ABSTRACT
Bone homeostasis is maintained by balanced osteoblast-mediated tissue production and osteoclast-mediated tissue destruction, and is disrupted in pathological conditions such as osteoporosis. The mechanisms underlying osteogenic differentiation of bone marrow mesenchymal stem cells, which is critical to bone homeostasis, are not completely clear, despite extensively studies. Long noncoding RNAs (lncRNAs) have recently emerged as novel therapeutic targets in various diseases. However, the expression pattern and biological function of lncRNAs in osteogenic differentiation remain unclear. In this study, we aimed to determine the role of lncRNAs in osteogenic differentiation of human bone marrow mesenchymal stem cells. We found high lncRNA MCF2L-AS1 expression in human bone marrow mesenchymal stem cells, and used bioinformatics analysis to analyze its function. MCF2L-AS1 knockdown induced inhibition of osteoblast differentiation. Silencing of MCF2L-AS1 increased the expression of miR-33a and subsequently inhibited Runx2 expression at the post-transcriptional level. Moreover, MCF2L-AS1 directly interacted with miR-33a, and downregulation of miR-33a efficiently reversed the suppression of Runx2 induced by MCF2L-AS1 short hairpin RNA (shRNA). Thus, MCF2L-AS1 positively regulated the expression of Runx2 by sponging miR-33a, and promoted osteogenic differentiation in BMSCs. Our results indicated that the lncRNA MCF2L-AS1 plays a critical role in the osteogenic differentiation of BMSCs, and targeting lncRNA MCF2L-AS1 could be a promising strategy to promote osteogenic differentiation.
KEYWORDS: MCF2L-AS1, miR-33a, Runx2, osteogenic differentiation
Introduction
The balance between osteoblast-mediated production and osteoclast-mediated destruction of bone tissue is critical to bone homeostasis [1,2]. Altered osteogenic differentiation of mesenchymal stem cells (MSCs) contributes to the imbalance in osteonecrosis and bone regeneration [3]. Bone marrow mesenchymal stem cells (BMSCs) are important members of the stem cell family, and can differentiate into multiple cell types such as adipocytes, chondrocytes, and osteoblasts [4–6]. BMSCs also play a vital role in maintaining the hematopoietic stem cell niche by providing modulatory signals [7]. Despite extensive studies, the mechanisms involved in the differentiation of BMSCs are not completely clear.
Long noncoding RNAs (lncRNAs) are non-protein-coding RNAs that are >200 nucleotides in length. They have been identified as novel regulators of various biological activities and play critical roles in the progression of several diseases [8,9]. LncRNAs interact with microRNAs (miRNAs) as competing endogenous RNAs (ceRNAs), and thereby regulate MSC differentiation and metabolism. Linc-ROR functions as a ceRNA for miR-138 and miR-145, and promotes osteogenic differentiation of mesenchymal stem cells [10]. The lncRNA MCF2L-AS1 suppresses bone formation of periodontal ligament stem cells by sponging miRNA-758 [11]. lncRNA-HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of the femoral head [12]. miRNAs play a role in multiple biological processes by regulating mRNA targets,such as osteogenic differentiation of BMSCs [13]and osteoclastogenesis [14].Therefore, we hypothesized that a lncRNA-miRNA-mRNA axis could play an important role in regulating the differentiation of MSCs.
Based on previous findings, we aimed to determine the role of the lncRNA MCF2L-AS1 in osteoblastic differentiation of bone-marrow-derived MSCs (BM-MSCs) and the relation of lncRNA MCF2L-AS1 and miRNA.
Materials and methods
Animals
Sprague-Dawley (SD) rats (80–120 g) were purchased from the Animal Experiment Center of Fujian Medical University (Fuzhou, China). Animals were maintained in a 12-h light/dark cycle, with free access to food and water. All experimental procedures and protocols were in accordance with the Chinese Council on Animal Care Guidelines, and were approved by the Animal Ethical Committee of Fujian Medical University. Efforts were made to minimize numbers and suffering of animals used.
MSC isolation
Rats were sacrificed, and the hindlimbs were aseptically dissected out, free of soft tissues. The femurs and tibias were removed, and both ends of the bones were cut off. The marrow cavities were flushed with culture medium (Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 [DMEM/F-12; Gibco, Grand Island, NY, USA], 10% fetal bovine serum [FBS; Gibco], 1% penicillin and streptomycin [Gibco]) by using a 25-gauge needle. The cells obtained were suspended in culture medium and seeded into 10 cm2 culture flasks, and incubated in a humidified atmosphere of 5% CO2 at 37°C. After incubating for 3 days, non-adherent cells were removed by frequent medium change. The remaining adherent cells (primary BMSCs) were passaged after digestion using 0.25% trypsin. Cells at passage 3 were used in our experiments.
For induction of osteogenic differentiation of BMSCs, cells were cultured for 7 and 14 days in osteogenic medium (OS medium), which consisted of normal culture medium supplemented with dexamethasone (100 nM) and β-glycerophosphate (2 mM) purchased from Sigma-Aldrich (St Louis, MO, USA), until they reached 70–80% confluence. The medium was changed every 3 days.
Plasmid generation and cell transfection
Total length of linc-MCF2L-AS1 was amplified by PCR from MSC cDNA and subcloned into pBabe vector for transient or stable expression of linc- MCF2L-AS1 in BM-MSCs. Runx2 3ʹ-UTR sequence as well as total length of linc-MCF2L-AS1 obtained before were subcloned into the pmiR-GLO vector. All of the cDNA sequences were obtained by database searching (lncRNAdb: http://www.lncrnadb.org; NCBI: https://www.ncbi.nlm.nih.gov). All miRNA mimics and antisense inhibitors were purchased from GenePharma Company (Shanghai, China). miRNA mimics and inhibitors as well as DNA plasmids were transfected using transfection reagent Lipofectamine 2000 (Invitrogen, USA) by following the manufacturer’s instructions.
ALP activity
Alkaline phosphatase (ALP) activity was used to evaluate the osteogenic differentiation capability of BMSCs after the different treatments. In brief, the cells were lysed with a lysis buffer containing 0.1% Triton X-100. The supernatant of the lysate was used to determine the ALP activity by incubating with p-nitrophenyl phosphate (pNPP) at 37°C for 15 min, and measuring absorbance at 405 nm by using a microplate reader. Total DNA concentration was determined by CyQUANT1 Cell Proliferation Assays (Thermo Fisher Scientific, USA). The ALP activity was normalized to total DNA concentration.
Alizarin red detection
MSCs were grown in osteogenic differentiation medium by using the StemPro osteogenesis differentiation kit (Invitrogen). For detecting mineralization, cells were induced for 2 weeks, fixed with 70% ethanol, and stained with 2% alizarin red (Sigma-Aldrich, St. Louis, MO, USA). To quantitatively determine calcium, alizarin red was destained with 10% cetylpyridinium chloride in 10 mM sodium phosphate for 30 min at room temperature. The concentration was determined by measuring the absorbance at 562 nm on a multiplate reader and comparing to a standard calcium curve from calcium dilutions in the same solution. The final calcium level in each group was normalized to the total protein concentration detected in a duplicate plate.
Luciferase assay
Dual-luciferase assay was performed to evaluate the luciferase activity of the reporter constructs according to manufacturers’ instructions (Promega, USA) with some modifications. In brief, HEK293 cells were seeded in 24-well culture plates at a density of 1 × 106 cells per well, and allowed to grow until they reached 80% confluency. Cells were then transfected with TOP Flash or pmiR-GLO luciferase reporter plasmids together with linc-MCF2L-AS1 expression vectors or miRNA mimics. The pRSVb-galactoside vector was co-transfected into HEK293 cells as an internal control for normalization. The plate was placed into a PerkinElmer VictorTM X2 2030 multilabel reader (Waltham, USA) to measure the firefly luciferase activity and β-galactosidase activity. The ratio of firefly luciferase to β-galactosidase activity in each sample was used as a measurement of the normalized luciferase activity. All experiments were performed in triplicate.
Microarray analysis
LncRNA expression during osteogenic differentiation of rat BMSCs was used for microarray analysis. BMSCs were treated with 10 mmol/L sodium beta-glycerophosphate + 0.1 μmol/L dexamethasone + 50 mg/L vitamin C for 7 days. Microarray analysis of lncRNA was performed with the LncRNA Human Gene Expression Microarray V4.0 expression profile, with the absolute value of Fold Change (FC) greater than or equal to 1.5, and P value less than or equal to 0.05 as the standard, screening out the weak differentially expressed lncRNA profile.
Quantitative polymerase chain reaction (q-PCR)
Total RNA of the tissues or cells were extracted using the TRIzol total RNA extraction kit (Tiangen, China) according to the manufacturer’s instructions. cDNA was synthesized using a reverse transcription PCR kit (Tiangen, China) according to the manufacturer’s instruction. Gene expression at the mRNA level was evaluated by quantitative polymerase chain reaction (q-PCR) using Exicycler™ 96 Real-Time Quantitative Thermal Block (Bioneer, China). Amplification conditions were as follows: 95°C for 10 min, 40 cycles of 95°C for 10 s, 60°C for 20 s and 72°C for 30 s. All samples were run in triplicate. The relative changes in transcript levels were analyzed using the ΔΔCt method and comparing Ct values of mRNA expression relative to the internal control.
Western blotting
The cell lysates were collected and centrifuged at 20,000 r.p.m for 5 min at 4°C. The supernatant was transferred to a clean tube and protein concentrations were measured using the BCA Kit (Pierce, Rockford, IL, USA). Proteins were separated by 10% SDS-PAGE with 5% stacking gel and 6, 8, 10, and 13% gradient separating gel, and blotted onto nitrocellulose membranes. Membranes were blocked with 5% skim milk for 1 h and incubated overnight at 4°C with primary antibody. The primary antibodies and dilutions used were anti-ALP 1:500, anti-OCN 1:500, anti-RUNX2 1:1000, and anti-GAPDH 1:1000. All antibodies were purchased from Abcam (Cambridge, UK). Membranes were washed six times in TBST for 5 min and incubated with secondary antibodies, HRP-conjugated goat anti-mouse/rabbit antibody (1:5,000; Invitrogen) for 2 h at room temperature. After washing in TBST, the protein bands were visualized using ECL reagents (EMD Millipore). The optical density of bands was analyzed by reflectance densitometry on a Gel-Pro Analyzer.
Statistical analysis
The χ2 test was used to compare clinicopathological parameters in patients with JUB protein expression. The SPSS 16.0 software (Chicago, USA) was used to analyze the enumeration data. Each experiment was performed at least 3 times, on independent passages, usually in triplicates. Data were analyzed by Newman–Keuls test using Statistica software as indicated and are presented as mean ± SEM. p < 0.05 was considered statistically significant. Results of time-lapse microscopy experiments were analyzed with Wilcoxon test in R software.
Results
MCF2L-AS1 expression is up-regulated after osteogenic induction of BMSCs
To identify lncRNA expressed during osteogenic differentiation of rat BMSCs, flow cytometry was first used for verifying successful BMSC isolation (Figure 1a–b). Osteogenic differentiation was confirmed by alizarin red detection (Figure 1c). The lncRNA microarray analysis identified 24 differentially expressed lncRNAs, of which 9 were up-regulated and 15 were down-regulated in the model group compared with the blank group (Figure 1d). We then chose lncRNA MCF2L-AS1 for further experiments and verified its expression by qRT-PCR. The results showed that the expression of lncRNA MCF2L-AS1 was consistent with the microarray results (Figure 1e).
Figure 1.
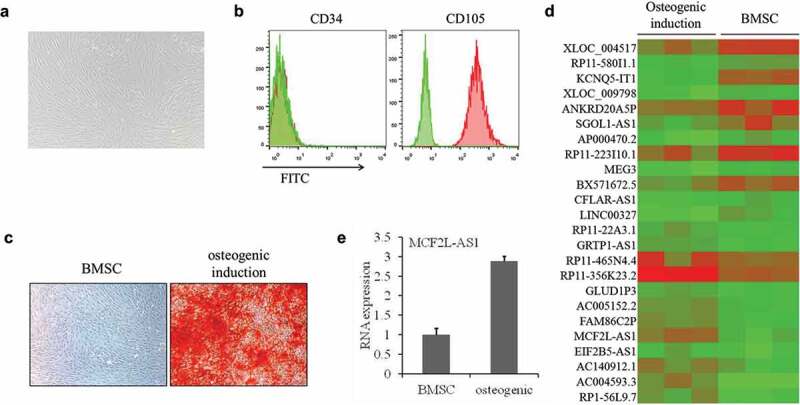
MCF2L-AS1 expression is up-regulated in BMSCs after osteogenic induction.
MCF2L-AS1 down-regulation inhibits osteogenic differentiation of rat BMSCs
To identify the role of linc-MCF2L-AS1 in osteogenesis, we examined its expression pattern during osteogenic differentiation. As shown in Figure 2a, we used lentiviral vectors to stably restore or silence the expression of linc-MCF2L-AS1 in rat BM-MSCs, and osteogenic differentiation was studied in the established stable cells. Expression levels of several osteogenic marker genes were measured at day 7, and ALP, OCN, and RUNX2 were downregulated by linc-MCF2L-AS1 knockdown (Figure 2b,c). Calcium nodules were evaluated by alizarin red S staining assays at day 14, and the results showed a significant reduction in calcium deposits in linc-MCF2L-AS1 knockdown cells (Figure 2d). Alkaline phosphatase (ALP) activity, an early marker of osteogenesis, was measured at day 3 with osteo-induction, and was decreased in linc-MCF2L-AS1-knockdown cells (Figure 2e). All of these data showed that linc-MCF2L-AS1 promotes osteogenesis of rat MSCs.
Figure 2.
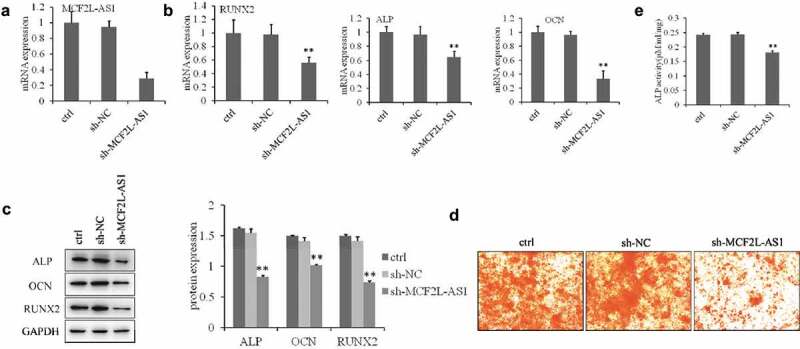
MCF2L-AS1 down-regulation inhibits osteogenic differentiation of rat BMSCs.
miR-33a is a direct target of lncRNA MCF2L-AS1
To elucidate the molecular mechanism by which lncRNA MCF2L-AS1 regulates osteogenic differentiation, predicted targets of lncRNA MCF2L-AS1 were analyzed using StarBase 2.0 (http://starbase.sysu.edu.cn/). As shown in Figure 3a, miR-33a was one of the targets of lncRNA MCF2L-AS1. To determine whether miR-33a is regulated by lncRNA MCF2L-AS1 directly, luciferase reporter constructs were constructed. The miR-33a mimics and lncRNA MCF2L-AS1 wild-type or mutant reporter were transferred into BMSC cells at the same time. The luciferase activity of lncRNA MCF2L-AS1 wild-type reporter decreased significantly; lncRNA MCF2L-AS1 was down-regulated by miR-33a (Figure 3b). miR-33a expression was up-regulated by treatment with sh-MCF2L-AS1 (Figure 3c). Furthermore, miR-33a could also suppress the lncRNA MCF2L-AS1 expression (Figure 3d). Taken together, these data showed that MCF2L-AS1 directly interacts with miR-33a.
Figure 3.
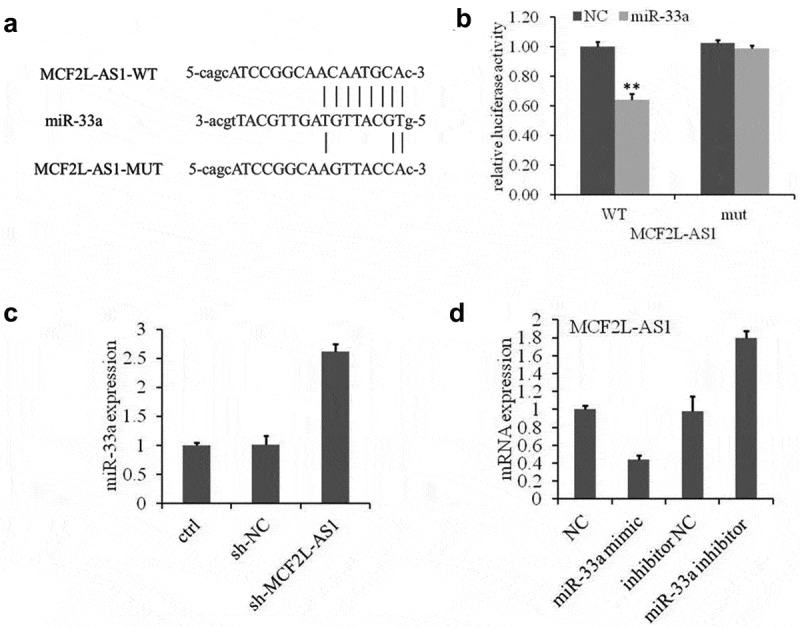
miR-33a is the direct target of lncRNA MCF2L-AS1.
miR-33a is a negative regulator of osteoblast differentiation
To elucidate the biological function of miR-33a in BMSCs, we examined the effects of miR-33a on osteogenesis by transfecting miRNA mimics into BMSCs. qRT-PCR showed that miR-33a expression was significantly higher in the miR-33a mimic group than in the other groups (Figure 4a). We measured Runx2, ALP, and OCN expression by qRT-PCR and western blot and found that the expression of Runx2, ALP, and OCN was down-regulated in the miR-33a inhibitor group compared with the osteogenic group (Figure 4b,c). Further, alizarin red S staining confirmed that miR-33a significantly repressed calcium nodule formation after 14 days of osteogenic differentiation (Figure 4d).
Figure 4.
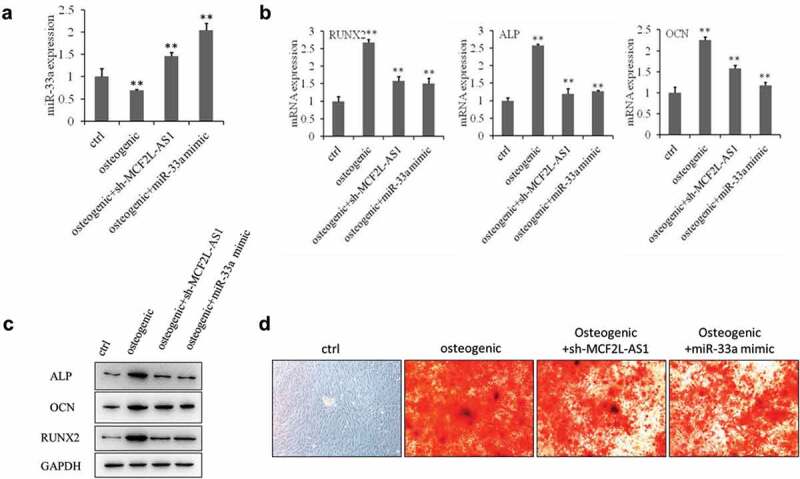
miR-33a is a negative regulator of osteoblast differentiation.
Discussion
The potential applications of BMSCs in tissue engineering, particularly of bone tissue, have attracted much attention in recent years. A thorough understanding of the molecular mechanisms associated with osteogenesis is essential to maintaining osteogenic capacity and realizing the therapeutic potential of BMSCs [15–17]. A number of critical signaling pathways, including Wnt, TGF-β/BMP, and Hedgehog, are involved in regulating the lineage commitment of MSCs [18–20]. lncRNAs regulate diverse cellular processes, and many lncRNAs play important roles in regulating adipogenesis and osteogenesis of MSCs [21]. lnc-NTF3e5 could enhance the expression of Runx2, ALP, and OSX, and further enhanced osteogenic differentiation potential of MSCs [22]. However, the role of lncRNAs in the lineage commitment of MSCs remains largely unknown.
In this study, lncRNA microarray analysis showed that the lncRNA MCF2L-AS1 was up-regulated during osteogenic induction of BMSCs. Down-regulation of MCF2L-AS1 decreased expression of the osteogenesis-associated genes Runx2, OCN, and ALP. These data indicated that MCF2L-AS1 partially promoted osteogenic differentiation induced by sodium beta-glycerophosphate + dexamethasone + vitamin C in rat BMSCs. Further, lncRNA MCF2L-AS1 likely acts as a natural miRNA “sponge” or RNA decoy; lncRNAs modulate target miRNAs and thereby regulate downstream gene expression. Based on the ceRNA hypothesis, we speculated that lncRNA MCF2L-AS1 influences the targets of miR-33a. Our results indicated that linc-ROR interacted with miR-33a as a miRNA sponge and participated in miRNA-mediated osteogenesis. We found that miR-33a was negatively regulated by MCF2L-AS1 in BMSCs and was associated with the effect of MCF2L-AS1 on BMSCs. Runx2 is an essential transcription factor for osteogenesis and activation of osteogenic differentiation marker genes, and plays a vital role in bone formation and metabolism. Transcription of the osteoblast-related genes ALP and OCN were directly stimulated by Runx2 [23]. These genes were significantly downregulated in the lncRNA-MCF2L-AS1 knockdown group, indicating that lncRNA-MCF2L-AS1 promoted the osteogenesis of BMSCs.
To our knowledge, this study is the first to describe the role and molecular mechanism of lncRNA MCF2L-AS1 in BMSC differentiation. Our results revealed that lncRNA-microRNA interaction of lncRNA MCF2L-AS1 is essential for promoting osteogenic differentiation. We could conclude that lncRNA MCF2L-AS1 influences bone formation by miRNA-33a and Runx2 via the lncRNA MCF2L-AS1/miR-33a/Runx2 axis. Our results could provide a potential target for the treatment of bone injury and related diseases. We also identified other lncRNAs related to osteogenesis, suggesting the involvement of other genes in the proliferation and osteogenesis of BMSCs, which we will study in the future.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Coughlin TR, Romero-Moreno R, Mason DE, et al. Bone: a fertile soil for cancer metastasis. Curr Drug Targets. 2017;18:1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Park D, Park CW, Choi Y, et al. A novel small-molecule PPI inhibitor targeting integrin alphavbeta3-osteopontin interface blocks bone resorption in vitro and prevents bone loss in mice. Biomaterials. 2016;98:131–142. [DOI] [PubMed] [Google Scholar]
- [3].Yu X, Li Z, Wan Q, et al. Inhibition of JAK2/STAT3 signaling suppresses bone marrow stromal cells proliferation and osteogenic differentiation, and impairs bone defect healing. Biol Chem. 2018;399:1313–1323. [DOI] [PubMed] [Google Scholar]
- [4].Li W, Wang C, Zhang M, et al. Young and old adipocytes have differential influence on the development of osteoblasts. Obes Res Clin Pract. 2018;12:520–527. [DOI] [PubMed] [Google Scholar]
- [5].Ruan H, Xiao R, Jiang X, et al. Biofunctionalized self-assembly of peptide amphiphile induces the differentiation of bone marrow mesenchymal stem cells into neural cells. Mol Cell Biochem. 2018;450:199–207. [DOI] [PubMed] [Google Scholar]
- [6].Yang F, Yang L, Li Y, et al. Melatonin protects bone marrow mesenchymal stem cells against iron overload-induced aberrant differentiation and senescence. J Pineal Res. 2017;63:e12422. [DOI] [PubMed] [Google Scholar]
- [7].Abbuehl JP, Tatarova Z, Held W, et al. Long-term engraftment of primary bone marrow stromal cells repairs niche damage and improves hematopoietic stem cell transplantation. Cell Stem Cell. 2017;21:241–255 e246. [DOI] [PubMed] [Google Scholar]
- [8].Ma Y, Bu D, Long J, et al. LncRNA DSCAM-AS1 acts as a sponge of miR-137 to enhance Tamoxifen resistance in breast cancer. J Cell Physiol. 2018;234:2880–2894. [DOI] [PubMed] [Google Scholar]
- [9].Sun W, Ma M, Yu H, et al. Inhibition of lncRNA X inactivate-specific transcript ameliorates inflammatory pain by suppressing satellite glial cell activation and inflammation by acting as a sponge of miR-146a to inhibit Nav 1.7. J Cell Biochem. 2018;119:9888–9898. [DOI] [PubMed] [Google Scholar]
- [10].Feng L, Shi L, Lu YF, et al. Linc-ROR promotes osteogenic differentiation of mesenchymal stem cells by functioning as a competing endogenous RNA for miR-138 and miR-145. Mol Ther Nucleic Acids. 2018;11:345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Peng W, Deng W, Zhang J, et al. Long noncoding RNA ANCR suppresses bone formation of periodontal ligament stem cells via sponging miRNA-758. Biochem Biophys Res Commun. 2018;503:815–821. [DOI] [PubMed] [Google Scholar]
- [12].Wei B, Wei W, Zhao B, et al. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS One. 2017;12:e0169097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Xiao Y, Guo Q, Jiang TJ, et al. miR-483-3p regulates osteogenic differentiation of bone marrow mesenchymal stem cells by targeting STAT1. Mol Med Rep. 2019;20:4558–4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li K, Chen S, Cai P, et al. MiRNA-483-5p is involved in the pathogenesis of osteoporosis by promoting osteoclast differentiation. Mol Cell Probes. 2019;6:101479. [DOI] [PubMed] [Google Scholar]
- [15].Gong X, Yu W, Zhao H, et al. Skeletal site-specific effects of zoledronate on in vivo bone remodeling and in vitro BMSCs osteogenic activity. Sci Rep. 2017;7:36129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li C, Wei GJ, Xu L, et al. The involvement of senescence induced by the telomere shortness in the decline of osteogenic differentiation in BMSCs. Eur Rev Med Pharmacol Sci. 2017;21:1117–1124. [PubMed] [Google Scholar]
- [17].Man Z, Li T, Zhang L, et al. E7 peptide-functionalized Ti6Al4V alloy for BMSC enrichment in bone tissue engineering. Am J Transl Res. 2018;10:2480–2490. [PMC free article] [PubMed] [Google Scholar]
- [18].Han W, Yu Y, Liu XY.. Local signals in stem cell-based bone marrow regeneration. Cell Res. 2006;16:189–195. [DOI] [PubMed] [Google Scholar]
- [19].Jing H, Su X, Gao B, et al. Epigenetic inhibition of Wnt pathway suppresses osteogenic differentiation of BMSCs during osteoporosis. Cell Death Dis. 2018;9:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang YD, Zhao SC, Zhu ZS, et al. Cx43- and smad-mediated TGF-beta/BMP signaling pathway promotes cartilage differentiation of bone marrow mesenchymal stem cells and inhibits osteoblast differentiation. Cell Physiol Biochem. 2017;42:1277–1293. [DOI] [PubMed] [Google Scholar]
- [21].Li Z, Jin C, Chen S, et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem. 2017;433:51–60. [DOI] [PubMed] [Google Scholar]
- [22].Lim TY, Wang W, Shi Z, et al. Human bone marrow-derived mesenchymal stem cells and osteoblast differentiation on titanium with surface-grafted chitosan and immobilized bone morphogenetic protein-2. J Mater Sci Mater Med. 2009;20:1–10. [DOI] [PubMed] [Google Scholar]
- [23].Kuo SW, Rimando MG, Liu YS, et al. Intermittent administration of parathyroid hormone 1-34 enhances osteogenesis of human mesenchymal stem cells by regulating protein kinase cdelta. Int J Mol Sci. 2017;18:2221. [DOI] [PMC free article] [PubMed] [Google Scholar]


