Abstract
Work on the filamentous fungus Neurospora crassa has contributed to or pioneered many aspects of research on circadian clock mechanism, a process that is functionally conserved across eukaryotes. Biochemical assays of the fungal circadian clock typically involve growth in liquid medium where Neurospora forms a spherical ball of submerged mycelium. Here, we revive a method for dispersed growth of Neurospora in batch culture using polyacrylic acid as an additive to the medium. We demonstrate that dispersed growth cultures utilize more carbon than mycelial balls, but nonetheless retain a functional circadian clock. This culturing method is suited for use in circadian experiments where uniform exposure to nutrients and/or increased biomass is required.
Keywords: circadian clock, glucose utilization, dispersed growth, Neurospora crassa
1. Introduction
Organisms use their circadian clock to anticipate daily changes in the environment. Neurospora crassa is a primary model organism for clock biology because the core feedback loop of the circadian clock is functionally conserved from fungi to animals (Loros, 2019). Neurospora became a leading clock model due to its genetic tractability and ease of circadian readout. The race tube assay, together with a ras-1 mutant background (which facilitates formation of distinct bands of conidiophores once per subjective night), is still widely used to determine circadian period length (Belden et al., 2007; Sargent et al., 1966). More recently, a high throughput assay for circadian period and phase determination was developed utilizing a core clock reporter gene driving luciferase (Gooch et al., 2008). The majority of assays used to determine circadian period length involve fungal growth on a solid substrate.
On the other hand, the bulk of circadian biochemistry experiments are performed by culturing mycelial discs in liquid (Aronson et al., 1994; Nakashima, 1981). The circadian clock runs normally in these conditions for up to 3 cycles, yet a limitation of liquid culturing is that the exposed edges of the ball arising from the disc have direct access to nutrients whereas the center can become nutrient limited leading to spatial heterogeneity. Neurospora mycelial balls grow uniformly from exposed edges (Emerson, 1950), and gas exchange is thought to reach the center of the mycelium (Marshall and Alexander, 1960). However, the density and morphology of the fungal ball’s center depends on the growth medium (Burkholder and Sinnott, 1945), and growth regimes likely exist where the center of the Neurospora ball is oxygen limited. The interplay of the circadian clock and metabolism is currently of great interest to the field (Reinke and Asher, 2019), and there is much reason to believe that nutritional differences may influence the core clock itself as well as the spectrum of clock-regulated processes. Because of this, a fungal growth regime in which mycelia have more uniform access to nutrients is needed for research to answer questions about cross-talk between metabolism and the clock.
Neurospora crassa has been successfully cultured in semi-dispersed growth in bioreactors (Cockrell et al., 2015; Havlik et al., 2017; Tralau et al., 2007), but there has been debate over whether the circadian clock functions under high mixing speeds. Germinated conidia have normal circadian function prior to cell fusion (Lindgren, 1994) and prior to fungal ball formation (Baker et al., 2009), suggesting that Neurospora does not require a highly connected mycelial network to run the clock. Dispersed growth in batch culture can be achieved by adding negatively charged polymers to the medium (Metz and Kossen, 1977). Data from Aspergillus niger suggest that the anionic polymers interact with fungal cell walls, repel other germinated conidia, and prevent fusion (Elmayergi et al., 1973; Jones et al., 1988). We have adapted this method for Neurospora dispersed growth. Our results are consistent with high metabolic output of dispersed cultures as observed in Aspergillus (Elmayergi et al., 1973) and with a functional circadian clock (Tralau et al., 2007). In future work, this culturing method will be deployed to validate differential metabolic programs in the subjective day versus night (Hurley et al., 2014, 2018) and to identify conserved rhythmic metabolites across eukaryotes (Krishnaiah et al., 2017).
2. Materials and Methods
2.1. Neurospora crassa strains and growth conditions
The strain used in all analyses was derived from a wild-type background (FGSC2489) transformed with a core clock transcriptional reporter for frequency (Hurley et al., 2014; Larrondo et al., 2015) at the csr-1 locus (Bardiya and Shiu, 2007). Bird medium (Metzenberg, 2004) with 1.3% or 1.8% glucose w/v as the carbon source was used. Where indicated, polyacrylic acid (Sigma # 181285; also referred to in the literature as “Junlon”) was included in Bird medium at 0.2% w/v to induce dispersed growth. Polyacrylic acid (PA) was fully dissolved in ddH2O under mixing and low heat, and pH was then adjusted to 6.5 using 10N sodium hydroxide. This solution was autoclaved before combining with filter sterilized 20X stocks of Bird Solutions 1 and 2 to 1X final concentrations (Metzenberg, 2004). Bird + PA medium can be filter sterilized after autoclaving, but the autoclave step is important to achieve dispersed growth (data not shown). Glassware was treated with Sigmacote (Sigma # SL2) per the manufacturer’s instructions to prevent mycelial growth above the liquid level (chips in older glassware can still seed fungal ball formation). Baffled flasks increased chaotic mixing and improved fully dispersed growth without ball formation. Dispersed growth was optimal at a 0.15 – 0.2 ratio of liquid medium to total flask volume, which could be due to increased aeration (Emerson, 1950) and/or chaotic mixing.
For liquid culturing, fresh conidia were isolated from 6–8 day old minimal medium slants and washed in sterile water. The concentration of conidia was determined by counting on a hemocytometer, and the inoculum at time 0 for each flask was 1x106 conidia / ml. Cultures were incubated in the light or dark at 25°C as indicated. Traditional liquid culturing (Chen et al., 2009; Nakashima, 1981) was performed in one assay for comparison to dispersed growth. 1x106 total conidia were inoculated into a Petri plate containing 25 ml of Bird 1.8% glucose medium. A mycelial mat formed after about 24 hours of stationary growth at 25°C in constant light. A disc was cut from this mycelial mat using a sterilized 6 mm diameter cork borer and transferred into a flask containing fresh Bird 1.8% glucose medium.
Glucose levels were determined from spent media using the Glucose (GO) Assay Kit (Sigma # GAGO-20) using to the standard curve method in the manufacturer’s instructions. Reaction volumes were decreased by a factor of 5 to accommodate Eppendorf tube volumes. Spent media was either measured immediately after isolation or flash frozen and thawed on ice before taking measurements (with equivalent results, data not shown).
Biomass was determined after isolating fresh Neurospora tissue by vacuum filtration, squeezing out excess moisture, and measuring the tissue mass (Mettler Toledo PB303-S).
2.2. RNA isolation and gene expression
Neurospora tissue was harvested by vacuum filtration and flash frozen in liquid nitrogen. Tissue was ground in liquid nitrogen with a mortar and pestle, and total RNA was extracted by adding 1 ml of TRIzol (Invitrogen # 15596026) directly to ~ 200-300 μl worth of ground tissue and processing as described previously (Chen et al., 2009). 3 μg of total RNA was used to synthesize cDNA with the Superscript IV First-Strand synthesis kit (Invitrogen # 18091050). RT-PCR was performed using SYBR green master mix (Qiagen # 204054) and a StepOne Plus Real-Time PCR System (Applied Biosystems) with run parameters as described (Hurley et al., 2015). Ct values were determined using StepOne software version 2.3 (Life Technologies) and normalized to the crp-43 gene (ΔCt), which does not cycle in expression level over circadian time (Hurley et al., 2015). The ΔΔCt method was used to determine mRNA levels relative to a reference time point. Primer sequences for the three sugar transporter genes were taken from a previous study (Wang et al., 2017).
Relevant RT-PCR primer sequences:
| CRP-43 (NCU08964): | 5′ CTGTCCGTACTCGTGACTCC 3′ and 5′ ACCATCGATGAGGAGCTTGC 3′ |
| GLT-1 (NCU01633): | 5′ CTACGTCAGTGGTCAAATTTCTG 3′ and 5′ CAGGGAGTAGGGCGACAATAG 3′ |
| HGT-1 (NCU10021): | 5′ AGATTTGCCCTCGCAAGGTC 3′ and 5′ TGGCGATAGGAATACGGTAAGC 3′ |
| HGT-2 (NCU04963): | 5′ TTGGGTATGACACTGGCACT 3′ and 5′ AGCAGACAGAATGGAGACGA 3′ |
2.3. Luciferase reporter detection with a CCD camera
Liquid cultures were prepared as in Section 2.1 with 25 μM luciferin (GoldBio # 115144-35-9) added to the medium. Dispersed conidia were germinated and entrained in constant light at 25°C and 100-125 RPM shaking for at least 6 hours. Cultures were then placed in individual containers lined with 6-ply black railroad board (to prevent light contamination between cultures), and flask openings were covered in plastic wrap with a ~ 3 mm hole for aeration. Flasks were transferred onto a shaker at 100 RPM inside a Percival incubator under constant darkness and 25°C temperature. Luminescence was recorded using a liquid nitrogen cooled CCD camera (Princeton Instruments & Roper Scientific, VersArray LN 1300 B) housed inside the incubator. Light signal was acquired for 15 minutes every hour using WinView software (Princeton Instruments, 32-bit version 2.5.25.2) to compile one frame per hour up to 73 hours.
For glucose spike experiments, cultures were prepared for luciferase detection as described above. Flasks were then removed at either 19.9 hours or 31.9 hours after transfer under the CCD camera. Glucose or water was added to each culture as indicated for a total of approximately 1 minute of manipulations under dim red lights, which do not reset the Neurospora clock (Chen et al., 2009). The glucose spike was 300 μl of a 40% w/v glucose stock solution (e.g. 6 mg/ml added), and mock treatment was 300 μl of sterile water.
Moving flasks generated blurred images of luminescent fungal cultures through the flask tops. The average intensity of each flask’s circular trajectory was determined using a custom ImageJ Macro (Larrondo et al., 2015), and background correction was performed for each frame. Background-corrected luminescence traces were then smoothed using a weighted sliding window over the center point (e.g. weighting of [1, 2, 1]) to de-noise). Linear trends were removed by fitting the smoothed data to a first degree polynomial (using the polyfit function in the R statistical computing environment) and subtracting the trend. Smoothed, detrended data were used to calculate peak phase for each trace. Periods were computed using the MESA (maximum entropy spectral analysis) method from custom MATLAB software (Dowse, 2013).
In order to compare peak times of the circadian luciferase reporter between experiments, a circadian time (abbreviated CT) axis was calculated for each experiment. The CT scale is conventionally used to compare rhythms with different period lengths on a common 24-hour timeline (Loros et al., 1989). CT values were calculated for Neurospora using the formula: [ (hours in constant dark / free running period length) x 24 + 12] mod 24. This CT formula applies to circadian experiments where experimental time 0 was a shift from light to dark conditions (i.e. CT12 is dusk).
2.4. Microscopy
Hyphae were sampled from a dispersed growth culture (25°C, constant light, 100 RPM shaking). The sample was placed on a polylysine-coated slide with a glass coverslip. Brightfield images were acquired using a Nikon Ti-E light microscope, equipped with a coolLED white light source, Andor Zyla 4.2 scMOS camera, and 10X CFI Plan Fluor objective. A scale bar was added using Fiji.
2.5. Data visualization
All figures were plotted in R, output as scalable vector graphics, and formatted using Inkscape. Data represent the mean of at least three biological replicates with standard deviation error bars unless otherwise indicated.
3. Results
3.1. Dispersed growth is achieved in N. crassa with the addition of anionic polymers
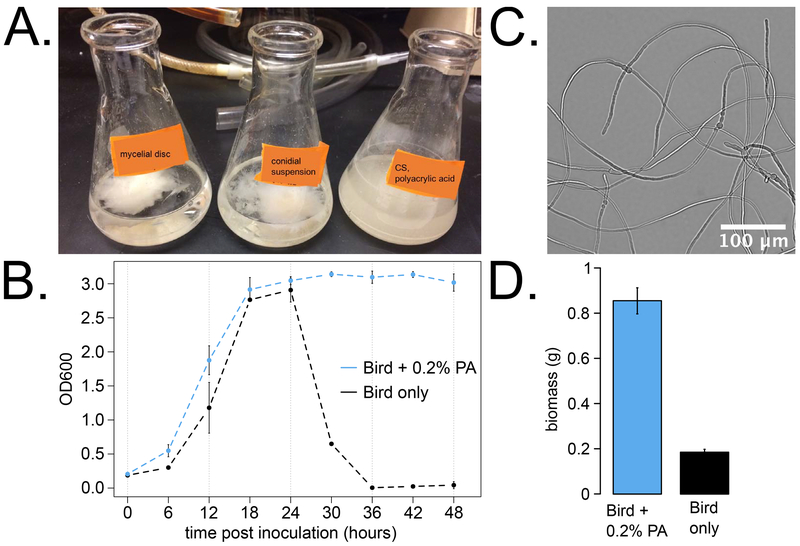
Negatively charged polyacrylic acid (PA) added into Bird medium induced dispersed growth in batch cultures of Neurospora, as described previously for Aspergillus and other fungi (Elmayergi et al., 1973; Jones et al., 1988). By eye, dispersed cultures grow as an opaque suspension in the medium compared to a fungal ball (Fig 1A). This qualitative observation was quantified using OD600 as a measure of medium opacity, and dispersed growth cultures maintained high OD opacity readings over 2 days of growth (Fig 1B). In standard Bird medium, inoculated conidia germinate, fuse, and coalesce into a fungal mass after approximately 24 hours of growth, which is indicated by low OD readings from clear medium (Fig 1B). Inoculating a suspension of conidia directly into liquid culture leads to formation of a mycelial ball having morphology similar to that of traditional mycelial disc cultures after about 24 hours.
Fig 1. Polyacrylic acid (PA) induced Neurospora dispersed growth.
Image of flasks at 3.5 days after inoculation with a 6 mm disc cut from a mycelial mat (A, left), with conidia into Bird 1.8% glucose medium (A, middle), or with conidia into Bird + PA medium (A, right). OD600 readings were recorded in 6 hour intervals over 2 days for dispersed growth cultures with PA versus fungal ball cultures grown in standard Bird medium (B, N = 1 – 6 replicates per time point). Hyphae from a dispersed culture were imaged at 22 hours post inoculation (C), at a time point when standard cultures without PA have begun to form a fungal ball (B). Biomass yield was quantified 2 days after inoculation with conidia into 50 ml of Bird 1.8% glucose + PA medium (D, left) or standard Bird 1.8% (D, right).
We observed that hyphae from dispersed growth cultures are in close proximity to each other but do not appear to fuse or branch frequently (Fig 1C). These microscopy observations are consistent with dispersed growth of Streptomyces in the presence of polyacrylic acid (Hobbs et al., 1989), Rhizopus arrhizus grown with polymers (Byrne and Ward, 1987), and Aspergillus niger with polyacrylic acid (Trinci, 1983).
Biomass of dispersed cultures was tracked for up to 3 days post inoculation, and the average yield of vacuum-filtered dry tissue was 0.51 ± 0.16 mg of per ml of liquid medium used per hour grown (N = 5). Biomass yield was directly compared between dispersed (+ PA) and fungal ball (− PA) cultures grown in constant light at 25°C for 2 days (N = 3 replicates). Dispersed growth yielded about four times more biomass than fungal ball cultures (Fig 1D). Given that Neurospora consumes 1 mg of glucose or more per mg of dry weight accumulated (Colvin et al., 1973), we next asked if dispersed cultures utilized more glucose from the medium.
3.2. Higher total glucose consumption in Neurospora dispersed growth cultures
The metabolic activity of dispersed cultures of A. niger was found to be higher than fungal ball controls by CO2 production and by glucose utilization (Elmayergi et al., 1973). Three models were proposed to explain these results (Metz and Kossen, 1977): 1) an equivalent mass of fungal ball is less metabolically active than dispersed hyphae due to nutrient and/or gas exchange limitations, 2) more conidia germinate in the presence of anionic polymers which leads to more metabolically active hyphae in dispersed cultures, 3) anionic polymers interact with the fungal cell wall and increase the uptake rate of potassium ions (Elmayergi et al., 1973). We posited that Neurospora dispersed growth would also be more metabolically active than mycelial ball growth.
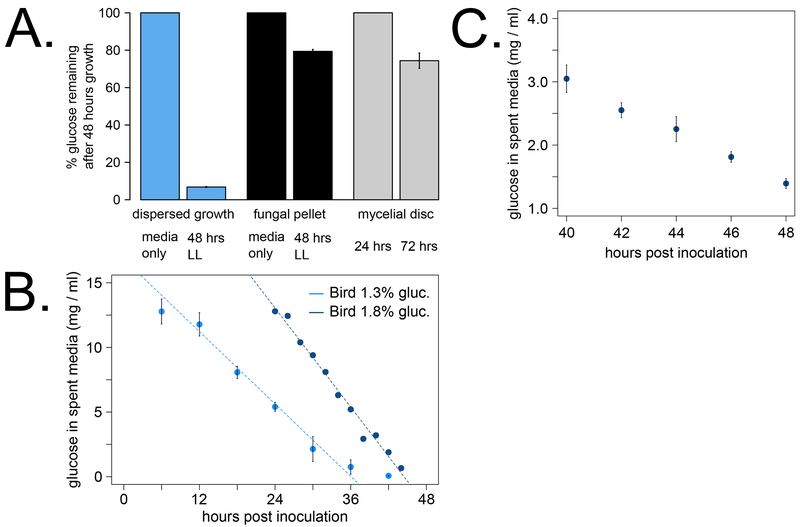
We found that dispersed cultures utilized more than 90% of the glucose after 2 days in culture (Fig 2A). On the other hand, fungal balls arising from conidia inoculum or from mycelial discs used only about 25% of the available glucose in 2 days (Fig 2A). The rate of glucose consumption in dispersed cultures was linear after about 12 hours growth, presumably due to a lag time during conidial germination (Fig 2B, R2 > 0.98). We finely mapped the glucose utilization rate of dispersed cultures to reach approximately 1 mg/ml glucose left in the medium at 2 days of growth (Fig 2C), which is an endpoint for many liquid culture experiments including non-circadian assays. Taken together, our data support previous findings that dispersed growth has significantly more active central carbon metabolism than a fungal ball and/or produces more tissue mass with active metabolism.
Fig 2. High glucose consumption of dispersed growth cultures.
Glucose levels from spent media were measured using a colorimetric assay to compare dispersed growth (A, blue bars) versus fungal balls (A, black bars) after 2 days grown in constant light. Percentages were calculated relative to the Bird culture medium with (A, blue) or without (A, black) polyacrylic acid (N = 3 replicates). Glucose levels from spent media were also determined at 24 and 72 hours after 6 mm mycelial disc transfer into Bird 1.8% glucose medium (A, gray bars, N = 4). The glucose consumption rate of dispersed cultures was estimated by sampling spent media from the same culture at regular intervals, measuring glucose levels, and fitting a linear model (B). In cultures starting at 1.3% glucose (N = 3), the slope of the linear fit from 12 – 36 hours was −0.467 ± 0.037 (dashed line, R2 = 0.981). In cultures starting at 1.8% glucose (N = 1), the slope of the linear fit from 24 – 44 hours was −0.638 ± 0.025 (dashed line, R2 = 0.986). Boxes below the x-axis indicate growth in constant light for 12 hours followed by transfer to darkness for the remainder of the experiment (B). The glucose consumption rate was finely sampled from 40 – 48 hours from dispersed growth cultures starting at 1.8% glucose (C, N = 3). The box below the x-axis denotes growth in constant light.
We next asked if dispersed growth cultures become starved for glucose as the primary carbon source. Sugar transport in Neurospora occurs through a low affinity, facilitated diffusion pathway in glucose replete conditions (System I) or through a high affinity, active transport mechanism that is induced upon glucose starvation (System II). Reported Km values for low affinity transport range from 8 mM (Scarborough, 1970) to 25 mM glucose (Schneider and Wiley, 1971). Thus, System I transport could continue to function down to 1 mg/ml (5.6 mM) of glucose in the environment (Fig 2C). High affinity glucose transport Km values range from 0.01 mM (Scarborough, 1970) to 0.04 mM (Schneider and Wiley, 1971). Of the 39 putative sugar transporter genes in the Neurospora genome, three genes were recently identified and characterized as major components of System I: glt-1 (NCU01633), and of System II: hgt-1 and hgt-2 (NCU10021 and NCU04963) (Wang et al., 2017). The Km values for all three transporters fall within the ranges of earlier work (Wang et al., 2017).
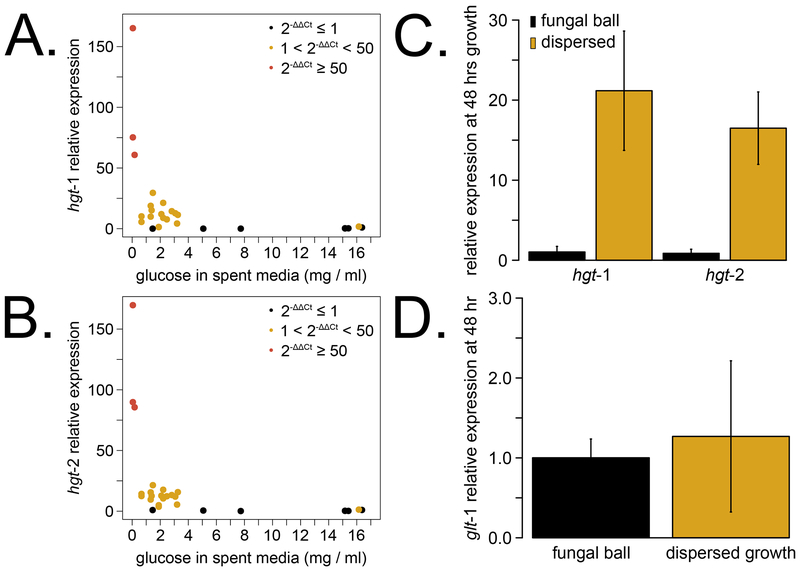
Gene expression levels of the glucose transporter genes were measured from a subset of the glucose utilization rate cultures (Fig 2B–C). We identified three expression patterns for the high affinity glucose transporter genes hgt-1 (Fig 3A) and hgt-2 (Fig 3B). Both genes were expressed at low levels when glucose concentrations in spent media were above 5 mg/ml (27.8 mM), which is well within the Km values for System I. Expression was primed at glucose levels from ~ 1 – 3 mg/ml (5.6 – 16.7 mM), and the highest expression was observed at glucose levels below ~ 0.5 mg/ml (2.8 mM). As glucose levels fall below the range of System I, the System II genes are highly induced. HGT-1 and HGT-2 were first identified in an RNA-Sequencing screen for genes differentially expressed at 20 mg/ml (2%) versus 0.5 mg/ml (0.05%) glucose (Wang et al., 2017), and our data are consistent with upregulation in low glucose concentrations. We also directly compared sugar transporter levels from dispersed versus ball cultures after 2 days of growth (Fig 3C–D). The high affinity transporters (Fig 3C) are intermediately expressed in dispersed growth but not active in fungal balls, which reflects differences in total glucose consumption. We observe similar levels of the low affinity glt-1 transporter expression between balls and dispersed cultures despite different concentrations of external glucose (Fig 3D). This result is consistent with previous findings that the System I transporter glt-1 is constitutively expressed down to levels as low as 0.05% or 2.8 mM glucose (Wang et al., 2017).
Fig 3. Dispersed growth cultures upregulate glucose starvation genes before 48 hours in culture.
Neurospora tissue was harvested at a subset of times post inoculation concurrent with glucose utilization rate experiments (Fig 2B–C, N = 5 time courses). Gene expression of sugar transporters was measured by RT-PCR, and relative expression levels of hgt-1 (A) and hgt-2 (B) were plotted against the spent medium glucose concentration for each sample. Relative expression values less than 1 (black dots) map to regions of glucose replete conditions, while 50-fold or more increases (red dots) correspond to glucose levels below 0.5 mg/ml. Intermediate values (yellow dots) cluster at 1 – 3 mg/ml of glucose in a primed expression state. Sugar transporter expression was directly compared between dispersed growth and fungal ball cultures harvested at 48 hours (1.4 ± 0.1 mg/ml and 15.9 ± 0.6 mg/ml glucose remaining in spent media, respectively; N = 3). The two high affinity transporters are intermediately expressed in dispersed compared to fungal ball growth (C), while the low affinity transporter is expressed at similar levels (D).
3.3. Circadian clock function in dispersed growth
The circadian clock functions in submerged mycelial balls, but a normal clock has not been shown in the dispersed growth regime supplemented with polyacrylic acid. Dispersed cultures of a Neurospora strain bearing a core clock luciferase reporter (Gooch et al., 2008; Larrondo et al., 2015) were grown in constant light at 25°C, and then transferred to constant darkness to set the circadian clock to subjective dusk. Cultures were observed via a CCD camera to monitor luciferase activity (see Materials & Methods). Rhythms were tracked in multiple flasks and compared to data from the same strain grown in standard 96-well plate solid medium assays (Fig 4).
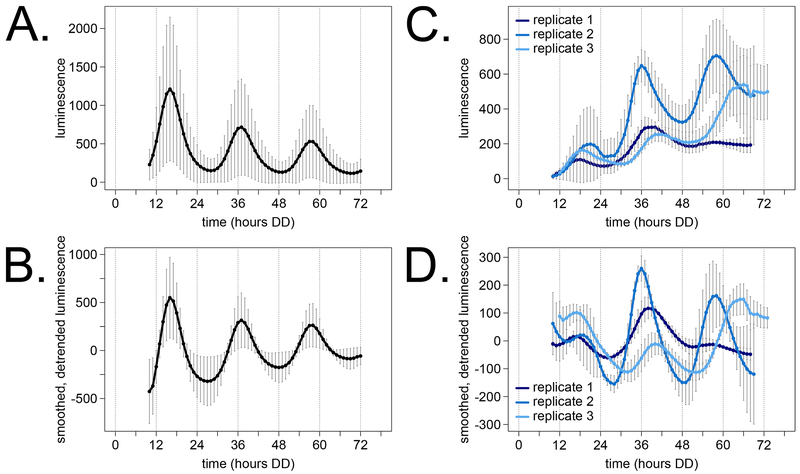
Fig 4. The circadian clock functions in dispersed growth cultures.
Luciferase reporter traces from 78 wells across 7 different 96-well plate experiments were compiled as a control for normal clock function and experimental variability (A). Carbon sources in the solid medium were 0.03% glucose, 0.05% arginine, and 0.01 M quinic acid as described previously (Larrondo et al., 2015). Traces from individual wells were smoothed and detrended (see Materials & Methods) before computing period and phase (B). Luminescence from dispersed cultures grown in Bird medium, 1.8% glucose + PA, was also measured from multiple flasks in three biological replicates (C). Replicate 1 was conducted in 50 ml liquid volume with a 12-hour growth in constant light prior to transfer to darkness (N = 2 flasks). Replicate 2 was carried out in 50 ml volumes with 6 hours growth in light (N = 4) prior to transfer to darkness. Replicate 3 used 20 ml volumes with 6 hours light (N = 6). Traces from individual flasks were smoothed and detrended for period and phase calculation (D).
The modern 96-well plate assay is a high throughput method for measuring the free running circadian period length and phase of various Neurospora strains and mutants (Hurley et al., 2014). Data from many technical and biological replicates were averaged together over the first 3 circadian days (Fig 4A). Although a wild-type rhythm is obvious, there is significant noise in reporter amplitude level between different 96-well plate experiments (i.e. error bars are larger at peaks than troughs). We implement smoothing and detrending on data from individual wells (see Materials & Methods) before measuring circadian period and phase from each trace (Fig 4B). The normalization method does not affect reporter peak times (Fig 4A versus Fig 4B).
We detected a circadian oscillation in all three biological replicates of dispersed growth cultures (Fig 4C – D). However, luciferase rhythms from dispersed growth cultures were much more variable than the solid medium assay between flasks of the same experiment and between biological replicates. One factor that could contribute to measurement noise is flask motion during camera image acquisition, which is not an issue for stationary solid medium assays. The linear trend from liquid cultures was increasing (Fig 4C) compared to the solid medium downward trend (Fig 4A). The major variable that contributes to this trend is accumulation of fungal biomass and reporter expression over time. The free running period length and reporter peaks were measured from each individual trace and averaged over technical replicates (Table 1). Consistent with visual inspection, the liquid culture measurements are noisier with higher standard deviations than solid medium. As expected, the transcriptional reporter peaks during the circadian day (CT0-12) in both growth regimes. The circadian clock appears to function normally in dispersed, fungal ball, and solid substrate growth conditions, where stationary solid medium plates are the simplest to track and quantify luminescence.
Table 1. Circadian period length and peak reporter phase measurements from liquid culture are variable, but consistent with, solid growth clock assays.
Normalized luminescence traces from solid medium controls (Fig 4B) and dispersed culture flasks (Fig 4D) were used to calculate an average free running period length (see Materials & Methods) for each experiment. We use reporter peak time as the phase reference point. Peaks were identified in silico as the time point corresponding to the local maximum luminescence value for each circadian day. Conventional circadian time (CT) was computed (see Materials & Methods) in order to compare phases between experiments with different free running period lengths.
| Experiment Label | Period (hours) | 1st Peak in DD (CT) | 2nd Peak in DD (CT) | 3rd Peak in DD (CT) |
|---|---|---|---|---|
| 96 well, solid medium | 21.3 ± 0.5 | 16.1 ± 0.5 (6.1) | 37.0 ± 0.7 (5.7) | 57.6 ± 1.0 (4.9) |
| replicate 1, dispersed | 19.5 ± 0.8 | 17.0 ± 0.0 (8.9) | 38.5 ± 2.1 (11.4) | 57.0 ± 2.8 (10.2) |
| replicate 2, dispersed | 20.1 ± 0.9 | 18.0 ± 1.7 (9.5) | 36.0 ± 0.0 (7.0) | 58.0 ± 2.2 (9.3) |
| replicate 3, dispersed | 24.2 ± 1.4 | 16.8 ± 0.8 (4.7) | 40.2 ± 1.2 (3.9) | 65.0 ± 0.6 (4.5) |
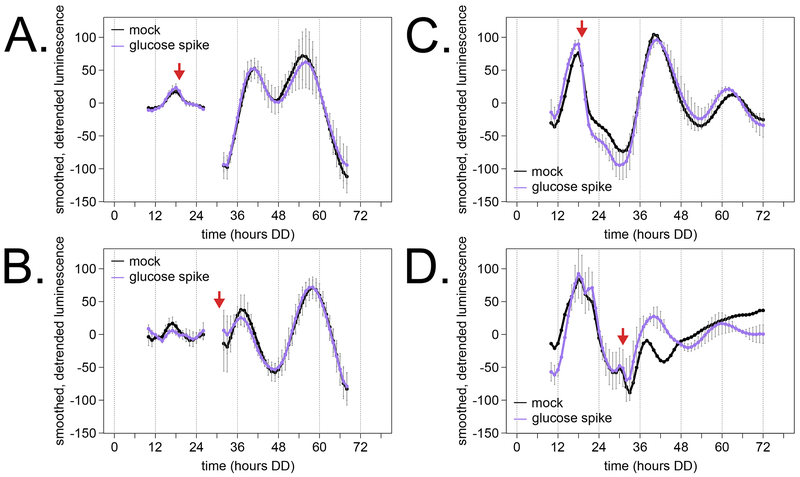
Light and temperature are two environmental signals that feed into the circadian clockwork and can reset clock phase with a strong cue (so called zeitgebers, or “time givers”). A 5-fold increase in environmental glucose concentration does not reset the Neurospora clock in disc cultures (Nakashima, 1981), suggesting that glucose levels do not act as a zeitgeber for fungal clocks. Dispersed cultures begin to transcriptionally prepare for glucose starvation after only about 40 hours of growth in Bird 1.8% glucose + PA medium (Fig 2C & Fig 3). We designed experiments to ask if a spike of additional glucose would reset the clock of dispersed cultures. We supplemented glucose at two different time points—DD20, just after the first transcriptional reporter peak, and DD32, during the rise of reporter expression in the second cycle.
Consistent with previous work, we found that a glucose spike does not reset the clock compared to mock controls in two biological replicates (Fig 5). As with unperturbed control experiments (Fig 4D, Table 1), we observed measurement noise in period and phase estimates after glucose addition (Table 2). Circadian day 2 peaks were within 2 CT-hours between glucose spike and mock cultures, demonstrating no phase shift in response to glucose. For circadian day 3, two phase advances and one phase delay were recorded after glucose addition relative to mock controls (Table 2). However, these data do not demonstrate a consistent, glucose-driven phase shift. Proper CT calculations depend on an accurate free running period estimate, and liquid culture traces have a higher variance in period calculation (Table 1 & 2). The raw peak time points in DD show that the three “advanced/delayed” phases are within 1.5 hours of their experimental counterparts (Table 2). Additionally, one replicate of a mock treatment flask did not show a clear peak in the third cycle (Fig 5D, Table 2). Despite the experimental and measurement variability, our results agree with previous work that glucose does not reset the clock in Neurospora.
Fig 5. Addition of glucose at two out-of-phase circadian time points does not reset the clock in dispersed growth cultures.
Dispersed cultures were prepared in 20 ml of Bird 1.8% glucose + PA medium for luciferase detection as described (Fig 4; Materials & Methods). Glucose (treatment) or water (mock) was added at the DD20 (A and C) or DD32 (B and D) time points indicated by red arrows. Replicate 1 included three technical replicate flasks per condition (A, B), and replicate 2 had two technical replicate flasks for glucose and one for mock treatment (C, D). At DD20, approximately 12 mg/ml glucose remains in spent media, and at DD32, about 3 mg/ml glucose remains (Fig 2B – C). Thus, the 6 mg/ml glucose spike represents a 1.5- to 3-fold change in the environmental glucose concentration. Missing data points from hours 27 – 31 are not shown due to a reporter detection artefact (A, B).
Table 2. Circadian phase of dispersed cultures is not affected by a glucose spike compared to mock treatment.
Normalized luminescence traces from dispersed culture flasks (Fig 5) were used to calculate an average free running period length and reporter peak times for each treatment as described in Table 1.
| Experiment Label | Period (hours) | 1st Peak in DD (CT) | 2nd Peak in DD (CT) | 3rd Peak in DD (CT) |
|---|---|---|---|---|
| Fig 5A, glucose at DD20 | 19.3 ± 1.7 | 18.0 ± 0.0 (10.4) | 40.5 ± 0.7 (14.4) | 56.0 ± 0.0 (9.6) |
| Fig 5A, mock at DD20 | 20.3 ± 0.7 | 18.0 ± 0.0 (9.3) | 41.0 ± 1.0 (12.5) | 55.0 ± 1.0 (5.0) |
| Fig 5B, glucose at DD32 | 21.1 ± 3.7 | 17.0 ± 0.0 (7.3) | 36.7 ± 1.5 (5.7) | 57.3 ± 0.6 (5.2) |
| Fig 5B, mock at DD32 | 20.2 ± 0.5 | 17.0 ± 0.0 (8.2) | 36.3 ± 1.2 (7.1) | 58.0 ± 1.0 (8.9) |
| Fig 5C, glucose at DD20 | 21.9 ± 3.4 | 18.0 ± 0.0 (7.7) | 41.0 ± 1.4 (8.9) | 61.5 ± 0.7 (7.4) |
| Fig 5C, mock at DD20 | 21.0 | 18.0 (8.6) | 40.0 (9.7) | 63.0 (12.0) |
| Fig 5D, glucose at DD32 | 19.9 ± 1.1 | 18.0 ± 0.0 (9.7) | 39.0 ± 1.4 (11.0) | 61.0 ± 2.8 (13.6) |
| Fig 5D, mock at DD32 | 20.2 | 18.0 (9.4) | 38.0 (9.1) | NA |
4. Discussion
Consistent with historical data from other fungi, Neurospora crassa grows as dispersed hyphae with high metabolic activity when anionic polymers are present in the medium (Metz and Kossen, 1977). This liquid culturing method is an equivalent alternative to mycelial disc growth for circadian biochemistry assays (Nakashima, 1981). Dispersed growth is optimized for biomass yield and for more uniform exposure to nutrients in the medium (Fig 1 & Fig 2). Dispersed growth yields more biomass than fungal ball cultures (Fig 1D), and therefore two models could explain the increased glucose consumption and mass production of dispersed hyphae (Fig 2 & Fig 3) (Metz and Kossen, 1977). In the first model, many more dispersed conidia germinate, but these cells have comparable metabolic activity to the medium-exposed edges of the fungal ball. In the second model, more conidia germinate, and the metabolic activity of dispersed hyphae are higher than all fungal ball hyphae. The current study cannot differentiate between these two possibilities but posits that the circadian clock functions equivalently under both growth regimes (Fig 4 & Fig 5).
Due to high metabolic activity, dispersed cultures are primed for glucose starvation when the environmental glucose levels drop below 3 mg/ml (16.7 mM), and high affinity glucose transporters are fully upregulated when glucose concentrations from spent media were 0.5 mg/ml (2.8 mM) or less (Fig 3). Previous work also showed that both low and high affinity glucose transporter messages are expressed when environmental glucose levels are low but not zero (0.5 mg/ml versus 20 mg/ml RNA-Seq data from (Wang et al., 2017)). Neurospora hyphae appear to express a mixed population of low affinity (Km ≥ 8 mM) and high affinity (Km ≤ 0.1 mM) glucose transporters in low environmental glucose concentrations. Interestingly, hgt-2 (NCU04963) is reported to be a clock-controlled gene from fungal discs cultured in Bird 1.8% glucose (Hurley et al., 2014). Single knockouts of hgt-2 (FGSC18806) or hgt-1 (FGSC22818) have a wild-type free running period length of 22.3 and 22.9 hours, respectively, and normal phase (data not shown). Sugar transport in Neurospora is complex with almost 40 putative transporter genes (Galagan et al., 2003), and circadian clock function does not depend on a specific transporter, carbon source, or environmental glucose level.
To our knowledge, this is the first report of densely sampled bioluminescence data for the clock from individual batch cultures (Fig 4). Solid medium luminescent assays continue to be the gold standard for estimating period and phase, as our data clearly show more noise in clock reporter detection from shaking liquid cultures (Fig 4, Table 1). Circadian parameters gleaned from 96-well plate assays represent an average luminescence rhythm from conidia, aerial hyphae, and mycelium developmental phases. Our data, together with previous work on germinating conidia (Baker et al., 2009; Lindgren, 1994), show that the circadian clock functions normally in germinating conidia and in hyphae. Thus, an interconnected mycelium is not required for synchronous rhythms in a population of dispersed hyphae. Lastly, for industrial applications, bioreactors with gentle mixing and medium containing anionic polymers should permit circadian function in Neurospora (Tralau et al., 2007).
Light and temperature are critical cues for organisms to maintain the proper time and phase of the circadian clock relative to the environment. 1.5- to 5-fold increases in the environmental glucose concentration do not affect the phase of the Neurospora clock (Fig 5 and (Nakashima, 1981)). This result is somewhat surprising given that serum shock or glucose alone are used to synchronize mammalian cells in culture (Hirota et al., 2002). Plants and animals partition feeding times based on time of day, and mammalian feeding out of phase with the host circadian clock exacerbates metabolic disease outcomes. Food partitioning is not as clearly physiologically relevant for the fungal life cycle because its chances of encountering a sugar source are equally likely over a circadian day. On the other hand, Neurospora does partition some of its metabolism because rhythmic genes related to catabolic reactions tend to peak in expression during the day, followed by anabolism at night (Hurley et al., 2014, 2018). A handful of mutants exist where the circadian period length is altered by varying glucose levels—prd-1, ras2, rco-1, and csp-1 (Emerson et al., 2015; Gyöngyösi et al., 2017; Olivares-Yanez et al., 2016; Sancar et al., 2012). Thus, the wild-type Neurospora clock is normally buffered against glucose levels in the environment by nutritional / metabolic compensation pathways. Glucose or other carbon sources may function as zeitgebers for only a subset of circadian organisms based on metabolic and temporal feeding demands (Reinke and Asher, 2019).
Future work with this dispersed growth system will query the population of rhythmic metabolites in Neurospora crassa with even access to nutrients. We will then be able to determine if any metabolites are rhythmically abundant from fungi to mammals (Krishnaiah et al., 2017) and perhaps in the common eukaryotic ancestor running its circadian clock. To further optimize monitoring circadian rhythms in liquid culture, we predict that more than 3 cycles of data could be obtained using a luciferase variant containing a PEST degron (Cesbron et al., 2013). Glucose spike experiments at time points beyond 48 hours growth (where starvation transporters are fully active) would be enabled with this PEST luciferase tool.
Acknowledgements
This work is supported by the National Institutes of Health (F32 GM128252 to CMK, R35 GM118021 to JCD, R35 GM118022 to JJL, and U01 EB022546-01 to co-PI JCD). We thank the Fungal Genetics Stock Center (Kansas City, Missouri, USA) for curating N. crassa strains. We thank Brad Bartholomai for assistance taking images of dispersed growth hyphae.
This article is part of the Fungal Adaptation to Hostile Challenges special issue for the third International Symposium on Fungal Stress (ISFUS), which is supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grant 2018/20571-6 and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) grant 88881.289327/2018-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Aronson B, Johnson K, Loros J, Dunlap J. 1994. Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science, 263(5153), 1578–1584. [DOI] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. 2009. Quantitative Proteomics Reveals a Dynamic Interactome and Phase-Specific Phosphorylation in the Neurospora Circadian Clock. Molecular Cell, 34(3), 354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya N, Shiu PKT. 2007. Cyclosporin A-resistance based gene placement system for Neurospora crassa. Fungal Genetics and Biology, 44(5), 307–314. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Larrondo LF, Froehlich AC, Shi M, Chen CH, Loros JJ, Dunlap JC. 2007. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes & Development, 21(12), 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkholder PR, Sinnott EW. 1945. Morphogenesis of Fungus Colonies in Submerged Shaken Cultures. American Journal of Botany, 32(7), 424–431. [Google Scholar]
- Byrne GS, Ward OP. 1987. Effects of polymers on pelleting of Rhizopus arrhizus. Transactions of the British Mycological Society, 89(3), 367–371. [Google Scholar]
- Cesbron F, Brunner M, Diernfellner ACR. 2013. Light-dependent and circadian transcription dynamics in vivo recorded with a destabilized luciferase reporter in Neurospora. PLoS ONE, 8(12), 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Ringelberg CS, Gross RH, Dunlap JC, Loros JJ. 2009. Genome-wide analysis of light-inducible responses reveals hierarchical light signalling in Neurospora. The EMBO Journal, 28(8), 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockrell AL, Pirlo RK, Babson DM, Cusick KD, Soto CM, Petersen ER, Davis MJ, Hong CI, Lee K, Fitzgerald LA, Biffinger JC. 2015. Suppressing the Neurospora crassa circadian clock while maintaining light responsiveness in continuous stirred tank reactors. Scientific Reports, 5, 10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin HJ, Sauer BL, Munkres KD. 1973. Glucose utilization and ethanolic fermentation by wild type and extrachromosomal mutants of Neurospora crassa. Journal of Bacteriology, 116(3), 1322–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowse HB. 2013. Maximum entropy spectral analysis for circadian rhythms: Theory, history and practice. Journal of Circadian Rhythms, 11(1), 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayergi H, Scharer J, Moo-Young M. 1973. Effects of polymer additives on fermentation parameters in a culture of A. niger. Biotechnology and Bioengineering, 15, 845–859. [Google Scholar]
- Emerson JM, Bartholomai BM, Ringelberg CS, Baker SE, Loros JJ, Dunlap JC. 2015. period-1 encodes an ATP-dependent RNA helicase that influences nutritional compensation of the Neurospora circadian clock. Proceedings of the National Academy of Sciences of the United States of America, 112(51), 15707–15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson S 1950. The growth phase in Neurospora corresponding to the logarithmic phase in unicellular organisms. Journal of Bacteriology, 60(3), 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature, 422(6934), 859–868. [DOI] [PubMed] [Google Scholar]
- Gooch VD, Mehra A, Larrondo LF, Fox J, Touroutoutoudis M, Loros JJ, Dunlap JC. 2008. Fully codon-optimized luciferase uncovers novel temperature characteristics of the Neurospora clock. Eukaryotic Cell, 7(1), 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyöngyösi N, Szöke A, Ella K, Káldi K. 2017. The small G protein RAS2 is involved in the metabolic compensation of the circadian clock in the circadian model Neurospora crassa. Journal of Biological Chemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlik D, Brandt U, Bohle K, Fleißner A. 2017. Establishment of Neurospora crassa as a host for heterologous protein production using a human antibody fragment as a model product. Microbial Cell Factories, 16(1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T, Okano T, Kokame K, Shirotani-Ikejima H, Miyata T, Fukada Y. 2002. Glucose down-regulates Per1 and Per2 mRNA levels and induces circadian gene expression in cultured rat-1 fibroblasts. Journal of Biological Chemistry, 277(46), 44244–44251. [DOI] [PubMed] [Google Scholar]
- Hobbs G, Frazer CM, Gardner DCJ, Cullum JA, Oliver SG. 1989. Dispersed growth of Streptomyces in liquid culture. Applied Microbiology and Biotechnology, 31(3), 272–277. [Google Scholar]
- Hurley JM, Dasgupta A, Andrews P, Crowell AM, Ringelberg C, Loros JJ, Dunlap JC. 2015. A Tool Set for the Genome-Wide Analysis of Neurospora crassa by RT-PCR. G3, 5(10), 2043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Dasgupta A, Emerson JM, Zhou X, Ringelberg CS, Knabe N, Lipzen AM, Lindquist EA, Daum CG, Barry KW, Grigoriev IV, Smith KM, Galagan JE, Bell-Pedersen D, Freitag M, Cheng C, Loros JJ, Dunlap JC. 2014. Analysis of clock-regulated genes in Neurospora reveals widespread posttranscriptional control of metabolic potential. Proceedings of the National Academy of Sciences of the United States of America, 111(48), 16995–17002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Jankowski MS, De Los Santos H, Crowell AM, Fordyce SB, Zucker JD, Kumar N, Purvine SO, Robinson EW, Shukla A, Zink E, Cannon WR, Baker SE, Loros JJ, Dunlap JC. 2018. Circadian Proteomic Analysis Uncovers Mechanisms of Post-Transcriptional Regulation in Metabolic Pathways. Cell Systems, 7(6), 613–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Moore D, Trinci APJ. 1988. Effects of Junlon and Hostacerin on the Electrokinetic Properties of Spores of Aspergillus niger, Phanerochaete chrysosporium and Geotrichum candidum. Journal of General Microbiology, 134, 235–240. [Google Scholar]
- Krishnaiah SY, Wu G, Altman BJ, Growe J, Rhoades SD, Coldren F, Venkataraman A, Olarerin-George AO, Francey LJ, Mukherjee S, Girish S, Selby CP, Cal S, Er U, Sianati B, Sengupta A, Anafi RC, Kavakli IH, Sancar A, Baur JA, Dang CV, Hogenesch JB, Weljie AM. 2017. Clock Regulation of Metabolites Reveals Coupling between Transcription and Metabolism. Cell Metabolism, 25(5), 1206. [DOI] [PubMed] [Google Scholar]
- Larrondo LF, Olivares-Yanez C, Baker CL, Loros JJ, Dunlap JC. 2015. Decoupling circadian clock protein turnover from circadian period determination. Science, 347(6221), 1257277-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren KM. 1994. Characterization of ccg-1, a clock-controlled gene of Neurospora crassa. Ph.D. thesis, Chapter 6, 159–186. Dartmouth College, Hanover, NH. [Google Scholar]
- Loros JJ. 2019. Principles of the animal molecular clock learned from Neurospora. European Journal of Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC. 1989. Molecular cloning of genes under control of the circadian clock in Neurospora. Science, 243(4889), 385–388. [DOI] [PubMed] [Google Scholar]
- Marshall K, Alexander M. 1960. Growth characteristics of fungi and actinomycetes. Journal of bacteriology, 80(473), 412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz B, Kossen N. 1977. The growth of molds in the form of pellets–a literature review. Biotechnology and Bioengineering, 19, 781–799. [Google Scholar]
- Metzenberg RL. 2004. Bird Medium: an alternative to Vogel Medium. Fungal Genetics Reports, 51, 19–20. [Google Scholar]
- Nakashima H 1981. A liquid culture method for the biochemical analysis of the circadian clock of Neurospora crassa. Plant & Cell Physiology, 22(2), 231–238. [Google Scholar]
- Olivares-Yanez C, Emerson J, Kettenbach A, Loros JJ, Dunlap JC, Larrondo LF. 2016. Modulation of circadian gene expression and metabolic compensation by the RCO-1 corepressor of Neurospora crassa. Genetics, 204(1), 163–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke H, Asher G. 2019. Crosstalk between metabolism and circadian clocks. Nature Reviews Molecular Cell Biology, 20(4), 227–241. [DOI] [PubMed] [Google Scholar]
- Sancar G, Sancar C, Brunner M. 2012. Metabolic compensation of the Neurospora clock by a glucose-dependent feedback of the circadian repressor CSP1 on the core oscillator. Genes & Development, 26(21), 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent ML, Briggs WR, Woodward DO. 1966. Circadian nature of a rhythm expressed by an invertaseless strain of Neurospora crassa. Plant Physiology, 41(8), 1343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough GA. 1970. Sugar transport in Neurospora crassa II. A second glucose transport system. The Journal of Biological Chemistry, 245(15), 3985–3987. [PubMed] [Google Scholar]
- Schneider RP, Wiley WR. 1971. Kinetic characteristics of the two glucose transport systems in Neurospora crassa. Journal of Bacteriology, 106(2), 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tralau T, Lanthaler K, Robson GD, Crosthwaite SK. 2007. Circadian rhythmicity during prolonged chemostat cultivation of Neurospora crassa. Fungal Genetics and Biology, 44(8), 754–763. [DOI] [PubMed] [Google Scholar]
- Trinci APJ. 1983. Effect of Junlon on morphology of Aspergillus niger and its use in making turbidity measurements of fungal growth. Transactions of the British Mycological Society, 81(2), 408–412. [Google Scholar]
- Wang B, Li J, Gao J, Cai P, Han X, Tian C. 2017. Identification and characterization of the glucose dual-affinity transport system in Neurospora crassa: pleiotropic roles in nutrient transport, signaling, and carbon catabolite repression. Biotechnology for Biofuels, 10(1), 17. [DOI] [PMC free article] [PubMed] [Google Scholar]