Abstract
Pollen development is critical to the reproductive success of flowering plants, but how it is regulated is not well understood. Here, we isolated two allelic male-sterile mutants of OsMYB80 and investigated how OsMYB80 regulates male fertility in rice. OsMYB80 was barely expressed in tissues other than anthers, where it initiated the expression during meiosis, reached the peak at the tetrad-releasing stage and then quickly declined afterward. The osmyb80 mutants exhibited premature tapetum cell death, lack of Ubisch bodies, no exine and microspore degeneration. To understand how OsMYB80 regulates anther development, RNA-seq analysis was conducted to identify genes differentially regulated by OsMYB80 in rice anthers. In addition, DNA affinity purification sequencing (DAP-seq) analysis was performed to identify DNA fragments interacting with OsMYB80 in vitro. Overlap of the genes identified by RNA-seq and DAP-seq revealed 188 genes that were differentially regulated by OsMYB80 and also carried an OsMYB80-interacting DNA element in the promoter. Ten of these promoter elements were randomly selected for gel shift assay and yeast one-hybrid assay, and all showed OsMYB80 binding. The 10 promoters also showed OsMYB80-dependent induction when co-expressed in rice protoplast. Functional annotation of the 188 genes suggested that OsMYB80 regulates male fertility by directly targeting multiple biological processes. The identification of these genes significantly enriched the gene networks governing anther development and provided much new information for the understanding of pollen development and male fertility.
Keywords: DAP-seq, Male sterility, Oryza sativa, OsMYB80, Pollen development, RNA-seq
Accession number: The nucleotide sequences reported in this article have been submitted to the NCBI Sequence Read Archive under accession numbers SRP058039 and SRP196452.
Introduction
Anther development involves a series of events from the formation of stamen primordium to the release of mature pollen during anther dehiscence (Zhang et�al. 2011). During early anther development, stamen primordial cells divide and differentiate into pollen mother cells (PMCs) surrounded by four layers of anther wall cells, from surface to interior the epidermis, endothecium, middle layer and tapetum (Zhang et�al. 2011). The PMC undergoes meiosis, forming four haploid microspores that are initially enclosed as a tetrad by callose wall (Ariizumi and Toriyama 2011). Subsequently, the callose wall is degraded by callase produced by the tapetum, and individual microspore is released into the anther locule (Wan et�al. 2011). During further development, microspores go through cell enlargement, vacuolation and two rounds of mitosis and eventually form two small sperm cells enclosed by a large vegetative cell full of nutrients, while outside of the microspore is covered by a wall that is gradually thickened till the pollen grain matures (Zhang et�al. 2011). In accompany with microspore development after meiosis, the tapetum goes through the programmed cell death (PCD) and actively synthesizes and secretes materials for pollen wall formation and pollen maturation (Shi et�al. 2015a).
The mature pollen wall generally has two structural layers: the inner intine and the outer exine (Ariizumi and Toriyama 2011). The intine is believed to be formed by the male gametophyte and consists of cellulose, hemicellulose and pectic polymers similar to the primary walls of common plant cells (Ariizumi and Toriyama 2011). The exine commonly comprises two layers, the outer sexine and the inner nexine. The sexine contains an outermost roof, the tectum, and radially directed rods, the bacula, which together determine the species-specific appearance of pollen grains. The nexine consists of layers of nexine I, on which the bacula is anchored, and nexine II, which is laid down on the intine (Ariizumi and Toriyama 2011). The spaces between the tectum and nexine I are usually filled with the tryphine derived from the tapetum degeneration (Ariizumi and Toriyama 2011). The main constituent of exine is the sporopollenin polymer, whose precursors are believed to be produced and secreted by the tapetum and loaded on the surface of pollen grains. It is mainly composed of long polyhydroxylated aliphatic chains and of small amounts of aromatic rings derived from the phenylpropanoid metabolism (Dominguez et�al. 1999).
Recent genetic studies in Arabidopsis and rice have identified a large number of evolutionally conserved genes that are required for pollen development and male fertility (Shi et�al. 2015a). Among them, a group of genes is involved in the biosynthesis of aliphatic lipids and phenolic compounds. For example, Arabidopsis CER1 and its rice ortholog OsCER1 (Aarts et�al. 1997, Ni et�al. 2018), Arabidopsis CER3/WAX2/YRE/FLP1 and its rice ortholog WDA (Jung et�al. 2006, Rowland et�al. 2007) and Arabidopsis CER6/CUT1 (Fiebig et�al. 2000) are involved in the biosynthesis of very-long-chain fatty acids and alkanes, which are precursors of aliphatic polymers. Arabidopsis AtGPAT1 and its rice homolog OsGPAT3 encode glycerol-3-phosphate acyltransferases that play key roles in regulating lipids and extracellular lipid polyesters by generating lysophosphatidic acids and acylating glycerol 3-phosphate at the sn-1 or sn-2 hydroxyl with acyl-CoA or acyl carrier protein (ACP) to alter glycerolipid triacylglycerol biosynthesis (Zheng et�al. 2003, Men et�al. 2017, Sun et�al. 2018). OsSTRL2 encodes an atypical strictosidine synthase that plays crucial roles in regulating anther development and pollen wall formation in rice (Zou et�al. 2017a). Rice DPW2 encodes a hydroxycinnamoyl-CoA:ω-hydroxy fatty acid transferase of the BAHD/HXXXD acyl-transferase family that plays a role in the biosynthesis of key components of the anther cuticle and pollen wall (Xu et�al. 2017). Arabidopsis MS2 and its rice ortholog DPW encode fatty ACP reductases that can catalyze the fatty ACP into fatty alcohols (Chen et�al. 2011, Shi et�al. 2011). Rice OsNP1 encodes a glucose-methanol-choline (GMC) oxidoreductase that probably has a role in catalyzing the conversion of long-chain fatty acid to ω-dicarboxylic fatty acid required for pollen exine formation (Chang et�al. 2016a). The product of a GMC oxidoreductase can serve as the substrate for cytochrome P450 proteins such as the Arabidopsis CYP704B1 and CYP703A2 and their rice orthologs OsCYP704B2 and OsCYP703A3 that can catalyze in-chain hydroxylation or ω-hydroxylation of medium- and long-chain fatty acids (Morant et�al. 2007, Dobritsa et�al. 2009, Li et�al. 2010, Yang et�al. 2014). Arabidopsis ACOS5 and its rice ortholog OsACOS12 encode acyl-CoA synthetases that can catalyze the medium- and long-chain fatty acids into fatty acyl-CoA esters (de Azevedo et�al. 2009, Li et�al. 2016), and CoA esters can be condensed to malonyl-CoA by polyketide synthases encoded by Arabidopsis PKSA/LAP6 and PKSB/LAP5 and their rice orthologs OsPKS1 and OsPKS2 (Dobritsa et�al. 2010, Kim et�al. 2010, Zhu et�al. 2017, Zou et�al. 2017b), which are then reduced by tetraketide reductases encoded by the Arabidopsis TKPR1 and TKPR2 and rice OsTKPR1 (Grienenberger et�al. 2010, Xu et�al. 2019). The polyhydroxyalkyl α-pyrone monomers together with the fatty acids and fatty alcohols are prone to form ether or ester linkages in sporopollenin polymer (Grienenberger et�al. 2010).
Another group of genes is involved in polysaccharide metabolisms. For example, Arabidopsis CalS5/AtGSL2, AtGSL10, AtGSL1 and AtGSL5 and rice OsGSL5 (AtGSL2 homolog) encode callose synthases required for the synthesis of callose wall surrounding meiocytes, tetrads and early microspores (Dong et�al. 2005, Enns et�al. 2005, Shi et�al. 2015b). Arabidopsis AtUGP1 and AtUGP2 and their rice orthologs OsUgp1 and OsUgp2 encode UDP-glucose pyrophosphorylases that are essential for callose wall formation during meiosis (Chen et�al. 2007, Park et�al., 2010). Rice SPG2/IRX9L encodes a family GT43 glycosyltransferase involved in xylan backbone biosynthesis, while UPEX1 encodes a family GT31 glycosyltransferase likely involved in the galactosylation of arabinogalactan proteins; both SPG2 and UPEX1 are required for primexine synthesis (Li et�al. 2017, Suzuki et�al. 2017). Rice Osg1 encodes β-1,3-glucanase for the degradation of callose wall surrounding the microspore (Wan et�al. 2011). QRT1 and QRT3 encode a pectin methylesterase (pectinase) and an endo-polygalacturonase, respectively, that act sequentially to degrade the cell wall surrounding PMCs, and mutation of either gene results in the formation of pollen grains that are not separated (Rhee et�al. 2003, Francis et�al. 2006). Other genes such as OsUAM3 encoding a UDP-arabinopyranose mutase (Sumiyoshi et�al. 2015), GTL1 encoding a glycosyl transferase (Moon et�al. 2013) and CAP1 encoding an arabinokinase-like protein (Ueda et�al. 2013) act in the microsporocyte.
Many genes with functions in transportation were also found to be required for pollen fertility. In the past few years, several plant ABCG proteins, such as AtABCG11 (PanikashviLi et�al. 2010), AtABCG26 (Quilichini et�al. 2014), AtABCG9, AtABCG31 (Choi et�al. 2014), AtABCG1 and AtABCG16 (Yadav et�al. 2014) in Arabidopsis and OsABCG26 (Zhao et�al. 2015, Chang et�al. 2016b), OsABCG15 (Qin et�al. 2013, Niu et�al. 2013a) and OsABCG3 (Chang et�al. 2018) in rice, have been shown to contribute to pollen wall development. In addition, RAFTIN1 associated with the Ubisch body, type III lipid transporter proteins (LTPGs) and glycosyl phosphatidylinositol-anchored nonspecific LTPGs were also found to be required for pollen wall development (Wang et�al. 2003, Kim et�al. 2012, Huang et�al. 2013). Additional transporters including sugar transporters (Hirose et�al. 2010), UDP-GlcNAc and UDP-GalNAc transporter ROCK1 (Niemann et�al. 2015) and magnesium transporters (Li et�al. 2015) are probably involved in the transportation of nutrients for pollen development.
In addition to the catalytic enzymes and transport proteins, many genes required for pollen development are involved in signal transduction and transcriptional regulation, including hormone receptors, receptor-like kinases and transcriptional regulator (Shi et�al. 2015a). The Arabidopsis transcription factors DYSFUNCTIONAL TAPETUM1 (DYT1), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1), ABORTED MICROSPORES (AMS), AtMYB80 (also named AtMYB103 or AtMYB188) and MS1 form a transcriptional cascade regulating genes of sporopollenin synthesis (Zhu et�al. 2011). DYT1 is the most upstream regulator that directly regulates TDF1, TDF1 directly regulates AMS, AMS regulates AtMYB80 and AtMYB80 regulates MS1 (Zhu et�al. 2011). Corresponding to these Arabidopsis transcription factors, rice UNDERDEVELOPED TAPETUM1 (UDT1/bHLH164) is orthologous to DYT1 (Jung et�al. 2005), TAPETUM DEGENERATION RETADATION (TDR/bHLH5) is orthologous to AMS (Li et�al. 2006, Zhang et�al. 2008a), OsMYB80 is orthologous to AtMYB80 (Phan et�al. 2012) and PERSISTENT TAPETAL CELL1 (PTC1/OsMS1) is orthologous to MS1 (Li et�al. 2011, Yang et�al. 2019a). In addition to these proteins, other transcription factors have also been found essential for pollen development in rice, including the MYB family transcription factor GAMYB (Aya et�al. 2009, Liu et�al. 2010), TDR-INTERACTING PROTEIN2 (TIP2/bHLH142) (Fu et�al. 2014, Ko et�al. 2014), ETERNAL TAPETUM 1/DELAYED TAPETUM DEGENERATION (EAT1/DTD/bHLH141) (Ji et�al. 2013, Niu et�al. 2013b), TDR-INTERACTING PROTEIN3 (TIP3) (Yang et�al. 2019b) and TGA transcription factor OsTGA10 (Chen et�al. 2018). GAMYB is involved in gibberellin-mediated regulation of pollen wall development (Aya et�al. 2009). GAMYB and UDT1 work in parallel to positively activate the expression of TDR (Liu et�al. 2010). TIP2/bHLH142 acts the downstream of UDT1 but the upstream of TDR and EAT1, and TIP2/bHLH142 can form a dimer with TDR to further activate EAT1 transcription by binding to its promoter (Fu et�al. 2014, Ko et�al. 2014). EAT1 and TIP3 also physically interact with TDR to affect the expression of genes related to tapetum PCD and pollen wall formation (Niu et�al. 2013b, Yang et�al. 2019b). OsTGA10 regulates tapetum development and pollen formation by interacting with TIP2 and TDR to affect the expression of AP25 and MTR (Chen et�al. 2018). TDR not only modulates tapetum development and degeneration but also functions in aliphatic metabolism and pollen formation by directly activating the expression of its target genes OsCP1, OsC6 and OsCYP703A3 (Li et�al. 2006, Zhang et�al. 2008a, Zhang et al. 2010, Yang et�al. 2014). PTC1/OsMS1 regulates tapetal development and pollen formation by affecting the expression of tapetum- and microspore-expressed genes (Li et�al. 2011).
AtMYB80 is an R2R3 transcription factor that plays a critical role in anther development (Phan et�al. 2012). AtMYB80 has been shown to bind the promoters of ACOS5, CYP703A2, CYP704B1, PKSA, PKSB, TKPR1 and MS2 that are involved in sporopollenin synthesis, the promoter of transcriptional regulator MS1, the promoter of A1 aspartic protease gene UNDEAD regulating tapetum PCD, and the promoters of two other genes (GLOX1 encoding a glyoxal oxidase and VANGUARD1 encoding a pectin methylesterase) of unknown functions in pollen development, either by itself or in association with AMS or other unknown transcription factors (Phan et�al. 2011, Wang et�al. 2018). OsMYB80 is homologous to AtMYB80 in sequence and can replace the function of AtMYB80 in regulating male fertility when expressed in Arabidopsis plant (Phan et�al. 2012), but how OsMYB80 functions in rice has not been studied. In this study, we isolated two male-sterile mutants of the OsMYB80 gene, which enabled us to analyze the function of OsMYB80 in rice development. Here, we report that OsMYB80 regulates male fertility in rice by directly targeting a large number of genes in different biological pathways.
Results
Isolation of OsMYB80 mutants and analysis of OsMYB80 gene expression
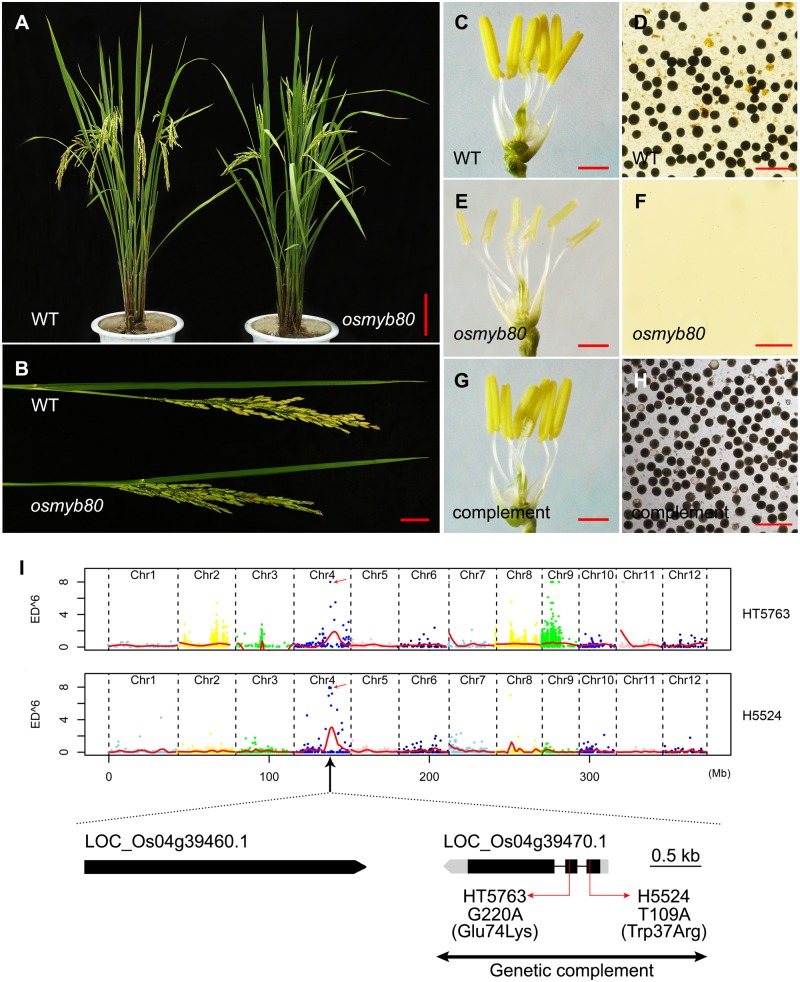
We screened an ethyl methanesulfonate (EMS) mutant library derived from the indica rice Huanghuazhan (HHZ) and isolated two complete male-sterile mutants (HT5763 and H5524). Both mutants exhibited normal vegetative and floral development (Fig.�1A, B), except for the anthers being smaller without pollen (Fig.�1C–F). Both mutant plants pollinated with the wild-type (WT) pollen exhibited normal seed set, indicating normal female fertility. The F1 plants were completely fertile, and the F2 progeny exhibited 3:1 segregation of fertile to sterile [232:80 for HT5763 (χ23:1 = 0.0339) and 224:61 for H5524 (χ23:1 = 1.0344)], suggesting that the male sterile phenotype was controlled by a single recessive gene for both mutants.
Fig. 1.
Mutation of OsMYB80 induced no-pollen male sterility in rice. (A–F) Whole plants, panicles, anthers and pollen grains stained with I2-KI were compared between the WT and osmyb80 mutant. (G, H) The anthers and pollen grains from osmyb80 mutant complemented with the OsMYB80 gene. Scale bars: 10 cm in (A); 2 cm in (B); 1 mm in (C), (E) and (G); 100 μm in (D), (F) and (H). (I) Identification of the causal gene in HT5763 and H5524 mutant plants. The top panel showed the distribution of Euclidean distance scores (ED6) of single nucleotide polymorphisms (SNPs) on chromosomes of HT5763 and H5524. The red lines represent the Loess regression curves of ED6 values. The colored dots indicate the mutations along the chromosomes. Arrow shows the position of OsMYB80 gene on chromosome 4. Gene structure and mutation sites in the OsMYB80 gene are shown. Black boxes represent exons, gray boxes represent untranslated regions (UTRs) and the lines between boxes represent introns. The DNA fragment for gene complementation is indicated by the line.
To identify the causal mutant genes, 30 sterile individuals in each F2 population mentioned above were selected for bulk sequencing (Supplementary Table S1). The sequenced data were analyzed using the Simultaneous Identification of Multiple Causal Mutations (SIMM) pipeline (Yan et�al. 2017), which identified two different mutation sites in LOC_Os04g39470 (OsMYB80). The G to A mutation in HT5763 caused Glu74 (GAG) substitution by Lys (AAG), and the T to A mutation in H5524 caused Trp37 (TGG) substitution by Arg (AGG) (Fig.�1I). High-resolution melting (HRM) analysis of the F2 individuals indicated that both mutations co-segregated with the male sterile phenotype. For simplicity, further analyses were conducted with the HT5763 mutant.
To confirm the mutation, a complementation experiment was performed by introducing a 2,512-bp genomic DNA fragment, including 1,203-bp upstream region and 1,309-bp gene body of OsMYB80, into the HT5763 mutant plant. Five independent transgenic lines of homozygous mutation background showed normal anther and pollen fertility (Fig.�1G, H), indicating that the mutations in OsMYB80 were responsible for the male sterile phenotype.
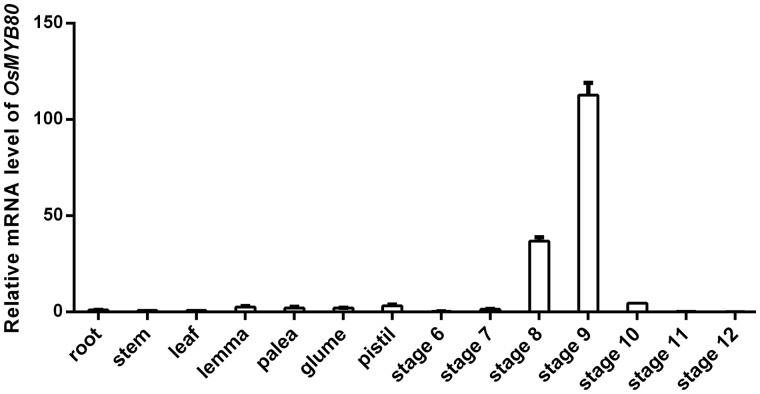
Because osmyb80 mutants exhibited defects only in male fertility, to understand the devoted role of OsMYB80, we analyzed the tissue specificity and developmental expression patterns of OsMYB80 using quantitative reverse transcription PCR (qRT-PCR). OsMYB80 was barely expressed in root, stem, leaf, lemma, palea, sterile lemma and pistil tissues. In anthers, however, the gene showed a developmentally regulated expression pattern (Fig.�2). OsMYB80 mRNA was very low in anthers before meiosis (stage 7), quickly increased during meiosis (stage 8), reached the highest level when microspores were released from the tetrad (stage 9), then declined to a very low level when microspores were vacuolated (stage 10) and became undetectable afterward (Fig.�2). The periods of active OsMYB80 expression overlapped with the process of meiotic division, microspore release from the tetrad, progress of tapetum PCD and active synthesis and secretion of sporopollenin precursors (Zhang et�al. 2011).
Fig. 2.
qRT-PCR analysis of OsMYB80 gene expression. Anthers were collected at developmental stages 6–12. Other tissues were harvested from plants at the flowering stage. OsACTIN1 was used as the internal control. Data are shown as mean � SD (n = 3).
Characterization of cellular defects in osmyb80 mutant anthers
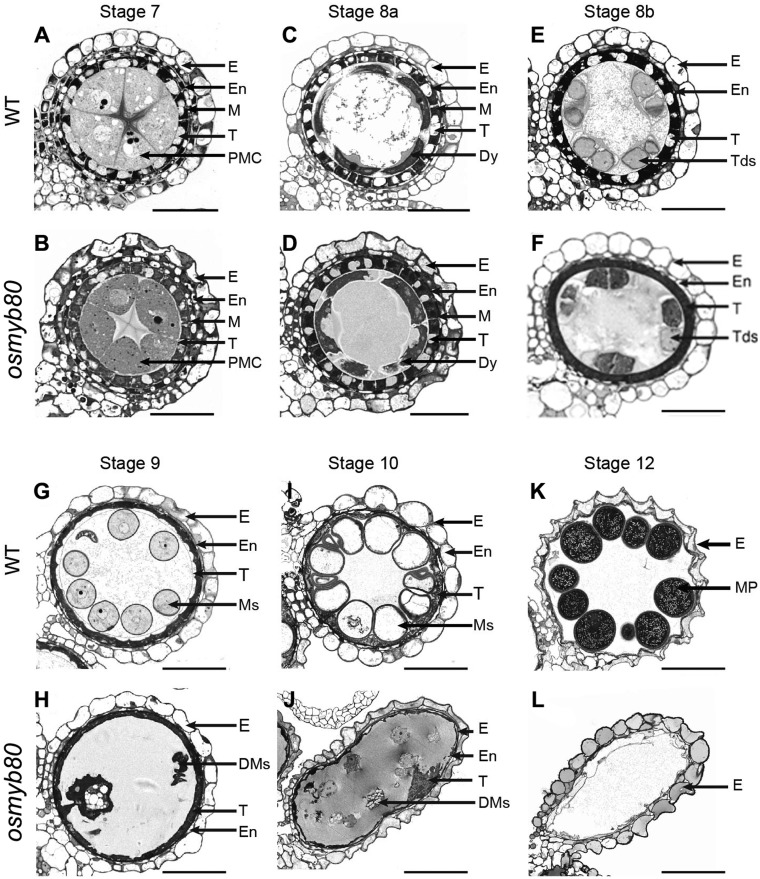
To understand how OsMYB80 affects pollen development, we analyzed the semi-thin sections of HT5763 mutant anthers. No clear difference between the WT and osmyb80 was detected before stage 8a when microspore mother cells (MMCs) finished meiosis I and formed dyads (Fig.�3A–D). At stage 8b, MMCs underwent meiosis II and formed tetrads of haploid microspores in the WT (Fig.�3E). In contrast, microspores in the mutant tetrads were more condensed and exhibited an irregular shape (Fig.�3F). At the same stage, the tapetal layer was more condensed and stained darker in the mutant than that in the WT (Fig.�3F), suggesting premature PCD. Microspores were released from the tetrads by stage 9 in the WT, but the mutant microspores degenerated, leaving debris in the locule (Fig.�3G, H). Subsequently, the WT microspores underwent vacuolation and two rounds of cell division, forming spherical pollen grains filled with cellular contents (Fig.�3I, K), but the mutant anther exhibited empty and shriveled locule lacking microspores (Fig.�3J, L).
Fig. 3.
Histological features of anther development in the WT and osmyb80 mutant. WT (A, C, E, G, I, K) and osmyb80 (B, D, F, H, J, L) anthers at developmental stages 7–12 are shown. Scale bar = 20 μm. DMs, degenerated microspores; Dy, dyad cell; E, epidermis; En, endothecium; M, middle layer; MP, mature pollen; Ms, microspores; T, tapetal layer; Tds, tetrads.
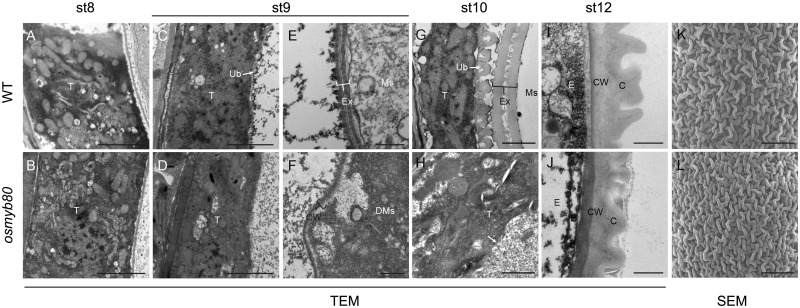
To more precisely characterize the cellular differences, transmission electron microscopy (TEM) analysis was performed on anthers at stages 8b–12. The WT tapetum underwent PCD from stage 8, became highly condensed afterward and almost completely degenerated by stage 11 (Zhang et�al. 2011). Discrete organelles were clearly visible in the WT tapetal cells at stage 8b (Fig.�4A). However, the organelles in the mutant tapetal cells were apparently disorganized (Fig.�4B), indicating a more advanced PCD. The WT tapetum started to form Ubisch bodies at the inner surface at stage 9 (Fig.�4C) that gradually grew into electron-dense orbicules facing the locule (Fig.�4G). However, no Ubisch bodies were formed by the mutant tapetum and tapetal cell membrane was apparently incomplete at stage 10 (Fig.�4D, H). Ubisch bodies are proposed to export the tapetum-produced materials for pollen exine formation and anther epidermis cuticle structure (Ariizumi and Toriyama 2011, Shi et�al. 2015a). The deposition of sporopollenin precursors on the microspore surface and thickening of the pollen cell wall were clearly visible in the WT (Fig.�4E, G). However, in osmyb80, deposition of sporopollenin precursors on the primary cell wall of microspore was not detected (Fig.�4F) and pollen exine was not formed before the microspore degeneration (Fig.�4H). At the mature stage, both the WT and osmyb80 anther epidermises were covered by grid-like cuticle structures that looked like fingers under the transmission electron microscope. However, the finger-like structures were slightly shorter in osmyb80 than in WT (Fig.�4I, J). Consistently, the WT anther surface exhibited deeper gullies than the mutant anther in the grid-like structure under a scanning electron microscope (SEM) (Fig.�4K, L).
Fig. 4.
TEM and SEM analyses of the WT and osmyb80 anthers. The transverse sections of the WT (A, C, E, G, I) and osmyb80 (B, D, F, H, J) anthers at stage 8 (A, B), stage 9 (C–F), stage 10 (G, H) and stage 12 (I, J) are compared. (K, L) SEM observation of the WT and osmyb80 anther epidermis. Scale bars: (A–D, G, H) 1 μm; (E, F, I, J) 500 nm; and (K, L) 10 μm.
Transcriptome analyses of genes regulated by OsMYB80
As a transcription factor, OsMYB80 is expected to affect anther development by regulating the expression of downstream genes. To identify genes that might be regulated by OsMYB80, we performed two sets of comparative transcriptome analyses using RNA-Seq. One set was to compare the transcriptomes in anthers at stage 9 between the WT and osmyb80 mutant. The other set was to compare the WT anthers at stages 9 and 7, because OsMYB80 was highly expressed at stage 9 and barely detectable at stage 7 (Fig.�2). Each sample had three biological replicates; thus, nine transcriptome libraries were sequenced. A total of 45.2–63.22 million raw reads were generated for each library (Supplementary Table S2). After the removal of low-quality reads, the clean reads were aligned to the Nipponbare reference genome (MSU v7) and the numbers of reads covering each gene were calculated.
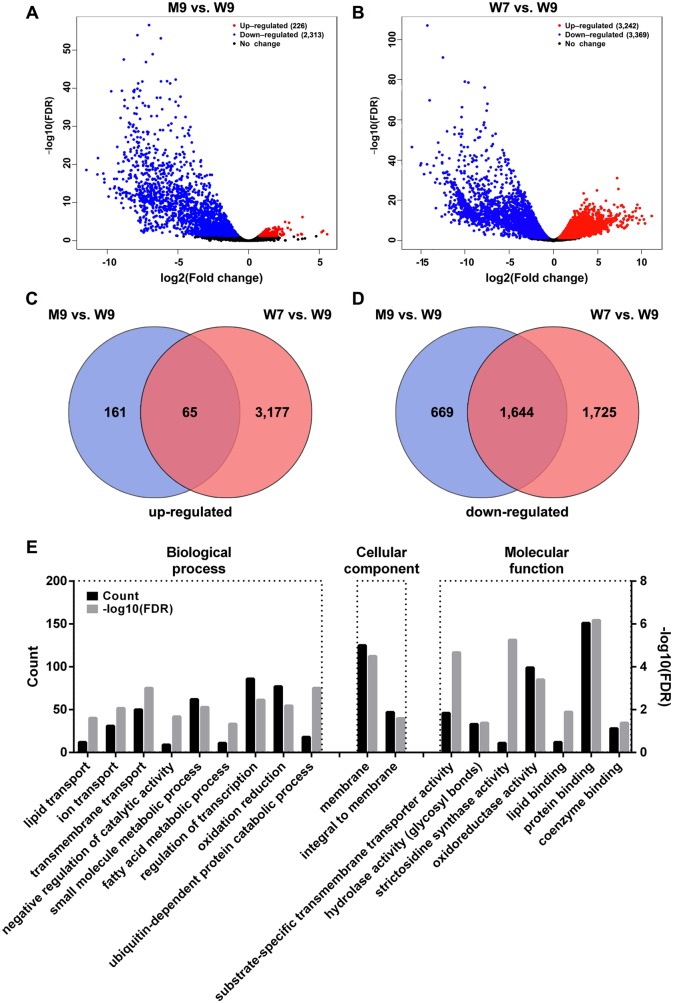
The expression patterns were highly consistent among the three replicates for each sample (Supplementary Fig. S1). The differentially expressed genes (DEGs) were extracted with fold change ≥2 and false discovery rate (FDR) <0.05. There were 226 upregulated genes and 2,313 downregulated genes in osmyb80 anther, compared with the WT anther at stage 9 (Fig.�5A and Supplementary Table S3). Comparison of DEGs in WT anthers at stages 7 and 9 resulted in 3,242 upregulated genes and 3,369 downregulated genes at stage 7 (Fig.�5B and Supplementary Table S4).
Fig. 5.
Identification of OsMYB80-related DEGs. (A) Volcano plot of DEGs between the osmyb80 and WT anthers at stage 9 (M9 vs. W9). (B) Volcano plot of DEGs between WT anthers at stages 7 and 9 (W7 vs. W9). Red dots and blue dots represent significantly up- and downregulated genes, respectively. Black dots represent no-change genes. (C) Overlap of upregulated genes identified in (A) and (B). (D) Overlap of downregulated genes identified in (A) and (B). (E) Significantly enriched GO analysis of OsMYB80-related DEGs. The count of genes (left y-axis) and −log10(FDR) (right y-axis) were shown for each enriched GO term.
qRT-PCR assays were performed to validate the DEGs identified by RNA-seq, using RNA samples that were collected at a different time from those for RNA-seq. We randomly chose 20 DEGs, 12 genes from the osmyb80 vs. WT anthers at stage 9 and 8 genes from the WT anthers at stage 7 vs. stage 9. The qRT-PCR and RNA-seq data showed close agreement (Supplementary Table S5), indicating that the transcriptome data were highly reliable.
Because the mutation of OsMYB80 caused multiple phenotypical defects in anther development, these defects (e.g. premature tapetum PCD) might trigger abnormal expression of certain genes. On the other hand, DEGs between the WT anthers at stages 7 and 9 also included those that were regulated by factors other than OsMYB80. Thus, to identify genes that were related to OsMYB80, we overlapped the DEGs between the two comparisons and extracted the shared genes with similar expression patterns, resulting in 1,644 upregulated and 65 downregulated genes associated with the OsMYB80 function (Fig.�5C, D and Supplementary Table S6). Importantly, the shared DEGs contained 22 known male sterility genes identified in rice (Supplementary Table S7).
AgriGO was used to identify enriched gene ontology (GO) terms of the OsMYB80-regulated genes. As shown in Fig.�5E, the DEGs were significantly enriched in transport functions, fatty acid and small-molecule metabolic processes, regulation of transcription, ubiquitin-dependent protein degradation process, oxidation and reduction and binding with proteins, lipids and coenzymes. Genes related to oxidoreductase activity, hydrolase activity specifically on glycosyl bonds and strictosidine synthase were also significantly enriched. The top 50 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the DEGs are shown in Supplementary Fig. S2. The enrichment of these genes was consistent with the active synthesis and transportation of metabolites by the tapetum at stage 9 and the lack of exine formation in osmyb80 mutant anthers.
Identification of OsMYB80-binding sequences by DNA affinity purification sequencing
It is perceivable that the DEGs identified by RNA-seq include genes that are regulated directly as well as indirectly by OsMYB80. To identify genes that are directly regulated by OsMYB80, we used DNA affinity purification sequencing (DAP-seq) for genome-wide identification of OsMYB80-binding sites in vitro (Bartlett et�al. 2017). The analysis identified 7,968 enriched 200-bp peaks with −log10(P-value) ≥ 5 (Supplementary Table S8). In total, 3,968 (49.80%) of these peaks were located within 1.5 kb upstream of the annotated open reading frames (ORFs), 1,960 (24.60%) were located within 1 kb downstream of putative ORFs and the remaining (25.60%) were distributed in intergenic regions in the genome (Supplementary Fig. S3A). The enriched peaks were almost uniformly distributed in the upstream regions but decreased with the distance to the stop codon in the downstream regions (Supplementary Fig. S3B). There were at least 34 known rice male sterile genes associated with a DAP- fragment (Supplementary Table S9).
Overlap of the genes identified by DAP-seq with the OsMYB80-regulated DEGs revealed 296 shared genes, of which 287 were upregulated and 9 were downregulated by OsMYB80 (Supplementary Table S10). Because most MYB proteins were known to regulate gene expression by binding to the promoters (Prouse and Campbell 2012), we therefore focused on the OsMYB80-regulated DEGs with the DAP-seq fragments in the promoter region, which identified 188 genes in total (Supplementary Table S11). Fourteen genes carried two DAP-seq fragments and three genes carried three DAP-seq fragments in their promoters, suggesting multiple OsMYB80-binding sites in these gene promoters (Supplementary Table S11).
Identification of putative OsMYB80-binding motifs in the promoters
OsMYB80 is orthologous to AtMYB80 (Phan et�al. 2012). Previously, Xu et�al. (2014) identified two motifs in the AtMYB80 promoter (ATGTTT and TTTGTTA) that were required for AtMYB80 autoregulation of its promoter activity in anthers. Although OsMYB80 and AtMYB80 promoters showed low identity overall, two conserved DNA motifs were identified at similar positions in the OsMYB80 promoter that differ from the AtMYB80 motifs of only one SNP in each motif (Fig.�6A). To test if these two motifs were functional for OsMYB80 binding, we conducted an electrophoretic mobility shift assay (EMSA). As shown in Fig.�6B, both motifs showed strong interaction with OsMYB80.
Fig. 6.
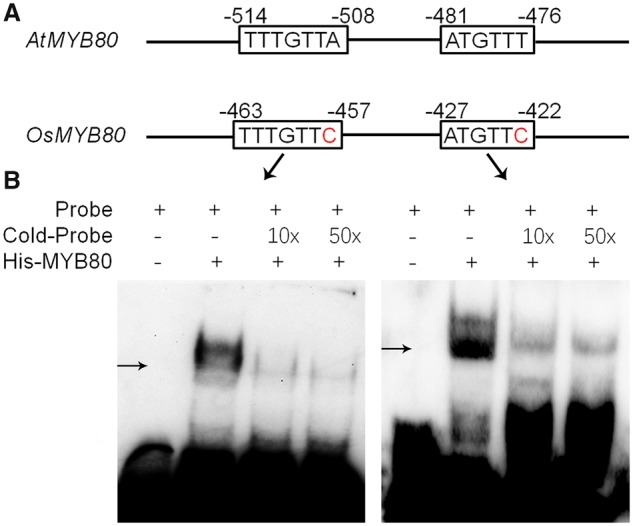
OsMYB80 interacts with two conserved DNA motifs in its promoter. (A) Distribution of the two conserved DNA motifs in the promoters of AtMYB80 and OsMYB80. (B) EMSA of OsMYB80 binding with the two DNA motifs.
In addition to the two DNA motifs in AtMYB80 promoter and the two motifs in OsMYB80 promoter, AtMYB80 was shown to bind three other motifs, including MYB1AT (A/T AACCA), MYB1LEPR (GTTAGTT) and MYBPZM (MYBPZM-1/2, CC A/T ACC; MYBPZM-1.1, CCAACCA) (Phan et�al. 2011). In addition, a series of conserved R2R3 MYB-binding motifs were identified in previous studies, including MYBCORE (CTGTTG), MYBST1 (GGATA), MYBGAHV (TAACAAA), MYBCOREATCYCB1 (AACGG), MYBHv1 (CAACGG), MRE (AACCTAA), MYBS (TATCCA), MBS1 (TA A/G CTG), MBS2 (CAACTG) and MBS3 (CGGTCA) (Lu et�al. 2002, Phan et�al. 2011). Hence, we analyzed if these cis-elements were presented in the DAP fragments on the promoters of DEGs in Supplementary Table S11. The search identified 154 DAP fragments in 140 gene promoters containing at least one putative MYB80- and/or R2R3 MYB-binding motif (Supplementary Table S12). The most abundant motif presented in the DAP fragments was MYBPZM (53), followed by MYB1AT (45), AtMYB80-1 (25), MBS1 (24) and MYBST1 (23) (Supplementary Fig. S4). The DAP fragments were also subjected to MEME assay, which revealed a number of other enriched DNA elements (Supplementary Fig. S5). The top four enriched motifs (ACC T/A A A/C, C/T/G C/T/A ACC T/A/C A/G A/C, G/T/A T/G T A/T GGT G/A/T and TAG G/C T A/G) were similar to the MRE motif or its complement (AACCTAA or TTAGGTT), and the fifth one (CCAAC T/C) is similar to the MYBPZM (CC A/T ACC and CCAACCA) motif. The other motifs had not been reported with a function (Supplementary Fig. S5).
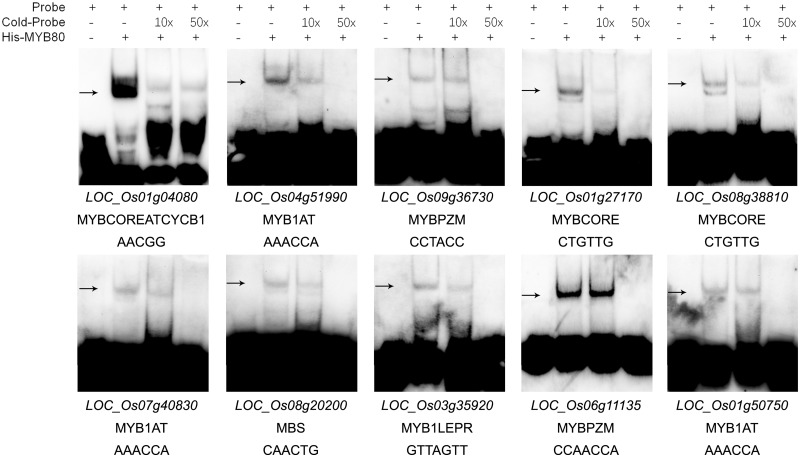
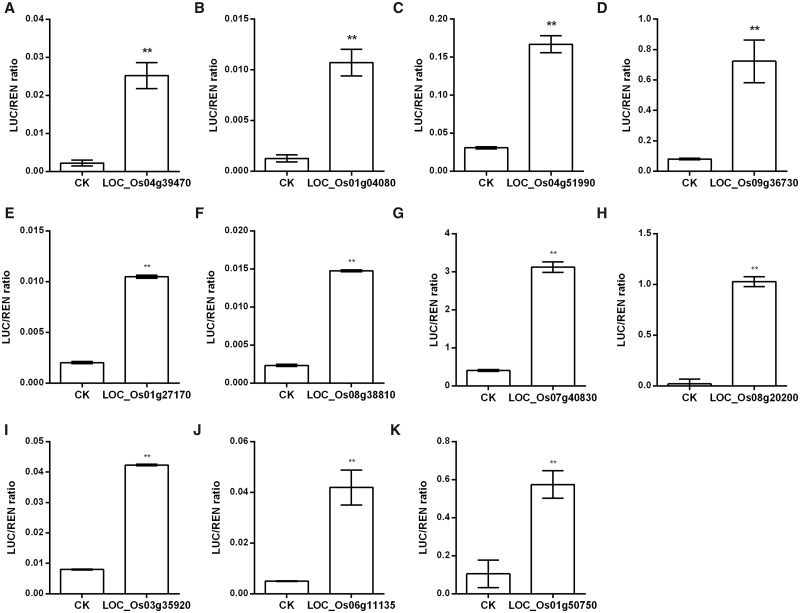
Genes in Supplementary Table S12 were likely to be the candidate genes directly regulated by OsMYB80, because they showed OsMYB80-dependent differential expression in RNA-seq assay, their promoters bound OsMYB80 in DAP-seq assay and there were putative MYB80-binding motifs presented in their promoters. To further test their candidacy, we randomly selected 10 genes and tested the binding of their putative promoter elements with OsMYB80 using EMSA (Table�1). As expected, all of these promoter elements bound OsMYB80 (Fig.�7). All these motifs were then tested for OsMYB80 binding using the yeast one-hybrid assay, and all showed positive results (Fig.�8). We further tested if OsMYB80 could regulate the promoters harboring these motifs in rice protoplast. As shown in Fig.�9, all these promoters showed higher activities in the presence of OsMYB80 than in the absence of OsMYB80, further suggesting that OsMYB80 directly regulates these promoters.
Table 1.
List of experimentally validated OsMYB80-binding promoters
| Gene | Description | Position | Motif |
|---|---|---|---|
| LOC_Os04g39470 | OsMYB80 | −463/−457 | OsMYB80-1 |
| −427/−422 | OsMYB80-2 | ||
| LOC_Os01g04080 | Expressed protein | −604/−600 | MYBCOREATCYCB1 |
| LOC_Os04g51990 | Transferase family protein | −820/−815 | MYB1AT |
| LOC_Os09g36730 | MYB family transcription factor | −322/−317 | MYBPZM |
| LOC_Os01g27170 | Potassium transporter | −705/−700 | MYBCORE, MYBCOREATCYCB1 |
| LOC_Os08g38810 | BURP domain-containing protein | −707/−702 | MYBCORE, AtMYB80-1 |
| LOC_Os07g40830 | App1 | −441/−436 | MYB1AT, MBS1 |
| LOC_Os08g20200 | Male sterility protein | −506/−501 | MBS2, MYBPZM |
| LOC_Os03g35920 | Expressed protein | −163/−157 | MYB1LEPR, AtMYB80-2 |
| LOC_Os06g11135 | Gibberellin receptor GID1L2 | −147/−141 | MYBPZM |
| LOC_Os01g50750 | Zinc finger, C3HC4-type domain-containing protein | −244/−239 | MYB1AT, MYBPZM |
Position represents distance of the validated motif to the start codon. The validated motifs are marked in bold.
Fig. 7.
EMSA validation of the OsMYB80-binding motifs in 10 selected DAP-seq fragments. The gene ID and the corresponding motif were listed below each experiment. The detected bands were indicated by arrows.
Fig. 8.
Validation of OsMYB80 binding with the 10 selected DAP-seq fragments using yeast one-hybrid assay. AD indicates the control yeast strain transformed with the empty pGADT7 vector without OsMYB80.
Fig. 9.
Activation of the 10 selected promoters by OsMYB80 in rice protoplast. (A) OsMYB80. (B-K) Ten randomly selected promoters. The selected gene promoters were constructed upstream of the LUC reporter gene and co-expressed with OsMYB80 in rice protoplast. The REN gene under 35S promoter was used as the internal control. The relative LUC/REN ratio was determined for each promoter. The ratio shown as mean � SD (n = 4). CK indicates a basal signal without OsMYB80. The corresponding gene promoters are listed in Table�1.
Discussion
In this study, we isolated two osmyb80 mutant plants of male sterile phenotype. Histological analysis of the mutant anther sections showed premature tapetum PCD, no-pollen exine formation and microspore cell death at the end of the tetrad stage. These phenotypic defects were similar to what in the Arabidopsis atmyb80 mutant anther (Zhang et�al. 2007). Our results showed that OsMYB80 was specifically expressed in rice anthers during the period of tetrad formation and microspore release from the tetrad, which was similar to the timing of AtMYB80 expression during the Arabidopsis anther development (Zhang et�al. 2007). By comparing gene expression profiles, we found 1,644 upregulated and 65 downregulated genes associated with the OsMYB80 function. GO and KEGG analyses indicated that functions related to fatty acid/lipid/sporopollenin biosynthesis, transmembrane transport, protein degradation and transcriptional regulation were significantly enriched in the OsMYB80-regulated DEGs. These enriched gene functions were also similar to what had been reported in the atmyb80 mutant (Zhu et�al. 2010, Phan et�al. 2011). These similarities were consistent with the conserved function of OsMYB80 and AtMYB80 in regulating anther development.
Our DAP-seq assay identified 7,968 enriched genomic fragments associated with OsMYB80 protein in vitro. Strikingly, at least 34 known rice male sterile genes were associated with a DAP fragment (Supplementary Table S9). However, most of these male sterile genes did not show an OsMYB80-dependent differential gene expression in our RNA-seq analysis. Whether these male sterile genes are indeed regulated directly by OsMYB80 remains to be verified.
Overlap of genes identified by DAP-seq with the OsMYB80-regulated DEGs identified 188 DEGs carrying DAP-seq fragment(s) in the promoter region (Supplementary Table S11). Most of these DEGs contain one or a few putative DNA motifs in the DAP-seq fragments that were known to interact with MYB80 or other R2R3 MYB proteins. Some of these 188 OsMYB80-regulated genes were also identified by a gene co-expression network analysis using sporopollenin synthesis and transport genes as a guide, but none showed co-expression with those genes involved in meiosis (Aya et�al. 2011). We randomly selected 10 genes and tested the binding of their putative DNA motifs with OsMYB80 using EMSA and yeast one-hybrid assays and found positive interaction of all these elements. In addition, these promoters showed OsMYB80-dependent induction in rice protoplast. These results suggested that most of these DEGs are directly regulated by OsMYB80. As to the DEGs that do not carry a known R2R3 MYB-binding DNA motif, they may carry novel OsMYB80-binding elements. Indeed, the MEME assay of the DAP fragments revealed a number of putative DNA elements of unknown function (Supplementary Fig. S5). Further characterization of these unknown elements will likely identify new MYB80-binding DNA motifs.
There are 11 Arabidopsis genes whose promoters were known to bind AtMYB80 (Phan et�al. 2011, Wang et�al. 2018). Only two of them (MS1 and MS2) have an ortholog in our candidate list (LOC_Os09g27620 and LOC_Os08g20200). Of the remaining nine reported AtMYB80-regulated genes, four of them, including AtCYP703A2 (ortholog of LOC_Os08g03682), AtCYP704B1 (ortholog of LOC_Os03g07250), AtACOS5 (ortholog of LOC_Os04g24530) and AtPKSA (ortholog of LOC_Os10g34360), had an ortholog displaying OsMYB80-dependent differential expression in our RNA-seq assay, but none was identified by DAP-seq. One possibility is that these orthologous genes are not subjected to direct regulation by OsMYB80. Another possibility is that these genes were missed by the DAP-seq assay.
Our DAP-seq assay generated 7,968 enriched fragments. However, most of the genes associated with a DAP-seq fragment were excluded from our candidate gene list because they did not meet the differential gene expression criteria. Overlap of the genes identified by DAP-seq with the OsMYB80-regulated DEGs resulted in only 296 shared genes (Supplementary Table S10). It is possible that some of the 7,968 DAP-seq fragments were derived from in vitro artificial interaction of the DNA with OsMYB80 protein. It is also possible that some of the excluded genes identified by DAP-seq were indeed regulated by OsMYB80, but they were dismissed by our stringent differential gene expression criteria, which required ≥2-fold differential expression and FDR ≤0.05 in both sets of the RNA-seq comparisons. Furthermore, it was reported recently that AtMYB80 binds the promoters of CYP704B1, ACOS5 and TKPR1, but these three genes did not show a differential gene expression in atmyb80 mutant (Wang et�al. 2018). It was proposed that AtMYB80 needs to team up with other transcription factors to regulate the expression of these genes (Wang et�al. 2018). This type of genes would also be identified by our DAP-seq assay but excluded by the RNA-seq assay.
Searching against published male sterility genes, we found only 3 of the 188 putative OsMYB80-regulated genes that had been reported with a function in pollen wall synthesis and pollen development in rice, including LOC_Os06g40550 (OsABCG15; Qin et�al. 2013), LOC_Os08g38810 (RAFTIN1; Wang et�al. 2003) and LOC_Os09g27620 (PTC1/OsMS1; Li et�al. 2011, Yang et�al. 2019a). Seven genes are orthologous to the Arabidopsis genes known to regulate pollen development and male fertility, but their functions have not been tested in rice, including LOC_Os09g36730 orthologous to AtMYB7/4/32 (Preston et�al. 2004); LOC_Os08g28820 and LOC_Os09g10260 orthologous to SKP1/ASK1 (Yang et�al. 1999), LOC_Os07g41650 orthologous to QRT1 (Francis et�al. 2006), LOC_Os01g03670 orthologous to TKPR2 (Grienenberger et�al. 2010), LOC_Os04g09520 orthologous to LTPG2 (Kim et�al. 2012) and a DPW paralog (LOC_Os08g20200) orthologous to MS2/FAR2 (Chen et�al. 2011). It will be interesting to test if mutations of these genes affect pollen development and male fertility in rice. The remaining genes have not been reported with a function related to pollen development.
Based on BlastP analysis, 24 of the 188 candidate genes directly regulated by OsMYB80 (Supplementary Table S11) appear to encode catalytic enzymes for aliphatic lipids, aromatics and phenolics metabolisms, including cytochrome P450 proteins, monoglyceride lipase, phospholipase C, 3-oxoacyl-synthase, acyl-CoA reductase, triacylglycerol lipase, acyl-CoA oxidase, myristoyl-ACP thioesterase, HXXXD-type acyl-transferase, dihydroflavonol-4-reductase, strictosidine synthase, laccase precursor protein, flavanone 3-hydroxylase, 2-hydroxyacid dehydrogenase, glyoxal oxidase, acyl-CoA thioesterase, cinnamoyl-CoA reductase, 3-oxoacyl-synthase, flavonoid O-methyltransferase, epoxide hydrolase and pantothenate kinase. Two genes were orthologous to the Arabidopsis genes required for pollen development, including LOC_Os08g20200 orthologous MS2 (Chen et�al. 2011) and LOC_Os01g03670 orthologous to TKPR2 (Grienenberger et�al. 2010); but none has been demonstrated with a function in pollen development in rice. Given the fact that pollen wall development is associated with the active synthesis of aliphatic lipids and phenolics and that OsMYB80 is required for pollen exine formation, many of these genes are expected to contribute to sporopollenin�synthesis and exine formation in rice.
Seven candidate genes are involved in polysaccharide metabolisms (Supplementary Table S11), including glycosyl hydrolase, pectinesterase, xyloglucan endotransglucosylase, endoglucanase and β-1,3-glucanase. One of them (LOC_Os07g41650) is orthologous to the Arabidopsis QRT1, a pectinesterase required for the separation of pollen tetrads (Francis et�al. 2006). As the osmyb80 mutant exhibited a defect in callose wall degradation, we speculate that LOC_Os07g43940 (encoding a β-1,3-glucanase) may play a role in degrading the callose wall surrounding the microspore. The other cell wall degradation enzymes may degrade the cell wall surrounding the tetrads or degrade the tapetum cell wall as a mean to recycle the tapetum cell wall components.
There are 27 candidate genes encoding proteins of various transportation functions (Supplementary Table S11). Two of them, OsABCG15 and RAFTIN1, were known to be essential for rice pollen wall development (Wang et�al. 2003, Qin et�al. 2013), while LOC_Os04g09520 is orthologous to Arabidopsis LTPG2 required for pollen wall development (Kim et�al. 2012). Other genes, including three different lipid transfer proteins, four different proteins involved in vesicle transportation, several transporters for sugar, sucrose, amino acids, nucleotide, malate, metal-nicotianamine, K+, Na+ and other metal ions, aquaporin, H+/Na+ antiporter, plasma membrane ATPase and protein secretion protein SEC61, have not been reported with a role in pollen development. It will be interesting to test if mutations of these genes impact pollen development.
At least 25 candidate genes encode components of the ubiquitination and proteasomal degradation pathway (Supplementary Table S11). Two of them, LOC_Os08g28820 and LOC_Os09g10260 were orthologous to the Arabidopsis gene SKP1/ASK1 required for pollen development (Yang et�al. 1999). The presence of so many components of the ubiquitination and proteasomal degradation pathway suggested active protein degradation during the OsMYB80-expression stage (stages 8–10), which may be associated with the active progression of PCD and exhaustion of the tapetum cells during this period (Zhang et�al. 2011).
At least 28 genes encode proteins in signal transduction and transcriptional regulation (Supplementary Table S11). Only LOC_Os09g27620 (OsMS1/PTC1) has been reported with a role in pollen development in rice (Li et�al. 2011, Yang et�al. 2019a). LOC_Os09g36730 is orthologous to AtMYB4/7/32 that is required for pollen development in Arabidopsis (Preston et�al. 2004). It is perceivable that some of these genes may directly regulate the DEGs identified by our RNA-seq assay. The presence of so many regulatory genes in this group explains why the number of OsMYB80-dependent DEGs was so much bigger than the number of candidate DEGs directly regulated by OsMYB80. Significantly, three genes (LOC_Os06g11135, LOC_Os07g06880 and LOC_Os09g28720) are homologous to GA receptor GID1 (Yoshida et�al. 2018). It is known that GA plays a critical role in anther development and male fertility (Aya et�al. 2009, Kwon and Paek 2016). Whether these GID1-like genes play a role in GA signaling during pollen development remains to be studied.
In addition to the above major functional groups, three genes encode putative pollen wall antigens (LOC_Os07g40830, LOC_Os07g40850 and LOC_Os07g40890), which are pollen cell wall proteins putatively involved in stress responses and metabolic processes during pollen development (Chen et�al. 2016). Five genes, including LOC_Os03g57200 (glutathione S-transferase), LOC_Os07g46570 (glutaredoxin family protein), LOC_Os01g15490 (PAPS reductase), LOC_Os03g24600 (methionine sulfoxide reductase) and LOC_Os01g12800 (peroxisomal membrane protein), may be related to the redox homeostasis of the tapetal cells (Traverso et�al. 2013). Three genes are probably related to cell death, including LOC_Os09g19710 (hypersensitive response-induced protein), LOC_Os02g07820 (senescence-associated protein) and LOC_Os02g33820 (abscisic stress-ripening protein ASR5). It is known that the production of reactive oxygen species is critical for tapetal PCD and pollen development in Arabidopsis (Smirnova et�al. 2014, Xie et�al. 2014). Miss regulation of the redox- and cell death-related genes may be associated with the abnormal PCD displayed by the osmyb80 mutant. Other than these genes, 2 proteases (LOC_Os04g45960 and LOC_Os01g04710), 2 ribosome-inactivating proteins (LOC_Os07g37090 and LOC_Os03g47460), 2 RNA-binding proteins (LOC_Os07g37090 and LOC_Os03g47460), 3 proteins that act on lysine metabolism (LOC_Os07g23190, LOC_Os04g48380 and LOC_Os07g40620), 3 heat shock proteins (LOC_Os08g43490, LOC_Os03g56540 and LOC_Os07g33350), 14 genes of various other functions and 38 genes of unknown functions were also identified as OsMYB80 directly regulated genes.
In summary, OsMYB80 directly regulates a large number of genes that probably act on different aspects of pollen formation. Based on the phenotypic defects exhibited by osmyb80 mutant and the putative functions of the OsMYB80-regulated genes, we propose a schematic model to summarize the biological processes regulated by OsMYB80 (Supplemental Fig. S6). These processes include the biosynthesis of precursors for pollen exine; transportation of pollen exine precursors for pollen wall formation; transportation of small nutrient molecules to nurture the pollen cell growth; degradation of the cell wall surrounding PMCs and the tetrads for microspore separation; massive protein degradation, redox homeostasis and cell death gene expression associated with the tapetum PCD; and signal transduction and transcriptional regulation that regulate downstream events for pollen development. Most of the OsMYB80-regulated genes we identified here were not known to be involved in pollen development. The identification of these genes significantly enriched the gene networks governing anther development and provided much new insight into the molecular mechanisms governing anther development and male fertility.
Materials and Methods
Plant materials and growth conditions
The HHZ mutant library generated by EMS treatment (Chen et�al. 2014) was used for the isolation of HT5763 and H5524 mutants. The mutants were cross-pollinated by the WT HHZ to produce the F1 plants, which further self-pollinated to generate the F2 populations. All plants were grown in the paddy field under natural conditions with regular care.
Characterization of the mutant phenotype
Plants and flowers at the mature stage were photographed using a Nikon D80 Digital Camera. For pollen fertility analysis, pollen grains at the mature stage were stained with I2-KI solution and photographed using a Nikon AZ100 microscope as described previously (Chang et�al. 2016a). Female fertility was tested by manual cross-pollination of the mutant plants with WT HHZ pollen, using osnp1-1 male sterile mutant plants as control (Chang et�al. 2016a). For transverse section and electron microscopic analyses of anthers, spikelets at different developmental stages were collected and treated as described by Chang et�al. (2016b).
Identification of the causal genes
The F2 plants were grown to the mature stage, and 30 individuals of the male sterile phenotype were harvested for DNA isolation. An equal amount of DNA from each F2 plant was mixed for genome re-sequencing using the Illumina HiSeq 2000 platform. The re-sequencing data were processed using the SIMM pipeline (Yan et�al. 2017).
Gene expression analysis
Rice tissues, including root, stem, leaf, lemma, palea, sterile lemma and pistil, were collected at the flowering stage. Anthers were harvested at different developmental stages as described by Chang et�al. (2016b). Total RNA was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) and then reverse-transcribed using PrimeScript™ RT Reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Dalian, China) according to the manufacturer’s instructions. qRT-PCR analysis was conducted according to the previous descriptions (Pan et�al. 2015). Primers for qRT-PCR are listed in Supplementary Table S13. The relative gene expression level was calculated using the comparative CT method (Livak and Schmittgen 2001) with OsActin1 as the internal control. Three biological replicates were analyzed for each sample.
Transgenic complementation of osmyb80 mutant
The 2,512-bp OsMYB80 genomic DNA, including 1,203-bp upstream region and 1,309-bp gene body, was PCR-amplified using the OsMYB80-com primer pair (Supplementary Table S13) and HHZ genomic DNA as a template. The PCR product was cloned into the pMD19-T vector (Takara) and confirmed by sequencing. The plasmid was then digested with BamHI and HindIII, and the segment was then ligated into the binary vector pCAMBIA1300 predigested with the same enzymes. The construct was introduced into Agrobacterium tumefaciens AGL10 strain and transformed into rice calli derived from the offspring of osmyb80 heterozygote plants. The positive transgenic lines were identified by PCR with primers Hyg-PCR (Supplementary Table S13) for the selection marker gene Hyg. To identify the background genotype of transgenic plants, primers com-background (Supplementary Table S13) flanking the 2,512-bp OsMYB80 DNA were used to amplify the genomic DNA and the PCR product was then diluted 1,000� for subsequent HRM analysis (Lochlainn et�al. 2011) using the primer pair OsMYB80-HRM (Supplementary Table S13).
Transcriptomic library construction and sequencing
Three replicates of osmyb80 mutant anthers at stage 9 (M9) and WT HHZ anthers at stage 7 (W7) and stage 9 (W9) were harvested from a number of plants, giving a total of nine samples (three tissues � three replicates). The samples were immediately frozen in liquid nitrogen prior to RNA exaction using TRIzol reagent. From each sample, 3 �g total RNA was used for RNA-seq library construction. The library was sequenced on Illumina HiSeq 2000 platform to ∼6 Gb PE150 raw data following the manufacturer’s instructions.
Bioinformatics analysis of transcriptomic data
The RNA-seq data were processed as previously described (Sun et�al. 2016). Genes with fold change ≥2 and the FDR of ≤0.05 were considered to be significantly differentially expressed. DEGs showing similar expression patterns between the two comparisons (M9 vs. W9 and W7 vs. W9) were extracted as candidate genes regulated by OsMYB80. The enriched GO terms of the overlapped DEGs were identified with AgriGO v2.0 (Tian et�al. 2017). Those with a FDR of <0.05 were defined as significantly enriched GO terms. The KEGG pathways for the DEGs were identified with KEGG automatic annotation server with the protein sequences as input (Moriya et�al. 2007).
DAP-seq analysis
The DAP-seq analysis was performed according to the protocol described by Bartlett et�al. (2017). Briefly, 5 μg HHZ genomic DNA was sheared into ∼200-bp fragments and then ligated with the Illumina-based sequencing adaptors to form a DNA library. The library was examined for adaptor-ligation frequency by quantitative PCR (qPCR) before applied to DAP-seq assay.
OsMYB80 ORF fused to the Halo affinity tag was expressed in vitro using TNT� SP6 Coupled Wheat Germ Extract System (Promega, Fitchburg, WI, USA). The HaloTag-OsMYB80 protein was then purified from nonspecific proteins in the expression system using the magnetic HaloTag ligand (chloroalkane) beads (Promega) and verified by Western blotting with the anti-HaloTag antibody (Promega). The purified protein was incubated with 500 ng adaptor-ligated genomic DNA library at 30�C for 2 h before washing away the unbound DNA fragments.
The samples were heated at 98�C for 10 min to release the OsMYB80-bound DNA, and the recovered DNA was then PCR-amplified with the indexed TruSeq primers (Illumina, San Diego, CA, USA). Indexed DNA samples were subsequently combined and size-selected to remove the residual adaptor dimers. Purified DNA libraries were then sequenced using the Illumina HiSeq sequencing platform.
Identification OsMYB80-binding sequences and the associated genes
To identify OsMYB80-binding sequences, the clean DAP-seq reads were aligned to the Nipponbare reference genome (MSU v7.0) with HISAT2 (Kim et�al. 2015). The enriched peaks were identified using MACS2 (Zhang et�al. 2008b) with −log10(P-value) ≥5. Then, the regions of the peaks were defined based on the annotations of the reference genome. Those located within 1.5 kb upstream of a gene were defined as peaks in promoters, and those located within 1 kb downstream of a gene were defined as peaks in 3′-UTR. Peaks located with >1.5 kb from the boundaries of a gene were defined as peaks in intergenic regions.
Electrophoretic mobility shift assay
The recombinant protein His-OsMYB80 was purified from Escherichia coli BL21 using Ni-NTA Superflow columns (Qiagen, Venlo, Netherlands) according to the manufacturer’s protocol. DNA probes were prepared by annealing a 5′-biotinylated oligonucleotide to a complementary unmodified oligonucleotide. The complementary oligonucleotides were diluted in annealing buffer (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM EDTA) to a final concentration of 20 μM, heated to 95�C for 5 min and cooled down at 25�C for 2–3 h. The same procedure was followed to generate unmodified dsDNA fragments for competition assays. Probes for selected genes are listed in Supplementary Table S14.
Gel shift assays were performed as described previously (Hendrickson, 1985). One microgram of purified recombinant protein was incubated with the biotinylated DNA probe in a buffer [5 ng μl−1 poly(dI�dC), 5% glycerol, 0.1% NP-40, 0.1 M KCl, 10 mM MgCl2, 20 mM EDTA] at 25�C for 0.5 h. Then, the samples were examined by gel electrophoresis using native 6% PAGE gel in ice-cold 0.5� TBE buffer (120 V, 55 min), followed by wet transfer of the gel to Biodyne B Nylon Membrane (Thermo Fisher Scientific, Waltham, MA, USA) in 0.5� TBE buffer (380 mA, 47 min). The LightShift Chemiluminescent DNA EMSA Kit (Thermo Fisher Scientific) was used for the detection of the biotinylated probes according to the manufacturer’s instructions.
The yeast one-hybrid assay
The Matchmaker� Gold Yeast One-Hybrid Library Screening System User Manual (Takara) was followed for the yeast one-hybrid assay. To construct the target-reporter plasmid (pBait-AbAi), three tandem copies of the DNA target element were synthesized (Supplementary Table S15) and inserted upstream of the AbAr reporter gene in pBait-AbAi plasmid. The insert DNA was synthesized as two antiparallel oligonucleotides with overhang sticky ends compatible with the sticky ends of the pBait-AbAi vector predigested with HindIII and SalI. After construction, the pBait-AbAi plasmids were digested with BstBI and the linearized plasmids were transformed into Y1HGold yeast strain (Takara). The transformants were selected on a synthetic dropout-Ura plate and confirmed by colony PCR analysis using the Matchmaker Insert Check PCR Mix 1 (Takara) for fragment integration into the yeast genome. The resulting bait-reporter yeast strains were then examined for the minimal inhibitory concentration of Aureobasidin A (AbA).
To confirm the interaction between the DNA fragments and OsMYB80, the full-length coding sequence of OsMYB80 was cloned into the pGADT7 vector (Takara). The OsMYB80 construct or empty vector (negative control) was transformed into the Y1H bait-reporter strain and selected on a synthetic dropout-Leu plate. Activation of the reporter gene was measured based on the AbA inhibition of yeast colony growth (50–250 ng/ml).
Protoplast transfection assay
For the transient transcriptional activity assay, ∼1,500-bp promoter sequences of the tested genes were PCR-amplified using primers in Supplementary Table S16 and then constructed upstream of the LUC gene in pGreen II 0800-LUC vector using In-Fusion cloning method (Hellens et�al. 2005). The renilla luciferase (REN) gene under the control of 35S promoter in the pGreen II 0800-LUC vector was used as the internal control (Yoo et�al. 2007). The CDS sequence of OsMYB80 was PCR-amplified using the primer set OsMYB80-effector (Supplementary Table S16), digested with BamHI and XbaI and cloned into the pGreen II 62-SK vector (Nova Tech, Singapore) under the control of 35S promoter. Rice protoplast was prepared and transfected as previously described (Yoo et�al. 2007). The firefly LUC and REN activities were measured using the Dual-Luciferase Reporter Assay System (Promega). The experiments were repeated at least four times, and each time with four independent measurements assessed.
Supplementary Material
Acknowledgments
We thank the Microscope Center in Life Science School of Sun Yat-sen University for using their facilities for microscopic analysis. We also thank Dr. Yanqiang Li for the analysis of RNA-seq data.
Funding
Major Program of Guangdong Basic and Applied Research [2019B030302006]; National Natural Science Foundation of China [31500254, U1704232 and U1901203]; Natural Science Foundation of Guangdong Province [2017A030310500 and 2018B030308008]; National Key Research and Development Plan Program [2016YFD0101801 and 2016YFD0100406]; Shenzhen Commission on Innovation and Technology Programs [JCYJ20180507181837997]; Guangzhou Science and Technology Innovation Commission [201804010034]; China Postdoctoral Science Foundation [2017M612685]; and Youth Innovation Promotion Association of Chinese Academy of Sciences [2017399].
Disclosures
The authors have no conflicts of interest to declare.
References
- Aarts M.G.M., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., et al. (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623. [DOI] [PubMed] [Google Scholar]
- Ariizumi T., Toriyama K. (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu. Rev. Plant Biol. 62: 437–460. [DOI] [PubMed] [Google Scholar]
- Aya K., Suzuki G., Suwabe K., Hobo T., Takahashi H., Shiono K., et al. (2011) Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS One 6: e26162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K., Ueguchi-Tanaka M., Kondo M., Hamada K., Yano K., Nishimura M., et al. (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett A., O'Malley R.C., Huang S.C., Galli M., Nery J.R., Gallavotti A., et al. (2017) Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12: 1659–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Chen Z., Wang N., Xie G., Lu J., Yan W., et al. (2016a) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc. Natl. Acad. Sci. USA 113: 14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z., Chen Z., Yan W., Xie G., Lu J., Wang N., et al. (2016b) An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 253: 21–30. [DOI] [PubMed] [Google Scholar]
- Chang Z., Jin M., Yan W., Chen H., Qiu S., Fu S., et al. (2018) The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Xu J., Devis D., Shi J., Ren K., Searle I., et al. (2016) Origin and functional prediction of pollen allergens in plants. Plant Physiol. 172: 341–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., Zhao X., Shao Z., Wei Z., Wang Y., Zhu L., et al. (2007) Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell 19: 847–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Yu X.H., Zhang K., Shi J., Oliveira S.D., Schreiber L., et al. (2011) Male Sterile 2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157: 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Lu J., Lu Q., Wang N., Wang C., Xie G., et al. (2014) Screening and analysis of male sterile mutants derived from elite indica cultivar Huanghuazhan. Guangdong Agri. Sci. 41: 1–4. [Google Scholar]
- Chen Z.S., Liu X.F., Wang D.H., Chen R., Zhang X.L., Xu Z.H., et al. (2018) Transcription factor OsTGA10 is a target of the MADS protein OsMADS8 and is required for tapetum development. Plant Physiol. 176: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Ohyama K., Kim Y.Y., Jin J.Y., Lee S.B., Yamaoka Y., et al. (2014) The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 26: 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Azevedo S.C., Kim S.S., Koch S., Kienow L., Schneider K., McKim S.M., et al. (2009) A novel fatty Acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 21: 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Lei Z., Nishikawa S., Urbanczyk-Wochniak E., Huhman D.V., Preuss D., et al. (2010) LAP5 and LAP6 encode anther-specific proteins with similarity to chalcone synthase essential for pollen exine development in Arabidopsis. Plant Physiol. 153: 937–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Shrestha J., Morant M., Pinot F., Matsuno M., Swanson R., et al. (2009) CYP704B1 is a long-chain fatty acid-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiol. 151: 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez E., Mercado J.A., Quesada M.A., Heredia A. (1999) Pollen sporopollenin: degradation and structural elucidation. Sex Plant Reprod. 12: 171–178. [Google Scholar]
- Dong X., Hong Z., Sivaramakrishnan M., Mahfouz M., Verma D.P. (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 42: 315–328. [DOI] [PubMed] [Google Scholar]
- Enns L.C., Kanaoka M.M., Torii K.U., Comai L., Okada K., Cleland R.E. (2005) Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol. Biol. 58: 333–349. [DOI] [PubMed] [Google Scholar]
- Fiebig A., Mayfield J.A., Miley N.L., Chau S., Fischer R.L., Preuss D. (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P. (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z., Yu J., Cheng X., Zong X., Xu J., Chen M., et al. (2014) The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Kim S.S., Lallemand B., Geoffroy P., Heintz D., Souza C.A., et al. (2010) Analysis of TETRAKETIDE α-PYRONE REDUCTASE function in Arabidopsis thaliana reveals a previously unknown, but conserved, biochemical pathway in sporopollenin monomer biosynthesis. Plant Cell 22: 4067–4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., et al. (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W. (1985) Protein-DNA interactions studied by the gel electrophoresis-DNA binding assay. BioTechniques 3: 198–207. [Google Scholar]
- Hirose T., Zhang Z., Miyao A., Hirochika H., Ohsugi R., Terao T. (2010) Disruption of a gene for rice sucrose transporter, OsSUT1, impairs pollen function but pollen maturation is unaffected. J. Exp. Bot. 61: 3639–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M.D., Chen T.L.L., Huang A.H.C. (2013) Abundant type III lipid transfer proteins in Arabidopsis tapetum are secreted to the locule and become a constituent of the pollen exine. Plant Physiol. 163: 1218–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C., Li H., Chen L., Xie M., Wang F., Chen Y., et al. (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol. Plant 6: 1715–1718. [DOI] [PubMed] [Google Scholar]
- Jung K.H., Han M.J., Lee D.Y., Lee Y.S., Schreiber L., Franke R., et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K.H., Han M.J., Lee Y.S., Kim Y.W., Hwang I., Kim M.J., et al. (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., Salzberg S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Lee S.B., Kim H.J., Min M.K., Hwang I., Suh M.C. (2012) Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 53: 1391–1403. [DOI] [PubMed] [Google Scholar]
- Kim S.S., Grienenberger E., Lallemand B., Colpitts C.C., Kim S.Y., de Souza C.A., et al. (2010) LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B encode hydroxyalkyl-pyrone synthases required for pollen development and sporopollenin biosynthesis in Arabidopsis thaliana. Plant Cell 22: 4045–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S.S., Li M.J., Ku M.S.B., Ho Y.C., Lin Y.J., Chuang M.H., et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C.T., Paek N.C. (2016) Gibberellic acid: a key phytohormone for spikelet fertility in rice grain production. Int. J. Mol. Sci. 17: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Pinot F., Sauveplane V., Werck-Reichhart D., Diehl P., Schreiber L., et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Yuan Z., Vizcay-Barrena G., Yang C., Liang W., Zong J., et al. (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol. 156: 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Huang Y., Tan H., Yang X., Tian L.F., Luan S., et al. (2015) An endoplasmic reticulum magnesium transporter is essential for pollen development in Arabidopsis. Plant Sci. 231: 212–220. [DOI] [PubMed] [Google Scholar]
- Li N., Zhang D.S., Liu H.S., Yin C.S., Li X.X., Liang W.Q., et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.L., Liu Y., Douglas C.J. (2017) Role of glycosyl transferases in pollen wall primexine formation and exine patterning. Plant Physiol. 173: 167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li D., Guo Z., Shi Q., Xiong S., Zhang C., et al. (2016) OsACOS12, an orthologue of Arabidopsis acyl-CoA synthetase 5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 16: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.H., Bao W.J., Liang W.Q., Yin J.Y., Zhang D.B. (2010) Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Biol. 52: 670–678. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lochlainn S.�., Amoah S., Graham N.S., Alamer K., Rios J.J., Kurup S., et al. (2011) High Resolution Melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.A., Ho T.H.D., Ho S.L., Yu S.M. (2002) Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14: 1963–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Men X., Shi J., Liang W., Zhang Q., Lian G., Quan S., et al. (2017) Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J. Exp. Bot. 68: 513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S., Kim S.R., Zhao G., Yi J., Yoo Y., Jin P., et al. (2013) Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 161: 663–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant M., J�rgensen K., Schaller H., Pinot F., M�ller B.L., Werck-Reichhart D., et al. (2007) CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. Plant Cell 19: 1473–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35: W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni E.D., Zhou L.Y., Li J., Jiang D.G., Wang Z.H., Zheng S.Y., et al. (2018) OsCER1 plays a pivotal role in very-long-chain alkane biosynthesis and affects plastid development and programmed cell death of tapetum in rice (Oryza sativa L.). Front. Plant Sci. 9: 1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann M.C.E., Bartrina I., Ashikov A., Weber H., Nov�k O., Sp�chal L., et al. (2015) Arabidopsis ROCK1 transports UDP-GlcNAc/UDP-GalNAc and regulates ER protein quality control and cytokinin activity. Proc. Natl. Acad. Sci. USA 112: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B.X., He F.R., He M., Ren D., Chen L.T., Liu Y.G. (2013a) The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice. J. Integr. Plant Biol. 55: 710–720. [DOI] [PubMed] [Google Scholar]
- Niu N., Liang W., Yang X., Jin W., Wilson Z.A., Hu J., et al. (2013b) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat. Commun. 4: 1445. [DOI] [PubMed] [Google Scholar]
- Pan X.Y., Hasan M.M., Li Y.Q., Liao C.S., Zheng H.Y., Liu R.Y., et al. (2015) Asymmetric transcriptomic signatures between the cob and florets in the maize ear under optimal- and low-nitrogen conditions at silking, and functional characterization of amino acid transporters ZmAAP4 and ZmVAAT3. J. Exp. Bot. 66: 6149–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D., Shi J.X., Bocobza S., Franke R.B., Schreiber L., Aharoni A. (2010) The Arabidopsis DSO/ABCG11 transporter affects cutin metabolism in reproductive organs and suberin in roots. Mol. Plant 3: 563–575. [DOI] [PubMed] [Google Scholar]
- Park J.I., Ishimizu T., Suwabe K., Sudo K., Masuko H., Hakozaki H., et al. (2010) UDP-glucose pyrophosphorylase is rate limiting in vegetative and reproductive phases in Arabidopsis thaliana. Plant Cell Physiol. 51: 981–996. [DOI] [PubMed] [Google Scholar]
- Phan H.A., Iacuone S., Li S.F., Parish R.W. (2011) The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23: 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan H.A., Li S.F., Parish R.W. (2012) MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 78: 171–183. [DOI] [PubMed] [Google Scholar]
- Preston J., Wheeler J., Heazlewood J., Li S.F., Parish R.W. (2004) AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40: 979–995. [DOI] [PubMed] [Google Scholar]
- Prouse M.B., Campbell M.M. (2012) The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 1819: 67–77. [DOI] [PubMed] [Google Scholar]
- Qin P., Tu B., Wang Y., Deng L., Quilichini T.D., Li T., et al. (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 54: 138–154. [DOI] [PubMed] [Google Scholar]
- Quilichini T.D., Samuels A.L., Douglas C.J. (2014) ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 26: 4483–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee S.Y., Osborne E., Poindexter P.D., Somerville C.R. (2003) Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 133: 1170–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O., Lee R., Franke R., Schreiber L., Kunst L. (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett. 581: 3538–3544. [DOI] [PubMed] [Google Scholar]
- Shi J., Cui M., Yang L., Kim Y.J., Zhang D. (2015a) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci. 20: 741–753. [DOI] [PubMed] [Google Scholar]
- Shi J., Tan H., Yu X.H., Liu Y., Liang W., Ranathunge K., et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Sun X., Zhang Z., Feng D., Zhang Q., Han L., et al. (2015b) GLUCAN SYNTHASE-LIKE 5 (GSL5) plays an essential role in male fertility by regulating callose metabolism during microsporogenesis in rice. Plant Cell Physiol. 56: 497–509. [DOI] [PubMed] [Google Scholar]
- Smirnova A.V., Matveyeva N.P., Yermakov I.P. (2014) Reactive oxygen species are involved in regulation of pollen wall cytomechanics. Plant Biol. J. 16: 252–257. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi M., Inamura T., Nakamura A., Aohara T., Ishii T., Satoh S., et al. (2015) UDP-Arabinopyranose Mutase 3 is required for pollen wall morphogenesis in rice (Oryza sativa). Plant Cell Physiol. 56: 232–241. [DOI] [PubMed] [Google Scholar]
- Sun C., Li Y., Zhao W., Song X., Lu M., Li X., et al. (2016) Integration of hormonal and nutritional cues orchestrates progressive corolla opening. Plant Physiol. 171: 1209–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Xiang X., Yang Z., Yu P., Wen X., Wang H., et al. (2018) OsGPAT3 plays a critical role in anther wall programmed cell death and pollen development in rice. Int. J. Mol. Sci. 19: 4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Narciso J.O., Zeng W., van de Meene A., Yasutomi M., Takemura S., et al. (2017) KNS4/UPEX1: A type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173: 183–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., et al. (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 45: W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso J.A., Pulido A., Rodr�guez-Garc�a M.I., Alch� J.D. (2013) Thiol-based redox regulation in sexual plant reproduction: new insights and perspectives. Front. Plant Sci. 4: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K., Yoshimura F., Miyao A., Hirochika H., Nonomura K., Wabiko H. (2013) Collapsed abnormal pollen1 gene encoding the Arabinokinase-like protein is involved in pollen development in rice. Plant Physiol. 162: 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Zha W., Cheng X., Liu C., Lv L., Liu C., et al. (2011) A rice beta-1,3-glucanase gene Osg1 is required for callose degradation in pollen development. Planta 233: 309–323. [DOI] [PubMed] [Google Scholar]
- Wang A., Xia Q., Xie W., Datla R., Selvaraj G. (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc. Natl. Acad. Sci. USA 100: 14487–14492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Guo Z.L., Zhou W.T., Zhang C., Zhang Z.Y., Lou Y., et al. (2018) The regulation of sporopollenin biosynthesis genes for rapid pollen wall formation. Plant Physiol. 178: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H.T., Wan Z.Y., Li S., Zhang Y. (2014) Spatiotemporal production of reactive oxygen species by NADPH oxidase is critical for tapetal programmed cell death and pollen development in Arabidopsis. Plant Cell 26: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Qu S., Tucker M.R., Zhang D., Liang W., Shi J. (2019) Ostkpr1 functions in anther cuticle development and pollen wall formation in rice. BMC Plant Biol. 19: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Shi J., Rautengarten C., Yang L., Qian X., Uzair M., et al. (2017) Defective pollen wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiol. 173: 240–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Iacuone S., Li S.F., Parish R.W. (2014) MYB80 homologues in Arabidopsis, cotton and Brassica: regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 14: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V., Molina I., Ranathunge K., Castillo I.Q., Rothstein S.J., Reed J.W. (2014) ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26: 3569–3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Chen Z., Lu J., Xu C., Xie G., Li Y., et al. (2017) Simultaneous identification of multiple causal mutations in rice. Front. Plant Sci. 7: 2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Hu Y., Lodhi M., McCombie W.R., Ma H. (1999) The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96: 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Wu D., Shi J., He Y., Pinot F., Grausem B., et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J. Integr. Plant Biol. 56: 979–994. [DOI] [PubMed] [Google Scholar]
- Yang Z., Liu L., Sun L., Yu P., Zhang P., Abbas A., et al. (2019a) OsMS1 functions as a transcriptional activator to regulate programmed tapetum development and pollen exine formation in rice. Plant Mol. Biol. 99: 175–191. [DOI] [PubMed] [Google Scholar]
- Yang Z., Sun L., Zhang P., Zhang Y., Yu P., Liu L., et al. (2019b) TDR INTERACTING PROTEIN 3, encoding a PHD-finger transcription factor, regulates Ubisch bodies and pollen wall formation in rice. Plant J. 99: 844–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Tanimoto E., Hirai T., Miyanoiri Y., Mitani R., Kawamura M., et al. (2018) Evolution and diversification of the plant gibberellin receptor GID1. Proc. Natl. Acad. Sci. USA 115: E7844–E7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Liang W., Yin C., Zong J., Gu F., Zhang D. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol. 154: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Luo X., Zhu L. (2011) Cytological analysis and genetic control of rice anther development. J. Genet. Genomics 38: 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang D.S., Liang W.Q., Yuan Z., Li N., Shi J., Wang J., et al. (2008a) Tapetum Degeneration Retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol. Plant 1: 599–610. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., et al. (2008b) Model-based Analysis of Chip-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.B., Zhu J., Gao J.F., Wang C., Li H., Li H., et al. (2007) Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52: 528–538. [DOI] [PubMed] [Google Scholar]
- Zhao G., Shi J., Liang W., Xue F., Luo Q., Zhu L., et al. (2015) Two ATP binding cassette g transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiol. 169: 2064–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Xia Q., Dauk M., Shen W., Selvaraj G., Zou J. (2003) Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. Plant Cell 15: 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Lou Y., Xu X.F., Yang Z.N. (2011) A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 53: 892–900. [DOI] [PubMed] [Google Scholar]
- Zhu J., Zhang G., Chang Y., Li X., Yang J., Huang X., et al. (2010) AtMYB103 is a crucial regulator of several pathways affecting Arabidopsis anther development. Sci. China Life Sci. 53: 1112–1122. [DOI] [PubMed] [Google Scholar]
- Zhu X., Yu J., Shi J., Tohge T., Fernie A.R., Meir S., et al. (2017) The polyketide synthase OsPKS2 is essential for pollen exine and Ubisch body patterning in rice. J. Integr. Plant Biol. 59: 612–628. [DOI] [PubMed] [Google Scholar]
- Zou T., Li S., Liu M., Wang T., Xiao Q., Chen D., et al. (2017a) An atypical strictosidine synthase, OsSTRL2, plays key roles in anther development and pollen wall formation in rice. Sci. Rep. 7: 6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T., Xiao Q., Li W., Luo T., Yuan G., He Z., et al. (2017b) OsLAP6/OsPKS1, an orthologue of Arabidopsis PKSA/LAP6, is critical for proper pollen exine formation. Rice 10: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.