Abstract
Background
In Europe, ticks are major vectors of both human and livestock pathogens (e.g. Lyme disease, granulocytic anaplasmosis, bovine babesiosis). Agricultural landscapes, where animal breeding is a major activity, constitute a mosaic of habitat types of various quality for tick survival and are used at different frequencies by wild and domestic hosts across seasons. This habitat heterogeneity, in time and space, conditions the dynamics of these host-vector-pathogen systems and thus drives acarological risk (defined as the density of infected ticks). The principal objective of the OSCAR project (2011-2016) was to examine the links between this heterogeneity and acarological risk for humans and their domestic animals. Here, we present the data associated with this project.
New information
This paper reports a database on the distribution and densities of I. ricinus ticks - the most common tick species in French agricultural landscapes - and the prevalence of three tick-borne pathogens (Anaplasma phagocytophilum, Borrelia spp. and Babesia spp.) in two sites in north-western (“Zone Atelier Armorique”: ZA site) and south-western (“Vallées et Coteaux de Gascogne”: VG site) France. The distribution and density of ticks along a gradient of wooded habitats, as well as biotic variables, such as the presence and abundance of their principal domestic (livestock) and wild hosts (small mammals), were measured from forest cores and edges to more or less isolated hedges, all bordering meadows. Ticks, small mammals and information on local environmental conditions were collected along 90 transects in each of the two sites in spring and autumn 2012 and 2013 and in spring 2014, corresponding to the main periods of tick activity. Local environmental conditions were recorded along each tick and small mammal transect: habitat type, vegetation type and characteristics, slope and traces of livestock presence. Samples consisted of questing ticks collected on the vegetation (mainly I. ricinus nymphs), biopsies of captured small mammals and ticks fixed on small mammals. In the VG site, livestock occurrence and abundance were recorded each week along each tick transect.
A total of 29004 questing ticks and 1230 small mammals were captured during the study across the two sites and over the five field campaigns. All questing nymphs (N = 12287) and questing adults (N = 646) were identified to species. Ticks from small mammals (N = 1359) were also identified to life stage. Questing nymphs (N = 4518 I. ricinus) and trapped small mammals (N = 908) were analysed for three pathogenic agents: A. phagocytophilum, Borrelia spp. and Babesia spp.
In the VG site, the average prevalence in I. ricinus nymphs for A. phagocytophilum, Borrelia spp. and Babesia spp. were, respectively 1.9% [95% CI: 1.2-2.5], 2.5% [95% CI: 1.8-3.2] and 2.7% [95% CI: 2.0-3.4]. In small mammals, no A. phagocytophilum was detected, but the prevalence for Borrelia spp. was 4.2% [95% CI: 0.9-7.5]. On this site, there was no screening of small mammals for Babesia spp. In ZA site, the average prevalence in nymphs for A. phagocytophilum, Borrelia spp. and Babesia were, respectively 2.2% [95% CI: 1.6-2.7], 3.0% [95% CI: 2.3-3.6] and 3.1% [95% CI: 2.5-3.8]. In small mammals, the prevalence of A. phagocytophilum and Borrelia spp. were, respectively 6.9% [95% CI: 4.9-8.9] and 4.1% [95% CI: 2.7-5.9]. A single animal was found positive for Babesia microti at this site amongst the 597 tested.
Keywords: Ticks, Ixodes ricinus, small mammals, Apodemus sylvaticus, Myodes glareolus, prevalence, Anaplasma , Borrelia , Babesia , France, forest, agricultural landscapes, livestock, zoonotic disease
Introduction
In agricultural landscapes, where livestock production occupies a large proportion of the surface area, pastures often adjoin different semi-natural ecosystems (forests, woods, hedges). This type of landscape mosaic implies that areas exploited by livestock are also frequently used by a diverse range of wild fauna. Many parasites and pathogens are shared amongst these animal species, even in the absence of direct contact and some may be transmitted between agricultural and semi-natural systems via common arthropod vectors. In France, ticks are major vectors for both human (e.g. Borrelia burgdorferi s.l., the agent of Lyme disease) and livestock pathogens (e.g. Anaplasma phagocytophilum, inducing granulocytic anaplasmosis or Babesia divergens, causing bovine babesiosis), with Ixodes ricinus being the most commonly-involved vector.
I. ricinus is a three-stage tick that feeds on a wide variety of vertebrate hosts (Sonenshine and Roe 2014, Bonnet et al. 2016). While larvae and nymphs may feed on a range of different-sized hosts, adult ticks require a bloodmeal from a larger host, like roe deer Capreolus capreolus or domestic animals (Ruiz-Fons et al. 2012). Host species are differently exploited by ticks and display variable susceptibilities to infection by different tick-borne infectious agents, exhibiting different levels of reservoir competence (Ostfeld et al. 2014). The abundance and diversity of different hosts thus influence the density of infected ticks (i.e. the “acarological risk”) and hence the probability of contact with humans and livestock (LoGiudice et al. 2003, Boyard et al. 2007, Takumi et al. 2019).
Agricultural landscapes constitute a mosaic of habitat types that vary in quality for tick survival and host use. The habitat composition of a given plot and its connection with other habitats will determine its use by wild vertebrates and will thus shape local tick-host interactions (Estrada-Peña 2002, Li et al. 2012, Werden et al. 2014, Heylen et al. 2019). Breeding practices and particularly, the management of animal grazing in different types of pastures, will also influence exposure risk of livestock to ticks and the pathogens they carry (Richter and Matuschka 2006, Boyard et al. 2007, Gassner et al. 2008, Agoulon et al. 2012, Ruiz-Fons et al. 2012). However, agricultural mosaics are not temporally fixed and can vary both seasonally and yearly. We are also currently witnessing rapid landscape modifications due to the influence of global changes and particularly those associated with land-use (i.e. relative proportions of breeding/crop surfaces, forest or hedge fragmentation) and climate change (i.e. tick population dynamics are tightly linked to temperature and humidity regimes) (Medlock et al. 2013, Agoulon et al. 2016).
The main goal of the OSCAR project (Outil de Simulation Cartographique à l’échelle du paysage Agricole du Risque acarologique / Simulation Tool for Mapping Acarological Risk in Agricultural Landscapes) was to explore the relationships between landscape structure and acarological risk. The study was carried out in two agricultural sites that are part of the International Long-Term Ecological Research (ILTER) network (Zones Ateliers network in France http://www.za-inee.org/en/node/804) and encompass the intrinsic diversity of agricultural landscape features: one LTER site - the “Zone Atelier Armorique” (“ZA site” hereafter) - in north-western France and the second in south-western France in the region of “Vallées et Coteaux de Gascogne” (“VG site” hereafter, belonging to the recently labelled “Zone Atelier PyGar"). Before conducting analyses, the initial task of the OSCAR project consisted of mapping the distribution of ticks, pathogens and the principal domestic (cattle) and wild (small mammals and roe deer) hosts, along a gradient of landscape fragmentation, from forest cores and edges to more or less isolated hedges, all bordering meadows. This paper describes the collected datasets (Fig. 1) (1) on questing tick and small mammal densities, (2) on local environmental conditions (habitat, vegetation and livestock densities) of sampled transects and (3) on pathogen prevalence in ticks and small mammals. Due to time and manpower constraints, we restricted our assessment of tick host species to small mammals, livestock and roe deer, the principal reservoir hosts implicated in disease for production animals. Additional datasets used in some analyses, such as roe deer presence, were not collected in the framework of this study (Fig. 1), but are available elsewhere as outlined in the text.
Figure 1.
Type of collected data used to study the relationships between landscape structure and acarological risk (i.e. density of infected ticks). Dataset origins: in bold, datasets presented in the datapaper; (*) collected in the field or analysed in the laboratory; (+) calculated from field data; (o) obtained from independent databases. Data uses: [1] response variables: pathogen prevalence in ticks, tick densities, tick population structure; [2] explanatory variables.
Project description
Personnel
Laboratories involved: § BIOEPAR, # CEFS, ¶ MIVEGEC, ‡ EPIA, | ECOBIO, 1 UMR CBGP Montpellier
Coordinator of the project: Plantard O. §
Task managers of the project: Vourc’h G. ‡ (Sampling, biological analyses and database constitution), McCoy K.D. ¶ (Empirical estimation of factors influencing acarological risk from field data), Hoch T. § (Simulating acarological risk maps according to environmental changes)
Site managers and contacts for samplings: Verheyden H. # for the VG (‘‘Vallées et Coteaux de Gascogne’’) LTER site and Butet A. | for the ZA (“Zone Atelier Armorique”) LTER site.
Data management and Geographic Information System (GIS): Agoulon A. §, Bastian S. §, Dorr N. ‡, Lebert I. ‡, Lourtet B. #, Mahé H. §, Rantier Y. |
Sample collection
VG site: Angibault J. #, Bailly X. ‡, Bard E. ‡, Bastian S. §, Cargnelutti B. #, Cebe N. #, Chastagner A. ‡, Delrue B. §, Lebert I. ‡, Léger E. ¶, Lourtet B. #, Mahé H. §, Masseglia S. ‡, McCoy K.D. ¶, Merlet J. #, Noël V. ¶, Perez G. §,|, Picot D. #, Pion A. ‡, Poux V. ‡, Quillery E. §, Toty C. ¶, Vaumourin E. ‡, Verheyden H. #, Vincent S. ‡, Vourc'h G. ‡
ZA site: Agoulon A. §, Al Hassan D. |, Armand F. §, Audiart J.-Y. §, Bastian S. §, Billon D. §, Bouju-Albert A. §, Boullot F. §,|, Bruneau A. §, Butet A. |, Daniel J. §, de la Cotte N. §, Delrue B. §, Faille F. §, Gonnet M. ‡, Hermouet A. §, Hoch T. §, Jambon O. |, Jouglin M. §, Lemine-Brahim M. §, Mahé H. §, Moreau E. §, Navarro N. §, Pavel I. §, Perez G. §,|, Plantard O. §, Quillery E. §, Rantier Y. |, Renaud J. §, Roy P. §
Identification of small mammals
VG site: Bastian S. §, Butet A. |, Cèbe N. #, Chastagner A. ‡, Cosson J. 1, Léger E. ¶, Masseglia S. ‡, McCoy K.D. ¶, Noël V. ¶, Perez G. §,|, Vaumourin E. ‡, Vourc'h G. ‡
ZA site: Butet A. |, Perez G. §,|, Agoulon A. §, Bastian S. §, Bouju-Albert A. §, Gonnet M. ‡, Hermouet A. §, Moreau E. §, Pavel I. §, Plantard O. §
Tick identification
VG site: Pion A. ‡, Poux V. ‡
ZA site: Agoulon A. §, Bouju-Albert A. §, Hermouet A. §, Plantard O. §
Laboratory analysis
VG site: Chastagner A. ‡, Masseglia S. ‡, McCoy K.D. ¶, Noël V. ¶, Léger E. ¶
ZA site: Bouju-Albert A. §, Daniel J. §, Faille F. §, Hermouet A. §, Jouglin M. §, Léger E. ¶, McCoy K.D. ¶, Noël V. ¶, Perez G. §,|, Quillery E. §
Livestock survey: (VG site only): Angibault J. #, Cargnelutti B. #, Lourtet B. #, Sevila J. #, Verheyden H. #
Study area description
LTER site “Vallées et Coteaux de Gascogne” (VG site)
The VG site is a Long Term Ecological Research (LTER) site (referenced as zone atelier Pyrénées Garonne - PYGAR since 2016, https://pygar.omp.eu/), located 75 km from Toulouse in south-western France (43°16'2.64"N, 0°51'51.00"E) (Fig. 2). The area is hilly (altitude 200–400 m above sea level) and dissected by north-south valleys with a mild oceanic climate and summer droughts. Woodland covers 24% of the area with two main forest patches of about 500 and 700 ha, many woods smaller than 50 ha and hedges dominated by Quercus spp. Areas dedicated to cultivated crops cover 32% of the main study site. Meadows cover another 40%, amongst which half are grazed by domestic animals (mostly cattle, horses, sheep, but sometimes goats and pigs), either individually or in mixed groups. The roe deer density has been estimated at around 6 roe deer/km2 in open areas and more than 30 roe deer/km2 in one of the forest areas (Hewison et al. 2007).
Figure 2.
Map of the two studied sites in France: the “Vallées et Coteaux de Gascogne” LTER site (VG) and the “Zone Atelier Armorique” LTER site (ZA). Landscape types: LH, Agricultural landscapes with a Low Hedgerow network density; HH, Agricultural landscapes with a High Hedgerow network density; FE, Forest Edge; FC, Forest Core. A single label per landscape type was drawn on the map (LH, HH, FE, FC), but corresponds to several sampling points in the field. For example, for the FE label, 20 sampling points were designated around the forest (see Fig. 3 for the number of points).
LTER site “Zone atelier Armorique” (ZA site)
The ZA site (https://osur.univ-rennes1.fr/za-armorique) (Fig. 2) is a labelled LTER area of the CNRS (Centre National de la Recherche Scientifique), where ecological studies have been conducted for over 25 years. It is an agricultural landscape situated in the vicinity of Rennes, which is south of the Mont-Saint-Michel’s Bay (north-east Brittany, Western France) (48°29'22.40"N, 1°33'41.48"W). The area includes a wide array of agricultural landscape features, a forest of about 1000 ha and many woods smaller than 50 ha. The southern part of the site is a fine-grain heterogeneous landscape with a complex network of hedgerows (160 m/ha) enclosing small fields. At the northern part of the site, agricultural intensification has led to a more homogeneous coarse-grain landscape with fewer hedgerows per hectare (70 m/ha) enclosing larger fields. The proportion of grassland is greater in the southern part, whereas fields of maize and cereal dominate the northern part. Small woods are disseminated within both northern and southern areas of the site (Hassan et al. 2012).
Sampling methods
Study extent
The study was performed in the two LTER sites (ZA and VG) from 2012 to 2014. Questing ticks and small mammals were sampled during five field campaigns: spring and autumn 2012, spring and autumn 2013 and spring 2014. The sampling design is presented in Fig. 3.
Figure 3.

A Schematic representation of single and associated sampling transects of ticks and small mammals in the different landscape types.
B Details of:
- questing tick transect-lines, where the drag transect was subdivided into sub-transects
- small mammal trap-lines, which contained 34 traps spaced 3 m apart across the initial part of a subset of tick transects
Landscape types:
LH, Agricultural landscapes with a Low Hedgerow network density
HH, Agricultural landscapes with a High Hedgerow network density
FE, Forest Edge
FC, Forest Core
The sampling zones (n = 60) were located in 4 landscape types: Agricultural landscapes with a Low Hedgerow network density (LH); Agricultural landscapes with a High Hedgerow network density (HH); Forest Edge (FE); and Forest Core (FC) (Fig. 3). Small mammals were sampled in 24 zones (amongst the 60 sampling zones), trap-lines being systematically paired with one or two questing tick transect-lines (Fig. 3). Small mammal trap-lines were distributed amongst the four landscape types as follows: six in LH, six in HH, six in FE and six in FC. For each trap-line, 34 traps were spaced 3 m apart along the 100 m line.
Questing ticks were sampled in all 60 zones (including the 24 zones for small mammal sampling). In each zone, one or two transect-lines were defined: 1) a single transect-line was sampled when found along hedgerows and in FC; 2) two transect-lines were run when situated at wood and forest edges (i.e. on either side of the ecotone: one in the meadow and one in the forest) (Fig. 3). This resulted in a total of 90 questing tick transect-lines which were distributed as follows: 30 in LH, 30 in HH, 20 in FE and 10 in FC. For each transect-line, ticks were collected along lines of 300 m, divided into 10 sub-transects of 10 m2 each (10 m length x 1 m width), with a space of 20 m between sub-transects (Fig. 3).
The design was fully applied (60 sampling zones) in four campaigns (spring and autumn 2012, spring 2013 and 2014), but only 36 transect lines from the 24 zones used to quantify small mammal presence were sampled during autumn 2013, corresponding to an optimisation of the sampling effort during a less favourable period of tick activity.
Sampling description
Recording local environmental conditions
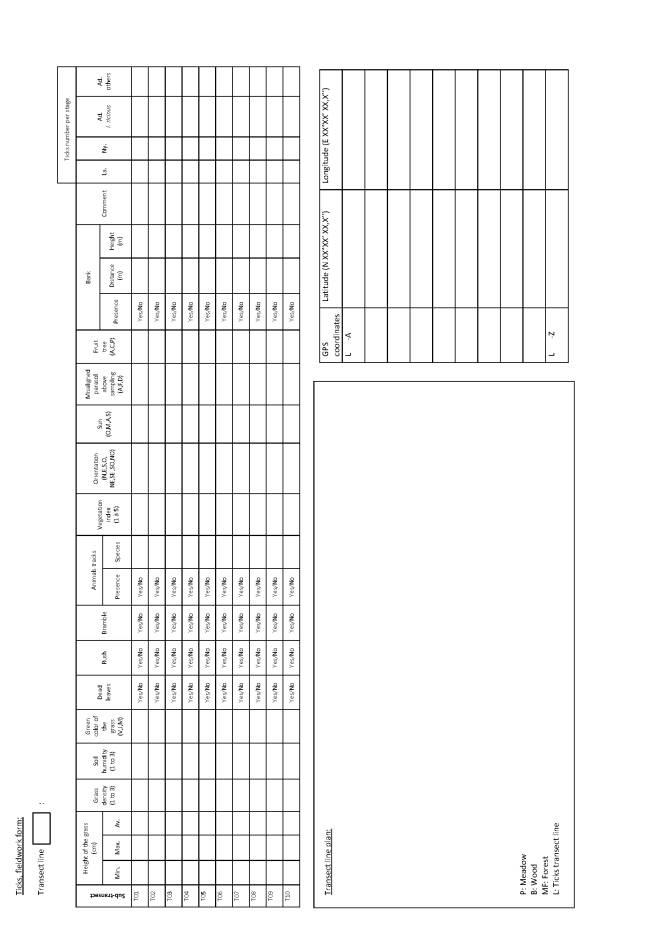
Georeferencing of sampling locations of ticks (Table 1) and small mammals (Table 2) was obtained in the field using a Trimble GNSS GeoExplorer XT 6000 receiver. A differential correction in post-processing made it possible to obtain decimetric precision. The points obtained were exported in a shape (shp) format and inserted into Geographic Information System (ArcGIS) software. Drawings of the sampling lines were performed on maps by the operators during sampling and were corrected with the GIS database with the help of orthophotos (BD ORTHO®, resolution 50 cm x 50 cm, IGN). During sampling, local environmental conditions were recorded for the questing tick transect-lines, the tick sub-transects and the small mammal trap-lines. The following variables were recorded in the field during tick sampling (Fig. 4) and small mammal sampling (Fig. 5): date and time of the day, habitat type, vegetation type and characteristics, slope, traces of use by livestock. In the VG site, livestock occurrence and abundance were also recorded each week along each tick transect. The livestock survey was only performed in the VG site in association with other research projects and these data were not collected in the ZA site. The data were entered into specific tables of the database (Tables 3, 4, 5, 6).
Table 1.
Field description for tick sub-transect locations. c., characters.
| Field | Description | Type |
| ECHT_ID | Identifier for tick sub-transect line: campaign - site - landscape type - transect line number - sub-transect line number | Text (50 c.) |
| X_CENTRE | X coordinate of the sub-transect centroid (RGF93_Lambert_93, EPSG 2154) | Real (19, 11) |
| Y_CENTRE | Y coordinate of the sub-transect centroid (RGF93_Lambert_93, EPSG 2154) | Real (19, 11) |
| ECHT_ECHLT | Identifier for the transect: campaign - site -landscape type - transect line number | Text (50 c.) |
| LENGTH | Length of the sub-transect (metres) | Real (13, 11) |
| LATITUDE | Decimal Latitude of the sub-transect centroid (WGS84; EPSG 4326) | Real (10, 7) |
| LONGITUDE | Decimal Longitude of the sub-transect centroid (WGS84; EPSG 4326) | Real (10, 7) |
Table 2.
Field description for small mammal trap-line locations. c., characters.
| Field | Description | Type |
| X_CENTRE | X coordinate of the trap-line centroid (RGF93_Lambert_93, EPSG 2154) | Real (18, 11) |
| Y_CENTRE | Y coordinate of the trap-line centroid (RGF93_Lambert_93, EPSG 2154) | Real (18, 11) |
| LENGTH | Length of the trap-line (metres) | Real (12, 11) |
| ECHLM_ID | Identifier of the trap-line: campaign - site - landscape type - trap-line number | Text (15 c.) |
| LATITUDE | Decimal Latitude of the sub-transect centroid (WGS84; EPSG 4326) | Real (10, 7) |
| LONGITUDE | Decimal Longitude of the sub-transect centroid (WGS84; EPSG 4326) | Real (10, 7) |
Tick form.
Figure 4a.
Page 1
Figure 4b.
Page 2
Small mammal form.
Figure 5a.

Page 1
Figure 5b.

Page 2
Figure 5c.
Page 3
Table 3.
Field description of the dataset including the characteristics of the tick transect lines. c., characters.
| Field | Description | Type |
| ZONE_ID | Identifier of the LTER site (VG or ZA) | Text (5 c.) |
| SECT_CODE | Identifier for the landscape type: forest core (FC, CF in table), forest edge (FE, LF in table), agricultural landscape with a high hedgerow network density (HH, BD in table), agricultural landscape with a low hedgerow network density (LH, BO in table) | Text (5 c.) |
| LTIQ_ID | Identifier for the transect line: site - landscape type - transect line number | Text (20 c.) |
| ECHLT_ID | Identifier for the transect line: campaign - site - landscape type - transect line number | Text (20 c.) |
| ECHT_ID | Identifier for tick sub-transect line: campaign - site - landscape type - transect line number - sub-transect line number | Text (30 c.) |
| ECHLT_DATE | Sampling date for a transect | Date/Time |
| ECHLT_SAISON | Identifier for campaign (1 = spring 2012, 2 = autumn 2012, 3 = spring 2013, 4 = autumn 2013, 5 = spring 2014) | Integer |
| ECHLT_HDEB | Starting hour of tick sampling in the transect | Date/Time |
| ECHLT_HFIN | Ending hour of tick sampling in the transect | Date/Time |
| ECHLT_SOL | Land use: 1 = meadow, 2 = wood, 3 = forest, 4 = meadow/hedge, 5 = meadow/wood, 6 = meadow/forest | Boolean |
| ECHLT_PHERBH | Average height of the grass in the meadow landscape (cm) | Integer |
| ECHLT_BHERBH | Average height of the grass in the wood landscape (cm) | Integer |
| ECHLT_FHERBH | Average height of the grass in the forest landscape (cm) | Integer |
| ECHLT_FTYPE | Forest type: 1 = deciduous, 2 = coniferous, 3 = mixed | Boolean |
| ECHLT_HHERB | Wet grass: 1 = yes, 0 = no | Boolean |
| ECHLT_ANIP | Presence of livestock on the pasture: 1 = yes, 0 = no | Boolean |
Table 4.
Field description of the dataset including characteristics of tick sampling in each tick sub-transect. c., characters.
| Field | Description of the sub-transect | Type |
| ECHT_ID | Identifier for the tick sub-transect | Text (30 c.) |
| ECHT_ECHLT_ID | Key to Table 3 | Text (20 c.) |
| ECHT_TIR | Identifier of sub-transect | Text (3 c.) |
| ECHT_HERB_MOY | Average height of the grass in the sub-transect (cm) | Boolean |
| ECHT_HERB_DENS | Grass in the sub-transect: 1 = none, 2 = sparse, 3 = dense | Boolean |
| ECHT_SOL_HUM | Soil humidity: 1 = dry, 2 = slightly wet, 3 = presence of water | Real |
| ECHT_HERB_VER | Green colour of the grass: V = green on 2/3 of the sub-transect, J = yellow on 2/3 of the sub-transect, M = mixed, NP = not relevant if no grass | Text (3 c.) |
| ECHT_PFEUIL | Presence of dead leaves: 1 = yes, 0 = no | Boolean |
| ECHT_JONC | Presence of rush: 1 = yes, 0 = no | Boolean |
| ECHT_RONC | Presence of bramble: 1 = yes, 0 = no | Boolean |
| ECHT_IND_VEG | Vegetation index (hedge or wood): 1 = no hedge, 2 = discontinuous hedge, 3 = continuous hedge not deeper than 2 m, 4 = deeper hedge, between 2 and 5 m, 5 = hedge deeper than 5 m or wood | Boolean |
| ECHT_PARASOL | Misaligned parasol above sampling: A = no branches (no parasol), F = dense branches over less than 2/3 of the sub-transect, D = dense branches over more than 2/3 of the sub-transect | Text (1 c.) |
| ECHT_TALU | Presence of a bank: 1 = yes, 0 = no | Boolean |
| ECHT_DT_TALU | Distance between the bank and the sub-transect (metres) | Real |
| ECHT_HT_TALU | Bank height (metres) | Real |
| ECHT_NB_LIRLA | Number of Ixodes ricinus larvae | Boolean |
| ECHT_NB_LIRNY | Number of Ixodes ricinus nymphs | Boolean |
| ECHT_NB_LIRADM | Number of Ixodes ricinus male adults | Boolean |
| ECHT_NB_LIRADF | Number of Ixodes ricinus female adults | Boolean |
| ECHT_NB_LIFNY | Number of Ixodes frontalis nymphs | Boolean |
| ECHT_NB_IRADND | Number of adult Ixodes ricinus ticks (male or female) | Boolean |
Table 5.
Field description of the dataset including characteristics of the small mammal trap-lines. c., characters.
| Field | Description | Type |
| ZONE_ID | Identifier of the LTER site (VG or ZA) | Text (5 c.) |
| SECT_CODE | Identifier of the landscape type: forest core (FC, CF in table), forest edge (FE, LF in table), landscape with high hedgerow network density (HH, BD in table), landscape with low hedgerow network density (LH, BO in table) | Text (5 c.) |
| ECHLM_ID | Identifier of the trap-line: campaign - site - landscape type - trap-line number | Text (30 c.) |
| ECHLM_DATE | Sampling date for placing the traps | Date/Time |
| ECHLM_SITLIG | Trap-line place (interface): 1 = meadow/hedge, 2 = meadow/wood, 3 = meadow/forest, 4 = forest | Boolean |
| ECHLM_TYP_PRAI | Meadow type: 1 = grasses, 2 = mowing meadow, 3 = other | Boolean |
| ECHLM_HCONT | Continuity of the hedge: 1 = continuous, 2 = not continuous | Boolean |
| ECHLM_HDENS | Hedge density: 1 = dense, 2 = slightly dense | Boolean |
| ECHLM_HBERB | Presence of herbaceous layer in hedge: 1 = yes, 0 = no | Boolean |
| ECHLM_HARBU | Presence of shrub layer in hedge: 1 = yes, 0 = no | Boolean |
| ECHLM_HARBO | Presence of arborescent layer in hedge: 1 = yes, 0 = no | Boolean |
| ECHLM_HLSOL | Width of the hedge at the level of the ground, in the hedge (metres) | Integer |
| ECHLM_HLCAN | Width of the canopy above the hedge (metres) | Boolean |
| ECHLM_BHERB | Presence of a herbaceous layer in the woods: 1 = yes, 0 = no | Boolean |
| ECHLM_BARBU | Presence of shrub layer in the woods: 1 = yes, 0 = no | Boolean |
| ECHLM_BARBO | Presence of arborescent layer in the woods: 1 = yes, 0 = no | Boolean |
| ECHLM_BDENS | Wood density: 1 = dense, 2 = slightly dense | Boolean |
| ECHLM_BTYPE | Wood type: 1 = deciduous, 2 = coniferous, 3 = mixed | Boolean |
| ECHLM_FHERB | Presence of herbaceous layer in forest: 1 = yes, 0 = no | Boolean |
| ECHLM_FARBU | Presence of shrub layer in forest: 1 = yes, 0 = no | Boolean |
| ECHLM_FARBO | Presence of arborescent layer in forest: 1 = yes, 0 = no | Boolean |
| ECHLM_FDENS | Forest density: 1 = dense, 2= slightly dense | Boolean |
| ECHLM_FTYPE | Forest type: 1 = deciduous, 2 = coniferous, 3 = mixed | Boolean |
| ECHT_ID | Identifier for small mammal trap-line and checking number | Text (30 c.) |
| ECHT_REL_COD | Identifier of trap checks: R1 = 24 h, R2 = 48 h | Text (5 c.) |
| ECHT_DATE | Day of trap check | Date/Time |
| ECHT_NUAGE | Cloud cover: 0 = blue sky, 1 = 1/4 cloud cover, 2 = half covered, 3 = 3/4 covered, 4 = completely covered | Integer |
| ECHT_VENT | Presence of wind: 0= no wind, 1 = light wind, 2 = discontinuous, 3 = strong | Boolean |
| ECHT_ANIM | Presence of livestock in the field: 1 = yes, 0 = no | Boolean |
| ECHT_ESP | Animal types: 1 = cattle, 2 = sheep, 3 = horse, 4 = other | Boolean |
| ECHT_NB_ANI | Number of animals in the field | Boolean |
| ECHT_PRES_MAM | Small mammal sign: 1 = yes, 0 = no | Boolean |
| ECHT_PIEGE_NOT_OK | Traps disturbed or closed without capture: 1 = yes, 0 = no | Boolean |
| ECHT_PIEGE_NB | Number of traps disturbed or closed without capture (between 1 and 34) | Integer |
Table 6.
Field description of the dataset concerning small mammal sampling and identification. c., characters.
| Field | Description | Type |
| MAM_ID | Identifier of the trapped small mammals: campaign - site - landscape type - trap-line number - small mammal number | Text (30 c.) |
| MAM_ECHM_ID | Identifier for small mammal trap-line and check number | Text (30 c.) |
| MAM_DATE | Autopsy day | Date |
| MAM_SEXE | Identifier for sex: 1 = Male, 2 = Female | Boolean |
| MAM_SANG | Blood sampling: 1 = yes, 0 = no | Boolean |
| MAM_SMETHO | Blood sampling method: IC = intracardiac, RO = retro-orbital | Text (2 c.) |
| MAM_PDSENT | Small mammal weight before autopsy (g) | Integer |
| MAM_STAD | Small mammal stage: 1 = juvenile, 2 = sub-young, 3 = adult | Boolean |
| MAM_LTEST | Testicule length | Boolean |
| MAM_GESTANT | Pregnant female: 1 = yes, 0 = no | Boolean |
| MAM_NB_F | If pregnant = yes, number of fœtuses | Boolean |
| MAM_ALLAIT | Lactating female: 1 = yes, 0 = no | Boolean |
| MAM_PRELEV_ORE | Ear sample: 1 = yes, 0 = no | Boolean |
| MAM_PRELEV_FOIE | Liver sample: 1 = yes, 0= no | Boolean |
| MAM_PRELEV_RNA | RNA sample from spleen: 1 = yes, 0 = no | Boolean |
| MAM_PRELEV_RATE | Spleen sample: 1 = yes, 0 = no | Boolean |
| MAM_CARC_PDIS | Carcass partially dissected and frozen: 1 = yes, 0 = no | Boolean |
| MAM_NB_TIK | Total number of ticks on the small mammal | Boolean |
| MAM_NB_TIK_LA | Total number of larvae on the small mammal | Boolean |
| MAM_NB_TIK_NY | Total number of nymphs on the small mammal | Boolean |
| MAM_NB_TIK_AD | Total number of adult ticks on the small mammal | Boolean |
| MAM_TYP_ECTO | Ectoparasitic species: fleas, mites, lice, fleas + mites, fleas + lice, mites + lice, fleas + mites + lice, ectoparasite species not specified, none | Text (50 c.) |
| LMAM_NOM_LAT | Species name (Latin) | Text (50 c.) |
| LMAM_NOM_FR | Species name (French) | Text (50 c.) |
| MAM_ID | Identifier of the trapped small mammals: campaign - site - landscape type - trap-line number - small mammal number | Text (30 c.) |
| MAM_ECHM_ID | Identifier for small mammal trap-line and check number | Text (30 c.) |
Sampling of questing ticks
Questing ticks (Fig. 3) were sampled by flagging (Boyard et al. 2007). In each sub-transect, a 1x1 m white flannel cloth (or ‘flag’) was slowly dragged (0.5 m/s) along 9 m (explored surface of 10 m2) across the lower vegetation and leaf-litter (Agoulon et al. 2012). Ticks were counted, collected from the flag and stored in 70% ethanol for later identification (life stage and species) and detection of infectious agents using molecular analyses (Fig. 6, Table 7). Tick identifications were performed using a binocular microscope, according to Pérez-Eid (2007).
Figure 6.
Molecular analyses of ticks; +ve, positive sample.
Table 7.
Field description of the dataset concerning the analyses of tick DNA for infectious agents. c.: characters
| Field | Description | Type |
| ECHLT_ID | Identifier of the transect: season-site-landscape-transect number - Identifier for campaign (1 = spring 2012, 3 = spring 2013) | Text (20 c.) |
| ECHLT_DATE | Sampling date for a transect | Date/Time |
| ECHT_ID | Identifier for the tick transect -subtransect: campaign - site - landscape - transect number - sub-transect number | Text (30 c.) |
| TIQ_ID | Identifier for a tick | Text (30 c.) |
| ANA_RESULT1 | Result method 1: detection of Anaplasma from tick DNA (yes = 1, no = 0) | Boolean |
| ANA_RESULT2 | Result method 2: detection of Anaplasma from tick DNA (yes = 1, no = 0) | Boolean |
| ANA_CO_SEQ | Sequencing analysis: obtained sequence for Anaplasma (yes = 1, no = 0) | Boolean |
| BOR_RESULT | Result: detection of Borrelia from tick DNA (yes = 1, no = 0) | Boolean |
| BOR_CO_SEQ | Sequencing analysis: obtained sequence for Borrelia (yes = 1, no = 0) | Boolean |
| BOR_REM | Remark: assignment to a species | Memo |
| BAB_RESULT | Result: detection of Babesia by PCR from tick DNA (yes = 1, no = 0) | Boolean |
| BAB_CO_SEQ | Sequencing analysis: obtained sequence for Babesia (yes = 1, no = 0) | Integer |
| BAB_CO_REM | Remark: assignment to a species | Memo |
Sampling of small mammals
The 100 m trap-line contained 34 INRAE live-traps, fitted with dormitory boxes and baited with a mixture of seeds and fresh apple. After placement, the traps were checked in the morning 24- and 48-hours after setup (Figs 3, 7). Captured small mammals were identified to species, sexed and weighed to 0.5 g in a field laboratory (Table 6). They were euthanised by authorised experimenters in accordance with French law and dissected. A blood sample and ear and spleen biopsies were performed for the detection and characterisation of infectious agents during the first four field campaigns. Blood sampling was performed on trapped animals using the retro-orbital method (Hoff 2000). Blood pellets were separated from serum by centrifugation. Serum samples were stored at −20°C and are available for supplementary analysis upon request. Ticks from small mammals were counted immediately after being euthanised in VG, but in ZA, due to the high number of captured mammals, dead animals were frozen and ticks were collected later during dissections. All collected ticks were stored in 70% ethanol for later identification and use for molecular analyses. The animals captured in spring 2014 were not euthanised, but were released at least 500 m away from the capture site to avoid recapture and ticks were quickly collected on these individuals.
Figure 7.
Molecular analyses of small mammals. +ve, positive sample.
Molecular analyses
In tick (Fig. 6) (Table 7): Amongst the 12287 nymphs collected during the five campaigns, 4518 I. ricinus nymphs were selected at random from the two major periods of tick activity, i.e. spring campaigns of 2012 and 2013. For each tick, DNA was extracted using the ammonia-based protocol described in Schouls et al. (1999). Borrelia detection was performed using the quantitative PCR (SYBRGreen) protocol outlined in Jacquot et al. (2016). To identify the infecting Borrelia species, positive samples were re-amplified using nested PCR protocols for the FlaB and OspC genes (Gómez-Díaz et al. 2011) and amplicons were directly sequenced using Sanger technology (Eurofins, France). Detection of A. phagocytophilum DNA was ascertained by real-time PCR by targeting msp2/p44 genes and genotypes were characterised by 454 sequencing of groEL, msp4 and ankA genes (GATC, Germany) (Chastagner et al. 2017). The detection of Babesia spp. was achieved by nested PCR of the 18S rRNA gene (Jouglin et al. 2017). Positive amplicons were purified using ExoSAP-IT (Ozyme, France) and sent for Sanger sequencing (GATC, Germany). Additional investigations were also conducted on the population genetics of some ticks (nymphs), using either microsatellite (d'Ambrioso 2016) or SNP loci (Quillery et al. 2014).
In small mammals (Fig. 7) (Tables 6, 8): Small mammals trapped in spring and autumn sessions of 2012 and 2013 were analysed for the three pathogenic agents (N = 300 small mammals in VG site and N = 608 in ZA site). However, a couple of individuals could not be tested for all pathogens because of insufficient DNA quantity. Spleens were stored at −20°C for detection of A. phagocytophilum (Chastagner et al. 2016) and Babesia (Jouglin et al. 2017). Ear biopsies were stored in 70% ethanol for detection of Borrelia spp. (Jacquot et al. 2016). DNA from spleen and ear samples were extracted using the NucleoSpinTissue kit (Macherey Nagel, Düren, Germany) (Chastagner et al. 2016, Perez et al. 2017). DNA of A. phagocytophilum was detected by real-time PCR targeting the msp2 gene, according to the protocol of Courtney et al. (2004). Detection of Babesia spp. was achieved by nested PCR of the 18S rRNA gene; different primers were used to amplify Babesia spp. from small mammals and from ticks because of high rates of false positive amplifications with small mammal DNA (Jouglin et al. 2017). Positive amplicons were purified using ExoSAP-IT (Ozyme, France) and sent for Sanger conventional sequencing (GATC, Germany). DNA of B. burgdorferi s.l. in ear samples was detected and typed as outlined for ticks.
Table 8.
Field description of the dataset concerning the analyses of infectious agents from small mammals. c.: characters.
| Field | Description | Type |
| ECHLM_ID | Identifier of the trap-line: campaign - site - landscape type - trap-line number | Text (30 c.) |
| ECHLM_DATE | Sampling date for the placement of traps | Date/Time |
| MAM_ID | Identifier of the trapped small mammals: campaign - site - landscape type - trap-line number - small mammal number | Text (30 c.) |
| LMAM_NOM_LAT | Species name | Text (50 c.) |
| BOOR_RESULT_PCR | Result: detection of Borrelia from small mammal ear DNA: 1 = yes, 0 = no | Boolean |
| BOOR_SEQ | Sequencing analysis of Borrelia: 1 = yes, 0 = no | Boolean |
| BOOR_SP | Species name of Borrelia | Memo |
| ANR_RESULT_QPCR | Result: detection of Anaplasma from spleen DNA: 1 = yes, 0 = no | Boolean |
| ANR_RA_SEQ | Sequencing analysis: obtained sequence for Anaplasma (1 = yes, 0 = no) | Integer |
Livestock survey in VG site
Livestock abundance was measured in the VG site on the pasture adjoining each tick transect-line in 2012 and 2013 (Table 9). The number of cattle, sheep, goats and horses grazing in each pasture was monitored on a weekly basis from autumn 2011 to spring 2013, excluding the winter (November to March). The number of individuals grazing in each pasture was then summed per season (spring: week 17 to 26, summer: week 27 to 35, autumn: week 36 to 44) to obtain a livestock abundance estimate, given as the number of head.day per season. When averaged per count day and summed across the whole VG site, the livestock mean density was 20.3 animals/km2 in the open landscapes (HH and LH).
Table 9.
Field description for livestock dataset. c., characters. Heads.day refers to the number of individual animals that were counted in a pasture on a given day.
| Field | Description | Type |
| LTIQ_ID | Identifier for the transect line: site - landscape type - transect line number | Text (20 c.) |
| BET_ID | Identifier for livestock | Text (30 c.) |
| BET_SAISON | Season: spring (week 17 to 26), summer (week 27 to 35), autumn (week 36 to 44) | Text (10 c.) |
| BET_ANNEE | Year | Integer |
| BET_CUMUL | Sum of livestock heads.day at pasture over the considered season (spring 70 days, summer 63 days, autumn 63 days) | Integer |
| LBET_ESPECE | Species name: bovine, caprine, equine, ovine | Text (20 c.) |
DataBase
All the data of Tables 1, 2, 3, 4, 5, 6, 7, 8, 9 were united in a single Access database. The relationship between the tables is given in Figs 8, 9.
Figure 8.
Relational model for ticks: relationships between tables concerning tick sampling and analyses. Similar colour corresponds to similar data present in two tables. Key is primary key. ECHLT_*, Identifier code for tick transect-line; ECHT_*, Identifier code for tick sub-transect line.
Figure 9.
Relational model for small mammals: relationships between tables concerning small mammal sampling and analyses. Similar colour corresponds to similar data present in two tables. Key is primary key. ECHLM_*, Identifier code for small mammal trap-line; MAM_*, Identifier code for captured small mammal.
The data presented in this dataset are detailed by campaign and by site in Table 10.
Table 10.
Summary of available data in the present dataset according to campaign and site. Identifier for campaigns: 1 = spring 2012, 2 = autumn 2012, 3 = spring 2013, 4 = autumn 2013, 5 = spring 2014.
| Site | VG | ZA | ||||||||
| Campaign | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 |
| Local environmental conditions | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Number of tick transect lines | 90 | 90 | 90 | 36 | 90 | 89 | 89 | 90 | 36 | 90 |
| Tick identification | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Pathogens analysis in ticks | yes | no | yes | no | no | yes | no | yes | no | no |
| Number of small mammal trap-lines | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 | 24 |
| Small mammal identification | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes |
| Pathogens analysis in small mammals | yes | yes | yes | yes | no | yes | yes | yes | yes | no |
| Identification of small mammals ticks | yes | yes | yes | yes | no | yes | yes | yes | yes | no |
| Livestock | yes | yes | yes | yes | no | no | no | no | no | no |
Variables not included in the datapaper
Information on the infection rate and movement of roe deer in some of the studied habitat types were recorded at the VG site (see, for exemple, Martin et al. 2018). They are available on http://eurodeer.org/ or upon request to the CEFS.
Weather data were obtained from Météo-France weather stations close to ZA (Broualan, Rennes-St Jacques, Pontorson) and VG (Boussan, Fabas, Palaminy) sites. Additional weather data were measured near the VG site at the meteorological weather station (INRAE in SAMAN), located at the UMR DYNAFOR (INRAE-INPT) in Saint-André (F-31420) or near the ZA site at the COSTEL meteorological weather station (CNRS in COSTEL), located in the LEGT RENNES. According to the location, the weather stations were equipped with sensors to measure air and ground temperatures, air humidity, pluviometry, wind speed and direction, relative humidity, atmospheric pressure and light intensity. The data (2011-2014) are available upon request to the corresponding author.
Additional variables were calculated to measure landscape heterogeneity around the sampling locations. These data and their production (ecotone length between wooded habitat and meadows, proportion of woodland cover, grassland cover and crops, mean distance between wooded patches, perimeter-area ratio of wooded patches, connectivity of wooded habitat patch) are presented in Perez et al. (2016) and Perez et al. (2020).
Geographic coverage
Description
VG site (19004 ha):
top left 43°22'11,59''N, 0°43'59,17''E;
bottom right: 43°11'41,25''N, 0°59'15,61''E
ZA site (14203 ha):
top left 48°34'20,83''N, 1°19'21,26''W;
bottom right: 48°25'20,46''N, 1°29'56,85''W
Usage rights
Use license
Other
IP rights notes
Creative Commons CC-BY 4.0
Data resources
Data package title
Data from ANR OSCAR Project
Resource link
Portail Data INRAE, https://data.inrae.fr/
Number of data sets
4
Data set 1.
Data set name
Field description of tick datasets
Data format
tab
Number of columns
1
Character set
UTF-8
Download URL
https://data.inrae.fr/dataset.xhtml?persistentId=doi: 10.15454/93LPP7
Description
The data concerning questing tick sampling are presented in the 3 following tables.
Table 3. Field description of the dataset, including the characteristics of the questing tick transect-lines. (Associated file: TickTransectData.tab).
Table 4. Field description of the dataset, including characteristics of questing tick sampling in each tick sub-transect. (Associated file: TickSamplingData.tab).
Table 7. Field description of the dataset concerning the analyses of tick DNA for infectious agents. (Associated file: TickAnalysisData.tab).
The date format ISO 8601 (YYYY-MM-DD) was used.
Data set 2.
Data set name
Description of small mammal datasets
Data format
tab
Number of columns
1
Character set
UTF-8
Download URL
https://data.inrae.fr/dataset.xhtml?persistentId=doi: 10.15454/93LPP7
Description
The data concerning small mammal sampling are presented in the 3 following tables.
Table 5. Field description of the characteristics of the small mammal trap-lines in the dataset. (Associated file: SmallMammalsTrapLineData.tab)
Table 6. Field description of the dataset concerning small mammal sampling and identification (Associated file: SmallMammalsSamplingData.tab)
Table 8. Field description of the dataset concerning the analyses of small mammal DNA for infectious agents (Associated file: SmallMammalsPathogenData.tab)
The date format ISO 8601 (YYYY-MM-DD) was used.
Data set 3.
Data set name
Description of the livestock dataset
Data format
tab
Number of columns
1
Character set
UTF-8
Download URL
https://data.inrae.fr/dataset.xhtml?persistentId=doi: 10.15454/93LPP7
Description
Field description for the livestock dataset (Table 9) (Associated file: LivestockData.tab)
Data set 4.
Data set name
Tick sub-transects and small mammal trap-line locations
Data format
shapefile
Number of columns
1
Download URL
https://data.inrae.fr/dataset.xhtml?persistentId=doi: 10.15454/93LPP7
Description
Two tables describing the sample locations for questing ticks (Table 1) and for small mammals (Table 2).
(Associated files: TickTransect.shp and SmallMammalsTrapLine.shp)
Additional information
We provide a quick description of the results in the following section. A total of 29004 questing ticks and 1230 small mammals were collected during the study at the two sites and over the five campaigns. All questing nymphal (N = 12311) and adult ticks (646) were identified to species. Ticks from small mammals (N = 1359) were also identified to the stage.
Sampled ticks
During the five campaigns (from spring 2012 to spring 2014), 16047 larvae, 12287 I. ricinus nymphs, 646 I. ricinus adults and 24 Ixodes frontalis nymphs were collected on the vegetation (Table 11).
Table 11.
Number of collected ticks per campaign and per site. No, number; IR, Ixodes ricinus; IF, Ixodes frontalis. Identifier for campaigns: 1 = spring 2012, 2 = autumn 2012, 3 = spring 2013, 4 = autumn 2013, 5 = spring 2014.
| Campaign | Site | No sampled transect-lines | No larvae | No IR nymphs | No IR adults | No IF nymphs |
| 1 | VG | 90 | 24 | 1588 | 59 | 1 |
| 1 | ZA | 89 | 5214 | 2622 | 109 | 7 |
| 2 | VG | 90 | 758 | 143 | 11 | 0 |
| 2 | ZA | 89 | 3649 | 277 | 22 | 7 |
| 3 | VG | 90 | 69 | 932 | 85 | 0 |
| 3 | ZA | 90 | 1508 | 3196 | 164 | 0 |
| 4 | VG | 36 | 27 | 16 | 8 | 0 |
| 4 | ZA | 36 | 867 | 330 | 20 | 4 |
| 5 | VG | 90 | 25 | 848 | 69 | 0 |
| 5 | ZA | 90 | 3906 | 2335 | 99 | 5 |
| Total | 16047 | 12287 | 646 | 24 |
Fig. 10 presents the density of I. ricinus nymphs, according to landscape type and field campaign. Densities were generally higher in the ZA site than in the VG site, regardless of the campaign or landscape type. However, large heterogeneities were found amongst the five campaigns in both sites.
Figure 10.

I. ricinus nymphal density in the two sites (VG and ZA), according to campaign and landscape type.
Landscape types:
LH: Agricultural landscapes with a Low Hedgerow network density
HH: Agricultural landscapes with a High Hedgerow network density
FE: Forest Edge
FC: Forest Core
Sampled small mammals
Over the study, 335 small mammals were trapped in the VG site (Table 12) and 895 in the ZA site (Table 13). Seven different species were found in VG against five in ZA. In both sites, wood mice (Apodemus sylvaticus) were the dominant species, accounting for 75% of the captured individuals. Bank vole (Myodes glareolus) was the second most frequently-encountered species in both sites (VG: 11% and ZA: 24%).
Table 12.
Small mammal species in the VG site over the 5 field campaigns
| Species name | Number of captured individuals |
| Apodemus sylvaticus | 250 |
| Myodes glareolus | 37 |
| Crocidura russula | 18 |
| Microtus arvalis | 14 |
| Sorex coronatus | 11 |
| Microtus agrestis | 4 |
| Microtus pyrenaicus | 1 |
| Total | 335 |
Table 13.
Small mammal species in the ZA site over the 5 field campaigns.
| Species name | Number of captured individuals |
| Apodemus sylvaticus | 668 |
| Myodes glareolus | 216 |
| Microtus agrestis | 4 |
| Sorex coronatus | 4 |
| Microtus subterraneus | 3 |
| Total | 895 |
Local environmental conditions
In the VG site, the forest type was mainly deciduous (N = 41) with one mixed forest (including coniferous trees). In the ZA site, collections were performed in 33 deciduous forest type and eight mixed forests. Table 14 presents some results of local environmental variables collected during tick sampling.
Table 14.
Summary values of local environmental conditions for transects and sub-transects in VG and ZA sites for the 5 field campaigns (1 to 5). Description of the fields are given in Tables 3, 4. NC: Not concerned (The field makes no sense for the landscape type in question. For example, there cannot be information in a field concerning meadows when the sub-transect line is in the forest); ND: Not documented (missing data).
| Transects and sub-transects | Site | VG | ZA | ||||||||
| Campaign | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Number of tick transect lines | 90 | 90 | 90 | 36 | 90 | 89 | 89 | 90 | 36 | 90 | |
| ECHLT_PHERBH | Median | 20 | 10 | 50 | 20 | 30 | 45 | 20 | 30 | 13,5 | 60 |
| Min | 5 | 5 | 15 | 10 | 5 | 10 | 10 | 10 | 0 | 0 | |
| Max | 60 | 120 | 105 | 50 | 50 | 110 | 50 | 160 | 100 | 110 | |
| ECHLT_BHERBH | Median | 20 | 15 | 30 | 20 | 30 | 20 | 10 | 10 | 7,5 | 20 |
| Min | 5 | 0 | 5 | 5 | 10 | 0 | 5 | 0 | 5 | 0 | |
| Max | 40 | 35 | 60 | 40 | 50 | 80 | 100 | 30 | 15 | 100 | |
| ECHLT_FHERBH | Median | 20 | 25 | 30 | 22,5 | 25 | 15 | 17,5 | 15 | 10 | 20 |
| Min | 0 | 5 | 15 | 5 | 10 | 5 | 5 | 0 | 0 | 5 | |
| Max | 30 | 30 | 55 | 40 | 60 | 20 | 20 | 50 | 30 | 30 | |
| Number of sub-transect | 900 | 900 | 900 | 360 | 900 | 890 | 890 | 900 | 900 | 900 | |
| ECHT_HERB_DENS | 1 | 172 | 161 | 93 | 83 | 64 | 291 | 293 | 254 | 117 | 176 |
| 2 | 304 | 311 | 282 | 105 | 178 | 193 | 129 | 226 | 97 | 231 | |
| 3 | 424 | 428 | 524 | 172 | 657 | 404 | 468 | 420 | 146 | 492 | |
| ND | 0 | 0 | 1 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | |
| ECHT_SOL_HUM | 1 | 282 | 721 | 133 | 344 | 189 | 684 | 807 | 685 | 331 | 731 |
| 2 | 514 | 141 | 665 | 15 | 634 | 156 | 71 | 195 | 27 | 154 | |
| 3 | 104 | 38 | 101 | 0 | 76 | 35 | 12 | 20 | 2 | 14 | |
| ND | 0 | 0 | 1 | 1 | 1 | 15 | 0 | 0 | 0 | 1 | |
| ECHT_HERB_VER | J | 31 | 224 | 2 | 43 | 0 | 0 | 78 | 61 | 15 | 23 |
| M | 147 | 339 | 15 | 59 | 54 | 7 | 134 | 89 | 64 | 34 | |
| ND | 0 | 0 | 1 | 0 | 1 | 14 | 0 | 0 | 0 | 1 | |
| NC | 3 | 14 | 24 | 26 | 1 | 79 | 92 | 103 | 89 | 63 | |
| V | 719 | 323 | 858 | 232 | 844 | 790 | 586 | 647 | 192 | 779 | |
| ECHT_PFEUIL | 0 | 321 | 171 | 433 | 108 | 318 | 385 | 404 | 388 | 118 | 497 |
| 1 | 579 | 729 | 467 | 252 | 581 | 488 | 473 | 512 | 242 | 403 | |
| ND | 0 | 0 | 0 | 0 | 1 | 17 | 13 | 0 | 0 | 0 | |
| ECHT_JONC | 0 | 878 | 887 | 892 | 354 | 879 | 809 | 789 | 798 | 327 | 761 |
| 1 | 22 | 13 | 7 | 5 | 20 | 59 | 90 | 102 | 33 | 139 | |
| ND | 0 | 0 | 1 | 1 | 1 | 22 | 11 | 0 | 0 | 0 | |
| ECHT_RONC | 0 | 679 | 627 | 544 | 169 | 574 | 669 | 571 | 684 | 265 | 659 |
| 1 | 211 | 273 | 353 | 190 | 322 | 200 | 289 | 214 | 94 | 241 | |
| ND | 10 | 3 | 1 | 4 | 21 | 30 | 2 | 1 | 0 | ||
| ECHT_IND_VEG | 1 | 6 | 7 | 7 | 1 | 7 | 22 | 13 | 23 | 1 | 16 |
| 2 | 65 | 69 | 23 | 5 | 22 | 101 | 66 | 99 | 17 | 72 | |
| 3 | 37 | 75 | 53 | 15 | 27 | 62 | 62 | 108 | 46 | 112 | |
| 4 | 119 | 73 | 98 | 21 | 83 | 68 | 75 | 37 | 8 | 31 | |
| 5 | 603 | 596 | 715 | 317 | 679 | 637 | 663 | 633 | 288 | 668 | |
| ND | 70 | 80 | 4 | 1 | 82 | 0 | 11 | 0 | 0 | 1 | |
| ECHT_PARASOL | A | 247 | 258 | 255 | 82 | 244 | 122 | 151 | 210 | 47 | 123 |
| D | 387 | 383 | 483 | 207 | 173 | 327 | 412 | 465 | 205 | 530 | |
| F | 266 | 119 | 162 | 71 | 201 | 370 | 224 | 225 | 104 | 246 | |
| ND | 0 | 140 | 0 | 0 | 282 | 71 | 103 | 0 | 4 | 1 | |
| ECHT_TALU | 0 | 819 | 817 | 828 | 349 | 774 | 698 | 526 | 561 | 169 | 420 |
| 1 | 81 | 82 | 70 | 10 | 126 | 191 | 295 | 338 | 189 | 477 | |
| ND | 0 | 1 | 2 | 1 | 0 | 1 | 69 | 1 | 2 | 3 |
The livestock survey was performed in the VG site: livestock occurred on 28 of the 90 questing tick transect-lines, cattle being the main species present in pastures (Table 15). Median heads.day values at pasture was 112 for the 3 seasons (min = 0, max = 1848). Caprine were present along two transect-lines, equines along three transect-lines and ovine along three transect-lines. One meadow along a transect-line (VG-BD-L002) was occupied by the four livestock species.
Table 15.
Results of livestock survey in the VG site: sum of heads.day by species at pasture over the considered season (spring = 70 days, summer = 63 days, autumn = 63 days). Transect name (site - landscape type - transect number). Identifier for the landscape type: BD (bocage dense) = agricultural landscape with a high hedgerow network density (HH), BO (bocage ouvert) = agricultural landscape with a low hedgerow network density (LH), LF (Lisière de forêt) = forest edge (FE)
| Livestock | Transect name | Spring | Summer | Autumn | Total |
| bovine | VG-BD-L002 | 0 | 322 | 413 | 735 |
| VG-BD-L004 | 0 | 0 | 56 | 56 | |
| VG-BD-L006 | 420 | 378 | 378 | 1176 | |
| VG-BD-L015 | 0 | 0 | 168 | 168 | |
| VG-BD-L020 | 0 | 0 | 112 | 112 | |
| VG-BD-L032 | 420 | 378 | 378 | 1176 | |
| VG-BD-L033 | 0 | 546 | 364 | 910 | |
| VG-BD-L034 | 0 | 567 | 637 | 1204 | |
| VG-BD-L035 | 112 | 168 | 77 | 357 | |
| VG-BD-L036 | 0 | 126 | 0 | 126 | |
| VG-BD-L044 | 56 | 224 | 56 | 336 | |
| VG-BD-L046 | 0 | 21 | 224 | 245 | |
| VG-BD-L048 | 140 | 77 | 56 | 273 | |
| VG-BD-L050 | 0 | 322 | 560 | 882 | |
| VG-BD-L069 | 147 | 147 | 56 | 350 | |
| VG-BO-L105 | 0 | 126 | 56 | 182 | |
| VG-BO-L109 | 0 | 0 | 182 | 182 | |
| VG-BO-L113 | 0 | 0 | 161 | 161 | |
| VG-BO-L136 | 0 | 0 | 182 | 182 | |
| VG-BO-L140 | 0 | 56 | 0 | 56 | |
| VG-BO-L142 | 0 | 112 | 56 | 168 | |
| VG-BO-L145 | 0 | 0 | 56 | 56 | |
| VG-LF-L201 | 1470 | 1260 | 1400 | 4130 | |
| VG-LF-L202 | 1848 | 567 | 0 | 2415 | |
| VG-LF-L206 | 0 | 0 | 21 | 21 | |
| VG-LF-L207 | 1274 | 742 | 1323 | 3339 | |
| VG-LF-L210 | 210 | 119 | 126 | 455 | |
| VG-LF-L215 | 1321 | 882 | 1358 | 3561 | |
| total | 7418 | 7140 | 8456 | 23014 | |
| caprine | VG-BD-L002 | 84 | 21 | 84 | 189 |
| VG-BO-L145 | 0 | 0 | 21 | 21 | |
| total | 84 | 21 | 105 | 210 | |
| equine | VG-BD-L002 | 0 | 42 | 63 | 105 |
| VG-BD-L033 | 0 | 42 | 63 | 105 | |
| VG-BO-L109 | 56 | 0 | 0 | 56 | |
| total | 56 | 84 | 126 | 266 | |
| ovine | VG-BD-L002 | 105 | 0 | 105 | 210 |
| VG-BO-L145 | 0 | 0 | 21 | 21 | |
| VG-LF-L207 | 56 | 0 | 0 | 56 | |
| total | 161 | 0 | 126 | 287 |
Pathogen results
A selected subset of questing nymphs (N = 4518 I. ricinus) and 908 trapped small mammals (N = 300 in VG site and N = 608 in ZA site) were analysed for the three pathogenic agents: A. phagocytophilum, Borrelia spp. and Babesia spp. (Table 16).
Table 16.
Results of A. phagocytophilum, Borrelia spp. and Babesia spp. in nymphs from field campaigns 1 to 3 and in small mammals from field campaigns 1 to 4. No Babesia-positive small mammals were found. n/N, number of positive samples/number of analysed samples; Prev, prevalence in %; 95% CI, in [], 95% Confidence Interval for prevalence.
| Questing nymphs | Small mammals | |||||
| Site | Pathogens | A. phagocytophilum | Borrelia spp. | Babesia spp. | A. phagocytophilum | Borrelia spp. |
| VG | n/N | 35/1891 | 47/1891 | 51/1891 | 0/300 | 6/143 |
| Prev 95%CI |
1.9 [1.2-2.5] |
2.5 [1.8-3.2] |
2.7 [2.0-3.4] |
0.0 | 4.2 [0.9-7.5] |
|
| ZA | n/N | 57/2627 | 78/2627 | 82/2627 | 42/608 | 26/606 |
| Prev 95%CI |
2.2 [1.6-2.7] |
3.0 [2.3-3.6] |
3.1 [2.5-3.8] |
6.9 [4.9-8.9] |
4.1 [2.7-5.9] |
|
Pathogen results in I. ricinus nymphs. A. phagocytophilum was detected, respectively in 1.9% and 2.2% of questing I. ricinus nymphs from VG and ZA. Six species of Borrelia (B. afzelii, B. burgdorferi sensu stricto, B. garinii, B. valaisiana, B. spielmani, B. turdi or B. lusitaniae) were identified in nymphs in the two sites (Table 17). Amongst the 51 positive I. ricinus nymphs for Babesia spp. in the VG site, 23 were identified as Babesia venatorum and 11 had non-specific sequences. Amongst the 82 positive I. ricinus nymphs in the ZA site, 13 were identified as B. venatorum, two as Babesia capreoli and eight had non-specific sequences.
Table 17.
Identification of Borrelia species in infected nymphs.
| Species | VG | ZA |
| Borrelia afzelii | 8 | 16 |
| Borrelia burgdorferi sensu stricto | 15 | 13 |
| Borrelia garinii | 6 | 20 |
| Borrelia valaisiana | 10 | 14 |
| Borrelia spielmani | 0 | 1 |
| Borrelia turdi or B. lusitaniae | 0 | 1 |
| Co-infection | 4 | 6 |
| Non exploitable sequence | 4 | 7 |
| Total | 47 | 78 |
Pathogen results in small mammals (Table 16). A. phagocytophilum was not found in VG, but showed a prevalence of 6.9% in small mammals of ZA (Chastagner et al. 2016). Small mammals were infected only by B. afzelii with respective prevalences of 4.2% and 4.1% in VG and ZA. Amongst the six small mammals infected by Borrelia in the VG site, five were A. sylvaticus and one was M. glareolus. In the ZA site, amongst the 26 infected small mammals, 14 were A. sylvaticus, 11 were M. glareolus and one Microtus subterraneus (Perez et al. 2017). In the VG site, small mammals were not screened for Babesia spp. In the ZA site, one small mammal (M. glareolus, 2-ZA-CF-LM092-M3) amongst 597 tested was positive for Babesia (Jouglin et al. 2017).
Acknowledgements
We thank the French National Research Agency, which funded the OSCAR project (ANR-11-AGRO-001-04). The OSCAR project website can be accessed at: https://www6.inrae.fr/oscar. This study benefited from the help of the “Zone Atelier Armorique” and the Zone Atelier “Vallées et Coteaux de Gascogne”. We are grateful to Dominique L’Hostis, Sylvie Cocaud, Nathalie Morcrette, Esther Dzalé, Anne-Sophie Martel, members of the pole DigitaIST (INRAE) and Nathalie Gandon, Unité Ingénierie Numérique en Recherche (INRAE), for their help preparing this data paper and all the colleagues who have participated to the OSCAR project (see the “Personnel” mentioned in the section “Project description”).
Publications, using the presented data, are available on the OSCAR project website: https://www6.inrae.fr/oscar/Reperes/Publications/Publications-Internationales-revues-a-comite-de-lecture.
Contributor Information
Isabelle Lebert, Email: isabelle.lebert@inrae.fr.
Alain Butet, Email: alain.butet@univ-rennes1.fr.
Karen D. McCoy, Email: karen.mccoy@ird.fr.
Hélène Verheyden, Email: helene.verheyden@inra.fr.
References
- Agoulon Albert, Malandrin Laurence, Lepigeon Florent, Vénisse Maxime, Bonnet Sarah, Becker Claire AM, Hoch Thierry, Bastian Suzanne, Plantard Olivier, Beaudeau François. A Vegetation Index qualifying pasture edges is related to Ixodes ricinus density and to Babesia divergens seroprevalence in dairy cattle herds. Veterinary Parasitology. 2012;185:101–109. doi: 10.1016/j.vetpar.2011.10.022. [DOI] [PubMed] [Google Scholar]
- Agoulon Albert, Butet Alain, Hoch Thierry, Perez Grégoire, Plantard Olivier, Verheyden Hélène, Vourc’h Gwenaël. Dynamique des populations de tiques et liaison avec les facteurs environnementaux. Tiques et Maladies à Tiques. 2016:85–112. doi: 10.4000/books.irdeditions.9027. [DOI]
- Bonnet Sarah, Huber Karine, Joncour Guy, René‑Martellet Magalie, Stachurski Frédéric, Zenner Lionel. 2. Biologie des tiques. Tiques et Maladies à Tiques. 2016:53–84. doi: 10.4000/books.irdeditions.9020. [DOI]
- Boyard C., Barnouin J., Gasqui P., Vourc'h G. Local environmental factors characterizing Ixodes ricinus nymph abundance in grazed permanent pastures for cattle. Parasitology. 2007;134(7):987. doi: 10.1017/s0031182007002351. [DOI] [PubMed] [Google Scholar]
- Chastagner A., Moinet M., Perez G., Roy E., McCoy K. D., Plantard O., Agoulon A., Bastian S., Butet A., Rantier Y., Verheyden H., Cèbe N., Leblond A., Vourc’h G. Prevalence of Anaplasma phagocytophilum in small rodents in France. Ticks and Tick-borne Diseases. 2016;7(5):988–991. doi: 10.1016/j.ttbdis.2016.05.005. [DOI] [PubMed] [Google Scholar]
- Chastagner Amélie, Pion Angélique, Verheyden Hélène, Lourtet Bruno, Cargnelutti Bruno, Picot Denis, Poux Valérie, Bard Émilie, Plantard Olivier, McCoy Karen D, Leblond Agnes, Vourc'h Gwenaël, Bailly Xavier. Host specificity, pathogen exposure, and superinfections impact the distribution of Anaplasma phagocytophilum genotypes in ticks, roe deer, and livestock in a fragmented agricultural landscape. Infection, Genetics and Evolution. 2017;55:31–44. doi: 10.1016/j.meegid.2017.08.010. [DOI] [PubMed] [Google Scholar]
- Courtney J. W., Kostelnik L. M., Zeidner N. S., Massung R. F. Multiplex real-time PCR for detection of Anaplasma phagocytophilum and Borrelia burgdorferi. Journal of Clinical Microbiology. 2004;42(7):3164–3168. doi: 10.1128/jcm.42.7.3164-3168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ambrioso Jonathan. Structuration génétique d'Ixodes ricinus au sein des paysages agricoles. Master Biodiversité-Ecologie-Evolution, Université de Montpellier, France 2016
- Estrada-Peña Agustín. Understanding the relationships between landscape connectivity and abundance of Ixodes ricinus ticks. Experimental and Applied Acarology. 2002;28:239–248. doi: 10.1023/a:1025362903620. [DOI] [PubMed] [Google Scholar]
- Gassner Fedor, Verbaarschot Patrick, Smallegange Renate C., Spitzen Jeroen, Van Wieren Sipke E., Takken Willem. Variations in Ixodes ricinus Density and Borrelia infections associated with cattle introduced into a woodland in The Netherlands. Applied and Environmental Microbiology. 2008;74(23):7138–7144. doi: 10.1128/aem.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Díaz Elena, Boulinier Thierry, Sertour Natacha, Cornet Muriel, Ferquel Elisabeth, Mccoy Karen D. Genetic structure of marine Borrelia garinii and population admixture with the terrestrial cycle of Lyme borreliosis. Environmental Microbiology. 2011;13:2453–2467. doi: 10.1111/j.1462-2920.2011.02515.x. [DOI] [PubMed] [Google Scholar]
- Hassan Diab Al, Georgelin Ewen, Delattre Thomas, Burel Françoise, Plantegenest Manuel, Kindlmann Pavel, Butet Alain. Does the presence of grassy strips and landscape grain affect the spatial distribution of aphids and their carabid predators? Agricultural and Forest Entomology. 2012;15(1):24–33. doi: 10.1111/j.1461-9563.2012.00587.x. [DOI] [Google Scholar]
- Hewison Mark A. J., Angibault Jean-Marc, Cargnelutti Bruno, Coulon Aurélie, Rames Jean-Luc, Serrano Emmannuel, Verheyden Hélène, Morellet Nicolas. Using radio-tracking and direct observation to estimate roe deer Capreolus capreolus density in a fragmented landscape: A pilot study. Wildlife Biology. 2007;13(3):313–320. doi: 10.2981/0909-6396(2007)13[313:uradot]2.0.co;2. [DOI] [Google Scholar]
- Heylen D., Lasters R., Adriaensen F., Fonville M., Sprong H., Matthysen E. Ticks and tick-borne diseases in the city: Role of landscape connectivity and green space characteristics in a metropolitan area. Science of the Total Environment. 2019;670:941–949. doi: 10.1016/j.scitotenv.2019.03.235. [DOI] [PubMed] [Google Scholar]
- Hoff J. Methods of blood collection in the mouse. Lab Animal. 2000;29(10) [Google Scholar]
- Jacquot Maude, Abrial David, Gasqui Patrick, Bord Severine, Marsot Maud, Masseglia Sébastien, Pion Angélique, Poux Valérie, Zilliox Laurence, Chapuis Jean-Louis, Vourc’h Gwenaël, Bailly Xavier. Multiple independent transmission cycles of a tick-borne pathogen within a local host community. Scientific Reports. 2016;6:31273. doi: 10.1038/srep31273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouglin M., Perez G., Butet A., Malandrin L., Bastian S. Low prevalence of zoonotic Babesia in small mammals and Ixodes ricinus in Brittany, France. Veterinary Parasitology. 2017;238:58–60. doi: 10.1016/j.vetpar.2017.03.020. [DOI] [PubMed] [Google Scholar]
- Li Sen, Heyman Paul, Cochez Christel, Simons Leopold, Vanwambeke Sophie O. A multi-level analysis of the relationship between environmental factors and questing Ixodes ricinus dynamics in Belgium. Parasites & Vectors. 2012;5(1) doi: 10.1186/1756-3305-5-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoGiudice K., Ostfeld R. S., Schmidt K. A., Keesing F. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences. 2003;100(2):567–571. doi: 10.1073/pnas.0233733100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin Jodie, Vourc’h Gwenaël, Bonnot Nadège, Cargnelutti Bruno, Chaval Yannick, Lourtet Bruno, Goulard Michel, Hoch Thierry, Plantard Olivier, Hewison A. J. Mark, Morellet Nicolas. Temporal shifts in landscape connectivity for an ecosystem engineer, the roe deer, across a multiple-use landscape. Landscape Ecology. 2018;33(6):937–954. doi: 10.1007/s10980-018-0641-0. [DOI] [Google Scholar]
- Medlock J. M., Hansford K. M., Bormane A., Derdakova M., Estrada-Pena A., George J. C., Golovljova I., Jaenson T. G., Jensen J. K., Jensen P. M., Kazimirova M., Oteo J. A., Papa A., Pfister KA., Plantard O., Randolph S. E., Rizzoli A., Santos-Silva M. M., Sprong H., Vial L., Hendrickx G., Zeller H., Van Bortel W. Driving forces for changes in geographical distribution of Ixodes ricinus ticks in Europe. Parasites & Vectors. 2013;6:1. doi: 10.1186/1756-3305-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostfeld R. S., Levi T., Jolles A. E., Martin L. B., Hosseini P. R., Keesing F. Life history and demographic drivers of reservoir competence for three tick-borne zoonotic pathogens. PLoS One. 2014;9:1–8. doi: 10.1371/journal.pone.0107387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Eid C. Les tiques - Identification, biologie, importance médicale et vétérinaire. Tec et Doc, Editions Médicales. Lavoisier; France: 2007. [Google Scholar]
- Perez Grégoire, Bastian Suzanne, Agoulon Albert, Bouju Agnès, Durand Axelle, Faille Frédéric, Lebert Isabelle, Rantier Yann, Plantard Olivier, Butet Alain. Effect of landscape features on the relationship between Ixodes ricinus ticks and their small mammal hosts. Parasites & Vectors. 2016;9:20. doi: 10.1186/s13071-016-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Grégoire, Bastian Suzanne, Chastagner Amélie, Agoulon Albert, Plantard Olivier, Vourc'h Gwenaël, Butet Alain. Ecological factors influencing small mammal infection by Anaplasma phagocytophilum and Borrelia burgdorferi s.l. in agricultural and forest landscapes. Environmental Microbiology. 2017;19(10):4205–4219. doi: 10.1111/1462-2920.13885. [DOI] [PubMed] [Google Scholar]
- Perez Grégoire, Bastian Suzanne, Chastagner Amélie, Agoulon Albert, Rantier Yann, Vourc’h Gwenaël, Plantard Olivier, Butet Alain. Relationships between landscape structure and the prevalence of two tick-borne infectious agents, Anaplasma phagocytophilum and Borrelia burgdorferi sensu lato, in small mammal communities. Landscape Ecology. 2020;35(2):435–451. doi: 10.1007/s10980-019-00957-x. [DOI] [Google Scholar]
- Quillery E., Quenez O., Peterlongo P., Plantard O. Development of genomic resources for the tick Ixodes ricinus: isolation and characterization of single nucleotide polymorphisms. Molecular Ecology Resources. 2014;14(2):393–400. doi: 10.1111/1755-0998.12179. [DOI] [PubMed] [Google Scholar]
- Richter Dania, Matuschka Franz-Rainer. Modulatory effect of cattle on risk for Lyme Disease. Emerging Infectious Diseases. 2006;12(12):1919–1923. doi: 10.3201/eid1212.051552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Fons Francisco, Fernández-de-Mera Isabel G., Acevedo Pelayo, Gortázar Christian, la Fuente José de. Factors driving the abundance of Ixodes ricinus ticks and the prevalence of zoonotic I. ricinus-borne pathogens in natural foci. Applied and Environmental Microbiology. 2012;78(8):2669–2676. doi: 10.1128/aem.06564-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouls L. M., Van De Pol I., Rijpkema S. G. T., Schot C. S. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. Journal of Clinical Microbiology. 1999;37(7):2215–2222. doi: 10.1128/JCM.37.7.2215-2222.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshine Daniel E., Roe R. Michael. Biology of ticks. http://public.eblib.com/choice/publicfullrecord.aspx?p=1538390 Oxford University Press, 2014;Volume 1:1–560. [Google Scholar]
- Takumi Katsuhisa, Sprong Hein, Hofmeester Tim R. Impact of vertebrate communities on Ixodes ricinus-borne disease risk in forest areas. Parasites & Vectors. 2019;12(1):434. doi: 10.1186/s13071-019-3700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden Lisa, Barker Ian K., Bowman Jeff, Gonzales Emily K., Leighton Patrick A., Lindsay L. Robbin, Jardine Claire M. Geography, deer, and host biodiversity shape the pattern of Lyme Disease emergence in the Thousand Islands Archipelago of Ontario, Canada. PLoS ONE. 2014;9(1) doi: 10.1371/journal.pone.0085640. [DOI] [PMC free article] [PubMed] [Google Scholar]