ABSTRACT
Peritoneal metastasis develops in more than half of patients with gastric cancer but influencing factors are poorly characterized. Exosomes are increasingly recognized as a new mediator in cancer directional metastasis through the transfer of nucleic acids or proteins to neighboring or distant cells. The role of exosomes in peritoneal metastasis and whether it could establish pre-metastatic milieu are largely unknown. Here, we assessed the migration of gastric cancer (GC) cells and identified that PKH26-labeled exosomes from GC cells can be ingested by peritoneal mesothelial cells (MCs). Additionally, miRNA (miR-106a) that highly enriched in GC-derived exosomes (GC-exos) and essential for destroying the mesothelial barrier was demonstrated through the observation of the injury of the MCs including migratory enhancement and imbalance of apoptosis and proliferation. Moreover, either stimulating miR-106a or treatment with GC-exos could inhibit the expression of Smad7, accompanied by the concurrent elevated α-SMA and fibronectin in MCs. Silencing of miR-106a abolished GC-exos-induced gene expression in MCs. The MCs regain the viability, apoptosis reduction and Smad7 expression after rescue experiment conducted in miR-106a-enriched GC-exos. Xenograft model suggested that exosomal miR-106a had a potential to promote tumor growth through targeting Smad7. Collectively, we revealed that the delivery of miR-106a from GC-exos plays a crucial role in gastric cancer peritoneal metastasis.
Abbreviations: MiR-106a: microRNA-106a; Smad7: small mothers against decapentaplegic 7; GC: gastric cancer; MCs: mesothelial cells; Exos: exosomes; HG: high-differentiated gastric cancer cells; LG: low-differentiated gastric cancer cells.
KEYWORDS: Gastric cancer, exosomes, microRNA, smad7, metastasis
Introduction
Advanced gastric cancer remains the leading cause of cancer mortality worldwide, largely due to metastasis and development of chemotherapeutic resistance, although the annual endoscopic screening saved some of the early populations [1]. Diffuse-subtype and hyper-mutated intestinal-subtype are the most common molecular subtypes of gastric cancer and most patients present with late-stage disease and poor survival prospects due to the occurrence of various types of metastasis [2]. In China, it is statistically stated that there are an estimated 679, 100 new cases and 498, 000 mortality rate among all malignant tumors according to the latest statistics in 2015 [3]. It is reported that more than 80% of gastric tumors are metastatic at initial diagnosis [4], the treatment of surgery showed only a 5-year overall survival (OS) rates of 21.35% [5], the use of postoperative chemotherapy can only improve the overall survival rates to 26%, and alarming 50% of this mortality is associated with peritoneal metastasis even after the standard radical resection [6,7]. Advanced non-metastatic gastric cancer could acquire a better prognosis with surgical resection and perioperative chemotherapy or chemo-radiotherapy [8], efforts to prolong survival in metastatic gastric cancer still show little improvement. Therefore, it is imperative to establish a strategy to explicate the causes of peritoneal metastasis and clarify the possible mechanism.
Recently, it has been demonstrated that exosomes are expendable for cancer organotropic metastasis but is deduced an essential step in the process of peritoneal metastasis of gastric cancer [9]. Exosomes play an important role in the intercellular communication by transferring bioinformatics from parent cells to receptor cells [10]. Exosomes are defined as nano-/micron-sized membrane-containing derivatives (30–100 nm) of the endosomal system and correspond to the intraluminal vesicles of multivesicular bodies (MVBs), upon which the plasma membrane is fused with, followed by the loading of the “cargo”, are released into the extracellular environment. Early in 1983, exosomes had initially been reported, research on the nano-sized extracellular vehicles (EVs) did not start only until the discovery that small EVs transport small RNAs, including microRNAs (miRNAs) [11]. Our previous studies have reported that miR-106a has ectopic expression during gastric carcinogenesis and development, especially for cancer metastasis [12,13]. However, it is not unfolded how gastric cancer cells linked to peritoneum and whether exosomes-transported miRNA has functions in this process.
It is known that gastric cancer peritoneal metastasis is a complicated process. Tumor cells that have metastatic ability must go through several steps before they can be implanted. According to the classical “seed and soil” theory [14], a combination of several factors lead to metastasis [15]. a. Gastric cancer cells infiltrate aggressively and come adrift from the serous layer of the gastric wall to form “seeds”. b. “Seeds” communicates with “soil” peritoneum. c. Intercellular communication mediates phenotypic changes of mesothelial cells. d. Mesothelial cells lift focally from the peritoneum with apoptosis. e. Pre-metastatic milieu is formed that is favorable to cancer cell colonization. It is inferred that intercellular communication between “seed” and “soil” is a key link for cancer cells metastasis. Perhaps exosomes could build a new bridge to the illumination of such information transmission. Exosomes promote tumor metastasis mainly through epithelial–-mesenchymal transition (EMT) [16]. Transforming growth factor beta (TGF-β) is one of the key molecules to induce EMT in tumor cells. Smad7 is a key inhibitor for TGF-β-induced signal. Exosomes, therefore, as a mediator, could be used to study the signal changes guided by miR-106a and Smad7, which is helpful to discover the new mechanism of peritoneal metastasis of gastric cancer.
In this study, we hypothesize that exosomes, secreted by primary gastric cancer cells could potentially transfer miRNA to peritoneal mesothelial cells. We assess the biological characteristics of exosomes from parent cells, analyze the differential expression of miR-106a in exosomes, detect the changes of mesenchymal markers and target gene Smad7 after the transportation of miR-106a by exosomes. By in vitro and in vivo studies, our work provides novel and important insights into how exosomes promote tumorigenesis and develop distant metastasis through altering the phenotype and gene expression of recipient cells.
Materials and methods
Cell lines and tissues
The human gastric cancer cells AGS, BGC-823, MKN-45, MKN-74, NCI-N87 and human immortalized gastric mucosal epithelial cell GES-1 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in complete medium and incubated at 37°C in 5% CO2 incubator. The culture medium was compounded by RPMI-1640 (Hyclone, South Logan, UT, USA), 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) and 1% double antibiotics (100 U/mL penicillin, 100 U/mL streptomycin, Hyclone, South Logan, UT, USA). The human immortalized peritoneal mesothelial cells HMrSV5 that purchased from Bena Culture Collection (Beijing, China) were cultured using DMEM medium (Hyclone, South Logan, UT, USA) containing the same ingredients and maintained in the same humidified atmosphere. Gastric cancer tissues were collected from General Hospital of Ningxia Medical University with the approval of local ethics committee. All the samples were subjected to pathological diagnosis and a matched group containing 40 cases of gastric cancer and 40 cases of adjacent non-tumor tissues (5 cm from the center of the cancer loci) was divided. These gastric cancer patients did not have a previous history of radiotherapy and chemotherapy.
Exosomes isolation
Exosomes were isolated with Ribo™ Exosome Isolation Reagent (Ribobio, Guangzhou, China). Briefly, cells were cultured for 72 h in RPMI-1640 supplemented with 10% exosome-deleted FBS, and then the supernatant conditioned media were collected and centrifuged at 2000 × g at 4°C for 30 min to remove floating cells, cellular debris and unwanted proteins. The cell supernatant was then transferred to a new tube and placed on ice until use. One-third volume of Ribo™ Exosome Isolation Reagent was added to the tube and pipette several times until the sample was completed mixed (the solution would be cloudy); then, the sample was kept in a refrigerator at 4°C for overnight. Next day the 2 mL mixed liquid was pipette and transferred to a centrifugal tube, followed by a centrifugation step of the mixture at 1500 × g at 4°C for 30 min, and a small part of exosomes was obtained by discarding the supernatant. Repeat such centrifugation step three to four times until all the mixture was transferred. Leaving the exosomes pellets undisturbed, the clear supernatant was carefully removed, and then the exosomes were eluted and resuspended in phosphate-buffered saline (PBS).
Transmission Electron microscopy (TEM)
Exosomes suspensions were dropped on the parafilm and the copper grids (400 meshes, Pacific Grid-Tech, San Francisco, CA, USA) were suspended on the droplets for 3 min. The copper grids were coated with carbon membrane as absorbent. This material has a good adsorption effect on exosomes. After removing the unnecessary solutions with filter paper, the copper grids were re-suspended on 1% phosphotungstic acid (Alfa Aesar, Heysham, UK) for 3 min to negative staining. The abundant liquids were removed again, and the samples were left for drying. Grids were observed under Tecnai G2 F20 transmission Electron microscope (FEI, Hillsboro, OR, USA) at rated voltage 120kV.
Nanoparticle tracking analysis (NanoSight)
Vesicles that extracted from gastric cancer cells were analyzed by nanoparticle tracking analysis (NTA 3.2 Dev Build 3.2.16), using the NanoSight LM10 system (Malvern, Great Malvern, UK) to produce high-resolution results for particle size distribution and concentration. Particles were tracked for 60 s using NTA software. Each sample was analyzed four times and the counts were averaged.
PKH26 labeling and confocal laser scanning microscopy (LSCM)
To track exosomes internalization and determine whether HMrSV5 cells had an ability to assimilate the exosomes derived from gastric cancer cells, exosomes were fluorescently labeled with PKH26 Red Fluorescent Cell Linker Mini Kit (Sigma-Aldrich, St. Louis, MO, USA) according to the protocol recommended by the manufacturer with minor modifications. Briefly, 100 μL of exosomes was suspended in PBS solutions. A quantity of 0.5 μL Diluent C was mixed with 1 μL PBS-exos and 2 μL PKH26 separately. Immediately the stain solution was mixed with exosomes and incubated at room temperature for 5 min. By adding 1.5 mL BSA (0.5%), the labeling reaction was stopped, and the exosomes were eluted from the mixture by ultracentrifugation. The labeled exosomes were finally re-suspended in PBS. Repeat this step second time completely removed the unbound dyes and dyes dissolved in the solution. Then, the labeled exosomes were added to HMrSV5 cells and incubated at 37°C for 12 h, after which the cells were fixed with 4% formaldehyde and visualized under confocal laser scanning microscope. 4’,6-diamidino-2-phenylindole (DAPI) nuclear staining was used for comparative observation.
RNA isolation and quantitative real-time PCR (qRT-PCR)
qRT-PCR was used to detect the expression of miR-106a in cells and in exosomes, Smad7 and mesenchymal markers α-SMA and fibronectin. RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was generated from the RNA samples in order to act as the template for PCR amplification. For miRNA expression analysis, 1 μg of total RNA was reverse transcribed using the corresponding RT primer and Bestar™ qPCR RT kit (DBI® Bioscience, Ludwigshafen, Germany). PCR was performed on 1 μL of RT products by adding the miRNA forward primers, universal reverse primers and Bestar® SybrGreen qPCR Master Mix (DBI® Bioscience, Ludwigshafen, Germany). U6 snRNA was used for normalization. For mRNA analysis, 1 μg of total RNA was subjected to retro-transcription using the same reverse transcription kit without primers. qRT-PCR of Smad7, α-SMA and fibronectin were performed using SYBR Green qRT-PCR (DBI® Bioscience, Ludwigshafen, Germany) with their corresponding forward and reverse primers. Data were normalized to GAPDH. All samples were tested triplicate and uploaded on Agilent Stratagene Mx3000P fluorescence quantitative PCR instrument (Santa Clara, CA, USA). Primer sequences are listed in Table 1. Relative qualification was performed with the 2−ΔΔCt method.
Table 1.
Primers in this study.
| Name | Gene ID | Accession number | Accession number sequences (5“-3”) |
Product Length (bp) |
|---|---|---|---|---|
| hsa-miR-106a RT | 406,899 | NC_000023.11 | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCTACCTG | |
| hsa-miR-106a F | ACACTCCAGCTGGGAAAAGTGCTTACAGT | |||
| U6 F | 108,353,825 | NC_015438.3 | CTCGCTTCGGCAGCACA | |
| U6 R | AACGCTTCACGAATTTGCGT | |||
| All R | CTCAACTGGTGTCGTGGA | |||
| Smad7 F | 4092 | NC_000018.10 | TTCCTCCGCTGAAACAGGG | 116 |
| Smad7 R | CCTCCCAGTATGCCACCAC | |||
| α-SMA F | 59 | NC_000010.11 | TGTCCGATCTACTTTCCC | 106 |
| α-SMA R | GAGTTCTCACTTTCATCTGTT | |||
| Fibronectin F | 2335 | NC_000002.12 | CGGTGGCTGTCAGTCAAAG | 130 |
| Fibronectin R | AAACCTCGGCTTCCTCCATAA | |||
| GAPDH F | 2597 | NC_000012.12 | TGTTCGTCATGGGTGTGAAC | 154 |
| GAPDH R | ATGGCATGGACTGTGGTCAT | |||
| Smad7-F | 4092 | NC_000018.10 | GGGGTACCATGTTCAGGACCAAACGATCTGC | 1281 |
| Smad7-R | CCCTCGAGCTACCGGCTGTTGAAGATGACCT | |||
| Smad7-WT-F | 4092 | NC_000018.10 | CCGCTCGAGCTCGTATGATACTTCGACACTGTTC | |
| Smad7-WT-R | ATTTGCGGCCGCACATTTTAAAAATCGTTTAATGGAA | |||
| Smad7-MUT-F | 4092 | NC_000018.10 | AAATAAAGAAAAGATCGGTCGAGCTTTAATATAAATG | |
| Smad7-MUT-R | CATTTATATTAAAGCTCGACCGATCTTTTCTTTATTT |
Wound healing migration assay
The wound healing assay was used to assess the migration of gastric cancer cells. An artificial “wound” was created on a confluent cell monolayer (5 × 105/mL in 6-well plates) of AGS, BGC-823, MKN-45, MKN-74 and NCI-N87 cells. After 48 h of serum-free cultivation, cells were photographed under the MOTIC inverted microscope (Fujian, China). Five preset fields were calculated with Image-Pro Plus Software (Version 6.0; Media Cybernetics, Rockville, MD, USA). The relative mobility was calculated as a percentage of wound healing 48 h vs. time 0. In addition, the migratory ability of HMrSV5 cells after exosomes treatment was also evaluated by wound healing migration assay.
Transwell migration assay
The transwell assay was used to further evaluate the migratory ability of HMrSV5 cells. Briefly, HMrSV5 cells were incubated with AGS-exos for 24 h, followed by the trans-well assay. The treated cells were seeded into the upper chambers in serum-free RPMI-1640 medium. The lower chamber was filled with culture media containing 10% FBS. The cells that passed the chambers were fixed with 4% paraformaldehyde, stained with 0.1% crystal violet, and counted under a microscope (Olympus, Takachiho, Japan).
Plasmids and transfection
Smad7 overexpression plasmid was synthesized from Vipotion Biotechnology (Guangzhou, China). Generally speaking, the primers were designed and synthesized by Sangon Biotech (Shanghai, China) according to the sequence of Smad7 gene and pcDNA3.0 vector (Table 1). RNA extraction was conducted using Trizol method from AGS cells. Reverse transcription was carried out by Bestar qPCR RT Kit (DBI® Bioscience, Ludwigshafen, Germany), followed by the PCR amplification which was operated by Phanta®Super-Fidelity products (Vazyme, Jiangsu, China). After that, the PCR products were retrieved and subjected to enzyme digestion with pcDNA 3.0 vectors, in which XhoI and NotI (ThermoFisher Scientific, Waltham, MA, USA) restriction sites were selected. The recovered PCR products and the recovered vectors were linked together by T4 DNA ligase (ThermoFisher Scientific, Waltham, MA, USA). The products were transformed into a competent cell from Escherichia coli (E. coli) DH5α. The PCR amplification was used again to identify the bacterial suspension that acquired by shaking flask cultures. Recombinant plasmids were eluted with E. Z. N. A. ™ Plasmid Mini Kit I (Omega Bio-tek, Norcross, GA, USA) and finally identified by double enzyme digestion. Three identified plasmids were sequenced by Sangon Biotech (Shanghai, China) and blast in the NCBI database. The indicated cells were transfected with Smad7 plasmid using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA).
Software prediction and dual-luciferase reporter assay
Dual-Luciferase reporter assay was applied to identify the direct target of miR-106a. Smad7 wild (WT)/mutant (MUT) vectors were constructed. The primers were designed according to the Smad7 3’-UTR region and psiCHECK-2 vector (Table 1). The steps can be referred to the plasmid construction section. GES-1 was adjusted to a cell density at 8 × 105/mL, co-transfection was operated 24 h later by transfection of a mixture of 0.8 μg psiCHECK-2 Smad7 WT or MUT report plasmid together with 50 nM hsa-miR-106a mimic or negative control (Genepharma, Shanghai, China) using Lipofectamine™ 2000 reagent. Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) provided an efficient means of performing two reporter assays (firefly luminescence as a reporter gene and renilla luciferase reaction as an internal control). Continuous culture for 48 h, the fluorescence detection was carried out and the activity was read by Glomax bioluminescence detector (Promega, Madison, WI, USA).
Cell proliferation and cell viability assay (CCK8)
Cell counting kit-8 (CCK8, Dojindo, Kumamoto, Japan) allows sensitive colorimetric assays for the determination of cell viability in cell proliferation assays. HMrSV5 cells with the density of 1 × 105/mL were seed in triplicates in 96-well plates on day 0 and proliferation was measured after 24, 48, and 72 h, respectively. A quantity of 1 μg of exosomes that needed for incubation with recipient cells and 1 ng of vector DNA that prepared for transfection were completed 4 h before adding CCK8 detection solution. The relative optical density (OD) value was determined at 450 nm by microplate reader, and data were represented as mean ± standard deviation from at least three independent experiments.
Flow cytometry
Flow cytometry was used to detect apoptosis of HMrSV5 cells that were stimulated by exosomes from donor. Cells with the density of 5×105/mL were seeded into 6-well plates to culture for 48 h. Then, cells were collected, and the Annexin V-FITC/PI apoptosis detection kit (Multi sciences, Zhejiang, China) was used for apoptosis assay. After centrifugation for 5 min at 1000rpm, 200 μL binding buffer was added to tubes, and then 5 μL Annexin V-FITC and 10 μL propidium iodide (PI) were added to these cells. The samples were protected from light for 15 min before counting the stained cells by flow cytometry (BD FACSCalibur, Franklin Lakes, NJ, USA). Experiments were performed in triplicate.
Western blotting
To verify whether the exosomes can influence the gene expression of target cells, western blot was performed to detect Smad7 and mesenchymal related proteins. HMrSV5 cells were seeded into 6-well plate with the cell density at 5 × 105/mL, proteins were extracted after incubation for 48 h with cell lysis buffer (Beyotime Biotechnology, Shanghai, China). Total protein concentration was measured by Pierce™ BCA Protein Assay Kit (ThermoFisher Scientific, Waltham, MA, USA). Proteins were resolved by SDS-PAGE, transferred to polyvinylidene fluoride (PVDF, Merck Millipore, Billerica, MA, USA) membranes, blocked in 5% nonfat powdered milk in PBS-T (0.5% Tween-20) and probed with antibodies (CD9/CD81/TSG101, 1∶200, Santa Cruz Biotechnology, Dallas, TX, USA; Smad7, 1∶1000, Proteinteck, Rosemont, IL, USA; α-SMA, 1:2000, Cell signaling Technology, Danvers, MA, USA; Fibronectin, 1∶1000, Abcam, Cambridge, MA, USA). Proteins were detected using X-ray film and Immobilon™ Western Chemiluminescent HRP substrate (Merck Millipore, Billerica, MA, USA). The protein bands were analyzed by Image-Pro Plus Software (Version 6.0; Media Cybernetics, Rockville, MD, USA).
In situ hybridization
The expression of miR-106a was examined by in situ hybridization (ISH) in tissue samples. The ISH was performed in accordance with the procedure of the miRCURY LNA™ microRNA ISH Optimization Kit (Exiqon, Vedbaek, Demark). Briefly speaking, the paraffin specimens were cut into 10 μm. After routinely dewaxing and hydration, the miRNA was demasked using 15 μg/mL proteinase K at 37°C for 10 min. The double-DIG labeled locked nucleic acid (LNA) miR-106a probe was as follows: 5’-CTACCTGCACTGTAAGCACTTTT-3’. The hybridization was carried on with 40 nM of LNA™ probe at 55°C for 1 h in a hybridizer (Iris international, Tokyo, Japan). An anti-Digoxigenin-AP antibody (Roche, Basel, Switzerland) was used to recognize the DIG at 1∶800 for 60 min at room temperature. BCIP/NBT reaction solution (Roche Basel, Switzerland) and Neutral Red Staining Solution (Sigma-Aldrich, St. Louis, MO, USA) were used to colorimetric reaction and counterstaining, respectively. The positive miR-106a signal was seen as dark blue particles.
Xenograft model
SPF grade male 6-week, 18–22 g old BALB/c nude mice were purchased from Laboratory Animal Center of Southern Medical University (SCXK 2016–0041, Guangzhou, China) and randomly assigned to three groups (8 in each group). The animal experiments were proved by the Institute’s Animal Care and Use Committee. The parental BGC-823 cells which were stably transfected with Smad7 overexpression vector or blank pcDNA 3.0 vector was used to establish a subcutaneous xenotransplanted tumor model. After transfection, the cells were selected by 800 ug/mL of G418 culture to establish the cell lines stably expression of Smad7 protein. Then, resistant clones were chosen and expanded in RPMI-1640 culture medium. The treated cells (1 × 107/mL) were injected into the subcutaneous tissue of hind legs of nude mice, the whole process was manipulated in light chloral hydrate anesthetized states. Exosomes treatment (5 μg/mouse·day) was given through vena caudalis with the concentration quantified by Pierce™ BCA Protein Assay Kit (ThermoFisher, Waltham, MA, USA). Tumor growth and sizes of tumor nodules were measured and recorded every 4 days until the end of the experiment. Thirty-two days after implantation, the mice were sacrificed; their tumor development and Smad7 expression were determined by pathological method. V = ab^2/2. V: Tumor volume, a: long diameter, b: short diameter. Besides, another 6-week-old male BALB/c nude mice (Vital River Experimental Animal Technology Co., Ltd, Beijing, China, SCXK 2012–0001) (10 in each group) were inoculated the same density of BGC-823 cells that transfected with miR-106a antagomir (Ribobio, Guangzhou, China) at cartilago ensiformis to visualize metastatic lesions scattered over the abdomen. The mice were scarified 14 days later and the nodules were excised for further analyses.
H&E Staining and Immunohistochemistry
The tumor nodules were incised from xenotransplanted mice. The histological sections of tumor nodules were dyed by hematoxylin and eosin (H&E) staining to observe the morphological changes. The expression of Smad7 was explored by immunohistochemical staining. Nodules were fixed in 10% formalin and manufactured into paraformaldehyde-fixed paraffin-embedded (FFPE) sections. A rabbit anti-Smad7 polyclonal antibody (1∶300, Proteintech, Rosemont, IL, USA) was used in the EnVision™ Detection Systems, Peroxidase/DAB, Rabbit/Mouse (Dako, Glostrup, Denmark). Smad7 was stained as yellow in the cytoplasm or nucleus.
Statistical analysis
SPSS statistical software program (Version 17.0; IBM, Armonk, NY, USA) was chosen to conduct the data analysis. All data were expressed as the mean ± standard deviation. The comparison of two samples was performed using Student’s t test. Multiple samples were compared by One-way AVONA. Statistical charts were drawn by GraphPad Prism software (Version 5.0; San Diego, CA, USA). A P-value <0.05 was considered to be statistically significant.
Results
Identification of the characteristics of exosomes
To investigate the function of GC-derived exosomes during peritoneal metastasis, we first identified its characteristics and estimated its effect on peritoneal MCs. Before the identification, five different kinds of GC cells were cultured; the result of wound healing assay is shown in Figure 1A, B. Their relative migrated indexes were as follows: MKN-45 (36.26 ± 0.90), BGC-823 (45.78 ± 1.61), AGS (68.10 ± 2.23), MKN-74 (25.20 ± 0.42), and NCI-N87 (10.84 ± 1.25). The result was different among the groups (F = 690.52, P = 0.000). We selected AGS (high differentiation) and NCI-N87 (low differentiation) as parent cells for the following experiments based on their maximum and minimum mobility.
Figure 1.
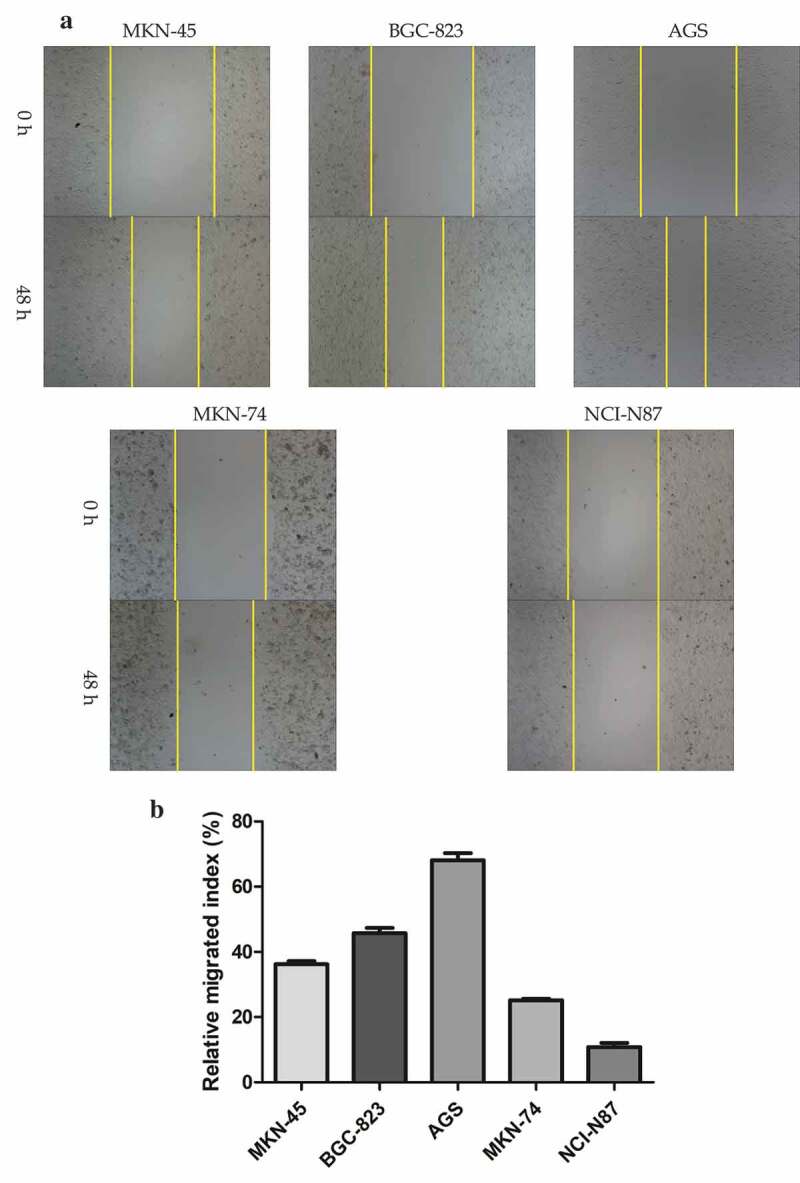
Identification of gastric cancer-derived exosomes. A. The basic migratory ability was assessed on five gastric cancer cell lines with different differentiated degrees using wound healing migration assay. AGS cells had the strongest migratory ability, followed by BGC-823, MKN-45 and MKN-74, NCI-N87 cells was the weakest. B. The relative migrated rate was counted out and significant differences among the five groups were observed (P < 0.001, as shown by Student’s t-test). C. Electron microscopy images of exosomes. The exosomes were characterized as a typical “cup-mouth” structure. D. Western blotting analysis of the exosomes marker proteins. Vesicles isolated from gastric cancer cells were positive for the exosomes markers CD9, CD81 and TSG101. Whereas equal amounts of proteins obtained from the cells were immunoblotted weaker. Galenxin was used as an internal reference. E. Nanosight graph showed that the majority of exosomes were at 97.5 nm (X = particle size, Y = count/mL).
Figure 1.
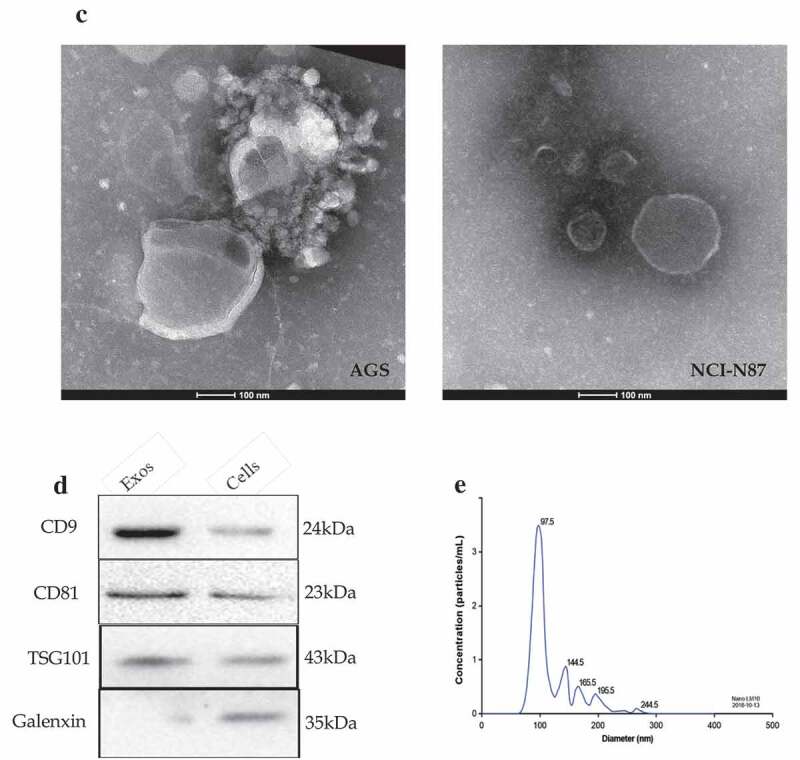
(Continued).
The exosomes from AGS and NCI-N87 cells were characterized by transmission electron microscope which is described in Figure 1C. The exosomes had a typical “cup-mouth” structure, and took on a round and ovoid shape that were wrapped in a bilayer membrane. The most widely accepted tetraspanin markers of exosomes, CD9, CD81 and TSG101, could be detected in exosomes by immunoblotting, while in the primary gastric cancer cell lysates, the expression of these markers was extremely weak. The calnexin was used as a negative control which was confirmed absent in exosomes but present in cells (Figure 1D). NTA system was used to measure the average size and estimate the number/mL of isolated nanoparticles from GC cells. The size distribution of nanoparticles isolated from AGS cell is demonstrated in Figure 1E. The curve demonstrated that the average number of nanoparticles/mL was 1.2e+008 ± 0.00e+000, exosomes showed a peak at 97.5 +/− 0.0 nm, mean at 109.6 ± 0.0 nm. Judging from these results, GC-secreted exosomes from the cell culture medium can be recognized to have been successfully extracted.
Internalization of GC-derived exosomes
To determine whether a particular selected miRNA species was over-represented in secreted exosomes, and confirm that the effect on recipient cell was derived from miRNA in exosomes rather than in cells, we examined the expression of exosomal miR-106a in different GC cells, and detect the expression of miR-106a in MCs to confirm GC-derived exosomes transfer miR-106a from donor cells to recipient cells. qRT-PCR result showed that the expression of exosomal miR-106a in five GC cells was different (F = 271.41, P = 0.000, Figure 2A), AGS-exos harbored the highest miR-106a (3.17 ± 0.10), whereas, NCI-N87-exos was the lowest (1.04 ± 0.11). The miR-106a expression in HMrSV5 co-cultured with AGS- or NCI-N87-exos was different (F = 121.07, P = 0.000, Figure 2B), AGS-exos transferred higher level of miR-106a to HMrSV5 (2.53 ± 0.18 vs. GES-1). In addition, we also found that the miR-106a expression in AGS-exos was much higher than that in the cells, the relative expression was 2.22 ± 0.19 (t = −5.14, P = 0.007), whereas, its expression for NCI-N87 cells was opposite (0.50 ± 0.02, t = 10.02, P = 0.001) (Figure 2C, D), suggesting that miR-106a was more enriched in the exosomes secreted from AGS cell. The relatively high abundance of miR-106a in exosomes and its low level in cells indicated that miR-106a was very selectively packaged and secreted via exosomes. Furthermore, we compared the expression of exosomal miR-106a from these two cells and obtained that the miR-106a in NCI-N87 cell was significantly lower than that in AGS cell (t = 25.03, P = 0.000, Figure 2E). The result indicated that exosomes secreted from AGS cell often packaged more miR-106a molecules.
Figure 2.
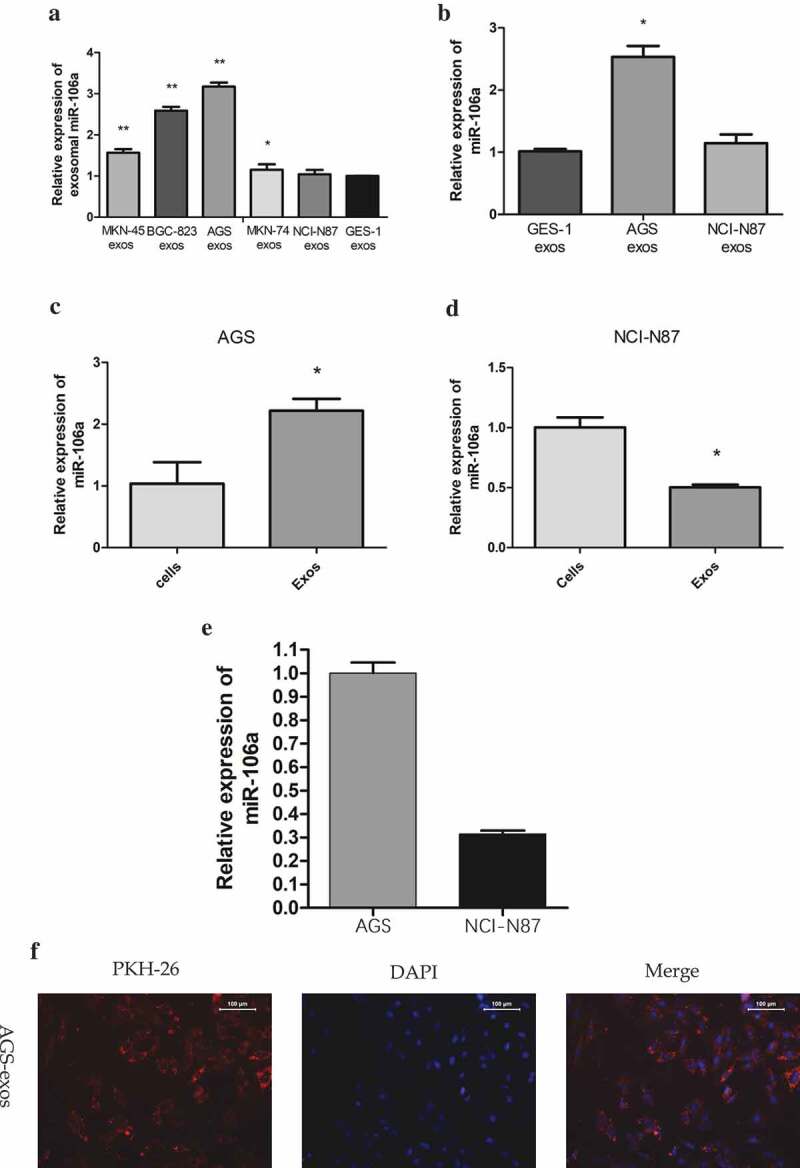
Quantitative analysis of miR-106a expression and the internalization of gastric cancer-derived exosomes. A. qRT-PCR detection of miR-106a expression in exosomes derived from five gastric cancer cells and GES-1 cells. GES-1 cells-derived exosomes were used as the negative control. **P < 0.001, *P < 0.05. B. qRT-PCR detection of miR-106a expression in HMrSV5 co-cultured with AGS/NCI-N87-derived exosomes and GES-1-derived exosomes (negative control). *P < 0.001. C. and D. qRT-PCR for determination of the relative expression of miR-106a in cells and in exosomes from AGS and NCI-N87 cells. *P < 0.01. E. Exosomal miR-106a expression in NCI-N87 and AGS cells. MiR-106a was mainly concentrated in the exosomes of AGS cells. Each experiment was repeated at least three times. F. Confocal laser scanning microscopy analysis of PKH26-labeled GC-derived exosomes (red) taken up by targeted peritoneal mesothelial cells after 12 h of co-culture. AGS cell was applied as donor cells and HMrSV5 cell was used as a recipient. A representative confocal microscopy image confirmed the internalization of exosomes within the cellular compartment. Left: PKH26-labeled exosomes (red fluorescence). Middle: DAPI-labeled cell nuclei (blue fluorescence). Right: Merge.
To justify whether exosomes could be taken up by target cells, we labeled GC derived-exosomes with PKH26, a red fluorescent dye, and then added the labeled exosomes to HMrSV5 cells. With long aliphatic tails, PKH26 dye could incorporate into the lipid membrane of exosomes [17]. From figure 2F, we found that HMrSV5 cells acquired positive PKH26 signals compared with control which presented no significant fluorescence at the same observation field. Besides, the fluorescence signals mainly dispersed on the cell cytoplasm. The observations suggested that the GC-derived exosomes could be internalized by HMrSV5 cells at metastatic sites.
Identification of Smad7 as adirect target of miR-106a
To gain insight into how exosome-transported miR-106a exerts its function in target cells during peritoneal metastasis, we used TargetScan, miRanda, DIANA-T to predict target genes of miR-106a. Smad7, which had a conservative site that could be complementary to miR-106a “seed” sequence, was identified (Figure 3A). Dual-luciferase reporter gene assay ascertained the direct combination of miR-106a and Smad7. From Figure 3B, we can see the luciferase ratio of Smad7-WT/miR-106a mimic was lower than Smad7-WT/miR-106a NC (falling by 30.82%; t = 5.54, P = 0.005), suggesting that Smad7 was significantly suppressed by miR-106a at post-transcriptional level. Whereas there was no significant difference between Smad7-MUT/miR-106a mimic and Smad7-MUT/miR-106a NC (t = 2.39, P = 0.075). Luciferase activity cannot be influenced when the binding site of Smad7 was mutated. qRT-PCR and western blotting revealed that the enhanced miR-106a could suppress both the expression of Smad7 mRNA level and protein level, but the Smad7 returned to its high expression when miR-106a inhibitor was arranged (Figure 3C. D). To further affirm their correlation, as illustrated in Figure 3E. F, the relative expression of miR-106a in gastric cancer was 2.28 ± 1.09 (t = −8.45, P = 0.000), corresponding to 0.65 ± 0.81 (t = 6.44, P = 0.000) for Smad7, and the positive expression of miR-106a was located in cancer tissues, but Smad7 was often strongly expressed in non-tumor tissues. There was a negative correlation between them (r = −0.056, P = 0.731, Figure 3G). The expression profile of miR-106a was illustrated in Figure 3H. The correlation between miR-106a/Smad7 expressions and the clinicopathological features of GC patients was analyzed in Table 2.
Figure 3.
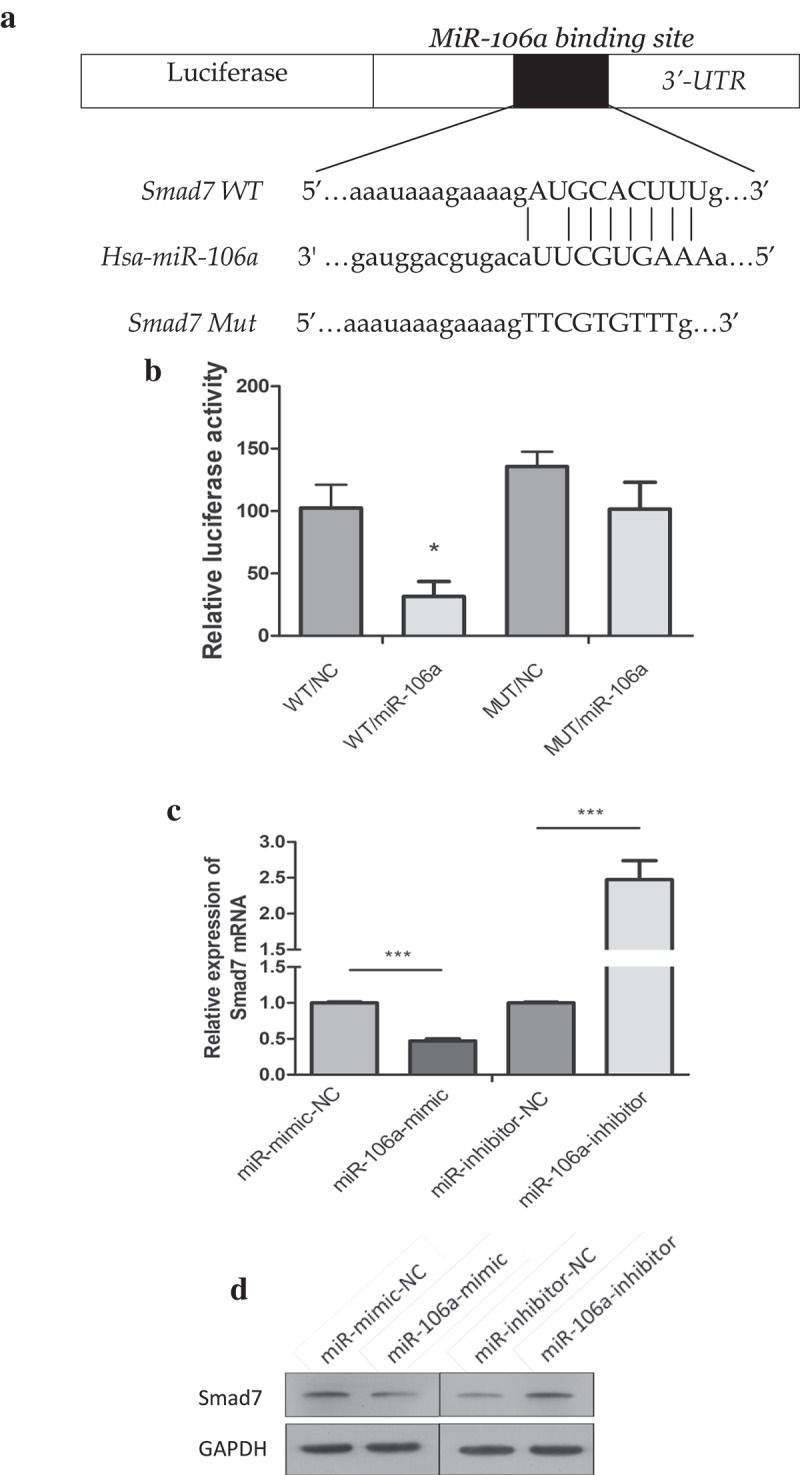
Correlation between miR-106a and Smad7. A. The wild type and mutant type of Smad7 3’-UTR regions were shown with the miR-106a sequence. B. Luciferase assay for detection of the direct regulation of miR-106a and Smad7. *P < 0.01. C. qRT-PCR detection of Smad7 mRNA in AGS cells transfected with miR-106a-mimic, mimic-NC, miR-106a-inhibitor and inhibitor-NC. ***P < 0.001. D. Western blot analysis for the protein level of Smad7. E. qRT-PCR detection of miR-106a or Smad7 in matched gastric cancer tissues and normal tissues. ***P < 0.001. F. The expression of miR-106a and Smad7 in gastric cancer and adjacent non-tumor tissues. The representative graphs of in situ hybridization and immunohistochemistry. G. The negative correlation between miR-106a and Smad7 in gastric cancer tissues. Expression values were expressed in 2−ΔΔCt. H. The expression trends of miR-106a in gastric cancer tissues. The normalized expression values were expression in log22−ΔΔCt.
Figure 3.
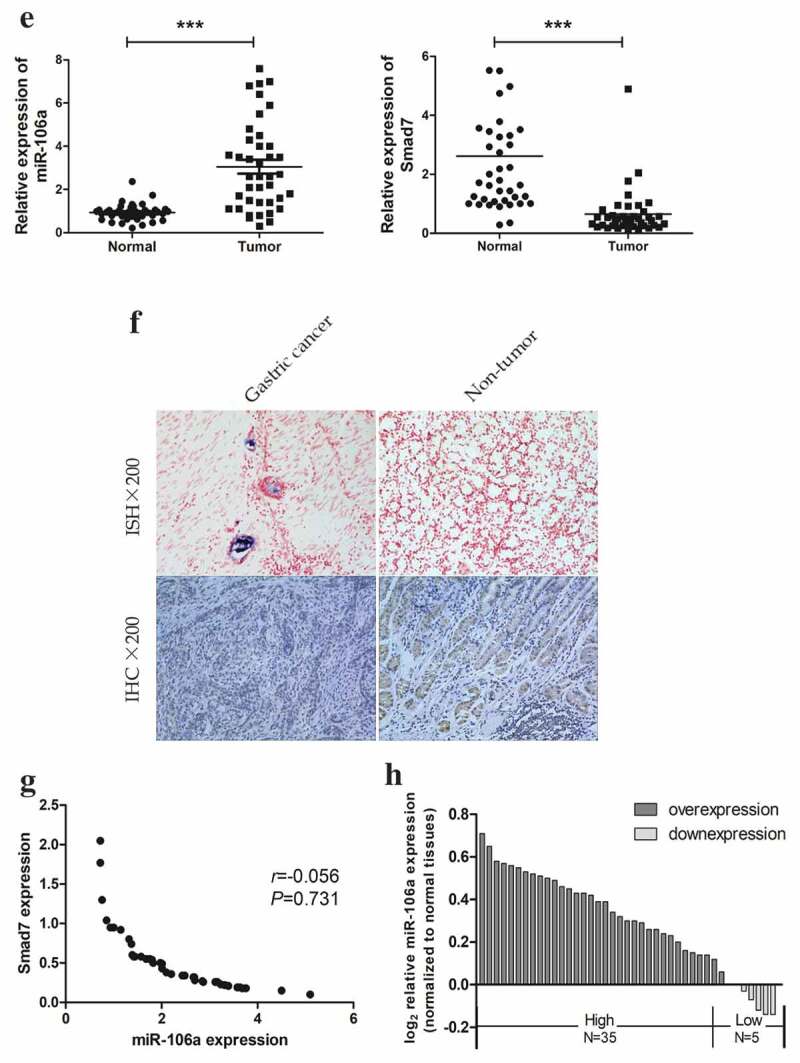
(Continued).
Table 2.
The correlation between miR-106a and Smad7 expression levels and the clinic pathological features of gastric cancer.
| miR-106a |
Smad7 |
||||
|---|---|---|---|---|---|
| Characteristics | Number | Fold change ± SD | P-value | Fold change ± SD | P-value |
| Sex | |||||
| Male | 22 | 2.5118 ± 1.27580 | 0.135 | 0.7862 ± 1.02471 | 0.255 |
| Female | 18 | 1.9917 ± 0.74372 | 0.4909 ± 0.38040 | ||
| Age (years) | |||||
| ≥60 | 23 | 2.4683 ± 1.08208 | 0.202 | 0.6995 ± 0.98368 | 0.680 |
| <60 | 17 | 2.0200 ± 1.07642 | 0.5910 ± 0.49821 | ||
| Tumor site | |||||
| Cardia | 7 | 2.4686 ± 1.20333 | 0.599 | 0.6204 ± 0.35941 | 0.971 |
| Body | 5 | 2.6400 ± 1.00055 | 0.7324 ± 0.80316 | ||
| Autumn | 28 | 2.1654 ± 1.09385 | 0.6474 ± 0.90220 | ||
| Size (cm) | |||||
| >5 | 9 | 3.3433 ± 1.05711 | 0.001# | 0.3257 ± 0.17514 | 0.014* |
| 3-5 | 26 | 2.0335 ± 0.94325 | 0.5908 ± 0.48357 | ||
| ≤3 | 5 | 1.6300 ± 0.55032 | 1.5693 ± 1.87072 | ||
| Histology grade | |||||
| Well | 5 | 1.1440 ± 0.33366 | 0.011* | 0.3842 ± 0.13025 | 0.432 |
| Moderate + poor | 35 | 2.4397 ± 1.06374 | 0.6918 ± 0.85567 | ||
| Lymph node | |||||
| Yes | 24 | 2.6200 ± 1.20633 | 0.013* | 0.7118 ± 0.98777 | 0.580 |
| No | 16 | 1.7644 ± 0.61991 | 0.5653 ± 0.42512 | ||
| Serosal invasion | |||||
| Yes | 27 | 2.5885 ± 1.13914 | 0.008# | 0.6977 ± 0.93522 | 0.621 |
| No | 13 | 1.6323 ± 0.61242 | 0.5608 ± 0.45211 | ||
| TNM stage | |||||
| Ⅰ + Ⅱ | 9 | 1.5900 ± 1.00169 | 0.029* | 1.2937 ± 1.50424 | 0.005# |
| Ⅲ + Ⅳ | 31 | 2.4774 ± 1.04471 | 0.4675 ± 0.28795 |
*P < 0.05, #P < 0.01.
GC-derived exosomes influence the phenotype and gene expression of MCs
MCs are considered as the first defense barrier of peritoneum and this barrier would be destroyed when GC cells are ready for implantation [15]. In order to determine whether GC-derived exosomes could destroy the mesothelial barrier, a series of functional assay in HMrSV5 cells was performed. CCK8 assay suggested that exosomes induced HMrSV5 cell damage in a time-dependent manner. Compared with HG-exos+NC, LG-exos decreased the proliferation of HMrSV5 cells significantly, whose trend was similar to HG-exos+miR-106a (Figure 4A).
Figure 4.
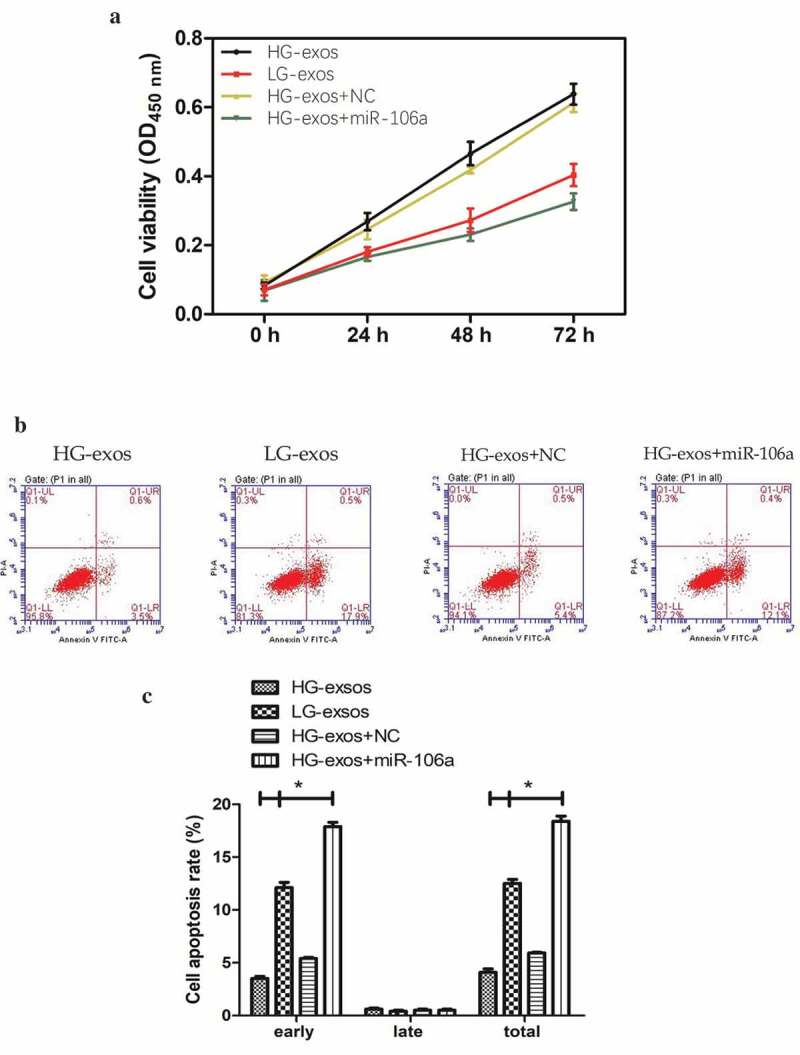
Phenotypical changes and gene expression of peritoneal mesothelial cells were analyzed in vitro. A. CCK8 assay for determination of HMrSV5 cells growth at indicated times. B. Flow cytometry for analysis of HMrSV5 cells apoptosis. Right lower quadrant: Annexin-positive/PI-negative, early apoptotic cells. Right upper quadrant: Annexin-positive/PI-positive, late apoptotic cells. Images indicated that LG-exos induced-apoptosis focus on early stage. C. The apoptosis rate of HMrSV5 cells. Compared with HG-exos+NC, cell apoptosis rate increased significantly in miR-106a group with the percentage at 12.5%, followed by LG-exos with the percentage at 6.6%, while the rate decreased in HG-exos group at −1.8%, *P < 0.001. D. HMrSV5 cells migratory ability was investigated by wound healing assay. E. Relative breadth was indicated as mean ± SD, *P < 0.001 compared with NC, as shown by the variance analysis. F. HMrSV5 cells migration was detected by transwell assay. G. The cell number from transwell assay, *P < 0.001. H. Exosomes and miR-106a mimic were added to HMrSV5 cells and the expression of Smad7, α-SMA and fibronectin were detected by qRT-PCR. Data were expressed as means ± SD. *P < 0.05, **P < 0.001. I. Western blotting analysis of the protein trend of these preparations in HMrSV5 cells. The panel above was representative of results which was repeated three times. These proteins showed the same trend as that the RNA level.
To further attest the destruction of MCs by exosomes, we conducted flow cytometry to decide the extent of apoptotic cells. As shown in Figure 4B, C, Annexin V-PI double staining exhibited that the cell apoptosis rate increased significantly in HG-exos+miR-106a group, and LG-exos can also promote the apoptosis of HMrSV5 cells, which was mainly manifested in the early apoptosis, HG-exos, however, did not have this function. The difference was statistically significant for early apoptosis (F = 1129.08, P = 0.000) and total apoptosis (F = 127.94, P = 0.000), but not for late apoptosis. The apoptosis rates of HMrSV5 cells exposed to HG-exos, LG-exos and HG-exos+miR-106a increased by −1.8%, 6.6% and 12.5%, respectively. These results indicated that exosomes purified from LG cells are more likely to exert the pro-apoptotic effects on HMrSV5 cells.
To make it easier to explore whether the potential role of exosomes-mediated transfer of miR-106a in the phenotypic changes of peritoneal MCs is involved in the formation of pre-metastastic microenvironment, we next validate the migratory ability of HMrSV5 cells in the presence of exosomes by wound healing assay. As illustrated in Figure 4D-G, the number of HMrSV5 migrating cells under the transfection of miR-106a mimic, LG-exos, HG-exos and NC was as follows: 36.68 ± 2.96, 55.32 ± 2.24, 21.34 ± 1.68 and 22.51 ± 2.00. The relative migrated breadth was different (F = 146.36, P = 0.000). The transwell assay confirmed the migration of HMrSV5 cells was increased with exosomes incubation (t = −8.54, P = 0.000).
Although we speculate that these changes above may be a preparation for better acceptance of tumor cells to be planted on MCs, the mechanism is still unclear. It should be noted that MCs need to lose apicobasal polarity, separate from each other, and detach from the peritoneal surface during the process of implantation [18,19]. Mesothelial-to-mesenchymal transition (MMT) may be acted as a pathway that makes the MCs to undergo these changes which finally lead to peritoneal function decline [19]. It is necessary to detect MMT-related markers to confirm whether MMT has appeared in this process. Mesenchymal indicator α-SMA and stromal components fibronectin together with Smad7 were all evaluated in Figure 4H, I. qRT-PCR results showed that the relative expression of Smad7 decreased, meanwhile, α-SMA and fibronectin increased to a great extent by LG-exos treatment but to a little extent by HG-exos. The difference was significant (FSmad7 = 460.75, FFibronectin = 502.60, Fα-SMA = 785.82, Pall = 0.000). Western blotting demonstrated that Smad7, α-SMA and fibronectin exhibited the same tendency as that of the RNA level. On the whole, these results hinted that miR-106a transported by GC-derived exosomes could induce MMT of HMrSV5 cells via targeting Smad7.
Figure 4.
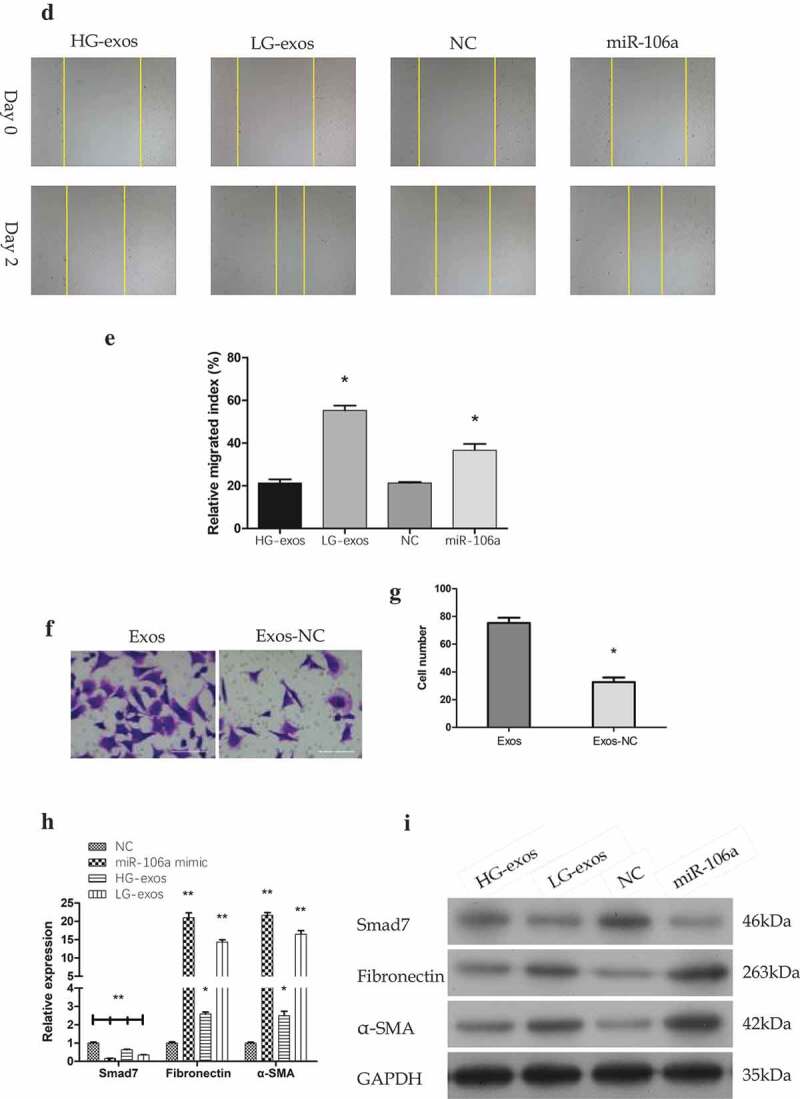
(Continued).
Rescue experiment confirmed the direct regulation of exosomal miR-106a on Smad7 in HMrSV5 cells
To further understand the effect of exosomes-transported miR-106a on MCs, we carried out rescue experiments to observe whether the cell phenotype is reversed and whether the gene expression is changed.
Cell treatment #1: LG-exo+NC, HG-exos, LG-exos, LG-exo+miR-106a inhibitor. As shown in Figure 5A, compared with LG-exos+NC, the relative value of miR-106a was 0.28 ± 0.01, 1.04 ± 0.02, 0.10 ± 0.01; Smad7 was 5.01 ± 0.18, 1.12 ± 0.08, 3.25 ± 0.10; Fibronectin was 0.27 ± 0.01, 1.06 ± 0.03, 0.54 ± 0.20, α-SMA was 0.31 ± 0.01, 1.01 ± 0.06, 0.62 ± 0.03. There was a significant difference among the groups (FmiR-106a = 559.88, FSmad7 = 804.54, FFibronectin = 356.92, Fα-SMA = 112.12, Pall = 0.000). The protein level exhibited the similar changes as the mRNA data (Figure 5B). Rescued experiments showed that when the inhibitor was given in LG-exos, the expression of miR-106a decreased, followed by the increase of Smad7 and the decrease of MMT markers.
Figure 5.
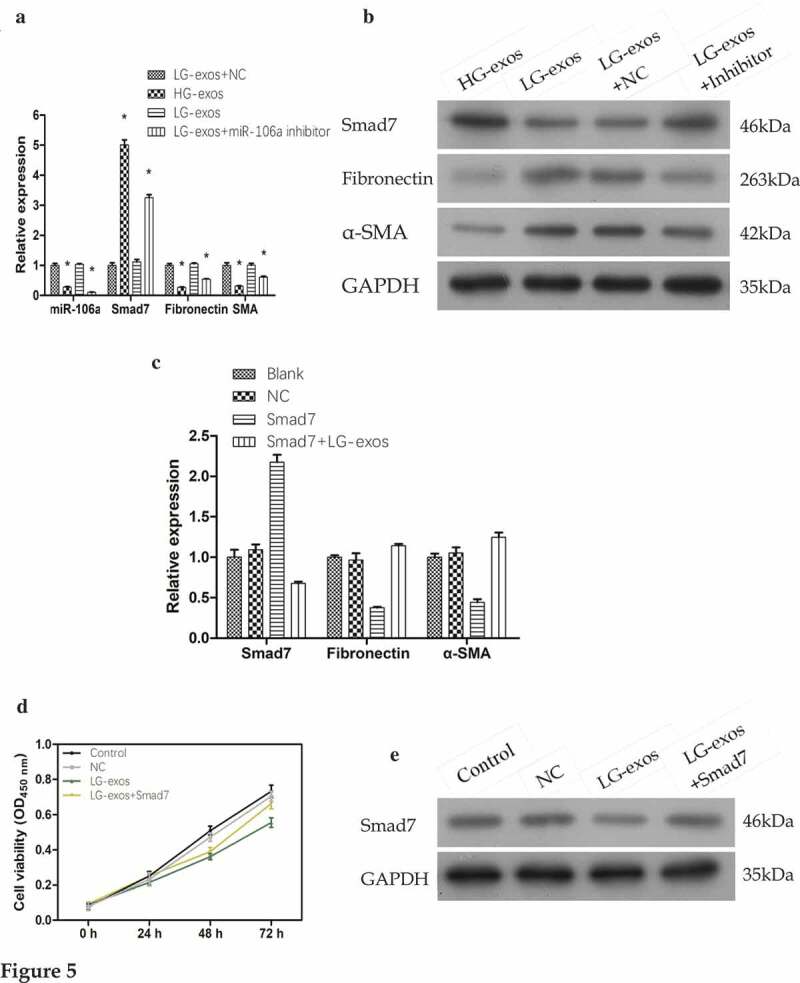
Rescue experiments for validating the gene expression and phenotypical changes of mesothelial cells. A. and B. Exosomes and miR-106a inhibitor were added to HMrSV5 cells and the expression of Smad7, α-SMA and fibronectin were detected by qRT-PCR and western blotting. Data were expressed as means ± SD. *P < 0.001. C. Smad7 overexpression plasmid was constructed into HMrSV5 cells and its role on MMT markers were validated by qRT-PCR. The constructed plasmid validly raised the expression of Smad7 in HMrSV5 cells. However, its expression was suppressed under the treatment of LG-exos. Moreover, the MMT markers in HMrSV5 cells were reversed. D. Cell proliferation from CCK8 assay showed that the inhibition of exosomes on HMrSV5 cells can be reversed by Smad7. E. The expression of Smad7 in HMrSV5 cells was checked by western blotting.
Cell treatment #2: Blank, NC, Smad7, LG-exos+Smad7. qRT-PCR results shown in Figure 5C was as follows: Smad7 (Smad7: 2.17 ± 0.10, Smad7+ LG-exos: 0.68 ± 0.02), fibronectin (Smad7: 0.38 ± 0.01, Smad7+ LG-exos: 1.14 ± 0.02), α-SMA (Smad7: 0.44 ± 0.04, Smad7+ LG-exos: 1.25 ± 0.06). The difference was significant (FSmad7 = 240.88, FFibronectin = 170.70, Fα-SMA = 124.53, Pall = 0.000). After using the overexpression plasmid of Smad7, its high expression was successfully detected when compared to blank and NC group, which suggested that the constructed vector was valid. Based on this, LG-exos could partially reverse the high expression of Smad7 in HMrSV5 cells so that fibronectin and α-SMA began to elevate.
Cell treatment #3: Control, NC, LG-exos, LG-exos+Smad7. CCK8 assay in Figure 5D manifested that the cell viability of HMrSV5 cells was inhibited after LG-exos treatment. However, when the overexpression plasmid of Smad7 acted, the inhibitory effect of exosomes on HMrSV5 cells proliferation was partly reversed. With the phenotype of HMrSV5 cells changed, the expression of Smad7 changed accordingly. Western blotting detection in Figure 5E displayed the Smad7 expression under the treatment.
Exosomal miR-106a influence GC growth by regulation of Smad7 in vivo
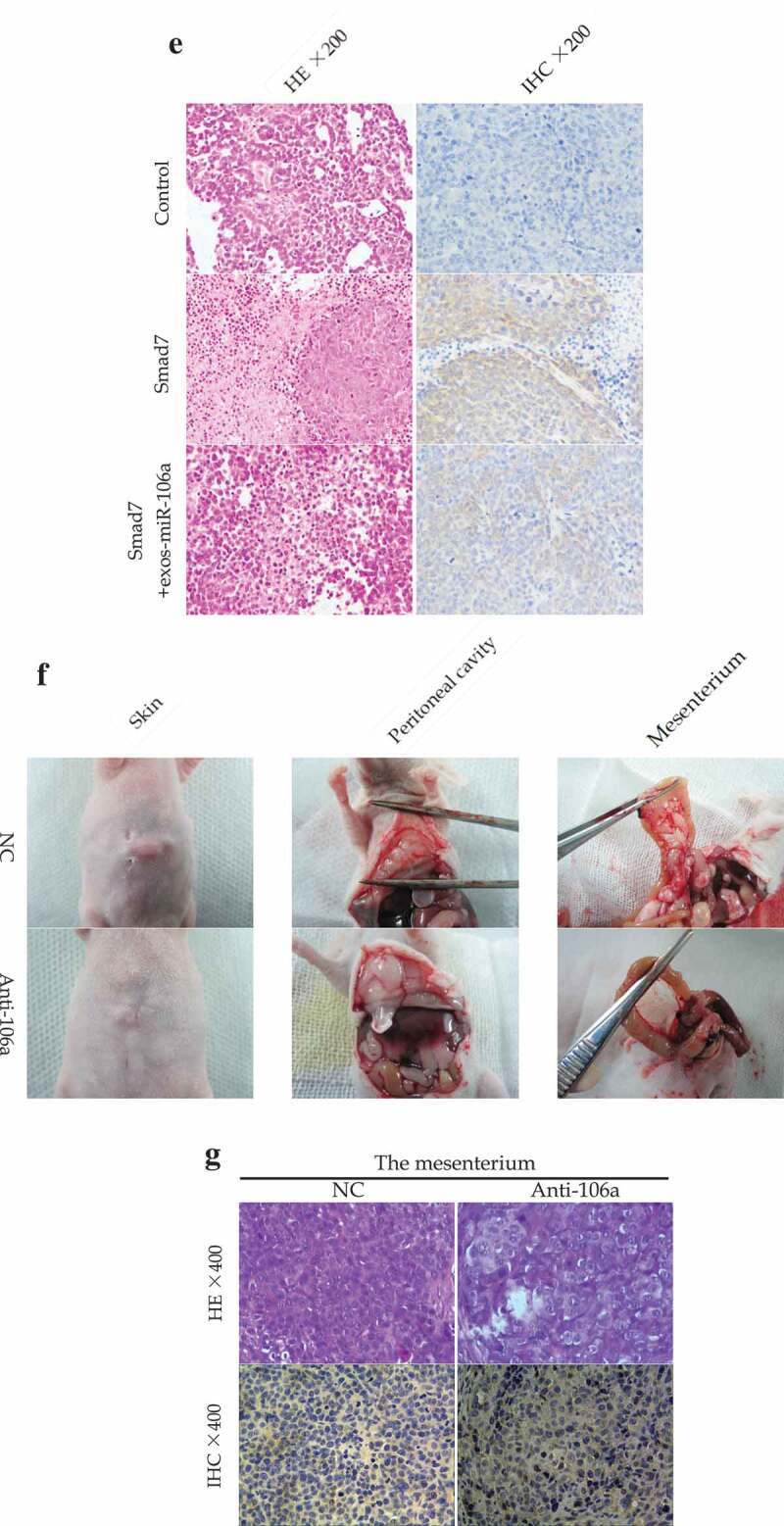
To further explore the potential influences of exosomal miR-106a in vivo, a subcutaneous tumor model was established. The mice were divided into three groups: Control, Smad7, Smad7+ exos-miR-106a. As can be observed in Figure 6A, tumor growth curve was a linear growth, suggesting that the tumor growth was slow in Smad7 group, but it would return to near normal growth level when treated with exosomes. As illustrated in Figure 6B, tumor volume in these groups was as follows: 585.00 ± 240.68, 106.00 ± 51.03, 555.17 ± 107.84. Statistical difference was established (F = 8.98, P = 0.016) and Smad7 group had the lowest tumor volume compared with the control group or Smad7+ exos-miR-106a group (P = 0.009, P = 0.012, respectively). Meanwhile, pathomorphological observation is shown in Figure 6C-E. Biopsy observation: tumors in control group were the largest, followed by the exosomes, and Smad7 group was the smallest. Under an optical microscope, the transplanted tumor cells arrayed as disorder and nest, differed in size and in shape, neoplastic cells with round, oval or irregular shape, the nuclear was big, deep-stained, thickened-karyotheca, chromatin granulated, and with many visible mitotic activities. In contrast, large necrosis appeared in Smad7 group, suggesting that Smad7 could induce cell death, but when combined with exosomes, this induction weakened by the regulation of miR-106a. Immunohistochemical staining showed that the Smad7 was positively located in tumor cells with uniform distribution in cytoplasm. The positively staining cells distributed diffusely with higher intensity. In exosomes group, however, staining cells were fewer and distributed focally and sporadically. This demonstrated that exosomal miR-106a influence GC growth by regulating Smad7.
Figure 6.
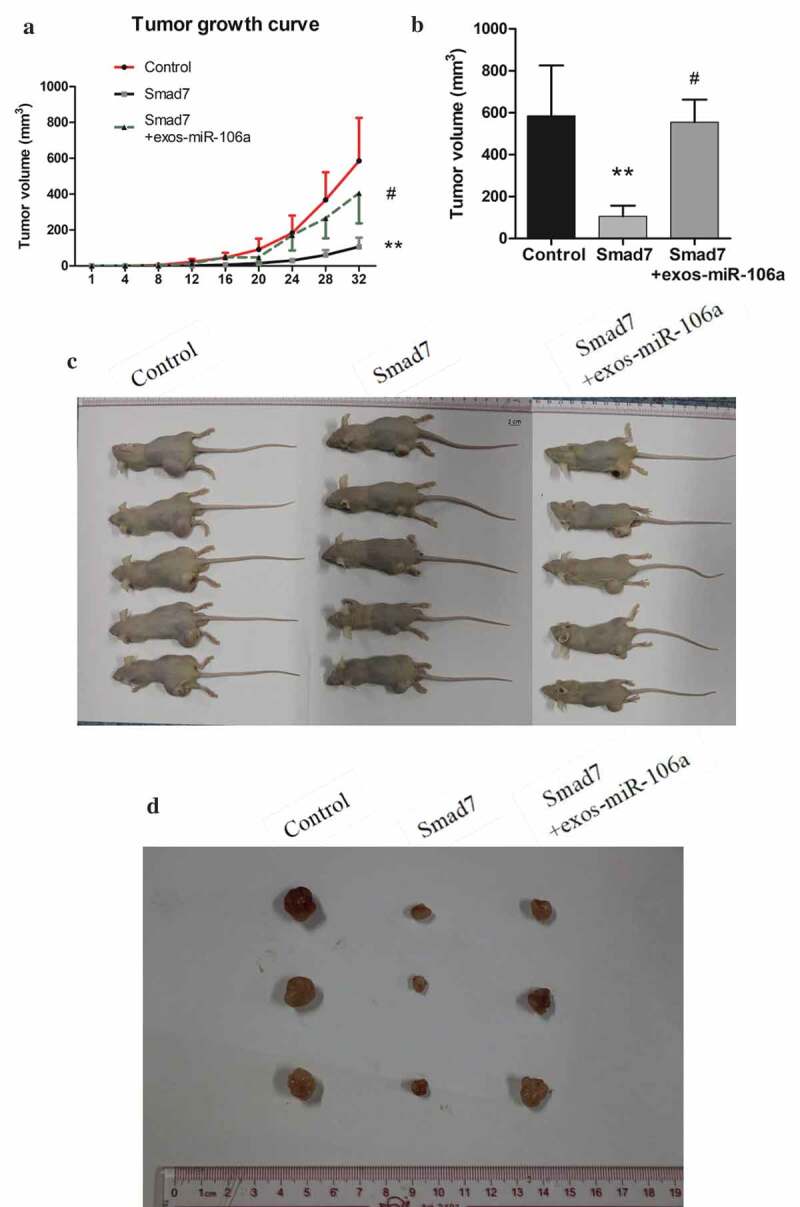
Exosomal miR-106a potentiates tumor growth in vivo. A. Subcutaneous xenotransplanted tumor model: the mice tumor growth curve was recorded after injection with mock-transduced BGC-823 cells, or BGC-823 cells transduced with Smad7 overexpression vector or exosomal miR-106a. B. Tumor volume of xenograft mice measured 32 days after injection. **P < 0.01, #P < 0.05. C. and D. Tumor tissues incised from the transplanted mice. Pathological studies including E. H&E and immunohistochemical staining for detection of the tumor cells growing on the limb of nude mice and the expression of Smad7 in tumor nodules. Magnification 200 ×. F. Abdominal xenotransplanted tumor model: miR-106a antagomir and antagomir NC transfected BGC-823 cells were injected into nude mice through a small incision at cartilago ensiformis of upper abdomen. Tumor nodules on the skin, peritoneal cavity, mesenterium were shown. G. H&E and immunohistochemical staining of the nodules on the mesenterium. Magnification 400 ×.
Figure 6.
(Continued).
To further assess the peritoneal implantation in mice, another abdominal tumor model was established. As found in figure 6F, miR-106a antagomir group harbored xenograft tumor nodules that significantly smaller than that of antagomir-control group no matter in quantity or size. Tumor tissues were confirmed by H&E staining, and IHC staining showed that Smad7 was positively expressed in anti-miR-106a group (Figure 6G). The phenomenon indicated that peritoneal dissemination had taken place in the peritoneum which might be regulated by miR-106a-Smad7 pathway.
Discussion
Small vehicles were originally thought to be trash bags that do not have important biological functions for cell life. It is now however, become increasingly clear as the integral role has been established in many types of diseases, especially in cancer [10,20]. Exosomes, as the representative of extracellular small vehicles, has emerged as a key regulator of multitudinous cellular processes, with its most fundamental role is to mediate intercellular communication that relying on the transportation of bioactive molecules (DNA, mRNAs, miRNAs, proteins, and lipids) from parent cells to receipt cells [21–23]. Based on the capability of travel between cell populations, exosomes allows reconstruction and reshape of recipient cells, and thus, extracellular environment may be impacted and with a number of papers demonstrated, exosomes produced by various cancers such as lung cancer, breast cancer, colon cancer and pancreatic cancer are able to promote the pre-metastatic niche formation or guide organ-oriented metastases [24–28].
Peritoneal metastasis, the most common way of dissemination for gastric cancer, the process that has been described by “seed and soil” theory, however, it is still not known what factors mediated the organotropic metastasis between gastric cancer cells and peritoneum. Exosomes attracted our great attention for its critical roles in the modification of “soil” suitable for “seed” cultivation and growth. We speculate that in this process, the important genetic substances from primary gastric cancer cells may reach the peritoneum through exosomes transportation. Peritoneum, on one hand, becomes the first barrier due to its extensive distribution [29]; on the other hand, it becomes the easiest place also due to the fulfillment of cellular components and blood vessels [30,31]. Genetic substances that carried by the exosomes are thought of as pioneers which could modify the original peritoneal structure to make it adapt for the metastasis of cancer cells.
With regards to the substances in the exosomes, based on the stability, miRNAs have been shown to be promising molecules for illuminating the underlying mechanisms during cancer development [32]. Previous studies raise an intriguing possibility that miR-106a had an ectopic expression in gastric cancer and could offer a promoting role to cancer metastasis [33–35]. MiR-106a belongs to miR-17 family, which has been widely studied [36,37]; however, in this study, we provide evidence of the oncogenic role of a single miRNA, miR-106a, in the peritoneal metastasis of gastric cancer, which has not yet been elucidated before.
Mesothelial cells are located on the most superficial layer of peritoneum, and their changes play a crucial role in the occurrence of peritoneal metastasis [38,39]. In this study, we assessed the migratory ability of gastric cancer cells and demonstrated that miR-106a was significantly enriched in low-differentiated GC-derived exosomes and could be transferred into MCs, which would be then transformed into pro-fibrotic myofibroblasts, as evidenced by the up-regulation of α-SMA and fibronectin expression, together with the down-regulation of Smad7 expression. It should be noted that with the changes of gene expression in MCs, the alternation of cell phenotype, including inhibition of proliferation, acceleration of apoptosis, was also observed in the meantime. Combined with previous reports, these changes in MCs are thought to be beneficial to the implantation of cancer cells [18,40]. The imbalance of cell proliferation and apoptosis result in the decrease of the number of MCs, and the loosening of intercellular junctions, followed by the exposure of hidden vascular and cellular components, which may provide a pro-metastatic microenvironment. In addition, we also examined the migratory ability of MCs. Similar to another study, ovarian cancer cells enhance the migration of MCs via cMet pathway [39], gastric cancer cells alter the migration of MCs through the exosomes-mediated transfer of miR-106a. Our results also suggested that exosomal miR-106a could induce MMT of MCs in vitro and influence GC growth in vivo, rescue experiments further confirmed that MMT transformation occurred in MCs. These morphological changes suggest that GC-derived exosomes have a capacity to destroy the mesothelial barrier at least to some extent. Combined with another study that in hepatocellular cancer, exosomal miR-1247 promoted the transformation of normal lung fibroblasts into cell-associated fibroblasts (CAFs) through targeting B4GALT3 to activate the NF-kB pathway [25], we, therefore, speculate that in gastric cancer, exosomes secreted from GC cells carry some important substances, like miRNAs (miR-106a), could also promote the transformation of MCs into cancer–associated MCs which in turn construct a space suitable for tumor growth and survival.
To further investigate the mechanisms underlying miR-106a induced changes in MCs, we used bioinformatics analysis to predict miR-106a’s putative targets. Then, Smad7 was chosen from the candidate as previous studies had demonstrated its close involvement in tumor progression [41,42]. In our study, we used luciferase reporter assays to confirm that miR-106a directly bound to Smad7 within 3’-UTR region and led to the degradation of its mRNA and protein level. Tissue samples defined the negative correlation between them. Additionally, results of proliferation, apoptosis, migration, nucleic acid and protein assays revealed that exosomal miR-106a induced phenotypic changes and MMT of peritoneal MCs via targeting Smad7.
According to previous studies, TGF-β signal pathway participates in tumor invasion and metastasis due to its influence on epithelial–mesenchymal transition (EMT), however, besides that, TGF-β-mediated MMT transition seemed to be equally important to facilitate the formation of metastatic niche [43]. Smad7, for our knowledge, is an inhibitor protein of TGF-β pathway for its effect on blocking or weakening signal transduction via recruiting E3-ubiquitin ligase SMURF2 to TβRⅠ for degradation, or blocking phosphorylation of Smad2 to hinder the polymerization of Smad2 and Smad4 [44,45]. Our studies revealed that LG-exos reduced Smad7 expression in MCs due to the transfer of high expression level of miR-106a. When Smad7 was suppressed, the TGF-β might be induced to a certain extent, and MMT transition might happen. Moreover, in vivo experiments verified that the tumor growth which was induced by GC-derived exosomes could be suppressed by the transduction of Smad7. Exosomal miR-106a interacted with Smad7 is an important factor in maintaining the malignant growth and peritoneal dissemination. So, it is valid to consider that exosomal miR-106a activates TGF-β/Smad pathway by mitigating the inhibitory action of Smad7 during gastric cancer peritoneal metastasis.
In conclusion, our study demonstrates that exosomal miR-106a transferred from GC cells could affect the structure and function of MCs and promote peritoneal metastasis through directly targeting Smad7. The data also indicate that GC-derived exosomes could induce MMT in MCs which is a necessary step in peritoneal metastasis. Based on these, our study opens a new gap in exploring the mechanism of gastric cancer peritoneal metastasis from the perspective of the combination of exosomes and miRNA.
Funding Statement
This work was supported by grant (81802416) from the National Natural Science Foundation of China, grant (XT2019007) from School level project of Ningxia Medical University, and grant (2018M633529) from China Postdoctoral Science Foundation.
Author contributions
Zhu Meng designed and performed the experiments, analyzed the data, wrote the manuscript; Zhang Ning performed the experiments; He Shuixiang and Lu Xinlan supervised the research.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388:2654–2664. [DOI] [PubMed] [Google Scholar]
- [2].Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. [DOI] [PubMed] [Google Scholar]
- [3].Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. [DOI] [PubMed] [Google Scholar]
- [4].Zong L, Abe M, Seto Y, et al. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. [DOI] [PubMed] [Google Scholar]
- [5].Chen JG, Chen HZ, Zhu J, et al. Cancer survival in patients from a hospital-based cancer registry, China. J Cancer. 2018;9:851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cho JM, Jang YJ, Kim JH, et al. Pattern, timing and survival in patients with recurrent gastric cancer. Hepato-gastroenterology. 2014;61:1148–1153. [PubMed] [Google Scholar]
- [7].Yoo CH, Noh SH, Shin DW, et al. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236–242. [DOI] [PubMed] [Google Scholar]
- [8].Orditura M, Galizia G, Sforza V, et al. Treatment of gastric cancer. World J Gastroenterol. 2014;20:1635–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ruivo CF, Adem B, Silva M, et al. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77:6480–6488. [DOI] [PubMed] [Google Scholar]
- [11].Tkach M, Thery C.. Communication by extracellular vesicles: where we are and where we need to go. Cell. 2016;164:1226–1232. [DOI] [PubMed] [Google Scholar]
- [12].Zhu M, Zhang N, He S, et al. MicroRNA-106a targets TIMP2 to regulate invasion and metastasis of gastric cancer. FEBS Lett. 2014;588:600–607. [DOI] [PubMed] [Google Scholar]
- [13].Zhu M, Zhang N, He S, et al. MicroRNA-106a functions as an oncogene in human gastric cancer and contributes to proliferation and metastasis in vitro and in vivo. Clin Exp Metastasis. 2016;33:509–519. [DOI] [PubMed] [Google Scholar]
- [14].Paget S. The distribution of secondary growths in cancer of the breast. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- [15].Mikula-Pietrasik J, Uruski P, Tykarski A, et al. The peritoneal “soil” for a cancerous “seed”: a comprehensive review of the pathogenesis of intraperitoneal cancer metastases. Cell Mol Life Sci. 2018;75:509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Li K, Chen Y, Li A, et al. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144:1486–1495. [DOI] [PubMed] [Google Scholar]
- [17].Yuyama K, Sun H, Mitsutake S, et al. Sphingolipid-modulated exosome secretion promotes clearance of amyloid-beta by microglia. J Biol Chem. 2012;287:10977–10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Deng G, Qu J, Zhang Y, et al. Gastric cancer-derived exosomes promote peritoneal metastasis by destroying the mesothelial barrier. FEBS Lett. 2017;591:2167–2179. [DOI] [PubMed] [Google Scholar]
- [19].Lopez-Cabrera M. Mesenchymal conversion of mesothelial cells is a key event in the pathophysiology of the peritoneum during peritoneal dialysis. Adv Med. 2014;2014:473134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jan AT, Rahman S, Khan S, et al. Biology, pathophysiological role, and clinical implications of exosomes: a critical appraisal. Cells. 2019;8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Valadi H, Ekstrom K, Bossios A, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- [22].Yu X, Odenthal M, Fries JW. Exosomes as miRNA carriers: formation-function-future. Int J Mol Sci. 2016;17:2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tai YL, Chen KC, Hsieh JT, et al. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109:2364–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang HG, Grizzle WE. Exosomes: a novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fang T, Lv H, Lv G, et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun. 2018;9:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Li XJ, Ren ZJ, Tang JH, et al. Exosomal microRNA miR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer. Cell Physiol Biochem. 2017;44:1741–1748. [DOI] [PubMed] [Google Scholar]
- [27].Valcz G, Galamb O, Krenacs T, et al. Exosomes in colorectal carcinoma formation: ALIX under the magnifying glass. Mod Pathol. 2016;29:928–938. [DOI] [PubMed] [Google Scholar]
- [28].Batista IA, Melo SA. Exosomes and the future of immunotherapy in pancreatic cancer. Int J Mol Sci. 2019;20:567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van Baal JO, Van de Vijver KK, Nieuwland R, et al. The histophysiology and pathophysiology of the peritoneum. Tissue Cell. 2017;49:95–105. [DOI] [PubMed] [Google Scholar]
- [30].Tobioka H, Sawada N, Zhong Y, et al. Enhanced paracellular barrier function of rat mesothelial cells partially protects against cancer cell penetration. Br J Cancer. 1996;74:439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Capobianco A, Cottone L, Monno A, et al. The peritoneum: healing, immunity, and diseases. J Pathol. 2017;243:137–147. [DOI] [PubMed] [Google Scholar]
- [32].Ge Q, Zhou Y, Lu J, et al. miRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang Z, Liu M, Zhu H, et al. miR-106a is frequently upregulated in gastric cancer and inhibits the extrinsic apoptotic pathway by targeting FAS. Mol Carcinog. 2013;52:634–646. [DOI] [PubMed] [Google Scholar]
- [34].Zhu M, Zhang N, Lu X, et al. Negative regulation of Kruppel-Like Factor 4 on microRNA-106a at upstream transcriptional level and the role in gastric cancer metastasis. Dig Dis Sci. 2018;63:2604–2616. [DOI] [PubMed] [Google Scholar]
- [35].Espinosa-Parrilla Y, Munoz X, Bonet C, et al. Genetic association of gastric cancer with miRNA clusters including the cancer-related genes MIR29, MIR25, MIR93 and MIR106: results from the EPIC-EURGAST study. Int J Cancer. 2014;135:2065–2076. [DOI] [PubMed] [Google Scholar]
- [36].Volinia S, Calin GA, Liu CG, et al. (2006) A microRNA expression signature of human solid tumors defines cancer gene targets, Proc Natl Acad Sci USA. 2006;103,2257–2261. [DOI] [PMC free article] [PubMed]
- [37].Concepcion CP, Bonetti C, Ventura A. The microRNA-17-92 family of microRNA clusters in development and disease. Cancer J. 2012;18:262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nakamura M, Ono YJ, Kanemura M, et al. Hepatocyte growth factor secreted by ovarian cancer cells stimulates peritoneal implantation via the mesothelial-mesenchymal transition of the peritoneum. Gynecol Oncol. 2015;139:345–354. [DOI] [PubMed] [Google Scholar]
- [39].Matte I, Lane D, Laplante C, et al. Ovarian cancer ascites enhance the migration of patient-derived peritoneal mesothelial cells via cMet pathway through HGF-dependent and -independent mechanisms. Int J Cancer. 2015;137:289–298. [DOI] [PubMed] [Google Scholar]
- [40].Yokoi A, Yoshioka Y, Yamamoto Y, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun. 2017;8:14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lamora A, Talbot J, Bougras G, et al. Overexpression of smad7 blocks primary tumor growth and lung metastasis development in osteosarcoma. Clin Cancer Res. 2014;20:5097–5112. [DOI] [PubMed] [Google Scholar]
- [42].Halder SK, Rachakonda G, Deane NG, et al. Smad7 induces hepatic metastasis in colorectal cancer. Br J Cancer. 2008;99:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rynne-Vidal A, Au-Yeung CL, Jimenez-Heffernan JA, et al. Mesothelial-to-mesenchymal transition as a possible therapeutic target in peritoneal metastasis of ovarian cancer. J Pathol. 2017;242:140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang S, Fei T, Zhang L, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007;27:4488–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yan X, Chen YG. Smad7: not only a regulator, but also a cross-talk mediator of TGF-beta signalling. Biochem J. 2011;434:1–10. [DOI] [PubMed] [Google Scholar]


