Abstract
Small RNAs (sRNAs) that act by base-pairing have been shown to play important roles in fine-tuning the levels and translation of their target transcripts across a variety of model and pathogenic organisms. Work from many different groups in a wide range of bacterial species has provided evidence for the importance and complexity of sRNA regulatory networks, which allow bacteria to quickly respond to changes in their environment. However, despite the expansive literature, much remains to be learned about all aspects of sRNA-mediated regulation, particularly in bacteria beyond the well-characterized Escherichia coli and Salmonella enterica species. Here we discuss what is known, and what remains to be learned, about the identification of regulatory base-pairing RNAs produced from diverse genomic loci including how their expression is regulated.
Keywords: small RNAs, sRNA biogenesis, UTR, RNA-seq
1. Introduction
Regulatory small RNAs (sRNAs) are typically transcripts of less than 300 nt that are diverse and complex in their means of action, which involve interactions with target mRNAs, other sRNAs, RNases, and RNA chaperones. While the best characterized organisms remain Escherichia coli and Salmonella enterica, there is evidence that regulatory RNAs play important roles in modulating cellular responses to environmental stress in nearly all bacteria taxa. This review provides a general overview of their mechanisms of action before a detailed discussion of sRNA prevalence, biogenesis, and regulation.
1.1. Types of sRNA regulators
In general, there are regulatory sRNAs that sequester proteins and those that act by base-pairing with other RNAs. Only a few sRNAs that function by protein sequestration have been described mechanistically. The 6S RNA binds directly to σ70-RNA polymerase, which inhibits its association with promoters [1], thus preventing their transcription. The regulatory sRNAs CsrB and CsrC function as molecular sinks to sequester the global regulators CsrA/RsmA away from target mRNAs (reviewed in [2]). As another example, the GlmY sRNA impacts the stability, and consequently the regulatory effect, of the GlmZ base-pairing sRNA. GlmY sequesters the RNA binding protein, RapZ, that otherwise would stimulate GlmZ turnover by recruiting the major endoribonuclease RNase E [3]. Additionally, some sRNAs that bind the RNA chaperone Hfq, which typically facilitates sRNA-target regulation (discussed in depth below), do not themselves base pair with other RNAs but instead sequester Hfq from sRNA base-pairing regulators (reviewed in [4]).
The most well-characterized sRNAs act by base-pairing. Some sRNAs are encoded antisense to their target RNA, and thus have perfect complementarity with this RNA. However, most sRNAs are transcribed at other locations on the genome (trans-encoded) and act by base-pairing with RNA targets with only limited complementarity. For both types of base-pairing sRNAs, the initial region of base-pairing on the sRNA is generally denoted as the “seed sequence.” The outcome of this base-pairing can vary, but ultimately leads to some regulatory effect on the target RNA.
This review is focused on base-pairing sRNAs, in particular on how they have been discovered, their various origins, and how they are regulated, but will first provide an overview of their mechanisms of action.
1.2. Regulation by antisense RNAs
Antisense RNAs (asRNAs) (reviewed in [5, 6]) were first discovered as regulators of E. coli ColE1 replication [7]. Subsequently, asRNAs were shown to control transposase expression. The first example was RNA-OUT of the E. coli Tn10/IS10 transposase system, which is transcribed antisense of the transposase mRNA to inhibit translation and thus downregulate transposition [8]. Other examples of asRNAs transcribed opposite transposase genes include two in S. enterica [9, 10], two in Caulobacter crescentus [11], and three in Listeria monocytogenes [12]. asRNAs also serve as the antitoxin of type I toxin-antitoxin (TA) systems (reviewed in [13]). In these systems, asRNAs base-pair with and prevent expression from the mRNA encoding the toxin by competing with ribosome binding [14], blocking translation of leader peptides [15], and/or providing RNA-double stranded sequences which can be degraded by RNase III [16].
Given that the asRNA and its corresponding target RNA are linked on the genome, transcription of the asRNA itself could serve as a regulatory mechanism. Possible transcriptional interference mechanisms include the simultaneous transcription and collision of RNA polymerases from two convergently elongating complexes, occlusion of one promoter due to elongating transcription from the other, and interference where an elongating polymerase collides with and displaces a “sitting duck” preinitiation polymerase (reviewed in [17, 18]). Some tests of these models have been carried out with asRNAs of coliphage 186 [19] and L. monocytogenes [12], and artificially tested in E. coli [20]. Other times both transcriptional interference and base-pairing could contribute to asRNA regulation, as has been suggested for the AmgR asRNA in S. enterica [21]. While deep sequencing indicates that there is an abundance of antisense RNAs, very few have been shown to have a physiological function. Repressing the synthesis of a potentially toxic protein is the overwhelming role in the limited documented examples of antisense sRNA function.
1.3. Regulation by trans-encoded base-pairing RNAs
trans-encoded base-pairing sRNAs can regulate mRNA targets at multiple levels. There are a few examples of sRNAs that affect transcription by base-pairing with the 5′ untranslated region (UTR) of an mRNA and modulating Rho-mediated premature termination (reviewed in [22]). However, the most prevalent mechanisms of sRNA-mediated regulation impact the stability and/or translation of an mRNA. This was the original mechanism described for the first characterized trans-encoded sRNAs like E. coli MicF [23] and Staphylococcus aureus RNAIII [24]. The base-pairing can block ribosomes from accessing the ribosome binding site (RBS), impede translation, or recruit RNases to process mRNAs or degrade them. In a more limited number of cases, base-pairing alters mRNA secondary structure and exposes a previously concealed RBS leading to increased translation as is the case for E. coli DsrA and RprA activation of rpoS translation [25, 26].
There are also more complex regulatory mechanisms. RNA-RNA interactions have now been observed to occur not only at 5′ UTR regions but also within coding sequences [27]. In some cases, base pairing leads to mRNA structural changes that conceal the upstream RBS and inhibit translation initiation as for Bacillus subtilis SR1 sRNA which binds 80 nt into the coding sequence of the ahrC mRNA [28]. It is also possible that coding sequence interactions could slow or inhibit elongating ribosomes. The internal base pairing can result in changes in the turnover of mRNAs. As an example, in S. enterica, the MicC sRNA binds 67 nt into the coding sequence of the ompD mRNA leading to RNase E-dependent ompD decay [29]. sRNAs additionally can interact with other sRNAs, serving as sponges to prevent the affected sRNAs from finding their mRNA targets, often by promoting their degradation [30]. While usually assumed to be non-coding, more and more examples of “dual-function” RNAs, encoding both a regulatory base-pairing region and a small regulatory protein are emerging (reviewed in [31]).
The region of base-pairing between the sRNA and the target is generally limited. Thus, in many bacteria, a protein chaperone has been found to mediate these RNA-RNA interactions. The best characterized bacterial RNA binding protein, is the homohexamer RNA matchmaker Hfq, which uses distinct surfaces to bind specific sequence motifs in sRNAs and their cognate targets (reviewed in [32]). In E. coli and S. enterica, deletion of Hfq results in the destabilization of sRNAs and decreased regulatory effects (reviewed in [33]). A relatively understudied RNA chaperone, ProQ, also has been shown to have RNA-RNA matchmaking capabilities in E. coli [30], S. enterica [34, 35], and Legionella pneumophila [36] (reviewed in [37]). However, ProQ also appears to have additional roles in RNA regulation. In both E. coli and S. enterica, ProQ was shown to block RNase-mediated degradation of both mRNAs and sRNAs [30, 38].
While much work has focused on Hfq and ProQ, which are found in a range of bacterial species (reviewed in [37, 39]), in some bacteria, such as many Gram-positive organisms, Hfq can bind sRNAs but is not required for sRNA stability or regulatory function. As an example, SR1 sRNA regulates ahrC mRNA independent of Hfq in B. subtilis [40]. sRNAs also have been detected in organisms such as Streptomyces [41], Chlamydia [42], Helicobacter [43], and Campylobacter [44], which do not have known Hfq or ProQ homologs. Either other RNA binding proteins serve RNA matchmaking roles or base-pairing RNA-mediated regulation occurs independently of a protein chaperone.
The identification of trans-encoded base-pairing sRNA targets has exploded in recent years with the development of several techniques to detect the targets on a genome-wide level (reviewed in [45]). Some of these methodologies, like RIL-seq (RNA Interaction by Ligation and Sequencing) [46] and CLASH (UV Cross-linking, Ligation, and Sequencing of Hybrids) [47] rely on the crosslinking of RNAs to proteins followed by protein immunoprecipitation and the ligation of RNA ends. The sequencing of these products reveals RNAs that are in proximity on the immunoprecipitated protein. Other approaches, such as GRIL-seq (Global sRNA Target Identification by Ligation and Sequencing) which relies on in vivo expression of an RNA ligase coupled with RNA-seq [48, 49], are independent of RNA binding proteins.
2. Methods to identify sRNAs
The annotation of regulatory RNAs in bacterial transcriptomes followed significantly after the annotation of mRNAs and housekeeping RNAs. This can, in part, be attributed to the lack of awareness about potential regulatory RNA loci in genetic screens and because the consequences of defects in sRNA-mediated regulation generally are more subtle than phenotypes associated with defects in protein factors. However, the methods to detect sRNAs have greatly expanded in recent years.
2.1. Direct detection
Initially, small transcripts in E. coli were detected by two-dimensional polyacrylamide gel electrophoresis and RNase T1 fingerprinting [50, 51]. Later, high-density microarrays revealed signals for intergenic sRNA candidates in E. coli [52] and other organisms such as Streptococcus pyogenes [53], Neisseria gonorrhoeae [54], Burkholderia thailandensis [55], to only name a few. The development of RNA-seq brought higher resolution for sRNA identification. However, the compactness of bacterial genomes with overlapping transcripts creates challenges in mapping sRNAs using standard RNA-seq methods.
Improvements to transcriptome annotation have come with strand-specific RNA-seq and the development of methodologies to define RNA boundaries. 5′ RNA-seq approaches distinguish transcriptional start sites (TSSs) from processed 5′ ends. For instance, differential RNA-seq (dRNA-seq) [56] utilizes a terminator 5′-phosphate-dependent exonuclease (TEX) to degrade RNA carrying a 5′ monophosphate (as a result of processing), leaving 5′ triphosphate-containing RNAs (indicative of transcripts with a native TSS) to be sequenced (reviewed in [57]). Similarly, treatment of RNA with tobacco acid pyrophosphatase (TAP) or RNA 5′ pyrophosphohydrolase (RppH), which both remove the pyrophosphate from the 5′ end of triphosphorylated RNA, can be used in RNA-seq approaches to distinguish TSSs from processing sites. In the Cappable-seq method, TSSs are labeled with a desthiobiotin cap for reversible binding to streptavidin and subsequent 5′ RNA-seq [58]. On the other end of the transcript, 3′ ends of RNAs can be identified by Term-seq through the ligation of 3′ sequencing adapters in the first step of RNA-seq library preparations [59]. This can be combined with bicyclomycin (BCM), an inhibitor of the termination factor Rho, to subclassify 3′ ends as Rho-termination-dependent [60]. SMRT (Single Molecule, Real-Time)-Cappable-seq utilizes single-molecule long read sequencing to map bacterial operons, concurrently identifying 5′ and 3′ ends [61]. Additionally, SEnd-seq (Simultaneous 5′ and 3′ End Sequencing) circularizes cDNA for simultaneous 5′ and 3′ end identification [62].
The identification of sRNAs in RNA-seq data sets is improved by approaches that enrich for these molecules. This can be via size selection for small transcripts [63, 64] or association with RNA binding proteins. A number of different methods have been developed for the sequencing of transcripts that bind proteins like Hfq and ribonucleases. Many of these such as CLIP-seq (Crosslinking Immunoprecipitation and Sequencing) [65], RIL-seq [27, 30] and CLASH [47, 66] rely on the crosslinking of RNAs to the proteins of interest followed by immunoprecipitation of either the native protein or tagged derivatives. In another approach termed Grad-seq (Gradient profiling by Sequencing), RNA binding proteins and their RNAs are fractionated on glycerol gradients and co-association is determined by sequencing all of the RNAs and deep mass-spectrometric analysis of the proteins in each fraction [34].
Collectively, these transcriptome datasets have revolutionized bacterial genetics in a variety of model and pathogenic bacteria. The datasets have challenged dogma by revealing an abundance of internal, antisense, and intergenic transcription comprising much more complex bacterial transcriptomes than first anticipated.
2.2. Computational detection
Initially there were certain criteria by which sRNAs were defined and computational predictions were developed to scan bacterial genomes to identify the best candidates (reviewed in [67, 68]). Early search programs focused on intergenic regions where sRNAs were first discovered. Many algorithms are based on comparative genomics, examining closely-related bacteria. In general, these programs search intergenic regions for sequence conservation, GC content, and other features such as promoter motifs, transcription factor binding sites, and intrinsic terminators. sRNA secondary structures should be of a thermodynamically favorable minimum free energy, with lower free energy than a random sequence. Therefore, RNA folding can also be helpful in the computational prediction of sRNAs. Programs that use such parameters include QRNA [69], RNAz [70], sRNAPredict [71], sRNAscanner [72], and RNASurface [73]. Machine learning based software also has been developed [74, 75]. The downside of these approaches is that they sometimes rely on conservation, though not all sRNAs are broadly conserved. Additionally, intergenic regions also harbor mRNA transcription elements such as promoters and transcription factor binding sites that can confound the data. Limiting search parameters to intergenic regions also misses sRNAs encoded elsewhere in the genome.
One way to avoid some of the issues mentioned above is to combine direct detection and computational predictions, which is the premise of programs such as ANNOgesic [76] and APERO [77]. These approaches integrate data from RNA-seq experiments to detect small transcript boundaries and subsequently predict sRNA features from these datasets. The identification of sRNAs by multiple approaches has led to overwhelming datasets, even for just one bacterial species. For example, in S. aureus, sRNAs have been detected by computational predictions [78], microarrays [79], and RNA-seq methods [80–83] to generate a list of over 500 candidate sRNAs. However, combining RNA-seq methods with the sRNA computational software DETR′PROK narrowed this number to 50 putative sRNAs [84]. While some sRNAs could be missed, this type of analysis could reveal promising sRNAs for further studies, especially for organisms where few sRNAs have functionally been characterized.
3. Prevalence of sRNAs across bacteria species
While the true number of sRNAs is not known for any organism, it is useful to consider the numbers of sRNAs reported in different bacterial species. With the surge in research on bacterial regulatory RNAs and the availability of RNA-seq technologies, there seems to be an abundance of small transcripts. However, careful mechanistic studies which document base-pairing and regulatory actions of these RNAs is limited for most bacteria.
We have assembled a table presenting the estimated numbers of detected sRNAs across species excluding asRNAs, excepting those asRNAs that have been functionally characterized. We also note key methods in identifying sRNAs, the approximate number of characterized sRNAs, and a representative base-pairing sRNA (Table 1). The number of reported sRNAs varies between species. For instance, ~20 have been reported in Chlamydia trachomatis, compared to ~100 in E. coli, and ~600 in Borrelia burgdorferi. Follow-up experiments which confirm the expression of these sRNAs are important before making sweeping conclusions about the existence of these regulators. Northern blot analysis is the gold standard for validating the presence of sRNAs. This approach provides information about both the abundance and size of a transcript, which cannot be accomplished with other methods, like Reverse-Transcriptase PCR, especially for sRNAs that overlap other transcripts. Of the ~500 putative sRNAs in Synechocystis sp., only 14 have been tested by northern blot analysis to document their presence and discrete sizes [85]. In comparison, nearly all ~100 E. coli sRNAs reported by various global approaches, including size-selection RNA-seq [86], have been confirmed by northern blot analysis and many have been shown to be functional. Most bacteria in Table 1 have hundreds of sRNAs detected but fewer than 5 examples of sRNAs documented to act by base-pairing ideally by mutating the sRNA seed region to disrupt regulation and then restoring the regulation with a compensatory mutation in the target RNA. Most of the representative sRNAs listed in Table 1 have been tested for direct base-pairing. In some phyla (Acidobacteria, Fusobacteria, Planctomycetes), we did not find any studies which specifically detected sRNAs, providing opportunities for future discoveries.
Table 1.
sRNAs detected across bacteria species.
| Phylum | Example Species | Estimated sRNAs1 | Example Methods2 | Example sRNA3 | Functionally Characterized4 |
|---|---|---|---|---|---|
| Acidobacteria | none | ||||
| Actinobacteria | Streptomyces coelicolor | ~100 | size selection RNA-seq [205] | scr5239 [206] | <5 |
| Mycobacterium tuberculosis | ~200 | size selection RNA-seq [207] | MrsI [207] | <5 | |
| Bacteroidetes | Porphyromonas gingivalis | ~30 | size selection microarray and RNA-seq [208] | none | <5 |
| Chlamydiae | Chlamydia trachomatis | ~20 | 5’ RNA-seq [42] | IhtA [209] | <5 |
| Cyanobacteria | Synechocystis sp. | ~500 | 5’ RNA-seq [210], size selection RNA-seq [211] | RblR [85] | <5 |
| Firmicutes | Bacillus subtilis | ~150 | 5’ RNA-seq [212] | FsrA [100] | <10 |
| Staphylococcus aureus | ~50 | total RNA-seq and computational pipeline [84] | RsaD [213] | <15 | |
| Listeria monocytogenes | ~150 | size selection RNA-seq [214] | LhrA [215] | <10 | |
| Alpha-proteobacteria | Caulobacter crescentus | ~150 | total RNA-seq [216] | CrfA [217] | <5 |
| Rickettsia prowazekii | ~40 | Total RNA-seq [218] | none | <5 | |
| Beta-proteobacteria | Neisseria meningitidis | ~70 | 5’ RNA-seq [130] | AniS [219] | <10 |
| Gamma-proteobacteria | Escherichia coli | ~100 | cDNA library [112], size selection RNA-seq [86], Hfq and ProQ RIL-seq [30] | OxyS [97] | <70 |
| Legionella pneumophila | ~90 | 5’ RNA-seq [220] | RocR [36] | <5 | |
| Pseudomonas aeruginosa | ~170 | 5’ RNA-seq [221] | RhlS [122] | <20 | |
| Vibrio cholerae | ~100 | 5’ RNA-seq [102] | Qrr1–4 [222] | <20 | |
| Yersinia pestis | ~220 | size selection RNA-seq [223] | Ysr141 [223] | <5 | |
| Delta-proteobacteria | Myxococcus xanthus | unknown | n/a5 | Pxr [193] | <5 |
| Epsilon-proteobacteria | Helicobacter pylori | ~60 | 5’ RNA-seq [56] | RepG [224] | <5 |
| Spirochaetes | Borrelia burgdorferi | ~600 | size selection RNA-seq [225] | DsrA [226] | <5 |
| Fusobacteria | none | ||||
| Planctomycetes | none |
Numbers of sRNAs (excluding uncharacterized asRNAs) for each example species were estimated using global approaches which directly reported sRNAs.
Example methods used for sRNA estimations.
One example of a characterized base-pairing sRNA.
Estimation of functional sRNAs where base-pairing has been experimentally documented.
Not applicable.
4. Sources of sRNAs
Abundant numbers of sRNAs have been reported across bacterial species for many years. However, the fact that the sRNAs themselves are encoded by nearly all parts of the bacterial genome (Figure 1) has only been appreciated more recently. The following sections will describe the unique genomic loci (independently transcribed, excised from mRNA 5′ and 3′ ends, processed from operons, and components of tRNAs) that produce small base-pairing RNA regulators.
Figure 1.
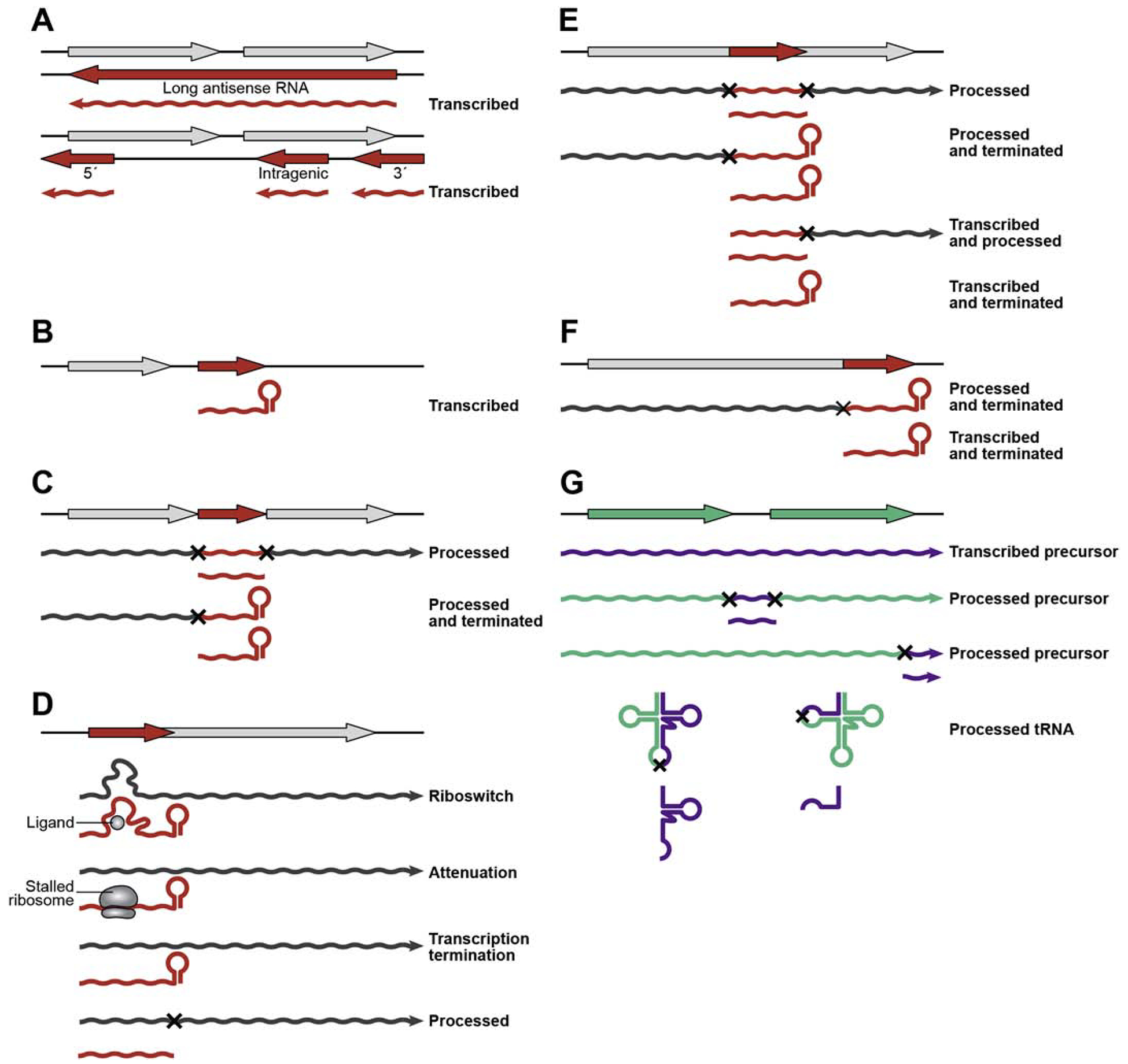
Origin of sRNAs from diverse genomic loci. (A) antisense (B) intergenic (C) operon- derived (D) 5′ derived (E) intragenic (F) 3′ derived (G) tRNA-derived. Coding sequences are denoted in gray, sRNA sequences in red, tRNA sequences in green, and regulatory tRNA sequences in purple. Processing cleavage sites are indicated with an “x”. Instances of known transcription termination are indicated by a hairpin. 3′ ends corresponding to known processed RNAs are blunt. An arrow is used if the mechanism of 3′ end generation is unknown.
4.1. Antisense sRNAs
Antisense sRNAs are encoded on the DNA strand opposite another gene, though base-pairing occurs in trans after both RNAs are transcribed (Figure 1A). Usually the target is the mRNA encoded on the opposite strand, though some asRNAs also have targets encoded elsewhere on the genome. RNA-seq data indicate extensive antisense transcription, with over 5,000 asRNAs detected in E. coli by RNA-seq [87], and up to 75% of all genes having antisense transcripts in Prochlorococcus sp. and S. aureus [88, 89]. asRNAs range in size from less than 100 nt to several thousand nt, even spanning several genes. One asRNA of ~7,000 nt spans 14 genes, comprising two adjacent operons in Prochlorococcus sp. [90], and a ~1,000 nt long asRNA is transcribed in opposition to an S-box riboswitch and the mccA gene in Clostridium acetobutylicum [91]. Others transcribed just opposite to the 5′ or 3′ ends of mRNAs such as SymR in E. coli [92] and SprA1 in S. aureus [93] are much smaller, just 77 nt and 60 nt in length, respectively. In Synechocystis sp., IsrR, a 177 nt asRNA completely internal to the gene encoded opposite, has also been shown to have a regulatory effect [94]. One would assume asRNAs can easily base-pair with the mRNA encoded on the opposite strand because of the perfect complementarity. However, as already mentioned, despite their abundance, few instances of bona fide regulatory roles have been documented.
4.2. Intergenic sRNAs
The largest number of functionally characterized base-pairing sRNAs are encoded as distinct transcripts in intergenic regions with their own promoters (Figure 1B). Most of these sRNAs contain an intrinsic terminator with a stable stem-loop and stretch of U residues. In E. coli ~50 intergenic sRNAs have been documented, ranging in size from ~50 to ~300 nt. A plethora of intergenic sRNAs also have been described in other bacteria. One study compared more than 23,000 intergenic regions and found ~900 intergenic sRNAs across 13 different bacterial species [95]. The authors also concluded that sRNA genes were significantly enriched in longer intergenic regions, which further implies that bacteria efficiently use genomic space.
One challenge is documenting the function for the numerous intergenic sRNAs in bacterial genomes. The genomic context and conservation of adjacent neighboring genes can be helpful in identifying gene functions, as these can be related. For instance, there are a number of examples, such as OxyR-OxyS and GcvA-GcvB in E. coli [96, 97], where the transcription factor that controls the expression of an sRNA is encoded adjacent to the sRNA gene.
Global evolutionary analysis of E. coli has indicated intergenic sRNAs in this organism are mostly conserved within the Enterobacteriales order [98]. However, gene rearrangements can result in the gain or loss of an intergenic sRNA, even among closely related species. For instance, the intergenic region of E. coli uspF and ompN contains the biofilm-inhibitor sRNA, EcsR1, which in S. enterica was fragmented due to a translocation event leading to the loss of the sRNA gene [99]. In general, the evolution of sRNAs continues to be poorly understood because sRNAs with the same function and targets have been described even when there is little sequence similarity between the two sRNAs. FsrA, one of the first intergenic sRNA characterized in B. subtilis, regulates mRNA targets involved in iron metabolism and storage [100], similar to Enterobacteriaceae RyhB [101] but has a completely different sequence and structure.
Since the characterization of the first E. coli sRNA [23], 30 years of research has documented regulatory mechanisms for ~30 E. coli intergenic sRNAs. As regulatory RNA- networks are being delineated in a range of bacteria, important physiological functions for intergenic sRNAs continue to be discovered. For example, VqmR in Vibrio cholerae regulates biofilm formation [102], B. subtilis RoxS regulates the NAD+/NADH equilibrium [103], RmaA regulates flagellar motility in Erwinia amylovora [104], S. aureus SprC regulates virulence and contributes to phagocytosis and murine infectivity [105]. While most functional characterization has been focused on intergenic-derived sRNAs, there has been increased discovery of base-pairing sRNAs in other genomic regions, as will be discussed in the following sections.
4.3. sRNAs processed from within operons
Some intergenic sRNAs have been found to be co-transcribed with other genes in operons and subsequently processed into functional small transcripts (Figure 1C). Generation of these regulators relies on transcription from the upstream genes. The 5′ end of the sRNA is always generated by cleavage events, but the 3′ end can result from transcription termination or cleavage. For instance, the sRNA sponge SroC, characterized in S. enterica, is derived from the gltIJKL mRNA encoding the glutamate/asparate ABC transporter. In this case, the 5′ end is generated by RNase E cleavage and the 3′ end is generated by transcription termination at an internal intrinsic terminator directly proceeding gltJ, which must allow some read-through for the polycistronic mRNA to be transcribed [96]. The mature SroC sRNA binds another sRNA, GcvB, as a sponge to prevent GcvB-mediated regulation. The uncharacterized sRNAs 3′ to fimA and glnA in E. coli [106] are also encoded within operons but less is known about their generation and activities.
4.4. 5′ derived sRNAs
The synthesis of sRNAs from 5′ mRNA regions is understudied, but these regions also have the potential for encoding trans-acting RNA regulators (Figure 1D). Classically, 5′ UTRs are hotspots for cis-acting RNA-regulatory elements, particularly riboswitches, RNA thermometers, and attenuators (reviewed in [107–109]). Riboswitches alter expression of their downstream genes through the binding of specific ligands, often metabolic byproducts from the gene’s enzymatic or signaling pathway. The secondary structures of RNA thermometers are changed with temperature to hide or expose ribosome binding sites. Finally, attenuators are structured leader sequences that encode a small upstream open reading frame (uORF) preceding the coding region of the mRNA. Ribosomes either translate or stall on the uORF, affecting the formation of a downstream terminator hairpin or Rho binding, which results in transcription termination and the release of an RNA corresponding to the attenuator.
In many of these regulatory scenarios, small, stable transcripts are generated. Indeed, small 5′ transcripts for the transcriptional riboswitches preceding mgtA [110] and lysC [111] were detected in an E. coli cloning-based screen, which captured RNAs of 30–65 nt [106]. Northern blot analysis verified that distinct fragments are detected at reasonable levels. Similarly, two uncharacterized E. coli sRNAs, named SroA and SroG [112], are produced from the thiamine riboswitch upstream of thiCOGE [113] and riboflavin riboswitch upstream of ribB [114], respectively.
While the functional properties of most 5′ UTR fragments have not been explored, there are some suggestions that 5′ sRNAs can have independent functions. For instance, two S- adenosylmethionine (SAM) riboswitches in L. monocytogenes leave behind stable, small 5′ RNAs denoted SreA and SreB, which have been shown to base-pair with and downregulate in trans the mRNA encoding the virulence regulator PrfA [115]. As another example, adenosyl cobalamine (B12) responsive riboswitches in Enterococcus faecalis and L. monocytogenes produce trans-acting sRNAs, EutX and Rli55 respectively [116, 117]. Both of these sRNAs bind the two-component response regulator EutV and thus prevent the protein from acting on its downstream eut target mRNAs. In Sinorhizobium meliloti, there are three tryptophan biosynthesis operons, but only one, trpE(G), encodes a known uORF. Interestingly, the attenuator RNA byproduct from this operon has been reported to function as a base-pairing sRNA. This sRNA, named rnTrpL, appears to base-pair with the trpDC mRNA from another tryptophan operon leading to its decay when tryptophan is abundant [118].
Outside of known cis-regulatory elements, other examples of small transcripts derived from 5′ UTR regions have been noted. In the same E. coli cloning based screen mentioned above, 15 others were detected [106]. Another study using RNA decay coupled with RNA-seq identified 12 fragments in 5′ UTRs [119]. One study examining the global consequences of deleting the 3′-to-5′ phosphorolytic exoribonuclease PNPase (pnp) revealed that several 5′ UTR fragments increased in a Δpnp strain, suggesting a subset of these transcripts could be regulated via this RNase [120]. Indeed stable 5′ derived small transcripts can arise both from cis-regulatory events and perhaps early termination or cleavage events at 5′ UTRs.
There are two characterized 5′ derived sRNAs for which there is no known cis-regulatory element. The E. coli SgrS RNA corresponds to the 5′ end of the setA mRNA and is generated by termination upstream of setA [121]. RhlS described in Pseudomonas aeruginosa provides another example [122]. This sRNA was discovered by mapping premature termination using Term-seq. A 3′ end was noted 34 nt upstream the start codon of rhlI, which encodes a protein involved in regulating N-acylhomoserine lactone quorum sensing. This region did not appear to encode a riboswitch when tested with N-butanoyl-homoserine lactone, and no other regulatory elements were predicted. RhlS contains a stem-loop followed by a polyuridine sequence, a hallmark of sRNAs. For both SgrS and RhlS, considering that they are part of the setA and rhlI 5′ UTR, respectively, there must be at least occasional transcriptional readthrough to fully transcribe the full-length mRNAs. As more studies demonstrate the abundance of 5′ mRNA fragments, the importance and biological relevance of these transcripts should be explored.
4.5. Intragenic sRNAs
Various RNA-seq studies have documented RNA regions within coding sequences that are more abundant than the rest of the mRNA (Figure 1E). Some bacteria seem to contain more intragenic transcription than others, although a subset could be due to misannotation. Nevertheless, 5′ RNA-seq mapped 63% of TSSs to internal coding regions in B. burgdorferi [123], 62% in Leptospira interrogans [124] and 61% in the haloarchaeon Haloferax volcanii [125]. Within E. coli, 37% of sequenced TSSs mapped to intragenic coding regions [87]. It is possible some of the transcripts initiating from internal promoters encode small proteins in the same or different frame as annotated gene [126]. However, an RNA decay experiment in E. coli using RNA-seq revealed 21 short, stabilized intragenic RNAs that the authors postulated could be base-pairing regulatory RNAs [119]. Furthermore, RIL-seq analysis indicated several are bound by Hfq and provided evidence that putative intragenic sRNAs base-pair with 5′ regions of mRNAs [27, 30]. Since no mechanistic studies to demonstrate base-pairing have been carried out, this class also deserves further study.
4.6. 3′ derived sRNAs
A quickly-expanding number of trans-acting sRNAs transcribed and/or processed from the 3′ end of mRNAs (Figure 1F) have been shown to have regulatory effects. Some 3′ UTR regions were first noticed as discrete small transcripts in early E. coli sRNA screens [106, 112]. However, their role as base-pairing sRNAs was first recognized in a S. enterica RNA-seq experiment [127] analyzing RNAs that co-immunoprecipitated with the RNA-RNA matchmaker, Hfq. Eight of these sRNAs were detected by northern blot analysis and one, DapZ, was shown to have its own promoter sequence internal to its overlapping mRNA, dapB. Other 3′ sRNAs can be processed from the larger mRNA, like SorX in Rhodobacter sphaeroides [128]. Even 3′ sRNAs transcribed as separate entities can be processed into a shorter form, like the 83 nt MicL-S derivative of the 300 nt E. coli MicL sRNA [129]. 3′ derived sRNAs can be identified by searches for 5′ ends in mRNA 3′ UTRs in RNA-seq data. For instance, 17 were identified and classified as transcribed or processed in Neisseria meningitidis using TEX treatment and dRNA-seq [130]. These sRNAs tend to be named based on their overlapping mRNA, keeping the first three letters of the mRNA and the fourth with a late letter of the alphabet, like CpxQ derived from the 3′ region of cpxP [131] or RbsZ which overlaps rbsB [30].
Given the known features of base-pairing sRNAs, 3′ UTR regions are ideal for evolution of these sRNAs, which require a stable stem-loop and stretch of poly-U’s to bind with Hfq. Thus, mRNA intrinsic terminators, which contain these features can be exploited. Co-evolution of 3′ sRNAs with their overlapping mRNAs eliminates evolving this structure de novo. In line with this suggestion, it has been noticed that mRNAs encoding 3′ derived sRNAs tend to have a longer stretch of poly-U’s than mRNAs without an sRNA [27]. Natural selection may favor rapid evolution in 3′ UTRs where mutations are less likely to be deleterious. The linked genetic location could imply similar physiological roles for the protein and sRNA encoded by the overlapping sequences. For example, the sRNA s-SodF is cleaved from the 3′ region of sodF (iron superoxide dismutase) and targets the nickel superoxide dismutase, sodN in Streptomyces coelicolor [132]. Therefore, understanding the genomic context of 3′ sRNAs may help to elucidate their functions.
Mechanistic studies have now documented that a number of 3′ derived sRNAs function as base-pairing regulators, and the association with Hfq strongly implies base-pairing potential for others. Consistent with this implication, ~300 E. coli RNA pairs were mapped to 3′ UTRs and other coding sequences in stationary phase using the RIL-seq approach, which detects Hfq-bound sRNAs and their target RNAs [27]. We expect more and more functional 3′ derived sRNAs to be discovered and characterized from numerous bacteria in the future.
4.7. tRNA precursors and fragments
tRNAs are either transcribed independently or in polycistronic tRNA transcripts. These tRNA precursors are then processed by a series of 5′ and 3′ end cleavages to complete their maturation (reviewed in [133]). Mature tRNAs also are subject to endonuclease cleavage. The byproducts of these cleavage products, tRNA-derived fragments (tRFs), range in size from 13 to 20 nt and have been identified in all domains of life (Figure 1G) (reviewed in [134]). tRFs are implicated in various biological functions in eukaryotes, but direct evidence for functional tRFs in bacteria is more limited. Here we present some indication that prokaryotic tRNA precursors and tRFs can function as base-pairing regulators.
Association of tRNA precursors with Hfq has suggested a regulatory function for these transcripts. Microarray analysis of Hfq co-immunoprecipitated RNA showed enrichment of the proM internal transcribed spacer (ITS) region from the argX-hisR-leuT-proM polycistronic tRNA precursor [135]. Subsequent northern blot analysis demonstrated proM tRNA precursors, but not mature tRNAPro, were enriched in Hfq-bound RNA samples. Similarly, Hfq co-immunoprecipitation coupled with RNA-seq and northern blot analysis showed the metZWV transcript, but not mature tRNAMet, binds Hfq [136]. Some mature tRNAs have been shown to bind Hfq in vitro [137]. Additionally, the sRNAs RybB and MicF were found to co-purify with the ITS regions on the metZWV tRNA precursor [138]. This interaction was only observed with full length metZWV and not the ITS fragments. However, the physiological relevance of these interactions is still unknown.
There is one example of a characterized tRNA precursor-derived fragment with a function. 3′ETSleuZ is processed from the 3′-external transcribed spacer (ETS) of the glyW-cysT- leuZ polycistronic tRNA precursor. 3′ETSleuZ functions as a base-pairing regulator by sponging and repressing the activity of the RyhB and RybB sRNAs [138]. Targets of these sRNAs include genes related to the TCA cycle and antibiotic sensitivity. Experiments assaying these regulons demonstrated 3′ETSleuZ sponges the sRNAs from their targets affecting cellular fitness. Given that sRNAs are powerful regulators, it may be beneficial to repress their activity via sponging from a constitutive tRNA transcript, such that specific induction of the sRNA must overcome the sponge to elicit effects. The RIL-seq approach found tRNA sequences in RNA duplexes with coding sequences, UTRs and sRNAs on the RNA chaperones Hfq and ProQ [27, 30], again implying base-pairing and perhaps new regulatory networks.
Some tRFs are generated by cleaving tRNAs into halves or quarters. In the archaeon H. volcanii, a stress-induced 26 nt tRF cleaved from tRNAVal reduces protein synthesis by directly binding to the small ribosomal subunit [139]. One study describes three tRFs cleaved from tRNAVal, tRNAGly, and tRNAGln in the Rhizobium Bradyrhizobium japonicum [140]. Intriguingly, the targets of these tRFs are not in the bacterium itself, but in its symbiotic partner the soybean, Glycine max. Overexpression and silencing of the Rhizobium tRFs affected root hair curling and nodule formation in the soybean via base-pairing with root hair and plant developmental gene targets. This emerging concept of RNA-mediated regulation across species and kingdoms is discussed in more detail below.
There are likely underexplored types of tRNA-linked regulatory elements. The cellular pool of tRNAs has been shown to be dynamic; for instance tRNAs are subject to degradation after amino acid starvation [141]. Conceivably, such byproducts could be recycled as regulators during such stress conditions. Other forms of tRNA precursors also could serve as regulators, like the tRNA-linked repeats, which are located immediately downstream tRNAs and share the last 18–19 nt of the 3′ end of a tRNA sequence [142]. Given the abundance of these transcripts, tRNA precursors and fragments could be a significant source of other regulatory RNAs.
5. Other potential sRNAs sources
There is now a greater appreciation of the diverse origins of base-pairing RNAs. Undoubtably, sRNA regulators will be found to be encoded by still other genomic locations, and we highlight a few conceivable sources below.
5.1. sRNAs from ribosomal RNAs
The three rRNAs (5S, 16S, and 23S) are co-transcribed in most bacteria and require processing into mature species through a series of cleavage and nucleotide/sugar modification events (reviewed in [143]). One can imagine that precursor or mature rRNA fragments (rRFs) could be functional, similar to tRFs. Fragments of mature E. coli rRNAs have been studied and shown to interact with translation factors [144], antibiotics [145, 146], mRNA/tRNA analogs [146], and to alter translation termination [147]. In eukaryotes small ribosome-derived RNAs have been functionally characterized (reviewed in [148]). For instance, the qiRNAs of the filamentous fungus Neurospora crassa are processed from 28S rRNA during DNA damage and mediate gene silencing in the DNA damage response pathway [149]. Such rRNA-derived regulators could also be active in bacteria.
5.2. sRNAs from other species
The concept of RNA-mediated crosstalk between bacteria, phage, and eukaryotic hosts is quite attractive. This is especially true for bacterial biofilms, microbiomes, bacteria-eukaryote symbionts, and host-pathogen interactions. Given that bacteria are excellent adaptors and manipulators of their environment, it is conceivable that molecules like regulatory RNAs participate in bacterial systems for hijacking host cells or for inter-organism communication.
Phage infections are likely instances of RNA-mediated crosstalk, in part because the virus can hijack the host genome for its replication. An obvious example of base-pairing crosstalk involving RNA is the CRISPR-Cas13 immunity system [150, 151]. Evidence is accumulating that phages also harbor other base-pairing sRNAs that modulate phage or host derived transcripts. Several putative phage sRNAs were detected during φR1–37 infection of Yersinia enterocolitica [152], and two sRNAs transcribed from the PAK_P3 phage during P. aeruginosa infection have been described [153]. Genomic regions containing bacterial prophage also have been shown to encode sRNAs, such as the AgvB sRNA found in pathogenic Enterohemorrhagic E. coli (EHEC) [154].
Bacterial-eukaryotic relationships are another likely environment for RNA-mediated crosstalk. The study described previously documenting regulatory RNAs from B. japonicum modulating its symbiotic host the soybean [140] provides one such example. L. monocytogenes, a Gram-positive intracellular bacterium, has been reported to secrete bacterial RNA/DNA which is detected by host cytosolic nucleic-acid sensing receptors [155]. It has also been reported that this bacterium secretes an RNA binding protein, Zea, which binds L. monocytogenes RNA extracellularly in the host cell cytoplasm and interacts with host proteins [156]. As detection methods improve to distinguish and categorize sRNAs from different species, we expect more examples of RNA-mediated crosstalk.
One question that arises is how sRNAs might be delivered from one species to another in their environment. The formation of extracellular vesicles to secrete molecules has been observed for both eukaryotes and prokaryotes. The most extensive characterization of bacterial vesicles is in Gram-negative bacteria, where they are termed outer membrane vesicles (OMVs), however, their biological significance is largely unknown. Several groups have shown OMVs play roles in pathogenesis and elicit host immune responses (reviewed in [157]). There has been much interest in the composition of OMVs, especially since it has been postulated that they harbor inter- and intra-kingdom “communication” molecules, like regulatory RNAs (reviewed in [158]). Some groups have sequenced extracellular RNA (exRNA) from OMVs and found them to be shorter than 250 nts and frequently derived from intergenic regions or fragments of tRNAs or rRNAs [159–161]. In one example, a tRNA-derived sRNA in P. aeruginosa has been reported to be transferred into human airway cells via OMVs to reduce the inflammatory response during infection [162]. The exRNA Rli32 of L. monocytogenes induces interferon-β production and promotes intracellular growth during macrophage infection [163]. Some caution is warranted given differences in vesicle isolation between groups and potential contamination of lysed cells or other secreted/extracellular factors. Given that the RNAs found in OMVs are so short, it is also conceivable that they just correspond to random, degraded RNA fragments. Nevertheless, there is considerable interest in the topic [164].
6. Regulation of sRNA synthesis
Like any other regulatory molecule, sRNAs themselves need to be expressed in a controlled manner to elicit targeted effects. Several mechanisms control sRNAs at the levels of transcription as well as processing/degradation.
6.1. Transcription initiation
The levels for sRNAs can be regulated by transcription from specific promoter sequences by an interplay of transcription activators and repressors. This is especially true for antisense and intergenic sRNAs.
Sigma factors (σ) compete with one another for binding to the RNA polymerase core subunits and direct transcription by interacting with promoter sequences. There are seven known sigma factors in E. coli, but the number in other bacteria varies from three in B. burgdorferi to 19 characterized/predicted in B. subtilis and more than 100 in the soil dwelling myxobacterium Sorangium cellulosum (reviewed in [165]). Some sigma factors like the stress response factor RpoS (σS) and envelope stress factor RpoE (σE), are relatively broadly conserved and affect sRNA expression in many species. Under membrane stress for instance, σE upregulates periplasmic proteases, protein chaperones, and specific sRNAs to counteract the misfolding of outer membrane proteins. In E. coli, the σE-dependent sRNAs MicA and RybB base-pair with and negatively regulate the mRNAs encoding membrane proteins, including ompA [166–169]. This is similar to V. cholerae, where σE directs increased transcription of the MicV and VrrA sRNAs, which share conservation with the E. coli RybB seed sequence and function to downregulate ompA and other outer membrane protein transcripts [170]. Sigma factors also can be species specific. In B. subtilis σK and σG direct transcription during late stages of sporulation and induce several sRNAs that have been postulated to regulate genes during sporulation [171]. Similarly, the alternative sigma factor SigB, regulates a subset of intergenic sRNAs in L. monocytogenes and S. aureus, such as Rli47 [172] and RsaA [78], respectively, in addition to some uncharacterized asRNAs [88, 173]. Feedback loops also exist, where an sRNA negatively regulates the synthesis of its own sigma factor, as for E. coli RybB and σE [168]. Through sigma factors, sRNAs can be targeted for expression under specific conditions where they are most needed.
Transcription factors can be used in conjunction with sigma factors to regulate the expression of sRNAs, either positively or negatively. They too can be broadly conserved, with the same transcription factor controlling sRNA transcription in many organisms. A classic example is the ferric uptake regulator (Fur). Under conditions of high iron, E. coli Fur represses synthesis of the sRNA RyhB, which targets mRNAs encoding iron-containing proteins during iron-limiting growth [174, 175]. Functional analogs of RyhB, such as PrrF in P. aeruginosa [176], NrrF in N. meningitidis [177], HrrF in Haemophilus influenzae [178] and FsrA in B. subtilis [100], are all transcriptionally regulated by Fur in these species. There are also species-specific transcription factors that regulate sRNA genes like ToxT, which activates expression of the TarA sRNA in V. cholerae [179]. As described for sigma factors, there are feedback loops involving transcription factors and sRNAs. For example, there are a number of two-component systems, where a membrane sensor kinase phosphorylates a transcription factor response regulator in response to environmental cues (reviewed in [180]) and thereby activates sRNA transcription. In a few examples, such as for the OmrA/B sRNAs and OmpR-EnvZ sensor kinase-response regulator in E. coli [181], the induced sRNAs base pair with and repress the mRNA encoding the two-component system, forming a negative feedback loop.
Often, sRNAs are the most strongly induced transcript in a specific stress response. For instance, the sRNAs OxyS and MicA are among the most highly-induced RNAs upon H2O2 and cell envelope stress, respectively [129, 182, 183]. Thus, sRNAs can serve as reporters of a particular cellular stress, and further understanding of the transcriptional regulation of sRNA genes can give insights into the response.
6.2. Transcription termination
Termination is a regulated process that can either result from formation of a stem-loop (hairpin) structure causing RNA polymerase pausing, and subsequent dissociation, or the binding of the termination factor Rho which recognizes specific sequences and dissociates the RNA polymerase elongation complex (reviewed in [184]). Given that sRNAs can be derived from 5′ UTRs, operons, and perhaps intragenic regions of mRNAs, there likely are mechanisms to favor production of the functional sRNA under some conditions and the functional mRNA under other conditions. Some evidence suggests that efficiency of sRNA termination is enhanced with the same stress conditions that induce transcription initiation of the sRNA [185]. It is also possible that the level of termination is constant in some cases with the basal level of termination sufficient to still elicit sRNA regulatory effects. The mechanisms by which transcription termination of sRNAs is regulated and also the role of the Rho protein are aspects that require further exploration.
6.3. Modification
One possible mechanism for controlling sRNAs could be RNA modifications. Post- transcriptional base modifications are widespread across the kingdoms of life and also have been detected for bacterial sRNAs (reviewed in [186–188]). Several sequencing studies have identified nicotinamide adenine dinucleotide (NAD) capped transcripts [189] and internal N6-methyladenosine (m6A) modifications in bacterial RNA [190]. Such modifications have been found in higher frequency for sRNAs than mRNAs, though only low percentages of the sRNAs are modified. This raises questions about the physiological roles of the modifications. It is possible that modifications contribute to processing, turnover and/or regulatory effects, but the biological consequences need to be further investigated.
6.4. Processing
Northern blot analysis of sRNAs frequently show multiple bands, indicative of precursors and/or processed forms of the sRNAs. Some sRNAs are cleaved from operons, 3′ UTRs, or tRNAs to produce functional regulators. However, even sRNAs that are transcribed as separate entities may be cleaved. 3′ UTR-derived sRNAs can have their own TSS intragenic to the protein coding sequence and also be processed from the mRNA, which contains the 3′ UTR. The E. coli sRNA RbsZ is likely made from its own promoter at the 3′ end of the ribose transporter rbsB, but is also processed from the mRNA itself and further cleaved into a shorter sRNA (RbsZ-S) [30]. The mechanisms for sRNA maturation can be complex and require the activity of multiple RNases. For example, the DicF sRNA in E. coli is believed to be cleaved from the dicBF operon by RNase III and then further processed by RNase E [191].
Sometimes different forms of an sRNA can have different regulatory consequences or be present at different levels throughout growth. Different forms of the E. coli SdsN sRNA have distinct mRNA targets, likely attributed to differences in secondary structure [192]. In Myxococcus xanthus, both the longer (Pxr-L) and shorter (Pxr-S) forms of the Prx sRNA are present in high amounts in vegetative cells, while Pxr-S levels decrease during starvation [193]. Conceivably a sRNA precursor could exist in the cell with some low level of activity, to then be processed under a specific stress condition to elicit a stronger effect. For example, compared to full-length E. coli ArcZ, the processed form of ArcZ binds much stronger to the rpoS target in the presence of Hfq in vitro [194]. The same may be true for other sRNAs. Processing results in a monophosphate at the 5′ end of the sRNA, which also can be advantageous. Indeed, RNase E-dependent cleavage has been shown to be stimulated by a 5′ monophosphate provided by an sRNA to activate degradation of the target RNA [195]. Thus, processing may provide an extra layer of regulation of sRNA activity beyond transcriptional synthesis.
The precise cues and players which contribute to such complex regulation of sRNA regulators continue to be discovered. RNA structures and specific sequences can dictate the cleavage of 3′ derived sRNAs [196]. Some of the cleavage sites for RNase E and III, which have been globally predicated by activating or deleting the RNase and performing RNA-seq in E. coli or S. enterica [197, 198], map to sRNA genes.
Obviously, another function of RNases is to turnover sRNAs. Across bacteria some of the key exo- and endoribonucleases in these processes are PNPase, RNase J, RNase E, RNase Y, and RNase III (reviewed [199, 200]). Control of degradation is crucial for regulating sRNA activity and can vary with conditions and the type of RNA. Some sRNAs are co-degraded with their base-pairing targets via RNase E or RNase III [201]. In this mechanism, one sRNA can only regulate one target transcript before it is eliminated. PNPase has also been shown to degrade some sRNAs, particularly those not bound by Hfq [202]. However, for other sRNAs this RNase can have a stabilizing effect, particularly for Hfq-bound sRNAs [120].
Other RNA binding proteins can affect sRNA turnover either generally or individually. In Gram-negative bacteria, Hfq is associated with the RNA degradosome promoting cleavage of bound targets and sRNAs. Paradoxically, however, deletion of Hfq results in the destabilization of many sRNAs. Thus, Hfq is required for both stabilizing the cellular pool of sRNAs as well as facilitating their turnover. The E. coli RapZ protein is an example of an RNA binding protein that specifically promotes turnover of just one sRNA, GlmZ, by acting as an adapter for RNase E [3].
Several transcripts that base-pair with and thus sponge sRNAs have been characterized. In some instances, an mRNA target sequesters and induces turnover of the sRNA, like the E. coli and S. enterica chb operon and ChiX [203, 204]. Other cases involve sRNA-sRNA pairing, where the sponging can result in RNase-mediated degradation. These types of sRNA sponges, like SroC [96], PspH [27], and RbsZ [30], have mainly been found in E. coli and S. enterica, but likely constitute an understudied mechanism to control sRNA-mRNA regulation in other bacteria.
7. Perspectives
Research in RNA biology is booming with more regulatory transcripts being discovered than ever before. The era of RNA-seq has facilitated the global identification of sRNAs from previously underappreciated genomic loci. However, finding functions for these sRNAs can be challenging. Moving forward it will be exciting to tease out new types of sRNA regulatory mechanisms and networks and to characterize sRNAs in bacteria which have not been extensively studied. Although they are small, sRNAs have big effects in the RNA world.
HIGHLIGHTS.
Varied numbers of regulatory sRNAs have been reported for many different bacteria
Most characterized sRNAs act via base-pairing with target RNAs
sRNAs are produced from diverse genomic loci including portions of mRNAs and tRNAs
sRNA levels are controlled by transcriptional and post-transcriptional mechanisms
ACKNOWLEDGMENTS
Thanks to K. Fröhlich, S. Brinsmade, L. Walling, and S. Melamed for providing helpful comments on the manuscript.
P.P.A. was supported by a Postdoctoral Research Associate (PRAT) fellowship from the National Institute of General Medical Sciences (NIGMS), award number 1Fi2GM133345-01. Research in the Storz laboratory was funded by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- [1].Wassarman KM, Storz G, 6S RNA regulates E. coli RNA polymerase activity, Cell, 101 (2000) 613–623. [DOI] [PubMed] [Google Scholar]
- [2].Romeo T, Babitzke P, Global regulation by CsrA and its RNA antagonists, Microbiol Spectr, 6 (2018) RWR-0009–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gopel Y, Papenfort K, Reichenbach B, Vogel J, Gorke B, Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA, Genes Dev, 27 (2013) 552–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kavita K, de Mets F, Gottesman S, New aspects of RNA-based regulation by Hfq and its partner sRNAs, Curr Opin Microbiol, 42 (2018) 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thomason MK, Storz G, Bacterial antisense RNAs: how many are there, and what are they doing?, Annu Rev Genet, 44 (2010) 167–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Georg J, Hess WR, Widespread antisense transcription in prokaryotes, Microbiol Spectr, 6 (2018) RWR-0029–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Itoh T, Tomizawa J, Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H, Proc Natl Acad Sci U S A, 77 (1980) 2450–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kittle JD, Simons RW, Lee J, Kleckner N, Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5’ end of the target RNA and a loop in the anti-sense RNA, J Mol Biol, 210 (1989) 561–572. [DOI] [PubMed] [Google Scholar]
- [9].Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J, Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq, PLoS genetics, 4 (2008) e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S, Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence, Nucleic Acids Res, 36 (2008) 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Landt SG, Abeliuk E, McGrath PT, Lesley JA, McAdams HH, Shapiro L, Small non-coding RNAs in Caulobacter crescentus, Mol Microbiol, 68 (2008) 600–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Regnault B, Coppee JY, Lecuit M, Johansson J, Cossart P, The Listeria transcriptional landscape from saprophytism to virulence, Nature, 459 (2009) 950–956. [DOI] [PubMed] [Google Scholar]
- [13].Wen J, Fozo EM, sRNA antitoxins: more than one way to repress a toxin, Toxins (Basel), 6 (2014) 2310–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Darfeuille F, Unoson C, Vogel J, Wagner EG, An antisense RNA inhibits translation by competing with standby ribosomes, Mol Cell, 26 (2007) 381–392. [DOI] [PubMed] [Google Scholar]
- [15].Thisted T, Gerdes K, Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene, J Mol Biol, 223 (1992) 41–54. [DOI] [PubMed] [Google Scholar]
- [16].Durand S, Gilet L, Condon C, The essential function of B. subtilis RNase III is to silence foreign toxin genes, PLoS genetics, 8 (2012) e1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shearwin KE, Callen BP, Egan JB, Transcriptional interference--a crash course, Trends Genet, 21 (2005) 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P, The excludon: a new concept in bacterial antisense RNA-mediated gene regulation, Nat Rev Microbiol, 11 (2013) 75–82. [DOI] [PubMed] [Google Scholar]
- [19].Dodd IB, Egan JB, Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch, Mol Microbiol, 45 (2002) 697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brophy JA, Voigt CA, Antisense transcription as a tool to tune gene expression, Mol Syst Biol, 12 (2016) 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee EJ, Groisman EA, An antisense RNA that governs the expression kinetics of a multifunctional virulence gene, Mol Microbiol, 76 (2010) 1020–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen J, Morita T, Gottesman S, Regulation of transcription termination of small RNAs and by small RNAs: molecular mechanisms and biological functions, Front Cell Infect Microbiol, 9 (2019) 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Andersen J, Forst SA, Zhao K, Inouye M, Delihas N, The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli, J Biol Chem, 264 (1989) 17961–17970. [PubMed] [Google Scholar]
- [24].Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S, Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule, EMBO J, 12 (1993) 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S, DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription, Proc Natl Acad Sci U S A, 95 (1998) 12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Majdalani N, Chen S, Murrow J, St John K, Gottesman S, Regulation of RpoS by a novel small RNA: the characterization of RprA, Mol Microbiol, 39 (2001) 1382–1394. [DOI] [PubMed] [Google Scholar]
- [27].Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H, Global mapping of small RNA-target interactions in bacteria, Mol Cell, 63 (2016) 884–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Heidrich N, Moll I, Brantl S, In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA, Nucleic Acids Res, 35 (2007) 4331–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J, Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation, Nat Struct Mol Biol, 16 (2009) 840–846. [DOI] [PubMed] [Google Scholar]
- [30].Melamed S, Adams PP, Zhang A, Zhang H, Storz G, RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles, Mol Cell, 77 (2020) 411–425 e417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Raina M, King A, Bianco C, Vanderpool CK, Dual-function RNAs, Microbiol Spectr, 6 (2018) RWR-0032–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Santiago-Frangos A, Woodson SA, Hfq chaperone brings speed dating to bacterial sRNA, Wiley Interdiscip Rev RNA, 9 (2018) e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Holmqvist E, Vogel J, RNA-binding proteins in bacteria, Nat Rev Microbiol, 16 (2018) 601–615. [DOI] [PubMed] [Google Scholar]
- [34].Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, Reinhardt R, Vogel J, Grad-seq guides the discovery of ProQ as a major small RNA-binding protein, Proc Natl Acad Sci U S A, 113 (2016) 11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smirnov A, Wang C, Drewry LL, Vogel J, Molecular mechanism of mRNA repression in trans by a ProQ-dependent small RNA, EMBO J, 36 (2017) 1029–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Attaiech L, Boughammoura A, Brochier-Armanet C, Allatif O, Peillard-Fiorente F, Edwards RA, Omar AR, MacMillan AM, Glover M, Charpentier X, Silencing of natural transformation by an RNA chaperone and a multitarget small RNA, Proc Natl Acad Sci U S A, 113 (2016) 8813–8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Olejniczak M, Storz G, ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers?, Mol Microbiol, 104 (2017) 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Holmqvist E, Li L, Bischler T, Barquist L, Vogel J, Global maps of ProQ binding in vivo reveal target recognition via RNA structure and stability control at mRNA 3’ ends, Mol Cell, 70 (2018) 971–982 e976. [DOI] [PubMed] [Google Scholar]
- [39].Updegrove TB, Zhang A, Storz G, Hfq: the flexible RNA matchmaker, Curr Opin Microbiol, 30 (2016) 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Heidrich N, Chinali A, Gerth U, Brantl S, The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism, Mol Microbiol, 62 (2006) 520–536. [DOI] [PubMed] [Google Scholar]
- [41].Vockenhuber MP, Sharma CM, Statt MG, Schmidt D, Xu Z, Dietrich S, Liesegang H, Mathews DH, Suess B, Deep sequencing-based identification of small non-coding RNAs in Streptomyces coelicolor, RNA Biol, 8 (2011) 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T, Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome, Nucleic Acids Res, 38 (2010) 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rieder R, Reinhardt R, Sharma C, Vogel J, Experimental tools to identify RNA-protein interactions in Helicobacter pylori, RNA Biol, 9 (2012) 520–531. [DOI] [PubMed] [Google Scholar]
- [44].Dugar G, Herbig A, Forstner KU, Heidrich N, Reinhardt R, Nieselt K, Sharma CM, High-resolution transcriptome maps reveal strain-specific regulatory features of multiple Campylobacter jejuni isolates, PLoS genetics, 9 (2013) e1003495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hor J, Gorski SA, Vogel J, Bacterial RNA biology on a genome scale, Mol Cell, 70 (2018) 785–799. [DOI] [PubMed] [Google Scholar]
- [46].Melamed S, Faigenbaum-Romm R, Peer A, Reiss N, Shechter O, Bar A, Altuvia Y, Argaman L, Margalit H, Mapping the small RNA interactome in bacteria using RIL-seq, Nature protocols, 13 (2018) 1–33. [DOI] [PubMed] [Google Scholar]
- [47].Iosub IA, Marchioretto M, Sy B, McKellar S, Nieken KJ, van Nues RW, Tree JJ, Viero G, Granneman S, Hfq CLASH uncovers sRNA-target interaction networks involved in adaptation to nutrient availability, bioRxiv, (2019) 481986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Han K, Tjaden B, Lory S, GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation, Nat Microbiol, 2 (2016) 16239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhang YF, Han K, Chandler CE, Tjaden B, Ernst RK, Lory S, Probing the sRNA regulatory landscape of P. aeruginosa: post-transcriptional control of determinants of pathogenicity and antibiotic susceptibility, Mol Microbiol, 106 (2017) 919–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ikemura T, Dahlberg JE, Small ribonucleic acids of Escherichia coli. I. Characterization by polyacrylamide gel electrophoresis and fingerprint analysis, J Biol Chem, 248 (1973) 5024–5032. [PubMed] [Google Scholar]
- [51].Ikemura T, Dahlberg JE, Small ribonucleic acids of Escherichia coli. II. Noncoordinate accumulation during stringent control, J Biol Chem, 248 (1973) 5033–5041. [PubMed] [Google Scholar]
- [52].Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S, Identification of novel small RNAs using comparative genomics and microarrays, Genes Dev, 15 (2001) 1637–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Perez N, Trevino J, Liu Z, Ho SC, Babitzke P, Sumby P, A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus, PLoS One, 4 (2009) e7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jackson LA, Pan JC, Day MW, Dyer DW, Control of RNA stability by NrrF, an iron-regulated small RNA in Neisseria gonorrhoeae, J Bacteriol, 195 (2013) 5166–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stubben CJ, Micheva-Viteva SN, Shou Y, Buddenborg SK, Dunbar JM, Hong-Geller E, Differential expression of small RNAs from Burkholderia thailandensis in response to varying environmental and stress conditions, BMC genomics, 15 (2014) 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermuller J, Reinhardt R, Stadler PF, Vogel J, The primary transcriptome of the major human pathogen Helicobacter pylori, Nature, 464 (2010) 250–255. [DOI] [PubMed] [Google Scholar]
- [57].Sharma CM, Vogel J, Differential RNA-seq: the approach behind and the biological insight gained, Curr Opin Microbiol, 19 (2014) 97–105. [DOI] [PubMed] [Google Scholar]
- [58].Ettwiller L, Buswell J, Yigit E, Schildkraut I, A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome, BMC genomics, 17 (2016) 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dar D, Shamir M, Mellin JR, Koutero M, Stern-Ginossar N, Cossart P, Sorek R, Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria, Science, 352 (2016) aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Dar D, Sorek R, High-resolution RNA 3’-ends mapping of bacterial Rho-dependent transcripts, Nucleic Acids Res, 46 (2018) 6797–6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Yan B, Boitano M, Clark TA, Ettwiller L, SMRT-Cappable-seq reveals complex operon variants in bacteria, Nat Commun, 9 (2018) 3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ju X, Li D, Liu S, Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria, Nat Microbiol, 4 (2019) 1907–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Gomez-Lozano M, Marvig RL, Molin S, Long KS, Identification of bacterial small RNAs by RNA sequencing, Methods Mol Biol, 1149 (2014) 433–456. [DOI] [PubMed] [Google Scholar]
- [64].Liu JM, Camilli A, Discovery of bacterial sRNAs by high-throughput sequencing, Methods Mol Biol, 733 (2011) 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Holmqvist E, Wright PR, Li L, Bischler T, Barquist L, Reinhardt R, Backofen R, Vogel J, Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo, EMBO J, 35 (2016) 991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Waters SA, McAteer SP, Kudla G, Pang I, Deshpande NP, Amos TG, Leong KW, Wilkins MR, Strugnell R, Gally DL, Tollervey D, Tree JJ, Small RNA interactome of pathogenic E. coli revealed through crosslinking of RNase E, EMBO J, 36 (2017) 374–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li W, Ying X, Lu Q, Chen L, Predicting sRNAs and their targets in bacteria, Genomics Proteomics Bioinformatics, 10 (2012) 276–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sridhar J, Gunasekaran P, Computational small RNA prediction in bacteria, Bioinform Biol Insights, 7 (2013) 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rivas E, Eddy SR, Noncoding RNA gene detection using comparative sequence analysis, BMC bioinformatics, 2 (2001) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Washietl S, Hofacker IL, Stadler PF, Fast and reliable prediction of noncoding RNAs, Proc Natl Acad Sci U S A, 102 (2005) 2454–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Livny J, Fogel MA, Davis BM, Waldor MK, sRNAPredict: an integrative computational approach to identify sRNAs in bacterial genomes, Nucleic Acids Res, 33 (2005) 4096–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Sridhar J, Sambaturu N, Sabarinathan R, Ou HY, Deng Z, Sekar K, Rafi ZA, Rajakumar K, sRNAscanner: a computational tool for intergenic small RNA detection in bacterial genomes, PLoS One, 5 (2010) e11970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Soldatov RA, Vinogradova SV, Mironov AA, RNASurface: fast and accurate detection of locally optimal potentially structured RNA segments, Bioinformatics, 30 (2014) 457–463. [DOI] [PubMed] [Google Scholar]
- [74].Barman RK, Mukhopadhyay A, Das S, An improved method for identification of small non-coding RNAs in bacteria using support vector machine, Sci Rep, 7 (2017) 46070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tran TT, Zhou F, Marshburn S, Stead M, Kushner SR, Xu Y, De novo computational prediction of non-coding RNA genes in prokaryotic genomes, Bioinformatics, 25 (2009) 2897–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yu SH, Vogel J, Forstner KU, ANNOgesic: a Swiss army knife for the RNA-seq based annotation of bacterial/archaeal genomes, Gigascience, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Leonard S, Meyer S, Lacour S, Nasser W, Hommais F, Reverchon S, APERO: a genome-wide approach for identifying bacterial small RNAs from RNA-Seq data, Nucleic Acids Res, 47 (2019) e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Geissmann T, Chevalier C, Cros MJ, Boisset S, Fechter P, Noirot C, Schrenzel J, Francois P, Vandenesch F, Gaspin C, Romby P, A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation, Nucleic Acids Res, 37 (2009) 7239–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mader U, Nicolas P, Depke M, Pane-Farre J, Debarbouille M, van der Kooi-Pol MM, Guerin C, Derozier S, Hiron A, Jarmer H, Leduc A, Michalik S, Reilman E, Schaffer M, Schmidt F, Bessieres P, Noirot P, Hecker M, Msadek T, Volker U, van Dijl JM, Staphylococcus aureus transcriptome architecture: from laboratory to infection-mimicking conditions, PLoS genetics, 12 (2016) e1005962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezee-Durant E, Barbet R, Jacquet E, Jacq A, Gautheret D, Felden B, Vogel J, Bouloc P, Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism, Nucleic Acids Res, 38 (2010) 6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Howden BP, Beaume M, Harrison PF, Hernandez D, Schrenzel J, Seemann T, Francois P, Stinear TP, Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure, Antimicrob Agents Chemother, 57 (2013) 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Broach WH, Weiss A, Shaw LN, Transcriptomic analysis of staphylococcal sRNAs: insights into species-specific adaption and the evolution of pathogenesis, Microb Genom, 2 (2016) e000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Carroll RK, Weiss A, Broach WH, Wiemels RE, Mogen AB, Rice KC, Shaw LN, Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus, mBio, 7 (2016) e01990–01915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Liu W, Rochat T, Toffano-Nioche C, Le Lam TN, Bouloc P, Morvan C, Assessment of bona fide sRNAs in Staphylococcus aureus, Front Microbiol, 9 (2018) 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hu J, Li T, Xu W, Zhan J, Chen H, He C, Wang Q, Small Antisense RNA RblR Positively Regulates RuBisCo in Synechocystis sp. PCC 6803, Front Microbiol, 8 (2017) 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Raghavan R, Groisman EA, Ochman H, Genome-wide detection of novel regulatory RNAs in E. coli, Genome Res, 21 (2011) 1487–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Thomason MK, Bischler T, Eisenbart SK, Forstner KU, Zhang A, Herbig A, Nieselt K, Sharma CM, Storz G, Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli, J Bacteriol, 197 (2015) 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penades JR, Valle J, Solano C, Gingeras TR, Genome-wide antisense transcription drives mRNA processing in bacteria, Proc Natl Acad Sci U S A, 108 (2011) 20172–20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Voigt K, Sharma CM, Mitschke J, Lambrecht SJ, Voss B, Hess WR, Steglich C, Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity, ISME J, 8 (2014) 2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Stazic D, Lindell D, Steglich C, Antisense RNA protects mRNA from RNase E degradation by RNA-RNA duplex formation during phage infection, Nucleic Acids Res, 39 (2011) 4890–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Andre G, Even S, Putzer H, Burguiere P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O, S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum, Nucleic Acids Res, 36 (2008) 5955–5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kawano M, Aravind L, Storz G, An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin, Mol Microbiol, 64 (2007) 738–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sayed N, Jousselin A, Felden B, A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide, Nat Struct Mol Biol, 19 (2011) 105–112. [DOI] [PubMed] [Google Scholar]
- [94].Duhring U, Axmann IM, Hess WR, Wilde A, An internal antisense RNA regulates expression of the photosynthesis gene isiA, Proc Natl Acad Sci U S A, 103 (2006) 7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Tsai CH, Liao R, Chou B, Palumbo M, Contreras LM, Genome-wide analyses in bacteria show small-RNA enrichment for long and conserved intergenic regions, J Bacteriol, 197 (2015) 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Miyakoshi M, Chao Y, Vogel J, Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA, EMBO J, 34 (2015) 1478–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G, A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator, Cell, 90 (1997) 43–53. [DOI] [PubMed] [Google Scholar]
- [98].Peer A, Margalit H, Evolutionary patterns of Escherichia coli small RNAs and their regulatory interactions, RNA, 20 (2014) 994–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Raghavan R, Kacharia FR, Millar JA, Sislak CD, Ochman H, Genome rearrangements can make and break small RNA genes, Genome Biol Evol, 7 (2015) 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gaballa A, Antelmann H, Aguilar C, Khakh SK, Song KB, Smaldone GT, Helmann JD, The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins, Proc Natl Acad Sci U S A, 105 (2008) 11927–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Masse E, Vanderpool CK, Gottesman S, Effect of RyhB small RNA on global iron use in Escherichia coli, J Bacteriol, 187 (2005) 6962–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Papenfort K, Forstner KU, Cong JP, Sharma CM, Bassler BL, Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation, Proc Natl Acad Sci U S A, 112 (2015) E766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Durand S, Braun F, Helfer AC, Romby P, Condon C, sRNA-mediated activation of gene expression by inhibition of 5’−3’ exonucleolytic mRNA degradation, Elife, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Schachterle JK, Zeng Q, Sundin GW, Three Hfq-dependent small RNAs regulate flagellar motility in the fire blight pathogen Erwinia amylovora, Mol Microbiol, 111 (2019) 1476–1492. [DOI] [PubMed] [Google Scholar]
- [105].Le Pabic H, Germain-Amiot N, Bordeau V, Felden B, A bacterial regulatory RNA attenuates virulence, spread and human host cell phagocytosis, Nucleic Acids Res, 43 (2015) 9232–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kawano M, Reynolds AA, Miranda-Rios J, Storz G, Detection of 5’- and 3’-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli, Nucleic Acids Res, 33 (2005) 1040–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Breaker RR, Riboswitches and translation control, Cold Spring Harb Perspect Biol, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Loh E, Righetti F, Eichner H, Twittenhoff C, Narberhaus F, RNA thermometers in bacterial pathogens, Microbiol Spectr, 6 (2018) RWR-0012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Turnbough CL Jr., Regulation of bacterial gene expression by transcription attenuation, Microbiol Mol Biol Rev, 83 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA, Riboswitch control of Rho-dependent transcription termination, Proc Natl Acad Sci U S A, 109 (2012) 5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR, An mRNA structure in bacteria that controls gene expression by binding lysine, Genes Dev, 17 (2003) 2688–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG, RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria, Nucleic Acids Res, 31 (2003) 6435–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Miranda-Rios J, Navarro M, Soberon M, A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria, Proc Natl Acad Sci U S A, 98 (2001) 9736–9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M, The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis, The FEBS journal, 282 (2015) 3230–3242. [DOI] [PubMed] [Google Scholar]
- [115].Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J, A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes, Cell, 139 (2009) 770–779. [DOI] [PubMed] [Google Scholar]
- [116].Mellin JR, Koutero M, Dar D, Nahori MA, Sorek R, Cossart P, Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA, Science, 345 (2014) 940–943. [DOI] [PubMed] [Google Scholar]
- [117].DebRoy S, Gebbie M, Ramesh A, Goodson JR, Cruz MR, van Hoof A, Winkler WC, Garsin DA, Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator, Science, 345 (2014) 937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Melior H, Li S, Madhugiri R, Stotzel M, Azarderakhsh S, Barth-Weber S, Baumgardt K, Ziebuhr J, Evguenieva-Hackenberg E, Transcription attenuation-derived small RNA rnTrpL regulates tryptophan biosynthesis gene expression in trans, Nucleic Acids Res, 47 (2019) 6396–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Dar D, Sorek R, Bacterial noncoding RNAs excised from within protein-coding transcripts, mBio, 9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Cameron TA, Matz LM, Sinha D, De Lay NR, Polynucleotide phosphorylase promotes the stability and function of Hfq-binding sRNAs by degrading target mRNA-derived fragments, Nucleic Acids Res, 47 (2019) 8821–8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Vanderpool CK, Gottesman S, Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system, Mol Microbiol, 54 (2004) 1076–1089. [DOI] [PubMed] [Google Scholar]
- [122].Thomason MK, Voichek M, Dar D, Addis V, Fitzgerald D, Gottesman S, Sorek R, Greenberg EP, A rhlI 5’ UTR-derived sRNA regulates RhlR-dependent quorum sensing in Pseudomonas aeruginosa, mBio, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Adams PP, Flores Avile C, Popitsch N, Bilusic I, Schroeder R, Lybecker M, Jewett MW, In vivo expression technology and 5’ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection, Nucleic Acids Res, 45 (2017) 775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Zhukova A, Fernandes LG, Hugon P, Pappas CJ, Sismeiro O, Coppee JY, Becavin C, Malabat C, Eshghi A, Zhang JJ, Yang FX, Picardeau M, Genome-wide transcriptional start site mapping and sRNA identification in the pathogen Leptospira interrogans, Front Cell Infect Microbiol, 7 (2017) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Babski J, Haas KA, Nather-Schindler D, Pfeiffer F, Forstner KU, Hammelmann M, Hilker R, Becker A, Sharma CM, Marchfelder A, Soppa J, Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq), BMC genomics, 17 (2016) 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Meydan S, Marks J, Klepacki D, Sharma V, Baranov PV, Firth AE, Margus T, Kefi A, Vazquez-Laslop N, Mankin AS, Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome, Mol Cell, 74 (2019) 481–493 e486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J, An atlas of Hfq-bound transcripts reveals 3’ UTRs as a genomic reservoir of regulatory small RNAs, EMBO J, 31 (2012) 4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Peng T, Berghoff BA, Oh JI, Weber L, Schirmer J, Schwarz J, Glaeser J, Klug G, Regulation of a polyamine transporter by the conserved 3’ UTR-derived sRNA SorX confers resistance to singlet oxygen and organic hydroperoxides in Rhodobacter sphaeroides, RNA Biol, 13 (2016) 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G, MicL, a new sigmaE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein, Genes Dev, 28 (2014) 1620–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Heidrich N, Bauriedl S, Barquist L, Li L, Schoen C, Vogel J, The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq, Nucleic Acids Res, 45 (2017) 6147–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Chao Y, Vogel J, A 3’ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response, Mol Cell, 61 (2016) 352–363. [DOI] [PubMed] [Google Scholar]
- [132].Kim HM, Shin JH, Cho YB, Roe JH, Inverse regulation of Fe- and Ni-containing SOD genes by a Fur family regulator Nur through small RNA processed from 3’UTR of the sodF mRNA, Nucleic Acids Res, 42 (2014) 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Shepherd J, Ibba M, Bacterial transfer RNAs, FEMS Microbiol Rev, 39 (2015) 280–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Raina M, Ibba M, tRNAs as regulators of biological processes, Front Genet, 5 (2014) 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S, Global analysis of small RNA and mRNA targets of Hfq, Mol Microbiol, 50 (2003) 1111–1124. [DOI] [PubMed] [Google Scholar]
- [136].Bilusic I, Popitsch N, Rescheneder P, Schroeder R, Lybecker M, Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation, RNA Biol, 11 (2014) 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Lee T, Feig AL, The RNA binding protein Hfq interacts specifically with tRNAs, RNA, 14 (2008) 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Masse E, A 3’ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise, Mol Cell, 58 (2015) 393–405. [DOI] [PubMed] [Google Scholar]
- [139].Gebetsberger J, Zywicki M, Kunzi A, Polacek N, tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii, Archaea, 2012 (2012) 260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Ren B, Wang X, Duan J, Ma J, Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation, Science, 365 (2019) 919–922. [DOI] [PubMed] [Google Scholar]
- [141].Svenningsen SL, Kongstad M, Stenum TS, Munoz-Gomez AJ, Sorensen MA, Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli, Nucleic Acids Res, 45 (2017) 793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Rudd KE, Novel intergenic repeats of Escherichia coli K-12, Res Microbiol, 150 (1999) 653–664. [DOI] [PubMed] [Google Scholar]
- [143].Deutscher MP, Maturation and degradation of ribosomal RNA in bacteria, Prog Mol Biol Transl Sci, 85 (2009) 369–391. [DOI] [PubMed] [Google Scholar]
- [144].Munishkin A, Wool IG, The ribosome-in-pieces: binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA, Proc Natl Acad Sci U S A, 94 (1997) 12280–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Fourmy D, Recht MI, Blanchard SC, Puglisi JD, Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic, Science, 274 (1996) 1367–1371. [DOI] [PubMed] [Google Scholar]
- [146].Purohit P, Stern S, Interactions of a small RNA with antibiotic and RNA ligands of the 30S subunit, Nature, 370 (1994) 659–662. [DOI] [PubMed] [Google Scholar]
- [147].Arkov AL, Mankin A, Murgola EJ, An rRNA fragment and its antisense can alter decoding of genetic information, J Bacteriol, 180 (1998) 2744–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Lambert M, Benmoussa A, Provost P, Small non-coding RNAs derived from eukaryotic ribosomal RNA, Noncoding RNA, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y, qiRNA is a new type of small interfering RNA induced by DNA damage, Nature, 459 (2009) 274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F, C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector, Science, 353 (2016) aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Meeske AJ, Nakandakari-Higa S, Marraffini LA, Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage, Nature, 570 (2019) 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Leskinen K, Blasdel BG, Lavigne R, Skurnik M, RNA-sequencing reveals the progression of phage-host interactions between phiR1–37 and Yersinia enterocolitica, Viruses, 8 (2016) 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Chevallereau A, Blasdel BG, De Smet J, Monot M, Zimmermann M, Kogadeeva M, Sauer U, Jorth P, Whiteley M, Debarbieux L, Lavigne R, Next-generation “-omics” approaches reveal a massive alteration of host RNA metabolism during bacteriophage infection of Pseudomonas aeruginosa, PLoS genetics, 12 (2016) e1006134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL, Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli, Mol Cell, 55 (2014) 199–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Abdullah Z, Schlee M, Roth S, Mraheil MA, Barchet W, Bottcher J, Hain T, Geiger S, Hayakawa Y, Fritz JH, Civril F, Hopfner KP, Kurts C, Ruland J, Hartmann G, Chakraborty T, Knolle PA, RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids, EMBO J, 31 (2012) 4153–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Pagliuso A, Tham TN, Allemand E, Robertin S, Dupuy B, Bertrand Q, Becavin C, Koutero M, Najburg V, Nahori MA, Tangy F, Stavru F, Bessonov S, Dessen A, Muchardt C, Lebreton A, Komarova AV, Cossart P, An RNA-binding protein secreted by a bacterial pathogen modulates RIG-I signaling, Cell Host Microbe, 26 (2019) 823–835 e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Ellis TN, Kuehn MJ, Virulence and immunomodulatory roles of bacterial outer membrane vesicles, Microbiol Mol Biol Rev, 74 (2010) 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Tsatsaronis JA, Franch-Arroyo S, Resch U, Charpentier E, Extracellular vesicle RNA: a universal mediator of microbial communication?, Trends Microbiol, 26 (2018) 401–410. [DOI] [PubMed] [Google Scholar]
- [159].Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, Wilmes P, The extracellular RNA complement of Escherichia coli, Microbiologyopen, 4 (2015) 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Blenkiron C, Simonov D, Muthukaruppan A, Tsai P, Dauros P, Green S, Hong J, Print CG, Swift S, Phillips AR, Uropathogenic Escherichia coli releases extracellular vesicles that are associated with RNA, PLoS One, 11 (2016) e0160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Sjostrom AE, Sandblad L, Uhlin BE, Wai SN, Membrane vesicle-mediated release of bacterial RNA, Sci Rep, 5 (2015) 15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA, Mielcarz DW, Demers EG, Dolben EL, Hammond JH, Hogan DA, Stanton BA, A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles, PLoS Pathog, 12 (2016) e1005672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Frantz R, Teubner L, Schultze T, La Pietra L, Muller C, Gwozdzinski K, Pillich H, Hain T, Weber-Gerlach M, Panagiotidis GD, Mostafa A, Weber F, Rohde M, Pleschka S, Chakraborty T, Abu Mraheil M, The secRNome of Listeria monocytogenes Harbors Small Noncoding RNAs That Are Potent Inducers of Beta Interferon, mBio, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Ainsztein AM, Brooks PJ, Dugan VG, Ganguly A, Guo M, Howcroft TK, Kelley CA, Kuo LS, Labosky PA, Lenzi R, McKie GA, Mohla S, Procaccini D, Reilly M, Satterlee JS, Srinivas PR, Church ES, Sutherland M, Tagle DA, Tucker JM, Venkatachalam S, The NIH extracellular RNA communication consortium, J Extracell Vesicles, 4 (2015) 27493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Bervoets I, Charlier D, Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology, FEMS Microbiol Rev, 43 (2019) 304–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Johansen J, Rasmussen AA, Overgaard M, Valentin-Hansen P, Conserved small non-coding RNAs that belong to the sigmaE regulon: role in down-regulation of outer membrane proteins, J Mol Biol, 364 (2006) 1–8. [DOI] [PubMed] [Google Scholar]
- [167].Udekwu KI, Wagner EG, Sigma E controls biogenesis of the antisense RNA MicA, Nucleic Acids Res, 35 (2007) 1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Thompson KM, Rhodius VA, Gottesman S, SigmaE regulates and is regulated by a small RNA in Escherichia coli, J Bacteriol, 189 (2007) 4243–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA, Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon, Proc Natl Acad Sci U S A, 108 (2011) 12875–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Peschek N, Hoyos M, Herzog R, Forstner KU, Papenfort K, A conserved RNA seed-pairing domain directs small RNA-mediated stress resistance in enterobacteria, EMBO J, 38 (2019) e101650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Silvaggi JM, Perkins JB, Losick R, Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis, J Bacteriol, 188 (2006) 532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Marinho CM, Dos Santos PT, Kallipolitis BH, Johansson J, Ignatov D, Guerreiro DN, Piveteau P, O’Byrne CP, The sigma(B)-dependent regulatory sRNA Rli47 represses isoleucine biosynthesis in Listeria monocytogenes through a direct interaction with the ilvA transcript, RNA Biol, 16 (2019) 1424–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Becavin C, Archambaud C, Cossart P, Sorek R, Comparative transcriptomics of pathogenic and non-pathogenic Listeria species, Mol Syst Biol, 8 (2012) 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Vassinova N, Kozyrev D, A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli, Microbiology, 146 Pt 12 (2000) 3171–3182. [DOI] [PubMed] [Google Scholar]
- [175].Masse E, Gottesman S, A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli, Proc Natl Acad Sci U S A, 99 (2002) 4620–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML, Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis, Proc Natl Acad Sci U S A, 101 (2004) 9792–9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Mellin JR, Goswami S, Grogan S, Tjaden B, Genco CA, A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis, J Bacteriol, 189 (2007) 3686–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Santana EA, Harrison A, Zhang X, Baker BD, Kelly BJ, White P, Liu Y, Munson RS Jr., HrrF is the Fur-regulated small RNA in nontypeable Haemophilus influenzae, PLoS One, 9 (2014) e105644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Richard AL, Withey JH, Beyhan S, Yildiz F, DiRita VJ, The Vibrio cholerae virulence regulatory cascade controls glucose uptake through activation of TarA, a small regulatory RNA, Mol Microbiol, 78 (2010) 1171–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Goulian M, Two-component signaling circuit structure and properties, Curr Opin Microbiol, 13 (2010) 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Brosse A, Korobeinikova A, Gottesman S, Guillier M, Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system, Nucleic Acids Res, 44 (2016) 9650–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Mancini S, Imlay JA, The induction of two biosynthetic enzymes helps Escherichia coli sustain heme synthesis and activate catalase during hydrogen peroxide stress, Mol Microbiol, 96 (2015) 744–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA, Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica, J Bacteriol, 191 (2009) 7279–7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Ray-Soni A, Bellecourt MJ, Landick R, Mechanisms of bacterial transcription termination: all good things must end, Annu Rev Biochem, 85 (2016) 319–347. [DOI] [PubMed] [Google Scholar]
- [185].Morita T, Ueda M, Kubo K, Aiba H, Insights into transcription termination of Hfq-binding sRNAs of Escherichia coli and characterization of readthrough products, RNA, 21 (2015) 1490–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Felden B, Gilot D, Modulation of bacterial sRNAs activity by epigenetic modifications: inputs from the eukaryotic miRNAs, Genes (Basel), 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Vasilyev N, Gao A, Serganov A, Noncanonical features and modifications on the 5’-end of bacterial sRNAs and mRNAs, Wiley Interdiscip Rev RNA, 10 (2019) e1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Marbaniang CN, Vogel J, Emerging roles of RNA modifications in bacteria, Curr Opin Microbiol, 30 (2016) 50–57. [DOI] [PubMed] [Google Scholar]
- [189].Cahova H, Winz ML, Hofer K, Nubel G, Jaschke A, NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs, Nature, 519 (2015) 374–377. [DOI] [PubMed] [Google Scholar]
- [190].Deng X, Chen K, Luo GZ, Weng X, Ji Q, Zhou T, He C, Widespread occurrence of N6-methyladenosine in bacterial mRNA, Nucleic Acids Res, 43 (2015) 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Faubladier M, Cam K, Bouche JP, Escherichia coli cell division inhibitor DicF-RNA of the dicB operon. Evidence for its generation in vivo by transcription termination and by RNase III and RNase E-dependent processing, J Mol Biol, 212 (1990) 461–471. [DOI] [PubMed] [Google Scholar]
- [192].Hao Y, Updegrove TB, Livingston NN, Storz G, Protection against deleterious nitrogen compounds: role of sigmaS-dependent small RNAs encoded adjacent to sdiA, Nucleic Acids Res, 44 (2016) 6935–6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Yu YT, Yuan X, Velicer GJ, Adaptive evolution of an sRNA that controls Myxococcus development, Science, 328 (2010) 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA, Positive regulation by small RNAs and the role of Hfq, Proc Natl Acad Sci U S A, 107 (2010) 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Bandyra KJ, Said N, Pfeiffer V, Gorna MW, Vogel J, Luisi BF, The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E, Mol Cell, 47 (2012) 943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Updegrove TB, Kouse AB, Bandyra KJ, Storz G, Stem-loops direct precise processing of 3’ UTR-derived small RNA MicL, Nucleic Acids Res, 47 (2019) 1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Chao Y, Li L, Girodat D, Forstner KU, Said N, Corcoran C, Smiga M, Papenfort K, Reinhardt R, Wieden HJ, Luisi BF, Vogel J, In vivo cleavage map illuminates the central role of RNase E in coding and non-coding RNA pathways, Mol Cell, 65 (2017) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Gordon GC, Cameron JC, Pfleger BF, RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA, mBio, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Mohanty BK, Kushner SR, Enzymes involved in posttranscriptional RNA metabolism in Gram-negative bacteria, Microbiol Spectr, 6 (2018) RWR-0011–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Durand S, Condon C, RNases and helicases in Gram-positive bacteria, Microbiol Spectr, 6 (2018) RWR-0003–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR, Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB, Nucleic Acids Res, 33 (2005) 1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Andrade JM, Pobre V, Matos AM, Arraiano CM, The crucial role of PNPase in the degradation of small RNAs that are not associated with Hfq, RNA, 18 (2012) 844–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L, Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target, Genes Dev, 23 (2009) 2004–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Overgaard M, Johansen J, Moller-Jensen J, Valentin-Hansen P, Switching off small RNA regulation with trap-mRNA, Mol Microbiol, 73 (2009) 790–800. [DOI] [PubMed] [Google Scholar]
- [205].Moody MJ, Young RA, Jones SE, Elliot MA, Comparative analysis of non-coding RNAs in the antibiotic-producing Streptomyces bacteria, BMC genomics, 14 (2013) 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Vockenhuber MP, Suess B, Streptomyces coelicolor sRNA scr5239 inhibits agarase expression by direct base pairing to the dagA coding region, Microbiology, 158 (2012) 424–435. [DOI] [PubMed] [Google Scholar]
- [207].Gerrick ER, Barbier T, Chase MR, Xu R, Francois J, Lin VH, Szucs MJ, Rock JM, Ahmad R, Tjaden B, Livny J, Fortune SM, Small RNA profiling in Mycobacterium tuberculosis identifies MrsI as necessary for an anticipatory iron sparing response, Proc Natl Acad Sci U S A, 115 (2018) 6464–6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Phillips P, Progulske-Fox A, Grieshaber S, Grieshaber N, Expression of Porphyromonas gingivalis small RNA in response to hemin availability identified using microarray and RNA-seq analysis, FEMS microbiology letters, 351 (2014) 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Grieshaber NA, Grieshaber SS, Fischer ER, Hackstadt T, A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis, Mol Microbiol, 59 (2006) 541–550. [DOI] [PubMed] [Google Scholar]
- [210].Kopf M, Klahn S, Scholz I, Matthiessen JK, Hess WR, Voss B, Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803, DNA Res, 21 (2014) 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Xu W, Chen H, He CL, Wang Q, Deep sequencing-based identification of small regulatory RNAs in Synechocystis sp. PCC 6803, PLoS One, 9 (2014) e92711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Irnov I, Sharma CM, Vogel J, Winkler WC, Identification of regulatory RNAs in Bacillus subtilis, Nucleic Acids Res, 38 (2010) 6637–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Augagneur Y, King AN, Germain-Amiot N, Sassi M, Fitzgerald JW, Sahukhal GS, Elasri MO, Felden B, Brinsmade SR, Analysis of the CodY RNome reveals RsaD as a stress-responsive riboregulator of overflow metabolism in Staphylococcus aureus, Mol Microbiol, (2019). [DOI] [PubMed] [Google Scholar]
- [214].Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T, The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages, Nucleic Acids Res, 39 (2011) 4235–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Nielsen JS, Larsen MH, Lillebaek EM, Bergholz TM, Christiansen MH, Boor KJ, Wiedmann M, Kallipolitis BH, A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes, PLoS One, 6 (2011) e19019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Schrader JM, Zhou B, Li GW, Lasker K, Childers WS, Williams B, Long T, Crosson S, McAdams HH, Weissman JS, Shapiro L, The coding and noncoding architecture of the Caulobacter crescentus genome, PLoS genetics, 10 (2014) e1004463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Landt SG, Lesley JA, Britos L, Shapiro L, CrfA, a small noncoding RNA regulator of adaptation to carbon starvation in Caulobacter crescentus, J Bacteriol, 192 (2010) 4763–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Schroeder CL, Narra HP, Sahni A, Rojas M, Khanipov K, Patel J, Shah R, Fofanov Y, Sahni SK, Identification and characterization of novel small RNAs in Rickettsia prowazekii, Front Microbiol, 7 (2016) 859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Fantappie L, Oriente F, Muzzi A, Serruto D, Scarlato V, Delany I, A novel Hfq-dependent sRNA that is under FNR control and is synthesized in oxygen limitation in Neisseria meningitidis, Mol Microbiol, 80 (2011) 507–523. [DOI] [PubMed] [Google Scholar]
- [220].Sahr T, Rusniok C, Dervins-Ravault D, Sismeiro O, Coppee JY, Buchrieser C, Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence, RNA Biol, 9 (2012) 503–519. [DOI] [PubMed] [Google Scholar]
- [221].Wurtzel O, Yoder-Himes DR, Han K, Dandekar AA, Edelheit S, Greenberg EP, Sorek R, Lory S, The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature, PLoS Pathog, 8 (2012) e1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL, The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae, Cell, 118 (2004) 69–82. [DOI] [PubMed] [Google Scholar]
- [223].Schiano CA, Koo JT, Schipma MJ, Caulfield AJ, Jafari N, Lathem WW, Genome-wide analysis of small RNAs expressed by Yersinia pestis identifies a regulator of the Yop-Ysc type III secretion system, J Bacteriol, 196 (2014) 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Pernitzsch SR, Tirier SM, Beier D, Sharma CM, A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori, Proc Natl Acad Sci U S A, 111 (2014) E501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Popitsch N, Bilusic I, Rescheneder P, Schroeder R, Lybecker M, Temperature-dependent sRNA transcriptome of the Lyme disease spirochete, BMC genomics, 18 (2017) 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Lybecker MC, Samuels DS, Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi, Mol Microbiol, 64 (2007) 1075–1089. [DOI] [PubMed] [Google Scholar]


