Abstract
Key points
Sleep restriction has previously been associated with the loss of muscle mass in both human and animal models.
The rate of myofibrillar protein synthesis (MyoPS) is a key variable in regulating skeletal muscle mass and can be increased by performing high‐intensity interval exercise (HIIE), although the effect of sleep restriction on MyoPS is unknown.
In the present study, we demonstrate that participants undergoing a sleep restriction protocol (five nights, with 4 h in bed each night) had lower rates of skeletal muscle MyoPS; however, rates of MyoPS were maintained at control levels by performing HIIE during this period.
Our data suggest that the lower rates of MyoPS in the sleep restriction group may contribute to the detrimental effects of sleep loss on muscle mass and that HIIE may be used as an intervention to counteract these effects.
Abstract
The present study aimed to investigate the effect of sleep restriction, with or without high‐intensity interval exercise (HIIE), on the potential mechanisms underpinning previously‐reported sleep‐loss‐induced reductions to muscle mass. Twenty‐four healthy, young men underwent a protocol consisting of two nights of controlled baseline sleep and a five‐night intervention period. Participants were allocated into one of three parallel groups, matched for age, , body mass index and habitual sleep duration; a normal sleep (NS) group [8 h time in bed (TIB) each night], a sleep restriction (SR) group (4 h TIB each night), and a sleep restriction and exercise group (SR+EX, 4 h TIB each night, with three sessions of HIIE). Deuterium oxide was ingested prior to commencing the study and muscle biopsies obtained pre‐ and post‐intervention were used to assess myofibrillar protein synthesis (MyoPS) and molecular markers of protein synthesis and degradation signalling pathways. MyoPS was lower in the SR group [fractional synthetic rate (% day–1), mean ± SD, 1.24 ± 0.21] compared to both the NS (1.53 ± 0.09) and SR+EX groups (1.61 ± 0.14) (P < 0.05). However, there were no changes in the purported regulators of protein synthesis (i.e. p‐AKTser473 and p‐mTORser2448) and degradation (i.e. Foxo1/3 mRNA and LC3 protein) in any group. These data suggest that MyoPS is acutely reduced by sleep restriction, although MyoPS can be maintained by performing HIIE. These findings may explain the sleep‐loss‐induced reductions in muscle mass previously reported and also highlight the potential therapeutic benefit of HIIE to maintain myofibrillar remodelling in this context.
Keywords: atrophy, high‐intensity interval exercise, protein synthesis, sleep loss
Key points
Sleep restriction has previously been associated with the loss of muscle mass in both human and animal models.
The rate of myofibrillar protein synthesis (MyoPS) is a key variable in regulating skeletal muscle mass and can be increased by performing high‐intensity interval exercise (HIIE), although the effect of sleep restriction on MyoPS is unknown.
In the present study, we demonstrate that participants undergoing a sleep restriction protocol (five nights, with 4 h in bed each night) had lower rates of skeletal muscle MyoPS; however, rates of MyoPS were maintained at control levels by performing HIIE during this period.
Our data suggest that the lower rates of MyoPS in the sleep restriction group may contribute to the detrimental effects of sleep loss on muscle mass and that HIIE may be used as an intervention to counteract these effects.
Introduction
Sleep plays a key role in many physiological and cognitive functions, and it is recommended that adults obtain between 7 and 9 h of sleep each night (Spiegel et al. 1999; Van Dongen et al. 2003; National Sleep Foundation, 2015). Sleep has also been causatively implicated in the maintenance of muscle mass (Nedeltcheva et al. 2010; Dattilo et al. 2012), with an increased likelihood of both sarcopenia and lower total skeletal muscle mass in those reporting insufficient (i.e. <7 h per night) or poor sleep quality (Chien et al. 2015; Buchmann et al. 2016; Hu et al. 2017). During periods of energy deficit, a disproportionately greater loss of muscle mass has been reported in humans following sleep restriction [5.5 h time in bed (TIB) each night for 14 days] compared to normal sleep (8.5 h TIB each night for 14 days) (Nedeltcheva et al. 2010). In animal models, muscle atrophy has also been observed following sleep deprivation (i.e. >24 h of extended wakefulness) (Dattilo et al. 2012; Monico‐Neto et al. 2015a ; Monico‐Neto et al. 2015b ; de Sa Souza et al. 2016); however, the potential mechanisms by which sleep restriction may lead to muscle atrophy have not been comprehensively studied in humans.
Changes in muscle mass are, in the short‐term (i.e. days to weeks), largely determined by the balance between the rates of muscle protein synthesis (MPS) and muscle protein breakdown (MPB) (Rennie, 1985; Gibson et al. 1987). Previous reports suggest that sleep deprivation may influence these processes. Indeed, protein catabolism in humans (determined by urea excretion) was increased following 72 h of sleep deprivation (Kant et al. 1984), although whether this was accompanied by changes in muscle protein turnover is unknown. Interventions known to induce loss of muscle mass (such as muscle disuse, limb immobilization and step reduction) suggest that reductions in MPS, and specifically myofibrillar protein synthesis (MyoPS), rather than increases in MPB, underlie these changes (Symons et al. 2009; Breen et al. 2013; Rudrappa et al. 2016). Accordingly, a reduction in MPS, and potentially MyoPS, may underpin reductions to muscle mass induced by sleep restriction. However, a potential catabolic influence of sleep restriction cannot be excluded given previous findings of increased autophagy and ubiquitination markers in rodent sleep deprivation studies (Monico‐Neto et al. 2015a ; de Sa Souza et al. 2016). Nonetheless, no study to date has investigated the effects of sleep restriction on MPS, in whole or fractionated human skeletal muscle, or the potential catabolic influence of sleep restriction on markers of MPB.
At the molecular level, protein synthesis is regulated via the activation of the AKT‐mTOR‐p70S6K signalling pathway, which subsequently leads to increases in strength and muscle mass over time (Bodine et al. 2001). However, little is known about how sleep restriction may influence these pathways. Following 96 h of sleep deprivation, either no change in p‐mTORser2448 and p‐p70S6Kthr389 (Monico‐Neto et al. 2015a ) or an increase in p‐mTORser2448 and p‐4EBP1ser65 (de Sa Souza et al. 2016) were observed in rodent skeletal muscle. Although MPS is considered the driving force underpinning changes in muscle mass, MPB may also contribute to these changes. Molecular markers of the ubiquitin proteasome system (UPS) (i.e. FOXO1/3, MuRF1 and MAFbx), as well as the autophagy signalling pathways (i.e. LC3 and p62/SQSTM1) that lead to protein degradation, are often measured as a proxy for protein degradation (Tipton et al. 2018). In rodents, 96 h of sleep deprivation elevated levels of ubiquitinated proteins, markers of protein degradation (e.g. p‐FOXO3Ser253) and autophagy pathways (e.g. LC3 and p62/SQSTM1) and also induced muscle atrophy (Monico‐Neto et al. 2015a ; de Sa Souza et al. 2016). However, such mechanisms in human models reflecting levels of sleep loss commonly experienced in society are yet to be investigated.
Although resistance exercise is regarded as the most potent stimulus to induce skeletal muscle protein synthesis, high‐intensity interval exercise (HIIE) is also capable of inducing muscle protein synthesis, and its associated signalling pathways (Miller et al. 2005; Di Donato et al. 2014; Bell et al. 2015). Given the increasing prevalence of inadequate sleep in modern society (Ford et al. 2015) and the important metabolic and structural roles of skeletal muscle, there are clear health implications to mitigating reductions in muscle mass resulting from inadequate sleep. Given the potency of the stimulus provided by HIIE to stimulate MyoPS (Bell et al. 2015) and its time‐efficient nature (Hood et al. 2011), as well as its other proven clinical benefits (such as improvements in insulin sensitivity and oxidative capacity) (Little et al. 2011; Saner et al. 2018), HIIE may be a potential therapeutic intervention to counteract the detrimental effects of sleep restriction on muscle quality.
The effects of sleep restriction and HIIE on MyoPS and its associated signalling pathways have not previously been investigated in humans. Consequently, the present study aimed to determine the potential mechanisms by which sleep restriction may contribute to previously reported reductions in muscle mass. Specifically, the study used a previously published model of sleep restriction (i.e. five nights, with 4 h TIB each night) (Reynolds et al. 2012) to investigate the effect of sleep restriction, with or without three sessions of HIIE, on the rate of MyoPS and associated molecular signalling pathways. We hypothesized that the rate of MyoPS would be lower in the sleep restriction group and this would be reflected by changes in the molecular signalling pathways associated with MPS and MPB, whereas performing three sessions of HIIE would attenuate this effect.
Methods
Ethical approval
All procedures involved conform to the standards set by the latest revision of the Declaration of Helsinki (except for registration in a database) and were approved by the Victoria University Human Research Ethics Committee (HRE15‐294).
Participants
Twenty‐four healthy, recreationally‐active men, aged between 18 and 40 years of age, volunteered to participate, having had all procedures and risks explained, and provided their written informed consent. Participants were screened to determine their eligibility, as well as to rule out any pre‐existing medical conditions that may have precluded their participation (e.g. cardiovascular, metabolic or musculoskeletal problems). Eligible participants included in the study were (i) not taking any medications before and during the study; (ii) not performing shift work (within the previous three months); (iii) had regular sleeping habits and no previously‐diagnosed sleep disorders; (iv) had not travelled overseas in the previous two months; and (v) had a body mass index between 19 and 30 kg m–2. Prior to commencing the study, 1 week of habitual sleep was monitored via wristwatch actigraphy and sleep diaries; participants who averaged <6 or >9 h sleep per night during the monitoring period were also excluded from the study.
Study setting
The study was conducted in a temperature‐controlled sleep laboratory with the capacity for three participants to complete the study at any one time, each within their own bedroom. Participants were monitored at all times throughout the study by a member of the research team to ensure adherence to the protocol.
Study overview
Following the initial screening procedures, eligible participants attended the exercise physiology laboratory for baseline assessments of anthropometric measurements (i.e. height and body mass), and aerobic fitness () determined from a graded exercise test (GXT). In a counterbalanced order, participants were then assigned to one of three experimental groups, matched for age, body mass index, habitual sleep duration, and : normal sleep (NS, n = 8), sleep restriction (SR, n = 8) or sleep restriction and exercise (SR+EX, n = 8). Baseline participant characteristics for the three groups are shown in Table 1. One week prior to beginning the study participants also provided a pre‐study, resting skeletal muscle biopsy, to determine baseline levels of deuterium oxide (D2O) enrichment.
Table 1.
Baseline characteristics of participants
| NS (n = 8) | SR (n = 8) | SR+EX (n = 8) | |
|---|---|---|---|
| Age (years) | 24 ± 4 | 25 ± 5 | 24 ± 4 |
| Height (cm) | 177 ± 8 | 179 ± 6 | 179 ± 7 |
| Mass (kg) | 78.7 ± 13.3 | 74.5 ± 11.7 | 80.2 ± 9.5 |
| BMI | 25.2 ± 3.6 | 23.3 ± 3.0 | 24.6 ± 2.5 |
| (mL kg−1 min−1) | 43.7 ± 9.7 | 47.2 ± 6.7 | 48.0 ± 5.0 |
| (W) | 319 ± 59 | 330 ± 44 | 362 ± 48 |
| Habitual sleep duration (min) | 457 ± 45 | 428 ± 44 | 437 ± 39 |
Values are the mean ± SD. There were no statistically significant differences between the three groups for any of the baseline characteristics. BMI, body mass index.
The experimental component of the study consisted of an eight‐night stay within the sleep laboratory. All groups completed two initial nights of baseline sleep (8 h TIB, 23.00 h to 07.00 h), followed by a five‐night intervention period, during which the NS group spent 8 h TIB (23.00 h to 07.00 h), whereas both SR and SR+EX spent 4 h TIB per night (03.00 h to 07.00 h). Between 23.00 h and 03.00 h, lighting was dimmed to below 15 lux to reduce the effect of lighting on circadian rhythms (Duffy & Wright, 2005). The SR+EX group also performed three exercise sessions during the intervention period on days 4, 5 and 6 at 10.00 h. Following the intervention period, all groups completed a final night of ad libitum recovery sleep. An overview of the study protocol is shown in Fig. 1.
Figure 1. Schematic representation of the study protocol.
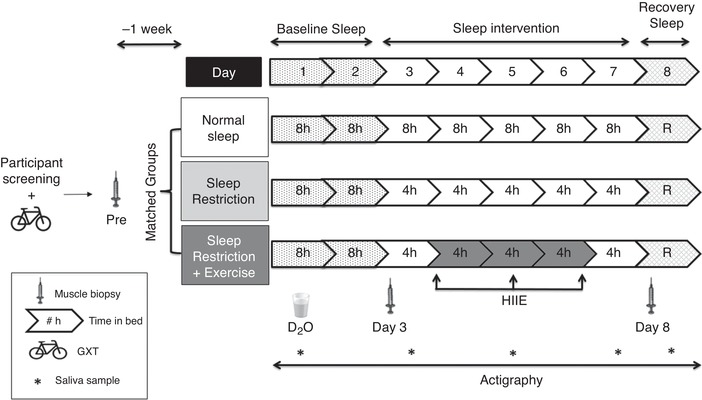
D2O – deuterium oxide ingestion; R, ad libitum recovery sleep; participant screening refers to medical questionnaires, exclusion criteria and habitual sleep, as well as physical activity monitoring.
To assess MyoPS and molecular markers of protein synthesis and degradation pathways, two additional resting skeletal muscle biopsies were sampled during the experimental testing sessions, at 10.00 h on both day 3 (following two nights of baseline sleep) and day 8 (following the final night of the sleep intervention). Throughout the study, participants were provided with a standardized diet consisting of fixed proportions (relative to body mass) of carbohydrates (4.5 g kg−1 day−1), protein (1.5 g kg−1 day−1) and fat (1 g kg−1.d−1). All meal times (six throughout the day) were kept constant throughout the study and participants were requested to eat all food provided. Dietary intake was replicated on the days prior to both experimental sessions. Caffeine intake was prohibited throughout the study. Participants were asked to match their habitual step counts by walking outside of the facility at designated periods throughout the day, accompanied by a member of the research staff. Waking hours were spent watching television, reading, working on a computer or talking to the staff.
Experimental procedures
Sleep monitoring
Sleep was assessed using wrist‐watch activity devices worn on the non‐dominant wrist (Actiwatch 2; Philips Respironics, Murrysville, PA, USA) with device activity sensitivity set to medium (40 activity counts per 30 s epoch) (Kosmadopoulos et al. 2014; Sargent et al. 2016), collected at 32 Hz and scored using a validated automated scoring algorithm (Oakley, 1997). These devices were used to objectively measure total sleep time (TST) obtained both habitually (prior to the study) and throughout the study. A member of the research team recorded the exact time that bedroom lights were switched on and off, allowing participants the designated sleep opportunity each night.
Physical activity monitoring
Throughout the study, and during 1 week prior (for habitual step‐count), daily step counts were monitored using commercially available and validated step‐counting applications on the participants’ personal mobile phone devices (i‐Health app; Apple Inc., Cupertino, CA, USA; and Samsung Health, Samsung Electronics Co., Ltd., Suwon, South Korea) (Hochsmann et al. 2018). Participants were encouraged to replicate their habitual daily step counts throughout the study protocol (Table 2). Participants were supervised at all times, and instructed to avoid any additional moderate or vigorous physical activity throughout the study.
Table 2.
Habitual and sleep study step counts for participants in each group
| Step count | NS | SR | SR+EX |
|---|---|---|---|
| Habitual | 12260 ± 3964 | 10965 ± 2136 | 11831 ± 919 |
| Study | 10652 ± 2476 | 10033 ± 1839 | 10953 ± 2316 |
Values are the mean ± SD. There were no significant differences between daily step counts, habitually and within the study protocol, in any of the groups.
GXT
A baseline assessment of aerobic fitness (i.e. peak oxygen uptake () and peak power (W) () was performed on an electronically‐braked cycle ergometer (Excalibur, V2.0; Lode, Groningen, The Netherlands). Following a standardized warm‐up (5 min at 30 W), participants cycled against an incremental ramp protocol where the resistance continually increased by 1 W every 2 s (30 W min−1) until volitional exhaustion (i.e. cadence fell below 60 rpm). Expired air was continuously analysed for O2 and CO2 concentrations via a gas analyser (Moxus 2010; AEI Technologies, Pittsburgh, PA, USA), which was calibrated immediately before each test.
HIIE
The HIIE protocol was adapted from a previous study (Little et al. 2010) and consisted of 10 × 60 s intervals performed on a cycle ergometer (RacerMate; Velotron, Seattle, WA, USA) at 90% of each participant's . Each interval was interspersed with 75 s of active recovery (at 60 W). Each session started with a 3 min warm up at 60 W. Heart rate (HR) (FT1; Polar, Kempele, Finland) and ratings of perceived exertion (RPE) (Borg, 1970) were recorded at the end of each interval. The mean power per interval was 318 ± 53 W, mean HR throughout the protocol was 156 ± 13 bpm, peak HR was 182 ± 12 bpm, and the average RPE per interval was 15 ± 2 AU.
Muscle biopsies
One week prior to commencing the study, and during the experimental sessions (day 3 and day 8), muscle biopsies were sampled from the vastus lateralis muscle of the same leg, using a suction‐modified Bergström needle, under local anaesthesia of the skin and fascia (1% lidocaine). The pre‐study biopsy was sampled at 10.00 h, following an overnight fast, and was used to determine background enrichment of D2O (prior to the ingestion of the D2O bolus) for the subsequent MyoPS analysis. The samples collected on days 3 and 8 were also obtained at 10.00 h, both following an overnight fast. All samples were immediately frozen in liquid nitrogen and stored at –80°C for subsequent analyses.
Assessment of MyoPS
D2O ingestion and saliva sample collection
The fractional synthetic rate (FSR) of MyoPS was assessed via the D2O tracer technique as previously reported (Wilkinson et al. 2014). At 18.00 h on day 1, each participant ingested 150 mL of D2O (70 atom %; Cambridge Isotope Laboratories, Tewksbury, MA, USA). Saliva samples were collected in salivettes prior to D2O ingestion and then on days 3, 5, 7 and 8 to determine body water enrichment. 2H enrichment of saliva was determined by a cavity ring‐down spectroscopy with the use of a liquid isotope analyser (Picarro L2130‐I analyser; Picarro, Santa Clara, CA, USA) with an automated injection system. Total body water 2H enrichment was used as a surrogate for plasma alanine 2H labelling, as previously described (Wilkinson et al. 2014). The mean body water enrichment [atomic percentage excess (APE), mean ± SD) throughout the intervention was 0.18 ± 0.03 APE on day 3, 0.15 ± 0.04 APE on day 5, 0.12 ± 0.03 APE on day 7 and 0.12 ± 0.03 APE on day 8.
Muscle preparation to determine myofibrillar fractional synthesis rate
Frozen muscle samples (40 to 60 mg) were homogenized in 500 µl of ice‐cold Tris homogenization buffer (25 mm Tris‐HCl, TritonX‐100 (0.5% final volume), protease inhibitor (catalogue no. 4693116001; Roche, Basel Switzerland) and phosphatase inhibitor (catalogue no. 4906845001; Roche). Metal beads were added to all samples and run at 20 Hz for 40 s in a tissue homogenizer (Tissue Lyser II; Qiagen, Valencia, CA, USA). The muscle homogenate was then centrifuged at 4500 rpm for 10 min at 4°C. The supernatant was removed and the pellet was used to prepare the myofibrillar fraction as previously described (Burd et al. 2010). Cation exchange chromatography was then performed on the myofibrillar samples using columns containing Dowex resin (Dowex 50wx8‐200 ion exchange resin; Sigma‐Aldrich, St Louis, MO, USA) to extract free amino acids from the myofibrillar fractions (Burd et al. 2010). The amino acid samples were then derivatized as their N‐acetyl‐n‐propyl‐esters, in accordance with previously reported protocols (Hector et al. 2018; McGlory et al. 2018).
FSR
The 2H/1H ratio of the myofibrillar samples were determined using gas chromatography pyrolysis isotope ratio mass spectrometry (Metabolic Solutions, Nashua, NH, USA), to assess the incorporation of deuterium into protein‐bound alanine. This was used to assess the FSR of myofibrillar proteins with the use of the enrichment of body water, corrected for the mean number of deuterium moieties incorporated per alanine (i.e. 3.7), as described previously (Wilkinson et al. 2014), as the surrogate precursor labelling between subsequent biopsies. Saliva enrichments were assessed using cavity ring‐down spectroscopy using methods, as described previously (Hector et al. 2018; McGlory et al. 2018).
The standard equation (Wilkinson et al. 2014) used to determine FSR was:
where FSR is the fractional synthetic rate, E t1 is APE day 8, E t0 is APE day 3, E p is average saliva APE, time is time between biopsies (days) and APE is the atomic percentage excess.
Preparation of whole‐muscle lysates for western blotting
Frozen muscle (10–20 mg) was homogenized as described previously (Granata et al. 2016) in an ice‐cold lysis buffer (dilution 1:20 w/v) containing 50 mm Tris, 150 mm NaCl, 1 mm EDTA, 1% IGEPAL, deionized water and a protease/phosphatase inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA), adjusted to pH 7.4. Protein concentration was determined in triplicate with a commercial colorimetric assay (Protein Assay kit‐II; Bio‐Rad, Gladesville, NSW, Australia), against bovine serum albumin standards (A9647; Sigma‐Aldrich).
Western blotting
Muscle homogenate was diluted in 4 × Laemmli buffer (0.25 m Tris, 4% SDS, 20% glycerol, 0.015% bromophenol blue and 10% 2‐mercaptoethanol) and equal amounts of total protein (15 or 20 µg) were loaded in different wells on Criterion™ 4–20% TGX Stain‐Free™ Precast Gels (Bio‐Rad). A stain‐free system (Bio‐Rad, Australia) was used as a loading control, with protein expression normalized to total protein loaded per lane. Each participant's samples were loaded into adjacent lanes on the same gel. Each gel also contained four to six internal standards of varying dilutions, made from a mixed homogenate of every sample in equal concentrations. These standards were used to form a calibration curve, with density plotted against protein content. Protein abundance was then calculated from the measured band intensity for each sample on the gel, using the linear regression equation from the calibration curve (Murphy & Lamb, 2013).
Gel electrophoresis was run for 20 min at 80 V and then for a further 60 to 90 min at 80 to 150 V. Transfer of proteins from the gel to 0.2 µm polyvinylidene fluoride membrane at 25 V for 10 min was performed via turbo transfer (Bio‐Rad). Membranes were then blocked in 5% non‐fat dry milk diluted in Tris‐buffered saline with 0.1% Tween‐20 (TBST) for 60 min. Membranes were washed in TBST and incubated overnight at 4°C with the appropriate primary antibody, prepared at a 1:1000 dilution in TBST with 5% bovine serum albumin and 0.02% sodium azide. The primary antibodies used were from Cell Signaling Technologies and include total AKT (CST9272), p‐AKTser473 (CST9271), total mTOR (CST2983), p‐mTORser2448 (CST5586), caspase‐3 (CST9662), total p70S6K (CST9202), total TSC2 (CST4308), p‐TSC2thr1462 (CST3617), LC3 (CST3868), p‐4EBP1thr37/46 (CST2855) and NFκB (CST8242). The measurement of p‐p70S6Kthr389 protein content could not be assessed because of issues with respect to obtaining reliable results with the available antibodies (i.e. CST9205 and CST9234). Following four TBST washes, the membranes were then incubated at room temperature with the appropriate host species‐specific secondary antibody for 60 min, before being exposed to a chemiluminescence solution (Clarity Western ECL; Bio‐Rad; or Supersignal West Femto Maximum Sensitivity Substrate; Thermo Fisher, Waltham, MA, USA). Images were taken with a ChemiDoc Imaging System fitted (Bio‐Rad). Densitometry was performed with Image Lab, version 5.0 (Bio‐Rad). Images are typically displayed with at least five bandwidths above and below the band of interest.
Quantitative RT‐PCR
RNA extraction
RNA was extracted from frozen muscle samples (10–20 mg) by adding 800 µL of TRIzol (15596‐026; Life Technologies, Grand Island, NY, USA) and a stainless steel metal bead to each sample and using a homogenizing instrument (Tissue Lyser II; Qiagen) (Kuang et al. 2018). Prior to storage, separate aliquots were taken for RNA quantification using a Nanodrop spectrophotometer (Thermo Fisher) and RNA integrity testing using an automated microcapillary electrophoresis system (Experion; Bio‐Rad Laboratories, Hercules, CA, USA). cDNA was synthesized from 1 µg of RNA using the iScript™ Reverse transcription supermix for RT‐PCR kit (Bio‐Rad Laboratories). DEPC water (180 µL) was added to all samples following cDNA synthesis and samples were stored at −20°C. As a result of small muscle sample collections for one of the participants, only samples from seven participants in the NS group were prepared for RT‐PCR. Relative mRNA expression was measured by qPCR (QuantStudio 7 Flex; Applied Biosystems, Foster City, CA, USA) using SsoAdvanced Universal SYBR Green Supermix (Bio‐Rad). Primers were designed using Primer‐BLAST (Ye et al. 2012) to include all splice variants, and were purchased from Sigma‐Aldrich. All reactions were performed in duplicate on 384‐well MicroAmp optical plates (4309849; Applied Biosystems) using an epMotion M5073 automated pipetting system (Eppendorf AG, Hamburg, Germany). The total reaction volume of 5 µL contained cDNA template, 2.5 µL of 2 × mastermix and 0.3 µm or 0.9 µm primers. All assays ran for 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C.
Primer design and data normalization
The expression of each target gene was normalized to the geometric mean of expression of the three most stably expressed reference genes, as described previously (Vandesompele et al. 2002; Kuang et al. 2018), and using the 2−ΔΔCt method (where Ct is the quantification cycle) (Schmittgen & Livak, 2008). A list of housekeeping and target gene primer sequences is provided in Table 3.
Table 3.
RT‐PCR primer sequences
| Primer name | Primer sequence | Product Size (bp) | Efficiency (%) | Accession number |
|---|---|---|---|---|
| Target genes | ||||
| Myostatin |
F – GGAGAAGATGGGCTGAATCCG R – GCATCGTGATTCTGTTGAGTGC |
111 | 98.9 | NM_005259 |
| Murf1 |
F – CCGTCGAGTGACCAAGGAGA R – CCAGGATGGCATACAACGTG |
80 | 98.6 | NM_032588 |
| Mafbx |
F – GCAGCTGAACAACATTCAGATCAC R – CAGCCTCTGCATGATGTTCAGT |
97 | 99.5 | NM_058229 |
| Foxo1 |
F – TGAGGGTTAGTGAGCAGGTTAC R – GGACTGCTTCTCTCAGTTCCT |
73 | 108.1 | NM_002015 |
| Foxo3 |
F –TTGGTTTGAACGTGGGGAAC R –TGTGTCAGTTTGAGGGTCTGC |
119 | 91.1 | NM_001455.4 |
| Mighty |
F – CCAACTCCGGAGCAAATTTTTCA R – TCCGAAGCACAAGCTTCACT |
106 | 94.7 | NM_024595 |
| p62/SQSTM1 |
F – AGAATCAGCTTCTGGTCCATCGG R – CTTTTCCCTCCGTGCTCCAC |
129 | 103.2 | http://NM_003900.5 |
| Housekeeping genes | ||||
| B2M |
F – TGCTGTCTCCATGTTTGATGTATCT R – TCTCTGCTCCCCACCTCTAAGT |
86 | 98 | NM_004048.2 |
| ACTB |
F – GAGCACAGAGCCTCGCCTTT R – TCATCATCCATGGTGAGCTGGC |
70 | 107 | NM_001101.3 |
| TBP |
F – CAGTGACCCAGCAGCATCACT R – AGGCCAAGCCCTGAGCGTAA |
205 | 99 | NM_003194.4 |
F, forward primer; R, reverse primer.
Statistical analysis
Statistical analyses were conducted using Prism, version 7.03 (GraphPad Software Inc., San Diego, CA, USA). Pre‐ to post‐intervention changes in gene expression and protein content were assessed for each group using a mixed ANOVA with one between‐subjects measure (group) and a within‐subjects measure (time). Significant effects of interaction (group × time), time (pre vs. post) and group (NS vs. SR vs. SR+EX) are reported where effects are seen. Where significant effects occurred, Bonferonni post hoc testing was performed to locate the differences. All statistical analyses of gene expression and protein content data were conducted using raw values. Gene expression data in‐text are reported as percent fold‐changes from pre‐intervention values (%, mean ± SD) with 95% confidence interval (CI) from fold‐change data (as a percentage), with the P value from the raw data reported. Data in the figures represent fold‐changes from pre‐intervention values, with individual responses. A one‐way ANOVA was used to assess differences between groups for the mean actigraphy sleep data and MyoPS data. All data in the text, figures and tables are presented as the mean ± SD, and 95% CI with P < 0.05 indicate statistical significance.
Results
Sleep data
There were no between‐group differences for baseline TST [mean ± SD TST (min) of nights 1 and 2, P value, NS; 448 ± 25 min, SR; 452 ± 17 min, and SR+EX; 459 ± 9 min, P = 0.50]. The NS group showed no difference in TST between the baseline and intervention periods [mean ± SD difference (min), 95% CI min, P value, 1 ± 28 min, 95% CI = –20 to 17 min, P > 0.99]. Both sleep‐restricted groups had a reduction in TST during their respective interventions compared to the baseline period (SR; –224 ± 20 min, 95% CI = −241 to −203 min, P < 0.001; SR+EX −223 ± 9 min, 95% CI = −242 to −204 min, P < 0.001). Compared to the NS group, TST during the intervention period was significantly reduced in both the SR group (−219 ± 8 min, 95% CI = −236 to −202 min, P < 0.001) and SR+EX group (−214 ± 8 min, 95% CI = −231.5 to −197 min, P < 0.001). There were no significant differences in TST between the SR and SR+EX groups during the intervention period (−5 ± 3 min, 95% CI = −23 to 12 min, P = 0.72).
MyoPS
There was a significant difference between groups for MyoPS throughout the intervention period (P < 0.001). Compared to the NS group, the myofibrillar protein FSR was significantly lower [mean ± SD between‐group difference FSR (% day–1), 95% CI, P value] in the SR group; –0.29 ± 0.08 FSR % day–1, 95% CI [−0.50, −0.09 FSR % day–1], P = 0.004) (Fig. 2). Furthermore, MyoPS was significantly lower in the SR group compared to the SR+EX group (−0.37 ± 0.09 FSR % day–1, 95% CI = −0.58 to −0.17 FSR % day–1, P < 0.001). There was no difference in MyoPS between the NS and SR+EX groups (0.08 ± 0.06 FSR % day–1, 95% CI = −0.29 to 0.12 FSR % day–1, P = 0.95).
Figure 2. FSR of myofibrillar proteins throughout the sleep intervention.
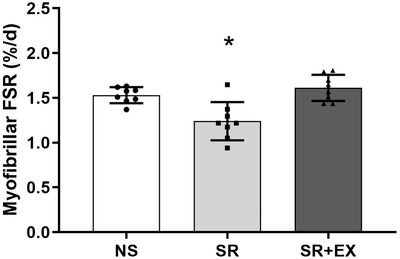
NS, SR and SR+EX groups. Bars are the mean ± SD. *Significant difference from both NS and SR+EX conditions; one‐way ANOVA (P < 0.05).
Protein synthesis and protein degradation related mRNA gene expression
There was no significant interaction effect for mRNA gene expression of Foxo1 (P = 0.69), Foxo3 (P = 0.74), Myostatin (P = 0.19), Mafbx (P = 0.24), Mighty (P = 0.81) or p62/SQSTM1 (P = 0.09) compared to pre‐intervention values. Despite there being a significant interaction effect for Murf1 mRNA gene expression (P = 0.026), post hoc analyses revealed no difference between groups from pre to post‐intervention (NS: 61 ± 81%, 95% CI = −114 to −9%, P = 0.25), (SR: −26 ± 33%, 95% CI = −23 to 75%, P = 0.27), (SR+EX: −29 ± 35%, 95% CI = −20 to 28%, P = 0.20) (Fig. 3).
Figure 3. Skeletal muscle mRNA expression of genes related to protein synthesis, degradation and autophagy signalling pathways, normalized to pre‐intervention values (i.e. values shown are fold change from pre‐intervention, calculated via the 2−ΔΔCt method). Foxo1 (A), Foxo3 (B), Myostatin (C), Murf1 (D), Mafbx (E), Mighty (F).
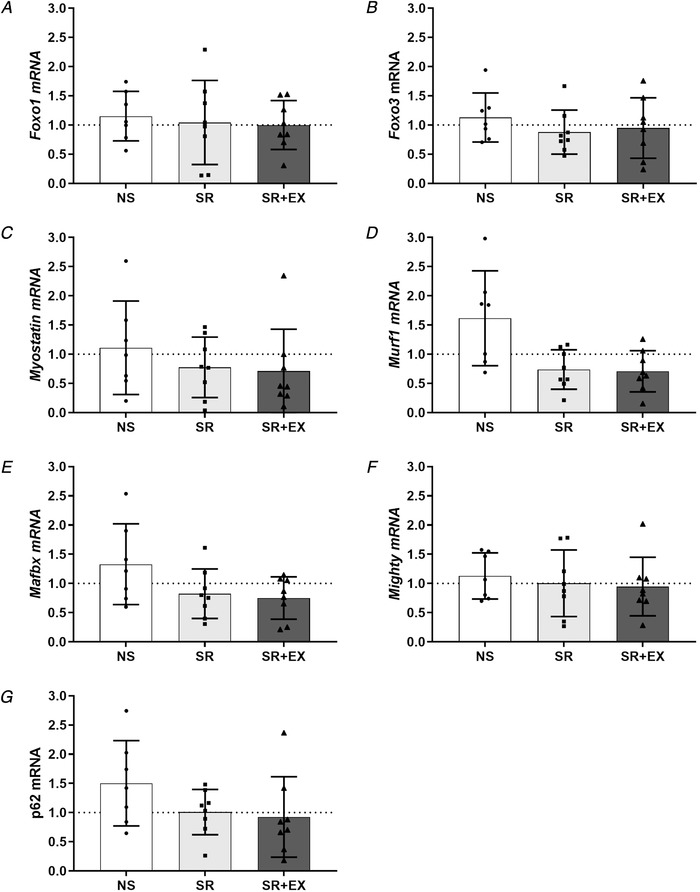
NS, SR and SR+EX groups. Bars are the mean ± SD; n = 7 in NS, n = 8 in both SR and SR+EX. Individual participant responses are indicated by individual symbols.
Protein synthesis and protein degradation related protein content
There were no changes in p‐AKTser473/AKT (total) (P = 0.15), p‐mTORser2448/mTOR (Total) (P = 0.15), p70S6K (P = 0.28), Caspase 3 (P = 0.19), p‐4EBP1thr37/46 (P = 0.67), p‐TSC2thr1462/TSC2 (Total) (P = 0.29), LC3BII:I ratio (P = 0.88) or NFκB (P = 0.72) protein content from pre‐ to post‐intervention (Fig. 4).
Figure 4. Skeletal muscle protein content of protein synthesis, autophagy and protein degradation pathways.
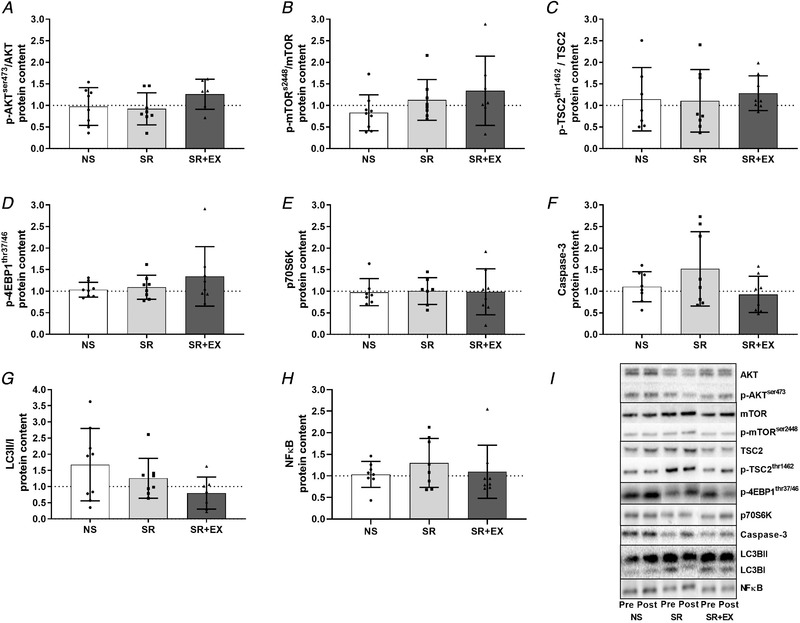
p‐AKTser473/AKT (total) (A), p‐mTORser2448/mTOR (total) (B), p‐TSC2thr462/TSC2 (total) (C), p‐4EBP1thr37/46 (D), p70S6K (total) (E), Caspase‐3 (F), LC3BII/LC3BI ratio (G), total NFκB (H) and representative western blots images (I). Data are presented as the fold‐change from pre‐intervention. NS, SR and SR+EX groups. Bars are the mean ± SD; n = 8 in each group. Individual participant responses are indicated by individual symbols.
Discussion
We observed a significantly lower rate of MyoPS following 5 nights of SR, whereas there was no difference in rates of MyoPS between the SR+HIIE and NS group. There were, however, no changes in molecular markers that are commonly used to indicate activation of the protein synthesis and degradation pathways. These results provide novel insights into mechanisms underlying the previously described decreases in muscle mass that have been attributed to inadequate sleep, and suggest HIIE as a potential therapeutic intervention to mitigate these changes.
Sleep restriction, exercise and MyoPS
Rates of protein synthesis are considered the primary driving mechanism for changes in muscle mass (atrophy and hypertrophy) (Rennie, 1985; Gibson et al. 1987; de Boer et al. 2007). Previous reports have shown reductions in muscle mass when obtaining insufficient sleep (Nedeltcheva et al. 2010; Dattilo et al. 2012; de Sa Souza et al. 2016); however, the present study is the first to demonstrate a lower rate of MyoPS in a SR group following 4 h TIB for five consecutive nights. This period of time was probably too short to detect any meaningful reductions in muscle mass or muscle fibre cross‐sectional area (CSA). However, persistent reductions in MyoPS are associated with the loss of muscle mass, as have been reported with a variety of muscle disuse conditions (i.e. step reduction, limb immobilization and bed rest) (de Boer et al. 2007; Breen et al. 2013). As such, the lower rate of MyoPS reported here aligns with observations of decreases in muscle fibre CSA, reported previously in rodents (following 96 h of sleep deprivation) (Dattilo et al. 2012; Monico‐Neto et al. 2015a ; de Sa Souza et al. 2016) and increased loss of muscle mass with sleep restriction in humans (Nedeltcheva et al. 2010). Nonetheless, further research is needed to determine the long‐term effects of sleep restriction on both MyoPS and the maintenance of muscle mass as there may be implications for those populations at risk of regularly experiencing inadequate or reduced sleep (e.g. elderly populations, shift‐workers) or those interested in the maintenance of rates of MyoPS (e.g. athletes) (Akerstedt & Wright, 2009; Chien et al. 2015; Fullagar et al. 2016). Previous interventions where MyoPS is reduced in response to step reduction have suggested the development of an ‘anabolic resistance’, such that the amino acid‐induced stimulation of MyoPS is blunted (Breen et al. 2013). Because the SR group had rates of MyoPS that were ∼19% lower than that of the NS or SR+HIIE groups, it may be proposed that sleep restriction can inhibit MyoPS. As such, further research should investigate the potential effect sleep restriction may have on the anabolic stimuli provided by interventions such as feeding and exercise.
Resistance exercise is well known to increase rates of MyoPS in skeletal muscle with subsequent increases in muscle mass suggesting this exercise modality warrants further investigation in the context of sleep loss (Konopka & Harber, 2014; Brook et al. 2015; Brook et al. 2016; Robinson et al. 2017). However, increases in MyoPS (which may also be associated with skeletal muscle remodelling) have also been observed with endurance exercise (and in particular, HIIE), although the findings appear to be somewhat protocol‐dependent (Di Donato et al. 2014; Konopka & Harber, 2014). The results of the present study demonstrate that the rate of MyoPS was higher in the SR+EX group compared to the SR group. The higher MyoPS observed in the SR+EX group compared to the SR group is probably explained by aerobic exercise induced increases in MyoPS (Miller et al. 2005; Di Donato et al. 2014; Bell et al. 2015). Indeed, Miller et al. (2005) reported increases in MyoPS following 1 h of single‐leg kicking (67% W max), which remained elevated at 48 and 72 h post‐exercise. An additional group that combined normal sleep (8 h TIB) with exercise (i.e. NS+EX) would have allowed us to determine whether sleep restriction may have attenuated additional exercise‐induced‐increases in MyoPS similar to those reported previously with HIIE interventions (Bell et al. 2015). However, the comparable rates of MyoPS in the SR+EX and NS groups support previous findings that implicate HIIE as a potent stimulus for MyoPS.
Molecular markers of protein turnover
In the present study, there were no detectable changes in mRNA expression or protein content of markers of either the protein synthesis or degradation signalling pathways (i.e. AKT‐mTOR‐p70S6K, UPS and autophagy pathways). Similar to the present study, no change in key regulators of the protein synthesis pathway (i.e. p‐mTORser2448 or p‐p70S6Kthr389) were reported following 96 h of sleep deprivation in rats, despite reductions in muscle fibre CSA (Monico‐Neto et al. 2015a ; de Sa Souza et al. 2016). By contrast to our findings, however, these same studies reported an increase in markers of the UPS and autophagy signalling pathways (i.e. p‐FOXO3, LC3 and p62/SQSTM protein content) (Monico‐Neto et al. 2015a ; de Sa Souza et al. 2016) indicating that sleep deprivation increases MPB in rats. Although some evidence supports corresponding changes in MyoPS and its associated signalling responses (Brook et al. 2015), dissociations similar to those reported in the present study are not uncommon (de Boer et al. 2007; Greenhaff et al. 2008; Atherton et al. 2010). Furthermore, the absence of changes to the MPS and MPB signalling pathways may be explained by the transient nature of their regulation and in relation to biopsy timing, which has previously been demonstrated in different contexts (de Boer et al. 2007; Atherton et al. 2010). Despite not detecting changes to markers of the MPS and MPB signalling pathways in the present study, the D2O method for assessing MyoPS allows for an integrated measure of MyoPS (i.e. it represents MyoPS in the fed, fasted, exercised, sleep and awaken states) throughout the intervention, and also encapsulates the influence of sleep restriction and exercise in a realistic, free‐living setting, over an extended duration.
In the context of exercise, several studies have reported increases in phosphorylation of AKTser473, mTORser2448 and decreases in eEF2thr56 protein, immediately following an endurance exercise session, with levels returning to baseline within 3–4.5 h (Mascher et al. 2007; Di Donato et al. 2014). Furthermore, although MPB increases following resistance exercise, it returns to resting values within 48 h (Phillips et al. 1997; Kumar et al. 2009). Given our post‐intervention biopsy occurred 48 h following the final exercise session, any changes in signalling markers may no longer have been detectable. Indeed, the present study showed that, even in the exercise group, there were no changes to MPS and MPB signalling. When taken together with previous literature, this probably means that changes in the molecular signalling pathways for MPS and MPB occur during the earlier stages of the sleep intervention or accumulated throughout the day as the homeostatic drive for sleep increased (rather than the hours immediately following sleep) and/or in the hours immediately following HIIE (de Boer et al. 2007; Greenhaff et al. 2008; Atherton et al. 2010). To fully determine the mechanisms that may underpin the lower rates of MyoPS observed in the SR group, and the comparable rates of MyoPS in the SR+EX and NS groups, further research is required with additional muscle sampling throughout the intervention to capture early signalling responses to exercise, feeding and changes as the duration of wakefulness is extended.
Given the capacity for HIIE to promote MyoPS (Di Donato et al. 2014; Bell et al. 2015) and the findings of the present study suggesting that HIIE may be capable of maintaining regular rates of MyoPS during periods of sleep restriction, collectively, these data may be of interest to populations for whom inadequate sleep is common, and the maintenance of muscle mass is of a particular concern (e.g. shift‐workers, athletic populations, military personnel) (Akerstedt & Wright, 2009; Fullagar et al. 2016; Mysliwiec et al. 2016). Furthermore, the prevalence of sarcopenia in older adults (>65 years of age) is increased in those who report sleeping <6 h per night (Chien et al. 2015; Buchmann et al. 2016; Hu et al. 2017). Although still speculative from the present acute data, the results of the present study suggest that regular HIIE may be used to preserve muscle mass and function in these populations and provides the foundation for future research in the area.
Limitations
Although previous research suggests that alterations to MyoPS underlie changes in muscle mass (Rennie, 1985; Gibson et al. 1987; de Boer et al. 2007), the present study did not employ direct measurements of changes in muscle mass. Previous research has observed dual‐energy X‐ray absorptiometry‐derived losses in fat‐free soft tissue mass across a 14‐day sleep restriction period, although the magnitude of this effect was probably enhanced by the calorie‐restricted diet employed (Nedeltcheva et al. 2010). Nevertheless, the current intervention duration was probably too short to detect changes in lean mass with dual‐energy X‐ray absorptiometry (given the variability of the measure) (Bone et al., 2017) and future investigations of similar durations should consider incorporating direct measures of muscle mass (i.e. with magnetic resonance imaging or CSA). Although our data report a significantly lower rate of MyoPS (∼19%) with sleep restriction, with the present study design, it is unclear how this compares to baseline rates of MyoPS. As such, future studies may incorporate a baseline measurement into the study as a comparison measurement. Furthermore, as eluded to previously, the addition of a normal sleep and exercise group may have provided further mechanistic insight into the effect of sleep restriction and HIIE on MyoPS. Finally, without further research that includes female and older participants, the generalizability of these findings for the wider population remains to be elucidated.
Conclusions
We discovered that, following five nights of sleep restriction (4 h TIB, each night), the rate of MyoPS was significantly lower (by 19%) compared to the NS and SR+EX groups. To our knowledge, this is the first observation of a sleep‐restriction‐induced change in MyoPS in humans. Importantly, we showed that participants in the SR+EX group had higher rates of MyoPS than the SR group and were similar to those in the NS group. However, there was a dissociation between the changes in MyoPS and the molecular signalling pathways that regulate protein synthesis and degradation. Although further investigations are needed, these findings suggest that sleep restriction can have detrimental effects on the processes that maintain muscle mass, via the potential suppression of MyoPS. Our findings may partly help to explain previous reports of reduced muscle mass in those experiencing insufficient sleep (Nedeltcheva et al. 2010). Our observation that those participants performing HIIE during the SR intervention were able to maintain their rates of MyoPS at levels comparable to the NS group could have important implications for a range of populations experiencing inadequate sleep and lends support to the idea that HIIE could be a viable intervention to mitigate certain detrimental effects associated with sleep loss.
Additional information
Competing interests
The authors declare that they have no competing interests.
Author contributions
NS, GR, SP, DB and JB were involved in the conception and design of the present study. NS, ML, NP, AG, TS, JK, SP, DB and JB were involved in the acquisition, analysis or interpretation of the data. NS, ML, NP, AG, GR, TS, JK, SP, DB and JB were involved in drafting the work and revising it critically for important intellectual content. All authors approved the final version of the manuscript submitted for publication. All authors agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This publication was supported in part by a Sports Medicine Australia (SMA) Research Foundation Grant and Australian Postgraduate Award PhD Scholarship to NS. SMP gratefully acknowledges support from the Canada Research Chairs program and the funding for this work provided by the National Science and Engineering Research Council (NSERC) of Canada. TS was supported by an NSERC of Canada graduate scholarship.
Supporting information
Statistical Summary Document
Acknowledgements
We thank all of the participants for their willingness to be involved with this study. We acknowledge the assistance of Dwight Marinkovic during data collection, as well as Conor Verbruggen and Dr Chris McGlory throughout the analysis process.
Biography
Nick J. Saner obtained his PhD in November 2019 from Victoria University, where he examined the relationship between sleep, exercise and skeletal muscle by specifically asking the question: can exercise mitigate the negative metabolic effects associated with sleep loss? He won a number of research awards, including Young Investigator Award for Exercise & Sport Science Australia (ESSA), 2018. He is currently Clinical Research Coordinator at the Baker Heart and Diabetes Institute in Melbourne, Australia. Long term, he hopes to continue this line of research; specifically, understanding the role exercise can play in maintaining skeletal muscle function in the context of sleep loss.

Edited by: Michael Hogan & Troy Hornberger
This is an Editor's Choice article from the 15 April 2020 issue.
Linked articles: This article is highlighted in a Journal Club article by Knowles. To read this article, visit https://doi.org/10.1113/JP279790.
References
- Akerstedt T & Wright KP, Jr (2009). Sleep loss and fatigue in shift work and shift work disorder. Sleep Med Clin 4, 257–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton PJ, Etheridge T, Watt PW, Wilkinson D, Selby A, Rankin D, Smith K & Rennie MJ (2010). Muscle full effect after oral protein: time‐dependent concordance and discordance between human muscle protein synthesis and mTORC1 signaling. Am J Clin Nutr 92, 1080–1088. [DOI] [PubMed] [Google Scholar]
- Bell KE, Seguin C, Parise G, Baker SK & Phillips SM (2015). Day‐to‐day changes in muscle protein synthesis in recovery from resistance, aerobic, and high‐intensity interval exercise in older men. J Gerontol A Biol Sci Med Sci 70, 1024–1029. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ & Yancopoulos GD (2001). Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol 3, 1014–1019. [DOI] [PubMed] [Google Scholar]
- Bone JL, Ross ML, Tomcik KA, Jeacocke NA, Hopkins WG & Burke LM (2017). Manipulation of muscle creatine and glycogen changes dual x‐ray absorptiometry estimates of body composition. Med Sci Sports Exerc 49, 1029–1035. [DOI] [PubMed] [Google Scholar]
- Borg G (1970). [Physical training. 3. Perceived exertion in physical work]. Lakartidningen 67, 4548–4557. [PubMed] [Google Scholar]
- Breen L, Stokes KA, Churchward‐Venne TA, Moore DR, Baker SK, Smith K, Atherton PJ & Phillips SM (2013). Two weeks of reduced activity decreases leg lean mass and induces “anabolic resistance” of myofibrillar protein synthesis in healthy elderly. J Clin Endocrinol Metab 98, 2604–2612. [DOI] [PubMed] [Google Scholar]
- Brook MS, Wilkinson DJ, Mitchell WK, Lund JN, Szewczyk NJ, Greenhaff PL, Smith K & Atherton PJ (2015). Skeletal muscle hypertrophy adaptations predominate in the early stages of resistance exercise training, matching deuterium oxide‐derived measures of muscle protein synthesis and mechanistic target of rapamycin complex 1 signaling. FASEB J. 29, 4485–4496 [DOI] [PubMed] [Google Scholar]
- Brook MS, Wilkinson DJ, Smith K & Atherton PJ (2016). The metabolic and temporal basis of muscle hypertrophy in response to resistance exercise. Eur J Sport Sci 16, 633–644. [DOI] [PubMed] [Google Scholar]
- Buchmann N, Spira D, Norman K, Demuth I, Eckardt R & Steinhagen‐Thiessen E (2016). Sleep, muscle mass and muscle function in older people. Dtsch Arztebl Int 113, 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd NA, West DW, Staples AW, Atherton PJ, Baker JM, Moore DR, Holwerda AM, Parise G, Rennie MJ, Baker SK & Phillips SM (2010). Low‐load high volume resistance exercise stimulates muscle protein synthesis more than high‐load low volume resistance exercise in young men. PLoS One 5, e12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien MY, Wang LY & Chen HC (2015). The relationship of sleep duration with obesity and sarcopenia in community‐dwelling older adults. Gerontology 61, 399–406. [DOI] [PubMed] [Google Scholar]
- Dattilo M, Antunes HK, Medeiros A, Monico‐Neto M, Souza Hde S, Lee KS, Tufik S & de Mello MT (2012). Paradoxical sleep deprivation induces muscle atrophy. Muscle Nerve 45, 431–433. [DOI] [PubMed] [Google Scholar]
- de Boer MD, Selby A, Atherton P, Smith K, Seynnes OR, Maganaris CN, Maffulli N, Movin T, Narici MV & Rennie MJ (2007). The temporal responses of protein synthesis, gene expression and cell signalling in human quadriceps muscle and patellar tendon to disuse. J Physiol 585, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sa Souza H, Antunes HK, Dattilo M, Lee KS, Monico‐Neto M, de Campos Giampa SQ, Phillips SM, Tufik S & de Mello MT (2016). Leucine supplementation is anti‐atrophic during paradoxical sleep deprivation in rats. Amino Acids 48, 949–957. [DOI] [PubMed] [Google Scholar]
- Di Donato DM, West DW, Churchward‐Venne TA, Breen L, Baker SK & Phillips SM (2014). Influence of aerobic exercise intensity on myofibrillar and mitochondrial protein synthesis in young men during early and late postexercise recovery. Am J Physiol Endocrinol Metab 306, E1025–E1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JF & Wright KP, Jr (2005). Entrainment of the human circadian system by light. J Biol Rhythms 20, 326–338. [DOI] [PubMed] [Google Scholar]
- Ford ES, Cunningham TJ & Croft JB (2015). Trends in self‐reported sleep duration among US adults from 1985 to 2012. Sleep 38, 829–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullagar HH, Skorski S, Duffield R, Julian R, Bartlett J & Meyer T (2016). Impaired sleep and recovery after night matches in elite football players. J Sports Sci 34, 1333–1339. [DOI] [PubMed] [Google Scholar]
- Gibson JN, Halliday D, Morrison WL, Stoward PJ, Hornsby GA, Watt PW, Murdoch G & Rennie MJ (1987). Decrease in human quadriceps muscle protein turnover consequent upon leg immobilization. Clin Sci (Lond) 72, 503–509. [DOI] [PubMed] [Google Scholar]
- Granata C, Oliveira RS, Little JP, Renner K & Bishop DJ (2016). Training intensity modulates changes in PGC‐1alpha and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J 30, 959–970. [DOI] [PubMed] [Google Scholar]
- Greenhaff PL, Karagounis LG, Peirce N, Simpson EJ, Hazell M, Layfield R, Wackerhage H, Smith K, Atherton P, Selby A & Rennie MJ (2008). Disassociation between the effects of amino acids and insulin on signaling, ubiquitin ligases, and protein turnover in human muscle. Am J Physiol Endocrinol Metab 295, E595–E604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector AJ, McGlory C, Damas F, Mazara N, Baker SK & Phillips SM (2018). Pronounced energy restriction with elevated protein intake results in no change in proteolysis and reductions in skeletal muscle protein synthesis that are mitigated by resistance exercise. FASEB J 32, 265–275. [DOI] [PubMed] [Google Scholar]
- Hochsmann C, Knaier R, Eymann J, Hintermann J, Infanger D & Schmidt‐Trucksass A (2018). Validity of activity trackers, smartphones, and phone applications to measure steps in various walking conditions. Scand J Med Sci Sports 28, 1818–1827. [DOI] [PubMed] [Google Scholar]
- Hood MS, Little JP, Tarnopolsky MA, Myslik F & Gibala MJ (2011). Low‐volume interval training improves muscle oxidative capacity in sedentary adults. Med Sci Sports Exerc 43, 1849–1856. [DOI] [PubMed] [Google Scholar]
- Hu X, Jiang J, Wang H, Zhang L, Dong B & Yang M (2017). Association between sleep duration and sarcopenia among community‐dwelling older adults: a cross‐sectional study. Medicine (Baltimore) 96, e6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant GJ, Genser SG, Thorne DR, Pfalser JL & Mougey EH (1984). Effects of 72 h sleep deprivation on urinary cortisol and indices of metabolism. Sleep 7, 142–146. [DOI] [PubMed] [Google Scholar]
- Konopka AR & Harber MP (2014). Skeletal muscle hypertrophy after aerobic exercise training. Exerc Sport Sci Rev 42, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmadopoulos A, Sargent C, Darwent D, Zhou X & Roach GD (2014). Alternatives to polysomnography (PSG): a validation of wrist actigraphy and a partial‐PSG system. Behav Res Methods 46, 1032–1041. [DOI] [PubMed] [Google Scholar]
- Kuang J, Yan X, Genders AJ, Granata C & Bishop DJ (2018). An overview of technical considerations when using quantitative real‐time PCR analysis of gene expression in human exercise research. PLoS One 13, e0196438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Atherton P, Smith K & Rennie MJ (2009). Human muscle protein synthesis and breakdown during and after exercise. J Appl Physiol (1985) 106, 2026–2039. [DOI] [PubMed] [Google Scholar]
- Little JP, Gillen JB, Percival ME, Safdar A, Tarnopolsky MA, Punthakee Z, Jung ME & Gibala MJ (2011). Low‐volume high‐intensity interval training reduces hyperglycemia and increases muscle mitochondrial capacity in patients with type 2 diabetes. J Appl Physiol (1985) 111, 1554–1560. [DOI] [PubMed] [Google Scholar]
- Little JP, Safdar A, Wilkin GP, Tarnopolsky MA & Gibala MJ (2010). A practical model of low‐volume high‐intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. J Physiol‐London 588, 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher H, Andersson H, Nilsson PA, Ekblom B & Blomstrand E (2007). Changes in signalling pathways regulating protein synthesis in human muscle in the recovery period after endurance exercise. Acta Physiol (Oxf) 191, 67–75. [DOI] [PubMed] [Google Scholar]
- McGlory C, von Allmen MT, Stokes T, Morton RW, Hector AJ, Lago BA, Raphenya AR, Smith BK, McArthur AG, Steinberg GR, Baker SK & Phillips SM (2018). Failed recovery of glycemic control and myofibrillar protein synthesis with 2 wk of physical inactivity in overweight, prediabetic older adults. J Gerontol A Biol Sci Med Sci 73, 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BF, Olesen JL, Hansen M, Dossing S, Crameri RM, Welling RJ, Langberg H, Flyvbjerg A, Kjaer M, Babraj JA, Smith K & Rennie MJ (2005). Coordinated collagen and muscle protein synthesis in human patella tendon and quadriceps muscle after exercise. J Physiol 567, 1021–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monico‐Neto M, Antunes HK, Lee KS, Phillips SM, Giampa SQ, Souza Hde S, Dattilo M, Medeiros A, de Moraes WM, Tufik S & de Mello MT (2015a). Resistance training minimizes catabolic effects induced by sleep deprivation in rats. Appl Physiol Nutr Metab 40, 1143–1150. [DOI] [PubMed] [Google Scholar]
- Monico‐Neto M, Giampa SQ, Lee KS, de Melo CM, Souza Hde S, Dattilo M, Minali PA, Santos Prado PH, Tufik S, de Mello MT & Antunes HK (2015b). Negative energy balance induced by paradoxical sleep deprivation causes multicompartmental changes in adipose tissue and skeletal muscle. Int J Endocrinol 2015, 908159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy RM & Lamb GD (2013). Important considerations for protein analyses using antibody based techniques: down‐sizing western blotting up‐sizes outcomes. J Physiol‐London 591, 5823–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysliwiec V, Walter RJ, Collen J & Wesensten N (2016). Military sleep management: an operational imperative. US Army Med Dep J, 128–134. [PubMed] [Google Scholar]
- National Sleep Foundation (2015). National Sleep Foundation Recommends New Sleep Times, ed. Duggan A. [Google Scholar]
- Nedeltcheva AV, Kilkus JM, Imperial J, Schoeller DA & Penev PD (2010). Insufficient sleep undermines dietary efforts to reduce adiposity. Ann Intern Med 153, 435–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley N (1997). Validation with polysomnography of the Sleepwatch sleep/wake scoring algorithm used by the Actiwatch activity monitoring system. Bend: Mini Mitter, Cambridge Neurotechnology. [Google Scholar]
- Phillips SM, Tipton KD, Aarsland A, Wolf SE & Wolfe RR (1997). Mixed muscle protein synthesis and breakdown after resistance exercise in humans. Am J Physiol Endocrinol Metab 273, E99–E107. [DOI] [PubMed] [Google Scholar]
- Rennie MJ (1985). Muscle protein turnover and the wasting due to injury and disease. Br Med Bull 41, 257–264. [DOI] [PubMed] [Google Scholar]
- Reynolds AC, Dorrian J, Liu PY, Van Dongen HP, Wittert GA, Harmer LJ & Banks S (2012). Impact of five nights of sleep restriction on glucose metabolism, leptin and testosterone in young adult men. PLoS One 7, e41218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MM, Dasari S, Konopka AR, Johnson ML, Manjunatha S, Esponda RR, Carter RE, Lanza IR & Nair KS (2017). Enhanced protein translation underlies improved metabolic and physical adaptations to different exercise training modes in young and old humans. Cell Metab 25, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa SS, Wilkinson DJ, Greenhaff PL, Smith K, Idris I & Atherton PJ (2016). Human skeletal muscle disuse atrophy: effects on muscle protein synthesis, breakdown, and insulin resistance – a qualitative review. Front Physiol 7, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saner NJ, Bishop DJ & Bartlett JD (2018). Is exercise a viable therapeutic intervention to mitigate mitochondrial dysfunction and insulin resistance induced by sleep loss? Sleep Med Rev 37, 60–68. [DOI] [PubMed] [Google Scholar]
- Sargent C, Lastella M, Halson SL & Roach GD (2016). The validity of activity monitors for measuring sleep in elite athletes. J Sci Med Sport 19, 848–853. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD & Livak KJ (2008). Analyzing real‐time PCR data by the comparative C‐T method. Nat Protoc 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Spiegel K, Leproult R & Van Cauter E (1999). Impact of sleep debt on metabolic and endocrine function. Lancet 354, 1435–1439. [DOI] [PubMed] [Google Scholar]
- Symons TB, Sheffield‐Moore M, Chinkes DL, Ferrando AA & Paddon‐Jones D (2009). Artificial gravity maintains skeletal muscle protein synthesis during 21 days of simulated microgravity. J Appl Physiol (1985) 107, 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton KD, Hamilton DL & Gallagher IJ (2018). Assessing the role of muscle protein breakdown in response to nutrition and exercise in humans. Sports Med 48, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dongen HP, Maislin G, Mullington JM & Dinges DF (2003). The cumulative cost of additional wakefulness: dose‐response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep 26, 117–126. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A & Speleman F (2002). Accurate normalization of real‐time quantitative RT‐PCR data by geometric averaging of multiple internal control genes. Genome Biol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DJ, Franchi MV, Brook MS, Narici MV, Williams JP, Mitchell WK, Szewczyk NJ, Greenhaff PL, Atherton PJ & Smith K (2014). A validation of the application of D(2)O stable isotope tracer techniques for monitoring day‐to‐day changes in muscle protein subfraction synthesis in humans. Am J Physiol Endocrinol Metab 306, E571–E579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S & Madden TL (2012). Primer‐BLAST: a tool to design target‐specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical Summary Document


