Abstract
Isotopically nonstationary metabolic flux analysis (INST-MFA) provides a versatile platform to quantitatively assess in vivo metabolic activities of autotrophic systems. By applying INST-MFA to recombinant aldehyde-producing cyanobacteria, we identified metabolic alterations that correlated with increased strain performance in order to guide rational metabolic engineering. We identified four reactions adjacent to the pyruvate node that varied significantly with increasing aldehyde production: pyruvate kinase (PK) and acetolactate synthase (ALS) fluxes were directly correlated with product formation, while pyruvate dehydrogenase (PDH) and phosphoenolpyruvate carboxylase (PPC) fluxes were inversely correlated. Overexpression of enzymes for PK or ALS did not result in further improvements to the previous best-performing strain, while downregulation of PDH expression (through antisense RNA expression) or PPC flux (through expression of the reverse reaction, phosphoenolpyruvate carboxykinase) provided significant improvements. These results illustrate the potential of INST-MFA to enable a systematic approach for iterative identification and removal of pathway bottlenecks in autotrophic host cells.
Keywords: Cyanobacteria, Isobutyraldehyde, Photoautotrophic metabolism, Metabolic flux analysis, Pyruvate, Phosphoenolpyruvate, Stable isotope
1. Introduction
The generation of efficient microbial cell factories is challenging due to the fact that metabolic networks have evolved to maintain homeostasis and to resist externally imposed changes (Nielsen and Keasling, 2016). Under nutrient replete conditions, the central carbon metabolism of cyanobacteria—i.e., the Calvin-Benson-Bassham (CBB) cycle, glycolysis, oxidative pentose phosphate (OPP) pathway, and the tricarboxylic acid (TCA) cycle—function to generate the precursors needed to sustain growth (Broddrick et al., 2016; Shastri and Morgan, 2005) and to accumulate storage reserves necessary for survival during periods of nutrient scarcity (Carr, 1988; Preiss and Romeo, 1989). These same precursors can be converted into useful bioproducts through metabolic engineering of cyanobacterial hosts, which can efficiently utilize renewable energy and carbon sources and can be cultivated in locations that are not suitable for food production. In order to achieve high product titers and specific productivities, carbon fluxes must be effectively siphoned away from their native metabolic routes and redirected into recombinant pathways leading to the desired product. Although advances in genetic tools continue to promote flexibility and ease of synthetic biology in cyanobacteria (reviewed in Sengupta et al. (2018)), current strain improvement efforts proceed largely by trial-and-error because methods to systematically assess metabolic bottlenecks are still maturing. As a result, there is a critical need to develop and test rational approaches of bottleneck identification that can become the central axis of an iterative metabolic engineering cycle (McAtee et al., 2015).
Isotopically nonstationary metabolic flux analysis (INST-MFA from hereon) is the premier method for assessing carbon fluxes within the central metabolism of photosynthetic organisms under autotrophic growth conditions (Abernathy et al., 2017; Ma et al., 2014; Wu et al., 2015; Young et al., 2011). Since the theoretical underpinnings of INST-MFA were first established more than a decade ago (Nöh and Wiechert, 2006; Shastri and Morgan, 2007), advances in computational algorithms (Young et al., 2008), design of experiments (Adebiyi et al., 2015; Antoniewicz, 2013; Jazmin et al., 2014), and development of publicly available software tools (Kajihata et al., 2014; Young, 2014) have spurred the application of INST-MFA in the fields of biotechnology and medicine (Cheah et al., 2017). In fact, the increasing accessibility of INST-MFA has enabled cyanobacterial metabolism to be studied across a wide variety of strains and environmental conditions (reviewed in Cheah and Young (2018)). However, few studies to date have leveraged INST-MFA to guide rational strain engineering of cyanobacteria. Hence, the goal of this study was to further test and validate the efficacy of INST-MFA toward systematically debottlenecking cyanobacterial metabolic pathways, using a recombinant isobutyraldehyde (IBA)-producing strain of S. elongatus PCC 7942 (denoted SA590), originally developed by Atsumi et al. (2009), as a model system.
We performed INST-MFA to quantify intracellular flux alterations induced by two of the most successful productivity enhancement strategies identified in our previous study (Jazmin et al., 2017): (i) overexpression of pyruvate kinase (PK) and (ii) media supplementation of thiamin (Fig. 1A). PK catalyzes the direct conversion of phosphoenolpyruvate (PEP) to pyruvate (the central metabolic precursor of IBA), while thiamin is a required cofactor for multiple enzymes in the CBB cycle, TCA cycle, and IBA synthesis pathway of SA590. We compared central carbon fluxes under three conditions with gradually increasing aldehyde productivities. Based on these results, we concluded that IBA production rate is sensitive to perturbations of fluxes entering and leaving the pyruvate node. More specifically, we found that aldehyde flux is (i) positively correlated with flux through PK and (ii) inversely correlated with flux through pyruvate dehydrogenase (PDH) and PEP carboxylase (PPC). We investigated the effects of further overexpression of PK or acetolactate synthase (ALS), the latter of which is considered a rate-limiting step in the IBA synthesis pathway, in our previous best-performing strain SA590-PK. We also examined the effects of attenuating flux into competing pathways through either inducible knockdown of PDH or overexpression of a non-native enzyme that promotes flux in the reverse direction of PPC. Through this process, we found that expressing genes that oppose net flux through PDH or PPC led to significant improvements in aldehyde titer and production rates. Our results demonstrate that INST-MFA offers a basis for rational metabolic engineering of cyanobacteria and other industrially relevant photosynthetic hosts, by providing a systematic strategy to identify and overcome metabolic bottlenecks in the pathways leading to target compounds of interest.
Fig. 1. Thiamin supplementation and PK overexpression increase aldehyde productivity.
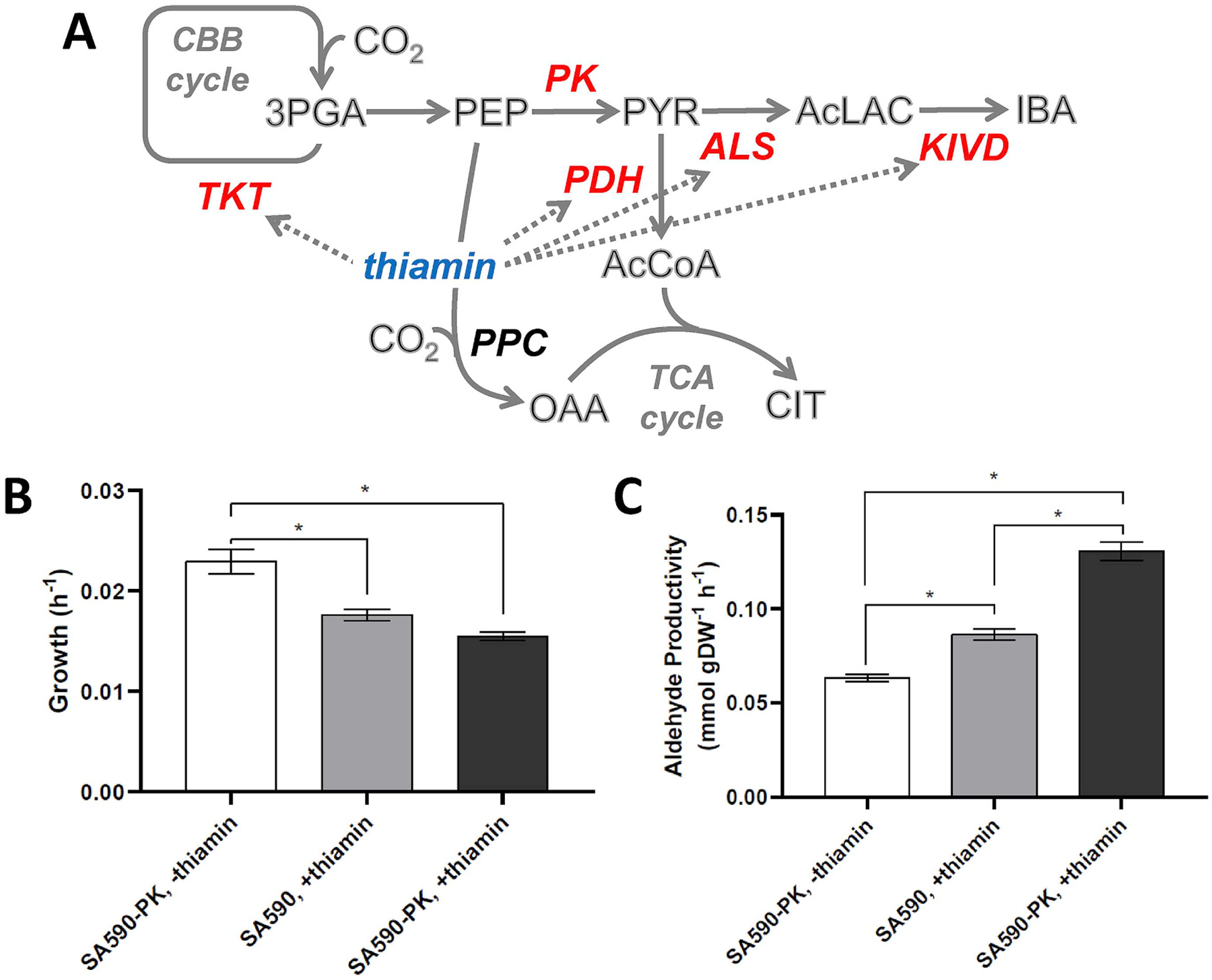
(A) Pathway diagram indicating targeted enzymatic steps (shown in red). Thiamin is a required cofactor for transketolase (TKT), ALS, KIVD, and PDH enzymes. Comparison of growth (B) and specific aldehyde productivity (C) in strains SA590 and SA590-PK with (+) or without (−) thiamin supplementation. Data ± SEM; n = 3. *p < 0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2. Materials and methods
2.1. Culture conditions and measurement of cell growth and aldehyde titer
All strains (listed in Table 1) were maintained on BG11 agar plates or cultured in BG11 liquid media supplemented with 50 mM NaHCO3. With the exception SA590-PK (−thiamin), 5 mg/L thiamin hydrochloride was supplemented in the liquid media. For isotope labeling experiments, 250-mL cultures were seeded to OD750 of ~1.1 in a 1-L Erlenmeyer flask. For experiments to compare numerous strains in parallel, 65-mL cultures were seeded to OD750 of ~1.1 in a 250-mL Erlenmeyer flask. Prior to sampling, all cultures were incubated in a water bath shaker at 130 rpm and 30 °C, and bubbled with humidified air. After a 12-h dark pulse (t = 0 h), air bubbling was stopped, IPTG was added to a final concentration of 1 mM, and the cultures were exposed to light at 120 μmol photons m−2 s−1. Cell density and aldehyde titers were monitored at t = 0, 6, 12, 24, and 30 h. In addition, experiments with strains overexpressing PK, ALS, or PCK under control of the PsmtA promoter were conducted with the same media supplemented with 1 μM ZnCl2, while ZnSO4 and CuSO4 were omitted from the media. SA590-PK was included in all experiments as a control. The correlation cell density (gDW/L) = 0.2504 × OD750 was used to estimate the biomass concentration on a dry-weight basis. Representative growth curves for SA590-PK, SA590-PK-αPDH, and SA590-PK-PCK are provided in Supplementary Information. In order to harvest sufficient biomass for analysis of intracellular metabolites, it was necessary to perform the experiments at a relatively high culture density. In all experiments, the highest OD750 attained at 30 h was ~1.8. Our experience is in agreement with previous work (Atsumi et al., 2009), which indicates that SA590 enters stationary growth phase at an OD750 of ~2.0. Therefore, the results we report in this work are reflective of cultures undergoing late exponential-phase growth. Growth rates and aldehyde productivities were regressed using the ETA software package (Murphy and Young, 2013).
Table 1.
Cyanobacterial strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| SA590 | IBA-producing parental PCC 7942 strain. Ptrc::kivd in NS1 (SpecR), PLlacO1::alsS-alvC-alvD in NS2 (KmR) | Atsumi et al. (2009) |
| SA590-PK | Plac::pk7942 in NS3 (CbR) of SA590 | Jazmin et al. (2017) |
| SA590-PK-αPDH | Plac::pk7942-αpdhB in NS3 (CbR) of SA590 | This work |
| SA590-PK-PK | PsmtA::pk7942 in NS4 (CmR) of SA590-PK | This work |
| SA590-PK-ALS | PsmtA::alsSL.lactis in NS4 (CmR) of SA590-PK | This work |
| SA590-PK-PCK | PsmtA::pckAE.coli in NS4 (CmR) of SA590-PK | This work |
| SA590-PK-PCK-FT | PsmtA::pckAE.coli::3×FLAG in NS4 (CmR) of SA590-PK | This work |
| 7942-LuxAB-FT | PsmtA::luxAB::3×FLAG in NS4 (CmR) of wild-type PCC 7942 | This work |
2.2. 13C labeling experiments, sample preparation, and derivatization
Isotope labeling experiments were conducted as previously described (Jazmin et al., 2017) with a few modifications: (i) culture volume was scaled down to 250 mL (from 500 mL), (ii) 25 mL of NaH13CO3 (Cambridge Isotope Laboratories, 98% isotopic purity) was introduced to the culture (down from 50 mL), and (iii) 20-mL aliquots were harvested at time points 1, 2, 5, 10, and 20min after tracer administration. L-norvaline (7 μg) was added to each sample as an internal standard prior to extraction of metabolites from cell pellets. Sample preparation and derivatization were conducted as previously described (Jazmin et al., 2017) with the following modification: metabolites were converted to methoxime tert-butyldimethylsilyl (MOXTBDMS) derivatives, instead of methoxime trimethylsilyl (MOX-TMS) derivatives, by reaction with 70 μL of MTBSTFA + 1% TBDMCS (Regis Technologies, Inc.). The rationale for this modification is provided in Supplementary Information. The resulting MOX-TBDMS derivatives were analyzed by GC-MS. Because all fluxes of interest were completely resolved based on the available GC-MS measurements, we did not perform LC-MS analysis of sugar phosphates as described in prior 13C flux studies of cyanobacterial metabolism (Jaiswal et al., 2018; Jazmin et al., 2017). Although sugar phosphate measurements are typically required to determine CBB cycle and OPP pathway fluxes (Adebiyi et al., 2015), the GC-MS measurements of MOX-TBDMS derivatives obtained in the current study were sufficient to fully constrain the flux solution.
2.3. GC-MS measurement of metabolite labeling and pool size
The GC-MS method was adapted from our previous study (Jazmin et al., 2017) with minor changes: analysis was performed on an Agilent 7890B/5977A GC-MS equipped with an Agilent DB-5ms column (30 m × 0.25 mm i.d. × 0.25 μm; Agilent J & W Scientific, Santa Clara, CA). The injection volume was 2 μL. Instrument parameters were identical to those of our previous study (Jazmin et al., 2017). Target ions to be used for mass isotopomer analysis were identified based on a list of known fragment ions (Wegner et al., 2014) and retention times. Raw ion chromatograms were integrated using a custom MATLAB mfile that applies consistent integration bounds and baseline correction to each ion (Antoniewicz et al., 2007) in order to calculate total ion counts (TICs) and mass isotopomer distributions (MIDs) of selected fragment ions. Relative changes in metabolite pool sizes were determined by normalizing the TICs from each sample to the L-norvaline (internal standard) peak area and to the cell density at the time of sample harvest. Atom percent enrichment (APE) of each fragment ion was calculated by first correcting the MID for natural abundance of stable isotopes (Fernandez et al., 1996) and then applying the formula , where N is the number of carbon atoms in the fragment ion and Mi is the fractional abundance of the ith mass isotopomer (Egnatchik et al., 2014).
2.4. Isotopically non-stationary metabolic flux analysis (INST-MFA)
The isotopomer model describing the central carbon metabolism of SA590 was adapted from previous studies (Jazmin et al., 2017) with the addition of intracellular lactate to the metabolic network. All measurements used for flux analysis are listed in Table 2. The INCA software package (Young, 2014) was applied to regress the model parameters (fluxes and pool sizes) to fit the measured MIDs of intracellular metabolites and extracellular rates of growth and aldehyde production. Flux estimations were performed on a cluster of 6 quad-core Linux computers (Intel Core i7 2600 processors with 4 GB of memory). Best-fit parameter estimates were obtained by running 50 flux estimations from random initial guesses. Parameter continuation was used to calculate 95% confidence intervals for each flux and pool-size estimate (Antoniewicz et al., 2006). The best-fit parameter estimates and their 95% confidence intervals are shown in Table S1−S3. The sum-of-squared residuals (SSR) and expected SSR ranges are tabulated for each flux solution in Table S4.
Table 2.
Isotope labeling measurements used for metabolic flux determination.
| Metabolite | Mass | Carbons | Composition |
|---|---|---|---|
| 3PGA | 585 | 123 | C23 H54 O7 Si4 P |
| SER | 390 | 123 | C17 H40 O3 N Si3 |
| PEP | 453 | 123 | C17 H38 O6 Si3 P |
| PYR | 174 | 123 | C6 H12 O3 N Si |
| ALA | 232 | 23 | C11 H26 O2 N Si2 |
| LAC | 233 | 23 | C10 H25 O2 Si2 |
| CIT | 465 | 123456 | C20 H39 O6 Si3 |
| SUC | 289 | 1234 | C12 H25 O4 Si2 |
| FUM | 287 | 1234 | C12 H23 O4 Si2 |
| MAL | 419 | 1234 | C18 H39 O5 Si3 |
Abbreviations: 3PGA, 3-phosphoglycerate; ALA, alanine; CIT, citrate; FUM, fumarate; LAC, lactate; MAL, malate; SER, serine; PEP, phosphoenolpyruvate; PYR, pyruvate; SUC, succinate. Measurements taken at t = 0, 1, 2, 5, 10, and 20 min were fitted to the INST-MFA model. Note that the MID of 3PGA could not be accurately quantified in the SA590-PK (−thiamin) experiment.
2.5. Generation of expression constructs and engineered strains
To further engineer the SA590 parental strain, we designed and synthesized the base expression vector pCX0104-LuxAB-FT (Fig. S1). This vector targets the expression cassette to a new integration site located between 236bp and 277bp downstream of the coding region of S. elongatus Synpcc7942_0104, which encodes a hypothetical protein with no currently known function. We have designated this new targeting site as “neutral site 4” (abbreviated here as “NS4”). Genes of interest were targeted into NS4 and expressed under control of the Zn2+-inducible PsmtA promoter (see Results section 3.2). Fig. S1 shows the base construct designed to express the Vibrio harveyi luciferase enzyme LuxAB. To generate constructs for Zn2+-inducible expression of other genes of interest, the luxAB::3×FLAG coding region of pCX0104-LuxAB-FT was excised with SpeI and either XbaI or BamHI, and replaced with genes of interest containing the same cut linkers. Replacing the luxAB::3×FLAG coding region with the coding region of pk (S. elongatus Synpcc7942_0098), alsS (L. lactis), or pckA (E. coli) resulted in plasmids pCX0104-PK, pCX0104-ALS, or pCX0104-PCK, respectively (Table S5). To generate a construct for Zn2+-inducible expression of the fusion protein PCK::3×FLAG, the luxAB coding region on pCX0104-LuxAB-FT was excised with SpeI and KpnI and replaced with the pckA coding sequence containing SpeI and KpnI linkers, generating plasmid pCX0104-PCK-FT (Table S5). The plasmid pPK-αPdhB was generated by modifying pPK/OX-NS3 (Jazmin et al., 2017) to simultaneously express pk (S. elongatus Synpcc7942_0098) and an antisense nucleotide sequence targeting the β subunit of pyruvate dehydrogenase complex E1 (abbreviated here as αPdhB). The αPdhB sequence is a 346bp reverse complement sequence of the pdhB gene (S. elongatus Synpcc7942_0143) spanning positions −213 to +133 relative to the translation start codon. These plasmids were transformed via double homologous recombination to generate the various engineered cyanobacterial strains listed in Table 1. Complete genome segregation of transgenes was confirmed by isolating single transformed colonies and verifying with PCR analysis. All of the DNA constructs and engineered strains were reconfirmed by DNA sequencing, restriction digestion, and PCR analysis. Corresponding primers used for sequencing and PCR are listed in Table S6.
2.6. Analysis of PsmtA-driven gene expression using LuxAB bioluminescent reporter
Cultures were grown in either regular BG11 liquid media with air bubbling or BG11 agar plates supplemented with 7.5 μg/mL chloramphenicol at 30 °C under continuous cool-white illumination (50 μE/m2s). For short-term studies of PsmtA induction in liquid 7942-LuxAB-FT cultures, fresh-grown agar cultures were adjusted to a cell density of OD750 = 0.1 in BG11 medium without ZnSO4 or CuSO4, and the cells were further grown for two days in continuous light (LL). Equal 50-mL aliquots of culture at OD750 = 0.2 were added to multiple flasks, and then ZnCl2 was added to achieve triplicate cultures with varying ZnCl2 concentrations. After Zn2+ addition, 100 μL of culture was mixed with 500 μL of deionized distilled water and 100 μL of 0.01% decanal. Luminescence was immediately measured as counts per second (cps) within 10 s after a 10-s delay in the luminometer. For long-term studies of PsmtA induction in 7942-LuxAB-FT agar cultures, tooth-picked colonies were grown on ZnSO4/CuSO4-free agar plates for 36 h, followed by a 12-h dark exposure to synchronize the circadian rhythms of the individual cells in the population. Then, different concentrations of ZnCl2 were added to the agar plates while lights were turned on. Luminescence levels and rhythms were measured under LL conditions as described previously (Xu et al., 2013).
2.7. RNA extraction and RT-qPCR of pdhA and pdhB expression
Extraction of total RNA from cyanobacterial cell pellets was performed with a Qiagen RNeasy kit (Catalog #74104) with the added step of using RNAprotect Bacteria Reagent (Catalog #76506). Purified mRNA was treated with DNase to remove any contaminating DNA. In brief, 1 μg of RNA was incubated with DNase I and DNase I buffer in a 10 μL reaction at 37 °C for 30 min. EDTA was added to a final concentration of 5 mM, and the reaction was incubated at 65 °C for 10 min to inhibit further DNase activity. cDNA was created from DNase-treated mRNA using the iScript cDNA synthesis kit (Bio-Rad, Hercules, California) in a 20-μL reaction according to the manufacturer’s protocol. Reactions not containing reverse transcriptase were run in parallel, to confirm the removal of DNA contamination.
Real-time quantitative PCR (RT-qPCR) was carried out in Bio-Rad Multiplate 96-well PCR plates, sealed with Microseal ‘B’ Seals, in 20-μL reactions. Each reaction contained 25 ng of cDNA, 250 nM forward and reverse primers, and 10 μL of 2X iQ SYBR Green Supermix (Bio-Rad, Hercules, California). Reaction plates were run on a Bio-Rad CFX96 thermocycler for 40 cycles; threshold cycle (Ct) was determined by Bio-Rad CFX Maestro software using the amplification curves. Primers for pdhB (SynnPCC7942_0143) and pdhA (SynnPCC7942_1944) were obtained based on the best designs generated by Primer3Plus (Table S6) (Nijveen et al., 2007). Target gene expression was normalized to the housekeeping gene rnpB (SynPCC7942_R0366) (Huang et al., 2016), and the ratio of normalized expression in SA590-PK-αPDH relative to SA590-PK was determined using the ΔΔCt method (Livak and Schmittgen, 2001).
2.8. Immunoblots for Zn2+-inducible PCK overexpression
Cultures of the strain SA590-PK-PCK-FT were inoculated from fresh agar plates into Zn2+/Cu2+-free BG11 liquid medium and grown at 30 °C in LL until OD750 = 0.2. Fifty-mL aliquots of culture were added to individual 125-mL flasks, to which varying concentrations of ZnCl2 were added. Following 6 h of Zn2+ induction, the cells were collected and total protein content was extracted as previously described (Xu et al., 2000). Extracts from strain SA590-PK grown in LL were included as a negative FLAG-free control. Seven μg of cell extract was loaded in each sample lane. Western blots for PCK::FLAG fusion protein were performed using mouse monoclonal anti-FLAG® M2 antibody (Sigma, cat. # F1804). Equal loading was confirmed by Coomassie blue staining.
2.9. Statistical analysis
Statistical tests were performed using the GraphPad Prism software. ANOVA was used to assess statistical significance for comparisons between three or more groups. The Student’s t-test was used to assess statistical significance for comparisons between two groups. In all experiments, p < 0.05 was considered statistically significant.
3. Results
3.1. 13C INST-MFA identifies flux alterations that correlate with increased IBA production
We previously observed that IBA production could be substantially increased through overexpression of PK and/or supplementation of thiamin in the culture medium (Jazmin et al., 2017). Thiamin pyrophosphate is a required cofactor for two enzymes in the IBA biosynthetic pathway (ALS and ketoisovalerate decarboxylase, KIVD), as well as transketolase (TKT) in the CBB cycle and PDH in the TCA pathway (Frank et al., 2007). Pyruvate is the central metabolic precursor of IBA, and our prior studies indicated that PK is a potential bottleneck that limits pyruvate availability in SA590 (Fig. 1A). We also previously identified isovaleraldehyde (IVA) as a minor byproduct of the IBA biosynthetic pathway. With thiamin supplemented in the culture medium, overexpression of PK led to ~50% improvement in total aldehyde productivity (IBA + IVA) without inhibiting growth (Fig. 1B,C; light gray vs. dark gray bars). The addition of thiamin to SA590-PK led to an approximate doubling in aldehyde specific productivity, but at the expense of growth (Fig. 1B,C; white vs. dark gray bars).
To further investigate the flux re-routing that occurs in response to increasing IBA production, we performed 13C labeling studies and INST-MFA on the three photoautotrophic cultures described in Fig. 1, which exhibit a range of aldehyde specific productivities. We calculated the atom percent enrichment (APE) of ten metabolites after administering NaH13CO3 tracer (Fig. 2). Steady-state labeling of the CBB cycle was achieved after ~20 min in SA590-PK (−thiamin), at which time PEP had the highest enrichment (62%) and succinate had the lowest enrichment (10%). Similar labeling trends were observed for both SA590 and SA590-PK when grown with thiamin supplementation (data not shown). The lower enrichment of 3-phosphoglycerate (3PGA) compared to its downstream product PEP, across all time points, is consistent with previous reports (Huege et al., 2011; Young et al., 2011). This counterintuitive result has been attributed to metabolic channeling of carboxysomal intermediates, which necessitates the introduction of metabolically inactive 3PGA and PEP pools in the INST-MFA model to accurately describe the enrichment time courses (Ma et al., 2014). Overall, CBB cycle and glycolytic intermediates approached higher final enrichments than TCA cycle metabolites. Substantial enrichments were also observed in the amino acids serine and alanine.
Fig. 2. 13C enrichment time courses of intracellular metabolites following addition of NaH13CO3 tracer to cultures of SA590-PK.
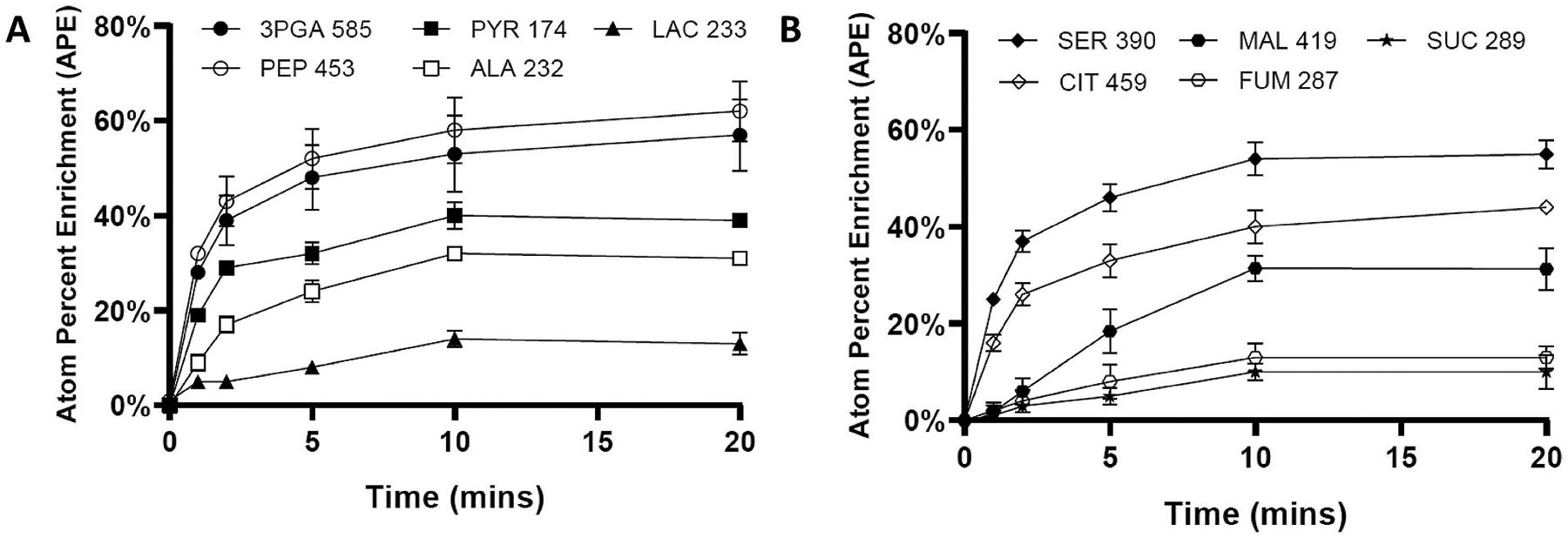
(A) Glycolytic/CBB cycle metabolites and (B) photorespiration/TCA cycle metabolites are shown for SA590-PK grown in the absence of supplemental thiamin. Data ± SEM; n = 3. Results have been corrected for natural abundance of stable isotopes (Fernandez et al., 1996).
Flux solutions obtained from INST-MFA provided valuable insights into the metabolic shifts associated with increasing IBA productivity. By arranging the flux estimates in order from lowest to highest aldehyde productivity (Fig. 3), we identified reactions that correlated with enhanced IBA productivity. ALS and PK fluxes were positively correlated with IBA production, while PDH and PPC fluxes were negatively correlated, and there was no significant correlation with net fluxes through citrate synthase (CS), malate dehydrogenase (MDH), or malic enzyme (ME). The results suggest that supplementation of thiamin effectively diverted carbon flux into aldehyde biosynthesis and away from competing pathways (i.e., PDH and PPC). In contrast, PK overexpression led to increased flux through PK and ALS without significantly altering flux through competing reactions. These results led to the hypothesis that SA590-PK could be further improved by targeting fluxes that strongly correlated with increasing aldehyde production. We therefore decided to test whether overexpression of ALS or PK, or downregulation of flux through PDH or PPC, would achieve further enhancements in IBA production within the genetic background of the SA590-PK strain.
Fig. 3. Pyruvate-associated fluxes under three experimental conditions arranged according to increasing aldehyde productivity.
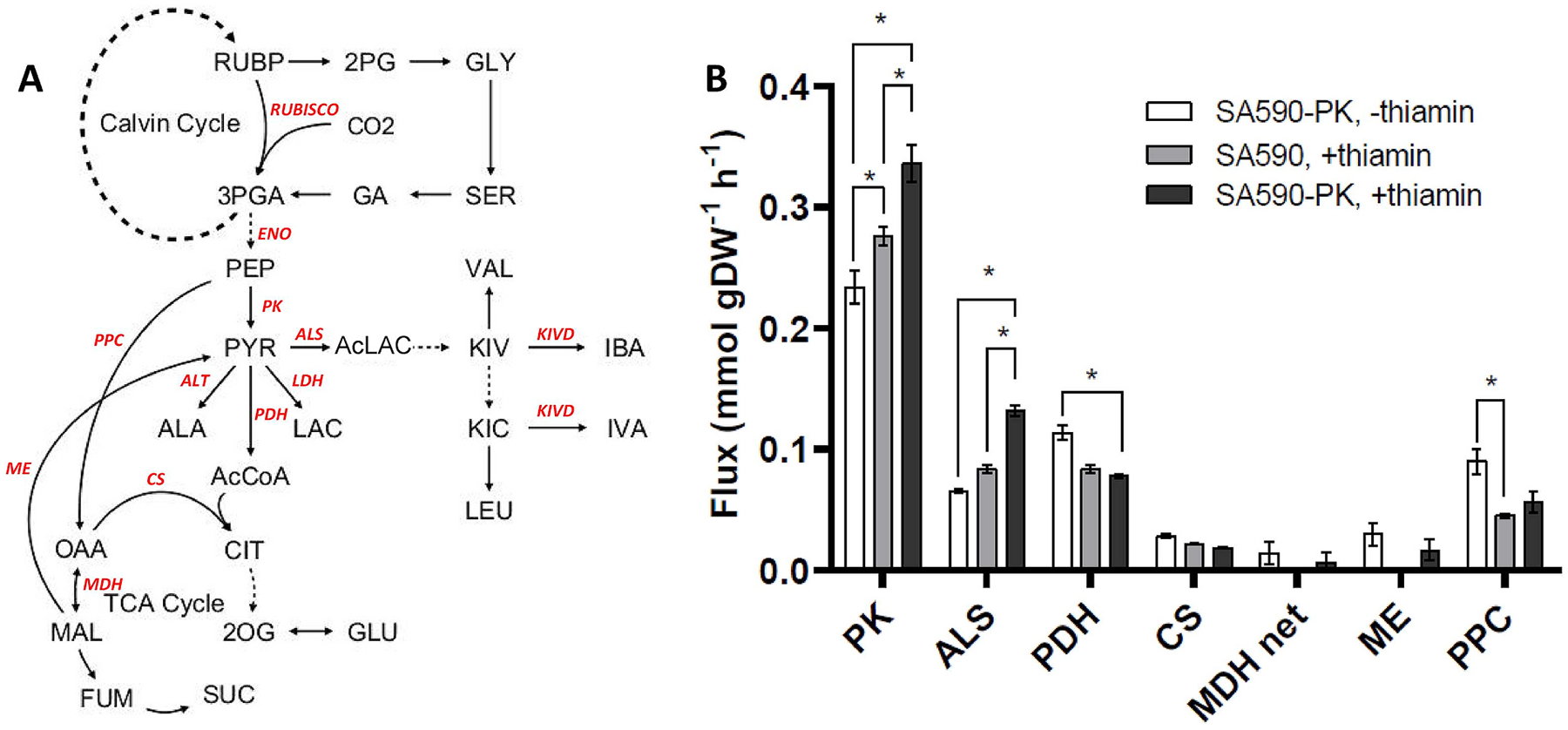
(A) Network diagram indicating key reactions and metabolites included in the INST-MFA model. (B) Fluxes estimated by INST-MFA. Abbreviations: ALS, acetolactate synthase; PK, pyruvate kinase; PDH, pyruvate dehydrogenase; CS, citrate synthase; MDH net, malate dehydrogenase (net flux from oxaloacetate to malate); ME, malic enzyme; PPC, PEP carboxylase. Data ± SEM, where SEM was calculated from the upper (UB) and lower bounds (LB) of the 95% flux confidence intervals (Table S1) using the formula SEM = (UB-LB)/3.92. n = 3. *p < 0.05.
3.2. A Zn2+-inducible cassette was designed for controlled expression of genes in PCC 7942
Engineering additional genes into SA590-PK was limited by the fact that its commonly used neutral sites (NS1, NS2, and NS3) were occupied by previously integrated constructs. Additionally, spectinomycin, kanamycin, and carbenicillin antibiotic resistance markers had already been utilized during prior steps of strain engineering. To circumvent these limitations, we selected a new targeting site, designated NS4, based on analysis of the genome database of S. elongatus PCC 7942 (see Materials and Methods). We designed a novel construct that integrates into this neutral site and uses a chloramphenicol antibiotic resistance marker (CmR). In addition, we designed the construct using the metal-inducible smtB/A-regulated expression system, which is native to PCC 7942, as we were motivated by a recent report that the PsmtA promoter is stronger and less leaky than Plac (Huang et al., 2016). To assess this new construct and evaluate its regulation of gene expression, we placed the bacterial luciferase (luxAB) genes under the control of the PsmtA promoter. A simplified schematic of our design (plasmid pCX0104-LuxAB-FT) is depicted in Fig. S1.
An earlier study that characterized PsmtA with the luxAB reporter system indicated a sensitive, dose-dependent response to ZnCl2 addition, and less-sensitive responses to CuSO4 or CdCl2 addition (Erbe et al., 1996). We tested and validated the ZnCl2 dose-dependent response of our novel construct in liquid cultures (Fig. S2A) and on agar plates (Fig. S2B). Though PsmtA-driven LuxAB luminescence exhibited a proportional dose-response to ZnCl2 addition up to 8 μM, we found that doses of ZnCl2 ≥5 μM were inhibitory to growth of SA590-PK under our culture conditions (Fig. S3). Hence, we restricted the inducer concentration to 1 μM ZnCl2 in all subsequent experiments.
3.3. Overexpression of PK or ALS does not impact SA590-PK growth and aldehyde production
As PK and ALS were overexpressed in SA590-PK using the weak Plac and PLlacO1 promoters, respectively, we hypothesized that stronger overexpression of these genes with PsmtA may lead to further improvements in IBA production. To test this hypothesis, we replaced luxAB in pCX0104-LuxAB-FT with the pk or alsS genes. The new plasmids, denoted pCX0104-PK and pCX0104-ALS (Table S5), were transformed separately into SA590-PK as described in Materials and Methods. The resulting strains, SA590-PK-PK and SA590-PK-ALS (Table 1), were compared to SA590-PK under photoautotrophic growth conditions. Further overexpression of PK or ALS did not impact growth or improve aldehyde productivity (Fig. 4), implying that these enzymes are not rate-limiting for IBA production in SA590-PK.
Fig. 4. Effects of manipulating target genes on growth and product formation in SA590-PK.
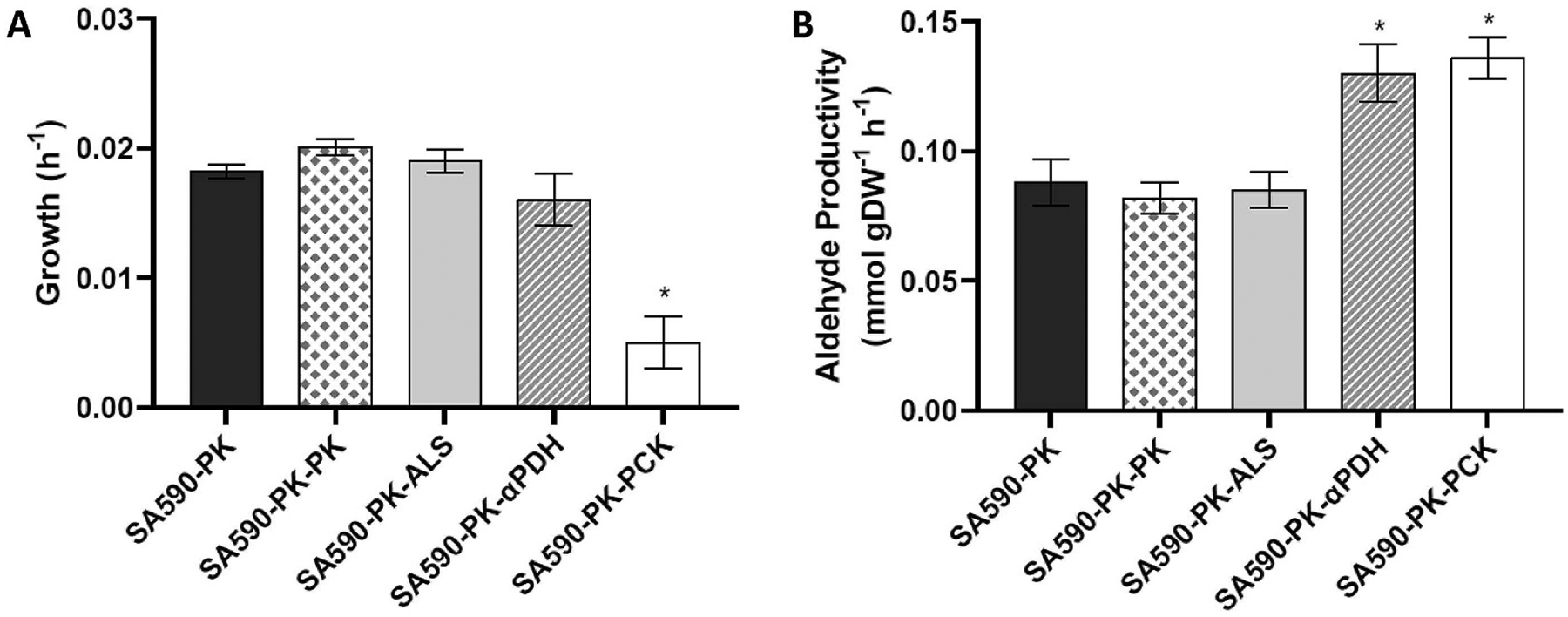
(A) Specific growth rate and (B) specific aldehyde productivity over 24 h (estimated from 6 to 30h time points in Fig. S5). Abbreviations indicate genes overexpressed in the control SA590-PK strain: ALS, acetolactate synthase (L. lactis); PK, pyruvate kinase (S. elongatus); αPDH, antisense RNA targeted to pyruvate dehydrogenase subunit B (S. elongatus); PCK, PEP carboxykinase (E. coli). Data ± SEM; n = 6 for SA590-PK, SA590-PK-αPDH, SA590-PK-PCK; n = 3 for SA590-PK-PK and SA590-PK-ALS. *p < 0.05 relative to control SA590-PK strain.
3.4. Downregulation of PdhB via antisense RNA increases aldehyde production of SA590-PK without inhibiting growth
The pyruvate dehydrogenase complex in S. elongatus is encoded by four genes: pdhA, pdhB, pdhC, and pdhD. Little is known about the regulation of PDH in S. elongatus, but genome annotations have designated pdhA and pdhB as encoding the E1 catalytic subunit of PDH, while pdhC and pdhD encode the E2 and E3 subunits, respectively. Although heterotrophic bacteria can survive without PDH activity (Kim et al., 2008; Schreiner et al., 2005; Wang et al., 2010), we hypothesized that a complete knockout of PDH would be lethal in PCC 7942 since there is no other major route to produce acetyl-CoA for lipid synthesis. Hence, we generated strain SA590-PK-αPDH by expressing both pk and an antisense sequence targeted to the pdhB gene under control of the IPTG-inducible Plac promoter in the SA590 background. Using RT-qPCR, we found that the relative mRNA expression of pdhB in SA590-PK-αPDH trended lower but was not significantly different from SA590-PK (Fig. S4A). Interestingly however, we found that the relative expression of pdhA was significantly downregulated, by as much as 48% (Fig. S4B), likely due to feedback repression of pdhA in response to pdhB knockdown. When SA590-PK and SA590-PK-αPDH were grown under similar conditions, we found that PDH knockdown led to a 47% increase in aldehyde productivity (Figs. 4B) and 37% increase in final aldehyde titer (Fig. S5B), without any negative impact on growth (Fig. 4A and Fig. S5A). Therefore, inhibiting flux through the competing PDH reaction was an effective strategy to increase IBA production, consistent with predictions derived from 13C INST-MFA.
3.5. Overexpression of PCK increases specific aldehyde productivity of SA590-PK at the expense of growth
Deletion of ppc in C. glutamicum has been reported to enhance production of α-ketoisovaleric acid (Buchholz et al., 2013) and L-valine (Hasegawa et al., 2013), both of which are derived from the same biosynthetic pathway used to produce IBA in SA590. However, PPC is an essential enzyme for growth of S. elongatus PCC 7942 under photoautotrophic conditions (Luinenburg and Coleman, 1990), and it cannot be readily complemented by expression of heterologous PPC isoforms (Luinenburg and Coleman, 1993). Rather than knocking down PPC expression as we did with PDH, we decided to heterologously express an opposing metabolic enzyme PEP carboxykinase (PCK) from E. coli. Two versions of the PCK overexpressing strain, SA590-PK-PCK and SA590-PK-PCK-FT (Table 1), were generated and used for functional evaluation and overexpression verification, respectively. As shown in Fig. 5, the PsmtA::PCK::3×FLAG protein exhibited Zn2+ dosage-dependent expression in strain SA590-PK-PCK-FT. Combined overexpression of PK and PCK in strain SA590-PK-PCK led to a 54% increase in aldehyde specific productivity (Figs. 4B) and 62% increase in final aldehyde titer (Fig. S5B) compared to PK overexpression alone, but dramatically reduced growth rate by as much as 72% (Fig. 4A and Fig. S5A). In addition, we observed a substantial redistribution of aldehyde products in response to PCK overexpression, with the ratio of IVA to IBA increasing from 9/91 in SA590-PK to 27/73 in SA590-PK-PCK (Fig. 6). While PCK overexpression shows promise as a strategy to divert flux into aldehyde biosynthesis, the unexpected effects on growth and product distribution motivated further examination into the nature of flux re-routing in SA590-PK-PCK.
Fig. 5. Zn2+ dosage-dependent overexpression of the PsmtA::PCK::3×FLAG protein in strain SA590-PK-PCK.
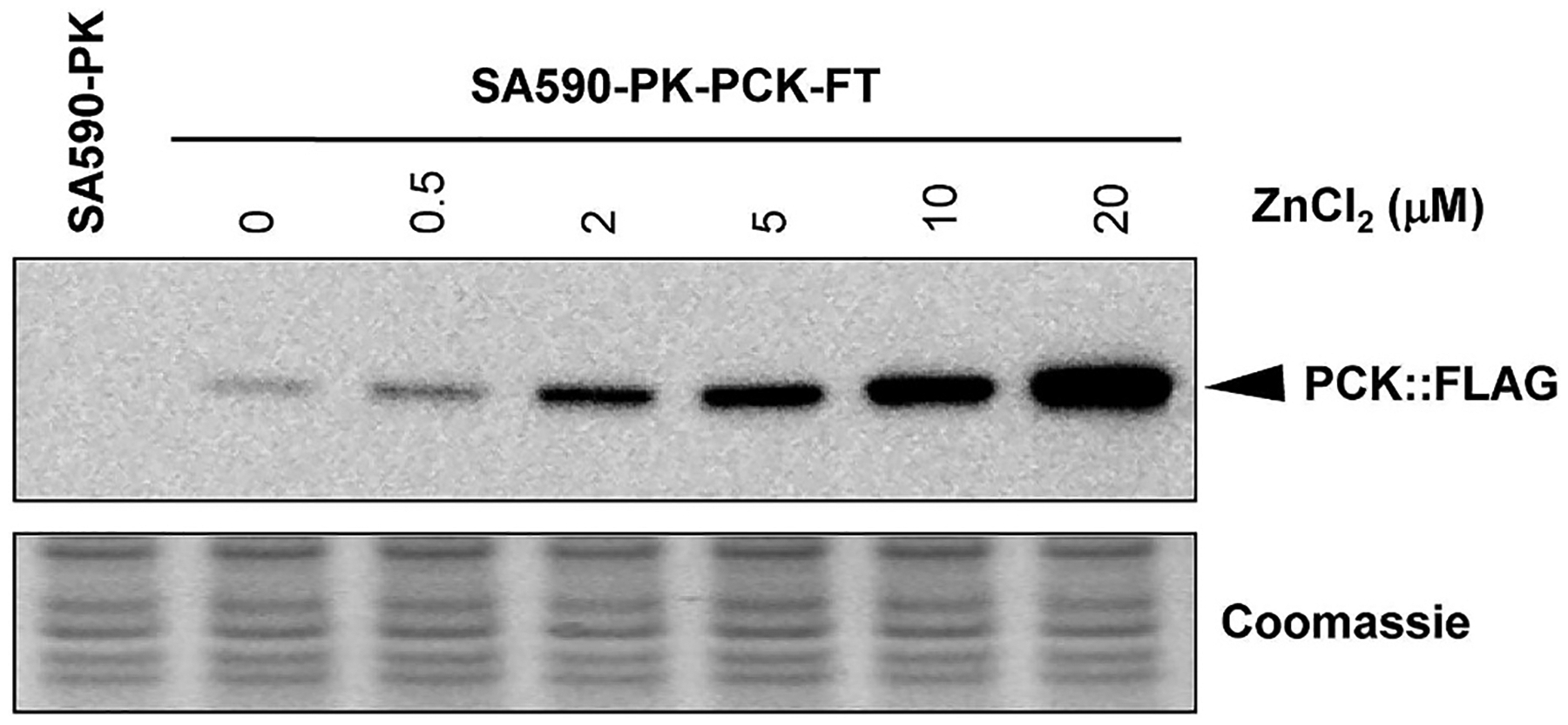
Expression of the PsmtA::PCK::3×FLAG protein was induced for 6 h with different concentrations of Zn2+ and assessed by Western blot. Seven micrograms of cell extract was loaded in each lane, and equal loading was confirmed by Coomassie blue staining. Extracts from strain SA590-PK grown in LL were included as a negative control.
Fig. 6. PCK expression alters aldehyde product composition in SA590-PK.
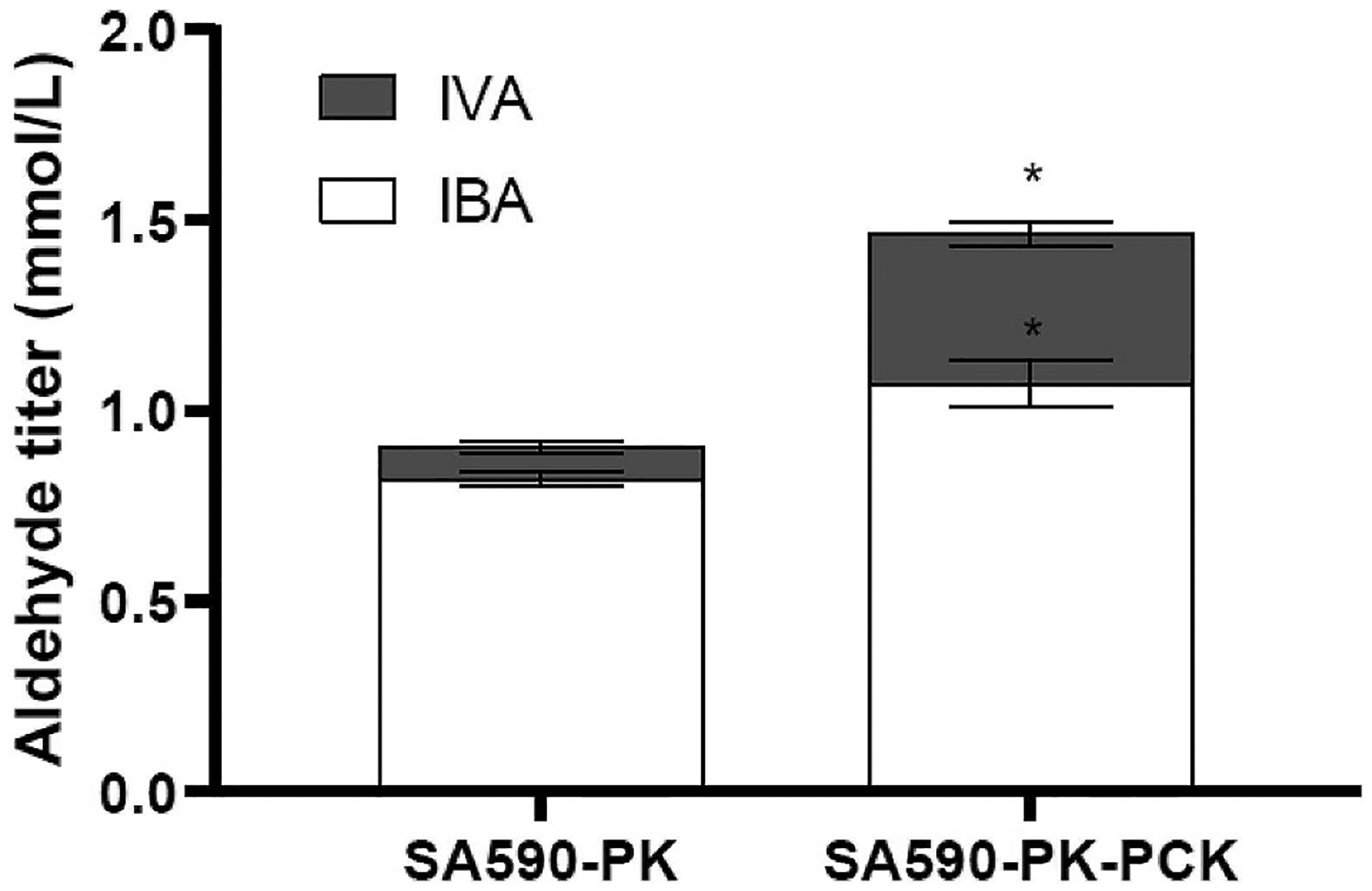
Medium concentrations of isobutyraldehyde (IBA) and isovaleraldehyde (IVA) at t = 30h of culture. *p < 0.05 relative to control SA590-PK strain.
3.6. Intracellular pool sizes of PCK overexpression strain indicate perturbations to pyruvate and amino acid metabolism
Because of the dramatic reduction in growth rate and the redistribution of flux from IBA to IVA observed in SA590-PK-PCK, we decided to profile intracellular pool sizes to assess whether metabolites were accumulating to inhibitory levels in response to PCK overexpression. Examination of pool sizes in the CBB cycle and glycolysis pathways revealed significant increases in pyruvate and lactate, suggesting carbon overflow at the pyruvate node (Fig. 7A). Interestingly, the alanine pool size was increased more than 18-fold in SA590-PK-PCK relative to SA590-PK (Fig. 7B), while several other amino acid levels were also perturbed, including aspartate, glutamate, serine, and valine. These changes in pyruvate and amino acid metabolism could be directly responsible for the observed shifts in growth and product distribution. In particular, synthesis of both IBA and IVA is catalyzed by the heterologous KIVD enzyme in SA590 (Fig. 3A), which has a primary substrate preference for α-ketoisovalerate (KIV, an intermediate in valine biosynthesis) and a secondary preference for α-ketoisocaproate (KIC, an intermediate in leucine biosynthesis) (de la Plaza et al., 2004). Although we could not measure the levels of KIV or KIC directly, the finding that valine was significantly elevated while leucine abundance was not affected in SA590-PK-PCK (Fig. 7B) could be an indication that the conversion of KIV into IBA had become rate-limiting while the conversion of KIC into IVA was still operating with excess capacity. Thus, alterations in amino acid metabolism in response to PCK overexpression could have been partially responsible for the redistribution of product flux from IBA to IVA in SA590-PK-PCK. In contrast, SA590-PK-αPDH exhibited less dramatic increases in lactate (Fig. 7C) and alanine (Fig. 7D) and maintained homeostatic levels of all other amino acids (Fig. 7D) following PDH knockdown, which likely explains the absence of any growth defect or product redistribution from IBA to IVA in this strain.
Fig. 7. Relative abundance of intracellular metabolites in SA590-PK-PCK and SA590-PK-αPDH.
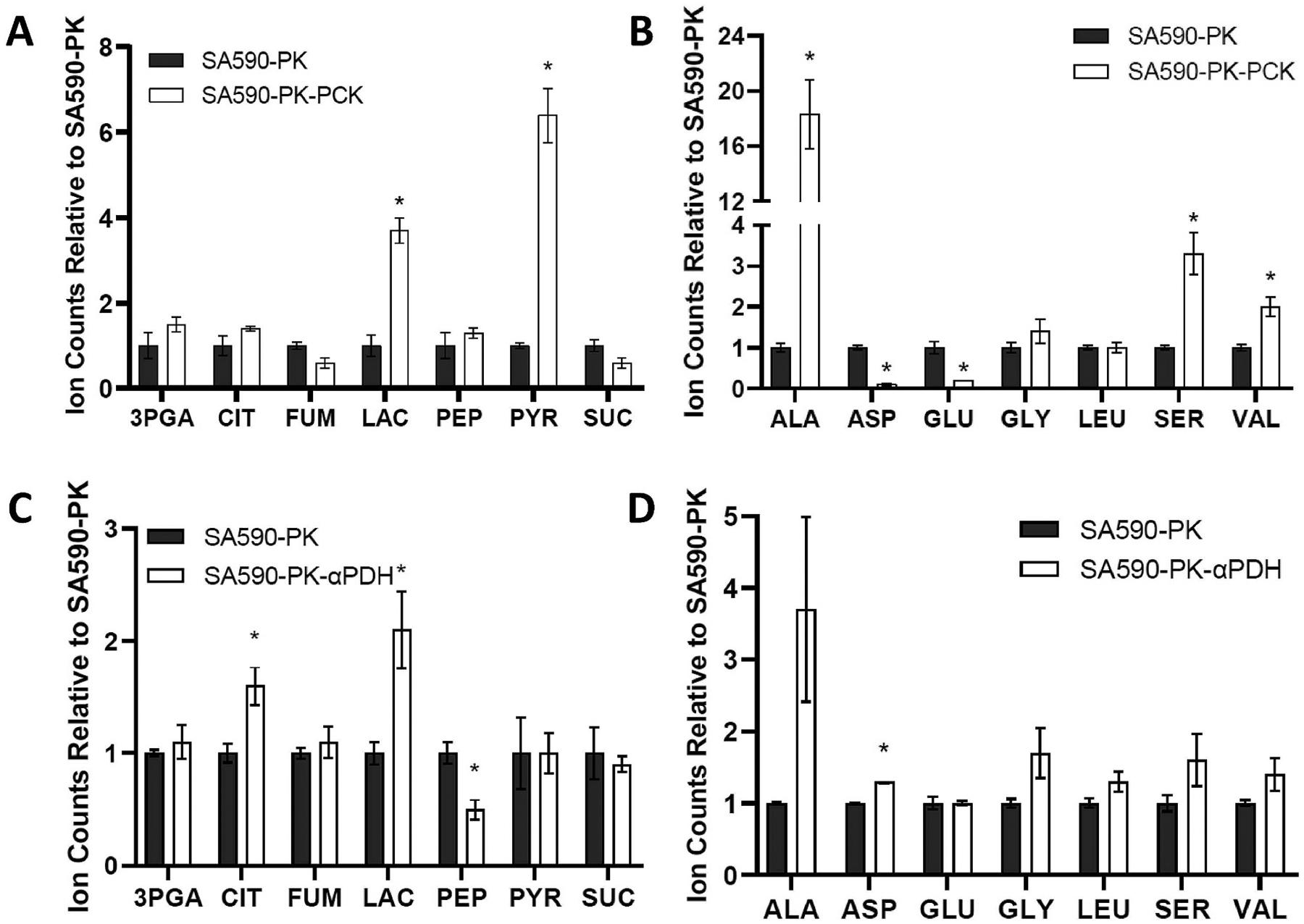
(A) CBB cycle and TCA cycle metabolites, and (B) amino acids in SA590-PK-PCK relative to SA590-PK. (C) CBB cycle and TCA cycle metabolites, and (D) amino acids in SA590-PK-αPDH relative to SA590-PK. Data shown are GC-MS ion counts normalized to an internal standard, L-norvaline, and also normalized to cell density at the time of sample collection. The normalized ion counts for both strains were then expressed relative to SA590-PK, which adjusts all relative counts for SA590-PK to unity. Data ± SEM; n = 3. *p < 0.05 relative to control SA590-PK strain.
4. Discussion
Rational selection of gene targets to improve strain performance is a major challenge in the field of metabolic engineering. Stoichiometric modeling approaches such as OptForce (Ranganathan et al., 2010) and OptKnock (Burgard et al., 2003) are frequently used to select target genes for overexpression or knockout in order to divert flux to a desired product. These methods rely on optimality assumptions to predict metabolic flux distributions based on an underdetermined system of mass balances and measurement constraints. 13C flux analysis, on the other hand, does not rely on such assumptions and therefore has the potential to identify unanticipated bottlenecks or unexpected metabolic phenotypes of engineered stains, as well as test the accuracy of theoretic predictions provided by stoichiometric models. In this study, we implemented a metabolic engineering strategy based on (i) application of 13C INST-MFA to assess metabolic flux alterations that correlate with increased strain productivity, followed by (ii) rational selection and validation of gene targets guided by these empirical flux correlations. Given that aldehydes (IBA and IVA) are derived from pyruvate in SA590, we optimized our analysis to precisely determine fluxes entering and leaving the pyruvate node. As a result, we pinpointed PK and ALS as fluxes that directly correlated with aldehyde production, and PDH and PPC as fluxes that varied inversely with aldehyde production (Fig. 3). These INST-MFA results provided a rational basis to generate and test new strategies for increasing aldehyde production in the best IBA-producing strain (SA590-PK) obtained in our prior study (Jazmin et al., 2017).
The positive correlation of PK and ALS fluxes with aldehyde productivity led us to hypothesize that further overexpression of these enzymes would provide additional improvements in SA590-PK. However, our experimental results showed that overexpression of these genes did not promote further increases in aldehyde productivity (Fig. 4). The most likely explanation for this result is that PK and ALS are already sufficiently expressed in SA590-PK under the IPTG-inducible Plac promoter and thus are no longer rate-controlling steps for aldehyde production. Alternatively, the PK reaction (which requires ADP) or other IBA pathway reactions may become cofactor-limited as more flux is re-routed to IBA. The recombinant IBA pathway in SA590 contains three cofactor-dependent enzymes: ALS and KIVD require thiamin, while ketol-acid reductoisomerase (IlvC) requires NADPH. Though we supplemented thiamin hydrochloride to the cultures in this study, we have not explored the effect of varying the supply of thiamin or other cofactors to maximize IBA production. Hence, efforts to optimize cofactor availability may be a worthwhile direction of future studies.
The inverse correlation of PDH and PPC fluxes with aldehyde production led us to hypothesize that attenuating these fluxes would augment aldehyde production in SA590-PK. Our experimental results support this hypothesis (Fig. 4). The observed increase in aldehyde productivity in response to PDH knockdown is in agreement with a previous study in C. glutamicum, where repression of PDH increased carbon flux towards the valine biosynthesis pathway (Buchholz et al., 2013). Opposing PPC flux by expression of its reverse reaction (PCK) caused an increase in aldehyde productivity, which is consistent with previous studies in E. coli where the simultaneous overexpression of PK and PCK led to increases in pyruvate-derived products (Chao and Liao, 1994; Hädicke et al., 2015). We also observed that the overexpression of PCK led to a dramatic inhibition of growth relative to the SA590-PK control strain. This could lend itself to an effective two-phase production strategy if PCK expression is driven from an inducible promoter, similar to the strategy developed by Shabestary et al. (2018) to divert carbon flux into bioproducts through inducible CRISPRi repression of cell growth in Synechocystis PCC 6803.
Due to the severe growth inhibition caused by PCK expression, we decided to investigate metabolite pool sizes of SA590-PK-PCK in further detail. We uncovered significant accumulation of pyruvate and pyruvate-derived products, most notably alanine, in comparison to SA590-PK (Fig. 7). This result suggests that the simultaneous expression of PK and PCK can be an effective approach to increase flux towards pyruvate-derived products. Furthermore, the two-fold increase in valine pool size in combination with the elevated ratio of IVA/IBA in SA590-PK-PCK suggests that KIVD, which is the final step in the IBA-producing pathway, may be saturated for its substrate KIV and could be targeted for overexpression to remove this potential bottleneck. It is likely that further optimization of the aldehyde biosynthetic pathway is needed to redirect the accumulated pathway intermediates toward the desired products. This may, in turn, relieve the growth inhibition induced by PCK overexpression through reducing the abundance of these intermediates back to homeostatic levels.
Several assumptions were made to simplify the metabolic network model used in this study. First, we assumed balanced flux through the recombinant aldehyde-producing pathway, and hence all flux through ALS was directed toward aldehyde products. This simplification disregards potential carbon losses due to secretion or excess accumulation of intermediates in the aldehyde biosynthetic pathway. Inclusion of these additional overflow products into the INST-MFA model is necessary to better predict bottlenecks to aldehyde production in future work. Second, the overflow of alanine and lactate in SA590-PK-PCK suggests that these metabolites may serve as additional carbon sinks under conditions of pyruvate excess. To accurately estimate fluxes through alanine transaminase and lactate dehydrogenase, the contributions of alanine to biomass (Nogales et al., 2012) and lactate overflow (Stal and Moezelaar, 1997) need to be more precisely quantified. However, these potential inaccuracies are likely small in comparison to the total flux passing through the pyruvate node. Third, we have assumed the same biomass composition (including the content of glycogen and other storage compounds) from our prior publication (Jazmin et al., 2017) for all SA590-derived strains. We previously found that net fluxes directly linked to the synthesis of biomass precursors were affected by changes in the yield coefficients of the growth equation, and therefore uncertainties in the biomass composition could have potential impacts on the INST-MFA results. However, we do not expect that these uncertainties would change the overall conclusions of our study, since the enzymes selected for manipulation are not directly involved in the synthesis of storage compounds or macromolecular constituents of biomass.
Lastly, out of the four predicted bottlenecks in this study, we have demonstrated that PDH and PPC are rate-controlling steps for aldehyde production by SA590 in vivo. We expect that higher productivities could be achieved through further optimization of the expression of αPdhB and PCK constructs. Induction of PCK resulted in severe growth inhibition (Fig. 4A) that could be potentially rescued by optimizing the inducer concentration. In contrast, it is surprising that knockdown of PDH flux by expression of αPdhB did not cause a growth defect as observed in previous studies (Buchholz et al., 2013). One possible explanation for this discrepancy is that our antisense strategy only provided ~50% reduction in PDH expression, while PDH activity was reduced to ≤20% of the wild-type level in the prior work of Buchholz et al. through a promoter replacement strategy. This notion is further supported by the relatively minor changes in intracellular pool sizes observed in our PDH knockdown strain (Fig. 7C,D). Therefore, further optimization of PDH knockdown in SA590 could potentially divert even more flux into aldehyde production.
5. Conclusion
Metabolic bottlenecks are not static features of biochemical networks, and therefore systematic strategies are needed to progressively identify and overcome pathway bottlenecks in producer strains of cyanobacteria and other host species. The current study presents one full iteration of metabolic engineering based on 13C flux analysis of the previous best IBA-producing strain: target identification, strain construction, and target validation. We found that PDH knockdown or PCK expression achieved 40–60% increases in final titer compared to the SA590-PK parental strain, which was already highly optimized compared to other cyanobacterial hosts (Angermayr et al., 2015). Experience has shown that there is rarely a “magic bullet” that can achieve large jumps in titer at later stages of strain development, since new bottlenecks inevitably emerge as existing bottlenecks are removed. Our work illustrates how application of flux analysis along with other metabolomics assays can provide a systematic approach for achieving iterative, stepwise improvements in strain performance. This process often begins by comparing a non-producing parent strain with an engineered producer strain to select initial targets for strain debottlenecking (Jazmin et al., 2017). Subsequent steps then involve comparing the metabolic phenotypes of several producer strains with varying titers/rates/yields in order to identify additional gene targets that can be manipulated to further enhance strain performance.
Starting from the IBA-producing strain SA590, we have now identified multiple strategies to increase aldehyde productivity by first overexpressing PK (Jazmin et al., 2017), and then simultaneously knocking down PDH or expressing PCK (this study). While flux to aldehydes in SA590 was initially limited by pyruvate availability, we have now engineered strains that produce a variety of overflow products at the pyruvate node. This leads us to hypothesize that redirecting fluxes from these accumulated intermediates into the IBA biosynthetic pathway could promote further increases in aldehyde production. In a broader sense, the strain engineering approaches we have employed here may also be beneficial to other cyanobacterial cell factories that use pyruvate as a metabolic precursor: e.g., lactate (van der Woude et al., 2014), 2,3-butanediol (Nozzi and Atsumi, 2015), or ethanol (Deng and Coleman, 1999). It would also be of interest to examine whether alternative approaches for increasing pyruvate availability, such as carboxysome knockout (Abernathy et al., 2019), might have similar effects on the production of IBA and other pyruvate-derived products.
Supplementary Material
Acknowledgements
This material is based upon work supported by the U.S. Department of Energy Genomic Science Program under award DE-SC0019404 and the National Institutes of Health under awards R37 GM067152 and R01 GM107434. The authors thank Irina Trenary and ZhiZhang Wang for maintaining GC-MS instrumentation, and Chi Zhao for kindly sharing cyanobacteria RNA extraction protocols and materials.
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymben.2020.03.007.
References
- Abernathy MH, Czajka JJ, Allen DK, Hill NC, Cameron JC, Tang YJ, 2019. Cyanobacterial carboxysome mutant analysis reveals the influence of enzyme compartmentalization on cellular metabolism and metabolic network rigidity. Metab. Eng 54, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy MH, Yu J, Ma F, Liberton M, Ungerer J, Hollinshead WD, Gopalakrishnan S, He L, Maranas CD, Pakrasi HB, Allen DK, Tang YJ, 2017. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. Biofuels 10, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adebiyi AO, Jazmin LJ, Young JD, 2015. 13C flux analysis of cyanobacterial metabolism. Photosynth. Res 126, 19–32. [DOI] [PubMed] [Google Scholar]
- Angermayr SA, Gorchs Rovira A, Hellingwerf KJ, 2015. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol 33, 352–361. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, 2013. 13C metabolic flux analysis: optimal design of isotopic labeling experiments. Curr. Opin. Biotechnol 24, 1116–1121. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G, 2006. Determination of confidence intervals of metabolic fluxes estimated from stable isotope measurements. Metab. Eng 8, 324–337. [DOI] [PubMed] [Google Scholar]
- Antoniewicz MR, Kelleher JK, Stephanopoulos G, 2007. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem 79, 7554–7559. [DOI] [PubMed] [Google Scholar]
- Atsumi S, Higashide W, Liao JC, 2009. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol 27, 1177–1180. [DOI] [PubMed] [Google Scholar]
- Broddrick JT, Rubin BE, Welkie DG, Du N, Mih N, Diamond S, Lee JJ, Golden SS, Palsson BO, 2016. Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. Unit. States Am 113, E8344–E8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz J, Schwentner A, Brunnenkan B, Gabris C, Grimm S, Gerstmeir R, Takors R, Eikmanns BJ, Blombach B, 2013. Platform engineering of Corynebacterium glutamicum with reduced pyruvate dehydrogenase complex activity for improved production of L-lysine, L-valine, and 2-ketoisovalerate. Appl. Environ. Microbiol 79, 5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgard AP, Pharkya P, Maranas CD, 2003. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng 84, 647–657. [DOI] [PubMed] [Google Scholar]
- Carr NG, 1988. Nitrogen Reserves and Dynamic Reservoirs in Cyanobacteria Univ. Warwick, Coventry, CV4 7AL (UK)). [Google Scholar]
- Chao YP, Liao JC, 1994. Metabolic responses to substrate futile cycling in Escherichia coli. J. Biol. Chem 269, 5122–5126. [PubMed] [Google Scholar]
- Cheah YE, Hasenour CM, Young JD, 2017. 13C Flux Analysis in Biotechnology and Medicine Systems Biology Wiley-VCH Verlag GmbH & Co. KGaA, pp. 25–70. [Google Scholar]
- Cheah YE, Young JD, 2018. Isotopically nonstationary metabolic flux analysis (INST MFA): putting theory into practice. Curr. Opin. Biotechnol 54, 80–87. [DOI] [PubMed] [Google Scholar]
- de la Plaza M, de Palencia PF, Pelaez C, Requena T, 2004. Biochemical and molecular characterization of alpha-ketoisovalerate decarboxylase, an enzyme involved in the formation of aldehydes from amino acids by Lactococcus lactis. FEMS Microbiol. Lett 238, 367–374. [DOI] [PubMed] [Google Scholar]
- Deng M-D, Coleman JR, 1999. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol 65, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egnatchik RA, Leamy AK, Noguchi Y, Shiota M, Young JD, 2014. Palmitate-induced activation of mitochondrial metabolism promotes oxidative stress and apoptosis in H4IIEC3 rat hepatocytes. Metab., Clin. Exp 63, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbe JL, Adams AC, Taylor KB, Hall LM, 1996. Cyanobacteria carrying an smt-lux transcriptional fusion as biosensors for the detection of heavy metal cations. J. Ind. Microbiol 17, 80–83. [DOI] [PubMed] [Google Scholar]
- Erbe JL, Taylor KB, Hall LM, 1995. Metalloregulation of the cyanobacterial smt locus: indentification of SmtB binding sites and direct interaction with metals. Nucleic Acids Res 23, 2472–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H, 1996. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom 31, 255–262. [DOI] [PubMed] [Google Scholar]
- Frank RAW, Leeper FJ, Luisi BF, 2007. Structure, mechanism and catalytic duality of thiamine-dependent enzymes. Cell. Mol. Life Sci 64, 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hädicke O, Bettenbrock K, Klamt S, 2015. Enforced ATP futile cycling increases specific productivity and yield of anaerobic lactate production in Escherichia coli. Biotechnol. Bioeng 112, 2195–2199. [DOI] [PubMed] [Google Scholar]
- Hasegawa S, Suda M, Uematsu K, Natsuma Y, Hiraga K, Jojima T, Inui M, Yukawa H, 2013. Engineering of Corynebacterium glutamicum for high-yield L-valine production under oxygen deprivation conditions. Appl. Environ. Microbiol 79, 1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C-H, Shen CR, Li H, Sung L-Y, Wu M-Y, Hu Y-C, 2016. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb. Cell Factories 15, 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huege J, Goetze J, Schwarz D, Bauwe H, Hagemann M, Kopka J, 2011. Modulation of the major paths of carbon in photorespiratory mutants of Synechocystis. PloS One 6, e16278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal D, Prasannan CB, Hendry JI, Wangikar PP, 2018. SWATH tandem mass spectrometry workflow for quantification of mass isotopologue distribution of intracellular metabolites and fragments labeled with isotopic 13C carbon. Anal. Chem 90, 6486–6493. [DOI] [PubMed] [Google Scholar]
- Jazmin LJ, O’Grady JP, Ma F, Allen DK, Morgan JA, Young JD, 2014. Isotopically nonstationary MFA (INST-MFA) of autotrophic metabolism. Methods Mol. Biol 1090, 181–210. [DOI] [PubMed] [Google Scholar]
- Jazmin LJ, Xu Y, Cheah YE, Adebiyi AO, Johnson CH, Young JD, 2017. Isotopically nonstationary 13C flux analysis of cyanobacterial isobutyraldehyde production. Metab. Eng 42, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajihata S, Furusawa C, Matsuda F, Shimizu H, 2014. OpenMebius: an open source software for isotopically nonstationary 13C-based metabolic flux analysis. BioMed Res. Int 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Ingram LO, Shanmugam KT, 2008. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol 190, 3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luinenburg I, Coleman JR, 1990. A requirement for phosphoenolpyruvate carboxylase in the cyanobacterium Synechococcus PCC 7942. Arch. Microbiol 154, 471–474. [Google Scholar]
- Luinenburg I, Coleman JR, 1993. Expression of Escherichia coli phosphoenolpyruvate carboxylase in a cyanobacterium (functional complementation of Synechococcus PCC 7942 ppc). Plant Physiol 101, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma FF, Jazmin LJ, Young JD, Allen DK, 2014. Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc. Natl. Acad. Sci. U.S.A 111, 16967–16972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAtee AG, Jazmin LJ, Young JD, 2015. Application of isotope labeling experiments and 13C flux analysis to enable rational pathway engineering. Curr. Opin. Biotechnol 36, 50–56. [DOI] [PubMed] [Google Scholar]
- Murphy TA, Young JD, 2013. ETA: robust software for determination of cell specific rates from extracellular time courses. Biotechnol. Bioeng 110, 1748–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Keasling Jay D., 2016. Engineering cellular metabolism. Cell 164, 1185–1197. [DOI] [PubMed] [Google Scholar]
- Nijveen H, Leunissen JAM, Geurts R, Bisseling T, Rao X, Untergasser A, 2007. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res 35, W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I, 2012. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc. Natl. Acad. Sci. Unit. States Am 109, 2678–2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nöh K, Wiechert W, 2006. Experimental design principles for isotopically in stationary 13C labeling experiments. Biotechnol. Bioeng 94, 234–251. [DOI] [PubMed] [Google Scholar]
- Nozzi NE, Atsumi S, 2015. Genome engineering of the 2,3-butanediol biosynthetic pathway for tight regulation in cyanobacteria. ACS Synth. Biol 4, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Preiss J, Romeo T, 1989. Physiology, biochemistry and genetics of bacterial glycogen-synthesis. Adv. Microb. Physiol 30, 183–238. [DOI] [PubMed] [Google Scholar]
- Ranganathan S, Suthers PF, Maranas CD, 2010. OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol 6, e1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner ME, Fiur D, Holatkoj J, Patek M, Eikmanns BJ, 2005. E1 enzyme of the pyruvate dehydrogenase complex in Corynebactetium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol 187, 6005–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A, Pakrasi HB, Wangikar PP, 2018. Recent advances in synthetic biology of cyanobacteria. Appl. Microbiol. Biotechnol 102, 5457–5471. [DOI] [PubMed] [Google Scholar]
- Shabestary K, Anfelt J, Ljungqvist E, Jahn M, Yao L, Hudson EP, 2018. Targeted repression of essential genes to arrest growth and increase carbon partitioning and biofuel titers in cyanobacteria. ACS Synth. Biol 7, 1669–1675. [DOI] [PubMed] [Google Scholar]
- Shastri AA, Morgan JA, 2005. Flux balance analysis of photoautotrophic metabolism. Biotechnol. Prog 21, 1617–1626. [DOI] [PubMed] [Google Scholar]
- Shastri AA, Morgan JA, 2007. A transient isotopic labeling methodology for 13C metabolic flux analysis of photoautotrophic microorganisms. Phytochemistry 68, 2302–2312. [DOI] [PubMed] [Google Scholar]
- Stal LJ, Moezelaar R, 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev 21, 179–211. [Google Scholar]
- van der Woude AD, Angermayr SA, Puthan Veetil V, Osnato A, Hellingwerf KJ, 2014. Carbon sink removal: increased photosynthetic production of lactic acid by Synechocystis sp. PCC6803 in a glycogen storage mutant. J. Biotechnol 184, 100–102. [DOI] [PubMed] [Google Scholar]
- Wang Q, Ou MS, Kim Y, Ingram LO, Shanmugam KT, 2010. Metabolic flux control at the pyruvate node in an anaerobic Escherichia coli strain with an active pyruvate dehydrogenase. Appl. Environ. Microbiol 76, 2107–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner A, Weindl D, Jäger C, Sapcariu SC, Dong X, Stephanopoulos G, Hiller K, 2014. Fragment formula calculator (FFC): determination of chemical formulas for fragment ions in mass spectrometric data. Anal. Chem 86, 2221–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Xiong W, Dai JB, Wu QY, 2015. Genome-based metabolic mapping and 13C flux analysis reveal systematic properties of an oleaginous microalga Chlorella protothecoides. Plant Physiol 167, 586–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ma P, Shah P, Rokas A, Liu Y, Johnson CH, 2013. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 495, 116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Johnson CH, 2000. Circadian clock-protein expression in cyanobacteria: rhythms and phase setting. EMBO J 19, 3349–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, 2014. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30, 1333–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Allen DK, Morgan JA, 2014. Isotopomer measurement techniques in metabolic flux analysis II: mass spectrometry In: Sriram G (Ed.), Plant Metabolism: Methods and Protocols Humana Press, Totowa, NJ, pp. 85–108. [DOI] [PubMed] [Google Scholar]
- Young JD, Shastri AA, Stephanopoulos G, Morgan JA, 2011. Mapping photoautotrophic metabolism with isotopically nonstationary 13C flux analysis. Metab. Eng 13, 656–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD, Walther JL, Antoniewicz MR, Yon H, Stephanopoulos G, 2008. An Elementary Metabolite Unit (EMU) based method of isotopically nonstationary flux analysis. Biotechnol. Bioeng 99, 686–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


