved by the local ethical board in November 2014 (INT 177/14) and subsequently reconfirmed on January 2018.
CTC enrichment and identification
Blood samples (10 ml) collected in K2EDTA tubes were subjected to CTC‐enrichment with Parsortix (Angle plc, Guildford, UK) within 1 hr from blood draw. Enriched cells were harvested according to the manufacturer's instructions and fixed for 20 min at room temperature (RT) with 2% PFA. Fixed samples were fluorescently stained with phycoerythrin (PE)‐labeled antibodies against epithelial markers EpCAM (clone HEA‐125, Miltenyi Biotec, Bergisch Gladbach, Germany, working dilution 1:11 for 10 min at 4°C), cytokeratins (pan‐cytokeratin clone C11, Abcam, San Francisco, CA, working dilution 1:10 for 10 min at RT) and EGFR (clone 423103, R&D Systems, Minneapolis, MN, working dilution 1:11 for 10 min at 4°C), and with allophycocyanin (APC)‐labeled antibodies recognizing leukocytes and monocytes: CD45 (clone 5B1, Miltenyi Biotec, working dilution 1:11 for 10 min at 4°C), CD14 (clone M5E2, BD Biosciences Pharmigen, San Diego, CA, working dilution 1:20 for 10 min at 4°C), CD16 (clone 3G8, BD Biosciences Pharmigen, working dilution 1:20 for 10 min at 4°C). Nuclei were stained with 1 μg/ml Hoechst 33342 (Sigma‐Aldrich, Saint Louis, MO) for 5 min at RT.
Labeled cells were analyzed using the DEPArray™ (Silicon Biosystems, Bologna, Italy), an automated platform to isolate and recover single cells, manually selected based on fluorescence labeling and morphology.24, 25 Selected single epithelial (PE+ve/APC−ve) or double‐negative (PE‐ve/APC‐ve) cells were recovered for downstream molecular analyses.
Molecular characterization of isolated CTCs
Recovered single cells and pools of white blood cells (WBC) were subjected to whole genome amplification employing the Ampli1™ WGA kit (Silicon Biosystems). Amplified DNA's quality was checked with the Ampli1™ QC kit (Silicon Biosystems) and a low‐pass whole‐genome sequencing (lpWGS) to detect copy number alterations (CNAs) was performed using the Ampli1™Low Pass kit (Silicon Biosystems) for barcoded libraries preparation, followed by sequencing with the IonTorrent Ion S5™ system (Thermo Fisher, Waltham, MA), using the Ion530 chip as for manufacturer's instructions.
Bioinformatics analyses
WGS sequences were aligned to the human reference genome (hg19) using tmap aligner tool on Torrent_Suite 5.4.0. CNAs were predicted by using Control‐FREEC 11.0 with the following settings: coefficientofVariation = 0.05, mateOrientation = 0, sex = XY or XX. Control FREEC produced different window size according to the sequencing depth in each sample. “Gain” and “loss” calls were filtered out when the Wilcoxon Rank‐Sum Test and Kolmogorov–Smirnov p values were greater than 0.05.
Comparison of single cells CNA profiles was performed considering copy number log‐ratio values evaluated for nonoverlapping equal regions size of 500 kb hierarchically clustered using Euclidean distance and Ward linkage method; as implemented in the R package. The most significantly and differentially altered regions between groups of responder and nonresponder CTCs were identified by GISTIC2.026 and subsequently validated using Fisher's exact test.
Large‐scale state transition scores27 were calculated using Genomic.Instability R package (https://github.com/SilvestriMR/Genomic.Instability).
Statistical analyses
The chi‐square or the Fisher exact tests were used to assess differences between groups as appropriate. Disease‐specific survival (DSS) was calculated from date of enrolment to the date of death or censored at the date of last follow‐up for living patients (median follow‐up = 20 months, range = 16–30 months). DSS was estimated using the Kaplan–Meier method and compared across the groups using the log‐rank test.
Results
Case series
The recruited patients (n = 24) were mostly middle‐aged and evenly distributed between those who presented at diagnosis with metastatic or with locoregional disease only. Similarly, the case series included a balanced number for intrahepatic and extrahepatic tumors and only two patients (8.3%) with gallbladder disease. Patients receiving previous treatments 6 or more months before recruitment were considered as untreated. Since 90% of patients were in the first‐line setting, the main treatment regimen was cisplatin + gemcitabine, as per guidelines. Two (10%) patients were enrolled at the beginning of the second line treatment. Only 11 patients have undergone previous surgery with curative intent. Patient characteristics are reported in Table 1 whereas Supporting Information Figure S1 shows the number of patients used in each specific type of below‐reported analyses.
Table 1.
Patient characteristics
| Number of cases | Percentage | |
|---|---|---|
| Gender | ||
| Female | 10 | 41.7 |
| Male | 14 | 58.3 |
| Age | ||
| ≤50 | 5 | 20.8 |
| 51–69 | 15 | 62.5 |
| ≥70 | 4 | 16.7 |
| Metastatic at diagnosis | ||
| Yes | 11 | 45.8 |
| No | 13 | 54.2 |
| Anatomical location | ||
| Intrahepatic | 13 | 54.2 |
| Extrahepatic | 9 | 37.5 |
| Gallbladder | 2 | 8.3 |
| Previous treatment | ||
| Yes | 6 | 25.0 |
| Cisplatin (CDDP) + Gemcitabine (GEM) | 3 | 50.0 |
| GEM | 2 | 33.3 |
| Radiotherapy + Capecitabine | 1 | 16.7 |
| No | 18 | 75.0 |
| Treatment regimen | ||
| First line | 22 | 92.2 |
| CDDP/GEM | 18 | 81.8 |
| FOLFOX | 04 | 18.2 |
| Second line | 2 | 8.3 |
| FOLFOX | 1 | 50.0 |
| FGFR inhibitor | 1 | 50.0 |
| Previous surgery | ||
| Yes | 11 | 45.8 |
| No | 13 | 54.2 |
Detection of epithelial and nonconventional CTCs
A CTC detection protocol developed in our laboratory to detect subpopulations of epithelial and nonepithelial CTCs was used.18 The protocol included an enrichment step based on size and deformability using the Parsortix rather than a marker‐based positive enrichment and was followed by a marker‐based positive selection for epithelial CTCs (eCTCs) and by a negative selection for nonepithelial CTCs. Selection and single‐cell recovery were performed using the DEPArray™.
In particular, during the DEPArray™ analysis, the combined expression of three epithelial (EpCAM, CK, EGFR) and three leukocytes (CD45, CD14, CD16) markers was evaluated, allowing the identification of both eCTCs (expressing any of the epithelial but none of the leukocyte markers) and of double‐negative cells (negative for all the evaluated markers).
The tumor origin of recovered double‐negative cells was then assessed by copy number alteration (CNA) analysis, which provides evidence for the malignant nature of each cell.28 Among double‐negative cells, those presenting an altered CNA profile were defined as bona fide nonconventional CTCs (ncCTCs), since confirmed as tumor cells by the presence of genomic alterations, although not fulfilling the conventional CTC identification criteria based on phenotype characteristics (Fig. 1). All recovered white blood cells presented as expected flat CNA profiles (Supporting Information Fig. S2).
Figure 1.
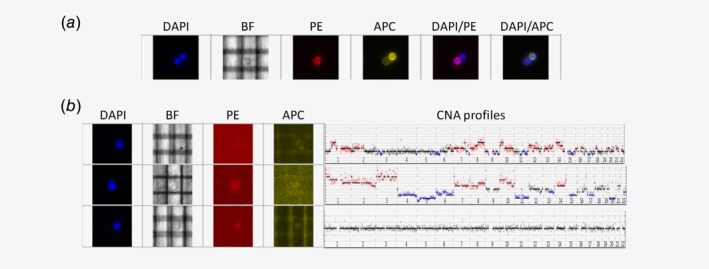
Image galleries showing morphologic and staining pattern of Parsortix‐enriched cells analyzed with the DEPArray platform. (a) Two nucleated (DAPI channel) cells, bottom/left and top/right: bottom/left cell is a classical eCTC, expressing epithelial markers (PE channel) and lacking leukocyte markers (APC channel); top/right cell is a leukocyte, showing the opposite staining pattern (PE−/APC+). (b) Three nucleated cells (DAPI+), negative for both epithelial and leukocyte markers (double‐negative, PE−/APC−). Although the three cells show a similar staining pattern and morphology (bright‐field channel, BF), the CNA profiles corresponding to each cell (reported on the right side) reveal the tumor origin only for the first two cells from the top, whereas the third cell has a normal diploid CNA profile. [Color figure can be viewed at http://wileyonlinelibrary.com]
Forty‐one blood samples collected at any time (before, during or after treatment) from the 21 patients of our case series were processed with the developed protocol. The numbers of eCTC and ncCTC for each of the analyzed blood samples are reported in Supporting Information Table S1.
Overall, we detected 18 eCTCs and 437 double negative cells. All the double‐negative cells were collected for CNA analysis: 70/437 cells could not be evaluated because of unsuccessful whole genome amplification (WGA), all the remaining 367 cells were analyzed by lpWGS and aberrant CNA profiles were identified for 73 cells, classifying them as bona fide ncCTCs (Supporting Information Table S1).
In order to test the impact of the newly identified subpopulation of ncCTC on the global CTC positivity rate, blood samples were categorized into four groups, according to their positivity for eCTC and ncCTCs (using 1CTC/10 ml as positivity threshold). The relative frequencies for the four CTC categories were (i) eCTCpos/ncCTCneg = 7%, (ii) eCTCpos/ncCTCpos = 12%, (iii) eCTCneg/ncCTCpos = 64% and (iv) eCTCneg/ncCTCneg = 17%.
Nonconventional CTCs were present in 31/41 samples (76%, median number = 2, range 1–7) either alone or in association with at least one eCTC. Conversely, eCTC alone was detectable only in 3/41 samples (7%). Thus, while considering eCTC only, the positivity rate was 19% (8/41 samples, median CTC number = 1.5, range 1–5), by adding also ncCTCs, an overall 4.3‐fold increase in CTC‐positivity was attained, leading to 83% of CTC+ve samples. The majority of CTC+ve samples (76%) would, therefore, have been missed with classic CTC‐detection methods.
eCTCs for prognosis
The prognostic role of baseline CTCs was investigated in 18 patients (three patients, for whom a baseline blood sample was not available, were excluded from the analysis).
We first assessed the association between baseline eCTC and disease‐specific survival (DSS). The median FU was 20 months and at least one eCTC was detectable in four patients (25%). All considered patients were in their first‐line treatment. No significant association between patients’ clinicopathological features and eCTC‐status was observed (Supporting Information Table S2).
Notwithstanding the small cohort of patients, Kaplan Meier's analyses supported a statistically significant (p = 0.03, log‐rank test) prognostic role of baseline eCTC status on DSS. The median survival time was 9 months for eCTC+ve patients and 19 months for eCTC−ve patients (Fig. 2).
Figure 2.
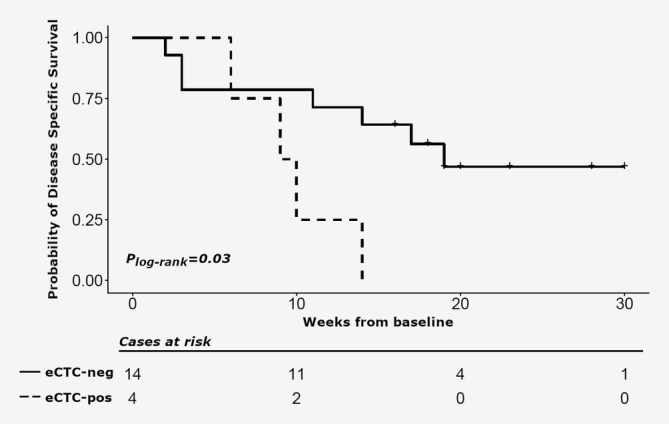
Kaplan–Meier estimates of disease‐specific survival (DSS) at baseline in cholangiocarcinoma patients. DSS estimates according to eCTC at baseline: ≥1CTC/10 ml of blood (dotted line) versus no CTC/10 ml of blood (solid line).
The same analysis performed on ncCTCs did not show an association with DSS (data not shown).
ncCTCs for treatment monitoring
Treatment can affect the presence of CTCs and their phenotype. To investigate this aspect, we compared e‐ and ncCTC‐positivity in the 38 blood samples drawn within a treatment line, in particular: at baseline in untreated patients (13 samples), at baseline in patients who already received previous treatment (6 samples) and during/at the end of treatment (19 samples).
We could detect CTCs (either eCTC or ncCTC) in all untreated patients at baseline (Fig. 3 a, first bar), with ncCTC being 2.75‐fold more frequent than eCTC. In baseline samples collected from patients who had received previous treatment (Fig. 3 a, second bar), we could never find any single eCTC alone, but only in association with ncCTCs (in 1/6 sample). Most samples (67%) contained only ncCTC and one sample only was negative for both CTC types.
Figure 3.
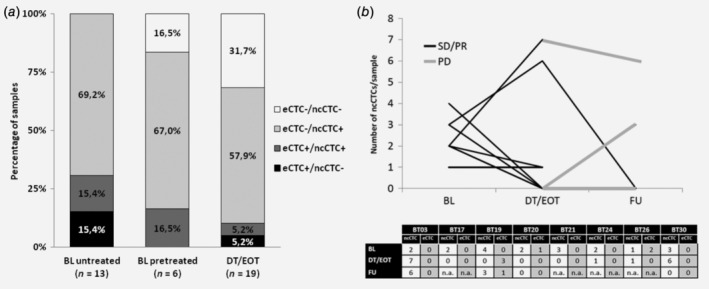
CTC subpopulations in cholangiocarcinoma patients as a function of treatment. (a) Bar chart reporting relative detection frequencies of eCTCs and ncCTCs in blood samples drawn at baseline (BL) from untreated patients (first bar), at BL from pretreated patients (second bar) and during/at the end of treatment (DT/EOT, third bar). (b) Line graph showing the changes of ncCTC numbers in samples longitudinally collected from eight patients undergoing treatment. ncCTC and eCTC counts for each blood sample are reported in the bottom table.
During and at the end of the treatment (DT/EOT), ncCTCs were 6.3‐fold more frequent than eCTCs, only 10% of samples were eCTC‐positive. Instead, 32% of the samples were CTC‐negative (Fig. 3 a, third bar).
The most relevant observed effect is, therefore, the increase of CTC‐negative samples collected DT/EOT versus baseline. Interestingly, all these CTC‐negative blood samples were drawn from patients that, at the time of CTC‐assessment, were classified as treatment‐responders, showing as the best response by RECIST criteria either a partial response (PR) or a stable disease (SD). These results suggest that, during treatment, CTC‐status and changes in the numbers of CTCs (in particular of ncCTCs, being the most frequently detected subpopulation) can reflect patients’ response/resistance to therapy.
To explore the possibility of monitoring treatment response with ncCTCs, we focused on eight patients for whom at least two blood samples, drawn at baseline and DT/EOT, were available (Fig. 3 b). All the patients were classified as responders (best response based on RECIST = PR or SD) at the time of the second CTC‐assessment (DT/EOT). For three of them, we also collected FU samples drawn 1–4 months after the end of therapy.
When comparing ncCTC counts at baseline versus DT/EOT, five patients (BT17, BT19, BT20, BT21, BT24) experienced a decrease in ncCTC numbers (with four becoming CTC‐negative) and one (BT26) remained unchanged with one ncCTC at both time points.
Two patients (BT03 and BT30) showed instead an increase in ncCTCs, passing from 2, 3 ncCTCs at baseline, to 7, 6 ncCTCs at the following assessment, respectively. However, patient BT30 became ncCTC‐ve 1 month after the end of treatment (FU blood draw), while patient BT03 experienced a rapid progression 1 month after the second CTC‐assessment. In fact, at the FU CTC‐assessment (1 month after progression) the patient still presented a high number of ncCTCs.
Patients BT19 and BT20 also had a disease progression after the end of treatment and were reevaluated for CTCs. While patient BT20 remained CTC‐negative also after disease progression, patient BT19 became ncCTC‐positive one month after PD (ncCTCs = 3). Interestingly, in this patient, three eCTC were detected during treatment.
Clinical and biological information conveyed by CNA profiles of single CTCs
Role of genomic instability of single CTCs
Genomic instability represents a hallmark of cancer, but is not easily assessable in tissue biopsies due to intrinsic heterogeneity and to contamination by normal cells. It is known to be associated with treatment resistance and clinical outcome, although sometimes in opposite directions29 and holds promise as a treatment predictive biomarker. Here we exploited the availability of WGS data on single‐CTCs collected during the treatment trajectory, to calculate the large‐scale state transition (LST) score, a surrogate score of genomic instability,27 computed as the number of chromosomal breaks between adjacent regions of 10 Mb. LST scores were then related to distinct clinical outcomes (PD vs. PR/SD).
Overall the median LST scores of single‐CTCs collected at baseline in patients developing PD (nonresponders) or achieving PR/SD (responders) as best response were slightly higher in the former group (12.5 vs. 8.5), although the difference was not statistically significant (p = 0.31). The complete list of LST scores for every single cell is reported in Supporting Information Table S3. Supporting Information Figure S3 reports LST values according to treatment response.
Identification of genomic regions involved in resistance mechanisms
To further investigate the informative content of CNA profiles obtained from single CTCs, a clustering analysis was performed including 88 cells (15 eCTCs and 73 ncCTCs) isolated from 23 patients (i.e., 3 more patients with respect to the 21 used for analyses of eCTC/ncCTC frequencies, for whom recovery of double‐negative cells has been subjected to a technical failure, hindering ncCTC classification; and removing one patient due to low WGS reads as reported in Supporting Information Figure S1). As shown in Figure 4 four distinct clusters (clu1–clu4) could be identified.
Figure 4.
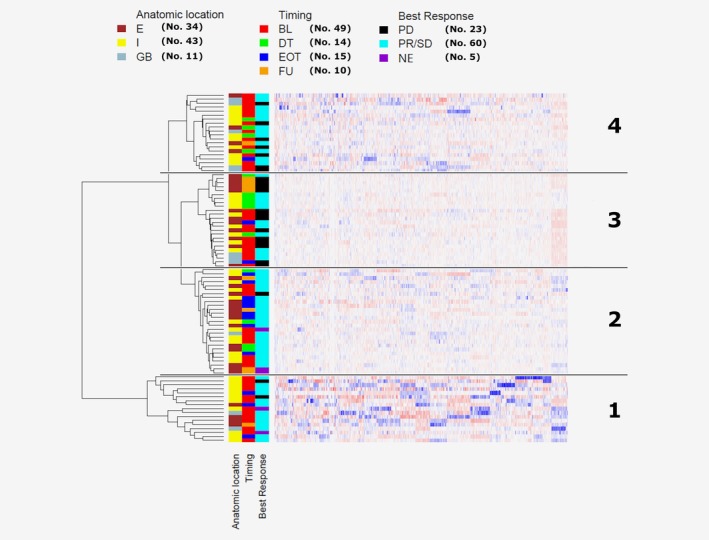
Clustering analysis of single‐CTCs’ CNA profiles. Hierarchical clustering analysis of 88 single CTCs (from 23 patients) using Euclidean distance and Ward linkage method. Reference bars on the top of the dendrogram indicate anatomic location (intrahepatic, I; entrahepatic, E; gallbladder, GB), blood draw timing (baseline, BL; during treatment, DT; end of treatment, EOT; follow‐up, FU) and treatment‐line best response (disease progression, PD; partial response/stable disease, PR/SD; not evaluable, NE). Numbers in brackets refer to the total number of CTCs within each category. The four main clusters are identified by numbers 1–4 (right side). [Color figure can be viewed at http://wileyonlinelibrary.com]
The clustering of genomic altered regions did not support a major role for patient individuality in the grouping of CTCs. The fact that not for all 23 patients similar numbers of CTCs were available affected this type of analysis, however when considering only the nine patients for whom at least four CTCs were available we observed that patient individuality seemed to drive the clustering in 5/9 cases.
The anatomic location of BTC defines different types of diseases with distinct molecular and clinical features.30 This seems to be only partially reflected in the altered chromosomal region clustering of CTCs (p = 0.090), likely due to the heterogeneity in blood drawing times (at baseline in either pretreated or not pretreated patients, during, at the end or after treatment). Nonetheless, when excluding pretreated patients from the analysis, the different blood drawing times were not a main driver of clustering (p = 0.216). The most striking separation in the four clusters occurred for CTCs derived from patients classified as responders (best response based on RECIST = PR or SD) versus nonresponders (best response = PD). Indeed 54.2 and 35% CTCs, respectively, falling into cluster 3 and 4, derived from patients whose best response to their treatment line was a progression (p = 0.00041).
Such a finding led us to hypothesize those specific genomic alterations associated with treatment resistance might be over‐represented in CTCs of cluster 3 and 4. This was tested by pairwise comparison of CNAs among the four clusters. Two chromosomal regions that are distinctly altered in cluster 2 versus 3 were identified (Figure 5), suggesting that when lacking tumor material, CTCs could be a surrogate for investigating CNAs and genes involved in treatment resistance.
Figure 5.
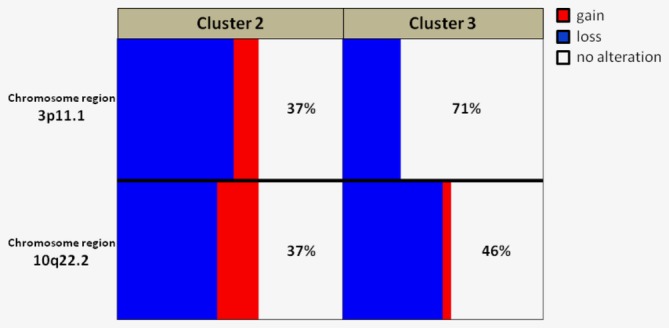
Comparison of alteration frequency in CTCs from cluster 2 and 3. Graph showing the frequency (%) of significantly different CNA in region 3p11.1 and 10q22.2 between CTCs included in cluster 2 (enriched in CTCs derived from patients responding to the treatment line) versus cluster 3 (enriched in CTCs derived from patients progressing during the treatment line). Clusters 2 and 3 refer to the heatmap reported in Figure 5. Gains, losses and no‐alteration are reported in red, blue and white, respectively. [Color figure can be viewed at http://wileyonlinelibrary.com]
Discussion
The diagnosis of BTC usually occurs at advanced stages, when surgical procedures are rarely feasible, biopsies are seldom performed and, as in the case of hilar BTC trans‐peritoneal biopsy, are even associated with disease dissemination.31
In such a scenario, a liquid biopsy approach is the only possible way to guide treatment in a precision medicine perspective. In particular, CTCs, albeit rare, may represent a unique source of biological material that can worthily substitute tissue biopsy.
In the past, the role of CTCs in patients with BTC has been addressed using the CellSearch™ which allows the identification and enumeration of classic eCTC only. In these patients, CTCs were reported to be associated with poor prognosis, but no studies are currently available on using CTCs to monitor treatment and to detect treatment‐associated molecular alterations. Here we provide proof‐of‐concept data to promote the use of the informative potential of CTCs on clinical outcome and resistance mechanisms, in the management of BTC patients.
To the best of our knowledge, this is the first study in BTC that does not use the CellSearch and also includes other CTC subpopulations besides the classic eCTCs. Nonetheless, it still confirms the prognostic role of eCTC on DSS, thus supporting the clinical validity of this newly developed approach. Moreover, when using this CTC detection protocol no single patient was classified as CTC negative at the time of diagnosis, therefore opening the way to actually exploit the information carried by these tumor cells to derive clinically and biologically useful data in the absence of tissue biopsies. However, we still acknowledge that the extremely low number of detected cells that might limit downstream analyses represents a possible weakness.
In patients with different tumor types, distinct CTC subpopulations, such as CTCs undergoing epithelial–mesenchymal transition (mCTCs), have been considered in addition to classic eCTCs.32 For instance, in women with metastatic breast cancer, mesenchymal traits in CTCs were associated with poor clinical outcomes especially during chemotherapy as reported by Yu et al.19 suggesting that the switching of CTC‐phenotype during treatment might play a role in the monitoring of treatment response. In a DEPArray study in women with stage‐IV breast cancer, Bulfoni et al. sorted single CTCs according to expression of either epithelial or mesenchymal markers or of both and attributed the worst clinical outcomes to those expressing mixed traits.33 At difference to our study, literature studies have not provided direct evidence for the malignant nature of such mesenchymal subpopulations, and were limited to phenotypic assessments, which may possibly be a reason for the failure in finding significant clinical associations even in some large and well‐planned studies.34
In our attempt toward the exploitation of CTCs in the clinical management of patients with BTC, a crucial step was testing the role of ncCTCs on treatment response monitoring. So far, only the study by Backen et al.17 had tried to exploit eCTC for treatment monitoring, but as commented by the authors themselves, the failure to observe a role of CTCs in treatment‐response monitoring suggested the need of using marker‐independent CTC platforms able to capture the entire CTC subpopulation heterogeneity. By considering the eight patients for who longitudinally collected samples were available, we report here that decreases/increases in the numbers of ncCTCs mirrored the treatment outcome even anticipating progression in two cases. Still acknowledging the limitations of our data with regard to the small size of the cohort, we feel that this represents a result highlighting the importance of a CTC subpopulation that was mostly neglected in previous studies. Indeed, with caution because of the preliminary nature of our data, we suggest here that treatment response may be associated with a decrease of ncCTCs during or at the end of the treatment, whereas ncCTC increase is associated with progression and can even anticipate it with respect to radiological assessments.
The analysis of single‐cell CNA profiles, not only provides a direct proof of CTC malignancy but also offers the opportunity to explore intra‐patient tumor heterogeneity in a more straightforward way than by bulk tissue analysis and to collect information on tumor evolution. We have exploited CNA data to investigate correlations between genomic instability and clinical outcome and to identify specific genome alterations associated with treatment resistance.
Genomic instability leading to copy number alterations is a hallmark of cancer35 and chromosomal instability measures such as the LST score can contribute to unravel the complex balance between the tumor evolution towards improved cellular fitness/treatment resistance or toward detrimental effects leading to cell death. Our data support the feasibility of performing chromosomal instability studies on single cells but are still too scanty to allow drawing any conclusions. In future studies with a larger number of patients, the LST score might represent an additional variable worth to be challenged. However, since LST is a rather rough score capturing the global instability of the genome, it could be probably refined by adding genome‐specific markers addressing genome regions/genes directly involved in treatment resistance/sensitivity.
Indeed, we report interesting data when exploiting CNA data to identify genomic regions involved in treatment response. Similarities among the CNA profiles in single CTCs derived from patients who were progressing during their treatment lines, allowed the identification of genome regions that might contain genes involved in drug‐resistance mechanisms, offering valuable hints to be challenged in future preclinical studies.
This latter finding gives further support to our hypothesis that CTC genomic analysis offers an opportunity for timely recognition of patients harboring deleterious alterations, but possibly also new treatment targets. Indeed a CNA signature in single CTCs able to distinguish patients likely to respond or not to chemotherapy treatments has already been published in small‐cell lung cancer36 confirming that CTCs’ molecular characterization holds promise.
Based on our results, proving that besides eCTC another CTC subpopulation (ncCTC) can be isolated in BTC and considering our preliminary data on its possible predictive role, we eventually envision the use of our liquid biopsy‐based approach to integrate the sometimes inadequate imaging evaluations in the therapeutic strategy design.
Conflict of interest
CR received two travel grants from Menarini Silicon Biosystems for attending the DEPArray User Meeting in Philadelphia, PA September 2018 and the 4th ACTC Meeting at Corfù, Greece October 2019.
Supporting information
Appendix S1: Supporting Information
Acknowledgements
We thank Mr F. Cascone, Dr A. Martinetti and Dr E. Sottotetti for helping in blood collection. We are in debt with Dr P. Miodini and Mrs R. Motta for skillful technical support in CTC enrichment, and with Dr S. Veneroni for the support of our study by the Institutional Blood Biobank. We thank the Integrated Biology Platform at the Fondazione IRCCS, Istituto Nazionale dei Tumori, Milano for performing WGS. The research leading to these results has received funding from AIRC under IG 2018—ID. 21694 project—P.I. Cappelletti Vera and Ministero della Salute RF‐2013‐02359692 P.I. Daidone Maria Grazia.
Contributor Information
Maria G. Daidone, Email: mariagrazia.daidone@istitutotumori.mi.it.
Vera Cappelletti, Email: vera.cappelletti@istitutotumori.mi.it.
Data availability
Data available on request from the authors.
References
- 1. Rizvi S, Khan SA, Hallemeier CL, et al. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat Rev Clin Oncol 2018;15(2):95–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet 2014;383(9935):2168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bridgewater J, Galle PR, Khan SA, et al. Guidelines for the diagnosis and management of intrahepatic cholangiocarcinoma. J Hepatol 2014;60(6):1268–89. [DOI] [PubMed] [Google Scholar]
- 4. Surveillance Research Program NCI . Cancer Stat Facts: Liver and Intrahepatic Bile Duct Cancer. Available from https://seer.cancer.gov/statfacts/html/livibd.html. Accessed January 12, 2018.
- 5. Jarnagin WR, Fong Y, Dematteo RP, et al. Staging, resectability, and outcome in 225 patients with hilar cholangiocarcinoma. Ann Surg 2001;234(4):507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valle J, Wasan H, Palmer DH, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362(14):1273–81. [DOI] [PubMed] [Google Scholar]
- 7. Lamarca A, Hubner RA, David Ryder W, et al. Second‐line chemotherapy in advanced biliary cancer: a systematic review. Ann Oncol 2014;25(12):2328–38. [DOI] [PubMed] [Google Scholar]
- 8. Simbolo M, Fassan M, Ruzzenente A, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget 2014;5(9):2839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakamura H, Arai Y, Totoki Y, et al. Genomic spectra of biliary tract cancer. Nat Genet 2015;47(9):1003–10. [DOI] [PubMed] [Google Scholar]
- 10. Abou‐Alfa GK, Andersen JB, Chapman W, et al. Advances in cholangiocarcinoma research: report from the third Cholangiocarcinoma Foundation Annual Conference. J Gastrointest Oncol 2016;7(6):819–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farshidfar F, Zheng S, Gingras MC, et al. Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH‐mutant molecular profiles. Cell Rep 2017;19(13):2878–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jusakul A, Cutcutache I, Yong CH, et al. Whole‐genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov 2017;7(10):1116–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cabel L, Proudhon C, Gortais H, et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol 2017;22(3):421–30. [DOI] [PubMed] [Google Scholar]
- 14. Al Ustwani O, Iancu D, Yacoub R, et al. Detection of circulating tumor cells in cancers of biliary origin. J Gastrointest Oncol 2012;3(2):97–104. 10.3978/j.issn.2078-6891.2011.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang JD, Campion MB, Liu MC, et al. Circulating tumor cells are associated with poor overall survival in patients with cholangiocarcinoma. Hepatology 2016;63(1):148–58. 10.1002/hep.27944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Valle JW, Wasan H, Lopes A, et al. Cediranib or placebo in combination with cisplatin and gemcitabine chemotherapy for patients with advanced biliary tract cancer (ABC‐03): a randomised phase 2 trial. Lancet Oncol 2015;16(8):967–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Backen AC, Lopes A, Wasan H, et al. Circulating biomarkers during treatment in patients with advanced biliary tract cancer receiving cediranib in the UK ABC‐03 trial. Br J Cancer 2018;119(1):27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reduzzi C, Motta R, Bertolini G, et al. Development of a protocol for single‐cell analysis of circulating tumor cells in patients with solid tumors. Adv Exp Med Biol 2017;994:83–103. [DOI] [PubMed] [Google Scholar]
- 19. Yu M, Bardia A, Wittner BS, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 2013;339(6119):580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gazzaniga P, Raimondi C, Gradilone A, et al. Circulating tumor cells, colon cancer and bevacizumab: the meaning of zero. Ann Oncol 2011;22(8):1929–30. [DOI] [PubMed] [Google Scholar]
- 21. Lowery MA, Ptashkin R, Jordan E, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: Potential targets for intervention. Clin Cancer Res 2018;24(17):4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wardell CP, Fujita M, Yamada T, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol 2018;68(5):959–69. [DOI] [PubMed] [Google Scholar]
- 23. Piantadosi S. Translational clinical trials: an entropy‐based approach to sample size. Clin Trials 2005;2(2):182–92. [DOI] [PubMed] [Google Scholar]
- 24. Fuchs AB, Romani A, Freida D, et al. Electronic sorting and recovery of single live cells from microlitre sized samples. Lab Chip 2006;6(1):121–6. [DOI] [PubMed] [Google Scholar]
- 25. Abonnenc M, Manaresi N, Borgatti M, et al. Programmable interactions of functionalized single bioparticles in a dielectrophoresis‐based microarray chip. Anal Chem 2013;85(17):8219–24. [DOI] [PubMed] [Google Scholar]
- 26. Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy‐number alteration in human cancers. Genome Biol 2011;12(4):R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greene SB, Dago AE, Leitz LJ, et al. Chromosomal Instability Estimation Based on Next Generation Sequencing and Single Cell Genome Wide Copy Number Variation Analysis. PLoSOne 2016;11(11):e0165089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Morrison G, Sero V, Xu Y, et al. Orthogonal identification of circulating tumor cells (CTCs) using single cell low pass whole‐genome sequencing (WGS) and copy‐number alteration (CNA) analysis [abstract]. In: Proceedings of the American Association for Cancer Research Annual Meeting 2017; 2017 Apr 1–5; Washington, DC. Philadelphia (PA): AACR. Cancer Res 2017;77:Abstract no. 1717. [Google Scholar]
- 29. Birkbak NJ, Eklund AC, Li Q, et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res 2011;71(10):3447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jain A, Kwong LN, Javle M. Genomic profiling of biliary tract cancers and implications for clinical practice. Curr Treat Options Oncol 2016;17(11):58. [DOI] [PubMed] [Google Scholar]
- 31. Heimbach JK, Sanchez W, Rosen CB, et al. Trans‐peritoneal fine needle aspiration biopsy of hilar cholangiocarcinoma is associated with disease dissemination. HPB (Oxford)HPB (Oxford) 2011;13(5):356–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Alix‐Panabières C, Mader S, Pantel K. Epithelial‐mesenchymal plasticity in circulating tumor cells. J Mol Med (Berl) 2017;95(2):133–42. [DOI] [PubMed] [Google Scholar]
- 33. Bulfoni M, Gerratana L, Del Ben F, et al. In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial‐to‐mesenchymal transition is associated with a poor prognosis. Breast Cancer Res 2016;18:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kasimir‐Bauer S, Hoffmann O, Wallwiener D, et al. Expression of stem cell and epithelial‐mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res 2012;14(1):R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 36. Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy‐number profiles in patients with chemosensitive and chemorefractory small‐cell lung cancer. Nat Med 2017;23(1):114–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Data Availability Statement
Data available on request from the authors.


