Abstract
In the present study, two isothermal molecular assays viz. reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) and reverse transcriptase recombinase amplification (RT-RPA) were developed to detect the cardamom vein clearing virus (CdVCV) infecting cardamom. Assays were optimized for parameters like duration, temperature and concentration of magnesium sulfate, and betaine in the case of RT-LAMP and magnesium acetate in the case of RT-RPA. Detection limits of both assays were determined and compared with conventional RT-PCR and SYBR Green-based real-time RT-PCR. RT-LAMP was found 10,000 times additional sensitive than RT-PCR and one-tenth that of real-time RT-PCR. RT-RPA was found 1000 times additional sensitive than RT-PCR and one-hundredth that of real-time RT-PCR. Both assays were specific, rapid, and sensitive for detecting CdVCV. Compared to real-time RT-PCR, these assays are economical and can be employed in large scale screening of cardamom plants against CdVCV for the selection of virus-free plants.
Keywords: Real-time RT-PCR, RT-LAMP, RT-PCR, RT-RPA, Sensitivity
Introduction
Cardamom (Elettaria cardamomum Maton) belongs to the family Zingiberaceae, is one of the most valued spice in the world grown mainly in India, Guatemala, Papua New Guinea, Sri Lanka, and Tanzania. Cardamom is an herbaceous perennial monocot plant that originated in the Western Ghats of Southern India (Ravindran and Madhusoodanan 2002). Three virus diseases are reported in cardamom namely, mosaic (katte), chlorotic streak, and vein clearing (kokke kandu). Among these mosaic and chlorotic streak diseases are caused by cardamom mosaic virus (CdMV) (genus: Macluravirus) and banana bract mosaic virus (BBrMV) (genus: Potyvirus), respectively (Jacob and Usha 2001; Siljo et al. 2012). The causal virus of vein clearing disease was unknown until recently. The recent study based on the small RNA sequencing and transcriptome sequencing identified a new virus species namely, cardamom vein clearing virus (CdVCV) (genus: Nucleorhabdovirus; family: Rhabdoviridae) as the cause of the disease (Bhat et al. 2020). Of the four plant infecting rhabdoviruses genera, Dichorhavirus and Varicosavirus have bipartite genome while Cytorhabdovirus and Nucleorhabdovirus have monopartite genomes (Jackson et al. 2005; Dietzgen et al. 2017). Differentiation of Cytorhabdoviruses and Nucleorhabdoviruses is done based on their site (cytoplasm or nucleus) of replication. Nucleorhabdoviruses have bacilliform enveloped virions, 45–100 nm in diameter, and 130–300 nm in length. CdVCV consists of a single molecule of negative-sense ssRNA genome of about 13 kb coding for six proteins namely, N (nucleocapsid), P (phospho), P3 (movement), M (matrix), G (glyco) and L (RNA-dependent RNA polymerase) (Bhat et al. 2020). Of these, G and M proteins constitute the major structural component of the virion envelope while N, P, and L proteins interact with genomic RNA to form the ribonucleoprotein core that is required for virus replication (Dietzgen et al. 2017). CdVCV is spread vegetatively and through the aphid Pentalonia caladii. Chlorosis of the veins is the first visible symptom of the disease followed by rosetting, loosening of leaf sheath and shredding of leaves. Another typical symptom of the disease is the formation of hook-like tillers wherein newly emerging leaves get intertwined in the older leaves hence the disease is locally known by the name kokke kandu. Other associated symptoms of the disease include mottling on the leaf sheath, light green streaks with shallow furrows on the capsules leading to cracking of fruits, and partial sterility of seeds (Venugopal 2002). The disease is prevalent in Hassan and Uttara Kannada Districts of Karnataka, India with incidence up to 60%. All ages of the plants are susceptible to the disease. New tillers arising out from the infected plants show typical symptoms of vein chlorosis and mottling on the leaf sheath (pseudostem). Of the three viral diseases of cardamom, vein clearing is the most severe as it leads to the complete degeneration of plants within 3–4 years of infection with yield reduction up to 62–84% in the first year of peak crop (Venugopal 2002).
Hence identification and propagation of healthy plants are the need of the hour to manage the spread of the virus. Common molecular detection methods like RT-PCR and real-time RT-PCR could be useful for the early detection of the virus in the field. Usage of these assays will be limited in large scale screening since these methods are both expensive and time consuming. In that context, other reliable molecular techniques namely, loop-mediated isothermal amplification (LAMP) and recombinase polymerase amplification (RPA) assays which are comparatively inexpensive and less time consuming are preferred. RT-LAMP refers to the LAMP technique employed with the use of reverse transcriptase for the detection of RNA viruses. RT-LAMP assay was reported for the rapid diagnosis of many RNA viruses at constant temperature (60–65 °C) and short incubation period (60 min) (Nie 2005; Le et al. 2010; Bhat et al. 2013; Banerjee et al. 2016; Congdon et al. 2019). RT-LAMP reaction includes four to six primers matching the DNA sequence and DNA is amplified by the strand-displacing activity of Bst polymerase. Positive RT-LAMP products can be viewed by color changes under UV light by adding certain dyes in the reaction mix or /and ladder-like arrangement on agarose gel electrophoresis. RT-LAMP assay for the detection of BBrMV in cardamom is reported previously (Siljo and Bhat 2014).
Recombinase polymerase amplification (RPA) developed by Piepenburg et al. (2006) is another novel technique to multiply DNA at isothermal condition by the action of primers, recombinase, single-stranded DNA binding proteins, and strand-displacing polymerase. RNA viruses can be easily detected through reverse transcriptase RPA (RT-RPA) with prior cDNA synthesis (Jiao et al. 2019). Exponential amplification of DNA fragments occurs in RT-RPA within 30–40 min at constant temperature (37–42 °C) results in amplicons of targeted sequence that can be visualized through gel electrophoresis. RT-RPA method was reported for the detection of RNA viruses like yam mosaic virus (Silva et al. 2015), rose rosette virus (Babu et al. 2017), maize chlorotic mottle virus (Jiao et al. 2019) and cucumber green mottle mosaic virus (Zeng et al. 2019). In the present study, we report the development and validation of RT-LAMP and RT-RPA assays for the specific and rapid testing of CdVCV. The sensitivity of the detection of these methods was compared with conventional and real-time RT-PCR.
Materials and methods
Virus isolates
For the initial standardization of both assays, RT-PCR-tested CdVCV-infected (positive control) and CdVCV-free (negative control) cardamom plants collected from Sakleshpur, Hassan District, Karnataka, India were used. Both assays were validated using field samples of cardamom with and without symptoms of the disease.
Primer design and synthesis
Based on the coat protein (nucleocapsid) gene nucleotide sequence of the CdVCV (GenBank accession number MN273311), primers were designed for the RT-LAMP and RT-RPA assays. All the RT-LAMP primers were designed using LAMP primer design software Primer Explorer version 5 (https://primerexplorer.jp/e/). The six primers designed for the RT-LAMP assay were: F3 (5′–GCATAAAAGATGAGTCAAATGCT–3′) and B3 (5′–AGTCCTGAAAAATTACTTCTTACC–3′) (external primers), FIP (5′–TAGAGGCAGAAACACTAGCCGCACAAAACTGAAACACTAGTCG–3′) and BIP (5′–AAGAAAACACAACCCCAATCTGTTTCTTTAGAGCGGATAGCG–3′) (internal primers), BL (5′–GATTTCGCCGATTTCTCGACA–3′) and FL (5′–TCTAGGTAAAACAGCGATCATGTC–3′) (loop primers). For RT-RPA, primers were designed as per the guidelines of the RPA reaction kit manufacturer (TwistDx, United Kingdom) (https://www.twistdx.co.uk). It comprised of forward primer (5′–CTATCCGCTCTAAAGAAGGTAAGAAGTAAT–3′) and revere primer (5′–GCTAGTGATTGTGTTGTAGTGGTTTATTATC–3′) to form an expected amplicon of 186 bp. Primers for RT-PCR (forward primer: 5′–TAAGATGAAAGATGAAGAGATAAT–3′ and reverse primer: 5′–ATATATGCAAGTGCGGCCA–3′) and SYBR Green-based real-time RT-PCR (forward primer: 5′–ACGTGTTCACAGAGGCAGTG–3′ and reverse primer: 5′–ATACAGGGTTGCGGACATTC–3′) were based on the nucleocapsid gene sequence of the CdVCV reported earlier (Bhat et al. 2020).
Total RNA isolation
RNA isolation from vein clearing disease affected cardamom plants was carried out using RNeasy Plant Mini kit (Qiagen, Germany). The yield of total RNA was determined using a spectrophotometer (Biophotometer plus, Eppendorf, Germany). Total RNA from the infected plant directly served as the template for RT-PCR, SYBR Green-based real-time RT-PCR, and RT-LAMP while for RT-RPA, cDNA was synthesized and used as a template. Eluted RNA was denatured by heating at 80 °C for 10 min and snap cooling in ice for 10 min before using it as a template.
cDNA synthesis
cDNA was synthesized using 10.5 μl of denatured total RNA, 4 μl of 5 × reaction buffer, 1 μM of CdVCV-specific primer (5′–ATATATGCAAGTGCGGCCA–3′), 2 μl of 10 mM dNTP mix, 20 U RNase inhibitor and 200 U of RevertAid reverse transcriptase, and the final volume made to 20 μl. The reaction was incubated at 42 °C for 60 min and then stopped by incubating at 70 °C for 10 min.
Development of RT-LAMP assay and optimization of reaction components, time and temperature
Initially, to check the correctness of the primers, RT-LAMP was done using the template from positive control, negative control, and water (no template) control. To check if primers cross-react with other viruses such as CdMV and BBrMV infecting cardamom, total RNA extracted from CdMV and BBrMV affected plants were also subjected to RT-LAMP using primers specific to CdVCV. The 25 μl RT-LAMP reaction mixture consisted 1 μl of denatured RNA template (about 60 ng), 2 × thermopol buffer, 4 mM MgSO4, 0.8 M betaine, 1.4 mM dNTP mix, 2 μM each of the internal primers (FIP and BIP), 200 nM each of external primers (F3 and B3), 1 μM each of loop primers (FL and BL), 1 mM MnCl2, 50 μM calcein, 8 U Bst polymerase, 1.5 U warm RTx, and sterile water. All three reactions were then incubated in an incubator (Thermo Scientific, USA) for 1 h at 65 °C followed by 5 min at 80 °C for inactivating Bst polymerase. The optimum concentration of MgSO4 and betaine for RT-LAMP was determined by testing different concentrations of MgSO4 (0–10 mM) and betaine (0.4–1.4 M). To determine the optimum concentration of MgSO4, betaine was kept at 0.8 M. Optimum concentration of betaine was determined by keeping MgSO4 concentration at 4 mM. To determine the optimum temperature, the RT-LAMP reaction was performed for 60 min at four different temperatures viz. 58 °C, 62 °C, 65 °C, 68 °C. Optimal incubation time for the reaction was determined by performing reactions for 15 min, 30 min, 45 min, 60 min, and 75 min at a constant temperature of 65 °C.
Visual detection of LAMP products was made possible by the addition of MnCl2 and fluorescent dye, calcein to the reaction mix, which results in the development of green fluorescence under UV light for positive reactions. RT-LAMP products were also run on 1.5% agarose gel electrophoresis and documented using a GelDoc system.
Development of RT-RPA and optimization of reaction components and time
To set the RT-RPA reaction, the supplied freeze-dried reaction pellet of the Twist Amp® basic kit was dissolved in 29.5 µl of rehydration buffer and 8.2 μl nuclease-free water. The resultant mixture was aliquoted equally (7.54 μl each) into five PCR tubes. This was followed by the addition of 0.48 µl each of forward and reverse primers (480 nM each) and 1 µl of template cDNA to the tubes. For initiating the RPA reaction, 0.5 μl of 14 mM magnesium acetate solution was added and incubated the tube for 40 min at 39 °C with moderate mixing and spinning briefly after 4 min. The RPA reaction was stopped by keeping the tube at 65 °C for 10 min. Initially, to check the specificity of designed primers, three RT-RPA reactions were set up using positive control, negative control, and water control (without template). RT-RPA products were run on 2% agarose gel and visualized under GelDoc system. Reaction conditions were optimized using different concentrations of magnesium acetate (12–20 mM) and different durations of incubation time (10–50 min). To check if primers cross-react with CdMV and BBrMV, total RNA isolated from CdMV and BBrMV infected plants were also subjected to RT-RPA using CdVCV-specific primers.
RT-PCR and SYBR-Green-based real-time RT-PCR
Conventional RT-PCR and SYBR Green-based real-time RT-PCR were performed using RNA isolated from infected cardamom plants as described in Bhat et al. (2020). Single tube RT-PCR reaction mix consisted 1 μl (about 60 ng) of total RNA, 1 × Taq polymerase buffer that contained 1.5 mM MgCl2, 10 mM dithiothreitol, 400 μM dNTP mix, 10 pM each of forward and reverse primers, 1 U of RNase inhibitor, 1.25 U of RevertAid reverse transcriptase, 0.75 U of Taq polymerase and RNase-free water to make 50 μl. The temperature profile in the thermocycler involved cDNA synthesis at 42 °C for 45 min followed by 35 cycles at 94 °C for 30 s, 50 °C for 40 s and 72 °C for 1 min and a final extension at 72 °C for 10 min. Products of RT-PCR was run on agarose gel electrophoresis and documented using a GelDoc system. SYBR-Green-based real-time RT-PCR assay carried out in Rotor-Gene Q system (Qiagen, Germany) contained 12.5 μl of 2 × QuantiFast™ SYBR Green PCR master mix, 1 μl of each primer (1 μM/μl), 50 U of RevertAid reverse transcriptase, 1 μl of RNA template and water to make 25 μl. Thermocycling steps involved 42 °C for 45 min, 95 °C for 5 min followed by 35 cycles of 95 °C for 20 s and 60 °C for 20 s. The real-time RT-PCR product was subjected to melt analysis to confirm its specificity (Bhat et al. 2020).
Determination of detection limits and comparison of sensitivity
To determine the detection limits of different assays, extracted total RNA from CdVCV-infected cardamom plant (cDNA in the case of RT-RPA) was subjected to serial dilution (from 100 to 10− 10) and each dilution was subjected RT-LAMP, RT-RPA, RT-PCR, and real-time RT-PCR. Products were run on agarose gel for comparing the sensitivity of the assays. The experiment was repeated three times to confirm the results.
Validation of RT-LAMP and RT-RPA assays
For validating the optimized RT-LAMP and RT-RPA assays, cardamom plants representing different geographical regions such as Hassan and Uttara Kannada Districts of Karnataka, India with and without symptoms were tested by RT-LAMP and RT-RPA assays along with real-time RT-PCR assay.
Results
Development and optimization of RT-LAMP assay
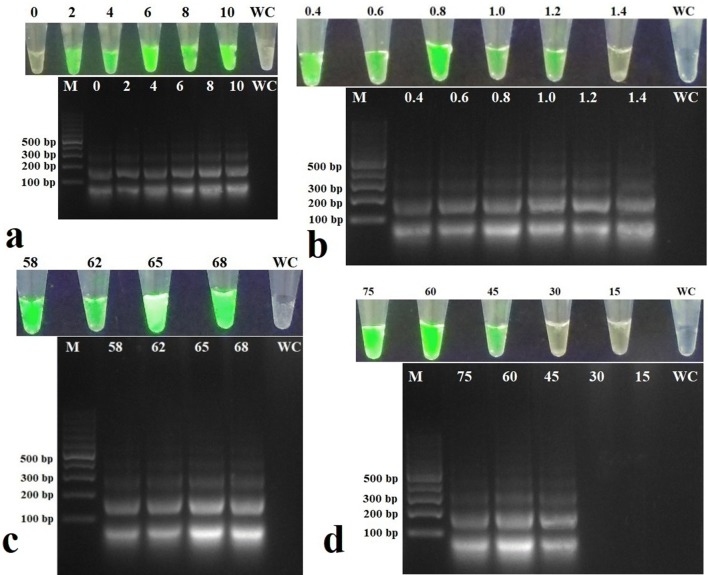
RT-LAMP product obtained with positive control showed green fluorescence when observed under UV light and a typical ladder-like arrangement when observed after agarose gel electrophoresis while no fluorescence or ladder-like arrangement was observed in the negative control and water control which indicated the specificity and proper design of primers. Further, RT-LAMP performed using template RNA extracted from CdMV- and BBrMV-infected cardamom plants did not show any fluorescence/ladder-like arrangement confirming the specificity of the primers for the detection of CdVCV (not shown). A magnesium sulfate concentration of 6 mM and betaine concentration of 0.8 M were found optimum for the detection of CdVCV through RT-LAMP (Fig. 1a, b). A temperature of 65 °C for 60 min was found better for the detection of CdVCV through RT-LAMP (Fig. 1c, d).
Fig. 1.
Effect of different concentrations of a MgSO4, b betaine, c temperature, and d duration on the detection of cardamom vein clearing virus (CdVCV) by reverse transcriptase loop-mediated isothermal amplification (RT-LAMP). Detection was done under UV light and agarose gel electrophoresis. a MgSO4. Lanes 0, 2, 4, 6, 8, and 10, are loaded with products of RT-LAMP carried out using different concentrations of MgSO4. b Betaine. Lanes 0.4, 0.6, 0.8, 1.0, 1.2, and 1.4 are loaded with products of RT-LAMP carried out using different concentrations of betaine. c Temperature. Lanes 58, 62, 65, and 68 are loaded with products of RT-LAMP carried out at different temperatures. d Duration. Lanes 15, 30, 45, 60, and 75 are loaded with products of RT-LAMP carried out over different durations. Lane M shows molecular size marker and Lane WC is water control
Development and optimization of RT-RPA assay
RT-RPA assay was found specific for the detection of CdVCV as the only positive control showed a positive reaction. Further, RT-RPA performed using extracted total RNA from CdMV- and BBrMV-infected cardamom plants did not show amplification indicating that the primers are specific for the detection of CdVCV (not shown). A magnesium acetate concentration of 16 mM was found optimum for RT-RPA (Fig. 2a). Optimization of incubation time revealed that RT-RPA can give expected amplicon within 10 min while the better intensity of bands was observed at 30, 40, and 50 min (Fig. 2b).
Fig. 2.
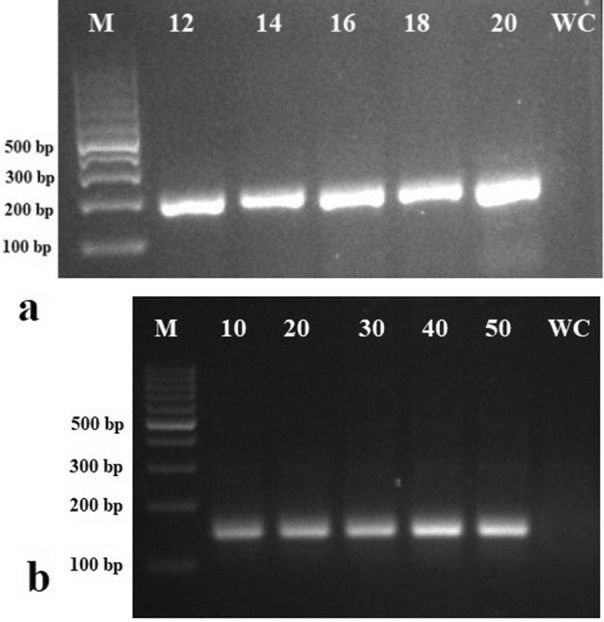
Agarose gel electrophoresis image showing the effect of different concentrations of a magnesium acetate and b duration on the detection of cardamom vein clearing virus (CdVCV) by reverse transcriptase-recombinase polymerase amplification (RT-RPA). a Magnesium acetate. Lanes 12, 14, 16, 18, and 20 are loaded with products of RT-RPA carried out at different concentrations of magnesium acetate. b Duration. Lanes 10, 20, 30, 40, and 50 are loaded with products of RT-RPA carried out at over different durations. Lane M shows molecular size marker and Lane WC is water control
Determination of detection limits of virus and comparison of the sensitivity of different assays
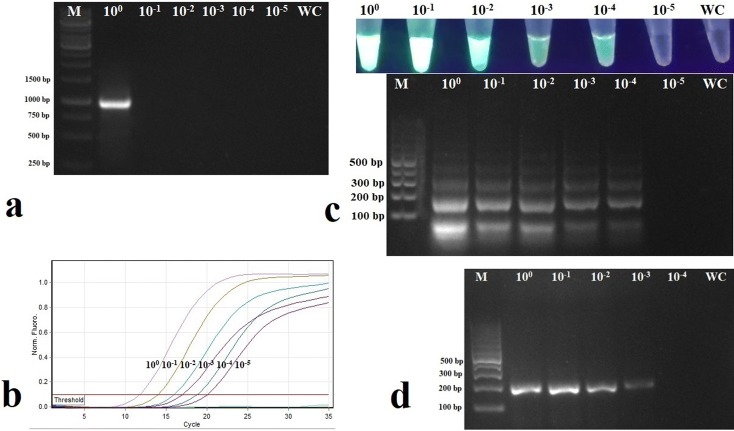
Detection limits of RT-LAMP and RT-RPA in detecting CdVCV revealed that RT-LAMP can detect the virus up to a 10− 4 dilution while the detection limit for RT-RPA was 10− 3 (Fig. 3c, d). The sensitivity of CdVCV detection using RT-LAMP and RT-RPA was compared with that of SYBR Green-based real-time RT-PCR and gel-based RT-PCR. RT-LAMP assay was 104 times higher sensitive than RT-PCR while it was 10 times less sensitive than real-time RT-PCR (Fig. 3a–c). On the other hand, RT-RPA was 103 times higher sensitive than RT-PCR and 102 times less sensitive than that of real-time RT-PCR (Fig. 3a, b, d). Among the two isothermal assays, RT-LAMP was ten times more sensitive than RT-RPA (Fig. 3c, d).
Fig. 3.
Comparison of the sensitivity of detection of cardamom vein clearing virus (CdVCV) by a reverse transcriptase-polymerase chain reaction (RT-PCR), b SYBR Green-based real-time RT-PCR, c reverse transcriptase loop-mediated isothermal amplification (RT-LAMP), and d reverse transcriptase recombinase polymerase amplification (RT-RPA). Lanes 100, 10− 1, 10− 2, 10− 3, 10− 4, 10− 5 show different dilutions of the original extracts of total RNA (cDNA in case of RT-RPA); Lane M shows molecular marker. Lane WC indicates water control. a Agarose gel electrophoresis of RT-PCR products. b Amplification curve obtained for different dilutions in SYBR Green-based real-time RT-PCR, c visual observation of RT-LAMP products under UV light and agarose gel electrophoresis, and d agarose gel electrophoresis of RT-RPA products
Validation of RT-LAMP and RT-RPA assays
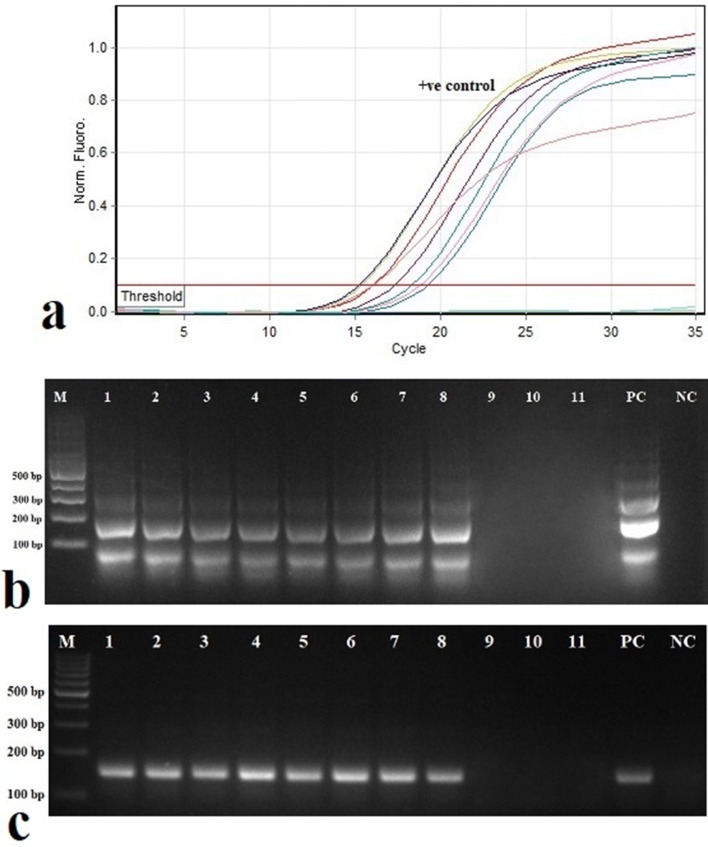
Validation of RT-LAMP and RT-RPA assays was carried out with field samples of cardamom plants. Out of 21 plants tested, 18 plants with symptoms were positive in RT-LAMP, and RT-RPA (Fig. 4b, c). Three non-symptomatic plants have shown a negative reaction to CdVCV. Similar results were obtained when samples were subjected to SYBR Green-based real-time RT-PCR (Fig. 4a).
Fig. 4.
Validation of reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) and reverse transcriptase recombinase polymerase amplification (RT-RPA) assays for the detection of cardamom vein clearing virus (CdVCV) in field samples of cardamom. a SYBR Green-based real-time RT-PCR showing amplification curves and Ct values obtained for different samples. b Agarose gel electrophoresis of RT-LAMP products. c Agarose gel electrophoresis of RT-RPA products. Lane 1–11 shows different samples. Lane M shows the molecular marker. Lane PC indicates positive control. Lane NC indicates negative control
Discussion
Cardamom, well-known as the queen of spices is affected by three important viral diseases. Being perennial and vegetatively propagated crops, the survival of viruses becomes easy in diseased cardamom plantations (Venugopal 2002). Thus the development of diagnostic assays is relevant for detecting these viruses in the early growth phase to eliminate and check further spread of viruses. Vein-clearing disease of cardamom although known to occur since 1993, the causal virus of the disease was identified only recently. The causal virus named as cardamom vein clearing virus (CdVCV) belongs to the genus, Nucleorhabdovirus under the family, Rhabdoviridae. A RT-PCR and SYBR Green-based real-time RT-PCR for the detection of CdVCV in cardamom was also reported (Bhat et al. 2020). In the current study, we developed, optimized, and validated assays such as RT-LAMP and RT-RPA for quick and sensitive detection of CdVCV in cardamom. The parameters such as the concentration of MgSO4 and betaine for RT-LAMP; magnesium acetate for RT-RPA; temperature and time for both RT-LAMP and RT-RPA were optimized. As reported by earlier researchers, the present findings also showed that RT-LAMP could be detected through green fluorescence or /and agarose gel electrophoresis with equal sensitivities (Tomita et al. 2008; Ravindran et al. 2012; Bhat et al. 2013; Siljo and Bhat 2014). Both RT-LAMP and RT-RPA assays developed in the current study were specific for the detection of CdVCV and did not cross-react with other viruses such as CdMV and BBrMV infecting cardamom. Assays such as conventional RT-PCR and real-time RT-PCR need more time and expensive in terms of equipment required to perform the tests which limit the wide usage of these assays for routine detection. Alternate techniques like RT-LAMP and RT- RPA are rapid and do not require expensive equipment like thermal cycler and hence can be performed in resource-poor laboratories. In the present study, sensitivity of these two isothermal assays was determined and compared with conventional RT-PCR and real-time RT-PCR assays. In general, sensitivity of these assays are in the order of real-time RT-PCR followed by RT-LAMP, RT-RPA, and conventional RT-PCR (Liu et al. 2010; Siljo and Bhat 2014; Jiao et al. 2019). In the current study also, similar results were observed where RT-LAMP and RT-RPA were 104 and 103 times more sensitive than that of conventional RT-PCR while they were slightly less sensitive (10− 1 and 10− 2) compared to real-time RT-PCR. Among the two isothermal assays optimized in the present study, RT-LAMP assay was ten times more sensitive than RT-RPA assay in the detection of CdVCV. Both RT-LAMP and SYBR Green-based real-time RT-PCR showed equal sensitivity in the detection of BBrMV infecting cardamom (Siljo and Bhat 2014). Similarly, Jiao et al. (2019) reported that RT-RPA is ten times more sensitive than RT-PCR in the detection of maize chlorotic mottle virus. When considering the cost of equipment and time required for the assays, both RT-LAMP and RT-RPA techniques are more practical methods to screen a large-field population of cardamom for CdVCV. These assays need only an incubator for setting up the reaction. However, among the two isothermal assays, RT-LAMP assay requires higher incubation temperature (about 65 °C), longer incubation time (about 1 h), a minimum of four primers for amplification while RT-RPA is a very simple assay that requires lower incubation temperature (about 37 °C), lesser incubation time (about 40 min), only two primers for amplification (Mohandas and Bhat 2020). In conclusion, in the present study, two molecular isothermal assays, RT-LAMP, and RT-RPA were optimized for the quick, specific and sensitive detection of CdVCV infecting cardamom. We believe that these assays will enable in the identification and production of virus-free cardamom plants and to check the spread of the virus. Besides, diagnostics can also be used to screen cardamom germplasm accessions for sources of resistance and to study the virus epidemiology, host range, and reservoir in nature.
Acknowledgement
Authors are thankful to Science and Engineering Research Board (SERB), Government of India for funding (EMR/2016/001135), Head, Division of Crop Protection, Director, ICAR-Indian Institute of Spices Research, Kozhikode, Kerala, India for facilities.
Author contribution
AIB conceptualized, designed experiments and finalized the manuscript; KPN performed all experiments and wrote the manuscript; both authors have read and approved the final manuscript.
Funding
Science and Engineering Research Board (SERB), Government of India (EMR/2016/001135).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
K. P. Naveen, Email: naveen916kp@gmail.com
A. I. Bhat, Email: aibhat65@gmail.com, Email: Ishwarabhat.a@icar.gov.in
References
- Babu B, Washburn BK, Miller SH, Poduch K, Sarigul T, Knox GW, Paret ML. A rapid assay for detection of Rose rosette virus using reverse transcription-recombinase polymerase amplification using multiple gene targets. J Virol Methods. 2017;240:78–84. doi: 10.1016/j.jviromet.2016.11.014. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Roy S, Sharma SK, Dutta SK, Chandra S, Ngachan SV. Reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for rapid diagnosis of chilli veinal mottle virus. Arch Virol. 2016;161:1957–1961. doi: 10.1007/s00705-016-2850-7. [DOI] [PubMed] [Google Scholar]
- Bhat AI, Pamitha NS, Naveen KP, Biju CN. Identification and characterization of cardamom vein clearing virus, a novel aphid-transmitted nucleorhabdovirus. Eur J Plant Pathol. 2020;156:1053–1062. doi: 10.1007/s10658-020-01958-2. [DOI] [Google Scholar]
- Bhat AI, Siljo A, Deeshma KP. Rapid detection of Piper yellow mottle virus and Cucumber mosaic virus infecting black pepper (Piper nigrum) by loop-mediated isothermal amplification (LAMP) J Virol Methods. 2013;193:190–196. doi: 10.1016/j.jviromet.2013.06.012. [DOI] [PubMed] [Google Scholar]
- Congdon BS, Kehoe MA, Filardo FF, Coutts BA. In-field capable loop-mediated isothermal amplification detection of Turnip yellows virus in plants and its principal aphid vector Myzus persicae. J Virol Methods. 2019;265:15–21. doi: 10.1016/j.jviromet.2018.12.014. [DOI] [PubMed] [Google Scholar]
- Dietzgen RG, Kondo H, Goodin MM, Kurath G, Vasilakis N. The family Rhabdoviridae: mono- and bipartite negative sense RNA viruses with diverse genome organization and common evolutionary origins. Virus Res. 2017;227:158–170. doi: 10.1016/j.virusres.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. Biology of plant rhabdoviruses. Ann Rev Phytopath. 2005;43:623–660. doi: 10.1146/annurev.phyto.43.011205.141136. [DOI] [PubMed] [Google Scholar]
- Jacob T, Usha R. 3′-Terminal sequence analysis of the RNA genome of the Indian isolate of Cardamom mosaic virus: a new member of genus Macluravirus of Potyviridae. Virus Genes. 2001;23:81–88. doi: 10.1023/A:1011191614839. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Jiang J, An M, Xia Z, Wu Y. Recombinase polymerase amplification assay for rapid detection of maize chlorotic mottle virus in maize. Arch Virol. 2019;164:2581–2584. doi: 10.1007/s00705-019-04361-3. [DOI] [PubMed] [Google Scholar]
- Le DT, Netsu O, Uehara-Ichiki T, Shimizu T, Choi IR, Omura T, Sasaya T. Molecular detection of nine rice viruses by a reverse-transcription loop-mediated isothermal amplification assay. J Virol Methods. 2010;170:90–93. doi: 10.1016/j.jviromet.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang Z, Qian Y, Mu J, Shen L, Wang F, Yang J. Rapid detection of tobacco mosaic virus using the reverse transcription loop-mediated isothermal amplification method. Arch Virol. 2010;155:1681–1685. doi: 10.1007/s00705-010-0746-5. [DOI] [PubMed] [Google Scholar]
- Mohandas A, Bhat AI. Recombinase polymerase amplification assay for the detection of piper yellow mottle virus infecting black pepper. VirusDis. 2020 doi: 10.1007/s13337-019-00566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X. Reverse transcription loop-mediated isothermal amplification of DNA for detection of Potato virus Y. Plant Dis. 2005;89:605–610. doi: 10.1094/PD-89-0605. [DOI] [PubMed] [Google Scholar]
- Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4:e204. doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran PN, Madhusoodanan KJ. Cardamom the genus Elettaria. London: Taylor and Francis; 2002. [Google Scholar]
- Ravindran A, Levy J, Pierson E, Gross DC. Development of a loop-mediated isothermal amplification procedure as a sensitive and rapid method for detection of 'Candidatus liberibacer solanacearum' in potatoes and psyllids. Phytopathology. 2012;102:899–907. doi: 10.1094/PHYTO-03-12-0055-R. [DOI] [PubMed] [Google Scholar]
- Siljo A, Bhat AI. Reverse transcription loop-mediated isothermal amplification assay for rapid and sensitive detection of Banana bract mosaic virus in cardamom (Elettaria cardamomum) Eur J Plant Pathol. 2014;138:209–214. doi: 10.1007/s10658-013-0318-0. [DOI] [Google Scholar]
- Siljo A, Bhat AI, Biju CN, Venugopal MN. Occurrence of Banana bract mosaic virus on cardamom. Phytoparasitica. 2012;40:77–85. doi: 10.1007/s12600-011-0193-1. [DOI] [Google Scholar]
- Silva G, Bömer M, Nkere C, Kumar PL, Seal SE. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J Virol Methods. 2015;222:138–144. doi: 10.1016/j.jviromet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Venugopal MN. Viral diseases of cardamom. In: Ravindran PN, Madhusoodanan KJ, editors. Cardamom The genus Elettaria. London: Taylor and Francis; 2002. pp. 143–159. [Google Scholar]
- Zeng R, Luo J, Gao S, Xu L, Song Z, Dai F. Rapid detection of Cucumber green mottle mosaic virus by reverse transcription recombinase polymerase amplification. Mol Cell Probes. 2019;43:84–85. doi: 10.1016/j.mcp.2018.12.005. [DOI] [PubMed] [Google Scholar]