Abstract
The mechanism by which CD4/CD8 lineage choice is coordinated with TCR specificity during positive selection remains an unresolved problem in immunology. The stochastic/selection model proposes that CD4/CD8 lineage choice in TCR-signaled CD4+CD8+ thymocytes occurs randomly and therefore is highly error-prone. This perspective is strongly supported by “coreceptor rescue” experiments in which transgenic CD4 coreceptors were ectopically expressed on thymocytes throughout their development and caused significant numbers of cells bearing MHC-II-specific TCR to differentiate into mature, CD8 lineage T cells. However, it is not known if forced coreceptor expression actually rescued positively selected thymocytes making an incorrect lineage choice or if it influenced developing thymocytes into making an incorrect lineage choice. We have now reassessed coreceptor rescue and the concept that lineage choice is highly error-prone with a novel CD4 transgene (referred to as E8I-CD4) that targets expression of transgenic CD4 coreceptors specifically to thymocytes that have already undergone positive selection and adopted a CD8 lineage fate. Unlike previous CD4 transgenes, the E8I-CD4 transgene has no effect on early thymocyte development and cannot itself influence CD4/CD8 lineage choice. We report that the E8I-CD4 transgene did in fact induce expression of functional CD4 coreceptor proteins on newly arising CD8 lineage thymocytes precisely at the point in thymic development that transgenic CD4 coreceptors would putatively rescue MHC-II-specific thymocytes that incorrectly adopted the CD8 lineage. However, the E8I-CD4 transgene did not reveal any MHC-II-selected thymocytes that adopted the CD8 lineage fate. These results demonstrate that CD4/CD8 lineage choice is neither error-prone nor stochastic.
How cell fate specification occurs in bipotential progenitor cells is an important problem in metazoan development. A mammalian example of such cell fate specification is the differentiation of bipotential CD4+CD8+ double-positive (DP)3 thymocytes into either CD4 or CD8 lineage T cells. In the thymus, DP thymocytes are signaled by their TCR to undergo positive selection and to differentiate into either CD4+CD8− or CD4− CD8+ single-positive (SP) T cells, which constitute the major T cell arm of the adaptive immune system. Importantly, coreceptor expression in SP T cells must be matched to TCR specificity and cellular function, such that CD4+ T cells possess helper function and express TCR specific for peptides presented by MHC-II molecules, while CD8+ T cells possess cytotoxic function and express TCR specific for peptides presented by MHC-I molecules (1–3). Understanding how TCR specificity, coreceptor phenotype, and cellular function are appropriately matched during T cell development in the thymus remains an unresolved issue.
The stochastic/selection model of CD4/CD8 lineage choice postulates that CD4/CD8 lineage choice by signaled DP thymocytes is entirely random, regardless of TCR specificity, and that signaled DP thymocytes differentiate into SP cells that then require additional TCR/coreceptor “rescue” signals to differentiate into long-lived T cells (4–9). Strong support for the stochastic/selection model has come from so-called “coreceptor rescue” experiments in which forced or persistent expression of one of the coreceptor proteins resulted in the appearance of mature T cells bearing TCR with inappropriate MHC recognition specificities for their T cell lineage (7, 8, 10, 11). In the case of CD4, forced expression of transgenic CD4 coreceptor proteins resulted in the appearance of mature CD8 lineage T cells bearing mismatched MHC-II-restricted TCR (7, 10). Furthermore, genetic deletion of the CD4 silencer element (12, 13) prevented down-regulation of CD4 expression on CD8 lineage T cells and caused the generation of mature CD8 lineage T cells bearing mismatched MHC-II-restricted TCR. Such experiments were interpreted as fulfilling the central premise of the stochastic/selection model that CD8 SP thymocytes arise during MHC-II-specific positive selection but that such cells normally fail to differentiate into long-lived CD8 lineage T cells because they cannot receive rescue signals from their MHC-II-specific TCR in the absence of CD4 coreceptors (12).
Because the CD4/CD8 lineage decision is central to our understanding of T cell development in the thymus, it was important to determine whether SP thymocytes that have randomly made an incorrect lineage choice in fact exist and whether they are in fact rescued by forced expression of the matching coreceptor protein. Previous reports of coreceptor rescue utilized experimental models in which forced expression of the rescuing coreceptor was initiated early and continuously throughout thymocyte development, so that the rescuing coreceptor might well have altered T cell selection and lineage choice in some way other than by rescuing putatively short-lived SP thymocytes. Consequently, the present study has addressed the issue of coreceptor rescue in a novel experimental model consisting of a CD4 coreceptor transgene (referred to as E8I-CD4) that targets CD4 coreceptors only to thymocytes that have already undergone positive selection and have committed to a CD8 lineage fate, precisely the point in thymocyte development that CD4 coreceptors would rescue MHC-II-specific CD8 lineage thymocytes. The present study demonstrates that the E8I-CD4 transgene does prevent the loss of CD4 coreceptor expression on CD8-committed thymocytes, but nevertheless fails to rescue or reveal significant numbers of MHC-II specific thymocytes that had made an incorrect CD8 lineage choice. Thus, the present study directly contradicts the concept of coreceptor rescue and directly contradicts the central premise of stochastic/selection.
Materials and Methods
Mice
C57BL/6 (B6), CD40, β2m0 (β2-microglobulin0), and OT-II TCR transgenic mice (14) were obtained from The Jackson Laboratory. 8DP4 transgenic mice bred to CD40β2m0 mice were generated and screened as previously described (15). All mice were maintained under pathogen-free conditions in accordance with National Institutes of Health guidelines.
Generation of E8I-CD4 transgenic mice
The E8I-CD4 transgenic vector consisted of the murine 7.6-kb E8I cis-Cd8 enhancer and Cd8α promoter (16) upstream of a murine CD4 cDNA. The construct was subcloned into a modified pIRES-2EGFP (Invitrogen) plasmid with the IRES-EGFP sequence replaced with a poly(A) fragment. To generate a transgenic mouse, the transgenic vector plasmid was digested with AfeI and NotI, and a 13-kb fragment comprised of the E8I-CD4 transgene (see Fig. 1B) was injected into C57BL/6 (B6) pronuclei. Founder offsprings were obtained and screened by Southern blot for transgene insertion and by flow cytometry of peripheral blood for CD4 expression on CD8+ T cells. E8I-CD4 transgenic mice were identified by the presence of a 1.3-kb product in PCR of tail DNA using the following primers (forward: 5′-GTCGAGCTCAAGCTTCGAATT-3′ and reverse: 5′-CAGTAGAC ACTGCCACAGCCC-3′) in a 32-cycle PCR reaction with an annealing temperature of 65°C.
FIGURE 1.
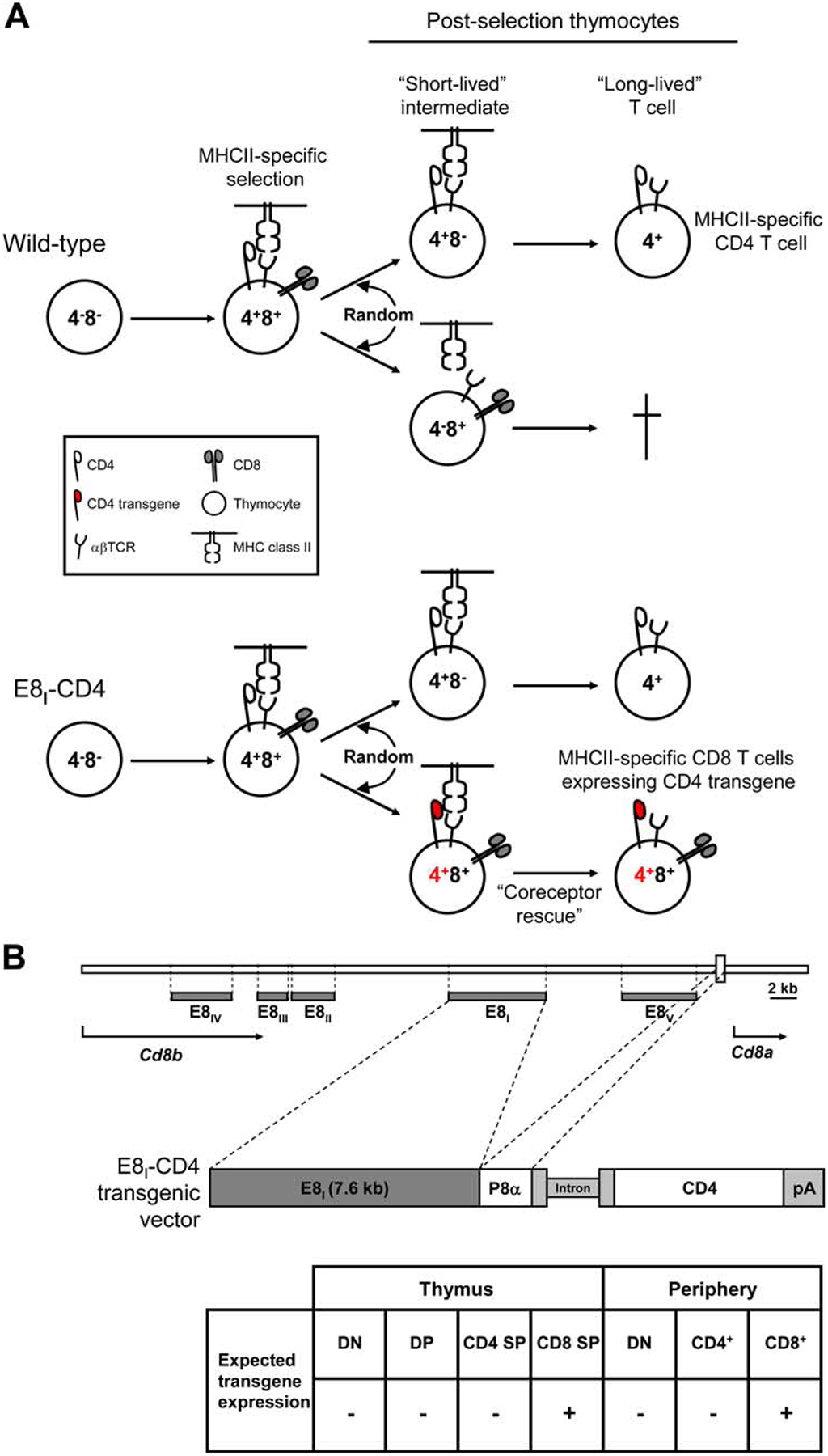
Experimental design. A, Schematic of the stochastic/selection model of lineage choice as applied to MHC-II-specific T cells. During positive selection, MHC-II-specific TCR signals induce DP thymocytes to randomly terminate expression of either CD4 or CD8 coreceptors, generating short-lived intermediate thymocytes bearing MHC-II-specific TCR that are destined to die if they fail to receive a TCR/coreceptor-mediated survival signal. Since CD4 is required for MHC-II-dependent TCR signaling, only short-lived MHC-II-selected thymocytes expressing CD4 coreceptors can receive a TCR-mediated survival signal and differentiate into long-lived T cells. In WT mice, MHC-II-specific thymocytes that have randomly adopted the CD8 lineage fate by terminating CD4 gene expression undergo cell death because they are unable to receive MHC-II-specific TCR survival signals (upper panel). In the present study, the E8I-CD4 transgene was designed to only induce transgenic CD4 coreceptor expression on those positively selected thymocytes that have adopted the CD8 lineage fate. Consequently, in E8I-CD4 transgenic mice, MHC-II-specific thymocytes that have randomly terminated CD4 gene expression would express transgenic CD4 coreceptors (shown in red) and so would receive MHC-II-specific TCR survival signals and differentiate into long-lived CD8 lineage T cells (lower panel). B, Design of the E8I-CD4 transgene. The E8I-CD4 transgene consisted of the mature CD8 T cell-specific 7.6-kb E8I transcriptional enhancer element (16) ligated to the CD8α promoter (P8α) and a murine CD4 intron splicing module (intron) driving CD4 cDNA expression (28). The origin of the E8I element and CD8α promoter are shown in a schematic representation of the Cd8 gene locus; horizontal gray bars correspond to regions of cis-enhancer elements (E8I-E8V) (29, 30).
Abs and flow cytometry
Cell suspensions were prepared from thymus and lymph nodes (LN) and stained with fluorochrome-conjugated Abs with the following specificities: CD4 (GK1.5 and RM4.5), CD69 (H1.2F3), Qa-2 (1-1-2), CD25 (7D4), TCRβ (H57–59), CD5 (53–7.3) (all from BD Pharmingen), CD8α (CT-CD8α, Caltag Laboratories), CD24 (M1/69, BD Pharmingen), and IL-7Rα (A7R34, eBioscience). OT-II transgenic TCR were identified by staining with Vα2 (B20.1, eBioscience) and Vβ5 (MH3–2, BD Pharmingen) Abs. For FACS analysis, 1–2 × 106 cells were incubated with 2.4G2 (antimouse FcγIII/II receptor; BD Pharmingen) and stained with fluorochrome-conjugated Abs and analyzed with a FACSVantage SE flow cytometer (BD Biosciences). Flow cytometry data were analyzed using software developed at the National Institutes of Health and displayed in 4-decade log scale.
Intracellular calcium flux
For calcium flux analyses, 2 × 106 LN cells were loaded with the calcium dye Indo-1 (Molecular Probes) at a final concentration of 1.8 μM and incubated for 30 min at 37°C. After washing, cells were incubated on ice for 40 min with 5 μg/ml biotinylated anti-TCRβ (H57–59) alone or in combination with 1 μg/ml biotinylated anti-CD4 (GK1.5) Abs, anti-CD4-FITC (RM4.5), and anti-CD8α-PE (53–6.72) (all from BD Pharmingen). Stained cells were then washed twice and prewarmed to 37°C for 2 min before being applied to the flow cytometer. Cells were signaled 30 s after being applied to the flow cytometer by cross-linking with avidin (4 μg/ml), and calcium mobilization was recorded for 4 min following cross-linking. Intracellular calcium concentrations were determined by the ratio of Indo-1 fluorescence at 405 vs 510 nm using FlowJo software (Tree Star).
Results
Experimental design and characterization of E8I-CD4 transgenic mice
The stochastic/selection model predicts that up to 50% of MHC-II-signaled DP thymocytes randomly shut off CD4 gene expression and make an incorrect lineage decision to become CD4−8+ thymocytes with mismatching TCR and coreceptors that cannot generate TCR-mediated rescue signals (4–9) (Fig. 1A, top panel). To determine whether significant numbers of MHC-II-selected thymocytes actually select a CD8 lineage fate and die as a result, we designed a CD4 transgene (referred to as E8I-CD4) that specifically targets transgenic CD4 coreceptors to thymocytes that have undergone positive selection and have recently adopted a CD8 lineage fate. We did so by utilizing the E8I-CD8 gene-enhancer element whose activity is restricted to postselection CD8 lineage T cells (16, 17). The E8I-CD4 transgene consists of CD4 cDNA whose expression is transcriptionally controlled by the E8I enhancer and CD8α promoter (Fig. 1B), and we introduced the E8I-CD4 transgene into C57BL/6 (B6) mice to generate E8I-CD4 transgenic mice. Consequently, the E8I-CD4 transgene would rescue any MHC-II-selected T cells that adopted a CD8 lineage fate (Fig. 1A, bottom panel).
Comparison of E8I-CD4 and nontransgenic (wild-type (WT)) littermate mice revealed that overall thymus cellularity and TCRβ expression were comparable in both mice (Fig. 2A). However, analysis of CD4 and CD8 expression revealed that CD4−CD8+ thymocytes that were present in WT mice (Fig. 2B, left panel, shown in blue) were largely absent in E8I-CD4 mice, which instead contained a population of CD8+ cells expressing CD4 at uniquely high levels (Fig. 2B, left panel, shown in red). Such CD4highCD8+ thymocytes in E8I-CD4 mice were equivalent to mature CD8 SP thymocytes in WT mice except for their expression of transgenic CD4, as both were identically CD24lowCD8+ cells (Fig. 2B, middle panel). Because CD24lowCD8+ cells in E8I-CD4 mice expressed transgenic CD4, we refer to them as “CD8 lineage cells” rather than referring to them as CD8 SP cells since these cells were not phenotypically CD4−CD8+. Indeed, CD8 lineage T cells in both the periphery (Fig. 2B, right panel) and thymus (Fig. 2B, left panel) of E8I-CD4 mice expressed high levels of transgenic CD4. However, with regard to expression of T cell markers other than CD4, CD8 lineage cells in E8I-CD4 mice were essentially identical to CD8+ T cells in WT mice (Fig. 2C).
FIGURE 2.
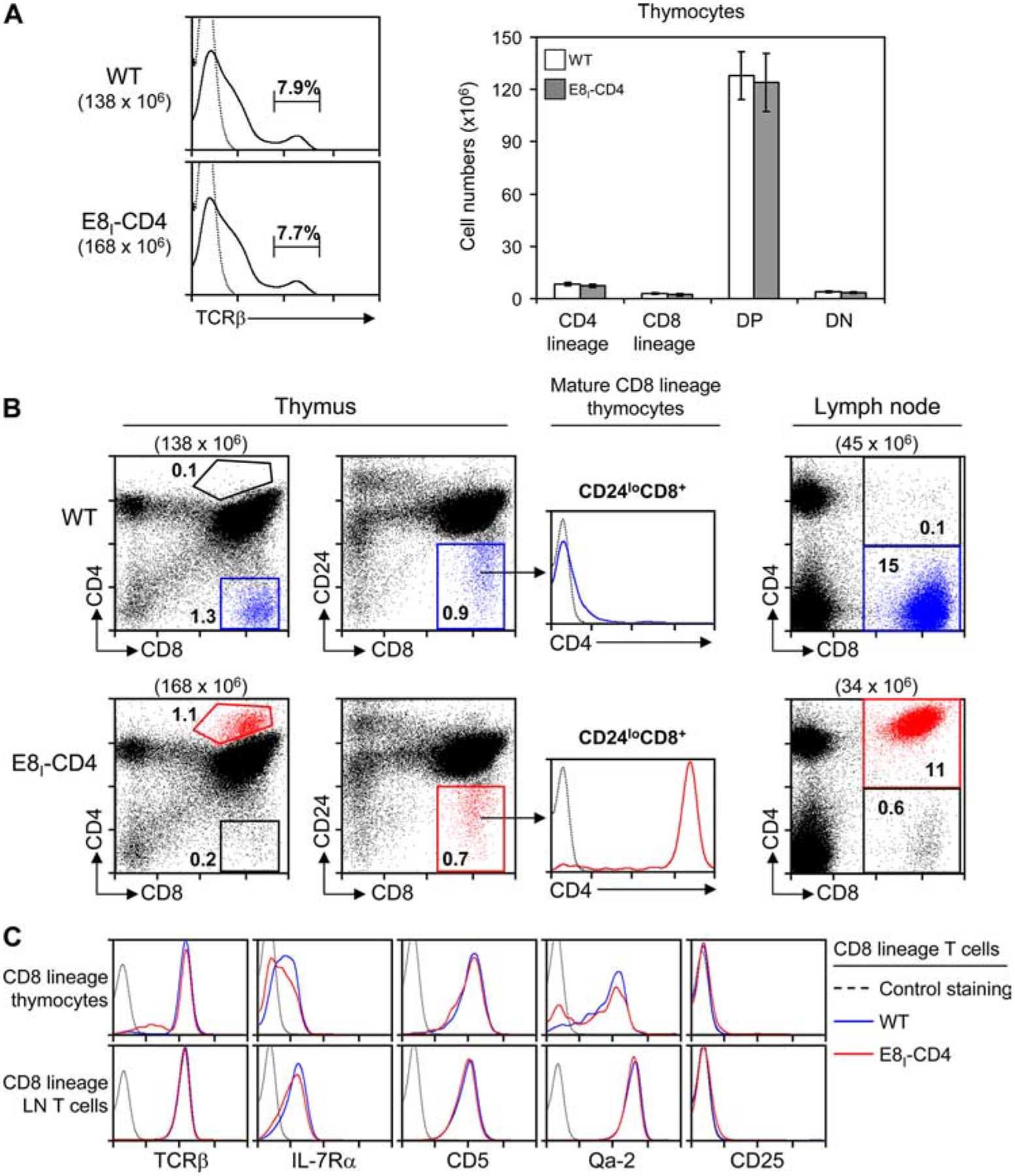
Characterization of E8I-CD4 transgenic mice. A, Single-cell suspensions of thymocytes from littermate (WT) and E8I-CD4 mice were stained with fluorochrome-conjugated Abs as indicated and analyzed by flow cytometry. Left, TCRβ expression and the frequency of TCRβhigh thymocytes; right, average number ±SEM) of thymocyte subsets from WT (n = 6) and E8I-CD4 (n = 6). CD4 lineage and CD8 lineage thymocytes were defined as CD24lowCD4+ and CD24lowCD8+, respectively. B, Thymocytes and LN cells from WT and E8I-CD4 transgenic mice were analyzed for CD4, CD8, and CD24 expression. Mature CD8 lineage cells in thymus and LN from WT mice are shown in blue, whereas mature CD8 lineage cells from E8I-CD4 mice are shown in red. Numbers in parentheses indicate total numbers of cells, while numbers in boxes indicate the frequency (percent) of cells in that box. Data are displayed on a 4-decade log scale and are representative of five independent experiments. C, Comparison of CD8 lineage T cells in WT and E8I-CD4 transgenic mice. Surface expression of the indicated proteins on CD8 lineage thymocytes and LN T cells from WT mice (blue line) and E8I-CD4 transgenic mice (red line) are shown. Data are displayed on a 4-decade log scale and are representative of five independent experiments.
Kinetics of E8I-CD4 transgene expression during thymocyte development
Potential rescue of mismatched MHC-II-specific CD8 lineage thymocytes requires that the E8I-CD4 transgene maintain CD4 protein expression despite down-regulation of endogenous CD4. Consequently, we assessed the ability of the E8I-CD4 transgene to maintain CD4 protein expression on developing CD8 lineage thymocytes. To relate thymocyte development to thymic selection, we defined progressive stages of CD8+ thymocyte maturation by their expression of TCRβ and CD24, as maturing thymocytes progressively increase surface expression of TCRβ and then down-regulate surface expression of CD24 (18–20) (Fig. 3, middle panels). As a result, pre-selection thymocytes are TCR−/loCD24high (Fig. 3A, middle panels, gates I and II), thymocytes undergoing positive selection are TCRintCD24high (gate III), postselection thymocytes are TCRhighCD24high (gate IV), and maturing thymocytes are TCRhighCD24low (gate V). In WT mice, it can be seen that surface CD4 expression is maintained on developing CD8+ thymocytes before and during positive selection (Fig. 3A, top right panel, gates I–III), after which surface CD4 expression progressively declines as postselection thymocytes differentiate into mature CD8 lineage T cells (Fig. 3A, top panel, gate V). In contrast, in E8I-CD4 mice, CD4 expression is maintained on developing CD8+ thymocytes throughout their selection and differentiation into mature CD8 lineage T cells, at which point it is dramatically increased (Fig. 3A, right panel, gates I–V). Thus, CD4 surface protein expression is never lost from developing CD8 lineage thymocytes in E8I-CD4 mice.
FIGURE 3.
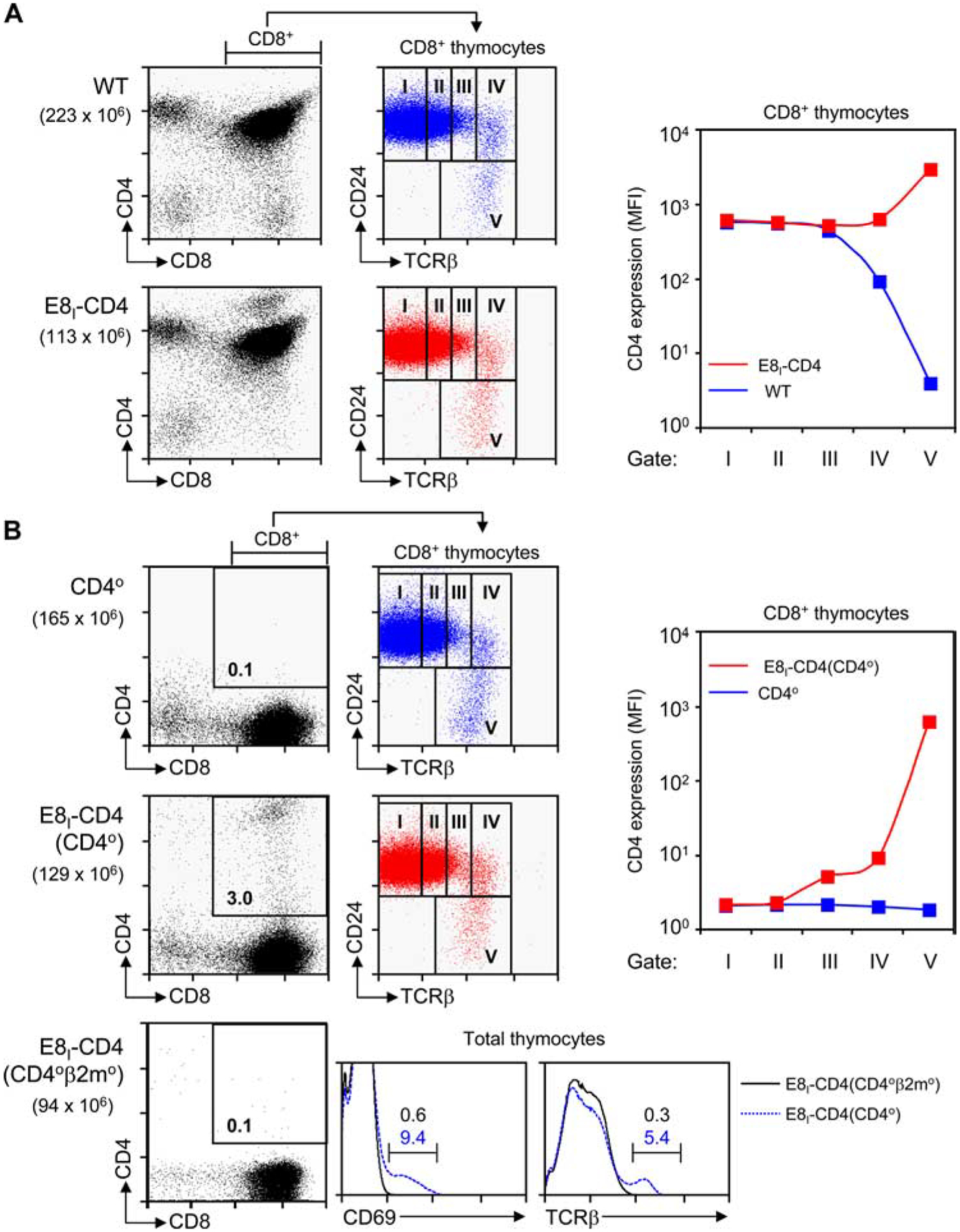
Kinetics of E8I-CD4 transgene expression. A, Induction of E8I-CD4 transgene upon down-regulation of endogenous CD4 expression. Thymocytes from WT mice (upper panel) and E8I-CD4 mice (lower panel) were stained for CD4, CD8, CD24, and TCRβ expression. To determine the developmental point during thymic selection that CD8+ cells express the E8I-CD4 transgene, we defined five progressive stages of thymic selection (I–V) by gating on CD24 vs TCRβ staining (middle column; blue indicates WT; red, E8I-CD4) on CD8+ thymocytes and then assessing transgenic CD4 expression on cells in each gate. Mean fluorescence intensity (MFI) of CD4 expression is plotted against the developmental stage (right panel; note that the y-axis of the graph is a log scale). Numbers in parentheses indicate total numbers of cells. Flow cytometry data are displayed on a 4-decade log scale and results are representative of three independent experiments. B, Timing of E8I-CD4 transgene expression during thymocyte development. The E8I-CD4 transgene was introduced into CD40 mice (middle panels) and CD40β2m0 mice (bottom panels) so that all CD4 expression was from the E8I-CD4 transgene. Thymocytes from CD40 mice (first row, blue) and E8I-CD4(CD40) mice (second row, red) were stained for CD4, CD8, CD24, and TCRβ expression. The developmental point during thymic selection that CD8+ cells express E8I-CD4 coreceptor transgene expression was assessed in thymocytes based on the same gates (I–V) as in A. MFI of CD4 expression is plotted against the developmental stage (right panel; note that the y-axis of the graph is a log scale). Thymocytes from E8I-CD4(CD40β2m0) mice (bottom panels) were stained for CD4, CD8, CD69, and TCRβ expression. Numbers in parentheses indicate total numbers of cells. Numbers in boxes represent the frequency of cells in that box. Flow cytometry data are displayed on a 4-decade log scale and results are representative of up to three independent experiments.
To clearly identify the point in thymocyte development that E8I-CD4 encoded CD4 proteins are expressed, we examined CD4 expression on thymocytes from E8I-CD4 transgenic mice that lacked endogenous CD4 expression. We introduced the E8I-CD4 transgene into CD4-deficient (CD40) mice to generate E8I-CD4(CD40) mice in which all CD4 expression was from the E8I-CD4 transgene (Fig. 3B). As expected from the transcriptional activity of the E8I enhancer element, transgenic E8I-CD4 expression was essentially limited to CD8+ thymocytes (Fig. 3B, second row). Significantly, transgenic E8I-CD4 expression was first detected on TCRβint CD24high thymocytes in gate III undergoing positive selection and steadily increased on postselection thymocytes in gates IV and V (Fig. 3B, right panel). These data confirm that the E8I-CD4 transgene drives surface CD4 protein expression on CD8 lineage thymocytes that are down-regulating expression of endogenous CD4.
To further document that the E8I-CD4 transgene was not expressed in preselection thymocytes, we introduced the E8I-CD4 transgene into CD40β2m0 mice, which contain only preselection thymocytes because MHC-I- and MHC-II-specific positive selection are both impaired. Notably, the E8I-CD4 transgene was not expressed in E8I-CD4(CD40β2m0) thymocytes, which remained preselection cells that were CD69− and TCRβlow (Fig. 3B, third row).
Taken together, these data demonstrate that the E8I-CD4 transgene is not expressed in preselection thymocytes, but it is first expressed in thymocytes that had undergone positive selection and adopted the CD8 lineage fate.
The E8I-CD4 transgene fails to reveal MHC-II-specific T cells adopting the CD8-lineage fate
To reveal MHC-II-selected thymocytes adopting the CD8 lineage fate, we bred the E8I-CD4 transgene into β2m0 mice to generate E8I-CD4(β2m0) mice. We found that thymocytes from E8I-CD4(β2m0) mice contained few, if any, cells identified by CD24lowCD8+ expression as CD8 lineage cells, and these were no greater in frequency or absolute number than in nontransgenic β2m0 littermates (Fig. 4A). Analysis of peripheral LN T cells similarly showed no differences in either frequency or absolute number of TCRβ+CD8+ cells in E8I-CD4(β2m0) mice compared with nontransgenic β2m0 littermates (Fig. 4B). Thus, the E8I-CD4 transgene did not reveal or rescue MHC-II-specific thymocytes that had adopted the CD8− lineage fate.
FIGURE 4.
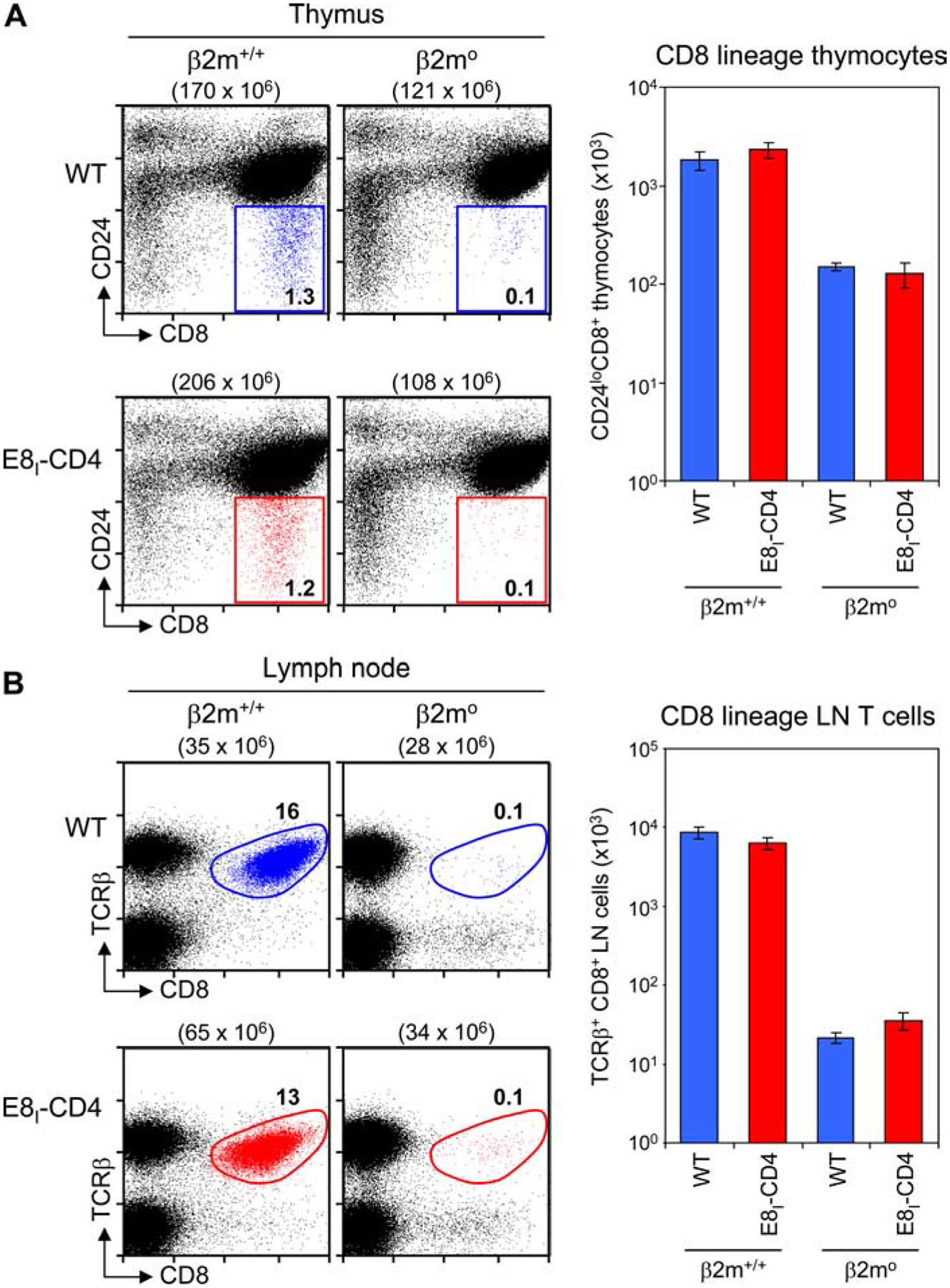
MHC-II-specific selection in E8I-CD4 transgenic mice. A, Thymocytes from WT and β2m-deficient B6 and E8I-CD4 transgenic mice were analyzed for CD4, CD8, and CD24 expression. Mature CD8 lineage thymocytes (i.e., CD8+CD24low) from WT mice are shown in blue, whereas mature CD8 lineage thymocytes from E8I-CD4 transgenic mice are shown in red. Numbers in parentheses indicate total numbers of cells, while numbers in boxes indicate the frequency (percent) of cells in that box. Data are displayed on a 4-decade log scale and are representative of four experiments. Bar graph depicts the average number ±SEM) of mature CD8 lineage thymocytes in four mice per group (right panel). Note that the y-axis of the bar graph is expressed as a log scale. B, LN T cells from WT and β2m-deficient B6 and E8I-CD4 transgenic mice were analyzed for TCRβ and CD8 expression. Mature CD8 lineage (i.e., CD8+ TCRβ+) T cells from WT mice are shown in blue, whereas mature CD8 lineage T cells from E8I-CD4 transgenic mice are shown in red. Numbers in parentheses indicate total numbers of cells, while numbers in boxes indicate the frequency (%) of cells in that gate. Bar graph depicts the average number ±SEM) of mature CD8 LN T cells in six mice per group (right panel). Note that the y-axis of the bar graph is expressed as a log scale.
Nevertheless, we further addressed this possibility with TCR transgenic mice. We introduced the E8I-CD4 transgene into mice expressing the OT-II TCR transgene that encodes Vα2+ Vβ5+ TCR that are positively selected by I-Ab thymic elements (14). In normal OT-II TCR transgenic mice, nearly all Vβ5+ T cells in the thymus and LN were CD4 lineage (i.e., CD8−) T cells, and very few Vβ5+ T cells were CD8 lineage (i.e., CD8+) T cells (Fig. 5). Notably, introduction of the E8I-CD4 transgene into OT-II TCR transgenic mice had no significant effect, as OT-II T cells in both the thymus and LN were still overwhelmingly CD4 lineage cells, with very few OT-II T cells becoming CD8 lineage T cells (Fig. 5). Thus, the E8I-CD4 transgene also failed to rescue or reveal CD8 lineage thymocytes bearing the OT-II transgenic TCR.
FIGURE 5.
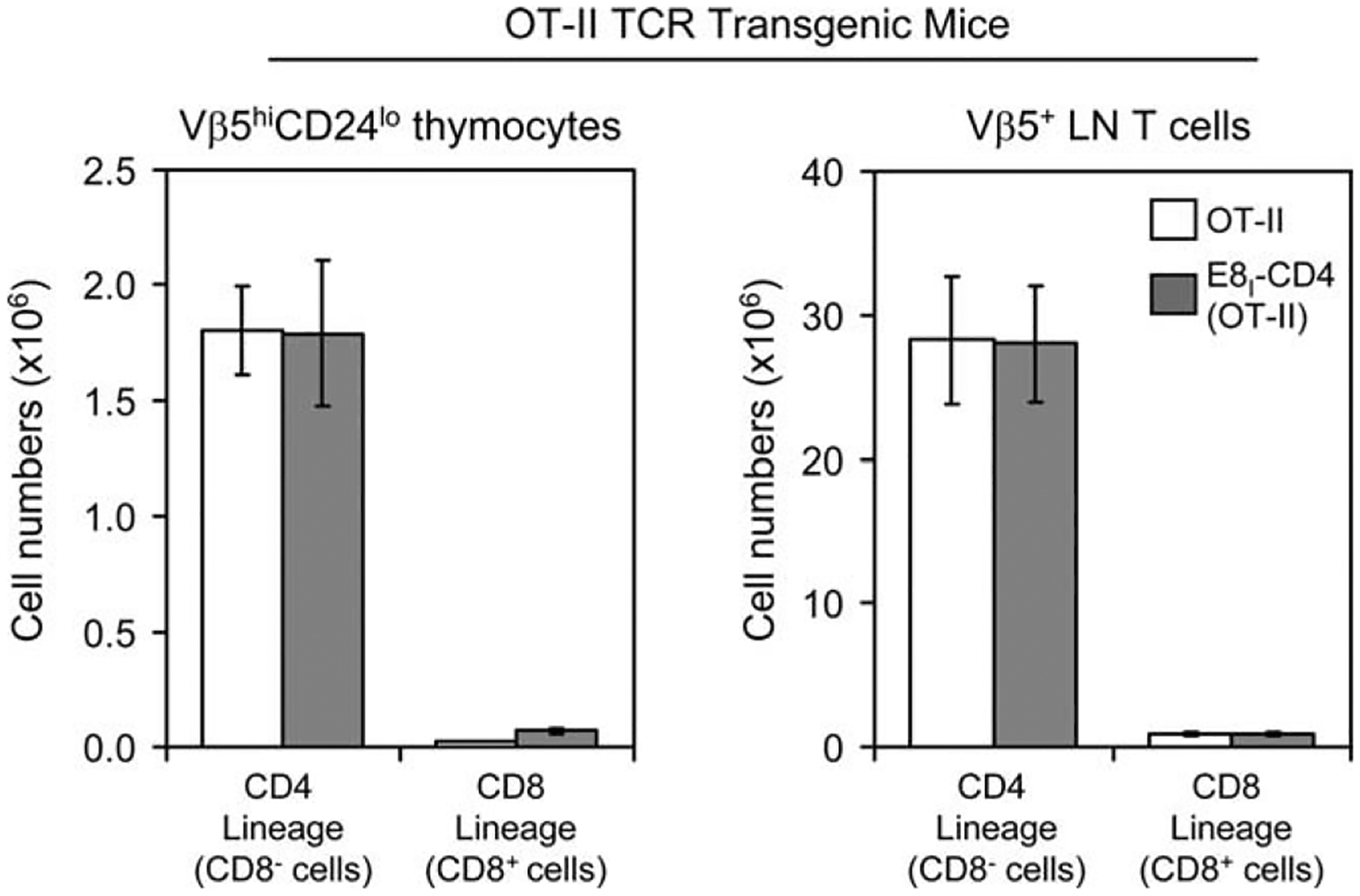
Effect of the E8I-CD4 transgene on development of OT-II transgenic T cells. We introduced the E8I-CD4 transgene into OT-II TCR transgenic mice (14) and analyzed clonotype-specific (Vβ5+) T cells in both thymus and LN of OT-II and E8I-CD4(OT-II) mice. The bar graphs display the average number ±SEM) of CD4 lineage (i.e., CD8−) and CD8 lineage (i.e., CD8+) mature T cells among thymocytes (left panel) and LN T cells (right panel) from OT-II (clear bars, n = 7) and E8I-CD4(OT-II) (shaded bars, n = 8).
E8I-CD4 transgene promotes TCR signaling in CD8 T cells
Because the E8I-CD4 transgene failed to reveal MHC-II-specific CD8 lineage thymocytes during thymic selection of either normal or TCR transgenic thymocytes, we considered that CD4 molecules encoded by the E8I-CD4 transgene might be signaling defective for unknown reasons and unable to augment TCR signal transduction. However, contrary to this possibility, E8I-CD4-encoded CD4 molecules, when coengaged with surface TCR, substantially augmented TCR signaled calcium mobilization in CD8 LN T cells from E8I-CD4 transgenic mice (Fig. 6A), confirming that E8I-CD4-encoded CD4 molecules were signaling competent.
FIGURE 6.
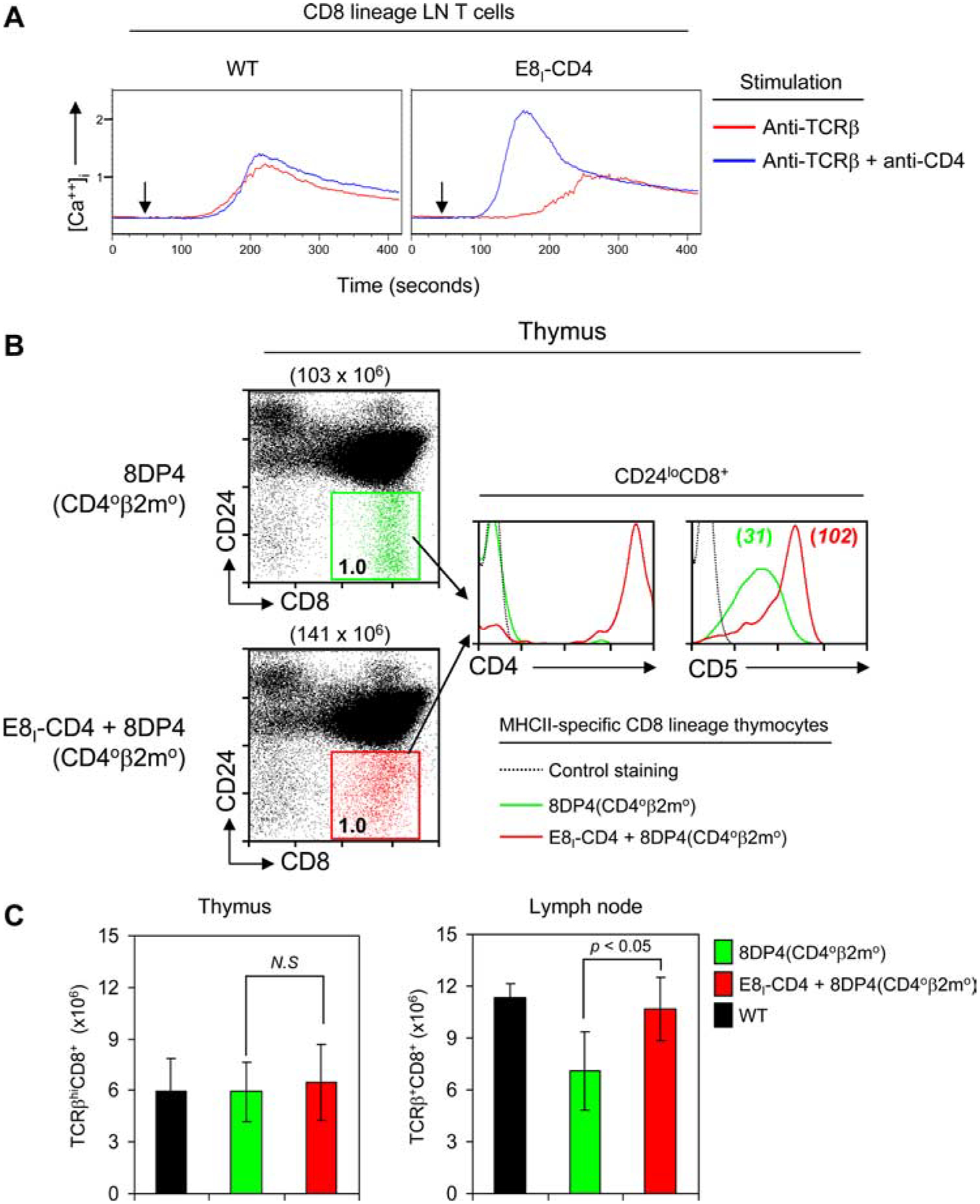
Expression of E8I-CD4 augments TCR signaling and improves the homeostasis of MHC-II-restricted CD8 T cells. A, Intracellular calcium flux in E8I-CD4 CD8 lineage T cells. Single-cell suspensions of LN T cells from WT and E8I-CD4 mice were loaded with Indo-1 dye, washed, and stained with fluorochrome-conjugated Abs to CD4 (RM4.5), CD8a (53–6.72), and 5 μg/ml of biotinylated anti-TCRβ (H57–59) with or without 1 mg/ml of biotinylated anti-CD4 (GK1.5). TCR signaling was initiated in stained cells by cross-linking with avidin after 30 s (arrowheads) of being loaded onto the flow cytometer at 37°C. Data are representative of two independent experiments. B, E8I-CD4 transgene expression on MHC-II-restricted CD8 lineage T cells generated in 8DP4 experimental mice. To assess E8I-CD4 transgene expression in MHC-II-specific CD8 T cells when such cells were generated during thymic selection, we introduced the E8I-CD4 transgene into 8DP4 experimental mice, which were on a CD40β2m0 background. As previously documented (15), MHC-II-restricted CD8 lineage T cells develop in 8DP4 experimental mice despite their expression of mismatching MHC-II TCR and CD8 coreceptors. Thymocytes were stained for CD4, CD8, CD24, and CD5. Mature CD8 lineage cells (i.e., CD8+CD24low) from 8DP4 mice are shown in green, while mature CD8 lineage cells from E8I-CD4 + 8DP4 mice are shown in red. Numbers above the two-color histograms indicate total numbers of cells, while numbers in boxes indicate the frequency (percent) of cells in that box (left panels). Numbers in the CD5 histogram (right panel) indicate CD5 mean fluorescent intensity (MFI). Data are displayed on a 4-decade log scale and are representative of three independent experiments. C, Absolute numbers of TCRβhighCD8+ thymocytes and TCRβ+CD8+ LN T cells in WT (black bar), 8DP4 experimental mice (green bar), and 8DP4 experimental mice expressing the E8I-CD4 transgene (red bar). Data are average ±SEM) of three mice per group and were compared with a two-tailed Student’s t test. N.S. indicates not significant (p > 0.05).
MHC-II-restricted CD8 thymocytes express the E8I-CD4 transgene
Next, we wanted to verify that the E8I-CD4 transgene would be expressed on MHC-II-specific CD8 lineage thymocytes when such mismatched cells were actually generated during thymic selection. To do so, we utilized “8DP4 experimental mice” that we have previously described in detail (15). Because 8DP4 experimental mice were on a β2m0CD40 genetic background, their thymus only expressed MHC-II selecting elements and their thymocytes only expressed transgenic CD4 molecules. 8DP4 experimental animals express the E8III-CD4 transgene that drives CD4 coreceptor expression only in preselection DP thymocytes so that CD4 expression and MHC-II-restricted TCR signaling are prematurely terminated during MHC-II-specific positive selection. As a result, MHC-II-signaled thymocytes in 8DP4 experimental mice uni3formly adopt the CD8 lineage fate (15). For the present study we introduced the E8I-CD4 transgene into 8DP4 experimental mice and analyzed CD4 expression on such MHC-II-selected CD8 lineage thymocytes (Fig. 6B). We confirmed that MHC-II-selected CD8 lineage (CD24lowCD8+) thymocytes were generated both in 8DP4 mice (Fig. 6B, left panel; shown in green) and in 8DP4 mice containing the E8I-CD4 transgene (Fig. 6B, left panel; shown in red). Most importantly, we found that the E8I-CD4 transgene was expressed in MHC-II-selected CD8 lineage cells, as nearly all such cells were CD4+ (Fig. 6B, middle histogram). Notably, expression of CD4 coreceptors encoded by the E8I-CD4 transgene dramatically increased CD5 expression on mismatched MHC-II-restricted CD8 lineage thymocytes in 8DP4 mice (Fig. 6B, right histogram), indicating that E8I-CD4 encoded CD4 coreceptors augmented in vivo signaling by MHC-II-specific TCR (21). These results demonstrate that CD4 coreceptors encoded by the E8I-CD4 transgene are indeed expressed on MHC-II-restricted CD8 lineage thymocytes when such cells are generated in the thymus, and that the transgene-encoded CD4 coreceptors are functional.
While MHC-II-specific CD8 lineage T cells are generated in the thymus of 8DP4 mice, the numbers of such MHC-II-specific CD8 lineage T cells present in the periphery are reduced compared with conventional B6 CD8+ T cells (Fig. 6C), because their TCR/coreceptor mismatch renders them unable to generate homeostatic TCR signals needed for long-term survival in vivo (15, 22). Thus, we asked if the E8I-CD4 transgene, by inducing expression of matching CD4 coreceptors, would increase the number of MHC-II-specific CD8 lineage T cells present in the periphery of 8DP4 experimental mice. In fact, expression of the E8I-CD4 transgene did significantly increase the number of MHC-II-specific CD8 lineage T cells present in the periphery of 8DP4 mice and increased them to levels comparable to conventional B6 CD8+ T cells (Fig. 6C). These results indicate that the E8I-CD4 transgene is capable of rescuing MHC-II-specific CD8 lineage T cells, but that TCR/coreceptor-mediated rescue signals are not required during development in the thymus but are only required in the lymphoid periphery.
Discussion
The present study documents that thymocytes that have been positively selected by MHC-II-specific TCR rarely adopt the incorrect CD8 lineage fate. Persistence of CD4 coreceptor expression on CD8-committed thymocytes was achieved by the E8I-CD4 transgene, which specifically targeted transgenic CD4 protein expression to CD8-committed thymocytes and prevented loss of cell-surface CD4 protein expression. However, despite persistent CD4 coreceptor expression, MHC-II-selected thymocytes only rarely adopted a CD8 lineage fate. Thus, the present study directly contradicts the concept of coreceptor rescue and directly contradicts the central premise of stochastic/selection. Instead, the present study documents that CD4/CD8 lineage choice in the thymus is neither stochastic nor significantly error-prone.
The stochastic/selection model postulates that CD4/CD8 lineage choice is highly error-prone, as signaled DP thymocytes randomly terminate expression of one or the other coreceptor to become short-lived SP thymocytes that require matching TCR and coreceptors to differentiate into mature T cells. Consequently, stochastic/selection predicts that ~50% of MHC-II-signaled thymocytes maintain CD4 coreceptor expression and differentiate into mature CD4+ T cells, while the other 50% of MHC-II-signaled thymocytes terminate CD4 coreceptor expression and die (4–7, 11, 23). To directly assess this prediction, the present study utilized the E8I-CD4 transgene, which encodes CD4 coreceptor molecules that are first expressed when positively selected thymocytes have adopted the CD8 lineage fate and begun down-regulating endogenous CD4 expression, precisely the developmental point at which CD4 coreceptor rescue of MHC-II-selected CD8 lineage thymocytes would occur. In fact, the E8I-CD4 transgene functioned as designed in that it successfully induced expression of competent CD4 coreceptors on MHC-I-selected CD8 lineage thymocytes and successfully induced expression of competent CD4 coreceptors on MHC-II-selected CD8 lineage thymocytes when such cells were generated in 8DP4 experimental mice by premature termination of intrathymic MHC-II signaling. Nevertheless, the E8I-CD4 transgene did not reveal MHC-II-selected thymocytes adopting the CD8 lineage fate during normal thymic selection, indicating that MHC-II-selected thymocytes do not normally make an incorrect lineage choice.
The present study differs importantly from previous CD4 coreceptor rescue (7, 10) experiments in the timing of CD4 transgene expression during thymocyte development and in the detection of MHC-II-specific CD8 lineage T cells. Previous CD4 coreceptor rescue experiments utilized CD4 transgenes whose expression was controlled by heterologous transcriptional control elements that initiated CD4 transgene expression early and continuously throughout thymocyte development, and resulted in the appearance of MHC-II-specific CD8 lineage T cells (7, 10). We do not think that the appearance of such MHC-II-specific CD8 lineage T cells experiments was due to coreceptor rescue of short-lived MHC-II-selected thymocytes, as has been thought (7, 10). Rather, we suggest that the appearance of MHC-II-specific CD8 lineage T cells was the result of a characteristic intrinsic to CD4 coreceptor transgenes driven by heterologous transcriptional control element, namely, that their expression is down-regulated during positive selection, causing premature disruption of MHC-II signaling during positive selection of thymocytes with low-affinity, coreceptor-dependent TCR and redirecting them to differentiate into CD8 lineage T cells (S. D. Sarafova, F. Van Laethem, T. Guinter, S. O. Sharrow, L. Feigenbaum, R. Bosselut, and A. Singer, manuscript in preparation). Indeed, differentiation of MHC-II-signaled thymocytes into CD4 lineage cells requires persistent MHC-II signaling, as premature disruption of MHC-II signaling during positive selection has been shown to redirect differentiation of MHC-II-specific thymocytes into CD8 lineage T cells (15, 24).
Appearance of MHC-II-restricted CD8 lineage T cells in CD4 silencer-deficient (CD4Δsil−/−) mice has a similar explanation (12). Targeted deletion of the Cd4 silencer element not only removed the transcriptional control element that silences CD4 expression on CD4− thymocytes, but it also removed a positive regulatory element (25) that augments CD4 coreceptor expression on signaled thymocytes during positive selection and differentiation into mature CD4+ T cells (12). Consequently, because CD4 coreceptor expression declined during positive selection, it is likely that MHC-II signaling was prematurely disrupted in CD4Δsil−/− thymocytes with low-affinity TCR, causing such MHC-II-signaled thymocytes to differentiate into CD8 lineage T cells.
We think that the CD4/CD8 lineage decision is best described by the kinetic signaling model (3, 26, 27). According to kinetic signaling, TCR-signaled DP thymocytes selectively terminate CD8 coreceptor transcription, regardless of the specificity of their TCR, to transcriptionally become CD4+CD8− intermediate thymocytes that may phenotypically appear as CD4+CD8low cells. It is in such intermediate thymocytes that CD4/CD8 lineage choice is made, and the choice is based on whether the TCR-mediated selection signal persists or ceases in the absence of CD8 coreceptor transcription. If diminished CD8 coreceptor expression does not disrupt TCR signaling, thymocytes develop into CD4 lineage T cells; however, if diminished CD8 coreceptor expression does disrupt TCR signaling, thymocytes develop into CD8 lineage T cells (3, 26, 27). Indeed, 8DP4 experimental mice were originally constructed to test a prediction of kinetic signaling (15); that is, that loss of CD4 coreceptor expression would disrupt MHC-II signaling in intermediate thymocytes and cause them to differentiate into CD8 lineage T cells. 8DP4 experimental mice also proved invaluable in the present study because they allowed us to experimentally confirm that the E8I-CD4 coreceptor transgene was expressed on MHC-II-selected CD8 lineage thymocytes when such cells were actually generated in the thymus.
In conclusion, the present study demonstrates that CD4/CD8 lineage choice in the normal thymus is not stochastic and is not significantly error-prone, contradicting the central premise of the stochastic/selection model and contradicting the results of previous coreceptor rescue experiments. We suggest that the appearance of T cells with mismatching TCR and coreceptors in experimental circumstances does not imply an underlying stochastic mechanism to CD4/CD8 lineage choice, but rather is due to redirected differentiation of positively selected thymocytes as described by kinetic signaling (3, 15, 27).
Acknowledgments
We thank Dr. Wilfried Ellmeier for the E8I and E8III regulatory elements and Dr. Naomi Taylor for critical reading of the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Abbreviations used in this paper: DP, double positive; β2m, β2-microglobulin; LN, lymph node; MFI, mean fluorescence intensity; SP, single positive; WT, wild type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Bosselut R 2004. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat. Rev. Immunol 4: 529–540. [DOI] [PubMed] [Google Scholar]
- 2.Kappes DJ, He X, and He X. 2005. CD4-CD8 lineage commitment: an inside view. Nat. Immunol 6: 761–766. [DOI] [PubMed] [Google Scholar]
- 3.Singer A, and Bosselut R. 2004. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol 83: 91–131. [DOI] [PubMed] [Google Scholar]
- 4.Chan S, Correia-Neves M, Benoist C, and Mathis D. 1998. CD4/CD8 lineage commitment: matching fate with competence. Immunol. Rev 165: 195–207. [DOI] [PubMed] [Google Scholar]
- 5.Chan SH, Benoist C, and Mathis D. 1994. In favor of the selective model of positive selection. Semin. Immunol 6: 241–248. [DOI] [PubMed] [Google Scholar]
- 6.Chan SH, Cosgrove D, Waltzinger C, Benoist C, and Mathis D. 1993. Another view of the selective model of thymocyte selection. Cell 73: 225–236. [DOI] [PubMed] [Google Scholar]
- 7.Davis CB, Killeen N, Crooks ME, Raulet D, and Littman DR. 1993. Evidence for a stochastic mechanism in the differentiation of mature subsets of T lymphocytes. Cell 73: 237–247. [DOI] [PubMed] [Google Scholar]
- 8.Itano A, Kioussis D, and Robey E. 1994. Stochastic component to development of class I major histocompatibility complex-specific T cells. Proc. Natl. Acad. Sci. USA 91: 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan S, Correia-Neves M, Dierich A, Benoist C, and Mathis D. 1998. Visualization of CD4/CD8 T cell commitment. J. Exp. Med 188: 2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron A, Hafen K, and von Boehmer H. 1994. A human CD4 transgene rescues CD4−CD8+ cells in β2-microglobulin-deficient mice. Eur. J. Immunol 24: 1933–1936. [DOI] [PubMed] [Google Scholar]
- 11.Corbella P, Moskophidis D, Spanopoulou E, Mamalaki C, Tolaini M, Itano A, Lans D, Baltimore D, Robey E, and Kioussis D. 1994. Functional commitment to helper T cell lineage precedes positive selection and is independent of T cell receptor MHC specificity. Immunity 1: 269–276. [DOI] [PubMed] [Google Scholar]
- 12.Leung RK, Thomson K, Gallimore A, Jones E, Van den Broek M, Sierro S, Alsheikhly AR, McMichael A, and Rahemtulla A. 2001. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat. Immunol 2: 1167–1173. [DOI] [PubMed] [Google Scholar]
- 13.Zou YR, Sunshine MJ, Taniuchi I, Hatam F, Killeen N, and Littman DR. 2001. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet 29: 332–336. [DOI] [PubMed] [Google Scholar]
- 14.Barnden MJ, Allison J, Heath WR, and Carbone FR. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol 76: 34–40. [DOI] [PubMed] [Google Scholar]
- 15.Sarafova SD, Erman B, Yu Q, Van Laethem F, Guinter T, Sharrow SO, Feigenbaum L, Wildt KF, Ellmeier W, and Singer A. 2005. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity 23: 75–87. [DOI] [PubMed] [Google Scholar]
- 16.Ellmeier W, Sunshine MJ, Losos K, Hatam F, and Littman DR. 1997. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7: 537–547. [DOI] [PubMed] [Google Scholar]
- 17.Hostert A, Tolaini M, Roderick K, Harker N, Norton T, and Kioussis D. 1997. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity 7: 525–536. [DOI] [PubMed] [Google Scholar]
- 18.Bendelac A, Matzinger P, Seder RA, Paul WE, and Schwartz RH. 1992. Activation events during thymic selection. J. Exp. Med 175: 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Punt JA, Suzuki H, Granger LG, Sharrow SO, and Singer A. 1996. Lineage commitment in the thymus: only the most differentiated (TCRhibcl-2hi) subset of CD4+CD8+ thymocytes has selectively terminated CD4 or CD8 synthesis. J. Exp. Med 184: 2091–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita I, Nagata T, Tada T, and Nakayama T. 1993. CD69 cell surface expression identifies developing thymocytes which audition for T cell antigen receptor-mediated positive selection. Int. Immunol 5: 1139–1150. [DOI] [PubMed] [Google Scholar]
- 21.Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, and Love PE. 1998. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med 188: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JH, Adoro S, Lucas PJ, Sarafova SD, Alag AS, Doan LL, Erman B, Liu X, Ellmeier W, Bosselut R, Feigenbaum L, and Singer A. 2007. “Coreceptor tuning”: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat. Immunol 8: 1049–1059. [DOI] [PubMed] [Google Scholar]
- 23.Correia-Neves M, Waltzinger C, Mathis D, and Benoist C. 2001. The shaping of the T cell repertoire. Immunity 14: 21–32. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, and Bosselut R. 2004. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nat. Immunol 5: 280–288. [DOI] [PubMed] [Google Scholar]
- 25.McCready PM, Hansen RK, Burke SL, and Sands JF. 1997. Multiple negative and positive cis-acting elements control the expression of the murine CD4 gene. Biochim. Biophys. Acta 1351: 181–191. [DOI] [PubMed] [Google Scholar]
- 26.Brugnera E, Bhandoola A, Cibotti R, Yu Q, Guinter TI, Yamashita Y, Sharrow SO, and Singer A. 2000. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13: 59–71. [DOI] [PubMed] [Google Scholar]
- 27.Singer A 2002. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr. Opin. Immunol 14: 207–215. [DOI] [PubMed] [Google Scholar]
- 28.Sawada S, Scarborough JD, Killeen N, and Littman DR. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77: 917–929. [DOI] [PubMed] [Google Scholar]
- 29.Ellmeier W, Sawada S, and Littman DR. 1999. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol 17: 523–554. [DOI] [PubMed] [Google Scholar]
- 30.Kioussis D, and Ellmeier W. 2002. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol 2: 909–919. [DOI] [PubMed] [Google Scholar]


