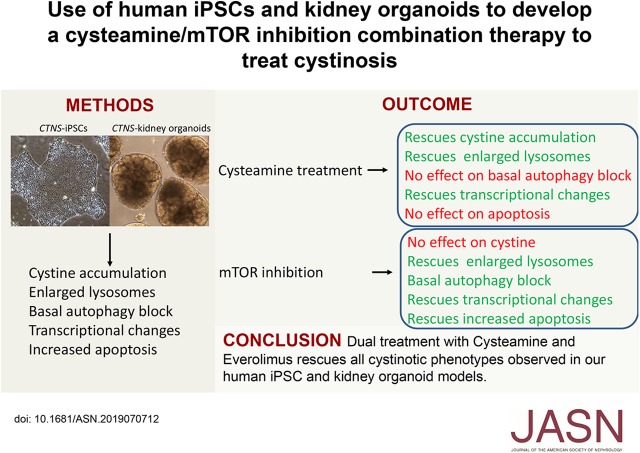
Significance Statement
In its severest form, the lysosomal storage disease cystinosis is characterized by accumulation of cystine; renal proximal tubule dysfunction; and kidney failure. Research has also implicated cystinosin in modulating the mammalian target of rapamycin (mTOR) complex 1 pathway. Use of the cystine-depleting drug cysteamine, the sole treatment option for cystinosis, only slows disease progression. The authors developed induced pluripotent stem cell and kidney organoid models of cystinosis that exhibit elevated cystine levels, enlarged lysosomes, increased apoptosis, and defective basal autophagy. Although the latter is not rescued by cysteamine treatment, mTOR inhibition with everolimus was able to restore basal autophagy to levels of healthy controls. Dual treatment of everolimus and cysteamine rescued all of the observed cystinotic phenotypes in the models, suggesting that a combination therapy may improve outcomes in patients with cystinosis.
Keywords: induced pluripotent stem cells, kidney organoids, new treatments, kidney disease, cystinosis
Visual Abstract
Abstract
Background
Mutations in CTNS—a gene encoding the cystine transporter cystinosin—cause the rare, autosomal, recessive, lysosomal-storage disease cystinosis. Research has also implicated cystinosin in modulating the mTORC1 pathway, which serves as a core regulator of cellular metabolism, proliferation, survival, and autophagy. In its severest form, cystinosis is characterized by cystine accumulation, renal proximal tubule dysfunction, and kidney failure. Because treatment with the cystine-depleting drug cysteamine only slows disease progression, there is an urgent need for better treatments.
Methods
To address a lack of good human-based cell culture models for studying cystinosis, we generated the first human induced pluripotent stem cell (iPSC) and kidney organoid models of the disorder. We used a variety of techniques to examine hallmarks of cystinosis—including cystine accumulation, lysosome size, the autophagy pathway, and apoptosis—and performed RNA sequencing on isogenic lines to identify differentially expressed genes in the cystinosis models compared with controls.
Results
Compared with controls, these cystinosis models exhibit elevated cystine levels, increased apoptosis, and defective basal autophagy. Cysteamine treatment ameliorates this phenotype, except for abnormalities in apoptosis and basal autophagy. We found that treatment with everolimus, an inhibitor of the mTOR pathway, reduces the number of large lysosomes, decreases apoptosis, and activates autophagy, but it does not rescue the defect in cystine loading. However, dual treatment of cystinotic iPSCs or kidney organoids with cysteamine and everolimus corrects all of the observed phenotypic abnormalities.
Conclusions
These observations suggest that combination therapy with a cystine-depleting drug such as cysteamine and an mTOR pathway inhibitor such as everolimus has potential to improve treatment of cystinosis.
Cystinosis is a rare, autosomal, recessive, lysosomal-storage disease caused by mutations in the CYSTINOSIN (CTNS) gene which encodes a cystine transporter.1,2 In the absence of CTNS, cystine accumulates within the lysosome where it causes lysosomal dysfunction. Nephropathic cystinosis is the most severe form of cystinosis and is initially associated with the renal proximal tubule failing to reabsorb essential metabolites from the urine (Fanconi syndrome). Kidney defects present between 6 and 18 months of age and progress to renal failure by the end of the first decade of life.1,3 Other complications include derangements in nonrenal tissues such as widespread cystine crystal formation (notably in the cornea), hypothyroidism, and neurologic and muscular symptoms.4–7
The current treatment for cystinosis is lifelong therapy with cysteamine, a molecule that cleaves the cystine disulphide bond to produce mixed disulphides that can escape the lysosome through alternative transporters.8,9 However, cysteamine only slows the progression of renal injury and kidney transplantation is inevitably required later in life.9,10 As a result, there remains a pressing need to develop more effective therapies for cystinosis.
Although there is not yet a complete understanding of the pathogenesis of cystinosis, several lines of evidence indicate that cystine loading causes lysosomal enlargement, impaired proteolysis, and delayed fusion with cargo-loaded vesicles.11–14 Other cellular features of cystinotic cells that are present in different cell types include reduced ATP and GSH levels, mitochondrial damage, oxidative stress, increased apoptosis, and proximal tubule cell dedifferentiation.9,15–27 In addition, cystinotic proximal tubule cells display decreased expression of the endocytic receptors megalin and cubulin as well as impaired megalin recycling.11,12
Defects in macroautophagy (herein called autophagy) are also found in cystinotic cells. Autophagy involves the sequestration of a portion of the cytoplasm by a double-layered membrane known as an autophagosome, followed by fusion with a lysosome to form an autolysosome.28 This final step can be modulated by bafilomycin A1 (BafA1), an inhibitor of autophagolysosome acidification that disrupts autophagosome-lysosome fusion.29 Under resting conditions, basal levels of autophagy are required in a “housekeeping” capacity to degrade long-lived and ubiquitinated proteins; N-linked glycans; damaged organelles, such as mitochondria; and to dampen certain pathways such as inflammatory, Notch, and Wnt signaling.30–32 Under stress conditions, such as starvation, autophagy is greatly upregulated to ensure metabolically useful molecules are recycled to maintain cellular homeostasis. Although autophagy can be induced in cystinotic cells by starvation,33 basal autophagy flux is reduced in a number of cystinotic cell lines, resulting in a build-up of autophagosomes that frequently contain mitochondria.26,33,34 In addition, the separate pathway of chaperone-mediated autophagy, in which specific cytosolic proteins are directly delivered across the lysosomal membrane for degradation, is also defective in fibroblasts from the Ctns-knockout (KO) mouse.33
More recently, CTNS has been implicated in modulating the mammalian target of rapamycin (mTOR) complex 1 (mTORC1) pathway, which integrates both intracellular and extracellular signals and serves as a core regulator of cellular metabolism, proliferation, survival, and autophagy.35 mTORC1 switches between active and inactive states in response to nutrient availability.36,37 Inhibition of mTORC1 by a class of drugs that include everolimus, which is used clinically as an immunosuppressant and anticancer agent, results in activation of autophagy.38,39 CYSTINOSIN physically interacts with mTORC1-binding partners that are necessary for mTORC1 activation by amino acids.40 Loss of CYSTINOSIN function in conditionally immortalized human and mouse proximal tubule cell lines leads to a reduction in mTORC1 activity as well as delayed reactivation after a return to amino acid–rich conditions.40,41
One of the challenges of the cystinosis field is a lack of good human-based cell culture models. To address this, we generated patient-specific and CRISPR/Cas9-edited cystinotic induced pluripotent stem cells (iPSCs). These cells have the advantages of being a renewable source of nonimmortalized cystinotic cells and can be differentiated into numerous tissues, including kidney organoids. Our analysis of CTNS iPSCs and kidney organoids revealed increased cystine levels, enlarged lysosomes, abnormal basal autophagy flux, increased apoptosis and altered gene expression compared with healthy controls, consistent with modeling key aspects of the cystinotic phenotype. We further discovered that some of these defects can be rescued by treatment with cysteamine or everolimus alone, but that combination therapy was the most efficacious. These results suggest that a cysteamine/everolimus combination therapy may have therapeutic potential to improve the treatment, and health outcomes, of individuals with cystinosis.
Methods
iPSC Lines and Maintenance
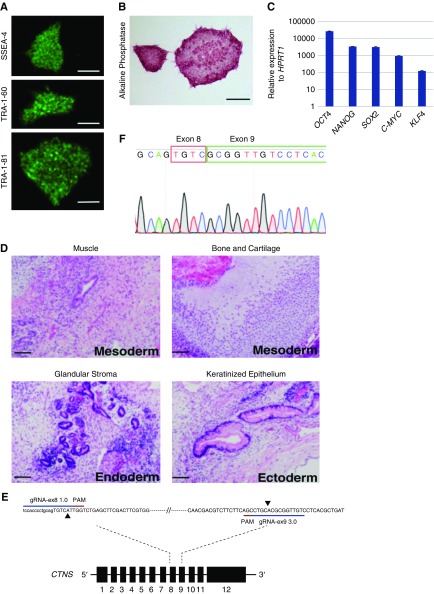
All work was carried out with the approval of the University of Auckland Human Participants Ethics Committee (UAHPEC 8712), Human and Disability Ethics Committee (17/NTA/204), and biosafety approval (GMO05). The CRL1502 clone C32 and the cystinosis iPSC lines were developed in Dr Wolvetang’s42 and Dr. Davidson’s laboratory, respectively. For the patient-derived cystinosis lines (CTNS−/−), adipose-derived mesenchymal cells were derived from a small fat sample (<1 g) taken from the kidney of an individual with nephropathic cystinosis undergoing renal transplantation. The sample was processed to obtain a stromal vascular fraction (SVF) by washing the fat sample in equal volumes of PBS and digesting extracellular matrix at 37°C for 30 minutes with 0.075% collagenase. Enzyme activity was neutralized with DMEM containing 10% FBS and centrifuged at 1200 × g for 10 minutes to obtain a high-density SVF pellet. The pellet was resuspended in 160 μl ammonium chloride and incubated at room temperature (RT) for 10 minutes to lyse contaminating red blood cells. The SVF was collected by centrifugation as above and filtered through a 100-μm nylon mesh to remove cellular debris and incubated overnight at 37°C with 5% carbon dioxide in DMEM with 10% FBS, 1% antibiotic/antimycotic solution. After incubation, plates were washed extensively with PBS to remove residual red blood cells. For transduction, 1×106 adipose-derived mesenchymal cells were seeded 24 hours prior in T75 flasks. Cells were infected with a lentiviral doxycycline-inducible polycistronic vector containing OCT4, SOX2, CMYC, KLF4, and NANOG. Five days after transduction, cells were passaged using trypsin and replated at different densities between 5×104 and 5×105 cells per 10 cm on MEF feeder layers. To induce reprogramming, culture medium was replaced 48 hours later by media supplemented with doxycycline (2 μg/ml). Human iPSC colonies were picked manually based on morphology between 4 and 8 weeks after doxycycline induction. Five CTNS−/− iPSC lines were generated, three of which (36, 108, 157) displayed a normal karyotype (determined by LabPLUS, Auckland, New Zealand). These lines were confirmed to be pluripotent based on immunostaining of cell surface markers (SSEA-4, TRA-1-60, TRA-1-81) and the formation of teratomas after transplantation of 1×106 cells under the kidney capsule of 8-week-old SCID mice (n=3 mice per line), according to established procedures.43
All iPSC lines were cultured on LDEV-free, hESC-qualified, Geltrex-coated tissue culture dishes in mTeSR1 (Stemcell Technologies) medium supplemented with 1% penicillin-streptomycin, and 2.5 μg/ml Plasmocin (InvivoGen). At approximately 70% confluence, cells were dissociated using 1/3 Accutase in Dulbecco PBS (DPBS). Cells were scraped from the dish, pelleted at 800 rpm for 5 minutes, and resuspended in mTeSR1 plus 5 μM Y27632 dihydrochloride (Stemcell Technologies) for the first 24 hours to facilitate cell survival. Unless otherwise stated, all drug treatments (100 nM everolimus, RAD001; Selleckchem; 1 mM cysteamine, 30 mM 3-methyladenine, 50 mM sucrose; Sigma) were added to cell culture medium and incubated with the cells for 24 hours. Cell lines were routinely tested for mycoplasma contamination using a MycoAlert Mycoplasma Detection Kit (Lonza) according to manufacturer’s instructions.
For starvation/refeeding experiments, cells were grown on 12-well culture plates until 70% confluent and incubated for 2 hours in fresh culture medium (basal condition). For starvation, cells were washed twice in PBS and incubated in HBSS for 1 hour. Refeeding was performed by incubating cells in normal culture medium for the indicated time points.
Generation of CTNS-KO Lines by Gene Editing
Guide RNA (gRNA) pairs targeted to introduce a 257-bp deletion in exon 8 and 9 of the CTNS gene were designed using RGEN (http://www.rgenome.net/cas-designer/) and COSMID (http://crispr.bme.gatech.edu/) online tools.44,45 KO efficiencies were first evaluated in HEK293 cells. Optimal gRNAs (gRNA_ex81.0, 5′-TCCACCCCCTGCAGTGTCATTGG-3′; gRNA_ex93.0, 5′-GCGTGAGGACAACCGCGTGCAGG-3′) were cloned into the pSPCas9(BB)-2A-green fluorescent protein (GFP) plasmid (48138; Addgene) and introduced into CRL1502 iPSCs by reverse transfection using TransIT-LT1 (Mirus Bio). GFP-positive cells were isolated 48 hours later by flow cytometric sorting and 8000 cells were plated on a 10-cm Geltrex-coated dish into prewarmed mTeSR1 containing 5 μM Y27632. Medium changes were carried out daily using mTeSR1 without Y27632. Single colonies were manually picked when they reached a suitable size (approximately 10 days postplating), clonally expanded, and screened for biallelic deletions using PCR primers flanking the deleted region (forward CTNS1_primer, 5′- CTCCACCCCCGCCAGTCCTC-3′; reverse CTNS_1primer, 5′-TCTGTGCACGGCTCTCAGC-3′). Homozygote deletions were verified by Sanger sequencing. Three clones, KO 15, 32, and 73 were expanded and karyotyped.
Generation of Cystinotic Kidney Organoids
We used a protocol developed in-house to convert iPSCs into kidney organoids.46 Briefly, iPSCs were cultured on 10-cm Geltrex-coated dishes to approximately 40%–50% confluency, then washed twice with 1× PBS, and treated with 1 mg/ml Dispase for 6 minutes at 37°C. Dispase was removed and cells were washed twice with 1× PBS. Using a cell scraper, cells are manually lifted from the dish; resuspended in BPEL medium47 containing 8 μM CHIR99021, 3 μM Y27632, and 1 mM β-mercaptoethanol; and evenly distributed into ultra-low attachment, 6-well plates (Corning). Half medium changes were carried out on day 2 with BPEL supplemented with 8 μM CHIR99021. On day 3, embryoid bodies (EBs) were allowed to sediment in a 50-ml tube and washed twice with PBS. EBs were returned to the ultra-low 6-well plate and transferred to stage II media (DMEM, 15% KnockOut Serum Replacement [Thermo Fisher], 1% nonessential amino acids, 1% penicillin/streptomycin, 1% HEPES, 1% GlutaMAX, 0.05% polyvinyl alcohol, 2.5 μg/ml Plasmocin) and grown for various periods of time (up to 2 weeks). Tubule formation was observed on day 7–8. Typically, 60%–80% of the EBs become kidney organoids. All drug treatments on organoids were administered on day 13 until day 14 when organoids were harvested for downstream analysis.
Immunostaining
Cells were washed with Tris-buffered saline (TBS) and fixed in 4% paraformaldehyde (PFA)/PBS (w/v) for 10 minutes at RT. After three washes, fixed cells were blocked at RT for at least an hour in blocking solution (TBS containing 2% BSA [w/v] and 10% normal horse serum with 0.1% Triton X-100 [v/v]). Cells were incubated with primary antibody (Supplemental Table 1) in the blocking solution overnight at 4°C in a humidified chamber. After 24 hours, cells were washed three times with 1× TBST (TBS containing 0.1% Triton X-100 [v/v]) and incubated with secondary antibodies (Supplemental Table 2) at 1:500 dilution in the blocking solution for 2 hours at RT. Cells were incubated with 10 μg/ml Hoechst 33258 for 5 minutes, washed twice with TBST, and mounted with ProLong Gold (Thermo Fisher) before imaging. Cells were imaged using a Zeiss LSM710 confocal microscope.
Immunochemistry of Organoids
Organoids were fixed in 4% PFA/PBS overnight at 4°C. After washing with 1× PBS plus 0.1% Tween 20, organoids were transferred into an embedding mold and filled with embedding medium (1% low-melting agarose, 0.9% agar, 5% sucrose). Once solidified, the blocks were transferred into 70% ethanol and incubated at 4°C overnight. Over the next 2 days, the blocks were transferred through a series of 95% and 2× 100% ethanol, 50:50 ethanol/xylol, 100% xylol, 1 hour each, rocking at RT, followed by 50:50 xylol/paraffin at 65°C overnight, and changes of paraffin every 4 hours. After embedding, the blocks were sectioned at 6 μm on a Leica microtome. Sections were air dried and then stored at 4°C. Immunohistochemistry was performed using standard procedures. Paraffin sections were deparaffinized at 65°C for 30 minutes, then incubated in two changes of xylol (10 minutes each). Antigen retrieval (10 mM sodium citrate buffer plus 0.05% Tween 20, pH 6.0, at 95°C for 30 minutes) was carried out for all antibodies. Immunostainings were imaged using a Zeiss LSM710 or Leica SP8 confocal microscope. For a list of antibodies see Supplemental Tables 1 and 2.
Magic Red-Cathepsin B Staining
At 48 hours before imaging, iPSC cells were seeded onto Geltrex-coated, 35-mm Fluro dishes (WPI). Before staining, cells were washed once with 1× DPBS. Cells were incubated for 1 hour with 26× Magic Red Cathepsin B (Bio-Rad, Hercules, CA) in mTeSR1. Hoechst 33258 was added for the final 15 minutes. Once staining was completed, the dyes were washed off with 1× DPBS and the cells were fixed with 4% PFA for 10 minutes. Cells were mounted with ProLong Gold and coverslips were added. Images were taken using a Zeiss LSM710 confocal microscope.
DQ-BSA Assay
iPSCs were seeded onto Geltrex-coated, 35-mm Fluro dishes 48 hours before imaging. On the day of the assay, cells were washed with 1× DPBS before incubation with 20 μg/ml working solution of DQ-BSA green (Invitrogen) in mTeSR1 for 3 hours. Hoechst 33258 was added for the final 15 minutes. After incubation, cells were washed with DPBS and replenished with fresh mTeSR1 media. Images were taken using a Zeiss LSM710 confocal microscope.
Apoptosis Assay
The ApopTag Plus Fluorescein In Situ Detection Kit (Millipore) was used to detect apoptosis following the manufacturer’s instructions. Briefly, cells were seeded onto Geltrex-coated, 35-mm Fluro dishes 48 hours before imaging. Cells were fixed with 1% PFA for 10 minutes at RT. Cells were washed twice with 1× PBS for 5 minutes. Precooled ethanol/acetic acid in a ratio of 2:1 was added for 5 minutes at −20°C to postfix cells. Cells were washed twice with 1× PBS. Equilibration buffer (75 μl) was added immediately for 10 seconds, followed by 55 μl working strength terminal deoxynucleotidyl transferase enzyme, and incubated for 1 hour at 37°C. Working strength stop/wash buffer was added and incubated for 10 minutes at RT. Cells were washed three times before applying anti-digoxigenin conjugate and incubating for 30 minutes in the dark at RT. Cells were washed four times and Hoechst 33258 was added for 15 minutes before mounting with ProLong Gold and adding coverslips. Images were taken using a Zeiss LSM710 confocal microscope.
Transmission Electron Microscopy
Samples (dissociated iPSCs or whole kidney organoids) were fixed in 2.5% glutaraldehyde and 0.1 M phosphate buffer, pH 7.4, at 4°C and kept in the fixative until processing. Samples were washed three times with 0.1 M phosphate buffer for 10 minutes, then fixed in 1% osmium tetroxide in 0.1 M phosphate buffer for an hour at RT, and washed twice in 0.1 M phosphate buffer for 5 minutes. The samples were then dehydrated in a graded series of ethanol washes for 10 minutes each at RT (50%, 70%, 90%, and twice at 100%), followed by two propylene oxide washes for 10 minutes at RT. The samples were then infiltrated with a graded series of propylene oxide/resin mix (2:1, 1:1, 1:2) for 30 minutes each, before being embedded in freshly made pure resin overnight. The next day, the samples were placed into molds and polymerized at 60°C for 48 hours. All washes were performed on a rocker. Sectioned samples were imaged using a Tecnai G2 Spirit Twin transmission electron microscope.
Transient Transfection of iPSCs and Kidney Organoids
For reverse transfection, 1 μg of plasmid was incubated with 2 μl TransIT-LT1 and 100 μl OptiMem (Gibco) for 15 minutes at RT. DNA complexes were added to Geltrex-coated, 35-mm Fluro dishes containing either mTeSR1 (iPSCs) or stage II media (organoids) for 15 minutes at RT. iPSCs were dissociated using 1/3 Accutase. Organoids were dissociated by incubating them in 100 μl TrypLE Express (gibco) at 37°C for up to 10 minutes. Once dissociated, cells were centrifuged at 800 × g for 5 minutes and resuspended in mTeSR1/stage II media. A total of 1×106 cells were then added to the dish containing DNA complexes and incubated at 37°C overnight. Cells were analyzed 24 hours post-transfection.
Immunoblotting
Cells were seeded onto 12-well plates at 2.5×105 cells per well 24–48 hours before the experiment. Cells were washed twice in ice-cold PBS and scraped on ice into 80 μl of ice-cold radioimmunoprecipitation assay buffer supplemented with protease (cOmplete Mini; Roche) and phosphatase inhibitors (1 mM sodium orthovanadate, 100 mM sodium fluoride, 1 mM β-glycerol phosphate, 2.5 mM sodium pyrophosphate). Samples were centrifuged at 12,000 × g for 10 minutes at 4°C. Protein content of the supernatant was determined using the Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). Equal amounts of protein were boiled in Laemmli buffer at 95°C for 5 minutes. A total of 20 μg of protein was separated by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad) using the semidry Trans-Blot Turbo device (Bio-Rad). Membranes were blocked in 5% BSA in TBST for 1 hour at RT, and probed using specific antibodies for LC3BII (1:1000), p-P70S6KThr389 (1:500), P70S6K (1:1000), p-RPS6Ser235–236 (1:1000), RPS6 (1:1000), 4EBP-1 (1:1000), p-EIF4ESer209 (1:1000), EIF4E (1:2000) (3868, 9205, 2078, 4856, 2317, 9644, 9741, and 9742 respectively; Cell Signaling Technologies), and β-actin (A2228, 1:10,000; Sigma). Primary antibodies were incubated overnight at 4°C with gentle agitation. The next morning, membranes were probed with either anti-rabbit or anti-mouse linked to horseradish peroxidase secondary antibodies (1:12,000 dilution; Supplemental Table 2) for 1 hour at RT. Membranes were exposed using enhanced chemiluminescence reagent (ECL Select Kit; GE HealthCare) and chemiluminescent signals were captured using the ChemiDoc image device (Bio-Rad). Densitometry analysis of protein bands were quantified using ImageJ software (National Institutes of Health, Bethesda, MD). The intensity of each band was recorded relative to a pooled control sample run on each gel.
4E-BP1 is a phosphoprotein that separates into multiple electrophoretic forms, therefore, to measure their phosphorylation state, the mobility-shift method was used.48,49 Phosphorylation results in the protein migrating at a higher apparent molecular mass. Total 4E-BP1 protein was recorded as the expression of all forms of 4E-BP1 (α-, β-, γ-, and δ-forms) and phosphorylation of 4E-BP1 was expressed as the ratio of the top bands relative to the total protein.
Image Analysis of Lysosomes and Fluorescent Puncta
DQ-BSA and Magic Red confocal raw images at a 63× magnification (approximately 10 random fields per condition) were analyzed using ImageJ analysis software. Nuclei were manually counted. To obtain a cross-sectional area of the enlarged vesicles, particle analysis was performed and the number of vesicles >10 μm2 were determined per field. Data were expressed as average number of enlarged vesicles per cell and statistically analyzed. For the measurement of autophagic puncta, cells were transfected with the LC3-mCherry-GFP vector and imaged by confocal microscopy (10 random fields per condition containing approximately one to three cells in three independent experiments) and analyzed using ImageJ. Nuclei and red and yellow puncta were manually counted using the ImageJ counting tool and the percentage of each puncta per cell was calculated.
ApopTag Plus–treated cells were viewed at 20× magnification (approximately 10 random fields per condition) and images analyzed using ImageJ. Nuclei and green puncta (apoptotic bodies) were manually counted using the ImageJ counting tool and the percentage of apoptotic bodies per cell was calculated for each condition.
HPLC–Tandem Mass Spectrometry for Cystine Measurements
A benchtop triple quadrupole mass spectrometer, Agilent 6140 (Agilent Technologies, Palo Alto, CA), operated in positive ionization mode with the multimode ionization probe source was used to determine the concentration of cystine. This was coupled to an Agilent 1200 HPLC system (Agilent Technologies). Samples were prepared as outlined by Jamalpoor et al.50 Briefly, frozen cell pellets were resuspended and thawed in 100 μl NEM (5 mmol/L in 0.1 M sodium phosphate buffer, pH 7.2) on ice. Cells were sonicated on ice three times for 10 seconds with 20 seconds cooling intervals (one cycle, 80% amplitude). Protein was precipitated by adding 50 μl sulfosalicylic acid (15% w/v) and sample was centrifuged at 20,000 rcf for 10 minutes at 4°C. Supernatant was recovered and diluted 1:10 in 0.1% formic acid. A volume of 5 μl of internal standard (20 μM cystine-D4) was added and the sample was pipetted into a glass vial. Chromatographic separation was achieved on a Thermo Scientific Hypercarb column (2.1×150 mm; Thermo Scientific) and was maintained at 30°C. The mobile phase consisted of water with 0.01% formic acid and acetonitrile (ACN) with 0.1% formic acid with fast gradient elution at a flow rate of 0.3 ml/min and run time of 5 minutes. The sample volume injected was 4 µl and the autosampler was set at 5°C. Instrument parameters of the mass spectrometer were: gas flow, 6 L/min; gas temperature, 300°C; vaporizer temperature, 250°C; nebulizer, 40 psi; and capillary voltage, 2500 V. Data were acquired and analyzed with Agilent MassHunter Software. A standard curve was plotted with the observed peak area ratio of analyte to the internal standard against the concentration of the analyte to extract the slope and intercept.
Mass Spectrometry for GSH Measurements
Cells were lysed on ice with cold 50% ACN and centrifuged at 16,000 × g for 10 minutes at 4°C. The supernatant was transferred to a cold Eppendorf tube and stored at −80°C until samples were ready to be processed. A volume of 2 μl of sample was added to 5 μl of a 50% solution of ACN and 50 mM ammonium bicarbonate (ABC). A volume of 3 μl of sample was treated with either 3 μl 1 mM tris(2-carboxyethyl) phosphine hydrochloride or 2 mM monobromobimane (MBrB) in 50% ACN/ABC and incubated at RT in the dark for 20 minutes. A volume of 3 μl of 2 mM MBrB and 3 μl 50 mM ABC was added to the tris(2-carboxyethyl) phosphine hydrochloride–treated samples. ACN (6 μl, 25%) was added to the MBrB-treated set. Samples were incubated at RT in the dark for 20 minutes. Following incubation, 950 μl 0.1% formic acid and 5 μl 4.292 μM GSH internal standard was added to all samples. 10 mg HLB SPE cartridges were conditioned with 0.5 ml methanol followed by 0.5 ml 0.1% formic acid. The entire sample was loaded onto the conditioned cartridge and washed with 1 ml 0.1% formic acid. Samples were eluted into clean tubes with 0.3 ml 10% ACN in 0.1% formic acid. Samples were spun in a SpeedVac until volumes were reduced to 50–100 μl. Samples were either injected neat or diluted 1:3 in 0.1% formic acid and run on a QStar XL LC-MS System and through an LC column (Zorbax SB-C18, 3.5 μm, 150×0.3 mm; Agilent Technologies).
ATP Assay
Cells were lysed using 1× lysis buffer for 10 minutes on ice. Lysate was centrifuged at 12,000 × g for 5 minutes at 4°C. Supernatant was collected and an ATP Determination Kit (Invitrogen) was used as per manufacturer’s directions. Samples were read using a VICTOR X Multilabel Plate Reader (PerkinElmer). The cell pellet was resuspended in 0.1 M sodium hydroxide and bicinchoninic acid assay performed 24 hours later to determine protein concentration.
Reabsorption Assay
A total of 20 μg/ml of 10 kDa Texas Red-dextran (Invitrogen) was added to stage II culture medium for 48 hours. Organoids were washed in stage II medium for 5 hours before fixation in 4% PFA, paraffin embedding, and sectioning as described above.
RNA Extraction, cDNA Synthesis, and Quantitative PCR Analysis
Cells were first washed with 1× PBS before being lysed in GENEzol for 5 minutes. RNA was extracted using GENEzol TriRNA Pure kit (Geneaid). cDNA was synthesized using qScript cDNA SuperMix (Quanta). For quantitative PCR (qPCR), PerfeCTa SYBR Green FastMix (Quanta) was used. qPCR was performed on a QuantStudio 6 Flex Real-Time PCR machine. Primers used are listed in Supplemental Table 3. Samples were normalized to HPRT1 and CREBP expression. Gene expression was calculated using the ddCt method.51 Error bars represent SD from technical triplicates.
RNA Sequencing and Analysis
Total RNA from four samples per iPSC line were prepared using the GENEzol TriRNA Pure kit. The quality (RNA integrity number), concentration, and purity (OD260/230 and OD260/280) of the RNA was determined on Bioanalyser (RNA nano chip), Qubit, and Nanodrop instruments. Library preparation and sequencing were performed commercially (New Zealand Genomics Limited, Otago, New Zealand). Libraries were prepared using the TruSeq standard total RNA kit with standard protocols (Illumina). Paired-end sequencing (2×125 bp) was performed on an Illumina HiSeq 2500 sequencer. Reads were adapter filtered and quality trimmed using BBDuk version 37.75 with a quality cutoff of phred=10 (trimq=10) and, to reduce the potential mapping errors, any reads <50 bp after trimming were removed. Quality control–filtered reads were mapped to the human genome (GRCh38) downloaded from ENSEMBL (www.ensembl.org/Homo_sapiens/Info/Index) using HISAT2 (version 2.0.5) in stranded mapping mode (–rna-strandness RF). Read counts were generated from the alignment files using HT-Seq (version 0.6.0) under the Union mode and strand option set to “reverse.” DESeq2 was used to generate differential expression calls and statistics for control versus KO comparison based on the observed read counts for each gene. Expression changes were declared significant for q-value <0.05. Heatmaps were generated in R using the pheatmap_1.0.8 package. Gene Ontology (GO) term enrichments were analyzed using the R package goseq (version 1.22). Enrichment was tested for all differentially expressed genes with a false discovery rate–corrected P value <0.05. The GO terms were also false-discovery-rate corrected at the same rate.
Statistical Analysis
Data are presented as the mean±SEM. GraphPad PRISM software version 7 (GraphPad Software) was used for all statistical analyses. The statistical significance of the differences between two groups was calculated using an unpaired t test. For between three or more groups, one-way ANOVA was used. A P value <0.05 was considered to be statistically significant.
Results
Generation of CTNS-iPSC Lines
The patient-specific CTNS iPSCs were generated as described in methods from adipose-derived mesenchymal cells grown from a fat sample from a patient with nephropathic cystinosis undergoing renal transplantation. Exon sequencing revealed compound heterozygosity for two described CTNS mutations: a 57-kb deletion and an L158P missense mutation in exon 8.2 Cells were reprogrammed into iPSCs and three CTNS−/− lines (36, 108, 157) with normal karyotypes (results not shown) were identified. All three lines stained positive for pluripotency markers alkaline phosphatase, SSEA-4, Tra-1-60, and Tra-1-81 and gave rise to teratomas containing tissues from all three germ layers (Figure 1, A, B and D, data for CTNS−/−36 shown). Re-expression of endogenous OCT4, NANOG, SOX2, CMYC, and KLF4 was confirmed by qPCR (Figure 1C, representative data for CTNS−/−36). Because all of these lines displayed similar phenotypes, line 36 was used for subsequent analyses (herein called CTNS−/−). In addition to patient-specific lines, we also generated independent CTNS-KO lines by performing CRISPR/Cas9 gene editing in CRL1502 iPSCs.42 gRNAs were used to introduce a 257-bp deletion in exon 8 and 9 of the CTNS gene, resulting in deletion of the second transmembrane domain (Figure 1, E and F). Three lines with homozygote deletions (KO 15, 32, and 73) were identified by Sanger sequencing. Because all three CTNS knockout (CTNSKO) lines displayed a similar phenotype (Supplemental Figure 1, A, B and C), CTNSKO line KO73 was used for subsequent experiments.
Figure 1.
Patient-derived CTNS iPSCs display markers of pluripotency. (A) CTNS−/− iPSCs stained for stem cell surface antigens SSEA-4, TRA-1-60, and TRA-1-81. Scale bar, 500 µm. (B) CTNS−/− iPSCs stained for alkaline phosphatase. Scale bar, 500 µm. (C) qPCR of endogenous genes relative to HPRT1 expression. Plotted data are mean±SD. (D) Hematoxylin and eosin–stained histologic sections of tumors derived from SCID mice after injection of CTNS−/− iPSCs under kidney capsule. All three germ layers were identified: mesoderm, endoderm, and ectoderm (n=3). Scale bar, 100 µm. (E) Schematic overview of the CRISPR-based strategy to disrupt the CTNS gene in wild-type iPSCs. The extent of the deletion in exon 8 and exon 9 is marked with black arrowheads. (F) Sanger sequencing chromatogram shows resulting sequence in CTNSKO iPSCs. PAM, protospacer adjacent motif.
CTNS iPSCs Load Cystine
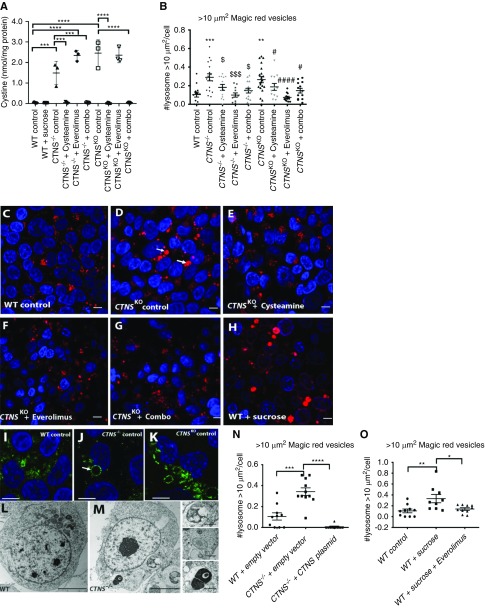
Cystine was measured in cystinotic and control iPSCs by HPLC–tandem mass spectrometry, revealing 33- to 54-fold higher levels in CTNS−/− and CTNSKO iPSCs compared with the CRL1502 control cells (Figure 2A, Supplemental Table 4). To assess whether cystine levels were responsive to cysteamine, CTNS iPSCs were treated with a dose range of cysteamine (10 µM, 100 µM, and 1 mM) for 1 or 24 hours. Whereas 10 µM cysteamine had no effect on cystine levels, 100 µM and 1 mM decreased cystine, with 1 mM being the most effective and significantly reducing cystine levels in CTNS−/− and CTNSKO iPSCs (Figure 2A, Supplemental Figure 1, D and E).
Figure 2.
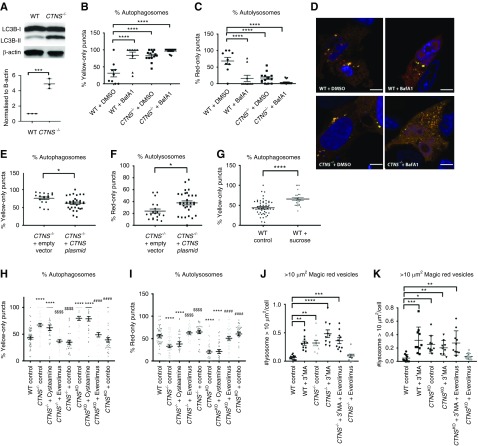
CTNS iPSCs display cystine accumulation and enlarged lysosmes. (A) Amount of cystine (nmol/mg of protein) in wild type (WT), CTNS−/− iPSCs, and CTNSKO iPSCs with various treatments (1 mM cysteamine; 100 nM everolimus; combo, 1 mM cysteamine and 100 nM everolimus; 24 hours). One-way ANOVA performed, data plotted as mean±SEM, three independent experiments. **P<0.01, ***P<0.001, ****P<0.0001. (B) Graph displaying quantification of the average number of Magic Red vesicles (lysosomes) per cell over 10 µm2. One-way ANOVA performed (n=600 cells from eight to ten random fields per condition, 20 cells per field, three independent experiments), data plotted as mean±SEM. ***P=0.001, WT control versus CTNS−/− control; **P<0.01, WT control versus CTNSKO control; $P<0.05, CTNS−/− control versus CTNS−/− 1 mM cysteamine and CTNS−/−combination; $$$P<0.001, CTNS−/− control versus CTNS−/−100 nM everolimus; #P<0.05, CTNSKO control versus CTNSKO 1 mM cysteamine and CTNS−/−combination; ###P<0.001, CTNSKO control versus CTNSKO 100 nM everolimus. (C–H) Representative images of fluorescent staining with Magic Red in WT control, CTNSKO control, and CTNSKO with treatments (1 mM cysteamine; 100 nM everolimus; combo, 1 mM cysteamine and 100 nM everolimus; 24 hours) and WT iPSCs treated with 50 mM sucrose for 24 hours. Scale bar, 10 µm. (I) Representative immunofluorescence staining with anti-LAMP1 (green) in WT iPSCs, and (J) CTNS−/− and (K) CTNSKO iPSCs, respectively. Arrow indicates enlarged vesicles. Scale bar, 10 µm. (L and M) Transmission electron micrograph of (L) WT and (M) CTNS−/− iPSCs showing enlarged vesicles. Scale bars, 5 µm in (L) and 1 µm in (M). (N) Graph displaying quantification of the average number of Magic Red vesicles per cell over 10 µm2 in WT iPSCs and CTNS−/− iPSCs overexpressing empty vector (pcDNA 3.1) or exogenous CTNS-GFP. One-way ANOVA performed (n=300 cells from five to eight random fields per condition, 20 cells per field, three independent experiments), data plotted as mean±SEM. ***P<0.001, ****P<0.0001. (O) Average number of Magic Red vesicles per cell over 10 µm2. WT iPSCs treated with 50 mM sucrose or sucrose and 100 nM everolimus for 24 hours. One-way ANOVA performed (n=300 cells from five random fields per condition, 20 cells per field, three independent experiments), all data are plotted mean±SEM. *P<0.05, **P<0.01. CTNSKO, CRISPR-generated cystinotic KO iPSCs. Nuclei counterstain in (C–H) and (I–K) was Hoechst.
CTNS iPSCs Display Enlarged Lysosomes
To assess the size and distribution of lysosomes in the cystinotic iPSCs and to functionally confirm their lysosomal identity, the cells were incubated with Magic Red, a substrate that is degraded by cathepsin B and fluoresces inside lysosomes and endolysosomes. Enlarged Magic Red+ puncta were observed more frequently in CTNS iPSCs compared with controls and tended to cluster in a perinuclear location (Figure 2, B, C and D). Quantification of the subset of enlarged lysosomes, defined as having an optical cross-sectional area of >10 µm2, showed the average number per cell was approximately 2.5-fold higher in CTNS iPSCs compared with controls (Figure 2B). To further confirm these structures are lysosomes, we examined cystinotic iPSCs with the lysosomal marker LAMP1 by immunofluorescence and at the ultrastructural level by electron microscopy. CTNS−/− and CTNSKO iPSCs were found to contain a mixture of small to enlarged LAMP1+ puncta, whereas control iPSCs show qualitatively fewer enlarged LAMP1+ puncta (Figure 2, I, J and K). Consistent with the LAMP1 and Magic Red data, we observed large degradative/storage-like bodies in CTNS−/− but not control iPSCs by electron microscopy (Figure 2, L and M). As expected for dysfunctional lysosomes, these bodies contain electron-dense material, intraluminal vesicles, and undigested membranes, and likely represent enlarged lysosomes and/or amphisomes (Figure 2M). To show this phenotype is due to loss of CYSTINOSIN, we performed rescue experiments by transfecting CTNS iPSCs with a plasmid encoding cystinosin-GFP and performed analysis 24 hours later.52 Overexpression of cystinosin-GFP, which colocalizes with Magic Red+ puncta (Supplemental Figure 1, G–J), reduced the number of enlarged lysosomes to levels below that seen in control iPSCs (Figure 2N).
To assess trafficking through the endocytic pathway, CTNS iPSCs and control iPSCs were incubated with DQ-BSA, a fluid-phase probe that becomes fluorescent when it reaches the lysosome, for 3 hours.53 Quantitation of positive puncta per cell showed that CTNS iPSCs had similar numbers of puncta compared with control iPSCs (average 5.6 total puncta per cell for CTNS iPSCs and 7.8 total puncta per cell for control iPSCs; Supplemental Figure 1, K–M). This result is consistent with the endocytic flux of fluid-phase cargos through the endocytic pathway being relatively normal in CTNS iPSCs, despite the presence of enlarged lysosomes.
We next sought to phenocopy the enlarged-lysosome phenotype of CTNS iPSCs in wild-type cells using sucrose, which accumulates within lysosomes of normal cells.54,55 Treatment of control iPSCs with 50 mM sucrose for 24 hours led to an increase in the number of enlarged lysosomes, similar to that seen in CTNS iPSCs, while having no effect on cystine loading (Figure 2, A, C, H and O). This observation supports the notion that the enlarged-lysosome phenotype of CTNS iPSCs is due to cystine accumulation. To confirm this, treatment of CTNS iPSCs with 1 mM cysteamine for 24 hours resulted in a reduction in the average number of enlarged lysosomes per cell, although it did not completely rescue to control levels (Figure 2, B and E). The total number of lysosomes per cell was not significantly affected by cysteamine treatment (Supplemental Figure 1F).
GSH, ATP, Apoptosis, and Mitochondria in CTNS iPSCs
We next examined GSH, ATP, apoptosis, and mitochondria because prior reports suggest these are altered in cystinotic cells.9,15–27 GSH levels (oxidized and reduced), ATP, and apoptosis were quantitated whereas mitochondria were assessed qualitatively by transmission electron microscopy and immunohistochemistry. No significant differences were seen with GSH (Supplemental Figure 1, N and O) or ATP (Supplemental Figure 1P). Apoptosis was quantified using the ApopTag detection assay and quantitation of positive puncta per cell revealed that CTNS iPSCs displayed >1.4-fold greater levels of apoptosis compared with controls (Supplemental Figure 1Q). Cysteamine treatment was unable to reduce the amount of apoptosis. No overt differences in mitochondrial morphology were observed in CTNS iPSCs compared with controls (Supplemental Figure 1, R–U).
RNA-Sequencing Analysis Reveals Differentially Regulated Genes in CTNSKO iPSCs
To gain further insights into the phenotype of CTNS iPSCs, we performed RNA-sequencing (RNA-seq) to identify differentially expressed genes between CTNSKO iPSCs and their isogenic control cells (n=4 biological repeats for each; National Center for Biotechnology Information BioProject accession number PRJNA591635). We found a total of 12,750 differentially expressed genes with 8792 significantly upregulated and 3958 significantly downregulated (P<0.05), compared with controls. Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed several significantly enriched pathways in CTNSKO iPSCs that include the ribosome, spliceosome, proteasome, oxidative phosphorylation, protein processing in the endoplasmic reticulum, and ubiquitin-mediated proteolysis (cutoff P<1×10−6; Table 1). Interestingly, pathways linked to Huntington, Parkinson, and Alzheimer disease were enriched.56 GO term enrichment analysis yielded a much more extensive list of gene sets that was more difficult to summarize (data not shown). However, in the “biological process” category, we found enrichment for pathways implicated in cystinosis including autophagy, vesicle trafficking, redox homeostasis, the mTOR pathway, and protein catabolism (Table 2).
Table 1.
Kyoto Encyclopedia of Genes and Genomes pathways significantly enriched in CTNSKO iPSCs (cutoff P<1×10−6)
| Entry | Name | Class | P Value | numDEInCat | numInCat |
|---|---|---|---|---|---|
| hsa03010 | Ribosome; Homo sapiens (human) | Genetic information processing; translation | p<0.001 | 81 | 87 |
| hsa05016 | Huntington disease; H. sapiens (human) | Human diseases; neurodegenerative diseases | p<0.001 | 134 | 182 |
| hsa05012 | Parkinson disease; H. sapiens (human) | Human diseases; neurodegenerative diseases | p<0.001 | 93 | 129 |
| hsa04110 | Cell cycle; H. sapiens (human) | Cellular processes; cell growth and death | p<0.001 | 105 | 124 |
| hsa03040 | Spliceosome; H. sapiens (human) | Genetic information processing; transcription | p<0.001 | 101 | 127 |
| hsa03008 | Ribosome biogenesis in eukaryotes; H. sapiens (human) | Genetic information processing; transcription | p<0.001 | 66 | 76 |
| hsa00190 | Oxidative phosphorylation; H. sapiens (human) | Metabolism; energy metabolism | p<0.001 | 90 | 131 |
| hsa04141 | Protein processing in endoplasmic reticulum; H. sapiens (human) | Genetic information processing; folding, sorting, and degradation | p<0.001 | 130 | 166 |
| hsa04120 | Ubiquitin-mediated proteolysis; H. sapiens (human) | Genetic information processing; folding, sorting, and degradation | p<0.001 | 111 | 135 |
| hsa03013 | RNA transport; H. sapiens (human) | Genetic information processing; translation | p<0.001 | 116 | 150 |
| hsa05010 | Alzheimer disease; H. sapiens (human) | Human diseases; neurodegenerative diseases | p<0.001 | 116 | 166 |
| hsa03050 | Proteasome; H. sapiens (human) | Genetic information processing; folding sorting, and degradation | p<0.001 | 37 | 44 |
Table 2.
GO Terms in biologic process category enriched in CTNSKO iPSCs
| GO Term | Term | P Value | numDEInCat | numInCat |
|---|---|---|---|---|
| GO:0016192 | Vesicle-mediated transport | p<0.001 | 769 | 1281 |
| GO:0016236 | Macroautophagy | p<0.001 | 202 | 299 |
| GO:0006914 | Autophagy | p<0.001 | 269 | 416 |
| GO:0045454 | Cell redox homeostasis | p<0.001 | 46 | 58 |
| GO:0000045 | Autophagosome assembly | p<0.001 | 57 | 74 |
| GO:0045022 | Early endosome to late endosome transport | p<0.001 | 32 | 36 |
| GO:0006900 | Membrane budding | p<0.001 | 56 | 72 |
| GO:0000422 | Mitophagy | p<0.001 | 132 | 200 |
| GO:0032006 | Regulation of Tor signaling | p<0.001 | 50 | 63 |
| GO:0006890 | Retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum | p<0.001 | 31 | 36 |
| GO:1901800 | Positive regulation of proteasomal protein catabolic process | p<0.001 | 67 | 93 |
| GO:0032008 | Positive regulation of TOR signaling | p<0.001 | 23 | 26 |
| GO:0032436 | Positive regulation of proteasomal ubiquitin-dependent protein catabolic process | p<0.001 | 59 | 80 |
| GO:0006623 | Protein targeting to vacuole | p<0.001 | 26 | 29 |
| GO:0045324 | Late endosome to vacuole transport | p<0.001 | 1 | 14 |
| GO:0031929 | TOR signaling | p<0.001 | 57 | 75 |
| GO:0070534 | Protein K63-linked ubiquitination | p<0.001 | 33 | 42 |
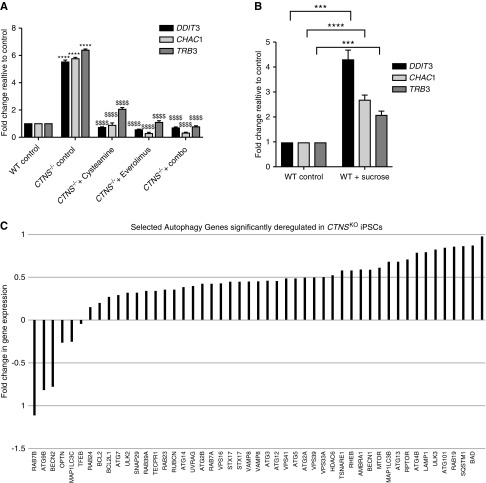
We next examined whether some of the differentially expressed genes would have utility as molecular biomarkers of the cystinotic phenotype. From the top 50 differentially expressed genes (Supplemental Figure 2), we focused on DDIT3 (also known as CHOP), which encodes a transcription factor belonging to the “integrated stress response” involved in cellular adaptation to stress.57 In addition, we identified two downstream targets of DDIT3: TRIB3, encoding a pseudokinase that acts as a negative feedback regulator of DDIT3,58 and CHAC1, encoding an enzyme that degrades GSH.59 Using qPCR we independently confirmed that DDIT3, TRIB3, and CHAC1 were significantly upregulated in CTNS iPSCs compared with control iPSCs (Figure 3A, Supplemental Figure 3A). To assess whether the expression of this gene triad is responsive to cysteamine, we treated CTNS iPSCs with 1 mM cysteamine for 24 hours and found that they significantly decreased to near-control levels. Incubation of control iPSCs with 50 mM sucrose for 24 hours also resulted in a significant upregulation of DDIT3, TRIB3, and CHAC1, indicating that these genes, although not specific biomarkers of cystinotic cells per se, may “read out” lysosomal dysfunction caused by the accumulation of substrates (Figure 3B).
Figure 3.
CTNS iPSCs display deregulation of genes. (A) qPCR of genes of interest—DDTI3, TRB3, and CHAC1—in wild-type (WT) and CTNS−/− iPSCs with various treatments (1 mM cysteamine; 100 nM everolimus: combo, 1 mM cysteamine and 100 nM everolimus; 24 hours) normalized to HPRT and CREEBP and expressed as fold change to WT. One-way ANOVA performed, data plotted as mean±SD. ****P<0.0001, compared with relative WT control; $$$$P<0.0001, compared with CTNSKO control. (B) qPCR analysis of target genes in WT iPSCs treated with 50 mM sucrose for 24 hours. Two-tailed unpaired t test performed, data plotted as mean±SD. ***P<0.001, ****P<0.0001. (C) Fold changes of selected autophagy genes from RNA-seq that are deregulated in CTNSKO iPSCs.
The mTORC1 Pathway Appears Unaffected in CTNS iPSCs
Closer scrutiny of the autophagy genes identified from the GO term analysis revealed that CTNSKO iPSCs show a slight upregulation of MTOR as well as two of its downstream targets (ULK1 and ATG13) compared with control cells (Figure 3C). To assess mTORC1 activity in CTNS iPSCs we performed Western blotting in triplicate for phosphorylated S6, RPS6, 4EBP-1, and EIF4e (all downstream targets of mTORC1), under basal conditions and after starvation for 60 minutes, followed by refeeding.60,61 We found no statistical difference in p-S6, p-RPS6, p-4EBP-1, and p-EIF4e levels between the cystinotic iPSCs and control cells under basal conditions (Supplemental Figure 3B) and, unlike prior reports,40,41 we did not detect a delay in the reactivation of mTORC1 at 2.5, 7, 12, or 15 minutes after refeeding (Supplemental Figure 3B and data not shown). Together, these observations indicate there are no significant defects in mTORC1 activity in CTNS iPSCs under basal conditions or after starvation.
Basal Autophagy Flux is Perturbed in CTNS iPSCs
Of the autophagy-related genes identified in the GO term analysis, there are genes involved in early through to late processes of autophagy including autophagosome formation (SQSTM1, BECLIN1, LC3B, RAB7A), movement (HDAC6), and tethering and fusion (RUBICON, UVRAG, VPS16, VAMP8, STX17, TSNARE1, STX17, SNAP29; these and others are shown in Figure 3C). In most cases, these genes are upregulated in CTNSKO iPSCs compared with controls (Figure 3C). Notably, an increase in SQSTM1/p62 can be indicative of a block in autophagy flux.
To explore basal autophagy levels, we first measured the levels of autophagosome-specific protein LC3B-II by Western blotting. Consistent with RNA-seq data, we found higher levels of LC3B-II in CTNS−/− iPSCs compared with control iPSCs, indicating either an increase in the number of autophagosomes or a decrease in autophagosome degradation (Figure 4A). To quantify autophagosome and autolysosome numbers, we transfected CTNS iPSCs and control iPSCs with a plasmid encoding the mCherry-LC3B-GFP sensor that fluorescently labels autophagosomes in yellow and autolysosomes in red.62 At 24 hours post-transfection, cells were analyzed and we found that under basal conditions CTNS−/− cells have approximately 2.6-fold higher levels of yellow puncta (autophagosomes) compared with control iPSCs (Figure 4, B and C).
Figure 4.
Basal autophagy is dysfuncational in CTNS iPSCs. (A) Representative Western blot against autophagosome marker LC3B-II and β-actin from wild-type (WT) and CTNS iPSCs and graph displaying quantification of three independent experiments. (B and C) Percentage of cells with yellow-only puncta (autophagosomes) and red-only puncta (autolysosomes) treated with DMSO or 400 nM BafA1 for 4 hours (representative of n=30 cells, from ten random fields per condition containing approximately one to three cells in three independent experiments). One-way AVOVA performed, the data are mean±SEM. ****P<0.0001 relative to WT. (D) Cells transfected with tandem mCherry-LC3B-GFP plasmid showing red and yellow puncta. Nuclei counter stain was 4′,6-diamidino-2-phenylindole. Scale bar, 10 µm. (E and F) Percentage of yellow-only and red-only puncta after exogenous expression of CTNS in CTNS−/− iPSCs. Two-tailed unpaired t test performed, the data are mean±SEM. *P<0.05. (G) Sucrose (50 mM) treatment for 24 hours on WT iPSCs to induce a cystinotic phenotype. Percentage of yellow-only puncta shown. Two-tailed unpaired t test performed, the data are mean±SEM. ****P<0.0001. (H and I) Effects of drug treatments on percentages of yellow and red puncta; CTNS iPSCs treated with 1 mM cysteamine alone, 100 nM everolimus alone, or a combination of both for 24 hours. One-way ANOVA performed, data plotted as mean±SEM (n=30 cells from ten random fields per condition containing approximately one to three cells in three independent experiments). ****P<0.0001, WT versus CTNS−/− and CTNSKO; $$$$P<0.0001, CTNS−/− versus CTNS−/− 100 nM everolimus and CTNS−/− combination; ####P<0.0001, CTNSKO versus CTNSKO 100 nM everolimus and CTNSKO combination. (J and K) Average number of Magic Red vesicles per cell over 10 µm2 in WT iPSCs and CTNS−/− or CTNSKO iPSCs treated with 3 mM 3-ΜΑ and 100 nM everolimus for 24 hours. One-way AVOVA performed, values are mean±SEM (n=300 cells from five to eight random fields per condition, 20 cells per field, three independent experiments). *P<0.05, **P<0.01, ***P<0.001.
To assess flux through the autophagy pathway, we treated CTNS iPSCs and control iPSCs expressing the mCherry-LC3B-GFP sensor with 400 nM BafA163 for 4 hours. Although BafA1 induced a 2.7-fold increase in the percentage of yellow puncta in control iPSCs compared with vehicle (DMSO)-treated cells, only a slight but nonsignificant increase was seen in CTNS iPSCs (Figure 4, B, C and D). To confirm the autophagy defect was specific to a loss of CYSTINOSIN, we cotransfected CTNS−/− iPSCs with a cystinosin-encoding plasmid, pCMV-CFP (to allow tracking of transfected cells) and mCherry-LC3B-GFP, resulting in an approximately 1.2-fold reduction in the percentage of yellow puncta (Figure 4, E and F). Taken together, these results indicate that loss of CYSTINOSIN in iPSCs causes an accumulation of autophagosomes under basal conditions due to reduced fusion of lysosomes with autophagosomes.
The basal autophagy block in CTNS iPSCs may be caused by the accumulation of cystine in the lysosome. To explore this, we treated control iPSCs with 50 mM sucrose for 24 hours and then transfected them with the mCherry-LC3B-GFP sensor. We found that the percentage of yellow puncta increased 1.5-fold in sucrose-loaded cells compared with control cells, indicative of a reduction in basal autophagic flux (Figure 4G). Given this, we next tested whether treatment with cysteamine would ameliorate the basal autophagy flux defect of CTNS iPSCs transfected with the mCherry-LC3B-GFP sensor. Unexpectedly, we found that cysteamine treatment did not greatly improve basal autophagy flux (Figure 4, H and I). We conclude that the basal autophagy defect in CTNS iPSCs is caused by a loss of CYSTINOSIN but this cannot be ameliorated by cysteamine treatment.
Basal Autophagy Flux Defects Are Rescued in CTNS iPSCs by mTORC1 Inhibition
The failure of cysteamine to restore basal autophagy flux in CTNS iPSCs may provide a rationale for why cysteamine therapy is not curative and led us to speculate that activating autophagy via mTORC1 inhibition may provide additional therapeutic benefit. To test this, we treated CTNS iPSCs for 24 hours with 100 nM everolimus and examined basal autophagy flux. We found that everolimus restores the number of yellow puncta (autophagosomes) to control levels and correspondingly increased the number of autolysosomes, in agreement with similar results reported using Ctns−/− mouse fibroblasts (Figure 4, H and I).33 Importantly, dual treatment of 1 mM cysteamine and 100 nM everolimus had similar effects as everolimus alone, without any sign of combination toxicity (Figure 4, H and I).
Cystine Levels Remain High in CTNS iPSCs after mTORC1 Inhibition
We then assessed cystine levels in everolimus and combined everolimus/cysteamine-treated cells. Everolimus alone had no significant effect on cystine levels in CTNSKO iPSCs but caused a 1.5-fold increase in CTNS−/− iPSCs (Figure 2A). Combination treatment decreased cystine in both cystinotic iPSC lines to levels similar to cysteamine treatment alone (Figure 2A), indicating that activation of the mTORC1 pathway does not interfere with the ability of cysteamine to deplete cystine.
mTORC1 Inhibition Reduces Enlarged Lysosomes in CTNS-iPSCs via Autophagy
Next we examined the effect of everolimus treatment on the enlarged lysosome phenotype. We found that everolimus reduces the average number of enlarged lysosomes to near-normal levels, making it more effective than cysteamine alone (Figure 2B). Qualitatively, we found that everolimus reduced perinuclear clustering of the lysosomes, but this could not be quantified due to the high cell density and small cytoplasmic volume of the iPSC cultures (Figure 2F).
Combined everolimus/cysteamine treatment yielded intermediate results with an approximately twofold reduction compared with untreated CTNS iPSCs, indicating that cysteamine interferes with the ability of everolimus to reduce the number of enlarged lysosomes (Figure 2, B and G). In addition, everolimus treatment reduced the number of enlarged lysosomes induced by sucrose loading, suggesting that its effects on the lysosome are not specific to cystinotic cells (Figure 2O). The total number of lysosomes was unaffected by treatments (Supplemental Figure 1F.
To determine if the action of everolimus on enlarged lysosomes was dependent on autophagy, we investigated the effects of 3-methyladenine (3-MA), an inhibitor of autophagy that acts downstream of mTORC1.64 First, we treated control and CTNS iPSCs for 24 hours with 30 mM of 3-MA alone. We observed an approximately four- to fivefold increase in the number of enlarged lysosomes per cell in control iPSCs whereas levels in CTNS−/− and CTNSKO iPSCs did not significantly increase further (Figure 4, J and K). Treating CTNS iPSCs with both 3-MA and everolimus failed to have any significant effect on the number of enlarged lysosomes (Figure 4, J and K), providing strong evidence that the effects of everolimus are mediated via stimulation of autophagy.
Everolimus Reduces the Levels of Apoptosis in CTNS iPSCs
Everolimus alone and combined treatment reduced the level of apoptosis in CTNS iPSCs (Supplemental Figure 1Q), indicating that cysteamine does not interfere with the ability of everolimus to decrease apoptosis.
Everolimus Reduces Expression of CHOP, TRB3, and CHAC1 in CTNS iPSCs
We then assessed the effects of everolimus alone and combined everolimus/cysteamine treatment on the expression of DDIT3, TRB3, and CHAC1. In both CTNS−/− and CTNSKO iPSCs, everolimus alone and combined treatment significantly reduced the expression levels of the gene triad to near-control levels, in keeping with the notion that these genes are providing a readout of lysosome dysfunction in iPSCs (Figure 3A, Supplemental Figure 3A).
Characterization of Cystinotic Kidney Organoids
Having established the potential therapeutic effects of combined everolimus/cysteamine treatment in CTNS iPSCs, we next assessed whether these compounds would show efficacy on human cystinotic kidney tissue, using a kidney organoid protocol we developed.46 Using this approach, we matured CTNS−/−, CTNSKO, and control cells into kidney organoids for 14 days.46 Similar to our results obtained with undifferentiated CTNS iPSCs, we found that the CTNS−/− and CTNSKO organoids also display cystine loading but show no differences in the ratio of GSH/GSH disulphide compared with isogenic control organoids (Figure 5A, Supplemental Table 4 and Figure 3C).
Figure 5.
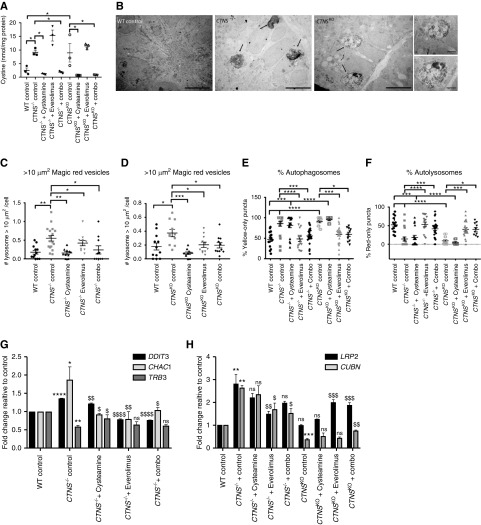
Cystinotic kidney organoids derived from CTNS iPSCs display increased cystine, enlarged lysosomes, basal autphagy defects and deregulation of genes. (A) Amount of cystine (nmol/mg of protein) in wild-type (WT), CTNS−/−, and CTNSKO organoids with various treatments. One-way ANOVA performed, data plotted as mean±SEM, n=30 organoids per experiment, three independent experiments. **P<0.01, ***P<0.001, ****P<0.0001. (B) Representative transmission electron microscope images of WT, CTNS−/−, and CTNSKO organoids displaying enlarged multivesicular bodies (black arrows). Scale bar, 5 µm in (A) and 2 µm in (B). (C) Average number of Magic Red vesicles per cell over 10 µm2 in WT, CTNS−/−, and (D) CTNSKO organoids. One-way AVOVA performed, the values are mean±SEM (n=300 cells from ten random fields per condition, ten cells per field, three independent experiments). *P<0.05, **P<0.01, ***P<0.001. (E and F) Effects of 1 mM cysteamine, 100 nM everolimus, and combination treatments for 24 hours on autophagy flux as determined by the percentage of yellow and red puncta in day 14 CTNS−/− and CTNSKO organoids. One-way ANOVA performed, all data plotted as mean±SEM (n=30 cells from ten random fields per condition containing approximately one to three cells in three independent experiments). *P<0.05, **P<0.01. (G) qPCR of genes of interest in control CTNS−/− organoids with various treatments expressed as fold change relative to control, data plotted as mean±SD. One-way ANOVA performed, data plotted as mean±SD. *P<0.5, **P<0.01, ****P<0.0001, compared with relative WT control; $P<0.5, $$P<0.01, $$$$P<0.0001, compared with CTNS−/− control. (H) qPCR of Megalin (LRP2) and Cubilin (CUBN) with various treatments expressed as fold change relative to control. One-way ANOVA performed, data plotted as mean±SD. **P<0.01, ***P<0.001, compared with relative WT control; $P<0.5, $$P<0.01, $$$P<0.001 compared with relative CTNS control.
At the level of light microscopy, both the CTNS−/− and CTNSKO kidney organoids appear equivalent to the control organoids and we see no evidence of abnormalities such as the characteristic cystinotic “swan-neck” lesion (data not shown). At the ultrastructural level, the mitochondria in CTNS organoids display similar numbers and morphology to controls (Supplemental Figure 4A). However, we qualitatively observed the presence of numerous enlarged vacuoles in the tubules of cystinotic organoids, reminiscent of the degradative/storage-like bodies seen in CTNS iPSCs, whereas these are rarely seen in control organoids (Figure 5B).
Defective endocytosis, another hallmark of cystinosis,11,12 was initially investigated by incubation of CTNSKO kidney organoids with 10 kDa Texas Red–labeled dextran, which is specifically taken up by LTL+ proximal tubules.65 No difference in dextran uptake was observed in cystinotic organoids compared with controls, suggesting that the absorptive function of the proximal tubule by fluid-phase endocytosis is intact (Supplemental Figure 4, B and C). Receptor-mediated endocytosis using fluorescently labeled albumin or transferrin were not technically successful in our hands, however, immunostaining of the endocytic receptor CUBN and the distal tubule marker CDH1 was consistent with unperturbed tubule segmentation and polarization in cystinotic organoids (Supplemental Figure 4D). Expression of CUBN and LRP2 was examined in the organoids by qPCR. iPSC-CTNSKO organoids showed normal LRP2 and significantly reduced CUBN expression compared with the isogenic control, whereas iPSC-CTNS−/− organoids displayed significantly higher levels of both genes compared with the control (Figure 5H).
To quantify the enlarged lysosomes in cystinotic organoids, we dissociated the tissue at day 12 into single cells and on day 14 incubated them in Magic Red for 1 hour. We found that CTNS organoids display approximately two- to threefold more enlarged lysosomes compared with control organoids (Figure 5, C and D, Supplemental Figure 4E).
To determine if basal autophagy flux is affected in CTNS kidney organoids, we reverse transfected dissociated single cells with the mCherry-LC3B-GFP sensor plasmid. We found that cystinotic kidney organoid cells display approximately 1.5- to twofold more autophagosomes compared with control organoid cells, consistent with a defect in basal autophagy (Figure 5, E and F, Supplemental Figure 4F).
We next assessed mRNA expression of DDIT3, TRB3, and CHAC1 in cystinotic and control kidney organoids using qPCR. We found that DDIT3 and CHAC1 were significantly increased in cystinotic kidney organoids compared with controls (Figure 5G, Supplemental Figure 3D). TRB3 was not upregulated, indicating that this gene may only be useful as a biomarker of lysosomal dysfunction in certain cell types.
We next examined the effects of single and combination treatment of cysteamine and everolimus on CTNS kidney organoids with respect to the phenotypes of cystine loading, CUBILIN and LRP2 expression, enlarged lysosomes, basal autophagy flux, and DDIT3, TRB3, and CHAC1. To this end, we treated day 13 CTNS and control kidney organoids with 1 mM cysteamine, 100 nM everolimus, or a combination of both drugs for 24 hours. In keeping with our observations in CTNS iPSCs, we found that cysteamine alone and when combined with everolimus reduced cystine levels whereas no effect was seen with everolimus treatment alone (Figure 5A). Treatments had no effects on dextran uptake or CUBN levels by immunostaining (Supplemental Figure 4, B and C) and no consistent trends were observed for CUBN and LRP2 transcripts by qPCR (Figure 5H). Cysteamine or everolimus alone and combined treatments reduced the numbers of enlarged lysosomes (Figure 5, C and D, Supplemental Figure 4E). Everolimus alone activated autophagy, as did combined treatment, but cysteamine alone had no effect (Figure 5, E and F, Supplemental Figure 4F). qPCR analysis of the treated kidney organoids showed that the expression levels of DDIT3 and CHAC1 were normalized to near-control values in response to cysteamine alone and combined treatments, as well as everolimus-alone treatment in the case of CTNS−/− organoids. However, everolimus-alone treatment significantly reduced DDIT3 but had no significant effect on CHAC1 in CTNSKO organoids (Figure 5G, Supplemental Figure 3D).
Discussion
In this report we generated new human models of cystinosis in the form of cystinotic iPSCs and kidney organoids and demonstrated a phenotype characterized by cystine loading, enlarged lysosomes, altered gene expression, increased apoptosis, and defective basal autophagy. Using this unique human-based platform, we tested the therapeutic effects of cysteamine and everolimus and found that a combination therapy was able to mitigate all of the observed cystinotic phenotypes in a beneficial and effective manner. Furthermore, these models have the potential to be used as a preclinical model for testing other therapeutics as well as a novel tool to deepen our understanding of the pathogenic mechanisms at play in cystinosis.
Of the defects observed in cystinotic iPSCs and kidney organoids, the block in basal autophagy flux appears most significant because cysteamine is unable to rescue it. The finding that the number of autophagosomes in CTNS iPSCs does not significantly increase in the presence of BafA1 may indicate an accumulation of autophagosomes under basal conditions due to a defect in lysosomal fusion. Although CYSTINOSIN is a hydrogen ion–driven transporter52 and lysosomal pH is important for lysosomal fusion,66 previous reports have shown that the pH of cystinotic lysosomes is normal.12 Thus, the underlying cause for the failure of lysosomes to fuse with autophagosomes remains unclear. There are a number of protein complexes that coordinate this fusion and have a requirement for the physical movement of both autophagosomes and lysosomes. Therefore, it is possible that one or more of these processes is affected in cystinotic cells.67 The observation that sucrose loading induces the equivalent phenotype in noncystinotic iPSCs and that other lysosomal-storage diseases show basal autophagy flux defects points to the intralysosomal accumulation of material as a generic cause of the flux defect.68–71 Our finding that CTNS iPSCs upregulate a number of genes in the autophagy pathway suggests that compromised basal autophagy leads to the activation of transcriptional feedback mechanisms to compensate for the reduced flux.
If cystine accumulation impairs autophagosome-lysosome fusion and reduces basal autophagy flux, then cysteamine treatment would have been expected to have rescued the basal autophagy phenotype of CTNS iPSCs. Interpreting the failure of cysteamine to restore basal autophagy is complicated by a report showing that cysteamine treatment of HeLa cells under basal conditions affects the autophagy pathway in two places: first by acting early to induce autophagosome formation but then acting later to inhibit autolysosome maturation.72 Thus, it is likely that although cysteamine may restore lysosomal functionality in cystinotic cells by depleting cystine, its independent inhibitory effects on autolysosome maturation induces an equivalent block in basal autophagy flux. This additional activity of cysteamine may be related to its antioxidant properties because reactive oxygen species are known to regulate autophagy via the redox modification of several autophagy components and antioxidants can inhibit basal autophagy.73,74
Several mouse studies have demonstrated a critical role for basal autophagy in maintaining proximal tubule function and support the notion that continued renal dysfunction in patients with cystinosis who are cysteamine treated is linked to a failure to restore basal autophagy. Specifically, it has been shown that blocking autophagy in kidney cells in vivo and in vitro under nutrient-replete conditions results in the accumulation of degradative vacuoles, deformed mitochondria, and p62- and ubiquitin-positive inclusion bodies.75–77 Similar observations have been reported for kidney biopsies and urinary cells from patients with cystinosis and in primary proximal tubule cells from the Ctns−/− mouse.26,27,75,78 Consistent with one of the functions of basal autophagy being the removal of damaged mitochondria, work using cystinotic mouse primary proximal tubule cells has led to a model in which defective autophagy-mediated clearance of damaged mitochondria (mitophagy) causes oxidative stress and triggers renal epithelial cell dysfunction.27 Although we did not detect an increased level of oxidative stress or deformed mitochondria in CTNS iPSCs, this may be due to iPSCs relying on glycolysis rather than oxidative phosphorylation for their energy needs.79 By contrast, proximal tubule cells contain a large quantity of mitochondria to drive membrane transport processes and it is reasonable to conclude that these cells would be highly dependent on efficient basal autophagy for mitochondrial quality control. Other tissues such as hepatocytes, neurons, heart, and skeletal muscle have also been found to be dependent on basal autophagy80 and, although neural and muscle tissues are eventually affected in individuals with cystinosis, it is the proximal tubule that is affected early in the disease. One explanation for this is that the lysosomal system of proximal tubule cells is under a high degradative load due to the uptake and degradation of albumin and other plasma proteins harboring disulphide bonds.81,82 Thus, the level of lysosomal cystine accumulation and the resulting basal autophagy dysfunction (assuming it is proportional to cystine load) would be expected to be pronounced in cystinotic proximal tubule cells in comparison to other tissues. We did not observe any signs of swan-neck lesions, defects in cell dedifferentiation, or mitochondria damage in our cystinotic kidney organoids. This is not unexpected given that kidney organoids are only cultured for a few weeks, represent a fetal stage of nephron differentiation, and lack filtration (limiting their degradative load).46
Prior studies using immortalized mouse and human proximal tubule cells found a transport-independent role for CYSTINOSIN that implicated it in the positive regulation of mTORC1.40,41 Despite detecting a slight upregulation of MTOR RNA and two of its downstream targets (ULK1 and ATG13) in CTNS iPSCs, we found no evidence to indicate that mTORC1 protein activity is altered. Similarly, no effect in mTORC1 activity was found in mouse cystinotic fibroblasts.33 At this stage, the cause for these discrepancies is not known but may be related to metabolic differences between cell lines. Alternatively, there may be confounding influences of the viral antigens used for immortalization because these can influence mTORC1 levels.83
We found that inhibiting mTORC1 with everolimus was capable of overcoming the basal autophagy block. Although the mechanism of this rescue is not known, a potential mediator is TFEB, a master transcriptional regulator of lysosomal biogenesis and autophagy.70 TFEB is downregulated in cystinotic cells84 but is activated after mTORC1 inhibition and can induce autophagy.85 Everolimus also reduced the frequency of enlarged lysosomes in CTNS iPSCs and because this effect was abrogated by 3-MA it is likely that these structures are being cleared by autophagy. This notion is in line with the discovery that autophagy is involved in the removal/recycling of aged and dysfunctional lysosomes (lysophagy) to prevent cell damage caused by the leakage of hydrolytic enzymes.86,87 At this stage it remains unclear if the basal autophagy block seen in cystinotic cells also results in a lysophagy defect. It is an attractive hypothesis because large lysosomes are particularly vulnerable to rupture and some cystinotic cells have been found to be sensitive to apoptosis.14,24,25,88,89 Interestingly, if dysfunctional lysosomes are not removed, the total number of lysosomes remains unchanged, even if some of them are nonfunctional.87 Thus, lysosome quality control is critical for maintaining cellular degradative capabilities and therefore lysophagy defects could contribute additional stress to cystinotic proximal tubule cells. Unlike previous reports, cysteamine treatment was unable to reduce the amount of baseline apoptosis in our cells.24,25 However, prior studies examined the effect of cysteamine pretreatment followed by the induction of apoptosis, rather than an effect on baseline levels, indicating that cysteamine may have a protective effect when cells are pretreated.
At present, mTOR inhibitors are used in the clinic as immunosuppressants and as treatments for some cancers.90,91 Our work suggests that everolimus, and related rapamycin derivatives, may also have therapeutic potential to treat cystinosis. This notion is consistent with studies of the lysosomal disorders mucopolysaccharidosis and Niemann–Pick disease, where overcoming a block in autophagic flux improves cell viability.68,92,93 In the case of Niemann–Pick disease type C, a combination therapy of low dose hydroxypropyl-β-cyclodextrin (which depletes the cholesterol accumulating in the lysosome) coupled with an autophagy stimulator has been proposed.94 We have arrived at the same conclusion with our findings and hypothesize that dual treatment of individuals who are cystinotic with cysteamine and an autophagy inducer such as everolimus will improve long-term outcomes. The next step in testing this hypothesis requires animal studies in either the zebrafish or rodent models.14,95,96 In considering these experiments, careful attention will need to be given to finding the lowest effective dosing schedule, because mTOR inhibitors have side effects that include dyslipidemia and impaired glucose homeostasis that may complicate their long-term use in patients with cystinosis.97 However, new mTOR pathway drugs continue to be developed and these may provide superior alternatives in the future.
Disclosures
Dr. Davidson reports grants from Cystinosis Research Foundation and grants from Cystinosis Foundation Ireland, during the conduct of the study. Dr. Harrison reports personal fees from Vertex, outside the submitted work. Dr. Holm reports grants from Cystinosis Research Foundation and grants from Cystinosis Foundation Ireland, outside the submitted work. Dr. D’Souza, Dr. Hollywood, Dr. Przepiorski, Mr. Sreebhavan, and Dr. Wolvetang have nothing to disclose.
Funding
This work was supported by the United States Cystinosis Research Foundation, Cystinosis Ireland, and Valrae Collins philanthropic support for Dr. Przepiorski.
Supplementary Material
Acknowledgments
We thank Adrian Turner, Jacqui Ross, and Ratish Kurian for help with electron microscopy and microscopy.
Dr. Davidson, Dr. Harrison, Dr. Hollywood, and Dr. Holm conceptualized the study; Dr. Davidson and Dr. Holm cosupervised the study; Dr. Davidson and Dr. Hollywood wrote the manuscript; Dr. D’Souza and Mr. Sreebhavan performed Western blot and HPLC experiments, respectively; Dr. Hollywood, Dr. Holm, and Dr. Przepiorski designed and performed experiments; Dr. Holm established the cystinotic patient lines; Dr. Davidson, Dr. Harrison, and Dr. Holm acquired funding; Dr. Wolvetang generated the CRL1502 iPSC line and reviewed manuscript.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2019070712/-/DCSupplemental.
Supplemental Table 1. Primary antibodies.
Supplemental Table 2. Secondary antibodies.
Supplemental Table 3. List of primers for qPCR.
Supplemental Table 4. Average numerical values of cystine measurements and protein. concentration of iPSCs and kidney organoids.
References
- 1.Gahl WA, Thoene JG, Schneider JA: Cystinosis. N Engl J Med 347: 111–121, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Kalatzis V, Nevo N, Cherqui S, Gasnier B, Antignac C: Molecular pathogenesis of cystinosis: Effect of CTNS mutations on the transport activity and subcellular localization of cystinosin. Hum Mol Genet 13: 1361–1371, 2004 [DOI] [PubMed] [Google Scholar]
- 3.Emma F, Nesterova G, Langman C, Labbé A, Cherqui S, Goodyer P, et al.: Nephropathic cystinosis: An international consensus document. Nephrol Dial Transplant 29[Suppl 4]: iv87–iv94, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scarvie KM, Ballantyne AO, Trauner DA: Visuomotor performance in children with infantile nephropathic cystinosis. Percept Mot Skills 82: 67–75, 1996 [DOI] [PubMed] [Google Scholar]
- 5.Trauner DA, Williams J, Ballantyne AO, Spilkin AM, Crowhurst J, Hesselink J: Neurological impairment in nephropathic cystinosis: Motor coordination deficits. Pediatr Nephrol 25: 2061–2066, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilmer MJ, Emma F, Levtchenko EN: The pathogenesis of cystinosis: Mechanisms beyond cystine accumulation. Am J Physiol Renal Physiol 299: F905–F916, 2010 [DOI] [PubMed] [Google Scholar]
- 7.Ivanova E, De Leo MG, De Matteis MA, Levtchenko E: Cystinosis: Clinical presentation, pathogenesis and treatment. Pediatr Endocrinol Rev 12[Suppl 1]: 176–184, 2014 [PubMed] [Google Scholar]
- 8.Gahl WA, Reed GF, Thoene JG, Schulman JD, Rizzo WB, Jonas AJ, et al.: Cysteamine therapy for children with nephropathic cystinosis. N Engl J Med 316: 971–977, 1987 [DOI] [PubMed] [Google Scholar]
- 9.Levtchenko EN, van Dael CM, de Graaf-Hess AC, Wilmer MJ, van den Heuvel LP, Monnens LA, et al.: Strict cysteamine dose regimen is required to prevent nocturnal cystine accumulation in cystinosis. Pediatr Nephrol 21: 110–113, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Gahl WA, Balog JZ, Kleta R: Nephropathic cystinosis in adults: Natural history and effects of oral cysteamine therapy. Ann Intern Med 147: 242–250, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Raggi C, Luciani A, Nevo N, Antignac C, Terryn S, Devuyst O: Dedifferentiation and aberrations of the endolysosomal compartment characterize the early stage of nephropathic cystinosis. Hum Mol Genet 23: 2266–2278, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Ivanova EA, De Leo MG, Van Den Heuvel L, Pastore A, Dijkman H, De Matteis MA, et al. : Endo-lysosomal dysfunction in human proximal tubular epithelial cells deficient for lysosomal cystine transporter cystinosin. Plos One 10: e0120998, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulman JD, Bradley KH, Seegmiller JE: Cystine: Compartmentalization within lysosomes in cystinotic leukocytes. Science 166: 1152–1154, 1969 [DOI] [PubMed] [Google Scholar]
- 14.Gaide Chevronnay HP, Janssens V, Van Der Smissen P, N’Kuli F, Nevo N, Guiot Y, et al.: Time course of pathogenic and adaptation mechanisms in cystinotic mouse kidneys. J Am Soc Nephrol 25: 1256–1269, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coor C, Salmon RF, Quigley R, Marver D, Baum M: Role of adenosine triphosphate (ATP) and NaK ATPase in the inhibition of proximal tubule transport with intracellular cystine loading. J Clin Invest 87: 955–961, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levtchenko E, de Graaf-Hess A, Wilmer M, van den Heuvel L, Monnens L, Blom H: Altered status of glutathione and its metabolites in cystinotic cells. Nephrol Dial Transplant 20: 1828–1832, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Wilmer MJ, de Graaf-Hess A, Blom HJ, Dijkman HB, Monnens LA, van den Heuvel LP, et al.: Elevated oxidized glutathione in cystinotic proximal tubular epithelial cells. Biochem Biophys Res Commun 337: 610–614, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Wilmer MJ, Christensen EI, van den Heuvel LP, Monnens LA, Levtchenko EN: Urinary protein excretion pattern and renal expression of megalin and cubilin in nephropathic cystinosis. Am J Kidney Dis 51: 893–903, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Wilmer MJ, Kluijtmans LA, van der Velden TJ, Willems PH, Scheffer PG, Masereeuw R, et al.: Cysteamine restores glutathione redox status in cultured cystinotic proximal tubular epithelial cells. Biochim Biophys Acta 1812: 643–651, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Chol M, Nevo N, Cherqui S, Antignac C, Rustin P: Glutathione precursors replenish decreased glutathione pool in cystinotic cell lines. Biochem Biophys Res Commun 324: 231–235, 2004 [DOI] [PubMed] [Google Scholar]
- 21.Mannucci L, Pastore A, Rizzo C, Piemonte F, Rizzoni G, Emma F: Impaired activity of the gamma-glutamyl cycle in nephropathic cystinosis fibroblasts. Pediatr Res 59: 332–335, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Laube GF, Shah V, Stewart VC, Hargreaves IP, Haq MR, Heales SJ, et al.: Glutathione depletion and increased apoptosis rate in human cystinotic proximal tubular cells. Pediatr Nephrol 21: 503–509, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Cherqui S, Sevin C, Hamard G, Kalatzis V, Sich M, Pequignot MO, et al.: Intralysosomal cystine accumulation in mice lacking cystinosin, the protein defective in cystinosis. Mol Cell Biol 22: 7622–7632, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park M, Helip-Wooley A, Thoene J: Lysosomal cystine storage augments apoptosis in cultured human fibroblasts and renal tubular epithelial cells. J Am Soc Nephrol 13: 2878–2887, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Park MA, Pejovic V, Kerisit KG, Junius S, Thoene JG: Increased apoptosis in cystinotic fibroblasts and renal proximal tubule epithelial cells results from cysteinylation of protein kinase Cdelta. J Am Soc Nephrol 17: 3167–3175, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Sansanwal P, Yen B, Gahl WA, Ma Y, Ying L, Wong LJ, et al.: Mitochondrial autophagy promotes cellular injury in nephropathic cystinosis. J Am Soc Nephrol 21: 272–283, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Festa BP, Chen Z, Berquez M, Debaix H, Tokonami N, Prange JA, et al.: Impaired autophagy bridges lysosomal storage disease and epithelial dysfunction in the kidney. Nat Commun 9: 161–177, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, et al.: Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mauvezin C, Neufeld TP: Bafilomycin A1 disrupts autophagic flux by inhibiting both V-ATPase-dependent acidification and Ca-P60A/SERCA-dependent autophagosome-lysosome fusion. Autophagy 11: 1437–1438, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Into T, Horie T, Inomata M, Gohda J, Inoue JI, Murakami Y, et al.: Basal autophagy prevents autoactivation or enhancement of inflammatory signals by targeting monomeric MyD88. Sci Rep 7: 1009, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casares-Crespo L, Calatayud-Baselga I, García-Corzo L, Mira H: On the role of basal autophagy in adult neural stem cells and neurogenesis. Front Cell Neurosci 12: 339, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seino J, Wang L, Harada Y, Huang C, Ishii K, Mizushima N, et al.: Basal autophagy is required for the efficient catabolism of sialyloligosaccharides. J Biol Chem 288: 26898–26907, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Napolitano G, Johnson JL, He J, Rocca CJ, Monfregola J, Pestonjamasp K, et al.: Impairment of chaperone-mediated autophagy leads to selective lysosomal degradation defects in the lysosomal storage disease cystinosis. EMBO Mol Med 7: 158–174, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieberman AP, Puertollano R, Raben N, Slaugenhaupt S, Walkley SU, Ballabio A: Autophagy in lysosomal storage disorders. Autophagy 8: 719–730, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Efeyan A, Zoncu R, Sabatini DM: Amino acids and mTORC1: From lysosomes to disease. Trends Mol Med 18: 524–533, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al.: The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM: Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.González E, Andrés A, Polanco N, Hernández A, Morales E, Hernandez E, et al.: Everolimus represents an advance in immunosuppression for patients who have developed cancer after renal transplantation. Transplant Proc 41: 2332–2333, 2009 [DOI] [PubMed] [Google Scholar]
- 39.Gude E, Gullestad L, Andreassen AK: Everolimus immunosuppression for renal protection, reduction of allograft vasculopathy and prevention of allograft rejection in de-novo heart transplant recipients: Could we have it all? Curr Opin Organ Transplant 22: 198–206, 2017 [DOI] [PubMed] [Google Scholar]
- 40.Andrzejewska Z, Nevo N, Thomas L, Chhuon C, Bailleux A, Chauvet V, et al.: Cystinosin is a component of the vacuolar H+-ATPase-Ragulator-Rag complex controlling mammalian target of rapamycin complex 1 signaling. J Am Soc Nephrol 27: 1678–1688, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanova EA, van den Heuvel LP, Elmonem MA, De Smedt H, Missiaen L, Pastore A, et al.: Altered mTOR signalling in nephropathic cystinosis. J Inherit Metab Dis 39: 457–464, 2016 [DOI] [PubMed] [Google Scholar]
- 42.Briggs JA, Sun J, Shepherd J, Ovchinnikov DA, Chung TL, Nayler SP, et al.: Integration-free induced pluripotent stem cells model genetic and neural developmental features of down syndrome etiology. Stem Cells 31: 467–478, 2013 [DOI] [PubMed] [Google Scholar]
- 43.Zhu F, Sun B, Wen Y, Wang Z, Reijo Pera R, Chen B: A modified method for implantation of pluripotent stem cells under the rodent kidney capsule. Stem Cells Dev 23: 2119–2125, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bae S, Park J, Kim JS: Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G: COSMID: A web-based tool for identifying and validating CRISPR/cas off-target sites. Mol Ther Nucleic Acids 3: e214, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Przepiorski A, Sander V, Tran T, Hollywood JA, Sorrenson B, Shih JH, et al.: A simple bioreactor-based method to generate kidney organoids from pluripotent stem cells. Stem Cell Reports 11: 470–484, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng ES, Davis R, Stanley EG, Elefanty AG: A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nat Protoc 3: 768–776, 2008 [DOI] [PubMed] [Google Scholar]
- 48.Zeng N, D’Souza RF, Figueiredo VC, Markworth JF, Roberts LA, Peake JM, et al.: Acute resistance exercise induces Sestrin2 phosphorylation and p62 dephosphorylation in human skeletal muscle. Physiol Rep 5: e13526, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan AH, D’Souza RF, Beals JW, Zeng N, Prodhan U, Fanning AC, et al. : The degree of Aminoacidemia after dairy protein ingestion does not modulate the postexercise anabolic response in young men: A randomized controlled trial. J Nutr 149: 1511–1522, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jamalpoor A, Sparidans RW, Pou Casellas C, Rood JJM, Joshi M, Masereeuw R, et al.: Quantification of cystine in human renal proximal tubule cells using liquid chromatography-tandem mass spectrometry. Biomed Chromatogr 32: e4238, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408, 2001 [DOI] [PubMed] [Google Scholar]
- 52.Kalatzis V, Cherqui S, Antignac C, Gasnier B: Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J 20: 5940–5949, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vázquez CL, Colombo MI: Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol 452: 85–95, 2009 [DOI] [PubMed] [Google Scholar]
- 54.Cohn ZA, Ehrenreich BA: The uptake, storage, and intracellular hydrolysis of carbohydrates by macrophages. J Exp Med 129: 201–225, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeCourcy K, Storrie B: Osmotic swelling of endocytic compartments induced by internalized sucrose is restricted to mature lysosomes in cultured mammalian cells. Exp Cell Res 192: 52–60, 1991 [DOI] [PubMed] [Google Scholar]
- 56.Beck M: The link between lysosomal storage disorders and more common diseases. J Inborn Errors Metab Screen 4: 1–8, 2016 [Google Scholar]
- 57.Ryoo HD: Long and short (timeframe) of endoplasmic reticulum stress-induced cell death. FEBS J 283: 3718–3722, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohoka N, Yoshii S, Hattori T, Onozaki K, Hayashi H: TRB3, a novel ER stress-inducible gene, is induced via ATF4-CHOP pathway and is involved in cell death. EMBO J 24: 1243–1255, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar A, Tikoo S, Maity S, Sengupta S, Sengupta S, Kaur A, et al.: Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep 13: 1095–1101, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, et al.: The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J 14: 5279–5287, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM: RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A 95: 1432–1437, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kimura S, Noda T, Yoshimori T: Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452–460, 2007 [DOI] [PubMed] [Google Scholar]
- 63.Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y: Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 266: 17707–17712, 1991 [PubMed] [Google Scholar]
- 64.Seglen PO, Gordon PB: 3-Methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A 79: 1889–1892, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Witzgall R, Kränzlin B, Gretz N, Obermüller N: Impaired endocytosis may represent an obstacle to gene therapy in polycystic kidney disease. Kidney Int 61[Suppl]: S132–S137, 2002 [DOI] [PubMed] [Google Scholar]
- 66.Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S: Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 3: 154–157, 2007 [DOI] [PubMed] [Google Scholar]
- 67.Nakamura S, Yoshimori T: New insights into autophagosome–lysosome fusion. J Cell Sci 130: 1209–1216, 2017 [DOI] [PubMed] [Google Scholar]
- 68.Sarkar S, Carroll B, Buganim Y, Maetzel D, Ng AHM, Cassady JP, et al.: Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Reports 5: 1302–1315, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo H, Zhao M, Qiu X, Deis JA, Huang H, Tang QQ, et al.: Niemann-Pick type C2 deficiency impairs autophagy-lysosomal activity, mitochondrial function, and TLR signaling in adipocytes. J Lipid Res 57: 1644–1658, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Awad O, Sarkar C, Panicker LM, Miller D, Zeng X, Sgambato JA, et al.: Altered TFEB-mediated lysosomal biogenesis in Gaucher disease iPSC-derived neuronal cells. Hum Mol Genet 24: 5775–5788, 2015 [DOI] [PubMed] [Google Scholar]
- 71.Seranova E, Connolly KJ, Zatyka M, Rosenstock TR, Barrett T, Tuxworth RI, et al.: Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem 61: 733–749, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan XM, Zheng F, Zhang L, Miao YY, Man N, Wen LP: Autophagy-mediated chemosensitization by cysteamine in cancer cells. Int J Cancer 129: 1087–1095, 2011 [DOI] [PubMed] [Google Scholar]
- 73.Pajares M, Jiménez-Moreno N, Dias IHK, Debelec B, Vucetic M, Fladmark KE, et al.: Redox control of protein degradation. Redox Biol 6: 409–420, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Underwood BR, Imarisio S, Fleming A, Rose C, Krishna G, Heard P, et al.: Antioxidants can inhibit basal autophagy and enhance neurodegeneration in models of polyglutamine disease. Hum Mol Genet 19: 3413–3429, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimura T, Takabatake Y, Takahashi A, Kaimori JY, Matsui I, Namba T, et al.: Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol 22: 902–913, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Isaka Y, Kimura T, Takabatake Y: The protective role of autophagy against aging and acute ischemic injury in kidney proximal tubular cells. Autophagy 7: 1085–1087, 2011 [DOI] [PubMed] [Google Scholar]
- 77.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al.: Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 169: 425–434, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang M, Wei Q, Dong G, Komatsu M, Su Y, Dong Z: Autophagy in proximal tubules protects against acute kidney injury. Kidney Int 82: 1271–1283, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA 4th, Ramalho-Santos J, et al.: Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One 6: e20914, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Levine B, Kroemer G: Autophagy in the pathogenesis of disease. Cell 132: 27–42, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu S, Hartleben B, Kretz O, Wiech T, Igarashi P, Mizushima N, et al.: Autophagy plays a critical role in kidney tubule maintenance, aging and ischemia-reperfusion injury. Autophagy 8: 826–837, 2012 [DOI] [PubMed] [Google Scholar]
- 82.Cherqui S, Courtoy PJ: The renal Fanconi syndrome in cystinosis: Pathogenic insights and therapeutic perspectives. Nat Rev Nephrol 13: 115–131, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yu Y, Kudchodkar SB, Alwine JC: Effects of simian virus 40 large and small tumor antigens on mammalian target of rapamycin signaling: Small tumor antigen mediates hypophosphorylation of eIF4E-binding protein 1 late in infection. J Virol 79: 6882–6889, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rega LR, Polishchuk E, Montefusco S, Napolitano G, Tozzi G, Zhang J, et al.: Activation of the transcription factor EB rescues lysosomal abnormalities in cystinotic kidney cells. Kidney Int 89: 862–873, 2016 [DOI] [PubMed] [Google Scholar]
- 85.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al.: A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Papadopoulos C, Meyer H: Detection and clearance of damaged lysosomes by the endo-lysosomal damage response and lysophagy. Curr Biol 27: R1330–R1341, 2017 [DOI] [PubMed] [Google Scholar]
- 87.Hung YH, Chen LM, Yang JY, Yang WY: Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 4: 2111, 2013 [DOI] [PubMed] [Google Scholar]
- 88.Ono K, Kim SO, Han J: Susceptibility of lysosomes to rupture is a determinant for plasma membrane disruption in tumor necrosis factor alpha-induced cell death. Mol Cell Biol 23: 665–676, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Galarreta CI, Forbes MS, Thornhill BA, Antignac C, Gubler MC, Nevo N, et al.: The swan-neck lesion: Proximal tubular adaptation to oxidative stress in nephropathic cystinosis. Am J Physiol Renal Physiol 308: F1155–F1166, 2015 [DOI] [PubMed] [Google Scholar]
- 90.Thomson AW, Turnquist HR, Raimondi G: Immunoregulatory functions of mTOR inhibition. Nat Rev Immunol 9: 324–337, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zaytseva YY, Valentino JD, Gulhati P, Evers BM: mTOR inhibitors in cancer therapy. Cancer Lett 319: 1–7, 2012 [DOI] [PubMed] [Google Scholar]
- 92.Bartolomeo R, Cinque L, De Leonibus C, Forrester A, Salzano AC, Monfregola J, et al.: mTORC1 hyperactivation arrests bone growth in lysosomal storage disorders by suppressing autophagy. J Clin Invest 127: 3717–3729, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, et al.: Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet 17: 3897–3908, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, et al.: Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports 2: 866–880, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nevo N, Chol M, Bailleux A, Kalatzis V, Morisset L, Devuyst O, et al.: Renal phenotype of the cystinosis mouse model is dependent upon genetic background. Nephrol Dial Transplant 25: 1059–1066, 2010 [DOI] [PubMed] [Google Scholar]
- 96.Elmonem MA, Khalil R, Khodaparast L, Khodaparast L, Arcolino FO, Morgan J, et al.: Cystinosis (ctns) zebrafish mutant shows pronephric glomerular and tubular dysfunction. Sci Rep 7: 42583, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morviducci L, Rota F, Rizza L, Di Giacinto P, Ramponi S, Nardone MR, et al.: Everolimus is a new anti-cancer molecule: Metabolic side effects as lipid disorders and hyperglycemia. Diabetes Res Clin Pract 143: 428–431, 2018 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.