Abstract
Cancer cells adapt to nutrient-deprived tumor microenvironment during progression via regulating the level and function of metabolic enzymes. Acetyl-coenzyme A (AcCoA) is a key metabolic intermediate that is crucial for cancer cell metabolism, especially under metabolic stress. It is of special significance to decipher the role acetyl-CoA synthetase short chain family (ACSS) in cancer cells confronting metabolic stress. Here we analyzed the generation of lipogenic AcCoA in bladder cancer cells under metabolic stress and found that in bladder urothelial carcinoma (BLCA) cells, the proportion of lipogenic AcCoA generated from glucose were largely reduced under metabolic stress. Our results revealed that ACSS3 was responsible for lipogenic AcCoA synthesis in BLCA cells under metabolic stress. Interestingly, we found that ACSS3 was required for acetate utilization and histone acetylation. Moreover, our data illustrated that ACSS3 promoted BLCA cell growth. In addition, through analyzing clinical samples, we found that both mRNA and protein levels of ACSS3 were dramatically upregulated in BLCA samples in comparison with adjacent controls and BLCA patients with lower ACSS3 expression were entitled with longer overall survival. Our data revealed an oncogenic role of ACSS3 via regulating AcCoA generation in BLCA and provided a promising target in metabolic pathway for BLCA treatment.
Subject terms: Cancer, Cell biology
Introduction
In cancer cells, considerable number of metabolic enzymes and intermediates are deregulated1. Acetyl-coenzyme A (AcCoA) is a key metabolic intermediate in anabolic and catabolic pathways, and plays a critical role in biomass production2,3. AcCoA homeostasis directly affects the level of histone acetylation, which contributes to transcriptional and posttranslational gene expression regulations4. Hence, AcCoA biosynthesis is crucial for cancer cell metabolism, especially under metabolic stress5.
AcCoA were basically generated from two pathways: one is mediated by ATP citrate lyase, which cleaved citrate into oxaloacetate and acetyl-CoA, and the other is mediated by acetyl-CoA synthetase short chain family (ACSS), which ligates acetate and CoA6. ACSS family contains three family members, ACSS1, ACSS2, and ACSS3. ACSS1 was originally found to be required for acetate uptake and cell survival in hepatocellular carcinoma7. In addition, ACSS1 was involved in cell survival and tumor growth of melanoma cells8. Most recently, it has been proposed that ACSS family proteins distinct heterogeneity of hepatocellular carcinoma in a metabolic manner9. Among the three ACSSs, ACSS2 was most extensively studied. Reports showed that ACSS2 precipitated in lipid metabolism and regulated carcinogenesis in hepatocellular carcinoma7,10, glioblastoma11, breast cancer6, prostate cancer6, and bladder cancer12. On the other hand, little is known about the function of newly identified ACSS member, ACSS3. It has been proposed that ACSS3 acts as an important prognosis biomarker in gastric cancer13 and ACSS3 serves as a biomarker to stratify subtypes of hepatocellular carcinoma9. However, the biological function of ACSS3 has not been verified in cancer cells yet.
As solid tumors usually confronted unfavorable environments where oxygen and nutrition were limited, alterative metabolic pathways were aberrantly activated to meet the demands for biomass production in cancer cells14. Under hypoxia status, AcCoA production from glucose was diminished and molecular tools have been developed to trace the metabolism of AcCoA15,16. It has been illustrated that acetate serve as an alternative carbon origin under metabolic stress conditions6,17. Therefore, it is of special significance to further validate the participation of ACSS proteins in tumorigenesis and evaluate the role ACSS proteins in alternative utilization of carbon sources.
Bladder urothelial carcinoma (BLCA) is the most common malignancy in the urinary tract, which lead to ~150,000 death worldwide each year18. Even though only ~25% of BLCA progressed into muscle invasive BLCA, half of the advanced cancers further metastasize19. What make the situation worse is that no well-defined prognostic marker for BLCA is currently available20. Hence, identification of proteins, especially metabolic enzymes, facilitates carcinogenesis of BLCA provides novel insights and potential therapeutic targets for BLCA.
Here we quantified the AcCoA metabolism in bladder cancer (BLCA) cells under normoxia and hypoxia, and evaluated the role ACSS3 in AcCoA metabolism during metabolic stress in bladder cancer cell lines. Moreover, we further dissected the function of ACSS3 in bladder cancer cell growth in vitro and in vivo. Importantly, we analyzed our clinical data and revealed an oncogenic role of ACSS3 in bladder cancer.
Material and methods
Cell culture
SV-HUC-1, UMUC3, T24 cells were purchased from the American Type Culture Collection. SV-HUC-1 cell line was maintained in F-12K Medium (HyClone). UMUC3 cell line was cultured in Eagle’s Minimum Essential Medium (HyClone). T24 cell line was cultured in McCoy’s 5a Medium (HyClone). All the cell lines were cultured in base medium supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) at 37 °C and 5% CO2.
13C Enrichment in lipogenic AcCoA detection
Fatty acid labeling assay was performed as previously described21. Briefly, cells were maintained in culture medium containing 2 mM glutamine. Cells were plated in six-well plates overnight before changing medium. Medium were changedto Dulbecco’s modified Eagle medium with 13C-labeled glucose (10 mM), 13C-glutamine (2 mM), and 13C-acetate for 48–72 h. Cells were washed with phosphate-buffered saline (PBS), add 0.75 mL of methanol : PBS (1 : 1), and place the plate at −20 °C for 10 min. To extract fatty acids, we scraped cells and transferred to glass tubes, added 0.5 mL cold chloroform, and vortexed for 1 min and dried under nitrogen gas. Then, add toluene (80 μl), methanol (600 μl), and methanolic-HCl (120 μl) to each tube, vortex, and incubate at 100 °C for 60 min. Water (0.4 ml) and hwxane (0.3 ml) were added to samples after reducing to room temperature, vortex and analyzed the top hexane phase by Gas Chromatography-Mass Spectrometer (GC-MS). The carbon sources of lipogenic AcCoA was determined by computed binomial distributions as previously described21.
Quantification of medium acetate
For acetate derivatization, 200 μl cell culture medium was collected into a 2 mL microfuge tube, followed by 40 μl 2H3-acetate (1 mM), 1-propanol (50 μl), and pyridine (50 μl). Tube was put on ice for 5 min and 100 μl NaOH (1 M), 30 μl methyl-chloroformate were added. After overtaxing for 20 s, add 300 μl of tert-butyl methyl ether (MTBE) to the tube and vortex again. The samples were then centrifuged at 10,000 × g for 5 min. The upper layer was transferred to GC-MS vials and analyzed with an Agilent 7890B GC system and 7000 Triple Quadrupole GC-MS system. Data were collected and analyzed as previously described22.
Histones and histone-bound acetate extraction
Cells were collected and washed with cold PBS supplemented with sodium butyrate (10 mM) and nicotinamide (50 mM). Extraction of nuclei was followed as described previously23. Histones were separated with SDS-polyacrylamide gel electrophoresis (PAGE) and detected with acetyl-histone-specific antibodies. Isolated histones were placed at 95 °C overnight with 10 M NaOH, then added with hydrochloric acid for GC-MS.
Transfection of small interfering RNA and small hairpin RNA
To construct inducible knockdown cell lines, two different small hairpin RNA (shRNA) sequences target ACSS3 and a control shRNA were cloned into pLKO-Puro plasmids. The sequences of shRNA are as follows: shACSS3-1: 5′-GGGTTACCTAAGGGTGTGGAAtt-3′, shACSS3-2: 5′-GAAAAGATATAAATGCAAGAAtt-3′. Lentivirus production and infection were generated as previously described in 293T cells. 293T cells were seeded at 105 cells per well and were transfected with plasmids. Viral supernatant was harvested 48 h after transfection. Cells were infected for 12 h and cultured for another 24 h and collected. Same sequences were synthesized as small interfering RNA (siRNA) and siRNAs were transfected with Lipofectamine 2000 (Invitrogen). Cells were collected 2 days post siRNA transfection.
Tumor xenografts
All animals were maintained in specific pathogen-free conditions according to the recommendation of Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Ethics Committee of First Affiliated Hospital of Zhengzhou University (Approval number 2019-KY-174).
For tumor xenograft model, 2 × 106 indicated UMUC3 and T24 cells were injected subcutaneously on the right side of the dorsum (n = 6 for each group). The tumor diameters were measured each week. Tumor volumes were calculated with the formula: V = 1/2 × A × B2. The mice were sacrificed after 6 weeks and tumors were analyzed.
Cell growth curve and colony-formation assay
Two thousand indicated cells were seeded in each well of a six-well plate and cultured for 10 days. The colonies were fixed with 4% paraformaldehyde and stained with crystal violet.
RNA extraction and quantitative reverse-transcriptase PCR
Cell total RNA was extracted using RNeasy kit (Qiagen) as manufacturer’s instructions. First strand cDNA was synthesis using QuantiTect Reverse Transcription kit (Qiagen). qRT-PCR was performed using SBBR GREEN PCR Master Mix (Applied Biosystems) and detected with ABI 7900 System. Relative mRNA expression was calculated using the ΔΔCt equations. The primers used for ACSS genes detection by RT-qPCR are as following:
ACSS3:
Forward primer: 5′-TGGACCAAAACGCTGGAGAAC-3′
Reverse primer: 5′-ACGATCAACGGCATTGTAACA-3′
ACSS2:
Forward primer: 5′-AAAGGAGCAACTACCAACATCTG-3′
Reverse primer: 5′-GCTGAACTGACACACTTGGAC-3′
ACSS1:
Forward primer: 5′-CACAGGACAGACAACAAGGTC-3′
Reverse primer: 5′-CCTGGGTATGGACGATGCC-3′
Western blotting
Western blot experiments was performed as previously described24. Briefly, protein was extracted with RIPA buffer (Byotime) supplemented with protease inhibitor cocktail (Roche). Total protein concentration was quantified with BCA Protein Assay Kit (Thermo). An equal amount of total proteins were separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membrane (Millipore). Then block the membrane with 5% non-fat milk, incubated with primary antibodies at 4 °C overnight and followed by secondary antibody for 1 h. The primary antibodies used were as follows: anti-ACSS1 (Invitrogen, PA5-59392), anti-ACSS2 (Cell Signaling Technology, 3568 S), anti-ACSS3 (Invitrogen, PA5-80305), anti-Tubulin (Proteintech, 66031–1), anti-acetyl-histone H4 (Millipore, 06–598), anti-histone H3 (CST, 4499), anti-acetyl-histone H3 (Millipore, 06–599), and anti-histone H4 (CST, 13919).
Clinical samples and immunohistochemistry
This project was approved by the Ethics Committee of First Affiliated Hospital of Zhengzhou University (Approval number 2019-KY-174) and patients enrolled were informed of the scientific usage of the samples. Serial sections of formalin-fixed, paraffin-embedded tissues from 60 patients were obtained between September 2014 and March 2017. The routine immunohistochemistry were performed as previously described25. Slices were immunostained using anti-ACSS3 (Invitrogen, PA5-80305) antibody. DAB (diaminobenzidine) stainings were performed with an HRP kit (UltraTek, USA) according to the manufacturer’s instructions. Stained specimens were visualized using a general optical microscope with a camera (Axiovert 100 M, Germany).
Immunofluorescence and confocal microscopy
Cells were planted on the glass coverslips a day in advance. After transfected with indicated plasmids, the cells were fixed with 4% paraformaldehyde. The cell membranes were p permeabilized with NP40, then blocked with blocking buffer for 1 h. After incubation with primary antibody against Myc-tag (Proteintech, 16286-1-AP) for overnight, cells were washed with Phosphate-Buffered Saline (PBS)-Tween and incubated with Alexa Fluor 488 donkey anti-rabbit secondary antibody (Invitrogen, R37118). Mitochondrial were stained with MitoTracker (Invitrogen, M22426) for 30 min and nuclei were stained with DAPI (Beyotime, C1002) for 10 min. The slides were observed using the Lecia Tcs SP8 confocal microscope.
Spheroid growth assay
Spheroid formation assay were performed as previously described26, cells were digested with trypsin, and cultured with 2% matrigel in 96-well ultralow attachment plates (Costar). Spheroid formation was initiated by centrifugation at 850 × g for 10 min and the spheroids were analyzed at the indicated time points. Spheroids were treated with ethanol (control) or doxycycline 48 h. At indicated time points, images were captured by a general optical microscope with a camera (Axiovert 100 M, Germany). Spheroid volume was calculated based on image analysis by area determination using image J software.
Statistical analysis
Statistical analysis was performed with GraphPad Prism 5.0. (GraphPad Software). Experiments were performed at least in triplicates and error bars stands for SD. Two-tailed Student’s t-test was performed to determine the significance of paired data. One-way analysis of variance for quantitative data from grouped DataSets. P-value < 0.05 was considered significant. *P < 0.05; **P < 0.01, and ***P < 0.001. A log-rank test was performed to compare tumor-free survival. P-values less than 0.05 were considered statistically significant.
Results
Lipogenic AcCoA metabolism is altered in BLCA under stress
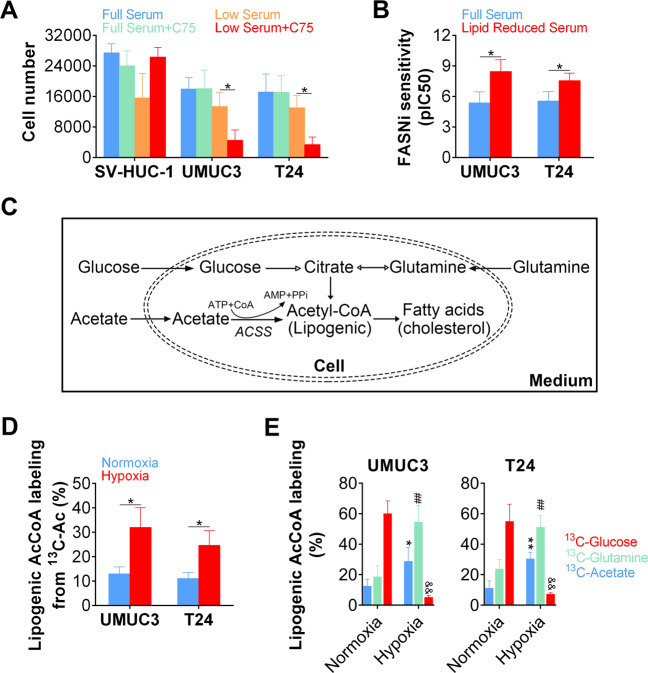
As FASN is closely related to the carcinogenesis, we therefore analyzed the role FASN in BLCA. We first analyzed the growth of BLCA cells treated with FASN inhibitor C75. Our data revealed that C75 did not affect the growth of normal bladder urothelial cells (SV-HUC-1) cultured with 10% serum (full serum) or 1% serum (low serum). Although the growth of BLCA cells (UMUC3 and T24) was not affected by C75 treatment in full serum, we observed that the growth of BLCA cells cultured in low serum was significantly inhibited by C75 administration (Fig. 1a). Moreover, we found that BLCA cells cultured with low serum were more sensitive to another FASN inhibitor, AZ22, as revealed by improvement of IC50 (Fig. 1b). As AcCoA is originated from alternative pathways (Fig. 1c) and hypoxic status mimics the in vivo lipid metabolic conditions5,6, we next evaluated the fatty acids metabolism in BLCA under hypoxia. We first analyzed the contribution of various precursors of lipogenic AcCoA and found that utilization of acetate (13C labeled) was dramatically increased in BLCA cells under hypoxia (Fig. 1d). Moreover, via isotopic labeling of glucose, glutamine, and acetate, our results revealed that the proportion of lipogenic AcCoA generated from glucose were largely reduced, whereas the proportion of lipogenic AcCoA generated from glutamine and acetate were increased significantly in BLCA cells (Fig. 1e).
Fig. 1. AcCoA is generated through alternative pathway in BLCA cells under metabolic stress.
a Sensitivity of normal bladder urothelial cells (SV-HUC-1) and bladder cancer cells to FASN inhibitor C75 in high (10%) or low (1%) serum levels. b Sensitivity of bladder cancer cells to FASN inhibitor AZ22 in high (10%) or low (1%) serum levels as revealed by pIC50. c Schematic of lipogenic AcCoA production. d Percentage of lipogenic AcCoA generation from acetate (labeled by 13C) under normoxia (21% O2) or hypoxia (1% O2) culture. e Sources of lipogenic AcCoA (labeled by 13C) under normoxia (21% O2) or hypoxia (1% O2) culture.
ACSS3 is responsible for lipogenic AcCoA synthesis in BLCA cells under metabolic stress
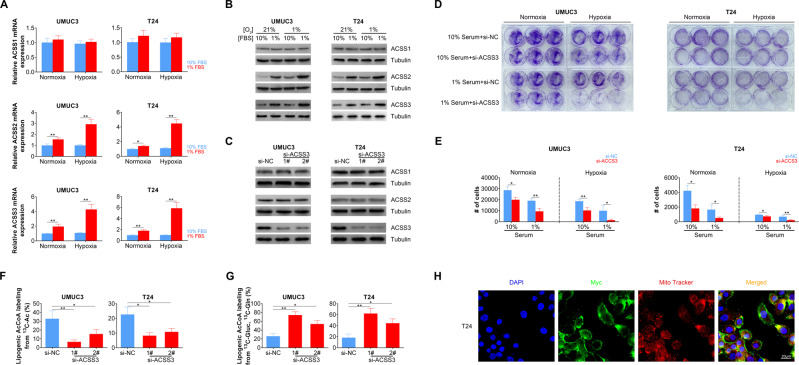
Previous study demonstrated that ACSS2 participated in lipid metabolism in breast cancer cells under hypoxic conditions5. Here we analyzed the role another ACSS family member plays, ACSS3, in BLCA. We first analyzed the expression levels of ACSS3 cultured with 10% serum or 1% serum under normal oxygen concentration, and found that the expression of all three members of ACSS family were not dramatically changed (Fig. 2a, b). However, when BLCA cells were cultured under hypoxia condition, the expression level of ACSS2 and ACSS3 in BLCA cells cultured with 1% serum were significantly increased in comparison with the expression level in BLCA cells cultured with 10% serum (Fig. 2a, b), indicating that metabolic stress, especially hypoxia, triggered the expression of ACSS2 and ACSS3 in BLCA cells. As ACSS2 has been reported to response to metabolic stress, we focused on the function of ACSS3. We utilized siRNA to downregulated ACSS3 expression in BLCA cells and both of the two siRNAs achieved considerable knockdown efficiency (Fig. 2c). Meanwhile, the protein level of ACSS1 and ACSS2 were not changed when ACSS3 was knocked down by siRNA (Fig. 2c). Colony-formation experiments revealed that downregulation of ACSS3 attenuated the number of colonies formed in both normoxia and hypoxia conditions in UMUC3 and T24 cells (Fig. 2d). Cell number counting revealed that ACSS3 knockdown significantly inhibited cell proliferation in both normoxia and hypoxia conditions (Fig. 2e), suggesting that ACSS3 was responsible for BLCA cell growth under metabolic stress. Importantly, a significant reduction was found in the lipogenic AcCoA from 13C-acetate (Fig. 2f). Meanwhile, labeling from 13C-glucose and 13C-glutamine were both increased (Fig. 2g) under indicated conditions. In order to clarify the localization of ACSS3, ACSS3-Myc plasmid was constructed and expressed in T24 cells. The expression of ACSS3 was detected through immunofluorescence staining assay using antibody against Myc-tag. As shown in Fig. 2h, myc signals were observed co-localized with MitoTracker, suggesting the colocalization with mitochondria.
Fig. 2. ACSS3 is induced under metabolic stress and facilitate BLCA cell growth.
a ACSS1, ACSS2, and ACSS3 mRNA expression levels in BLCA cells under normoxia (21% O2) or hypoxia (1% O2) culture with 10% or 1% serum levels. b ACSS1, ACSS2, and ACSS3 protein expression levels in BLCA cells under normoxia (21% O2) or hypoxia (1% O2) culture with 10% or 1% serum levels. c ACSS1, ACSS2, and ACSS3 protein expression levels in BLCA cells transfected with control of ACSS3 siRNA. d Colony formed from BLCA cells transfected with control of ACSS3 siRNA under indicated culture situations. e Cell number of BLCA cells transfected with control of ACSS3 siRNA under indicated culture situations after 48 h incubation. f Percentage of lipogenic AcCoA generation from acetate (labeled by 13C) in BLCA cells transfected with control of ACSS3 siRNA under hypoxia (1% O2) culture. g Percentage of lipogenic AcCoA generation from glutamine or glucose (labeled by 13C) in BLCA cells transfected with control of ACSS3 siRNA under hypoxia (1% O2) culture. h Subcellular localization of ACSS3-Myc expressed in T24 cells. ACSS3 was detected with anti-myc antibody. Mitochondria was stained with MitoTracker.
ACSS3 is crucial for histone acetylation in BLCA cells
To determine the net uptake of acetate by cancer cells, a delicate measurement has been developed previously5 and we utilized this measurement to study the role ACSS3 in lipid metabolism in BLCA cancer cells. Our results showed that ACSS3 knockdown attenuated acetate consumption from the medium (Fig. 3a). Meanwhile, more endogenous acetate was released to the medium suggesting that ACSS3 (Fig. 3a) is required for the utilization of endogenous acetate in cancer cells. We next calculated the ratio of acetate release and consumption in BLCA cells transfected with control siRNA or ACSS3 siRNA and found that this ratio was markedly increased in BLCA cells transfected with ACSS3 siRNA (Fig. 3b), further suggesting that ACSS3 facilitated acetate metabolism in cancer cells. We then studied the role of ACSS3 under metabolic stress. Interestingly, under normal culture status, downregulation of ACSS3 did not affect the levels of acetylated histone H3 and H4 (Fig. 3c). However, under metabolic stress, ACSS3 was required for maintaining the levels of acetylated histone H3 and H4 (Fig. 3c). Moreover, our data also revealed that metabolic stress induced a significant increase in histone acetylation in BLCA cells, which was aborted in ACSS3 knockdown cells (Fig. 3c), further indicating that ACSS3 is crucial for histone acetylation in BLCA cells.
Fig. 3. ACSS3 is required for acetate utilization and histone acetylation.
a Consumption of medium acetate and release of cytoplasmic acetate in BLCA cells transfected with control of ACSS3 siRNA under hypoxia (1% O2) culture. b Ratio of acetate release and consumption in BLCA cells transfected with control of ACSS3 siRNA under hypoxia (1% O2) culture. c Level of histone H3, acetylated histone H3, histone H4, and acetylated histone H4 in BLCA cells cultured in indicated conditions. Right panel are quantified results of protein expression levels.
ACSS3 silencing inhibits spheroid and tumor growth
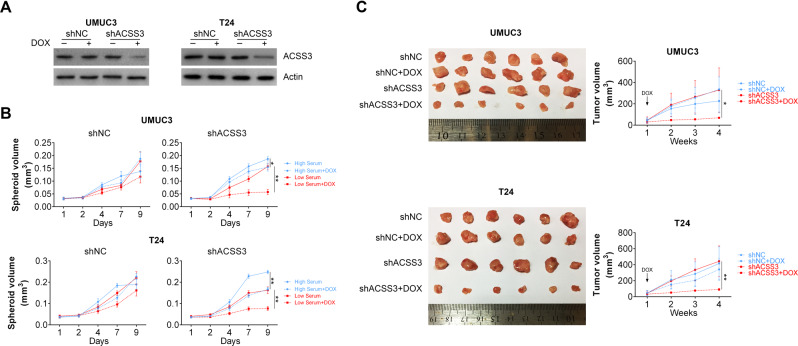
To further analyze the role ACSS3 in BCLA, we generated stable cell lines in which ACSS3 could be silenced by doxycycline administration. We first validated that doxycycline was able to induce ACSS3 silence in the stable cell lines (Fig. 4a). We then cultured these cells as spheroids in 10% or 1% serum with or without the administration of doxycycline. Importantly, our results showed that only in low serum culture, downregulation of ACSS3 resulted in significant impairment of spheroids formation in UMUC3 and T24 cells (Fig. 4b). In addition, BLCA cells were inoculated subcutaneously into nude mice and half of the mice were treated with doxycycline on the 7th day post inoculation. The tumor growth of the xenografts formed by BLCA cells expressing ACSS3 shRNA was significantly inhibited in comparison with xenografts formed by BLCA cells expressing control shRNA or xenografts formed by BLCA cells without doxycycline treatment (Fig. 4c). Hence, our data indicated an oncogenic role of ACSS3 in BLCA.
Fig. 4. ACSS3 is required for spheroid and xenografts formation.
a Induction of ACSS3 knockdown by doxycycline in BLCA cells. b Spheroid formation in BLCA cells stably transfected with control of ACSS3 shRNA with or without doxycycline administration under high serum or low serum culture. c Xenografts formed by BLCA cells stably transfected with control of ACSS3 shRNA with or without doxycycline administration under high serum or low serum culture. Right panels are measurements of tumor volumes at indicated time points.
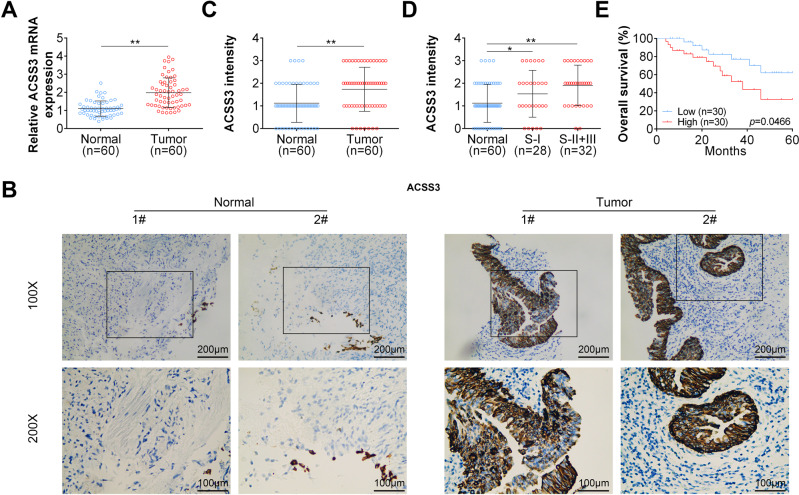
We then analyzed the expression pattern of ACSS3 in clinical samples (Table 1). We found that the both mRNA and protein levels of ACSS3 were significantly upregulated in BLCA samples compared with adjacent controls (Fig. 5a, b). We analyzed the staining intensity of the immunochemistry staining of ACSS3 and found that the staining intensity of ACSS3 was significantly higher in samples from BLCA patients in both stage I and stage II plus stage III (Fig. 5c, d). Moreover, BLCA patients with lower ACSS3 expression were entitled with longer overall survival time (Fig. 5e). Taken together, our analysis illustrated that the expression of ACSS3 was statistically elevated in BLCA patients and high ACSS3 level predicted poor outcome of BCLA prognosis.
Table 1.
Correlation between ACSS3 expression and clinicopathological characteristics of bladder cancer patients.
| Parameters | Total | ACSS3 expression | P-value | |
|---|---|---|---|---|
| Low (n = 30) | High (n = 30) | |||
| Gender | 0.194 | |||
| Male | 33 | 14 | 19 | |
| Female | 27 | 16 | 11 | |
| Age (years) | 0.519 | |||
| <59 | 12 | 7 | 5 | |
| ≥59 | 48 | 23 | 25 | |
| Tumor stage | 0.793 | |||
| Ta,T1 | 11 | 5 | 6 | |
| T2–T4 | 49 | 25 | 24 | |
| TNM stage | 0.010* | |||
| S-I | 28 | 19 | 9 | |
| S-II + III | 32 | 11 | 21 | |
| Grade | 0.004* | |||
| G1 | 33 | 22 | 11 | |
| G2–G3 | 27 | 8 | 19 | |
| Tumor size | 0.781 | |||
| <3 cm | 19 | 9 | 10 | |
| ≥3 cm | 41 | 21 | 20 | |
| Lymph node metastasis | 0.000* | |||
| Absent | 32 | 9 | 23 | |
| Present | 28 | 21 | 7 | |
| Distant metastasis | 0.028* | |||
| Absent | 48 | 21 | 27 | |
| Present | 12 | 9 | 3 | |
Fig. 5. ACSS3 is elevated in BLCA patient samples.
a ACSS3 mRNA expression levels in BLCA patient samples and adjacent normal tissues. b Representative immunohistochemistry images of ACSS3 staining in BLCA patient samples and adjacent normal tissues. c ACSS3 protein intensities in BLCA patient samples and adjacent normal tissues. d ACSS3 protein intensities in stage I or stage II + III BLCA patient samples and adjacent normal tissues. e Overall survival of BLCA patients with higher or low ACSS3 mRNA expression levels.
Discussions
Cancer cells adapt to nutrient-deprived tumor microenvironment during progression via adjusting the level and function of metabolic enzymes26. Several metabolic altered pathways are involved in bladder tumorigenesis27. With the advancement of multi-omics analysis, de novo identification of oncogenes have been carried out in various types of cancers, including BLCA28. A most recent multi-omics study has shown that fatty acid metabolism plays a central role in facilitating the progress of BLCA29. Moreover, another report showed that fibroblast growth factor receptor 3 promotes the cleavage and activation of sterol regulatory element-binding protein 1, which is the key regulator of lipogenesis, which in turn facilitates the BLCA cell growth30. Here we first analyzed the utilization of acetate in BLCA confronting environmental stress and found that BLCA cell lines altered the way of generation, which is in line with previous reports showing that cancer cells adapted to nutrient-deprived tumor microenvironment5,17,31.
As it has been shown that ACSS family members were critical for alternative utilization of acetate in cancer cells5,26, we suspected the role ACSS3 in BLCA cells under metabolic stress. As a member of ACSS enzyme family, ACSS3 is the least studied. ACSS1 is the main mitochondrial enzyme and ACSS2 mainly functions in nucleus and cytosol. Here we demonstrated that in BLCA cells, ACSS3 is localized in mitochondria and functions as an oncogene to promote lipid synthesis and histone acetylation through the use of acetate.
Importantly, our data showed that the expression level of ACSS3 were significantly upregulated in BLCA cell lines upon metabolic stress, suggesting an oncogenic role of ACSS3 in BLCA. Downregulation of ACSS3 significantly suppressed the growth of BLCA cells in vitro and in vivo. Indeed, previous report uncovered a higher expression of ACSS3 in tumor comparing to normal parental lesions and downregulation of ACSS3 inhibits progression of gastric cancer13. In hepatocellular carcinoma, ACSS3 was expressed in mitochondria and specifically recognized a subtype of hepatocellular carcinoma (namely iHCC2, which represents poor clinical survival rate)9, suggesting that the presence of ACSS3 may be noxious in hepatocellular carcinoma as well. Taken together, these data indicated an oncogenic role of ACSS3 even under routine culture conditions. Interestingly, methylation level of ACSS3 was found to be associated with prognosis of neuroblastoma patients32. It is of special significance to further determine the role of methylated ACSS3 in the progression of BLCA.
Interestingly, we found an upregulation of histone acetylation under low serum and hypoxic conditions in BLCA, which is consistent with the notion that fatty acid biosynthesis is significantly elevated in hypoxia and low serum conditions in breast cancer cell lines5, and the majority of histone acetylation is derived from fatty acid carbon33. Importantly, our results showed that the upregulated acetylation of histone H3 and histone H4 were significantly impaired in BLCA cells with downregulated ACCS3 level. Our data revealed that ACSS3 is required for the utilization of both environmental and intracellular acetate, which is identical to the role of ACSS2 in breast cancer cells5. As ACSS3 is mainly localized in mitochondria, the mechanisms of using acetate to promote histone acetylation should be different from that of ACSS2, which could directly converts the acetate to nuclear acetyl-CoA. The possible cause might be recycling of endogenous acetate, which needs further exploration.
In addition to the data illustrating that inhibition of ACSS3 suppressed xenograft formation in mice, we also analyzed clinical samples and found that the expression of ACSS3 was significantly increased in BLCA samples compared with adjacent normal tissues. Moreover, patients with higher ACSS3 suffered from poorer overall survival. As the correlation of high ACSS3 expression with poor survival can be validated in multiple online database (data not shown), it is of huge potential to further analysis of the relationship between ACSS3 expression and overall BLCA patient’s survival has great potential. Therefore, ACSS3 might be a promising prognosis marker for BLCA patients.
Taken together, our data demonstrated that fatty acid metabolism is altered in BLCA cells confronting nutrient stress in an ACSS-dependent manner. More importantly, ACSS3 facilitates the acetate utilization and histone acetylation in metabolic stressed BLCA cells and promotes BLCA cell growth. Our data revealed an oncogenic role of ACSS3 and fatty acid in BLCA, and provide a promising target in metabolic pathway for BLCA treatment.
Funding
This work was supported by Natural Science Foundation of China (81100464, 81200883, and 81570685), the Natural Science Foundation of Henan (2018061), the Medical Key Technologies R&D Program of Henan (201702031 and 201702015), and Key Scientific Research Foundation of the Higher Education of Henan Province, China (Grant Numbers 20A320032 and 20A320044).
Author contributions
C.H.G. and J.H.Z. conceived and designed the experiments. H.J.D. analyzed and interpreted the results of the experiments. Z.P.F. and X.W.H. performed the experiments
Data availability
All data generated or analyzed during this study are included in this published article.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The animal use protocol listed below has been reviewed and approved by the Animal Ethical and Welfare Committee. Approval number 2019-KY-174.
Footnotes
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41389-023-00484-0
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jianhao Zhang, Hongjian Duan
Change history
7/26/2023
This article has been retracted. Please see the Retraction Notice for more detail: 10.1038/s41389-023-00484-0
References
- 1.Corbet C, Feron O. Tumour acidosis: from the passenger to the driver’s seat. Nat. Rev. Cancer. 2017;17:577–593. doi: 10.1038/nrc.2017.77. [DOI] [PubMed] [Google Scholar]
- 2.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 5.Bulusu V, et al. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schug ZT, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun M, et al. The importance of acetyl coenzyme A synthetase for 11C-acetate uptake and cell survival in hepatocellular carcinoma. J. Nucl. Med. 2009;50:1222–1228. doi: 10.2967/jnumed.109.062703. [DOI] [PubMed] [Google Scholar]
- 8.Lakhter AJ, et al. Glucose-independent acetate metabolism promotes melanoma cell survival and tumor growth. J. Biol. Chem. 2016;291:21869–21879. doi: 10.1074/jbc.M115.712166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidkhori G, et al. Metabolic network-based stratification of hepatocellular carcinoma reveals three distinct tumor subtypes. Proc. Natl Acad. Sci. USA. 2018;115:E11874–E83. doi: 10.1073/pnas.1807305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X, et al. Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 2016;7:11960. doi: 10.1038/ncomms11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee MY, et al. Reprogrammed lipid metabolism in bladder cancer with cisplatin resistance. Oncotarget. 2018;9:13231–13243. doi: 10.18632/oncotarget.24229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang WC, et al. Mitochondrial acetyl-CoA synthetase 3 is biosignature of gastric cancer progression. Cancer Med. 2018;7:1240–1252. doi: 10.1002/cam4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rohrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016;16:732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 15.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc. Natl Acad. Sci. USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab. 2014;2:23. doi: 10.1186/2049-3002-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knowles MA, Hurst CD. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat. Rev. Cancer. 2015;15:25–41. doi: 10.1038/nrc3817. [DOI] [PubMed] [Google Scholar]
- 19.Stenzl A, et al. Treatment of muscle-invasive and metastatic bladder cancer: update of the EAU guidelines. Eur. Urol. 2011;59:1009–1018. doi: 10.1016/j.eururo.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 20.Schwentner C, Stenzl A, Gakis G. Monitoring high-risk bladder cancer. Curr. Opin. Urol. 2012;22:421–426. doi: 10.1097/MOU.0b013e3283555d04. [DOI] [PubMed] [Google Scholar]
- 21.Tumanov S, Bulusu V, Kamphorst JJ. Analysis of fatty acid metabolism using stable isotope tracers and mass spectrometry. Methods Enzymol. 2015;561:197–217. doi: 10.1016/bs.mie.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 22.Tumanov S, Bulusu V, Gottlieb E, Kamphorst JJ. A rapid method for quantifying free and bound acetate based on alkylation and GC-MS analysis. Cancer Metab. 2016;4:17. doi: 10.1186/s40170-016-0157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X, et al. USP17 suppresses tumorigenesis and tumor growth through deubiquitinating AEP. Int. J. Biol. Sci. 2019;15:738–748. doi: 10.7150/ijbs.30106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen X, et al. Bcl-3 regulates TGFbeta signaling by stabilizing Smad3 during breast cancer pulmonary metastasis. Cell Death Dis. 2016;7:e2508. doi: 10.1038/cddis.2016.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cairns RA, Mak TW. An alternative sugar fuels AML. Cancer Cell. 2016;30:660–662. doi: 10.1016/j.ccell.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Massari F, et al. Metabolic phenotype of bladder cancer. Cancer Treat. Rev. 2016;45:46–57. doi: 10.1016/j.ctrv.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Li R, Choi W, Ferguson Rd JE, Metcalfe MJ, Kamat AM. New discoveries in the molecular landscape of bladder cancer. F1000Res. 2016;5:2875. doi: 10.12688/f1000research.10031.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vantaku V, et al. Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin. Cancer Res. 2019;25:3689–3701. doi: 10.1158/1078-0432.CCR-18-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du X, et al. FGFR3 stimulates stearoyl CoA desaturase 1 activity to promote bladder tumor growth. Cancer Res. 2012;72:5843–5855. doi: 10.1158/0008-5472.CAN-12-1329. [DOI] [PubMed] [Google Scholar]
- 31.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2011;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decock A, et al. Genome-wide promoter methylation analysis in neuroblastoma identifies prognostic methylation biomarkers. Genome Biol. 2012;13:R95. doi: 10.1186/gb-2012-13-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McDonnell E, et al. Lipids reprogram metabolism to become a major carbon source for histone acetylation. Cell Rep. 2016;17:1463–1472. doi: 10.1016/j.celrep.2016.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.