ABSTRACT
Numerous researches show that MicroRNAs (miRNAs) participate in tumorigenesis, progression, recurrence and drug resistance of malignant tumors, including laryngocarcinoma. miR-552 works as an oncogene in both colorectal cancer and liver cancer. However, the potential role of miR-552 in laryngocarcinoma is unknown. Herein, we for first found that miR-552 expression was upregulated in laryngocarcinoma tissues compared with their normal controls. Moreover, miR-552 expression was also increasing in the laryngocarcinoma cells. miR-552 interference inhibited the proliferation and metastasis of laryngocarcinoma cells in vitro and in vivo. Mechanically, bioinformatics and luciferase reporter analysis identified p53 as a direct target of miR-552. miR-552 knockdown upregulated the p53 mRNA and protein expression in laryngocarcinoma cells. miR-552 expression was negatively associated with p53 expression in laryngocarcinoma tissues. More importantly, the p53 siRNA or p53 overexpression virus abrogated the discrepancy of growth and metastasis capacity between miR-552 interference laryngocarcinoma cells and control cells.
KEYWORDS: Laryngocarcinoma, miR-552, p53, proliferation, metastasis
Introduction
Laryngocarcinoma is one of the most common head and neck cancer worldwide [1]. There are more than 80 000 patients dying due to laryngocarcinoma each year [2]. Most laryngocarcinoma are diagnosed at an advanced stage for lacking effective early diagnosis biomarkers [3]. The 5-years survival rate of laryngocarcinoma is less than 50% [4]. Therefore, it is urgent to research the underlying mechanism of laryngocarcinoma and find the new therapeutic target for improving the clinical outcome of patients suffering from laryngocarcinoma.
MicroRNAs (miRNAs) belong to a group of small non-coding RNA molecule, containing about 22 nucleotides [5,6]. miRNAs regulate protein expression via its post-transcription by targeting molecules 3ʹUTR [7]. Increasing researchers found that miRNAs are involved in tumor initiation, proliferation, metastasis, recurrence and drug resistance [8,9]. For instance, miR-365 regulates liver cancer stem cells via RAC1 pathway [10]. MicroRNA-383-5p inhibits the progression of gastric carcinoma via targeting HDAC9 expression [11]. In human laryngocarcinoma, numerous miRNAs have been reported to be abnormally expressed during its progression, including miR-9, miR-29a-3p, miRNA-221, miR-210, etc [6,12–14]. miR-552 is a newly discovered miRNA, its function and mechanism of action in biological processes and diseases are not completely understood. Previous studies showed that m miR-552-5p facilitates osteosarcoma cell proliferation and metastasis by targeting WIF1 [15]. Moreover, MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression [16]. However, the role of miR-552 in laryngocarcinoma was unclear.
In the present study, we first found that miR-552 was upregulated in human laryngocarcinoma tissues and cell lines. Biological function study demonstrated that miR-552 promoted laryngocarcinoma cells' proliferation and metastasis in vitro and in vivo. Further mechanism study revealed that p53 was a direct target of miR-552 in laryngocarcinoma cells. In conclusion, our results highlighted the importance of miR-552 in promoting the proliferation and metastasis of laryngocarcinoma cells via p53 pathway.
Materials and methods
Human tissue samples
Laryngocarcinoma tissues and their normal control tissues were obtained from laryngocarcinoma patients who under surgical remove at the First Affiliated Hospital of China Medical University (Shenyang, Liaoning, China). Human laryngocarcinoma tissues and normal tissues were immediately snap-frozen in liquid nitrogen and stored at −80°C. Informed consent was obtained from each patient, and the research protocols were approved by the Ethics Committee of the First Affiliated Hospital of China Medical University.
Cell lines and cell culture
M2E, M4E, TU212 and Hep-2 cells were purchased from the Chinese Academy of Sciences, Shanghai, China. The laryngocarcinoma cells were cultured in Eagle’s Minimum Essential Medium supplemented with 10% fetal calf serum (FCS; Invitrogen, Carlsbad, CA, USA). LV3-has-miR-552 sponge (5ʹ- TTGTCTAACCAGTCACCTGTT −3ʹ) virus was purchased from Shanghai GenePharma (Shanghai, China). M4E and Hep-2 cells were dissociated with 0.5% trypsin and seeded into six-well plates. Then, the cells were infected with miR-552 sponge virus or miR-552 mimic virus and their control virus. The stable infectants were screened by using puromycin as before [17].
M4E miR-552 sponge or Hep-2 cells miR-552 sponge and their control cells were seeded into a six-well plate until they reached 60–70% confluence. Transfection of si-p53 or its negative control was performed in each well in the absence of serum with siRNA transfection reagent according to the manufacturer’s instructions (Polyplus, Illkirch, France). The sequence of si-p53 is as follows: 5ʹ- GCAUGAACCGGAGGCCCAU −3ʹ. Then, the above cells were subjected to CCK8 assay, transwell assay and matrigel invasion chamber assay.
Cell proliferation assays
For CCK8 assay, M4E miR-552 sponge/mimic or Hep-2 cells miR-552 sponge/mimic and their control cells were seeded in 96-well plates (3 × 103 cells per well). ATP activity was measured using a Cell Counting Kit-8 at indicated time points. ATP activity was measured using a Cell Counting Kit-8 at indicated time points. The procedure was as follows: The cell suspension (100 μl/well) was inoculated in a 96-well plate, and the plate was pre-incubated in a humidified incubator at 37°C for 1 h. This was followed by the addition of 10 μl of the CCK-8 solution to each well of the plate, and incubation of the plate for 1 h in the incubator. Finally, the absorbance was measured at 450 nm using a microplate reader (Synergy H1; BioTek Instruments, Inc., Winooski, VT, USA) [18].
For colony formation assay, M4E miR-552 sponge/mimic or Hep-2 cells miR-552 sponge/mimic and their control cells were cultured in 12-well plates (3x103 cells/well). The cells were incubated at 37°C for 7 days and then fixed with 10% neutral formalin for >4 h. The cells were dyed with crystal violet (Beyotime, Haimen, China). The cells were photographed under a microscope (Olympus, Tokyo, Japan).
For cell EdU immunofluorescence staining, M4E miR-552 sponge or Hep-2 cells miR-552 sponge and their control cells were seeded into 96-well plates and performed using the EdU Kit (RiboBio) at 48 h. The results were quantified with a Zeiss axiophot photomicroscope (Carl Zeiss) and Image-Pro plus 6.0 software.
Animal models
For xenograft formation assay, Hep-2 cells miR-552 sponge and its control cells (2 × 106) were injected subcutaneously into nude mice. Nude mice were sacrificed 6-weeks post inoculation and tumors were collected and examined.
For tail vein lung metastasis assay, Hep-2 cells miR-552 sponge and its control cells (2 × 106) were intravenously injected into male nude mice through the tail vein (Chinese Science Academy, Shanghai, China). After 3 months, lung metastasis was measured.
Cell migration assays
For cell migration experiments, 2 × 105 M4E miR-552 sponge/mimic or Hep-2 cells miR-552 sponge/mimic and their control cells were seeded into the upper chamber of a polycarbonate transwell in serum-free DMEM medium. The lower chamber was added with DMEM medium containing 20% FBS as chemoattractant. The cells were incubating for 12 h and the chamber was fixed with 10% neutral formalin for 12 h. The cells were dyed with crystal violet (Beyotime). The cells were then counted under a microscope (Olympus) and the cell number is expressed as the average number of the cells in each field.
Cell invasion assays
For cell invasion experiments, 2 × 105 M4E miR-552 sponge or Hep-2 cells miR-552 sponge and their control cells were seeded into the upper chamber of a polycarbonate transwell in serum-free DMEM medium. The lower chamber was added with DMEM medium containing 20% FBS as a chemoattractant. The cells were incubating for 24 h and the chamber was fixed with 10% neutral formalin for >4 h. The cells were dyed with crystal violet (Beyotime). The cells were then counted under a microscope (Olympus) and the cell number is expressed as the average number of the cells in each field.
Luciferase reporter assays
The wide type p53 3ʹ-untranslated region (UTR) containing miR-552 targeting sequence and the mutated type was amplified and cloned into the luciferase reporter plasmid pGL4.13 vector (Promega, Madison, WI). All the constructs were verified by sequencing. Briefly, laryngocarcinoma cells were co-transfected with miR-552 sponge or miR-control and pMIR-reporter luciferase vector containing a specific sequence of wild-type or mutant p53 fragment, using siRNA transfection (Invitrogen, NY, USA). Cells were collected and lysed for luciferase detection 48 h after transfection. The relative luciferase activity was normalized against the Renilla luciferase activity [19].
Real-time PCR
For the detection of mature miR-552, total RNA was subjected to reverse transcription using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems). qRT-PCR analysis of miR-552 expression was carried out using TaqMan MicroRNA assay kits (Applied Biosystems). Results were normalized to U6 snRNA using the comparative threshold cycle (Ct) method. The miR-552 primer sequences were forward: 5ʹ AACAGGTGACTGGTTAGACAA 3ʹ, U6 primer sequences were forward: 5ʹ ATTGGAACGATACAGAGAAGATT 3ʹ.
The total cells' RNA was extracted by using Trizol reagent (Invitrogen, 15596-018). Total cDNAs were synthesized by the ThermoScript TM RT-PCR system (Invitrogen, 11146-057). The total mRNA amount presented in the cells was measured by RT-PCR using the ABI PRISM 7300 sequence detector (Applied Biosystems). The p53 primer sequences were forward: 5ʹ ACCTATGGAAACTACTTCCTGAAA 3ʹ, reverse: 5ʹ CTGGCATTCTGGGAGCTTCA 3ʹ. The β-actin was used as a reference for relative expression calculation and its primer sequences were forward: 5ʹ GGCCCAGAATGCAGTTCGCCTT 3ʹ, reverse: 5ʹ AATGGCACCCTGCTCACGCA 3ʹ.
Western blotting assays
Twenty-five micrograms of proteins were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred to the nitrocellulose membrane. The membrane was blocked with 5% nonfat milk and incubated with the primary antibody for 2 h. The protein band, specifically bound to the primary antibody, was detected using an IRDye 800CW-conjugated secondary antibody and LI-COR imaging system (LI-COR Biosciences). The primary antibodies were p53 (1:1000; # 2524, Cell Signaling Technology) and GAPDH (1:5000; #5174, Cell Signaling Technology).
Statistical analysis
All statistical analyses were performed using GraphPad Prism (GraphPad Software, Inc. La Jolla, USA). Statistical analysis was carried out using t-test or Bonferroni Multiple Comparisons Test: *p < 0.05. A p-value of less than 0.05 was considered statistically significant.
Results
miR-552 expression was upregulated in laryngocarcinoma and cells
In order to explore the biological role of miR-552 in laryngocarcinoma, a large set of human laryngocarcinoma tissues was used to check the expression of miR-552. As shown in Figure 1(a), the miR-552 level was dramatically upregulated in laryngocarcinoma tissues compared with their normal controls. Laryngocarcinoma tissues were classified into laryngocarcinoma tissues than the normal tissues and the analyzed result showed that the levels of miR-552 in cancer tissues were higher than the normal group (Figure 1(b)). Furthermore, the expression level of miR-552 was higher in laryngocarcinoma patients at stage3/4 than that in laryngocarcinoma patients at stage1/2 (Figure 1(c)). Next, four human laryngocarcinoma cell lines were selected to detect miR-552 expression. We found that miR-552 expression in four laryngocarcinoma cell lines was significantly higher than the normal cell line (Figure 1(d)). Taken together, the above results demonstrated that miR-552 might play an important function in laryngocarcinoma progression.
Figure 1.
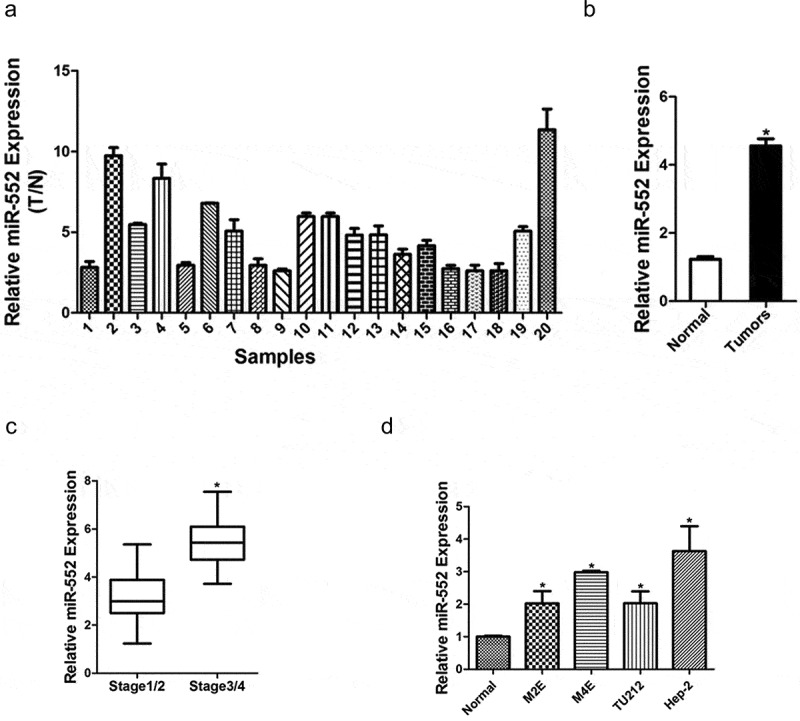
Expression of miR-552 was upregulated in human laryngocarcinoma tissues.
(a) Relative expression of miR-552 in human laryngocarcinoma patients’ tissues and their corresponding adjacent normal tissues (n = 20) (p < 0.05). (b) Average expression level of miR-552 in human laryngocarcinoma specimens (n = 20) and normal tissues (n = 20) (p < 0.05). (c) The expression of miR-552 in laryngocarcinoma patients with different stage tumors was investigated via real-time PCR analysis (stage1/2, n = 28; stage3/4, n = 28) (p < 0.05). (d) The expression of miR-552 in four laryngocarcinoma cells and normal cells was investigated via real-time PCR analysis (p < 0.05).
miR-552 promoted laryngocarcinoma cells' proliferation
To further explore the potential effect of miR-552 on laryngocarcinoma cells' behavior, Hep-2 and M4E cells were infected with miR-552 sponge virus and the knockdown effect was checked by real-time PCR (Figure 2(a)). Hep-2 and M4E cells were also infected with miR-552 mimic virus and the overexpress effect was checked by real-time PCR (Figure S2A). CCK8 assay was used to examine the cell growth; the result showed that miR-552 interference inhibited cell growth in laryngocarcinoma cells and ectopic miR-552 expression promoted laryngocarcinoma cells' growth (Figure 2(b)&Figure S2B). Next, the colony formation assay used to measure cell proliferation, the data showed that miR-552 knockdown laryngocarcinoma cells formed less and smaller colonies (Figure 2(c)). Conversely, miR-552 overexpression laryngocarcinoma cells formed much more colonies compared with control cells (Figure S2C). In addition, 5-ethynyl-2ʹ-deoxyuridine (EdU) staining showed that miR-552 interference suppressed laryngocarcinoma cells' proliferation (Figure 2(d)). More importantly, the in vivo experiments also confirmed that miR-552 knockdown inhibited laryngocarcinoma cells' growth (Figure 2(e,f)). Collectively, the above results showed that miR-552 promoted laryngocarcinoma cells' proliferation.
Figure 2.
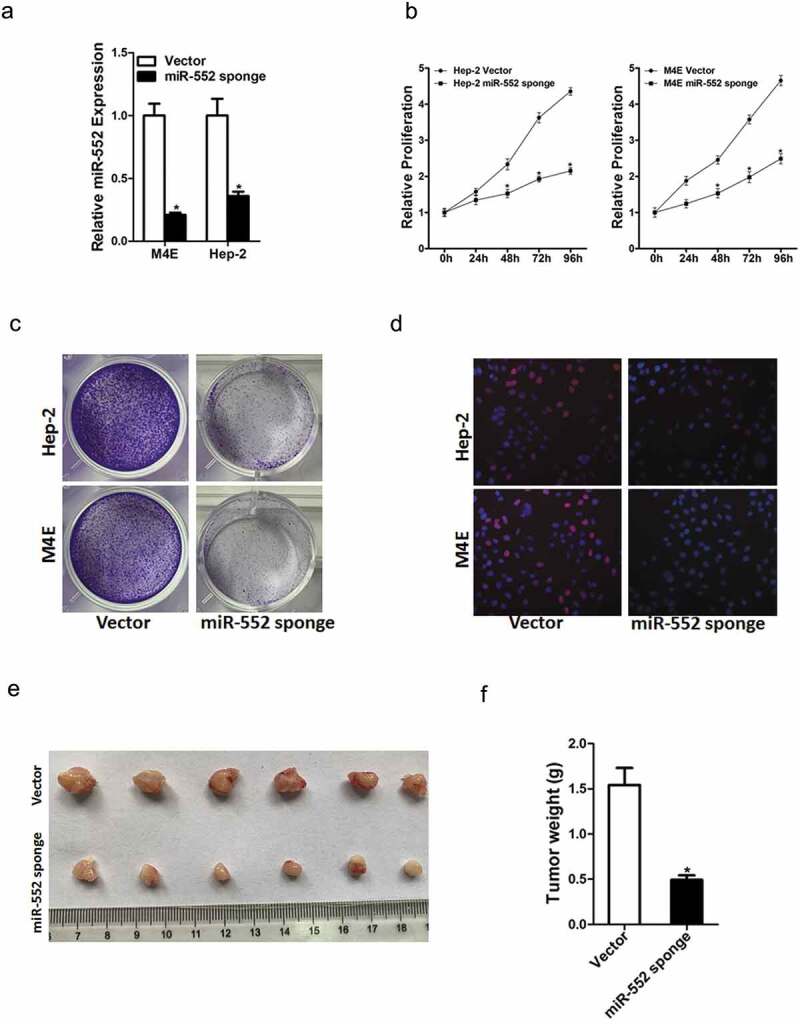
Interference of miR-552 suppresses laryngocarcinoma cells' proliferation.
(a) The interference effect of miR-552 in M4E and Hep-2 cells was checked by real-time PCR assay. (b) Cell proliferation was measured using CCK-8 assays in M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells. (c) Colony formation assays of M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells. (d) Cell proliferation was assessed using EdU immunofluorescence staining (red) in M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells. Nuclei were stained with Hoechst 33342 (blue). (e) Hep-2 miR-552 sponge and its control cells were implanted subcutaneously to induce xenograft tumor in nude mice. Mice were sacrificed at fourth weeks after inoculation; tumor was excised, pictured and weighed.
miR-552 promoted laryngocarcinoma cells' metastasis
Previous study showed that miR-552 could induce EMT of cancer, here, we want to know whether miR-552 could promote metastasis of laryngocarcinoma cells. To explore the biological function of miR-552 in laryngocarcinoma cells' metastasis, we used transwell assay and matrigel invasion chamber assay. The transwell assay results showed that miR-552 knockdown attenuated the migration ability of laryngocarcinoma cells (Figure 3(a,b)). Conversely, miR-552 overexpression enhanced the migration ability of laryngocarcinoma cells (Figure S3A&B). Moreover, matrigel invasion chamber assay revealed that the invasion ability was impaired in miR-552 interference laryngocarcinoma cells (Figure 3(c,d)). More importantly, miR-552 knockdown laryngocarcinoma cells formed much less metastasis foci in nude mice lung tissues (Figure 3(e,f)). Taken together, our data demonstrated that miR-552 promoted laryngocarcinoma cells' metastasis.
Figure 3.
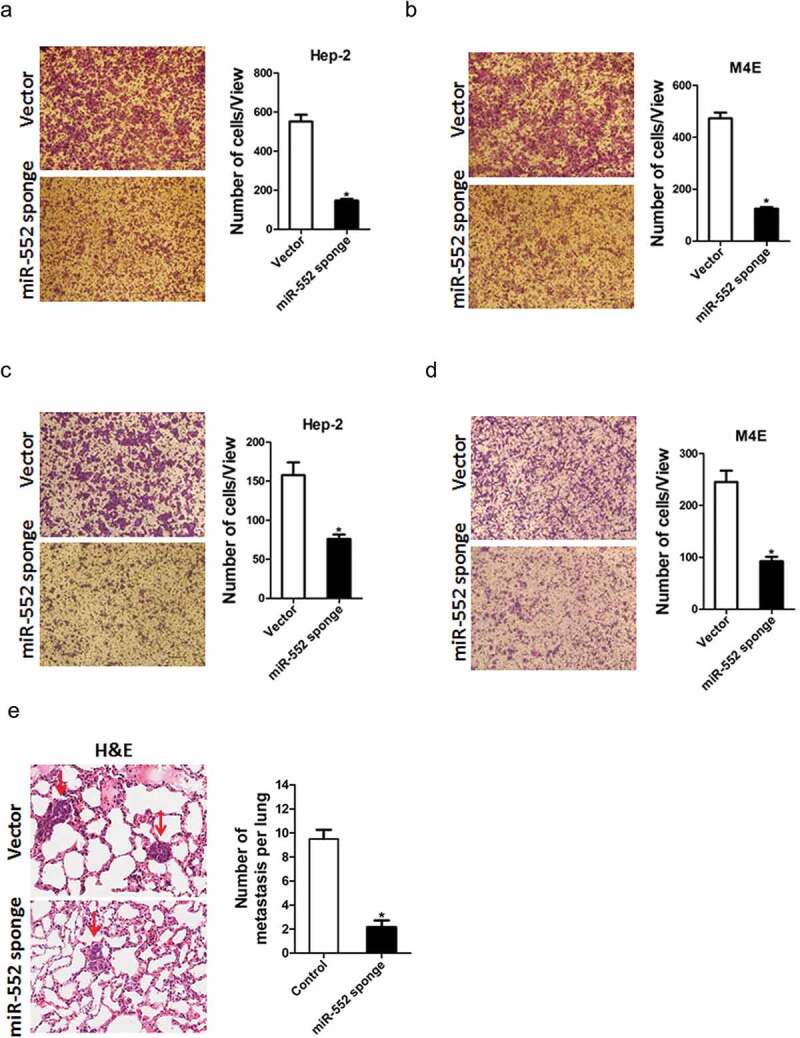
miR-552 depletion suppresses laryngocarcinoma cells' migration and invasion.
(a) The migration ability of Hep-2 miR-552 sponge and its control cells was performed utilizing polycarbonate membrane inserts in a 24-well plate. (b) The migration ability of M4E miR-552 sponge and its control cells was performed utilizing polycarbonate membrane inserts in a 24-well plate. (c) The invasive capacity of Hep-2 miR-552 sponge and its control cells was analyzed using Matrigel-coated Boyden chamber. (d) The invasive ability of M4E miR-552 sponge and its control cells was analyzed using Matrigel-coated Boyden chamber. (e) H&E staining of nude mice inoculated Hep-2 mir-552 sponge or control cells via tail vein for 12 weeks. The number of lung metastatic foci in each group (n = 7) was also calculated (p < 0.05).
miR-552 directly targeted p53 to promote laryngocarcinoma cells' progression
Next, we attempted to identify the potential target genes of miR-552 in laryngocarcinoma cells. Bioinformatics analysis found that miR-552 has a putative binding site in p53 mRNA 3ʹ-UTR (Figure 4(a)). To further explore whether miR-552 directly regulates p53 expression via interaction with its 3ʹ-UTR, the wild-type or mutant p53 3ʹ-UTR reporter plasmids were transfected into miR-552 sponge laryngocarcinoma cells and their control cells. The luciferase activity of the wild-type reporter was significantly upregulated in the absence of miR-552 (Figure 4(b)). However, miR-552-mediated repression of the reporter expression was abolished by mutation of the miR-552 binding site in the p53 3ʹ-UTR. Moreover, p53 mRNA and protein expression were also increased in miR-552 interference laryngocarcinoma cells (Figure 4(c,d)). There was a significant negative correlation between miR-552 and p53 mRNA expression in human laryngocarcinoma tissues (Figure 4(e)).
Figure 4.
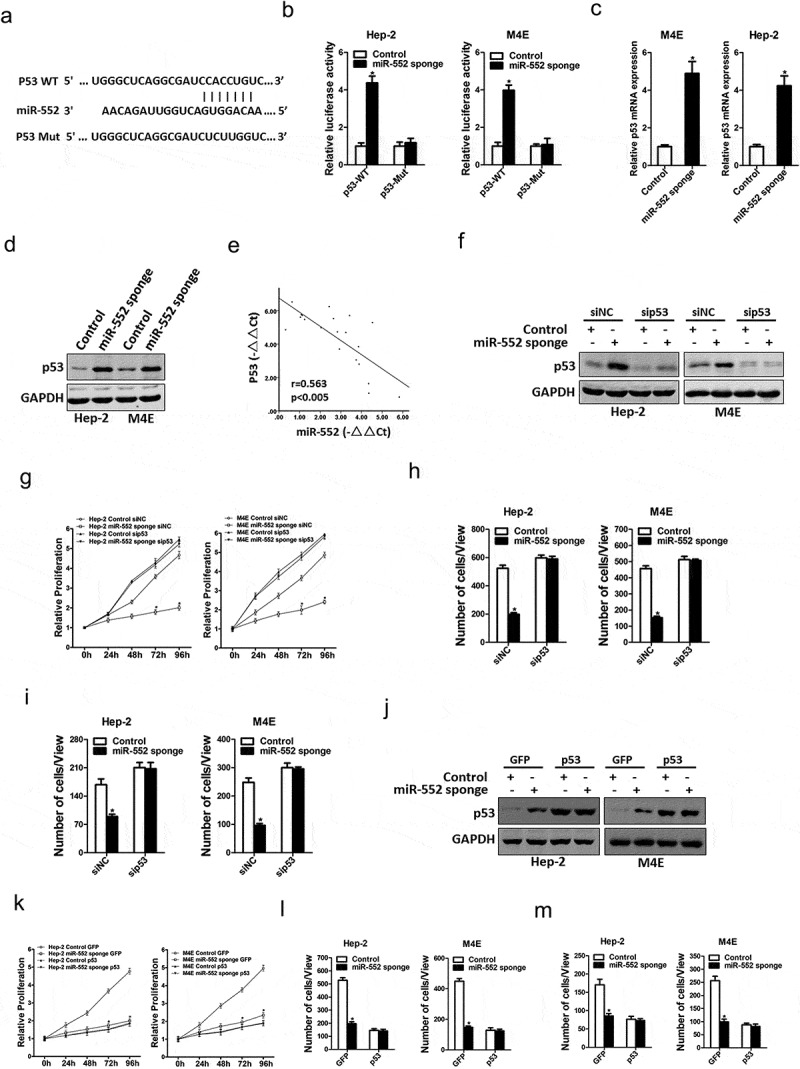
p53 was a direct target of miR-552 in laryngocarcinoma cells.
(a) A potential target site for miR-552 in the 3ʹ-UTR of human p53 mRNA, as predicted by the program Targetscan. To disrupt the interaction between miR-552 and p53 mRNA, the target site was mutated. (b) Luciferase reporter assays performed in M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells transfected with wild-type or mutant p53 3ʹ-UTR constructs. (c) The mRNA expression of p53 was checked in M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells by real-time PCR. (d) The protein expression of p53 was checked in M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells by western blot. (e) Significant correlation was observed between miR-552 and p53 expression in human laryngocarcinoma tissues (n = 20). (f) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were transfected with p53 siRNA and then checked by western bolt assay. (g) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were transfected p53 siRNA and then subjected to CCK8 assay. (h) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were transfected p53siRNA and then subjected to migration assay. (i) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were transfected p53siRNA and then subjected to Invasion assay. (j) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were infected with p53 overexpression virus and then checked by western bolt assay. (k) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were infected with p53 overexpression virus and then subjected to CCK8 assay. (l) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were infected with p53 overexpression virus and then subjected to migration assay. (m) M4E miR-552 sponge or Hep-2 miR-552 sponge and their control cells were infected with p53 overexpression virus and then subjected to Invasion assay.
To further investigate the role of p53 in miR-552-mediated proliferation and metastasis of laryngocarcinoma cells, special p53 siRNA was transfected into miR-552 knockdown laryngocarcinoma cells and control cells (figure 4(f)). As shown in Figure 4(g), p53 siRNA abolished the distinct growth capacity between miR-552 interference laryngocarcinoma cells and control cells (Figure 4(g)). Consistently, p53 siRNA also diminished the discrepancy of metastasis between miR-552 interference laryngocarcinoma cells and their control cells (Figure 4(h,i)). Next, miR-552 knockdown laryngocarcinoma cells and control cells were infected with p53 overexpression virus (Figure 4(j)). p53 overexpression also abolished the distinct growth and metastatic capacity between miR-552 interference laryngocarcinoma cells and control cells (Figure 4(k–m)). Collectively, the results suggested that miR-552 promoted laryngocarcinoma cells' proliferation and metastasis by directly targeting p53.
Discussion
Increasing evidence showed that miRNAs are discovered in laryngocarcinoma and played important roles in genes regulation at post-transcriptional levels [20]. But there are still many unknown miRNAs. So, it is urgent to explore new functional miRNAs in laryngocarcinoma. Previous studies demonstrated that miR-552 was involved in numerous tumors regulation. In the present study, we found that miR-552 was dramatically upregulated in human laryngocarcinoma tissues and cell lines. Interference of miR-552 suppressed laryngocarcinoma cells' proliferation and metastasis by targeting p53.
It was reported that miRNAs participated in the regulation of laryngocarcinoma. For instance, enhanced miR-9 promotes laryngocarcinoma cell survival via down-regulating PTEN [12]. MiR-29a-3p suppresses cell proliferation in laryngocarcinoma by targeting prominin 1 [13]. Existing research reported that miR-552 functions as an oncogene in liver cancer, colorectal cancer and osteosarcoma [21]. Our study identified that miR-552 expression in laryngocarcinoma tissues was upregulated compared to their matched normal adjacent tissues, which was also confirmed in laryngocarcinoma cells. In the face of cellular functions, miR-552 increased cell viability, colony formation, proliferation, and promoted migration and invasion in laryngocarcinoma cells both in vivo and in vitro. So, our results suggest that miR-552 acts as a promoting miRNA in laryngocarcinoma.
p53 tumor suppressor proteins play an important role in cell responses to DNA damage and other genomic aberrations [22]. Activation of p53 leads to cell cycle arrest, DNA repair, or apoptosis [23–25]. Its loss or mutation has been implicated as a cause of numerous tumors initiation, progression, recurrence and drug resistance [26,27]. However, the exact mechanism beneath p53 inactivation in laryngocarcinoma remains vague. We hereby revealed that p53 is a direct target of miR-552 in laryngocarcinoma cells. miR-552 sponge increased p53 mRNA and protein expression in laryngocarcinoma cells. Moreover, we also found that miR-552 directly regulates p53 expression via interaction with its 3ʹ-UTR. More importantly, p53 siRNA or p53 overexpression virus could diminish the distinct growth capacity or metastasis ability between miR-552 knockdown laryngocarcinoma cells and control cells. Herein, we for first revealed that miR-552 promotes laryngocarcinoma proliferation and metastasis via directly regulating p53. These findings of the present study not only shed a new light on the mechanism of laryngocarcinoma but suggest a potential therapeutic target against laryngocarcinoma patients.
Most of the previous studies indicate that miR-552 functions as an oncomiRNA in liver cancer, colorectal cancer, osteosarcoma and other types of cancer. It was reported that miR-552 facilitates osteosarcoma cell proliferation and metastasis by targeting WIF1 [15]. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression [16]. MiR-552 promotes colorectal cancer cells' proliferation and migration by directly targeting DACH1 [28]. However, our results showed that miR-552 promotes laryngocarcinoma proliferation and metastasis via directly regulating p53. This may be due to heterogeneity between different tumors. Different signaling pathways play different roles in different tumors. This also makes our research more meaningful.
Funding Statement
This work was supported by grants from Wu Jieping Medical Foundation [No. 320.6750.19021] and China Medical University young backbone support program [No. QGZ2018011].
Acknowledgements
We thank the Key Laboratory of Immunodermatology, Ministry of Health, Ministry of Education, No.1 Hospital of China Medical University for technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
The supplemental data for this article can be accessed here.
References
- [1].Miller KD, Goding Sauer A, Ortiz AP, et al. Cancer statistics for Hispanics/Latinos, 2018. CA Cancer J Clin. 2018;68:425–445. [DOI] [PubMed] [Google Scholar]
- [2].McGuire S. World cancer report 2014. Geneva, Switzerland: World Health Organization, international agency for research on cancer, WHO press, 2015. Adv Nutr. 2016;7:418–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lampri ES, Chondrogiannis G, Ioachim E, et al. Biomarkers of head and neck cancer, tools or a gordian knot? Int J Clin Exp Med. 2015;8:10340–10357. [PMC free article] [PubMed] [Google Scholar]
- [4].He FY, Liu HJ, Guo Q, et al. Reduced miR-300 expression predicts poor prognosis in patients with laryngeal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2017;21:760–764. [PubMed] [Google Scholar]
- [5].Zhong G, Xiong X. miR-205 promotes proliferation and invasion of laryngeal squamous cell carcinoma by suppressing CDK2AP1 expression. Biol Res. 2015;48:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zuo J, Wen M, Lei M, et al. MiR-210 links hypoxia with cell proliferation regulation in human Laryngocarcinoma cancer. J Cell Biochem. 2015;116:1039–1049. [DOI] [PubMed] [Google Scholar]
- [7].Wang H, Gou X, Jiang T, et al. The effects of microRNAs on glucocorticoid responsiveness. J Cancer Res Clin Oncol. 2017;143:1005–1011. [DOI] [PubMed] [Google Scholar]
- [8].Bach DH, Hong JY, Park HJ, et al. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int J Cancer. 2017;141:220–230. [DOI] [PubMed] [Google Scholar]
- [9].Ma L. MicroRNA and metastasis. Adv Cancer Res. 2016;132:165–207. [DOI] [PubMed] [Google Scholar]
- [10].Jiang ZB, Ma BQ, Liu SG, et al. miR-365 regulates liver cancer stem cells via RAC1 pathway. Mol Carcinog. 2019;58:55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Xu G, Li N, Zhang Y, et al. MicroRNA-383-5p inhibits the progression of gastric carcinoma via targeting HDAC9 expression. Braz J Med Biol Res. 2019;52:e8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu E, Su J, Zeng W, et al. Enhanced miR-9 promotes laryngocarcinoma cell survival via down-regulating PTEN. Biomed Pharmacother. 2016;84:608–613. [DOI] [PubMed] [Google Scholar]
- [13].Su J, Lu E, Lu L, et al. MiR-29a-3p suppresses cell proliferation in laryngocarcinoma by targeting prominin 1. FEBS Open Bio. 2017;7:645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yilmaz SS, Guzel E, Karatas OF, et al. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope. 2015;125:E377–381. [DOI] [PubMed] [Google Scholar]
- [15].Cai W, Xu Y, Yin J, et al. miR-552-5p facilitates osteosarcoma cell proliferation and metastasis by targeting WIF1. Exp Ther Med. 2019;17:3781–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Qu W, Wen X, Su K, et al. MiR-552 promotes the proliferation, migration and EMT of hepatocellular carcinoma cells by inhibiting AJAP1 expression. J Cell Mol Med. 2019;23:1541–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Xiang D, Cheng Z, Liu H, et al. Shp2 promotes liver cancer stem cell expansion by augmenting beta-catenin signaling and predicts chemotherapeutic response of patients. Hepatology. 2017;65:1566–1580. [DOI] [PubMed] [Google Scholar]
- [18].Xiang DM, Sun W, Ning BF, et al. The HLF/IL-6/STAT3 feedforward circuit drives hepatic stellate cell activation to promote liver fibrosis. Gut. 2018. Sep;67(9):1704–1715. [DOI] [PubMed] [Google Scholar]
- [19].Xiang DM, Sun W, Zhou T, et al. Oncofetal HLF transactivates c-Jun to promote hepatocellular carcinoma development and sorafenib resistance. Gut. 2019;68:1858–1871. [DOI] [PubMed] [Google Scholar]
- [20].Chen L, Liu S, Li K, et al. Evaluation of microRNA expression profiling in highly metastatic laryngocarcinoma cells. Acta Otolaryngol. 2018;138:1105–1111. [DOI] [PubMed] [Google Scholar]
- [21].Wang N, Liu W. Increased expression of miR-552 acts as a potential predictor biomarker for poor prognosis of colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22:412–416. [DOI] [PubMed] [Google Scholar]
- [22].Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. [DOI] [PubMed] [Google Scholar]
- [23].Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 2018;25:114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dhar D, Antonucci L, Nakagawa H, et al. Liver cancer initiation requires p53 inhibition by CD44-enhanced growth factor signaling. Cancer Cell. 2018;33:1061–1077 e1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sabapathy K, Lane DP. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat Rev Clin Oncol. 2018;15:13–30. [DOI] [PubMed] [Google Scholar]
- [26].Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 2015;25:486–495. [DOI] [PubMed] [Google Scholar]
- [27].Ruiz de Galarreta M, Bresnahan E, Molina-Sanchez P, et al. Beta-catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019;9:1124–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cao J, Yan XR, Liu T, et al. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH1 via the Wnt/beta-catenin signaling pathway in colorectal cancer. Oncol Lett. 2017;14:3795–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


