Although the respiratory tract is implicated as the primary portal of entry of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), gastrointestinal involvement is well-reported, associated with nausea, vomiting, diarrhea, and highly persistent viral particle shedding in feces.1 , 2
There is critical need to establish factors determining susceptibility to Coronavirus Disease 2019 (COVID-19) in patients with inflammatory bowel disease (IBD). Age, comorbidity, disease activity, and exposure to immuno-modulatory and biological therapies provide the basis for new guidelines for risk stratification and shielding.3
We hypothesize that expression levels of the SARS-CoV-2 spike protein receptor, angiotensin-converting enzyme 2 (ACE2),4 may also determine susceptibility to SARS-CoV-2-inflicted damage. Transmembrane serine protease 2 (TMPRSS2) primes the viral spike protein,5 allowing for the potent binding of ACE2. Both are known to be highly expressed in healthy ileal epithelium, with lower levels in epithelial cells in the colon. We report dysregulated mucosal ACE2 and TMPRSS2 expression in the colon and ileum in IBD, and identify the critical determinants of altered expression.
Methods
We compared RNA expression of ACE2 and TMPRSS2 in blood (paired-end sequencing), ileal, and colonic mucosal biopsies (microarray) from 138 patients with treatment-naïve IBD (cases) and 154 controls, predominantly with functional gastrointestinal disorders (Supplementary Table 1). They were recruited at 6 European centers, between 2012 and 2015, as part of the IBD-Character program (EU Character reference no. 305676). Demographics and further details are given in the supplementary information.
Results
ACE2 expression in the terminal ileum in controls was 25-fold higher than in the colon (P = 7.0 × 10–14; Supplementary Table 2), consistent with previous reports.
In IBD, expression in the terminal ileum was increased 10-fold compared with the colon (P = 7.9 × 10–14). In contrast, TMPRSS2 expression in the terminal ileum was lower than in the colon, both in controls (P = 3.6 × 10–16), and in IBD overall (P = 6.0 × 10–19).
Dysregulated Ileal Gene Expression
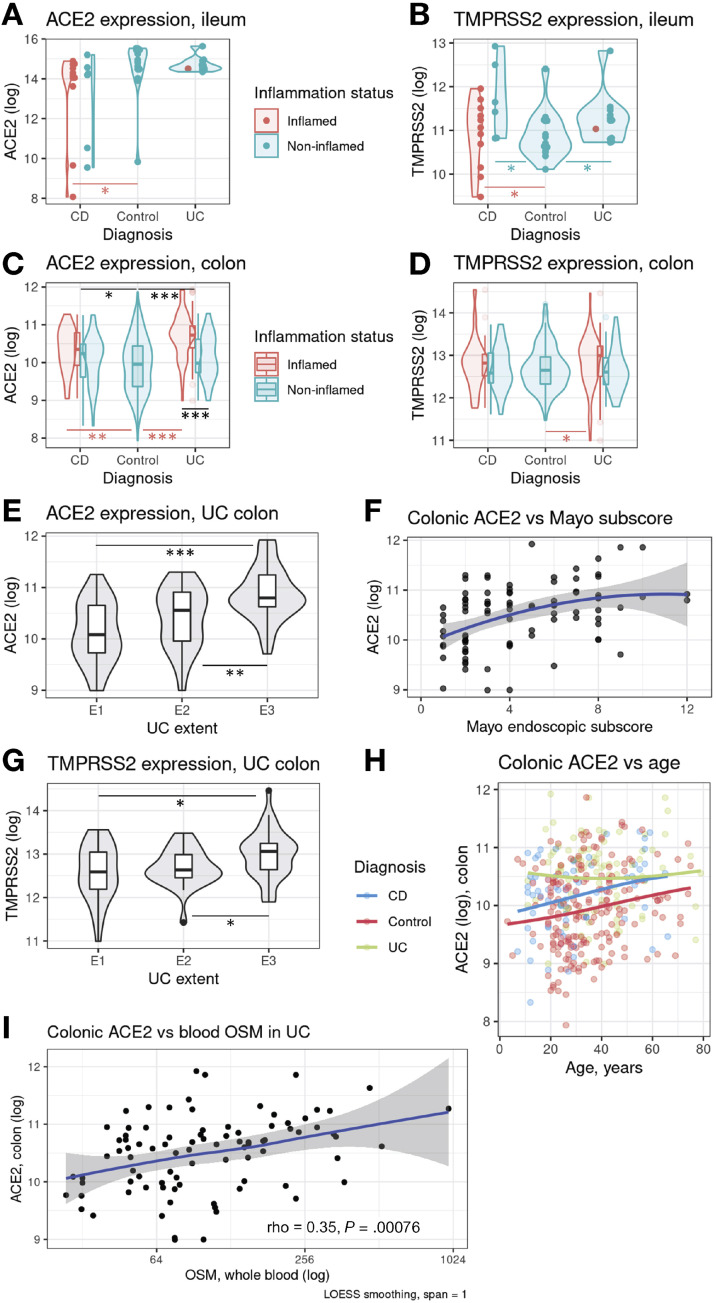
The expression of ACE2 in inflamed Crohn’s disease (CD) ileum was 60% lower (P = .0175) than in controls (Figure 1 A). Ileal TMPRSS2 was higher in CD noninflamed tissue than in controls (by 70%, P = .023, Figure 1 B). Ileal ACE2 did not differ between patients with ulcerative colitis (UC) and controls, but ileal TMPRSS2 was 30% higher (P = .023, Figure 1 B).
Figure 1.
ACE2 and TMPRSS2 expression in ulcerative colitis (UC) and Crohn's disease (CD); shown as log2(16+intensity). OSM – oncostatin M.
Dysregulated Colonic Expression
In CD, colonic ACE2 expression was increased by 30% relative to control (P = .006; Figure 1 C). TMPRSS2 expression in CD colon was similar to controls (Figure 1 D). In UC, the inflamed colonic mucosa expressed 70% more (P = 2.1 × 10–11) ACE2 transcript copies. UC mucosal ACE2 was 50% higher in inflamed vs noninflamed sites (P = 6.3 × 10–5).
Univariate Analyses of Factors Influencing Gene Expression
Colonic ACE2 levels associated with Montreal disease extent (Figure 1 E) and the Mayo endoscopic subscore (rho = 0.43, P = 3.2 × 10–5, Figure 1 F). Colonic TMPRSS2 was upregulated by inflammation (P = .0179, Figure 1 D), and extent (E1 vs E3: 150%, P = .0002, Figure 1 G), and was greater by 20% in men (P = .03).
Among patients with IBD, colonic ACE2 expression correlated weakly with high-sensitivity C-reactive protein (hsCRP) (rho = 0.23, P = .0043), age (rho = 0.19, P = .014, Figure 1 H) and serum albumin (rho = −0.17, P = .037). In controls, colonic ACE2 expression correlated with fecal calprotectin (n = 136, rho = 0.39, P = 2.7 × 10–6), hsCRP (n = 180, rho = 0.25, P = .00083), and age (rho = 0.20, P = .0066). In the control ileum tissue, ACE2 increased with age (rho = 0.64, P = .0099) and was 130% greater in men (P = .0256). The colonic expression of TMPRSS2, but not ACE2, was 20% higher in smokers from the control group (P = .0034). We found no important differences in ACE2 and TMPRSS2 expression with regard to recruitment centers.
We examined the relationship between mucosal ACE2 or TMPRSS2 and blood expression of TNF, OSM, IL10, TGFB1, GATA3, and STAT6 in IBD. Colonic (and also ileal) ACE2 correlated with blood OSM in patients with UC (rho = 0.35, P = .00076, Figure 1 I).
Dysregulation of the Renin-Angiotensin System
Mucosal angiotensinogen correlated with tissue inflammation (P = 4.7 × 10–11 in UC colon, P = 2.0 × 10–5 in CD colon) and disease severity (Mayo subscore rho = 0.58, P = 5.3 × 10–22; Froslie score rho = 0.39, P = 3.4 × 10–9). ACE expression in the blood associated negatively with IBD status (in UC P = 2.4 × 10–6, in CD P = 8.2 × 10–5) but ACE expression was greater in inflamed UC colon (P = .019). Renin was detectable in biopsies only, where it was reduced in colonic IBD compared with controls (UC P = 1.4 × 10–5, CD P = .0034).
Multivariable Analyses
Colonic expression
Multivariable analysis of ACE2 and TMPRSS2 expression were performed (Supplementary Methods). ACE2 gene expression in the colon was associated with increasing age in controls (β = 0.17, 95% confidence interval [CI] 0.02–0.32); with inflammation at biopsy site (β = 0.30, 95% CI 0.10–0.49) and E3 extent in UC (β = 0.40, 95% CI 0.18–0.62) and with endoscopic inflammatory subscore in CD (β = 0.23, 95% CI 0.00–0.46). Colonic TMPRSS2 expression was associated with hsCRP (β = 0.22, 95% CI 0.07–0.37) and smoking status in controls (β = 0.17, 95% CI 0.02–0.32), with Mayo subscore in UC (β = 0.23, 95% CI 0.01–0.44) and none of the parameters in IBD and CD.
Ileal expression
On multivariable analysis, ACE2 gene expression in the ileum was lower in the few patients with CD with B2 or B3 disease (β = −1.09, 95% CI −1.57 to −0.61). No other factors were implicated.
Discussion
We demonstrate that age, the presence of inflammation, and anatomic location are key determinants of expression of ACE2 and TMPRSS2 in patients presenting with IBD. These findings have potential implications for disease management, as well for mechanistic studies. Thus, the inflammation-related increase in ACE2 expression in the colon is consistent with recent mechanistic data highlighting the influence of cytokines on ACE2 expression in the respiratory epithelium.6 These data raise the possibility that active IBD may enhance viral particle production and uptake in the colon; and, furthermore, that infection and consequent inflammatory activation may exacerbate colitis.
The apparent reduction in ACE2 in the ileum in active CD also bears further investigation: this alteration may relate to the loss of epithelial surface in active ulceration, reduced ACE2 production by maturating epithelium, and consequent sampling effects; or may indeed be directly relevant to CD pathogenesis.
In summary, we identify age, smoking, and active disease as potential additional risk factors of vulnerability to COVID-19 in patients with IBD, through alterations of receptor expression. Our findings lend support to registry initiatives such as SECURE-IBD, which are necessary to monitor the possible impact of COVID-19 on IBD, and to the ongoing translational research program characterizing sites for therapeutic intervention in the molecular pathways of SARS-CoV-2 recognition.
Acknowledgments
IBD-Character Consortium: Alex T. Adams, MBChB, PhD, Erik Andersson, Ian D. Arnott, MD, Monica Bayes, PhD, Daniel Bergemalm, MD, PhD, Ferdinando Bonfiglio, PhD, Ray K. Boyapati, MD, Adam Carstens, MD, Christina Casén, MSc, Ewa Ciemniejewska, MSc, Mauro D'Amato, PhD, Fredrik A. Dahl, PhD, Trond Espen Detlie, MD, Hazel E Drummond, BSc, Gunn S. Ekeland, MSc, Daniel Ekman, MSc, Anna B. Frengen, PhD, Fernando Gomollón, MD, PhD, Mats Gullberg, PhD, Ivo G. Gut, PhD, Marta Gut, PhD, Simon C. Heath, PhD, Fredrik Hjelm, PhD, Henrik Hjortswang, MD, PhD, Gwo-Tzer Ho, PhD, Jørgen Jahnsen, MD, PhD, Daisy Jonkers, PhD, Åsa V. Keita, PhD, Nicholas A. Kennedy, MBBS, PhD, FRACP, Charles W. Lees, PhD, Torbjørn Lindahl, MSc, Mårten Lindqvist, PhD, Angelika Merkel, PhD, Eddie Modig, Bsc, Aina E.F. Moen, PhD, Hilde Nilsen, PhD, Elaine R. Nimmo, PhD, Colin L. Noble, MD, Niklas Nordberg, PhD, Kate R. O’Leary, MSc, Anette Ocklind, PhD, Christine Olbjørn, MD, Erik Pettersson, PhD, Marieke Pierik, MD, PhD, Dominique Poncelet, PhD, Dirk Repsilber, PhD, Céline Sabatel, PhD, Renaud Schoemans, PhD, Alan G. Shand, MD, Johan D. Söderholm, MD, PhD, Janne Sølvernes, MS, Mikael Sundell, BSc, Tone M. Tannæs, PhD, Leif Törkvist, MD, PhD, Morten H. Vatn, MD, PhD, Simen Vatn, MD, Anne-Clémence Veillard, PhD, Nicholas T. Ventham, MRCS(Eng) PhD, MBBS, David C. Wilson, MD, MRCPCH, Panpan You, MS.
CRediT Authorship Contributions
Jan Krzysztof Nowak, MD, PhD (Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Resources: Supporting; Visualization: Lead; Writing – original draft: Lead). Jonas Christoffer Lindstrøm, Msc, PhD (Data curation: Lead; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Equal; Writing – original draft: Equal). Rahul Kalla, MD, PhD (Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Writing – original draft: Equal; Writing – review & editing: Equal). Petr Ricanek, MD, PhD (Data curation: Supporting; Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Project administration: Equal; Resources: Equal; Writing – review & editing: Supporting). Jonas Halfvarson, MD, PhD (Formal analysis: Equal; Funding acquisition: Equal; Investigation: Equal; Project administration: Equal; Resources: Equal; Writing – review & editing: Equal). Jack Satsangi, MD, PhD (Conceptualization: Lead; Formal analysis: Equal; Funding acquisition: Lead; Investigation: Equal; Methodology: Equal; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Lead).
Footnotes
Conflict of interest These authors disclose the following: Jan Krzysztof Nowak reports personal fees from Norsa Pharma and nonfinancial support from Nutricia outside the submitted work. Rahul Kalla has served as a speaker for Ferring and has received support for research from IBD-Character (EU FP7 2858546). Jonas Halfvarson has received personal fees as speaker, consultant, and/or advisory board member for AbbVie, Aqilion AB, Celgene, Celltrion, Dr. Falk Pharma and the Falk Foundation, Ferring, Hospira, Janssen, MEDA, Medivir, MSD, Olink Proteomics, Pfizer, Prometheus Laboratories, Sandoz/Novartis, Shire, Takeda, Thermo Fisher Scientific, Tillotts Pharma, Vifor Pharma, and UCB and received grant support from Janssen, MSD, and Takeda, outside the submitted work. Jack Satsangi has served as a speaker, a consultant, and an advisory board member for MSD, Ferring, AbbVie, and Shire, consultant with Takeda, received speaking fees from MSD, travel support from Shire, and has received research funding from AbbVie, Wellcome, CSO, MRC, and the EC grant IBD-BIOM. The remaining authors disclose no conflicts.
Funding EU FP7 grant IBD-Character (2858546).
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2020.05.030.
Supplementary Methods
This inception cohort from IBD Character was recruited in the years 2012 to 2015 at 6 European centers: Oslo, Örebro, Edinburgh, Linköping, Zaragoza, and Maastricht. The inflammation in the endoscopically inspected segments was assessed using the Mayo endoscopic subscore in UC, the Froslie score in CD, qualitatively at the biopsy site (inflamed or noninflamed), and systemically (hsCRP). The disease was characterized with the Montreal classification. Self-declared current smoking status was noted. Study participants provided informed written consent; approval from local institutional review boards was obtained (including South Eastern Norway Bioethical Committee, reference number S-04209).
Mucosal biopsies were collected at index colonoscopy. They were collected to vials with a stabilizing solution (Allprotect; Qiagen, Hilden, Germany). Following homogenization (RLT Pluss Buffer, 5 mm steal bead at 4 m/s for 30 seconds, FastPrep-24 Classic Instrument from MP Biomedicals, Santa Ana, CA) and RNA isolation (QIAsymphony RNA Kit according to the protocol RNA CT 400; Qiagen), its purity and quality were assessed (Agilent 2100 Bioanalyser, Agilent 2100 Expert software, Agilent RNA 6000 Nano Kit; Agilent, Santa Clara, CA). Cutoff RNA Integrity Number and RNA concentration were 7.2 and 67 ng/μL, respectively. A total of 100 ng of messenger RNA (mRNA) was amplified, labeled with Cy3 cyanine, and hybridized according to the Agilent One-Color Microarray-Based Gene Expression Analysis workflow (version 6.9). Agilent SurePrint G3 human Gene Expression 8x60k v2 Microarrays were randomly filled with 600 ng of fragmented complementary RNA to compensate for diagnosis, sex, age, smoking status, and center. Hybridization occurred at 65°C and took 17 hours; after washing, the microarrays were scanned using the Agilent Microarray Scanner and the data were processed with Agilent Feature Extraction Software. The R limma package was used for array normalization, background correction (offset of 16), and quantile normalization.
Paired whole blood mRNA sequencing was performed. The whole blood RNA was stabilized in the PAXgene RNA tubes, isolated, and then quantified. Sequencing was conducted with the Ion AmpliSeq Human Gene Expression Core Panel. Ion Library TaqmanTM Quantitation was used for quality check. Torrent Software v. 4.6 with the ampliSeqRNA plugin v. 4.6.0.1 was used for alignment to hg19. The counts table was normalized with DESeq2.
Patients with the diagnoses of possible IBD or unspecified IBD were excluded. The data were analyzed using the R language (3.6.0; R Foundation for Statistical Computing, Vienna, Austria) and Statistica 13.3 (TIBCO Software, Palo Alto, CA), which was used for nonparametric statistical testing. Due to the lack of normality (Levene’s P < .05) in some of the subgroups, the Mann-Whitney U test was applied for unpaired comparisons and the Spearman’s correlation coefficient was calculated.
The forward stepwise regression was used for exploratory analysis of factor contribution to gene expression variability; the Q–Q plots were visually inspected to check for residual normality.
Post hoc power calculations (G∗Power, University of Dusseldorf, Germany) reveal that given the parameters for ACE2 expression in IBD and symptomatic non-IBD controls (Supplementary Table 2) and the assumed α level of 0.05 (2-tailed), the achieved power was 0.9998; in the case of ACE2 in inflamed UC colonic tissue (n = 27) vs controls (n = 191) the power was 0.8036. On the other hand, the comparison of inflamed IBD ileal mucosa with the control must be considered underpowered (β = 0.6513).
Negligible ACE2 expression was detected in whole blood (data not shown). The following genes were included in the additional analyses of the renin-angiotensin system: ACE, in the blood; AGT and REN in the mucosa.
Supplementary Table 1.
Group Characteristics
| CD | UC | Controls | |
|---|---|---|---|
| n | 64 | 74 | 154 |
| Age, y | 30.2 ± 15.9 | 38.6 ± 15.5 | 33.6 ± 14.4 |
| Sex, % female, % (n) | 46.9 (30) | 40.5 (30) | 55.8 (86) |
| Smoker, % (n) | 19.0 (12) | 5.4 (4) | 19.7 (30) |
| CRP, mg/L | 22.7±36.9 | 33.6±58.7 | 4.82±12.5 |
| Characteristics, % (n) | L1 25.0 (16) L2 31.2 (20) L3 43.7 (28) L4 21.8 (14) A1 25.0 (16) A2 50.0 (32) A3 25.0 (16) B1 82.8 (53) B2 9.4 (6) B3 4.7 (3) P 7.8 (5) |
E1 25.7 (19) E2 29.7 (22) E3 44.6 (33) |
|
| Endoscopic assessment | Mayo subscore 4.74 ± 2.90 | Froslie score 6.73 ± 4.78 |
Supplementary Table 2.
Expression of ACE2 and TMPRSS2 in the Intestinal Mucosa of Patients With CD, UC, and Controls
| IBD | CD | UC | Controls | ||
|---|---|---|---|---|---|
| Colon | All | 163 | 74 | 89 | 191 |
| ACE2 | 10.35 ± 0.66∗∗∗ | 10.17 ± 0.63∗ | 10.50 ± 0.64∗∗∗ | 9.94 ± 0.72 | |
| TMPRSS2 | 12.75 ± 0.56 | 12.72 ± 0.54 | 12.77 ± 0.58 | 12.69 ± 0.51 | |
| Noninflamed | 78 | 47 | 31 | ||
| ACE2 | 10.10 ± 0.62 | 10.07 ± 0.64 | 10.12 ± 0.58 | ||
| TMPRSS2 | 12.67 ± 0.50 | 12.67 ± 0.51 | 12.64 ± 0.50 | ||
| Inflamed | 85 | 27 | 58 | ||
| ACE2 | 10.58 ± 0.61∗∗∗,††† | 10.32 ± 0.58∗∗ | 10.69 ± 0.59∗∗∗,††† | ||
| TMPRSS2 | 12.82 ± 0.60∗,† | 12.80 ± 0.59 | 12.83 ± 0.61∗ | ||
| T. ileum | All | 29 | 17 | 12 | 15 |
| ACE2 | 13.84 ± 1.86∗ | 13.24 ± 2.24 | 14.68 ± 0.35 | 14.61 ± 1.43 | |
| TMPRSS2 | 11.21 ± 0.75∗ | 11.18 ± 0.87 | 11.25 ± 0.57 | 10.89 ± 0.55 | |
| Noninflamed | 17 | 6 | 11 | ||
| ACE2 | 14.12 ± 1.59 | 13.05 ± 2.38 | 14.70 ± 0.36 | ||
| TMPRSS2 | 11.42 ± 0.70∗∗ | 11.69 ± 0.87∗ | 11.27 ± 0.59∗ | ||
| Inflamed | 12 | 11 | 1 | ||
| ACE2 | 13.44 ± 2.19∗ | 13.34 ± 2.27∗ | 14.51 | ||
| TMPRSS2 | 10.91 ± 0.74 | 10.91 ± 0.78 | 11.03 |
NOTE. CD and UC data are compared with controls with ∗ denoting P < .05, ∗∗ < .01, ∗∗∗ < .001. Inflamed and noninflamed tissue is compared within subgroups defined by group and location with † denoting P < .05, †† < .01, ††† < .001. Values significantly different from control are printed in bold. Expression is represented as log2(16+intensity). The Mann-Whitney U test was used.
References
- 1.Xiao F., et al. Gastroenterology. 2020;158:1831–1833.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin X., et al. Gut. 2020;69:1002–1009. doi: 10.1136/gutjnl-2020-320926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danese S., et al. Nat Rev Gastroenterol Hepatol. 2020;17:253–255. doi: 10.1038/s41575-020-0294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wrapp D., et al. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann M., et al. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ziegler C., et al. https://papers.ssrn.com/abstract=3555145 Available at: