Abstract
Laryngeal cancer (LCa), a neoplasm of the head and neck region, is a leading cause of death worldwide. Surgical intervention remains the mainstay of LCa treatment, but a crucial point is represented by the possible nodal involvement. Therefore, it is urgently needed to develop biomarkers and therapeutic tools able to drive treatment approaches for LCa. In this study, we investigated deregulated microRNAs (miRNAs) in tissues from LCa patients with either lymph node metastases (N+) or not (N−). miRNA expression profiling was performed by a comprehensive PCR array and subsequent validation by RT-qPCR. Results showed a significant decrease of miR-449a expression in N+ compared to N− patients, and miR-133b down-modulation in LCa tissues compared to paired normal ones. Receiver operating characteristic (ROC) curve analysis revealed the potential diagnostic power of miR-133b for LCa detection. According to the validation results, we selected miR-449a for further in vitro studies. Ectopic miR-449a expression in the LCa cell line Hep-2 inhibited invasion and motility in vitro, slowed cell proliferation, and induced the downregulation of Notch1 and Notch2 as direct targets of miR-449a. Collectively, this study provides new promising biomarkers for LCa diagnosis and a new opportunity to use miR-449a for the treatment of nodal metastases in LCa patients.
Keywords: microRNA, laryngeal cancer, metastases, miR-449a, miR-133b, migration, invasion, Notch1, Notch2, lymph nodes
Graphical Abstract
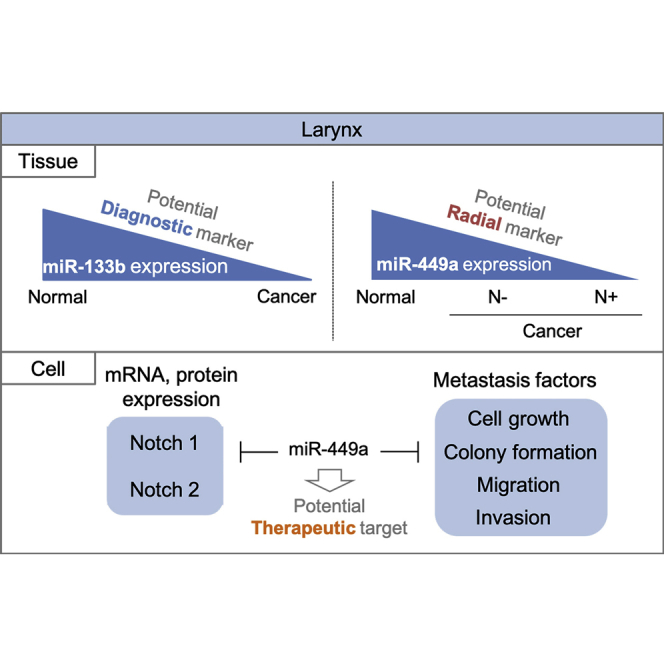
The need to identify molecular markers for early detection of laryngeal cancer prompted Kawasaki et al. to define a miRNA signature of tumor transformation and spreading. They showed the diagnostic and predictive potential of miR-133b and miR-449a, respectively, and demonstrated miR-449a-mediated suppression of the metastatic factors Notch1 and Notch2.
Introduction
Laryngeal cancer (LCa), known to be one of the most common cancers of the head and neck area, accounts for approximately 2% of newly diagnosed tumors worldwide.1,2 Histologically, more than 90% of LCa cases are categorized as squamous cell carcinomas.3 Therapeutic options, including surgical intervention, radiation, and combination therapies, are effective treatments for LCa at an early clinical stage. However, the treatment for advanced LCa cases still needs additional improvements, although significant advances have been obtained in the past few decades.4, 5, 6 The diffusion of LCa cells in perineural space is often associated with poor prognosis and, therefore, it requires surgical therapeutic treatment associated with adjuvant radiotherapy.7 Total laryngectomy is often required as a therapeutic intervention for the progressed LCa, but such surgical modality strongly adversely affects patient’s quality of life. For example, the dissection causes loss of voice and appearance deterioration. Furthermore, another severe problem is the possibility of cancer recurrence even after laryngectomy. Therefore, we urgently need to develop promising methods for early detection and effective clinical management of LCa to reduce disease morbidity and mortality and to improve quality of life among patients.
Metastases, which are tumor cells traveling from a primary site to distant organs, leading to the formation of new tumor colonies, are often responsible for a cancer patient’s unfavorable outcome.8,9 LCa can spread to cervical lymph nodes, which represent the first site of metastasization, and change the therapeutic approach to LCa patients who, in this case, require a more aggressive surgical strategy. The propensity of LCa to cause node metastases is correlated to both the anatomic site of origin and the molecular characteristics of the tumor. In this light, metastatic nodal involvement represents a crucial prognostic factor of LCa.10 In fact, the overall survival rate in LCa patients has a stronger association with node metastases compared to tumor extension.11 Conventional imaging analyses, such as computed tomography (CT), positron-emission tomography (PET), and MRI, generally used for diagnostic purposes, are able to confirm the occurrence of metastases, whereas micrometastases (not visible with conventional imaging) detection is still a challenge. Additionally, reliable molecular markers have not been found to accurately predict the presence of malignant metastases in patients. Likewise, effective treatment protocols also need further improvement.
MicroRNAs (miRNAs) belong to small, highly conserved non-coding RNA molecules (approximately 22 nt) that have a pivotal role in the regulation of gene expression at the post-transcriptional level. miRNAs work as either oncogenes or tumor suppressors, depending on their target messenger RNAs (mRNAs) and on the precise cell context. One miRNA negatively regulates multiple genes through imperfect binding to complementary sites in 3′ untranslated regions of target mRNAs, leading to mRNA degradation (does not occur in humans) or translational repression.12, 13, 14 Accumulated evidence has shown that miRNAs are closely associated with diverse pathological processes, such as cell proliferation, differentiation, apoptosis, and metastases in different malignancies.15,16 Numerous studies have demonstrated that miRNA expression patterns are aberrantly altered in different types of cancers, including LCa.17, 18, 19, 20, 21, 22, 23, 24, 25, 26 Due to these intriguing findings, definition of unique miRNA signatures in each cancer could contribute to a deeper understanding of molecular pathogenesis mechanisms and to the development of new powerful diagnostic and therapeutic approaches.
Several previous studies carried out by us and other authors have demonstrated the association between deregulated miRNAs and malignant progression, recurrence, and clinical aggressiveness of LCa.23, 24, 25, 26 Nevertheless, metastases-associated miRNAs in LCa remain to be defined. Therefore, further investigations aimed at identifying aberrant miRNA signatures in LCa appear necessary to lead to the elucidation of pathological mechanisms driving nodal involvement.
In the present study, we investigated miRNA expression patterns in LCa tissues in order to develop new reliable biomarkers for the definition of LCa prognosis, as well as for the prediction of nodal metastases and also for the identification of new therapeutic candidates for either replacement therapy or miRNA inhibition. The pathological characterizations of LCa and the available tissue biomarkers are still limited and do not allow a deep understanding of both molecular characteristics and spreading propensity. Based on these considerations, and to identify a nodal miRNA signature in LCa patients, we performed a comprehensive miRNA screening assay, followed by a validation test employing quantitative reverse transcription PCR (RT-qPCR) in LCa tissues affected by either lymph node metastases (N+) or not (N−). We found significant downregulation of miR-133b and miR-449a in LCa tissues with diagnostic potential for the disease and predictive potential for its nodal metastasis, respectively. Some previous reports also showed the tumor-suppressive role of miR-449a in other cancer types,27, 28, 29, 30, 31, 32 so we attempted to determine the biological functions of miR-449a in LCa. Through both gain- and loss-of-function studies in LCa cells, we observed the ability of miR-449a in suppressing metastatic factors such as cell proliferation, migration, and invasion in vitro assays. Moreover, we found that miR-449a inhibits, as direct target genes, the expression of Notch1 and Notch2, known as oncogenes in LCa.33,34 Collectively, our findings suggest that miR-449a works as an anti-tumor gene in LCa with potential for use as a therapeutic weapon for the prevention of LCa metastases.
Results
Profiling of miRNA Signatures in LCa
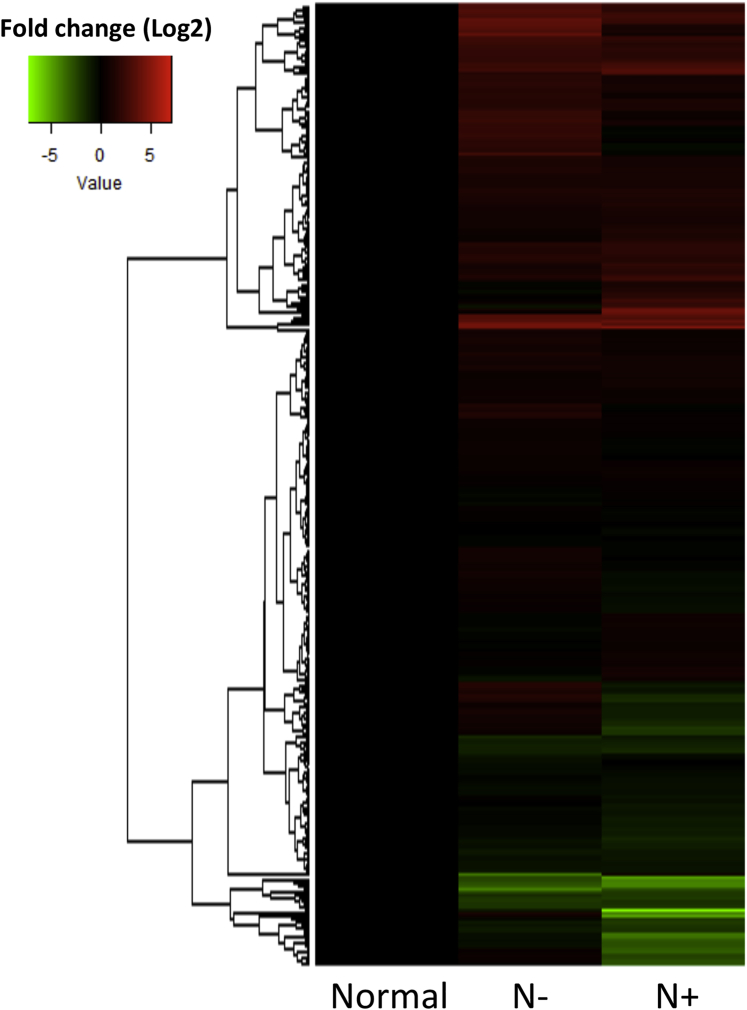
We performed a comprehensive PCR array-based screening, as described in Materials and Methods, to determine miRNA signatures in LCa tissues. Clinical parameters of enrolled subjects are summarized in Table 1. For miRNA profiling, clinical LCa tissue samples, collected from LCa patients with lymph node metastases (N+) (n = 23) or without (N−) (n = 23) and their adjacent normal counterparts (n = 30), were divided into five pools as mentioned in Materials and Methods. The PCR array analysis showed 309 miRNAs with either commonly or differentially detectable patterns across each group (N+, N−, and normal group) (Figures S1A–S1C). In addition, a hierarchical clustered heatmap exhibited different miRNA expression profiling among the groups (Figure 1), suggesting a miRNA dysregulation depending on laryngeal tissue context.
Table 1.
Clinical Information for All LCa Patients Enrolled
| Characteristics | n (%) | |
|---|---|---|
| Age (year) | ≤60 | 45 (59.2) |
| >60 | 31 (40.8) | |
| Sex | male | 63 (82.9) |
| female | 13 (17.1) | |
| T classification | T1–T2 | 15 (19.7) |
| T3–T4 | 61 (80.3) | |
| LNM | N0 | 38 (50.0) |
| N1–N4 | 38 (50.0) | |
LNM, lymph node metastasis.
Figure 1.
Hierarchical Clustering Demonstrating the Global and Distinct Expression Levels of miRNA across the Screening Set
The heatmap summarizes the differential patterns of miRNA expressions across the groups, profiled by the results of PCR array assays. The clustering analysis was performed by package R (version 3.4.3).
According to the expression pattern, 30 miRNAs were statistically significantly deregulated between cancers (N+ and N−) and normal tissues (p < 0.05) (Table 2), and, among them, 7 miRNAs (miR-133b, miR-184, miR-181a, miR-1, miR-140-3p, miR-198, and miR-885-5p) were drastically changed in their levels (p < 0.01). Furthermore, the expression of eight miRNAs was also highly altered in N+ when compared to N− (p < 0.05) (Table 3). Among them, four miRNAs (miR-545, miR-449b, miR-449a, and miR-652) were more significantly deregulated (p < 0.01).
Table 2.
Significantly Deregulated miRNAs among Laryngeal Cancer Tissues Compared to Their Paired Noncancerous Counterparts
| MicroRNA | ΔΔCt | 2−ΔΔCt (FC) | 95% CI for FC | Regulation | p Value | FDR-Adjusted p Value |
|---|---|---|---|---|---|---|
| miR-133b | 3.55 | 0.08 | −0.07–2.69 | down | 0.0004 | 0.06 |
| miR-184 | −3.61 | 12.1 | 5.44–41.87 | up | 0.0004 | 0.06 |
| miR-181a | −2.14 | 4.29 | 2.70–6.59 | up | 0.0016 | 0.17 |
| miR-1 | 3.46 | 0.09 | −0.03–1.47 | down | 0.0029 | 0.23 |
| miR-140-3p | 1.30 | 0.41 | 0.32–0.62 | down | 0.0060 | 0.34 |
| miR-198 | −3.76 | 13.9 | −0.95–36.12 | up | 0.0069 | 0.34 |
| miR-885-5p | 4.06 | 0.06 | 0.05–0.55 | down | 0.0087 | 0.35 |
| miR-139-5p | 1.79 | 0.29 | 0.22–0.44 | down | 0.0124 | 0.35 |
| miR-590-5p | −1.18 | 2.30 | 1.47–2.52 | up | 0.0126 | 0.35 |
| miR-324-3p | −0.86 | 1.87 | 0.98–2.02 | up | 0.0138 | 0.35 |
| miR-886-3p | −1.81 | 3.48 | 0.84–6.82 | up | 0.0145 | 0.35 |
| miR-196b | −2.91 | 7.46 | 5.93–15.58 | up | 0.0166 | 0.35 |
| miR-449b | 1.60 | 0.33 | 0.10–2.31 | down | 0.0170 | 0.35 |
| miR-185 | −2.34 | 4.92 | 2.85–7.06 | up | 0.0174 | 0.35 |
| miR-155 | −1.10 | 2.14 | 1.58–2.93 | up | 0.0180 | 0.35 |
| miR-28 | −1.49 | 2.83 | 1.82–3.56 | up | 0.0192 | 0.35 |
| miR-142-3p | −2.23 | 4.59 | 3.59–5.53 | up | 0.0194 | 0.35 |
| miR-490 | −1.56 | 3.03 | 1.46–4.37 | up | 0.0216 | 0.35 |
| miR-331-5p | −4.40 | 21.1 | 6.02–32.57 | up | 0.0241 | 0.35 |
| miR-20b | −0.90 | 2.00 | 1.26–2.23 | up | 0.0244 | 0.35 |
| miR-21 | −2.90 | 8.00 | 5.16–11.52 | up | 0.0248 | 0.35 |
| miR-425-5p | −1.31 | 2.46 | 1.73–4.56 | up | 0.0250 | 0.35 |
| miR-147b | −1.81 | 3.48 | 1.67–7.65 | up | 0.0256 | 0.35 |
| miR-133a | 5.17 | 0.03 | −0.07–0.96 | down | 0.0265 | 0.35 |
| miR-301 | −2.71 | 6.50 | 4.96–10.23 | up | 0.0312 | 0.39 |
| miR-539 | 1.73 | 0.31 | −0.24–2.89 | down | 0.0337 | 0.41 |
| miR-375 | 3.92 | 0.07 | 0.11–0.49 | down | 0.0367 | 0.42 |
| miR-132 | −1.01 | 2.00 | 1.54–2.55 | up | 0.0380 | 0.42 |
| miR-138 | −1.75 | 3.25 | 2.05–6.96 | up | 0.0381 | 0.42 |
| miR-182 | −1.88 | 3.73 | 2.42–5.10 | up | 0.0396 | 0.42 |
The t test was used for the calculation of p values. The 95% CI was calculated from fold change of each miRNA level.
Table 3.
Significantly Deregulated miRNAs in N+ Compared to N−
| MicroRNA | ΔΔCt | 2−ΔΔCt (FC) | 95% CI for FC | Regulation | p Value | FDR-Adjusted p Value |
|---|---|---|---|---|---|---|
| miR-545 | −4.93 | 29.9 | −3.73–150.39 | up | 0.003 | 0.74 |
| miR-449b | 3.41 | 0.09 | 0.06–0.19 | down | 0.006 | 0.74 |
| miR-449a | 2.80 | 0.14 | 0.01–0.49 | down | 0.009 | 0.74 |
| miR-652 | 1.78 | 0.29 | 0.20–0.45 | down | 0.009 | 0.74 |
| miR-96 | −3.26 | 9.8 | 7.27–14.93 | up | 0.012 | 0.76 |
| miR-512-3p | −4.12 | 17.1 | −127.45–501.93 | up | 0.028 | 0.99 |
| miR-627 | 6.32 | 0.01 | 0.01–0.02 | down | 0.044 | 0.99 |
| miR-372 | 2.75 | 0.14 | 0.14–0.39 | down | 0.044 | 0.99 |
The t test was used for the calculation of p values. The 95% CI was calculated from fold change of each miRNA level.
These results showed significant deregulation of several miRNAs in malignant laryngeal tissues, thus implicating their relevance to carcinogenesis and malignant metastasis as well as their potential use for both diagnostic and therapeutic purposes.
Criteria for Selection of Candidate miRNAs
Below are shown the criteria defined to carefully select tissue miRNA biomarker candidates, which may be significantly correlated to cancer pathogenesis and metastatic potential in LCa, with an appropriately detectable expression level: (1) mean Ct value ≤35.0 for each group (N+, N−, and normal group), and (2) Ct ≤ 35.0 in at least 50% of pools. We assumed that a mean Ct value higher than 35.0 across all pools is inappropriately determined due to its low expression level. For example, miR-545 was significantly deregulated in our screening (Table 3), but this miRNA was excluded from the next validation assessment because not only did its mean Ct value exceed 35.0, but it was also lower than 35.0 for less than 50% of the samples. These criteria provided us with reliable potential tissue biomarker candidate miRNAs for the subsequent validation set. Based on these premises, we selected three miRNAs (miR-133b, miR-449a, and miR-652), which were significantly deregulated between the cancer group and normal ones, or between N+ and N−, individually. Among the candidates, miR-133b was downregulated in the cancer cohort compared to the normal one with a fold change (FC) = 0.08 (p = 0.0004) (Table 2). Alternatively, miR-449a and miR-652 were down-modulated in N+ compared to N− with FC = 0.14 (p = 0.009) and FC = 0.29 (p = 0.009), respectively (Table 3). Based on the results, these three candidates, which were considered to be closely related to pathogenesis and metastatic capacity of LCa, were evaluated in a validation test to appropriately determine their expression levels and to select further studies for the analysis of biological functions, with particular reference to the metastatic potential.
Validation Study of Candidate miRNAs in LCa
To confirm the findings obtained from PCR array screening, we performed RT-qPCR analysis with a TaqMan miRNA assay as a validation set. As described above, the expression of three miRNAs (miR-133b, miR-449a, and miR-652) was examined. Sixty-four patients, including all samples used for the screening setting, were selected for the validation: 32 N+, 32 N−, and 64 paired normal tissues. For each patient, cancerous and non-cancerous tissues were considered.
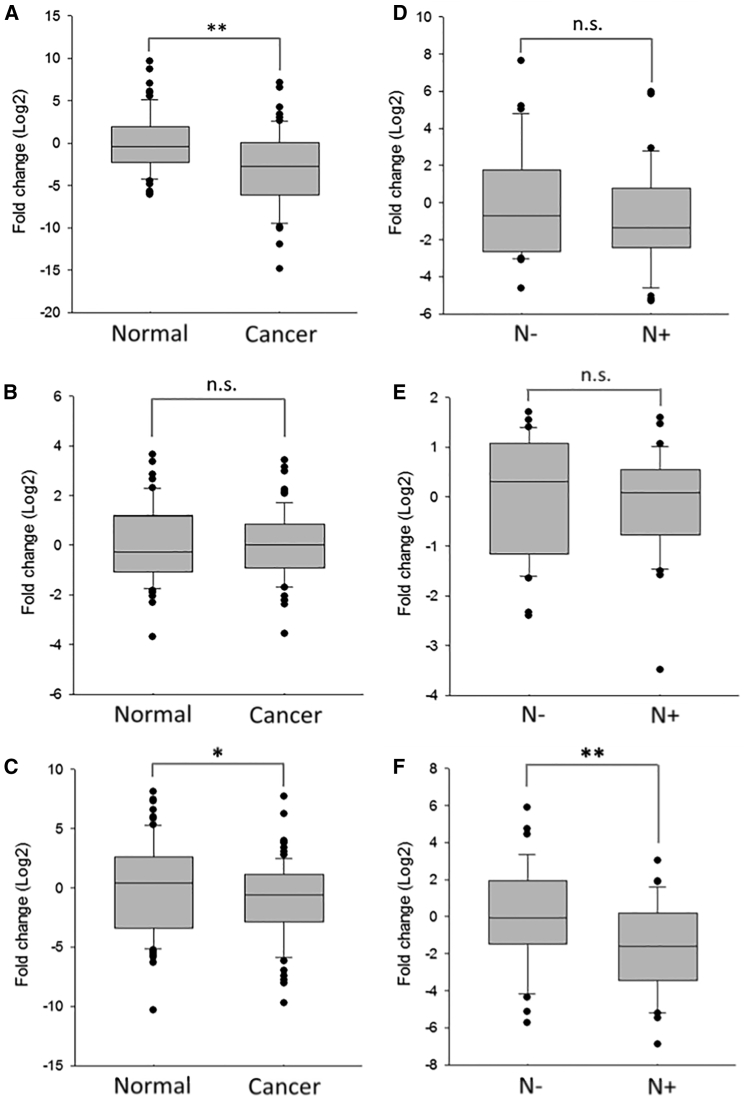
The validation setting confirmed upregulation/downregulation and the FC of each miRNA candidate. We used the median value of miRNA expression level to determine its regulation. As a result, the level of miR-133b was remarkably decreased in LCa tissues compared to normal ones (median log2 FC = −2.67 [95% confidence interval (CI) = −3.82 to −1.64], p < 0.01) (Figure 2A), indicating its potential as a malignancy predictor of LCa. In contrast, the slight change of miR-652 (median log2 FC = 0.16 [95% CI = −0.19–0.51], p = 0.78) (Figure 2B) and miR-449a (median log2 FC = −0.57 [95% CI = −4.19–3.06], p = 0.10) (Figure S2A) showed no statistical difference between the cancer cohort and the normal one, in agreement with the screening assessment. Importantly, miR-449a levels in N+ were significantly deregulated compared to N−, even more consistently than the high-throughput screening (median log2 FC = −1.96 [95% CI = −2.70 to −1.21], p < 0.01) (Figure S2B). Alternatively, miR-133b was altered but without reaching statistical significance (median log2 FC = −1.33 [95% CI = −2.30 to −0.43], p = 0.16) (Figure 2D). Although the screening test showed statistically significant aberrant expression of miR-652, this difference was not confirmed in the validation phase (median log2 FC = −0.04 [95% CI = −0.26–0.43], p = 0.34) (Figure 2E). Since miR-449a has great potential to distinguish between N+ and N−, we added 12 pairs of clinical samples (6 N+, 6 N−, and 12 paired noncancerous tissues) to more accurately determine the expression levels of miR-449a in LCa patients. The results of this additional validation unveiled significant differential miR-449a expression in LCa tissues when compared to normal counterparts (median log2 FC = −0.57 [95% CI = −1.30–0.17], p < 0.05) (Figure 2C) and confirmed that miR-449a was much more under-expressed in N+ compared to N− (median log2 FC = −1.61 [95% CI = −2.36 to −0.86], p < 0.01) (Figure 2F).
Figure 2.
Validation of Candidate miRNAs Expression in LCa Tissues
(A and B) The expression levels of (A) miR-133b and (B) miR-652 were validated in LCa tissues (n = 64) and the paired normal ones (n = 64) using RT-qPCR. (C) miR-449a expression level in LCa tissues (n = 76) and normal adjacent ones (n = 76). (D and E) The expression levels of (D) miR-133b and (E) miR-652 in LCa tissues with lymph node metastasis positive (N+) (n = 32) were compared to N− cases (n = 32). (F) miR-449a expression level was compared between N+ (n = 38) and N− (n = 38). All results are shown as a box-and-whisker plot. For the normalization, U6 snRNA was used as the endogenous control. Each sample was run in triplicate. Error bars show mean ± SD. A t test was used for the calculation of p values. ∗p < 0.05, ∗∗p < 0.01.
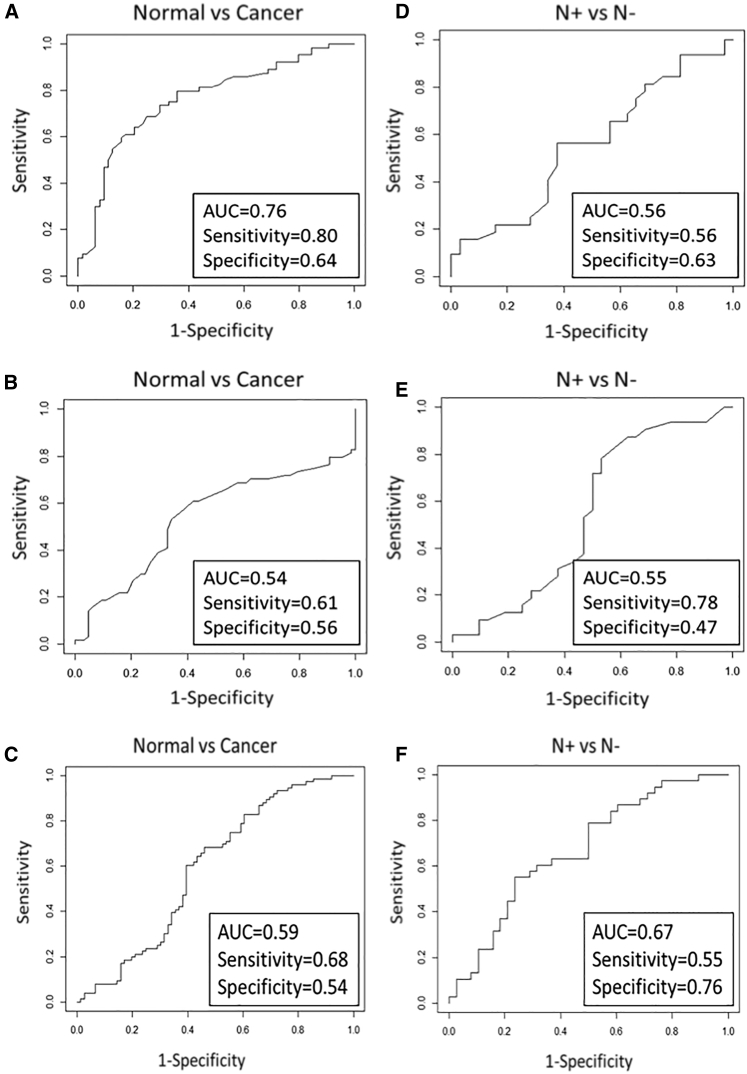
To estimate the diagnostic power of each miRNA assessed for the detection of LCa or for the involvement of nodal metastases, we used the receiver operating characteristic (ROC) curve analysis showing the sensitivity, specificity, and area under the curve (AUC). Notably, miR-133b was clearly deregulated but with mild diagnostic power to distinguish between LCa and normal status (sensitivity = 0.80, specificity = 0.64, AUC = 0.76) (Figure 3A), whereas it was not suitable to predict nodal involvement (sensitivity = 0.56, specificity = 0.63, AUC = 0.56) (Figure 3D). Alternatively, the increase of miR-652 expression was not enough for either detecting LCa (sensitivity = 0.61, specificity = 0.56, AUC = 0.54) (Figure 3B) or predicting lymph node metastasis (sensitivity = 0.78, specificity = 0.47, AUC = 0.55) (Figure 3E). Although miR-449a was not suitable as a diagnostic marker (sensitivity = 0.68, specificity = 0.54, AUC = 0.59) (Figure 3C), it showed potential performance as a predictive marker of nodal involvement (sensitivity = 0.55, specificity = 0.76, AUC = 0.67) (Figure 3F).
Figure 3.
ROC Curve Analysis of Each Validated miRNA for Evaluation of Biomarker Potential
(A–F) ROC curve evaluated (A) miR-133b in LCa tissues compared to the paired normal counterparts (sensitivity = 0.80, specificity = 0.64, AUC = 0.76); (B) miR-652 in LCa tissues compared to the paired normal counterparts (sensitivity = 0.61, specificity = 0.56, AUC = 0.54); (C) miR-449a in LCa tissues compares to the paired normal counterparts (sensitivity = 0.68, specificity = 0.54, AUC = 0.59); (D) miR-133b in N+ compared to N− (sensitivity = 0.56, specificity = 0.63, AUC = 0.56); (E) miR-652 in N+ compared to N− (sensitivity = 0.78, specificity = 0.47, AUC = 0.55); and (F) miR-449a in N+ compared to N− (sensitivity = 0.55, specificity = 0.76, AUC = 0.67).
Collectively, miR-133b has good potential as a powerful biomarker for the determination of LCa, and miR-449a is a potential predictive biomarker of nodal involvement.
miR-449a-Enforced Expression Inhibited Cell Proliferation and Colony Formation Ability of LCa Cells
According to the above results and based on the remarkable potential as a metastasis-associated miRNA, we selected miR-449a for functional studies in an LCa cell line in vitro.
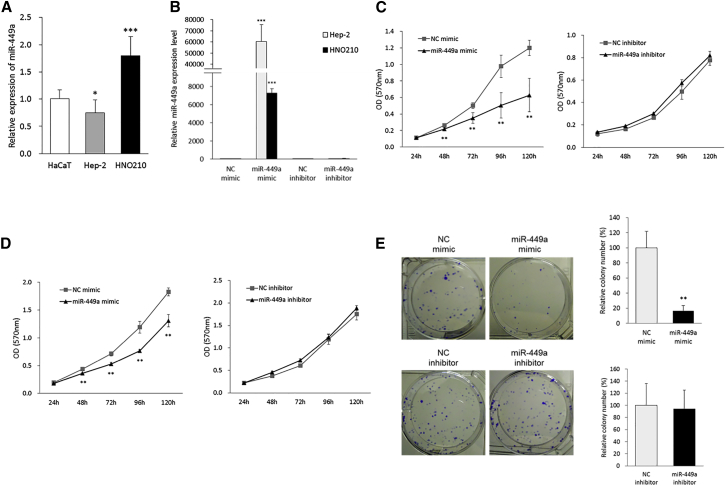
First, we evaluated the basal miR-449a expression level in two LCa cell lines (Hep-2 and HNO210) compared to a normal cell model (HaCaT) using RT-qPCR. The result showed that miR-449a basal expression levels were downregulated in Hep-2 (FC = 0.75 [95% CI = 0.59–0.90], p < 0.05) and upregulated in HNO210 (FC = 1.79 [95% CI = 1.56–2.03], p < 0.001) compared to the HaCaT cell line (Figure 4A).
Figure 4.
miR-449a Suppresses Cell Proliferation and Colony Formation Capacity of LCa Cells
(A) Basal miR-449a expression levels in two LCa cell lines (Hep-2 and HNO210) and HaCaT cells. U6 snRNA was used as a housekeeping gene. (B) miR-449a expression in transfected Hep-2 and HNO210 cells with either miR-449a mimic or inhibitor, or the corresponding NC. U6 snRNA was used as a housekeeping gene. (C and D) Cell proliferation was determined after the transfection of either miR-449a mimic or inhibitor, or the corresponding NC in (C) Hep-2 and (D) HNO210 cells. (E) Colony formation was determined in Hep-2 cells transfected with either miR-449a mimic or inhibitor, or the corresponding NC. Each sample was run in triplicate. Error bars show mean ± SD. The t test was used for the calculation of p values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To investigate biological functions of miR-449a in LCa cells, we performed both gain- and loss-of-function studies through transient transfection by Lipofectamine 2000 of either miR-449a mimic or inhibitor, as well as their respective negative controls (NCs) in Hep-2 and HNO210 cell lines. Mimic and inhibitor NCs are, respectively, a double-stranded and a single-stranded RNA molecule with scramble sequence, used to confirm the specific biological effect induced by the active nucleotides. First, we confirmed and determined the ectopic expression of miR-449a in the two LCa cell lines by RT-qPCR analysis. As a result, we observed the expected miR-449a upregulation after miRNA mimic transfection, compared to NC treatment (p < 0.001, respectively) (Figure 4B). In addition, miR-449a overexpression was more significant in Hep-2 with respect to the HNO210 cell model. Alternatively, miR-449a inhibitor did not change the miR-449a expression compared to the NC inhibitor in both cell lines (Figure 4B).
In order to analyze the antitumor effects of miR-449a, we assessed the inhibition of both cell viability and proliferation by an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide) assay, in a time-course experiment, and colony formation assays, as described in Materials and Methods. In particular, our results showed that miR-449a ectopic expression significantly suppressed cell proliferation of both Hep-2 (52%, p < 0.01) (Figure 4C) and HNO210 (72%, p < 0.01) cells (Figure 4D) at 120 h.
Starting from the above results, Hep-2 cells, which showed low miR-449a basal levels and were remarkably affected by miR-449a transfection, were selected as an optimal cell model for further studies based on miRNA replacement.
Clonogenic assays displayed a significant decrease of Hep-2 cell colony formation capacity to 16.5% upon miR-449a transfection, as visually observed by ImageJ analysis (p < 0.01) (Figure 4E). In contrast, the transfection of miR-449a inhibitor did not affect the clonogenic ability of Hep-2 cells.
In conclusion, miR-449a ectopic expression in Hep-2 cells caused a strong inhibition of cell proliferation, clearly showing that miR-449a plays a tumor suppressive role in Hep-2 cells.
miR-449a Overexpression Suppressed Cell Migration and Invasion in LCa Cells
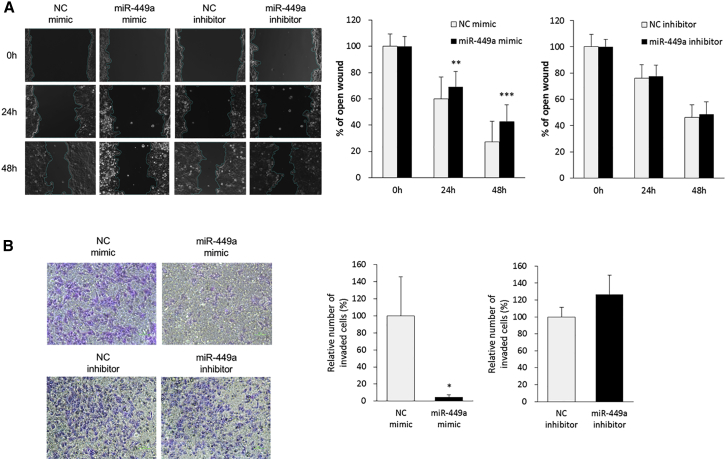
The metastatic ability of miR-449a in both cell migration and invasion was assessed via wound healing and transwell invasion assays, respectively, in Hep-2 cells transfected with miR-449a mimic or inhibitor. A wound-healing assay showed that miR-449a overexpression suppressed cell migration at 24 (p < 0.01) and 48 h (p < 0.001) from the scratch (Figure 5A). Moreover, a transwell invasion assay clearly demonstrated that overexpressed miR-449a strongly inhibited cell invasion to 5% at 24 h (p < 0.05) (Figure 5B). However, there was no significant difference between NC and miR-449a inhibitor treatments in both cell migration and invasion abilities.
Figure 5.
miR-449a Suppresses Cell Migration and Invasion in LCa Cells
(A) Cell migration was assessed by a wound-healing assay in Hep-2 transfected with miR-449a mimic or inhibitor, or the corresponding NC. (B) Cell invasion was evaluated in Hep-2 transfected with miR-449a mimic or inhibitor, or the corresponding NC employing a transwell invasion assay. The purple region indicates invaded cells, and black dots show pores of transwell chamber. Each sample was run in triplicate. Error bars show mean ± SD. The t test was used for the calculation of p values. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Overall, high expression of miR-449a suppressed metastatic potential of Hep-2 cells in vitro, suggesting its availability as a therapeutic agent in treating metastatic LCa.
Notch1 and Notch2 Are miR-449a Target Genes in LCa cells
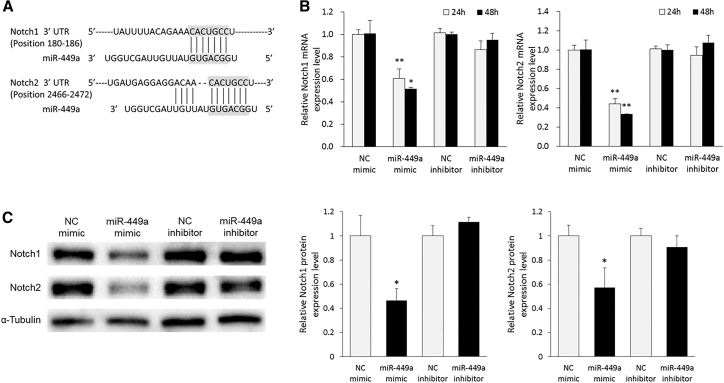
A further study of the molecular mechanisms underlying the metastatic potential of LCa has been carried out through the identification of miR-449a target genes. We first used online available in silico tools, with four different algorithms (TargetScan 7.1, DIANA-microT-CDS 5.0, miRANDA-mirSVR, and miRmap). According to the prediction, we focused on both Notch1 and Notch2 genes, which were considered to be linked with metastases in LCa33,34 and were commonly predicted by the tools as putative miR-449a targets. As shown in Figure 6A, miR-449a possesses complementary sites at 180–186 positions of the 3′ UTR of Notch1 mRNA and 2466–2472 positions of the 3′ UTR of Notch2 mRNA (predicted by TargetScan). On these bases, we assessed, by RT-qPCR, the expression of both Notch1 and Notch2 mRNA in transfected Hep-2 cells with miR-449a mimic or inhibitor and compared it to the mRNA levels in each corresponding NC-transfected ones. As a result, a significant decrease in Notch1 (at 24 h, FC = 0.60 [95% CI = 0.51–0.70], p < 0.001; at 48 h, FC = 0.52 [95% CI = 0.51–0.53], p < 0.01) and Notch2 (at 24 h, FC = 0.44 [95% CI = 0.39–0.50], p < 0.001; at 48 h, FC = 0.33 [95% CI = 0.33–0.34], p < 0.001) mRNA was found in Hep-2 cells overexpressing miR-449a (Figure 6B). In addition, decreased levels of Notch1 (FC = 0.46 [95% CI = 0.37–0.56], p < 0.01) and Notch2 proteins (FC = 0.57 [95% CI = 0.41–0.73], p < 0.01) were observed by western blot analysis (Figure 6C). Alternatively, miR-449a inhibitor did not affect both mRNA and protein expression of these Notch genes. Thus, it was demonstrated that miR-449a strongly suppressed Notch molecules at both the translational and transcriptional levels.
Figure 6.
miR-449a Negatively Regulates Notch1 and Notch2 in LCa Cells
(A) The figure shows representative interaction models between miR-449a and Notch molecules (Notch1 and Notch2). The bindings, predicted by TargetScan, show that Notch1 and Notch2 are putative target genes of miR-449a. (B) The Notch1 and Notch2 mRNA levels were measured in Hep-2 cells transfected with miR-449a mimic or inhibitor, or the corresponding NC using RT-qPCR. HPRT1 mRNA was used as a normalizer. (C) The protein expression levels of Notch1 and Notch2 were determined in Hep-2 cells at 48 h after the transfection of miR-449a mimic or inhibitor, or the corresponding NC by western blot analysis. α-Tubulin was used as a loading control of each western blot (WB). Each sample was run in triplicate. Error bars show mean ± SD. The t test was used for the calculation of p values. ∗p < 0.01, ∗∗p < 0.001.
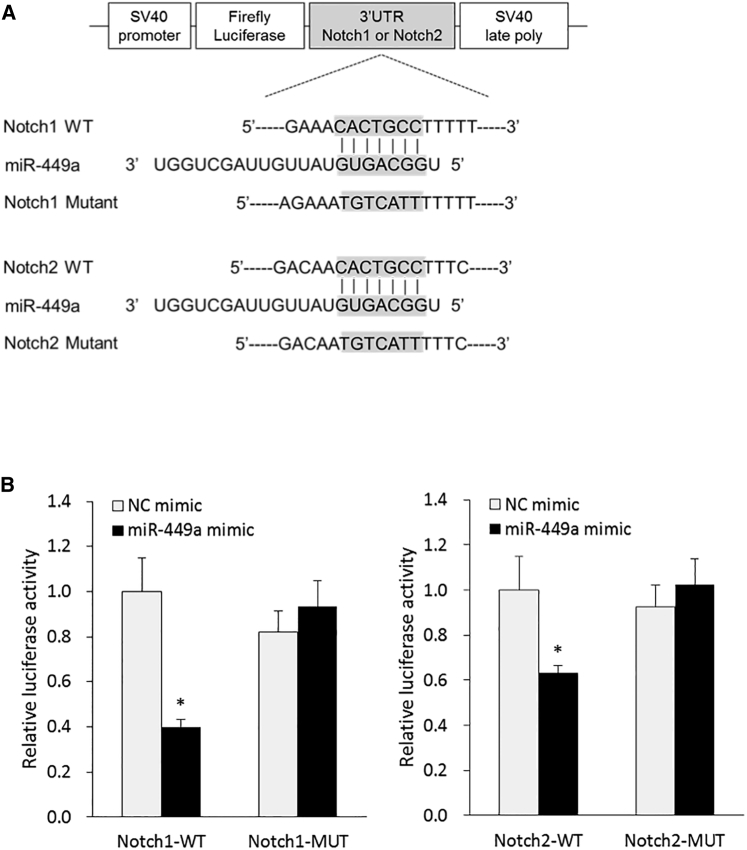
To confirm whether miR-449a directly binds to the 3′ UTR of Notch1 and Notch2, we performed a luciferase reporter assay using co-transfected Hep-2 cells with a miR-449a mimic or NC and a corresponding luciferase reporter plasmid containing a wild-type or mutated target 3′ UTR sequence of Notch1 and Notch2 genes, respectively (Figure 7A). As a result, the luciferase activities for wild-type 3′ UTR of both Notch1 and Notch2 were significantly reduced by 60% (p < 0.01) and 37% (p < 0.01), respectively, compared to the NC treatment conditions (Figure 7B). As expected, the corresponding mutated 3′ UTR sequences were not able to suppress luciferase activity induced by miR-449a mimic.
Figure 7.
miR-449a Directly Binds to 3′ UTR of Notch1 and Notch2 in LCa Cells
(A) Schematic diagram of luciferase reporter plasmid containing wild-type or mutated 3′ UTR target sequence of Notch1 and Notch2 for miR-449a. (B) The relative luciferase activity was determined using co-transfected Hep-2 cells with miR-449a mimic or NC and each corresponding luciferase reporter plasmid. Statistical significance was calculated in comparison between the wild-type group and the mutation group. Each sample was run in triplicate. Error bars show mean ± SD. The t test was used for the calculation of p values. ∗p < 0.01.
In conclusion, miR-449a targets both Notch1 and Notch2 through direct interaction between miR-449a and each gene, leading to the repression of their expression.
Discussion
Despite a growing body of studies, treatment and diagnosis approaches for metastatic LCa still remain to be further investigated. Trying to tackle this urgent need, miRNAs, key regulators of numerous genes at post-transcriptional levels, have emerged as new players modulating physiologic and pathologic events. Therefore, miRNAs have been considered to be potential diagnostic and therapeutic tools. In the present study, we mainly aimed to accumulate scientific evidence for the identification of candidate miRNAs associated with cancer metastatic potential in LCa patients. To achieve this aim, we have performed a comprehensive screening assay to determine a global miRNA profile in tissues collected from LCa patients with a subsequent validation to confirm the modulation of selected candidate miRNA (miR-133b, miR-449a, and miR-652) levels. Based on the results, we focused on miR-449a, which was significantly downregulated in LCa tissues affected by lymph node metastases (N+), for additional studies to understand its biological roles, particularly metastatic capacity.
The miR-449 family, consisting of miR-449a, miR-449b, and miR-449c, is frequently underexpressed in malignancies and may act as a tumor suppressor via the inhibition of a broad variety of oncogenes.27, 28, 29, 30, 31, 32 The miR-449 family shares a sequence similar to the miR-34 family, which is well studied as an onco-suppressor in various cancers;35 therefore, predictable overlap of biological roles could be found through the identification of the common regulatory genes. In our profiling study, miR-449a, which was much more strongly downregulated in N+ tissues compared to N− ones, demonstrated a good potential to diagnose metastasized LCa.
Using the results of the validation set, we analyzed the diagnostic potential of miR-449a as a biomarker. ROC curve analysis showed the predictive power of miR-449a as a molecular marker for the diagnosis of LCa patients with node metastases owing to its significant deregulation in N+.
Based on the results, we hypothesized that miR-449a is closely involved in molecular machinery of LCa progression and metastasis. To test this hypothesis, we examined biological functions of miR-449a. Our study showed that miR-449a significantly suppressed Hep-2 cell proliferation, evaluated by both MTT and clonogenic assays, thereby indicating the tumor-repressive effects of miR-449a in LCa. More importantly, scratch and transwell invasion assays revealed that miR-449a also had suppressive roles on cell migration and, especially, invasion. This demonstrates that miR-449a can inhibit both spreading and metastasizing growth and could be useful for therapeutic approaches in LCa patients affected by nodal metastases. These data support other reports that showed suppressive effects of miR-449a in various tumors.27, 28, 29, 30, 31, 32 In contrast, the loss-of-function experiments did not show the expected reversion of miR-449a-induced cellular effects, although they have traced the results observed upon transfection with the corresponding NC. The latter result could be ascribed to the low basal miR-449a levels in Hep-2 cells.
The most commonly used in silico prediction tools (see Materials and Methods) showed that both Notch1 and Notch2 are possible target genes of miR-449a. We demonstrated that miR-449a mimic transfection in LCa cells strongly decreased the expression of both genes. In addition, through a luciferase reporter assay, we showed direct interaction between miR-449a and the 3′ UTR of both Notch genes in LCa cells. The Notch family consists of four receptors, Notch1–4, widely expressed in human cells, where they play essential roles in the control of embryonic development and cell homeostasis. Notch signaling aberration is commonly found in the oncological microenvironment where it is strongly associated with divergent pathological events, including tumorigenesis, cancer development, invasion, and metastasis.36, 37, 38, 39 High expression of Notch genes in LCa tissues was already found in previous studies, which also linked it to metastases.33,34 In addition, other studies reported both direct and indirect interaction between Notch signaling and miR-449a in various diseases.31,40, 41, 42, 43
However, despite advances in investigations, the precise oncogenic or tumor suppressive role of Notch genes is still controversial, even within the same disease.36, 37, 38, 39 Furthermore, the biological implications of miR-449a and its correlation with Notch expression in LCa are still largely undefined. Because Notch is overexpressed in metastatic LCa, miR-449a may be used as an anti-cancer weapon used alone or in combination therapy with Notch inhibitors. Interestingly, our study revealed that miR-449a triggered repression of Notch1 and Notch2 expression as direct targets, in part resulting in the decrease of Hep-2 cell proliferation, migration, and invasion. To our knowledge, no detailed study including the biological functions of miR-449a in LCa has been reported. Therefore, our study provides new insights into the relevance of both Notch and miR-449a in LCa.
In the reported tissue miRNA screening and validation set, the miR-133b level was drastically changed (underexpressed) in LCa when compared to an adjacent normal counterpart, suggesting its possible use as diagnostic marker in LCa patients and also its potential therapeutic function for replacement therapy. Most importantly, the ROC curve analysis showed that miR-133b has a moderate potential diagnostic value to detect LCa.
miR-133b, which is a member of miR-133 family, belongs to a cluster composed of miR-1, miR-133, and miR-206 (cluster miR-1/133/206). The miR-133 family was initially thought to be a class of canonical muscle-specific miRNAs that was enriched in heart and skeletal muscle. All cluster members modulate myogenesis, muscle development, and remodeling.44 Apart from miR-206, which is not included in the array card, the expression of the other miR-133 family members was significantly reduced in LCa in the screening set.
Many studies have demonstrated that miR-133b is downregulated in different tumors and has anti-tumor activity.44, 45, 46, 47, 48 miR-133b was already negatively correlated with lymph node metastases in gastric cancer patients, and its exogenous overexpression markedly suppressed metastases in gastric carcinoma cells both in vitro and in vivo.45 Saito et al.22 showed that miR-133b expression levels were lower in LCa when compared to matched non-cancerous tissues. Notably, Sousa et al.46 reported that miR-133b was downregulated, but without statistical significance, and was not associated with lymph node metastases in head and neck cancer samples, including LCa specimens. At least two papers, investigating head and neck cancers,22,46 agree with our findings about miR-133b downregulation in tumor samples and no correlation with lymph node metastases in LCa patients.
To summarize this part of our study, miR-133b downregulation may be a powerful indicator of oncogenic laryngeal events and also a therapeutic tool for LCa treatment.
Since the validation did not show significant miR-652 modulation in LCa samples, probably its aberration does not have metastatic potential in LCa patients. Conversely, previous studies suggested that miR-652 could be associated with a metastatic role in other types of tumors, and miR-652 deregulation might be a therapeutic target for several diseases.49, 50, 51 Among them, one report revealed that miR-652 directly interacts with Jagged1 (a Notch1 ligand), and miR-652 inhibition by antisense oligonucleotides induces Jagged1 upregulation.49
Our present work is unlikely to open a scenario on the crucial importance of miR-652 in LCa, but the study of miR-652 biological function could be important for a better understanding of molecular pathological mechanisms of the neoplasm.
In conclusion, our comprehensive gene profile provides intriguing information on miRNA deregulated signatures associated with nodal involvement in LCa patients and significantly contributes to the development of therapeutic opportunities for the disease. Our results clearly showed that miR-449a works as a tumor suppressor and strongly reduces metastatic capacity of LCa, at least in part, via the inhibition of Notch1 and Notch2 molecules as direct targets. To our knowledge, our study provides the first evidence of Notch genes’ direct targeting by miR-449a in LCa. Taken together, the present data provide accumulative evidence about metastasis-associated miRNAs in LCa patients and demonstrate that miR-449a could become a new powerful tool to effectively treat malignant metastases in LCa patients.
Materials and Methods
Clinical Samples
Tissue samples were collected from patients enrolled at the Ear, Nose and Throat Division at the University of Naples “Federico II,” the University of Campania “L. Vanvitelli,” the “Monaldi” Hospital, and the “Antonio Cardarelli” Hospital. Informed consent was obtained from all patients.
Forty-six LCa tissues taken from 23 patients suffering from lymph node metastases (N+) or the same number of patients without metastases (N−), and 30 adjacent noncancerous samples were enrolled in the screening set. Seventy-six cancerous samples (38 N+ and 38 N−) and their counterparts, including all subjects in the screening, were used in the validation set. Clinical information on the patients is summarized in Table 1. Tissue specimens were introduced into RNAlater (Ambion, Life Technologies, Carlsbad, CA, USA) and were kept at 4°C until RNA extraction.
Cell Culture and Transfection
Immortalized human keratinocytes (HaCaT) were available within our research network. The LCa Hep-2 cell line was kindly provided by Dr. Alfredo Budillon (Department of Experimental Oncology, National Institute, Italy). The LCa cell line HNO210 was purchased from CLS Cell Lines Service (Eppelheim, Germany). Both cancer cell lines and human keratinocytes were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Lonza, Switzerland), 50 U/mL penicillin, 500 mg/mL streptomycin, and 4 mM glutamine (Gibco, Life Technologies, Carlsbad, CA, USA) in a humidified atmosphere of 5% CO2 at 37°C. The LCa cells were placed at a density of 4.0 × 105 cells per well in Opti-MEM media (Gibco, Life Technologies, Carlsbad, CA, USA) in a six-well plate. After incubation for 24 h, the cells were transfected with mimic miR-449a (mirVana miRNA mimic) (Life Technologies, Carlsbad, CA, USA), or scramble negative control (mirVana miRNA mimic, negative control #1) (Life Technologies, Carlsbad, CA, USA), or antago-miR-449a (mirVana miRNA inhibitor) (Life Technologies, Carlsbad, CA, USA), or scramble negative control (mirVana miRNA inhibitor, negative control #1) (Life Technologies, Carlsbad, CA, USA) using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA), according to the provided instructions. After incubating the transfected cells in culture for 24 h, the medium was replaced into fresh complete medium.
RNA Extraction
Total RNA, including miRNA, was extracted from either approximately 50 mg of clinical tissue specimens or the culture cells using a mirVana PARIS kit (Ambion, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol with the following modification: RNA was finally eluted into 50 μL of pre-heated elution buffer in order to concentrate the extract. RNA purity and quantity were measured by a spectrophotometer using the 260/280 ratio with a NanoDrop ND-1000 (Thermo Scientific, Wilmington, NC, USA). RNA samples were stored at −80°C until further processing.
PCR Array Screening Assay
As described above, 46 laryngeal tumors (23 N+ and 23 N−) and 30 of their adjacent normal tissues were selected in the screening study. Each type of sample (N+, N−, and normal) was divided into five pools, respectively, for statistical analysis. In detail, with regard to the N+ and N− groups, one pool including seven RNA samples and four pools individually containing four RNA samples were prepared. Alternatively, for the paired normal cohort, one pool including 14 RNA samples and four pools individually consisting of 4 RNA samples were prepared. 3 μL of each RNA sample (20 ng/μL) was mixed for preparing pool samples, resulting in a total of 15 samples prepared.
Starting from 60 ng, each pool sample was then subjected to reverse transcription steps. In this experiment, cDNA was synthesized with Megaplex RT primers, Human Pool A v2.1 (Applied Biosystems, CA, USA) following the Megaplex Pool A protocol, and stored at −20°C when not used immediately. The miRNA expression profiling was subsequently performed using TaqMan Array Human MicroRNA A Cards v2.0 (Applied Biosystems, CA, USA), which allows simultaneous determination of 377 mature miRNA signatures in a single experiment, and TaqMan Universal PCR Master Mix (Applied Biosystems, CA, USA) following the manufacturer’s manuals. The assay was run on a ViiA 7 real-time PCR system (Applied Biosystems, CA, USA).
The Ct value of every miRNA was determined using ViiA 7 software (Applied Biosystems, CA, USA) and setting a threshold of 0.2. Ct values of miRNAs undetermined by the instrument were set up at 40.0 in order to use them for the statistical analysis. For calculating the ΔCt of miRNAs of interest, Ct values of every miRNA were normalized by U6 small nuclear RNA (snRNA) as an endogenous control, and mean Ct values of a miRNA across five pools were used. The relative miRNA expression was calculated with the ΔΔCt method. FC was calculated using the 2−ΔΔCt method.
RT-qPCR for miRNA and mRNA Expression
For the validation of miRNAs, 32 N+ and the same number of N− and 64 of their neighboring normal tissues, which were included in all samples in the screening, were used in the validation set. In the validation of miR-449a, 12 pairs of samples were also used for an additional validation test.
cDNA synthesis was performed using a TaqMan miRNA reverse transcription kit (Applied Biosystems, CA, USA) for three miRNA candidates (miR-133b, miR-449a, and miR-652), and using a QuantiTect reverse transcription kit (QIAGEN, Hilden, Germany) for Notch1 and Notch2 mRNAs in accordance with the manufacturers’ instructions. The expression levels of miRNA candidates were detected using TaqMan Fast universal PCR master mix (Applied Biosystems, CA, USA), whereas expression levels of Notch1 and Notch2 were measured using SYBR Green master mix (Thermo Fisher Scientific, Waltham, MA, USA) via the ViiA 7 real-time PCR system (Applied Biosystems, CA, USA) with each primer for TaqMan miRNA primers (Applied Biosystems, CA, USA) or a QuantiTect primer assay (QIAGEN, Hilden, Germany), respectively.
Real-time PCR was also performed on a ViiA 7 real-time PCR system. Likewise, Ct, ΔCt, and FC were calculated according to the above-mentioned steps in the PCR array session. For evaluation of miRNA, U6 snRNA was also used as a control, while HPRT1 was applied to a reference gene to evaluate mRNA level. Each sample was run in triplicate.
MTT Assay
Transfected cells were seeded into 96-well plates at 2,000 cells/well and incubated at 37°C in 5% CO2. After 24, 48, 72, 96, and 120 h, the cells were stained using MTT reagent (Sigma-Aldrich, St. Louis, MO, USA) for 4 h. Acidified isopropanol was added to dissolve MTT into each well and mixed for 20 min by shaking. Absorbance at 570 nm was measured by an iMark microplate absorbance reader (Bio-Rad, Hercules, CA, USA).
Colony Formation Assay
Transfected cells were seeded into six-well plates at 2,000 cells/well and cultured for 9 days in DMEM (Gibco, Life Technologies, Carlsbad, CA, USA) with 20% FBS (Lonza, Switzerland). Cells were then fixed by 70% ethanol and stained with 0.25% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). After staining, the area of colonies was measured using ImageJ software.
Wound-Healing Assay
Transfected cells were cultured to confluence in 24-well plates. A sterile 10-μL tip was used to scratch through the cultured cells. Then, the medium was replaced with fresh serum-free DMEM (Gibco, Life Technologies, Carlsbad, CA, USA). The wound area was observed by a microscope (EVOS FL cell imaging system) (Life Technologies, Carlsbad, CA, USA) at each time point (0, 24, and 48 h). The closure of wound was calculated using ImageJ.
Transwell Invasion Assay
For the transwell assay, 5 × 104 transfected cells in serum-free medium were applied into the upper transwell chamber (Corning Life Sciences, NY, USA) coated with 0.2 mg/mL Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), and then 600 μL of DMEM (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% FBS (Lonza, Switzerland) was added into the lower chamber. After 24 h, each transwell chamber was washed with PBS buffer and the non-invasive cells were carefully removed using cotton swabs. The invasive cells were fixed in 100% methanol and stained with 0.25% crystal violet (Sigma-Aldrich, St. Louis, MO, USA), imaged, and quantified in five random fields using an inverted microscope (Eclipse microscope) (Nikon, Tokyo, Japan). The number of invaded cells was counted using ImageJ.
In Silico miRNA Target Gene Prediction
A combination of the four online available bioinformatics tools, that is, TargetScan (version 7.1) (http://www.targetscan.org/), DIANA-microT-CDS (version 5.0) (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index), and miRANDA-mirSVR (released 2010) (http://www.microrna.org/microrna/home.do), and miRmap (https://mirmap.ezlab.org/), was used to determine putative predicted targets of miR-449a.
Western Blot Analysis
Cultured cells were lysed with Pierce IP lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) plus 10 ng/mL aprotinin, 10 ng/mL leupeptin, 1 mM PMSF, 1 mM sodium orthovanadate, and 25 mM sodium fluoride on ice for 10 min.52 Cell lysates were collected by centrifugation at 13,000 rpm for 10 min, and then the protein concentrations were determined using Bradford reagent (SERVA Electrophoresis, Heidelberg, Germany). Protein samples (20 μg/lane) were separated on a 12% sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred onto polyvinylidene fluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA). Proteins were detected using Notch1, Notch2, and α-tubulin antibodies (1:1,000) (Cell Signaling Technology, Danvers, MA, USA), followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000) (ImmunoReagents, Raleigh, NC, USA). The immunoreactive protein bands were visualized using an enhanced chemiluminescence substrate kit (Bio-Rad, Hercules, CA, USA).
Luciferase Reporter Assay
Hep-2 cells were harvested into a 96-well plate at 5,000 cells/well and cultured overnight. Thereafter, cells were co-transfected with miRNA mimic or negative control together with each luciferase plasmid. In detail, miR-449a mimic (mirVana miRNA mimic) (Life, Technologies, Carlsbad, CA, USA) or scramble negative control (mirVana miRNA mimic, negative control #1) (Life Technologies, Carlsbad, CA, USA) was transfected using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA), while pEZX-MT06 plasmids containing either wild-type or mutated 3′ UTR of Notch1 or Notch2 (GeneCopoeia, Rockville, MD, USA) were transfected using Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA), following the provided manual. After a 24-h incubation, cell medium was exchanged with fresh DMEM containing 10% FBS. Twenty-four hours later, a dual-luciferase reporter assay was conducted using a Luc-Pair duo-luciferase HS assay kit (GeneCopoeia, Rockville, MD, USA), according to the manufacturer’s instructions. Luciferase activity was detected under the control of a Tecan Infinite M200 (Tecan, Männedorf, Switzerland). For normalization of firefly luciferase activity, the luminescence intensity of Renilla luciferase was used as an internal control of transfected cells.
Statistical Analysis
In the PCR array screening study, differential expression was assessed by linear regression models with empirical Bayes moderated t statistics using Bioconductor package limma v.3.32.8.53 To account for multiple testing, false discovery rate (FDR)-adjusted p values were computed according to the Benjamini-Hochberg method.54 Comparison between two groups was analyzed with Student’s t tests. A clustered heatmap was constructed by heatmap.2 functional analysis in the g plots package of the statistical computing tool R (version 3.4.3). ImageJ software was applied to estimate the results of colony formation and wound-healing assays, respectively. ROC curve analysis was conducted using R (version 3.4.3) to evaluate biomarker potential, including AUC, sensitivity, and specificity of tissue miRNA candidates. A 95% CI was calculated using data of fold change. Data are expressed as the mean ± SD. p < 0.05 was considered statistically significant.
Author Contributions
M.C., G. Misso, H.K., and T.T. designed the study, and H.K. and T.T. wrote the manuscript. F.R., A.L., M.M., Giovanni Motta, Gaetano Motta, D.T., S.D.L., F.O., T.A., and S.M. collected data of clinical trials. E.B., M.F., D.D.B., A.M.C., M.S., R.C., and M.F. performed data analysis. A.M.C. and M.F. contributed to manuscript revision. M.C. and G. Misso coordinated and edited the final version of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by POR Campania FESR 2014/2020 project SENSORMIRCIRCOLAR and by FSN 2014-2015-2016 Linea 5/11.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.04.006.
Contributor Information
Gabriella Misso, Email: gabriella.misso@unicampania.it.
Michele Caraglia, Email: michele.caraglia@unicampania.it.
Supplemental Information
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2013. CA Cancer J. Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Succo G., Crosetti E., Bertolin A., Lucioni M., Caracciolo A., Panetta V., Sprio A.E., Berta G.N., Rizzotto G. Benefits and drawbacks of open partial horizontal laryngectomies, part A: early- to intermediate-stage glottic carcinoma. Head Neck. 2016;38(Suppl 1):E333–E340. doi: 10.1002/hed.23997. [DOI] [PubMed] [Google Scholar]
- 3.Marioni G., Marchese-Ragona R., Cartei G., Marchese F., Staffieri A. Current opinion in diagnosis and treatment of laryngeal carcinoma. Cancer Treat. Rev. 2006;32:504–515. doi: 10.1016/j.ctrv.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Mirisola V., Mora R., Esposito A.I., Guastini L., Tabacchiera F., Paleari L., Amaro A., Angelini G., Dellepiane M., Pfeffer U., Salami A. A prognostic multigene classifier for squamous cell carcinomas of the larynx. Cancer Lett. 2011;307:37–46. doi: 10.1016/j.canlet.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 5.Belcher R., Hayes K., Fedewa S., Chen A.Y. Current treatment of head and neck squamous cell cancer. J. Surg. Oncol. 2014;110:551–574. doi: 10.1002/jso.23724. [DOI] [PubMed] [Google Scholar]
- 6.Misso G., Giuberti G., Lombardi A., Grimaldi A., Ricciardiello F., Giordano A., Tagliaferri P., Abbruzzese A., Caraglia M. Pharmacological inhibition of HSP90 and ras activity as a new strategy in the treatment of HNSCC. J. Cell. Physiol. 2013;228:130–141. doi: 10.1002/jcp.24112. [DOI] [PubMed] [Google Scholar]
- 7.Mesolella M., Iorio B., Misso G., Luce A., Cimmino M., Iengo M., Landi M., Sperlongano P., Caraglia M., Ricciardiello F. Role of perineural invasion as a prognostic factor in laryngeal cancer. Oncol. Lett. 2016;11:2595–2598. doi: 10.3892/ol.2016.4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta G.P., Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 10.Akman F.C., Dag N., Ataman O.U., Ecevit C., Ikiz A.O., Arslan I., Sarioglu S., Ada E., Kinay M., Dokuz Eylül Head and Neck Tumour Group (DEHNTG) The impact of treatment center on the outcome of patients with laryngeal cancer treated with surgery and radiotherapy. Eur. Arch. Otorhinolaryngol. 2008;265:1245–1255. doi: 10.1007/s00405-008-0664-2. [DOI] [PubMed] [Google Scholar]
- 11.Sessions D.G. Surgical pathology of cancer of the larynx and hypopharynx. Laryngoscope. 1976;86:814–839. doi: 10.1288/00005537-197606000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 13.Zamore P.D., Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 14.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimaldi A., Zarone M.R., Irace C., Zappavigna S., Lombardi A., Kawasaki H., Caraglia M., Misso G. Non-coding RNAs as a new dawn in tumor diagnosis. Semin. Cell Dev. Biol. 2018;78:37–50. doi: 10.1016/j.semcdb.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 16.Misso G., Zarone M.R., Grimaldi A., Di Martino M.T., Lombardi A., Kawasaki H., Stiuso P., Tassone P., Tagliaferri P., Caraglia M. Non coding RNAs: a new avenue for the self-tailoring of blood cancer treatment. Curr. Drug Targets. 2017;18:35–55. doi: 10.2174/1389450117666160606104208. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S., Calin G.A., Liu C.G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obayashi M., Yoshida M., Tsunematsu T., Ogawa I., Sasahira T., Kuniyasu H., Imoto I., Abiko Y., Xu D., Fukunaga S. microRNA-203 suppresses invasion and epithelial-mesenchymal transition induction via targeting NUAK1 in head and neck cancer. Oncotarget. 2016;7:8223–8239. doi: 10.18632/oncotarget.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanaihara N., Caplen N., Bowman E., Seike M., Kumamoto K., Yi M., Stephens R.M., Okamoto A., Yokota J., Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 20.de Leeuw D.C., van den Ancker W., Denkers F., de Menezes R.X., Westers T.M., Ossenkoppele G.J., van de Loosdrecht A.A., Smit L. MicroRNA profiling can classify acute leukemias of ambiguous lineage as either acute myeloid leukemia or acute lymphoid leukemia. Clin. Cancer Res. 2013;19:2187–2196. doi: 10.1158/1078-0432.CCR-12-3657. [DOI] [PubMed] [Google Scholar]
- 21.Kojima S., Chiyomaru T., Kawakami K., Yoshino H., Enokida H., Nohata N., Fuse M., Ichikawa T., Naya Y., Nakagawa M., Seki N. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br. J. Cancer. 2012;106:405–413. doi: 10.1038/bjc.2011.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito K., Inagaki K., Kamimoto T., Ito Y., Sugita T., Nakajo S., Hirasawa A., Iwamaru A., Ishikura T., Hanaoka H. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS ONE. 2013;8:e71480. doi: 10.1371/journal.pone.0071480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X., Li Z. The role of microRNAs expression in laryngeal cancer. Oncotarget. 2015;6:23297–23305. doi: 10.18632/oncotarget.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ricciardiello F., Capasso R., Kawasaki H., Abate T., Oliva F., Lombardi A., Misso G., Ingrosso D., Leone C.A., Iengo M., Caraglia M. A miRNA signature suggestive of nodal metastases from laryngeal carcinoma. Acta Otorhinolaryngol. Ital. 2017;37:467–474. doi: 10.14639/0392-100X-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki H., Zarone M.R., Lombardi A., Ricciardiello F., Caraglia M., Misso G. Early detection of laryngeal cancer: prominence of miRNA signature as a new tool for clinicians. Transl. Med. Rep. 2017;1:6502. [Google Scholar]
- 26.Gao W., Zhang C., Li W., Li H., Sang J., Zhao Q., Bo Y., Luo H., Zheng X., Lu Y. Promoter methylation-regulated miR-145-5p inhibits laryngeal squamous cell carcinoma progression by targeting FSCN1. Mol. Ther. 2019;27:365–379. doi: 10.1016/j.ymthe.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lizé M., Pilarski S., Dobbelstein M. E2F1-inducible microRNA 449a/b suppresses cell proliferation and promotes apoptosis. Cell Death Differ. 2010;17:452–458. doi: 10.1038/cdd.2009.188. [DOI] [PubMed] [Google Scholar]
- 28.Bou Kheir T., Futoma-Kazmierczak E., Jacobsen A., Krogh A., Bardram L., Hother C., Grønbæk K., Federspiel B., Lund A.H., Friis-Hansen L. miR-449 inhibits cell proliferation and is down-regulated in gastric cancer. Mol. Cancer. 2011;10:29. doi: 10.1186/1476-4598-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noonan E.J., Place R.F., Basak S., Pookot D., Li L.C. miR-449a causes Rb-dependent cell cycle arrest and senescence in prostate cancer cells. Oncotarget. 2010;1:349–358. doi: 10.18632/oncotarget.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo W., Huang B., Li Z., Li H., Sun L., Zhang Q., Qiu X., Wang E. MicroRNA-449a is downregulated in non-small cell lung cancer and inhibits migration and invasion by targeting c-Met. PLoS ONE. 2013;8:e64759. doi: 10.1371/journal.pone.0064759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niki M., Nakajima K., Ishikawa D., Nishida J., Ishifune C., Tsukumo S.I., Shimada M., Nagahiro S., Mitamura Y., Yasutomo K. MicroRNA-449a deficiency promotes colon carcinogenesis. Sci. Rep. 2017;7:10696. doi: 10.1038/s41598-017-10500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandbothe M., Buurman R., Reich N., Greiwe L., Vajen B., Gürlevik E., Schäffer V., Eilers M., Kühnel F., Vaquero A. The microRNA-449 family inhibits TGF-β-mediated liver cancer cell migration by targeting SOX4. J. Hepatol. 2017;66:1012–1021. doi: 10.1016/j.jhep.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y., Fang F., Ding Y.J., Dai M.Y., Yi X., Chen C., Tao Z.Z., Chen S.M. Notch 2 signaling contributes to cell growth, anti-apoptosis and metastasis in laryngeal squamous cell carcinoma. Mol. Med. Rep. 2016;14:3517–3524. doi: 10.3892/mmr.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai M.Y., Fang F., Zou Y., Yi X., Ding Y.J., Chen C., Tao Z.Z., Chen S.M. Downregulation of Notch1 induces apoptosis and inhibits cell proliferation and metastasis in laryngeal squamous cell carcinoma. Oncol. Rep. 2015;34:3111–3119. doi: 10.3892/or.2015.4274. [DOI] [PubMed] [Google Scholar]
- 35.Misso G., Di Martino M.T., De Rosa G., Farooqi A.A., Lombardi A., Campani V., Zarone M.R., Gullà A., Tagliaferri P., Tassone P., Caraglia M. Mir-34: a new weapon against cancer? Mol. Ther. Nucleic Acids. 2014;3:e194. doi: 10.1038/mtna.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap L.F., Lee D., Khairuddin A., Pairan M.F., Puspita B., Siar C.H., Paterson I.C. The opposing roles of NOTCH signalling in head and neck cancer: a mini review. Oral Dis. 2015;21:850–857. doi: 10.1111/odi.12309. [DOI] [PubMed] [Google Scholar]
- 37.Lobry C., Oh P., Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J. Exp. Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahlgren C., Gustafsson M.V., Jin S., Poellinger L., Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ranganathan P., Weaver K.L., Capobianco A.J. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer. 2011;11:338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 40.Marcet B., Chevalier B., Luxardi G., Coraux C., Zaragosi L.E., Cibois M., Robbe-Sermesant K., Jolly T., Cardinaud B., Moreilhon C. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Cell Biol. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 41.Capuano M., Iaffaldano L., Tinto N., Montanaro D., Capobianco V., Izzo V., Tucci F., Troncone G., Greco L., Sacchetti L. MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS ONE. 2011;6:e29094. doi: 10.1371/journal.pone.0029094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng J., Wu Q., Lv R., Huang L., Xu B., Wang X., Chen A., He F. MicroRNA-449a inhibition protects H9C2 cells against hypoxia/reoxygenation-induced injury by targeting the Notch-1 signaling pathway. Cell. Physiol. Biochem. 2018;46:2587–2600. doi: 10.1159/000489686. [DOI] [PubMed] [Google Scholar]
- 43.Poddar S., Kesharwani D., Datta M. miR-449a regulates insulin signalling by targeting the Notch ligand, Jag1 in skeletal muscle cells. Cell Commun. Signal. 2019;17:84. doi: 10.1186/s12964-019-0394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nohata N., Hanazawa T., Enokida H., Seki N. microRNA-1/133a and microRNA-206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget. 2012;3:9–21. doi: 10.18632/oncotarget.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y., Huang J., Zhang L., Qu Y., Li J., Yu B., Yan M., Yu Y., Liu B., Zhu Z. miR-133b is frequently decreased in gastric cancer and its overexpression reduces the metastatic potential of gastric cancer cells. BMC Cancer. 2014;14:34. doi: 10.1186/1471-2407-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sousa L.O., Sobral L.M., Matsumoto C.S., Saggioro F.P., López R.V., Panepucci R.A., Curti C., Silva W.A., Jr., Greene L.J., Leopoldino A.M. Lymph node or perineural invasion is associated with low miR-15a, miR-34c and miR-199b levels in head and neck squamous cell carcinoma. BBA Clin. 2016;6:159–164. doi: 10.1016/j.bbacli.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crawford M., Batte K., Yu L., Wu X., Nuovo G.J., Marsh C.B., Otterson G.A., Nana-Sinkam S.P. MicroRNA 133B targets pro-survival molecules MCL-1 and BCL2L2 in lung cancer. Biochem. Biophys. Res. Commun. 2009;388:483–489. doi: 10.1016/j.bbrc.2009.07.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kano M., Seki N., Kikkawa N., Fujimura L., Hoshino I., Akutsu Y., Chiyomaru T., Enokida H., Nakagawa M., Matsubara H. miR-145, miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int. J. Cancer. 2010;127:2804–2814. doi: 10.1002/ijc.25284. [DOI] [PubMed] [Google Scholar]
- 49.Bernardo B.C., Nguyen S.S., Winbanks C.E., Gao X.M., Boey E.J., Tham Y.K., Kiriazis H., Ooi J.Y., Porrello E.R., Igoor S. Therapeutic silencing of miR-652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J. 2014;28:5097–5110. doi: 10.1096/fj.14-253856. [DOI] [PubMed] [Google Scholar]
- 50.Xuan J., Guo S.L., Huang A., Xu H.B., Shao M., Yang Y., Wen W. miR-29a and miR-652 attenuate liver fibrosis by inhibiting the differentiation of CD4+ T cells. Cell Struct. Funct. 2017;42:95–103. doi: 10.1247/csf.17005. [DOI] [PubMed] [Google Scholar]
- 51.Nam R.K., Amemiya Y., Benatar T., Wallis C.J., Stojcic-Bendavid J., Bacopulos S., Sherman C., Sugar L., Naeim M., Yang W. Identification and validation of a five microRNA signature predictive of prostate cancer recurrence and metastasis: a cohort study. J. Cancer. 2015;6:1160–1171. doi: 10.7150/jca.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sirangelo I., Iannuzzi C., Vilasi S., Irace G., Giuberti G., Misso G., D’Alessandro A., Abbruzzese A., Caraglia M. W7FW14F apomyoglobin amyloid aggregates-mediated apoptosis is due to oxidative stress and AKT inactivation caused by Ras and Rac. J. Cell. Physiol. 2009;221:412–423. doi: 10.1002/jcp.21871. [DOI] [PubMed] [Google Scholar]
- 53.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:e3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 54.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.