Abstract
MTA2 is involved in tumor proliferation and metastasis. However, the role of MTA2 in cervical cancer thus far has not been identified. In this study, we report that elevated expression of MTA2 negatively correlates with Kallikrein-10 (KLK10) expression and poor prognosis of cervical cancer patients. Knockdown of MTA2 substantially inhibited tumor cell migration and invasion, and it enhanced KLK10 expression of the cervical cancer cells in vitro and in vivo. Functionally, shMTA2-mediated suppression of cell mobility was significantly restored by knockdown of KLK10. We also found that Sp1 (transcription factor specificity protein 1) is critical for shMTA2-induced transcriptional upregulation of KLK10 and subsequent biological functions. Furthermore, we found that the expression of miR-7 is elevated by MTA2 silencing and then by direct inhibition of Sp1 expression. Knockdown of Sp1 additively enhanced KLK10 expression in MTA2-knocked down cervical cancer cells, suggesting that the miR-7/Sp1 axis acts as an effector of MTA2 to impact KLK10 levels and mobility of cervical cancer cells. Taken together, our findings provide new insights into the physiological relationship between MTA2 and KLK10 via regulating the miR-7/Sp1 axis, and they provide a potential therapeutic target in cervical cancer.
Keywords: MTA2, KLK10, Sp1, miRNA-7, cervical cancer
Graphical Abstract

In vitro and in vivo studies revealed that knockdown of MTA2 inhibits metastasis of cervical cancer through induction of miR-7 targeting Sp1/KLK10 expression. These finding provide novel molecular mechanistic insight on the MTA2/miR-7/Sp1/KLK10 axis and its potential therapeutic target in cervical cancer.
Introduction
The mortality and incidence of cervical cancer are expected to decrease in the future with application of human papilloma virus (HPV) vaccines; however, cervical cancer remains a major public health issue in developed countries and the fourth leading cause of cancer death.1 Therefore, continuously studying the carcinogenesis of cervical cancer and developing novel therapeutic strategies for this disease are still an urgent need.
The metastasis-associated protein family (MTA1, MTA2, and MTA3) has been shown to be closely associated with the Mi2/NuRD nucleosome remodeling complexes.2 MTA2 has been reported to overexpress in a number of cancers and is involved in tumor progression and metastasis.3 MTA2 enhances metastatic behavior and is associated with poor outcomes in estrogen receptor-negative breast cancer.4 One study demonstrated that overexpression of MTA2 in non-small-cell lung cancer (NSCLC) inhibits epithelial cell adhesion molecule (Ep-CAM) and E-cadherin to promote NSCLC metastasis.5 These results showed that MTA2 might play a critical role in the progression of cancer cells.
Kallikrein-10 (KLK10) is a member of the kallikrein family.6 Recently, KLK10 was recognized as a novel putative tumor suppressor in breast cancer and prostate cancer. The mechanism of this tumor-specific loss characteristic was reportedly related to exon 3 hypermethylation and loss of KLK10 mRNA expression.7 It has been shown that expression of KLK10 was lower in breast cancer cells than in normal breast epithelial cells, and that overexpression of the KLK10 gene inhibited tumor growth in breast cancer cells.8 Another study also indicated that overexpression of KLK10 inhibits cell growth and increases the sensitivity to cisplatin treatment of esophageal cancer.9 However, knowledge on the function of KLK10 in cervical cancer is still unclear.
In recent years, more and more studies have indicated that miRNAs act as tumor suppressors or oncogenes and have been shown to be involved in the progression and metastasis of cancer cells.10 Studies have demonstrated that miR-7 was poorly expressed and also played a tumor-suppressive role in several types of tumors. For example, overexpressed miR-7 resulted in suppression of osteosarcoma proliferation, invasion, and migration whereas it induced apoptosis in vitro and in vivo.11 miR-7 inhibited the KLF4-VEGF signaling axis and played an important role in the regulation of angiogenesis.12 Previous studies indicated that miR-7 might have anti-tumor effects on cervical cancer cell proliferation and metastasis.13,14 However, another study reported that miR-7 modulated cellular energy homeostasis to promote cisplatin resistance of cervical cancer cells,15 suggesting that the role of miR-7 in cervical cancer progression is still unclear.
In this study, we found that the expression of MTA2 is negatively correlated with KLK10 in cervical cancer tissue and cell lines. Furthermore, we showed that downregulation of MTA2 induced miR-7-mediated transcriptional regulation of KLK10 expression via transcription factor specificity protein 1 (Sp1), and then inhibited cell migration and the invasion ability of cervical cancer cells. To our knowledge, these results are the first to indicate that MTA2 and KLK10 are functionally involved in the metastasis of cervical cancer, and they provide new insights regarding cervical cancer treatment.
Results
Expression of MTA2 Was Positively Associated with Advanced Tumor Grade and Poor Survival of Patients with Cervical Cancer
In the past, many studies have indicated that MTA2 is involved in the proliferation, migration, and poor prognosis of cervical cancer.16,17 The expression levels of MTA2 in human cervical tumor tissues were detected by immunohistochemistry (IHC) analysis. As shown in Figures 1A and 1B, expression of MTA2 was upregulated in grade III cervical tumor tissues compared with tumor grades I and II (p < 0.01). Further bioinformatics data from The Cancer Genome Atlas (TCGA) database supported this finding. As indicated in Figure 1C, expression of MTA2 in tumor tissue was significantly higher than in normal tissue (p < 0.01). Moreover, Kaplan-Meier analysis demonstrated that the patients with high expression of MTA2 had poor survival as compared with those with low expression of MTA2 (Figure 1D). Based on the Oncomine database analysis, we found that a high level of MTA2 expression was associated with cervical cancer compared to normal cervix uteri tissues (p < 0.01; Figure 1E). These results indicated that expression of MTA2 positively correlated with the tumor grade of cervical cancer.
Figure 1.
MTA2 Is Highly Expressed in Tumor Tissue and Correlated with Poor Prognosis of Cervical Cancer Patients
(A) Representative IHC staining of MTA2 from tumor grade I to grade III. Scale bars, 100 μm. (B) MTA2 protein amounts of the tissue sections from cervical cancer patients were examined by IHC staining and scored for their color density. The expression of MTA2 was positively correlated with tumor grade. ∗∗p < 0.01. (C) MTA2 expression levels of cervical cancer patients were analyzed by TCGA database. ∗∗p < 0.01, compared with normal tissue. (D) Kaplan-Meier curves for overall survival for human cervical cancer related to MTA2 expression. (E) MTA2 expression levels of cervical cancer patients were statistically higher than those for normal cervix uteri. ∗∗p < 0.01, compared with cervix uteri.
MTA2 Knockdown Attenuated Cervical Cancer Cell Lung Metastasis
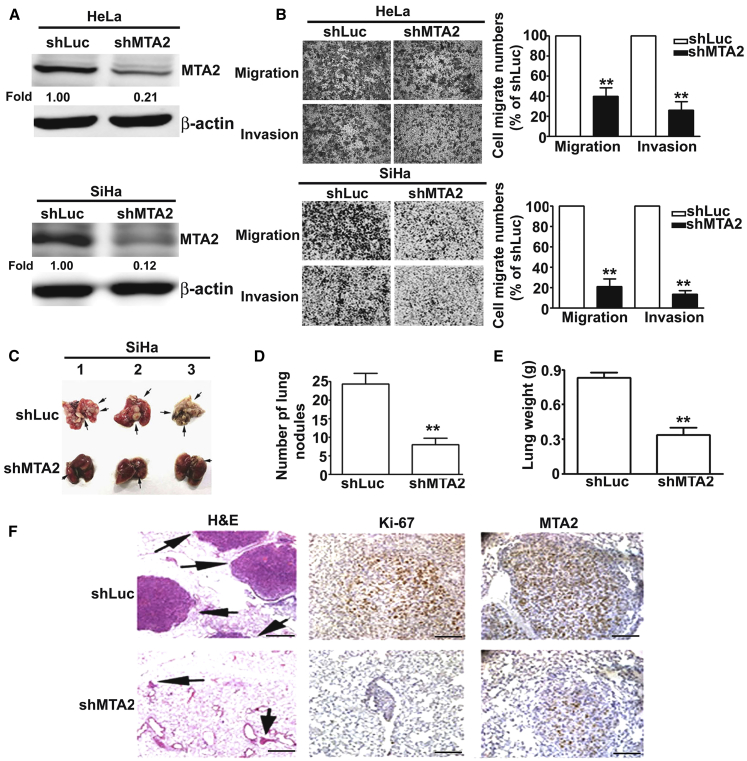
To investigate the biological function of MTA2 in human cervical cancer tumorigenesis, we knocked down the expression of MTA2 by specific shMTA2 (MTA2 short hairpin RNA [shRNA]) in HeLa and SiHa cells (Figure 2A) and found that inhibition of MTA2 suppressed the migratory and invasion activity of human cervical cancer cells (Figure 2B). To further define the role of MTA2 in an in vivo model, we injected shLuc and shMTA2 cervical cancer cells into female BALB/c mice via tail vein injection. Two months after injection, we found that knockdown of MTA2 significantly reduced the lung metastatic nodules in the shMTA2 group (Figures 2C and 2D). The lung weight of shMTA2 groups was also lighter than that in the shLuc groups (Figure 2E). Hematoxylin and eosin staining of lung sections revealed that the shLuc groups showed more invading tumor cells (Figure 2F, left panel). We also found that the expression levels of MTA2 and Ki-67 were significantly decreased in the shMTA2 group compared with the shLuc group by IHC staining (Figure 2F, right panel). These results demonstrated that inhibition of MTA2 suppressed the metastatic ability of human cervical cancer in vitro and in vivo.
Figure 2.
Inhibition of MTA2 Suppressed Cervical Cancer Cell Metastasis In Vitro and In Vivo
(A) HeLa and SiHa cells were infected with shLuc or shMTA2 and then treated with puromycin (2 μg/mL) for 5 days. Then, total lysates were isolated and analyzed by western blotting. (B) The migration and invasion abilities of shLuc and shMTA2-HeLa and shMTA2-SiHa cells were determined by a migration assay and Matrigel-invasion assay. Cells in the lower surface of the Borden chamber were stained and imaged under a light microscope at ×200 original magnification. Quantification of migrated cells is shown as a histogram chart. (C) Mouse lungs show the metastatic nodules of the shLuc group and shMTA2 group. (D) Quantification of metastatic nodules. (E) The lung weights of mice injected with shLuc and shMTA2 cells were detected. (F) The lungs were fixed in 4% paraformaldehyde and sectioned for H&E staining and IHC staining with anti-Ki-67 and anti-MTA2 antibodies. ∗∗p < 0.01, compared with the shLuc group. Scale bars, 20 μm.
KLK10 Is Critical for MTA2-Mediated Cell Migration/Invasion and Acts as a Prognosis Marker for Cervical Cancer Patients
To define the downstream effectors of MTA2, we performed a human protein array assay and found that knockdown of MTA2 significantly increased KLK10 protein expression in human cervical cancer cells (Figures 3A and 3B). To further clarify the results from the protein array assay, we knocked down the expression of MTA2 in SiHa cells and HeLa cells and found that the expressions of KLK10 were significantly increased in mRNA and in protein levels by quantitative RT-PCR (qRT-PCR) analysis (Figure 3C), a western blot assay (Figure 3D), and an ELISA assay (Figure 3E), respectively. We also analyzed the expressions of MTA2 and KLK10 among three cervical cancer cell lines with differential invasive abilities. We found that the expressions of KLK10 were inversely correlated with the expressions of MTA2 in mRNA and in protein levels of cervical cancer cell lines (Figures 3F and 3G). Transfection of the MTA2-expressing construct markedly abolished shMTA2-induced KLK10 induction (Figures 3H–3J). A previous study indicated that KLK10 was specifically downregulated in breast and cervical cancer cell lines.18 To further investigate the functional role of KLK10 in the progression of human cervical cancer cells, we suppressed the expression of KLK10 by siKLK10 (KLK10 small interfering RNA [siRNA]) in HeLa/shMTA2 cells and found dramatic restorations of the shMTA2-reduced migratory and invasive abilities (Figures 3K–3M). In the clinical tissues, we found that the expression of KLK10 in cervical tumor tissue was lower than that in normal tissue (p < 0.05; Figure 3N). The expression levels of KLK10 in cervical tumor tissues were inversely correlated with tumor grade (p < 0.05; Figure 3O and 3P) and positively correlated with overall survival of patients (p < 0.01; Figure 3Q). Collectively, these results indicated that KLK10 was negatively regulated by MTA2 and functionally involved in MTA2-mediated cervical cancer metastasis.
Figure 3.
KLK10 Is Critical for MTA2-Mediated Cell Migration/Invasion and Acts as the Prognosis Marker for Cervical Cancer Patients
(A) The total protein lysates of shLuc and shMTA2 stably transfected cells were collected for a human protein array assay. (B) Quantification of the protein expression of KLK10 was presented in terms of percentage of shLuc is shown as a histogram. (C–E) The expression levels of MTA2 and KLK10 in mRNA and protein levels of shLuc and shMTA2 cells were examined by a qRT-PCR assay (C), western blotting (D), and ELISA (E), respectively. β-Actin acted as an internal control. ∗∗p < 0.01, compared with shLuc cells. (F and G) The protein and mRNA expression levels of MTA2 and KLK10 of three cervical cancer cell lines were examined by western blotting (F) and qRT-PCR (G). β-Actin acted as the loading control. (H–J) Expression of MTA2 and KLK10 in protein and mRNA levels were examined by a western blot assay (H), ELISA (I), and qRT-PCR (J) with ectopic expression of MTA2. β-Actin acted as the protein loading control. GAPDH was used for normalization of the MTA2 and KLK10 mRNA levels. ∗p < 0.05, ∗∗p < 0.01, compared with shLuc cells transfected with Neo plasmid; #p < 0.05 compared with shMTA2 cells transfected with Neo plasmid. (K and L) The expression levels of MTA2 and KLK10 were detected by western blotting (K) and qRT-PCR assays (L) of shMTA cells transfected with KLK10 siRNA. (M) Effects of migratory and invasive abilities of KLK10 siRNA on shMTA2 cells were measured by a migration assay and invasion assay. ∗∗p < 0.01, compared with shLuc cells transfected with si-Control; #p < 0.05, compared with shMTA2 cells transfected with si-Control. (N) KLK10 expression levels of cervical tumor tissue and normal tissue were analyzed using TCGA database. ∗p < 0.05, compared with normal tissue. (O) Representative IHC staining of KLK10 from tumor grade I to grade III. Scale bars, 100 μm. (P) Expression of KLK10 correlated with tumor grade. ∗p < 0.05, grade III compared with grade I+II. (Q) Kaplan-Meier curves for overall survival of human cervical cancer patients.
Sp1 Is Involved in MTA2-Mediated Transcriptional Regulation of KLK10 and Subsequent Cell Migration and Invasion
Previous studies have reported that Sp1 negatively regulated the expression of KLK10 protein.19 To determine whether silencing MTA2 promoted KLK10 expression via suppressing transcriptional activity of Sp1, we performed western blotting analysis in shMTA2-SiHa and shMTA2-HeLa cells and found that MTA2 silencing inhibited total protein and nuclear expression of Sp1 in SiHa and HeLa cells (Figure 4A). In order to further clarify the role of Sp1 in MTA2-mediated KLK10 expression and metastatic ability, we used the si-Sp1 to specifically knock down the expression of Sp1. Western blotting analysis showed that Sp1 silencing increased the expression of KLK10 in both cells (Figure 4B). We also found that Sp1 silencing significantly increased the KLK10 expression in shMTA2-HeLa cells by a western blot assay (Figure 4C), as well as by an immunofluorescence staining assay (Figure 4D). We also found that treatment with si-Sp1 exhibited lower numbers of migrating and invasive cells in shMTA2-HeLa cells (Figure 4E). In addition, we found that the expression of Sp1 in cervical tumor tissue was higher than that in normal tissue from TCGA database (Figure 4F).
Figure 4.
Sp1 Is Involved in MTA2-Mediated Transcriptional Regulation of KLK10 and Subsequent Cell Migration and Invasion
(A) The protein extracted from shLuc and shMTA2 cells was separated into the nuclear part and total lysate and then subjected to western blotting to analyze the expression of MTA2 and Sp1. β-Actin was used as an internal control. Nuclear proteins were resolved and the levels of Sp1 were analyzed. Lamin B and α-tubulin were used as internal controls for nuclear and cytosolic fractions, respectively. (B) Detected expression of Sp1 and KLK10 in HeLa and SiHa cells after siRNA transfection. (C) MTA2, Sp1, and KLK10 expression levels were analyzed by western blotting in indicated cells. (D) Sp1 and KLK10 expression levels after siRNA transfection were analyzed by immunofluorescence staining. Scale bars, 50 μm. (E) The migratory and invasive abilities of indicated cells were analyzed by migration and invasion assays. (F) Expression status of Sp1 in Oncomine and TCGA databases. (G) The luciferase reporter constructs of the truncated KLK10 promoters were introduced into shLuc and shMTA2 cells. Luciferase activity was measured 48 h post-transfection. (H) Predicted Sp1 binding sites in the promoter region of KLK10. (I) The association between Sp1 and KLK10 in shMTA2 cells was examined by a chromatin immunoprecipitation (ChIP) assay with anti-Sp1 antibody, as well as PCR analysis of the genes encoding the promoter regions of KLK10 (−206/−6 and −692/−493). (J) Ectopic expression of MTA2 in shMTA2 cells and detection of the binding activity of Sp1 to the KLK10 promoter by a ChIP assay. ∗∗p < 0.01, compared with control.
Since KLK10 was regulated by MTA2 in both mRNA and protein levels, we further investigated whether MTA2-regulated KLK10 suppression is through transcriptional regulation. It has been reported that Sp1 is critically involved in transcriptional suppression of several important genes via binding to the 5′-flank promoter region.20,21 Therefore, we hypothesized that Sp1 binding to the KLK10 promoter inhibits its expression. We constructed serially deleted KLK10 5′-flank promoter region constructs (−1000 to −1, −1000 to −500, and −500 to −1 bp) and inserts to the pGL3-basic vector and evaluated the transcriptional activity by a Dual-Luciferase activity assay. As shown in Figure 4G, the region of −500 to −1 bp of the KLK10 5′-flank promoter is required for MTA2-mediated regulation of KLK10 promoter activity. There are two putative Sp1-binding sites (nucleotides −692/−493 and nucleotides −206/−6) on the KLK10 5′-flank promoter region (Figure 4H). To identify whether Sp1 binds to the KLK10 5′-flank promoter, a chromatin immunoprecipitation (ChIP) assay was performed. Consistent with the luciferase assay, the results from ChIP assay showed that knockdown of MTA2 decreased the activity of Sp1 binding to the Sp1 response element on the KLK10 5′-flank promoter region −206/−6, but not on the KLK10 5′-flank promoter region −692/−493 (Figure 4I). We transfected full-length MTA2-expressing vector in the shMTA2 cells and found that they significantly recovered the activity of Sp1 binding to the Sp1 response element on the KLK10 5′-flank promoter region (Figure 4J). These results showed that Sp1 is important for MTA2-mediated KLK10 expression and subsequent biological functions.
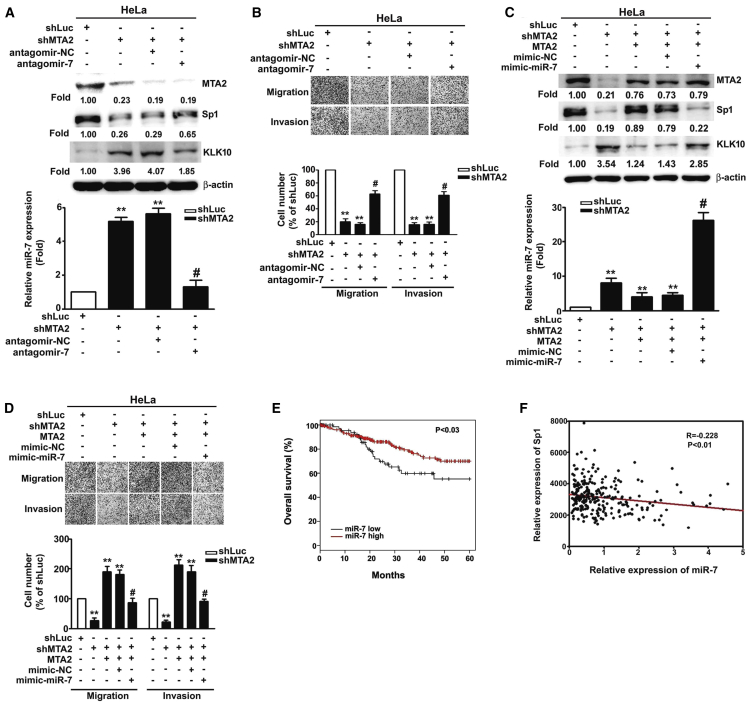
miR-7 Suppressed shMTA2-Mediated Induction of Sp1
miRNAs have been shown to play a critical role in cancer progression, including cancer metastasis.22 Therefore, the shMTA2 cells were subjected to miRNA microarray analysis (Figure 5A) and the results from the miRNA microarray were confirmed by qRT-PCR. We found that miR-7-5p, miR-10b, miR-320d, and miR-486-3p were increased in MTA2-silenced cells (Figure 5B). Furthermore, we also found the potential miR-7 targeting sites on the 3′ UTR regions of Sp1 by TargetScan, miRCode, and miRDB software (Figure 5C). To determine whether Sp1 is regulated by miR-7, the 3′ UTR of Sp1 containing the wild-type (WT) miR-7 binding site and mutant (Mut) binding site were constructed and inserted into the pmirGLO luciferase reporter plasmid. The binding of miR-7 to the 3′ UTR of Sp1 was further demonstrated by a luciferase reporter assay. The results showed that shMTA2-HeLa or shMTA2-SiHa cells significantly decreased the luciferase activity of the WT Sp1 3′ UTR, but not the miR-7 binding site Mut Sp1 3′ UTR (Figure 5D), suggesting that MTA2 regulates Sp1 mainly through induction of miR-7. In addition, overexpression of MTA2 significantly induced the luciferase activity of the Sp1 3′ UTR reporter (Figure 5E).
Figure 5.
miR-7 Suppressed shMTA2-Mediated Induction of Sp1
(A) Human miRNA microarray analysis to detect the miRNA expression profile of MTA2-silenced stable cells. (B) Expression levels of candidate miRNAs were determined by a qRT-PCR assay. (C) Identification of miRNA target gene. The miRbase, miRDB, and TargetScan programs were used to predict putative miRNA binding sites on the 3′ UTR of human Sp1. The schematic presentation includes the constructed Sp1 3′ UTR luciferase plasmids (WT and Mut) that were used in this study. (D) The WT and Mut of Sp1 3′ UTR luciferase reporters were transfected into MTA2-silenced stable cells, followed by detection with a luciferase reporter assay. (E) The Sp1 3′ UTR luciferase levels were analyzed in the indicated cells. ∗∗p < 0.01, compared with shLuc cells transfected with Neo plasmid; #p < 0.05, compared with shLuc stable cells transfected with MTA2 plasmid.
miR-7 Regulates the Sp1/KLK10 Axis and Inhibits MTA2-Induced Cell Metastasis
To clarify the regulatory effects of miR-7 on cervical cancer metastasis, antagomiR-7 was performed to suppress the expression of miR-7 in shMTA2 cells. Transfection with antagomiR-7 into MTA2-silenced HeLa cells significantly suppressed shMTA2-induced miR-7 expression and then recovered Sp1 expression as well as cell migratory and invasive abilities compared to shMTA2-HeLa cells (Figures 6A and 6B) and shMTA2-SiHa cells (Figures S1A–S1C). Consistently, overexpression of MTA2 in shMTA2-HeLa cells inhibited shMTA2-mediated miR-7 and KLK10 expression, enhanced Sp1 expression, and functionally restored the migratory and invasive abilities (Figures 6C and 6D). The result is similar to shMTA2 stable SiHa cells (Figures S2A and S2B). We also found that co-transfection of the miR-7 mimic with the MTA2-expressing vector into shMTA2-HeLa or shMTA2-SiHa cells abolished MTA2-regulated Sp1 and KLK10 as well as metastatic functions (Figures 6C and 6D; Figure S2C). In addition, we analyzed TCGA database and found that patients with low levels of miR-7 predicted poor survival probability compared with high levels of miR-7 (p < 0.03; Figure 6E). The expression of miR-7 was inversely correlated with the expression of Sp1 in cervical cancer tissue (p < 0.01; Figure 6F). These data suggested that miR-7 inhibits Sp1 and induces KLK10 expression to suppress migration and invasion of cervical cancer cells.
Figure 6.
miR-7 Regulates the Sp1/KLK10 Axis and Inhibits MTA2-Induced Cell Metastasis
(A) The MTA2-silenced stable HeLa cells were transfected with the antagomiR-NC or antagomiR-7 for 48 h followed by determination of the expression of miR-7 by qRT-PCR, as well as the expressions of MTA2, Sp1, and KLK10 by western blotting. (B) The migratory and invasive abilities of indicated cells were measured by migration and invasion assays. (C) The MTA2-silenced stable HeLa cells were co-transfected with MTA2 plasmid and miR-7 mimic for 48 h, followed by determination of the expression of miR-7 by qRT-PCR, the expressions of Sp1 and KLK10 by western blotting, and (D) the migratory and invasive ability were determined by migration and invasion assays. (E) Kaplan-Meier curves for overall survival of human cervical cancer patients. (F) Analysis of the correlation between miR-7 expression and Sp1 expression in human cervical cancer tissues from TCGA (p < 0.01). ∗∗p < 0.01, compared with shLuc cells; #p < 0.05, compared with shMTA2 stable cells transfected with antagomiR-NC or MTA2 plasmid.
Discussion
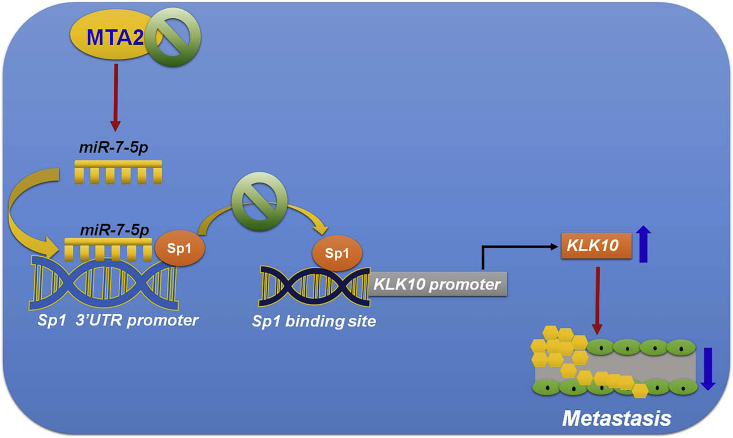
To date, studies on breast cancer, lung cancer, and colorectal cancer have found that expression of MTA2 is associated with tumor growth, invasion, and poor prognosis.3 However, the biological function and regulated mechanism of MTA2 in human cervical cancer remain to be clarified. Our current data revealed that MTA2 protein expressions were significantly increased in cervical cancer tissues compared with normal tissues, and that MTA2 mRNA and protein expression levels were upregulated in invasive cells (HeLa and SiHa) compared with non-invasive C33a cells. Furthermore, inhibition of MTA2 suppresses cancer cell metastasis in vitro and in vivo. Tissue arrays and TCGA database indicated that KLK10 expression was inversely correlated with MTA2 in cervical cancer tissue. In addition, proteinase arrays demonstrated that inhibition of MTA2 upregulated KLK10 expression. These data suggested that MTA2 is a key regulator for cervical cancer metastasis through regulating KLK10 expression. We additionally demonstrated that inhibition of MTA2 induced miR-7 expression and target to Sp1 3′ UTR, and Sp1 negatively regulated KLK10 transcription (Figure 7). These results show that MTA2 and KLK10 are involved in cell mobility and cancer metastasis. They also provide a potential anti-metastatic candidate for intervention in cervical cancer.
Figure 7.
Schematic Representation of the Inhibition of MTA2-Induced miR-7 Expression and the Target to Sp1 3′ UTR, Leading to Upregulated KLK10 Transcription, Then Suppression of the Metastatic Ability of Cervical Cancer
KLK10 expression is abundantly upregulated in several cancers, is positively correlated with the degree of tumor deterioration, and is not conducive to patient prognosis, including ovary cancer, pancreatic cancer, colon cancer, and gastric cancer.23,24 However, downregulated expression of KLK10 occurs in breast cancer and hepatocellular carcinoma.8,25 Upregulated KLK10 inhibits esophageal cancer proliferation and enhances cisplatin sensitivity in vitro.9 In NSCLC, forced expression of KLK10 remarkably suppressed cellular proliferation, migration in vitro, and oncogenicity in vivo.26 Our present study demonstrated that KLK10 was downregulated in cervical cancer tissue and highly invasive cancer cells. Through inhibition of MTA2, KLK10 expression was induced and cervical cancer cell migration and invasion abilities were inhibited, suggesting that KLK10 might acts as a tumor suppressor in cervical cancer.
It has been reported that inhibition of transcription factor Sp1 leads to upregulation of kallikrein-related peptidases, especially KLK10.19 Therefore, we wanted to know whether Sp1 is a negative regulator of KLK10 in cervical cancer. To confirm this hypothesis, we constructed a reporter plasmid that contained the 5′-flank region promoter of KLK10 with luciferase activity. Our results indicated that KLK10 5′-flank promoter activities were increased by MTA2 silencing in cervical cancer cells, suggesting that Sp1 indeed negatively regulated the transcription of KLK10. Li et al.18 revealed that deleted exon 1 and intron 1 of the KLK10 promoter is increased by about 40- to 50-fold, compared with the full length of the KLK10 promoter, suggesting that the KLK10 gene is a transcriptional suppressor and that hypermethylation of the KLK10 gene is a potential marker for breast cancer. The downregulation of KLK10 is often associated with hypermethylation in CpG-rich exon 3 in several cancers.26 Although the NuRD complex is mainly responsible for the deacetylation of gene transcription, the complex also contains MBD (methyl-CpG-binding)-related proteins and interacts with methyltransferases 7 and 8, indicating that the NuRD complex has not only a deacetylation function but also a methylation function.27 Based on the above findings and our results, it is concluded that inhibition of MTA2 expression might result in loss of the stability and methylation function of the NuRD complex, and subsequently abolish the suppression of KLK10 expression, but the detailed mechanisms require further exploration.
miR-7 has been shown to play a role as a tumor suppressor in many cancers.28 Inhibition of CKS1 protein expression by miR-7 inhibits proliferation and metastasis of thyroid cancer.29 It has also been found that transfection with the miR-7 mimic in cervical cancer inhibits focal adhesion kinase and suppresses cervical cancer metastasis.14 Our results indicated that inhibition of MTA2 induced miR-7 expression, suggesting that miR-7 is indeed involved in cervical cancer metastasis. Through analyzing the miRbase, miRDB, and TargetScan programs, it was found that miR-7 not only binds to FAK but also targets the Sp1 3′ UTR. Therefore, inhibiting the expression of Sp1 by miR-7 after knockdown of MTA2 may be one of the mechanisms involved in MTA2-regulated cancer cell metastasis. In addition, miR-1275 was found to be inhibited in shMTA2 stable cells and to bind to the 3′ UTR of the KLK10 promoter. Therefore, whether MTA2 affects KLK10 expression by regulating miR-1275 is worthy of further investigation.
To the best of our knowledge, this is the first study to demonstrate the role and possible mechanism of MTA2 on the metastasis of human cervical cancer cells. Inhibition of MTA2 significantly suppresses the migration and invasion, as well as the aggressiveness, of human cervical cancer cells in vitro and in vivo by controlling the expression of Sp1 and KLK10, via enhancing the levels of miR-7. In conclusion, although the precise mechanism by which MTA2 exerts its effects on the progression of cervical cancer remains to be determined, our present findings suggest that MTA2 and KLK10 could modulate the metastasis of cervical cancer and provide a new target candidate for its treatment.
Materials and Methods
Cell Culture
The human cervical cancer cell line C33A (BCRC no. 60554) and HeLa cells (BCRC no. 60005) were purchased from the Bioresources Collection and Research Center of the Food Industry Research and Development Institute (Hsinchu, Taiwan). SiHa cells (ATCC no. HTB35) were obtained from the American Type Culture Collection (Rockville, MD, USA). C33A cells were maintained in minimum essential medium (MEM), HeLa cells were cultured in DMEM/F12, and SiHa cells were maintained in DMEM. All culture media were supplemented with 10% fetal bovine serum (FBS; Gibco/Invitrogen, Waltham, MA, USA) and 1% penicillin/streptomycin (HyClone, Logan, UT, USA) in a CO2 humidified atmosphere at 37°C.
shRNA and siRNA
To inhibit the expression of human MTA2, KLK10, and Sp1, siRNA oligonucleotides or shRNA knockdown vectors were used. To construct the MTA2 shRNA plasmid (MTA2-shRNA-pLKO.1), materials were purchased from the RNAi (RNA interference) core of Academia Sinica (Taipei, Taiwan). The MTA2 targeting sequence was 5′-AGGGAGTGAGGAGTGAATTAA-3′, with pLKO.1-Luc used as a scrambled control. Puromycin (2 μg/mL) was used to select stably transduced cells. The cells were transfected with 100 nM siRNA or negative control by using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the instructions provided by the manufacturer. The sequences of siSp1, siKLK10, and negative control siRNA oligonucleotides are as follows: siControl, sense, 5′-UUCUCCGAACGUGUCACGTUTT-3′, antisense, 5′-ACGUGACACGUUCGGAGAATT-3′; siSp1, sense, 5′-CCAUUAACCUCAGUGCAUUTT-3′, antisense, 5′-AAUGCACUGAGGUUAAUGGTT-3′; siKLK10, sense, 5′-CUGUUGUCCAUCCCAAGUATT-3′, antisense, 5′-UACUUGGGAUGGACAACAGTT-3′
Western Blot Analysis
Western blot analysis was performed as described previously.30 Antibodies against β-actin (sc-69879, 1:5,000), MTA2 (sc-55566, 1:1,000), and KLK10 (sc-100551, 1:1,000) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against Sp1 (07645, 1:1,000) and horseradish peroxidase (HRP)-conjugated anti-mouse (AP124P, 1:10,000) were purchased from Merck Millipore (Burlington, MA, USA).
Human Protein Array Expression Profiling Analysis
Protease expression profiling analysis was performed as described previously.31 Briefly, the membranes coated with antibodies against multiple proteins were incubated with the mixture containing cell lysate. After the incubation, the membrane was reacted with streptavidin-HRP in array buffer and then incubated with the chemiluminescent reagent. The signals were analyzed by using an image analysis system (Fujifilm, Tokyo, Japan)
qRT-PCR
qPCR was performed as described previously.32 Briefly, total RNA was isolated from cells using TRIzol reagent (Invitrogen, Waltham, MA, USA) and quantified. cDNA was synthesized from 1 μg of RNA using the GoScript reverse transcription system (Promega, Madison, WI, USA) according to the manufacturer’s instructions. GAPDH or RUN6B was used as an internal control and determination of the difference in threshold cycle (Ct) between treated and untreated cells using the 2−ΔΔCt method.
Migration and Invasion Assay
A migration and invasion assay was performed as described previously.31 In brief, this assay uses 48-well modified Boyden chambers containing membrane filter inserts with 8-μm pores. These filter inserts were coated with Matrigel prior to the invasion assay. The lower compartment was filled with DMEM containing 10% FBS. Cells were placed in the upper part of the chamber, which contained serum-free medium, and incubated for 16–24 h. Cell migration and invasion were determined by counting cells on the filter at a magnification of ×100.
Transfection Assay
HeLa or SiHa cells were seeded into the 6-cm culture dish (3 × 105 cells) or six-well plate (1 × 105 cells/well) overnight, then transfected with siRNAs (100 nM), plasmid (3 μg), miRNA mimic (30 nM), or antagomiRNA (100 nM) in Opti-MEM medium (Thermo Fisher Scientific, Waltham, MA, USA) and added to 6 μL of TurboFect transfection reagent or 3 μL of Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, Waltham, MA, USA). After transfection for 48 h, the cells were lysed with extraction buffer for further cell experiments. The mimic miRNA, antagomiRNA, and siRNA were all synthesized by GenDiscovey Biotechnology (New Taipei City, Taiwan). Construction of MTA2 plasmid was purchased from Genewiz (Takeley, UK).
Immunofluorescence Staining
Cells were grown in Lab-Tek eight-well chamber slides (Thermo Fisher Scientific, Waltham, MA, USA) and then fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in PBS for 10 min. The cells were next incubated with anti-Sp1 (1:200) or anti-KLK10 (1:200) overnight at 4°C, followed by incubation with Cy3-conjugated anti-rabbit or fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin G (IgG) for 1 h. Fluorescence was monitored and imaged with a confocal microscope.
miRNA Prediction
Identification of the miRNA target gene was accomplished by using the miRBase (http://www.mirbase.org), miRDB (http://www.mirdb.org/), and TargetScan (http://www.targetscan.org/vert_61/) programs to predict putative miRNAs binding sites in the 3′ UTR of the human Sp1 promoter.
ChIP Assay
ChIP was performed as previously described.33 Primers for the ChIP assay were as follows: KLK10 (−692/−493), forward, 5′-ACGGTACAATTAGACAGAGG-3′, reverse, 5′-TCACCTGGGAGTCGCCGATA-3; KLK10 (−206/−6), forward, 5′-GGCGTCATTAGGGTAATTGA-3′, reverse, 5′-GGATCTGCTGGGGTGTGTGC-3′. Enrichment was calculated as the ratio between the net intensity of each bound sample divided by the input, and the vehicle control sample divided by the input, or (bound/input)/(control/input).
Luciferase Activity Assay
Cells were cotransfected with pGL4.13-Sp1-3′ UTR-WT/-Mut and miR-7 inhibitor using transfection reagent. The 3′ UTR mRNA sequences of Sp1 containing the miR-7 binding site were amplified by a PCR assay. The amplified DNA sequences were inserted into pmirGLO Dual-Luciferase miRNA target expression vector (Promega, Madison, WI, USA) to generate the pmirGLO-Sp1-3′ UTR (containing the binding site of miR-7) luciferase vectors. The pmirGLO-Sp1-3′ UTR plasmid was amplified by PCR using the primer pairs 5′-CTCGAGACCTTCTTGGCGTGGAGAGTCAA-3′ followed by an XhoI site and 5′-GTCGACTTCCCTTAGGAACCACCCAAGAAC-3′ followed by a SalI site. Site-directed mutagenesis of the miR-7 target site in the Sp1 3′ UTR (pGL4.13-Sp1-3′ UTR-Mut) was performed using the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). Luciferase activity was measured with a Dual-Luciferase reporter assay system (Promega E1910) according to the manufacturer’s protocol.
Experimental Metastasis Assay
Animal work was approved under the Institutional Animal Care and Use Committee of the Chung-Shan Medical University Experimental Animal Center. Animal experiments used 5-week-old female BALB/c mice (National Laboratory Animal Center, Taipei, Taiwan). All mice in group 1 (n = 6) were injected with 1 × 106 shLuc stable cells. Group 2 mice (n = 6) were injected with 1 × 106 shMTA2 stable cells. After 2 months, all mice were sacrificed, and the lungs of the mice were removed to analyze the status of invading tumors. Sections were stained with hematoxylin and eosin (H&E) to detect metastases. Immunohistochemical staining was performed to evaluate the protein expression of the invading tumor.
Statistical Analysis
Statistical analyses were performed with SPSS 12.0 (SPSS, Chicago, IL, USA). Results are presented as means ± standard errors, and statistical analyses were performed by Student’s t test and one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test. The overall survival curve of cervical cancer patients was calculated using the Kaplan-Meier method. The relationship between the Sp1 and miR-7 was assessed by Spearman rank correlation. Each experiment was performed at least three times. Significance was regarded as a p value below 0.05 or 0.01.
Author Contributions
Conception and design: C.-L.L., T.-H.Y., S.-F.Y., J.-J.L., and Y.-H.H. Development of methodology: S.-F.Y., S.-W.W., and Y.-H.H.. Acquisition of data: C.-L.L., T.-H.Y., and Y.-H.H. Analysis and interpretation of data: S.-W.W., S.-P.C., J.-J.L., and Y.-H.H. Writing, review, and/or revision of the manuscript: C.-L.L., T.-H.Y, S.-F.Y., S.-W.W., S.-P.C., J.-J.L., and Y.-H.H. Administrative, technical, or material support: C.-L.L., T.-H.Y., J.-J.L., and Y.-H.H.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Chung Shan Medical University Hospital (CSH-2019-D-007)
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.04.009.
Contributor Information
Jie-Jen Lee, Email: jjlee918@mmc.edu.tw.
Yi-Hsien Hsieh, Email: hyhsien@csmu.edu.tw.
Supplemental Information
References
- 1.Liontos M., Kyriazoglou A., Dimitriadis I., Dimopoulos M.A., Bamias A. Systemic therapy in cervical cancer: 30 years in review. Crit. Rev. Oncol. Hematol. 2019;137:9–17. doi: 10.1016/j.critrevonc.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Li D.Q., Pakala S.B., Nair S.S., Eswaran J., Kumar R. Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer Res. 2012;72:387–394. doi: 10.1158/0008-5472.CAN-11-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Covington K.R., Fuqua S.A. Role of MTA2 in human cancer. Cancer Metastasis Rev. 2014;33:921–928. doi: 10.1007/s10555-014-9518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covington K.R., Brusco L., Barone I., Tsimelzon A., Selever J., Corona-Rodriguez A., Brown P., Kumar R., Hilsenbeck S.G., Fuqua S.A. Metastasis tumor-associated protein 2 enhances metastatic behavior and is associated with poor outcomes in estrogen receptor-negative breast cancer. Breast Cancer Res. Treat. 2013;141:375–384. doi: 10.1007/s10549-013-2709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang B., Zhang H., Shen G. Metastasis-associated protein 2 (MTA2) promotes the metastasis of non-small-cell lung cancer through the inhibition of the cell adhesion molecule Ep-CAM and E-cadherin. Jpn. J. Clin. Oncol. 2015;45:755–766. doi: 10.1093/jjco/hyv062. [DOI] [PubMed] [Google Scholar]
- 6.Yousef G.M., Obiezu C.V., Luo L.Y., Magklara A., Borgoño C.A., Kishi T., Memari N., Michael P., Sidiropoulos M., Kurlender L. Human tissue kallikreins: from gene structure to function and clinical applications. Adv. Clin. Chem. 2005;39:11–79. doi: 10.1016/s0065-2423(04)39002-5. [DOI] [PubMed] [Google Scholar]
- 7.Hu J., Lei H., Fei X., Liang S., Xu H., Qin D., Wang Y., Wu Y., Li B. NES1/KLK10 gene represses proliferation, enhances apoptosis and down-regulates glucose metabolism of PC3 prostate cancer cells. Sci. Rep. 2015;5:17426. doi: 10.1038/srep17426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kioulafa M., Kaklamanis L., Stathopoulos E., Mavroudis D., Georgoulias V., Lianidou E.S. Kallikrein 10 (KLK10) methylation as a novel prognostic biomarker in early breast cancer. Ann. Oncol. 2009;20:1020–1025. doi: 10.1093/annonc/mdn733. [DOI] [PubMed] [Google Scholar]
- 9.Li L., Xu N., Fan N., Meng Q., Luo W., Lv L., Ma W., Liu X., Liu L., Xu F. Upregulated KLK10 inhibits esophageal cancer proliferation and enhances cisplatin sensitivity in vitro. Oncol. Rep. 2015;34:2325–2332. doi: 10.3892/or.2015.4211. [DOI] [PubMed] [Google Scholar]
- 10.Bouyssou J.M., Manier S., Huynh D., Issa S., Roccaro A.M., Ghobrial I.M. Regulation of microRNAs in cancer metastasis. Biochim. Biophys. Acta. 2014;1845:255–265. doi: 10.1016/j.bbcan.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Zhao M., Wang G. Upregulation of microRNA-7 contributes to inhibition of the growth and metastasis of osteosarcoma cells through the inhibition of IGF1R. J. Cell. Physiol. 2019;234:22195–22206. doi: 10.1002/jcp.28787. [DOI] [PubMed] [Google Scholar]
- 12.Li Y.Z., Wen L., Wei X., Wang Q.R., Xu L.W., Zhang H.M., Liu W.C. Inhibition of miR-7 promotes angiogenesis in human umbilical vein endothelial cells by upregulating VEGF via KLF4. Oncol. Rep. 2016;36:1569–1575. doi: 10.3892/or.2016.4912. [DOI] [PubMed] [Google Scholar]
- 13.Liu S., Zhang P., Chen Z., Liu M., Li X., Tang H. MicroRNA-7 downregulates XIAP expression to suppress cell growth and promote apoptosis in cervical cancer cells. FEBS Lett. 2013;587:2247–2253. doi: 10.1016/j.febslet.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 14.Hao Z., Yang J., Wang C., Li Y., Zhang Y., Dong X., Zhou L., Liu J., Zhang Y., Qian J. MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int. J. Clin. Exp. Med. 2015;8:480–487. [PMC free article] [PubMed] [Google Scholar]
- 15.Yang F., Guo L., Cao Y., Li S., Li J., Liu M. MicroRNA-7-5p promotes cisplatin resistance of cervical cancer cells and modulation of cellular energy homeostasis by regulating the expression of the PARP-1 and BCL2 genes. Med. Sci. Monit. 2018;24:6506–6516. doi: 10.12659/MSM.910969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S.L., Han Y., Zhang Y., Xie C.Y., Wang E.H., Miao Y., Li H.Y., Xu H.T., Dai S.D. Expression of metastasis-associated protein 2 (MTA2) might predict proliferation in non-small cell lung cancer. Target. Oncol. 2012;7:135–143. doi: 10.1007/s11523-012-0215-z. [DOI] [PubMed] [Google Scholar]
- 17.Ding W., Hu W., Yang H., Ying T., Tian Y. Prognostic correlation between MTA2 expression level and colorectal cancer. Int. J. Clin. Exp. Pathol. 2015;8:7173–7180. [PMC free article] [PubMed] [Google Scholar]
- 18.Li B., Goyal J., Dhar S., Dimri G., Evron E., Sukumar S., Wazer D.E., Band V. CpG methylation as a basis for breast tumor-specific loss of NES1/kallikrein 10 expression. Cancer Res. 2001;61:8014–8021. [PubMed] [Google Scholar]
- 19.Bin L., Kim B.E., Hall C.F., Leach S.M., Leung D.Y. Inhibition of transcription factor specificity protein 1 alters the gene expression profile of keratinocytes leading to upregulation of kallikrein-related peptidases and thymic stromal lymphopoietin. J. Invest. Dermatol. 2011;131:2213–2222. doi: 10.1038/jid.2011.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F., Jiang Z., Wang K., Guo J., Hu G., Sun L., Wang T., Tang X., He L., Yao J. Transactivation of the human NME5 gene by Sp1 in pancreatic cancer cells. Gene. 2012;503:200–207. doi: 10.1016/j.gene.2012.04.088. [DOI] [PubMed] [Google Scholar]
- 21.Kong L.M., Liao C.G., Fei F., Guo X., Xing J.L., Chen Z.N. Transcription factor Sp1 regulates expression of cancer-associated molecule CD147 in human lung cancer. Cancer Sci. 2010;101:1463–1470. doi: 10.1111/j.1349-7006.2010.01554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao X.W., Xiao J.Q., Li Z.Y., Zheng Y.C., Zhang N. Effects of microRNA-135a on the epithelial-mesenchymal transition, migration and invasion of bladder cancer cells by targeting GSK3β through the Wnt/β-catenin signaling pathway. Exp. Mol. Med. 2018;50:e429. doi: 10.1038/emm.2017.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexopoulou D.K., Papadopoulos I.N., Scorilas A. Clinical significance of kallikrein-related peptidase (KLK10) mRNA expression in colorectal cancer. Clin. Biochem. 2013;46:1453–1461. doi: 10.1016/j.clinbiochem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Luo L.Y., Katsaros D., Scorilas A., Fracchioli S., Bellino R., van Gramberen M., de Bruijn H., Henrik A., Stenman U.H., Massobrio M. The serum concentration of human kallikrein 10 represents a novel biomarker for ovarian cancer diagnosis and prognosis. Cancer Res. 2003;63:807–811. [PubMed] [Google Scholar]
- 25.Lu C.Y., Hsieh S.Y., Lu Y.J., Wu C.S., Chen L.C., Lo S.J., Wu C.T., Chou M.Y., Huang T.H., Chang Y.S. Aberrant DNA methylation profile and frequent methylation of KLK10 and OXGR1 genes in hepatocellular carcinoma. Genes Chromosomes Cancer. 2009;48:1057–1068. doi: 10.1002/gcc.20708. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Song H., Miao Y., Wang R., Chen L. Frequent transcriptional inactivation of Kallikrein 10 gene by CpG island hypermethylation in non-small cell lung cancer. Cancer Sci. 2010;101:934–940. doi: 10.1111/j.1349-7006.2009.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F., Sun L., Li Q., Han X., Lei L., Zhang H., Shang Y. SET8 promotes epithelial-mesenchymal transition and confers TWIST dual transcriptional activities. EMBO J. 2012;31:110–123. doi: 10.1038/emboj.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M., Pan M., You C., Dou J. The therapeutic potential of miR-7 in cancers. Mini Rev. Med. Chem. 2019;19:1707–1716. doi: 10.2174/1389557519666190904141922. [DOI] [PubMed] [Google Scholar]
- 29.Hua K., Jin J., Zhang H., Zhao B., Wu C., Xu H., Fang L. MicroRNA-7 inhibits proliferation, migration and invasion of thyroid papillary cancer cells via targeting CKS2. Int. J. Oncol. 2016;49:1531–1540. doi: 10.3892/ijo.2016.3660. [DOI] [PubMed] [Google Scholar]
- 30.Lin C.L., Lee C.H., Chen C.M., Cheng C.W., Chen P.N., Ying T.H., Hsieh Y.H. Protodioscin induces apoptosis through ROS-mediated endoplasmic reticulum stress via the JNK/p38 activation pathways in human cervical cancer cells. Cell. Physiol. Biochem. 2018;46:322–334. doi: 10.1159/000488433. [DOI] [PubMed] [Google Scholar]
- 31.Wu M.H., Lin C.L., Chiou H.L., Yang S.F., Lin C.Y., Liu C.J., Hsieh Y.H. Praeruptorin A inhibits human cervical cancer cell growth and invasion by suppressing MMP-2 expression and ERK1/2 signaling. Int. J. Mol. Sci. 2017;19:10. doi: 10.3390/ijms19010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiang K.C., Lai C.Y., Chiou H.L., Lin C.L., Chen Y.S., Kao S.H., Hsieh Y.H. Timosaponin AIII inhibits metastasis of renal carcinoma cells through suppressing cathepsin C expression by AKT/miR-129-5p axis. J. Cell. Physiol. 2019;234:13332–13341. doi: 10.1002/jcp.28010. [DOI] [PubMed] [Google Scholar]
- 33.Lin C.L., Chen C.M., Lin C.L., Cheng C.W., Lee C.H., Hsieh Y.H. Norcantharidin induces mitochondrial-dependent apoptosis through Mcl-1 inhibition in human prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2017;1864:1867–1876. doi: 10.1016/j.bbamcr.2017.07.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.