Abstract
Background
Many health facilities in malaria endemic countries are dependent on Rapid diagnostic tests (RDTs) for diagnosis and some National Health Service (NHS) hospitals without expert microscopists rely on them for diagnosis out of hours. The emergence of P. falciparum lacking the gene encoding histidine-rich protein 2 and 3 (HRP2 and HRP3) and escaping RDT detection threatens progress in malaria control and elimination. Currently, confirmation of RDT negative due to the deletion of pfhrp2 and pfhrp3, which encodes a cross-reactive protein isoform, requires a series of PCR assays. These tests have different limits of detection and many laboratories have reported difficulty in confirming the absence of the deletions with certainty.
Methods
We developed and validated a novel and rapid multiplex real time quantitative (qPCR) assay to detect pfhrp2, pfhrp3, confirmatory parasite and human reference genes simultaneously. We also applied the assay to detect pfhrp2 and pfhrp3 deletion in 462 field samples from different endemic countries and UK travellers.
Results
The qPCR assay demonstrated diagnostic sensitivity of 100% (n = 19, 95% CI= (82.3%; 100%)) and diagnostic specificity of 100% (n = 31; 95% CI= (88.8%; 100%)) in detecting pfhrp2 and pfhrp3 deletions. In addition, the assay estimates P. falciparum parasite density and accurately detects pfhrp2 and pfhrp3 deletions masked in polyclonal infections. We report pfhrp2 and pfhrp3 deletions in parasite isolates from Kenya, Tanzania and in UK travellers.
Interpretation
The new qPCR is easily scalable to routine surveillance studies in countries where P. falciparum parasites lacking pfhrp2 and pfhrp3 are a threat to malaria control.
Keywords: pfhrp2, pfldh, qPCR, RDT, Malaria
Research in context.
Evidence before this study
Rapid diagnostic tests (RDT) play a crucial role in malaria case management, control and elimination programs. RDTs allow malaria diagnosis on the spot in the clinic or in the field and do not require infrastructure or highly trained personnel. Most commercially available RDTs detect a protein unique to Plasmodium falciparum, histidine-rich protein 2 (HRP2). Some HRP2-based RDTs cross-react with another histidine-rich protein, HRP3. The emergence of Plasmodium falciparum parasites lacking pfhrp2 and pfhrp3 genes threatens the utility of RDTs and hampers malaria control and elimination efforts. Currently, identifying such parasites involves laborious multi-step PCR methods that are prone to error and are time consuming. Efforts to develop better methods to detect such parasites face challenges such as false deletion calls particularly at low parasite density due to inclusion of a multi-copy parasite reference gene; absence of a human normalizer gene and cross-binding of primers between pfhrp2 and pfhrp3.
Added value of this study
Our high-throughput multiplex qPCR assay accurately detects pfhrp2 and pfhrp3 deletion genotypes in single- and multi-clone infections and simultaneously estimates parasite density. The qPCR assay has superior performance to existing methods in speed, cost and ease of interpretation in detecting pfhrp2/3-deleted P. falciparum parasites from DNA derived from whole blood or filter-paper bloodspots. This was made possible by three unique features: the choice of a single copy parasite reference gene; the inclusion of a human normalizer gene and the modification of primers to improve specificity. We also report pfhrp2 and pfhrp3 deletions, for the first time, in UK travelers returned from Eritrea, Ethiopia, Kenya, Somalia, South Sudan, Sudan, Tanzania and Uganda. This is the first time pfhrp2/3 deletions have been reported in South Sudan and Somalia.
Implications of all the available evidence
The World Health Organization recommends monitoring the prevalence of pfhrp2 and pfhrp3 in countries where sporadic reports of deletions occur and in neighbouring areas. Based on our data and elsewhere in the literature, pfhrp2 and pfhrp3 deletions are present in 31 countries but the scale and scope is not well elucidated. The multiplex qPCR method can accurately and efficiently support surveillance efforts so that endemic countries have the data required to guide policy on RDT procurement and avert a serious public health threat.
Alt-text: Unlabelled box
1. Background
Malaria is caused by infecting protozoan parasites of the genus Plasmodium. P. falciparum continues to be the predominant species with an estimated global incidence of more than 228 million cases and about 405,000 deaths reported in 2018 [1]. Immunochromatographic rapid diagnostic tests (RDTs), which use membrane-bound antibodies to detect parasite proteins in finger-prick blood samples, play a crucial role in malaria control successes in disease endemic countries. Early diagnosis is critical to malaria elimination and eradication programs and RDT deployment is an important component of the strategy. As a result, the global availability and scale of use of RDTs has increased dramatically over the last 10 years [2]. Most RDTs used worldwide detect P. falciparum histidine-rich protein 2 (pfHRP2) and/or Plasmodium lactate dehydrogenase (pLDH) antigens. Some studies have shown that at least some pfHRP2-based RDTs also detect P. falciparum histidine-rich protein 3 (pfHRP3) due to a shared antigenic epitope [2], [3], [4], [5]. In sub-Saharan Africa, which bears 90% of the global malaria burden, RDTs accounted for 74% of diagnostic testing among suspected malaria cases in 2015, and pfHRP2-based tests were the most widely used [2].
Parasites with pfhrp2 and/or pfhrp3 gene (pfhrp2/3) deletions were first observed in South America and increasing reports of false-negative RDT results due to these parasites have now emerged from selected regions of Africa and Asia [6], [7], [8]. In some countries, a high proportion of RDT false-negative results due to these gene deletions has led to changes in national diagnostic guidelines [9,10]. However, before undertaking any drastic changes in diagnostic testing policies or deploying less sensitive, less heat stable RDTs that detect alternative antigens, malaria programs need robust epidemiological data about local pfhrp2/3 deletion prevalence. The World Health Organization (WHO) has prioritized studies of these parasites and developed a protocol for pfhrp2/3 deletion surveillance [11]. However, confirmation of pfhrp2/3 deletions using current techniques is challenging and time consuming. Most studies of pfhrp2 and pfhrp3 deletions deploy conventional nested PCR (nPCR) amplification of several genes followed by gel-electrophoresis [12]. In this genotyping approach, at least three independent genes are used to ascertain the quality of DNA and the presence of P. falciparum parasites to avoid unintentional misclassification of pfhrp2/3 deletions in samples with low-concentration or degraded DNA [13,14]. The nPCR approach requires several rounds of PCR for each gene and running the gel-electrophoresis for each PCR product. The nested-PCR genotyping approach is labor-intensive, time consuming and is prone to contamination, particularly when deployed in large-scale surveillance studies. The various nPCR methods used differ in limit of detection, and this can cause type I and type II errors. Further, performance of the reported pfhrp2 and pfhrp3 PCR methods is variable, with wide ranging limits of detection and the risk of cross-reactivity in some assays. In addition, gel-electrophoresis approaches do not detect deletions masked in polyclonal infections. Deletions in such infections contribute to the overall frequency of deletions in the parasite population, which has implications for the determination of deletion prevalence and RDT guideline policy [6,15].
In this study, we report the development of a multiplex qPCR assay which simultaneously detects DNA from the human host, a single-copy parasite house-keeping gene, and the pfhrp2 and pfhrp3 genes, including in polyclonal P. falciparum infections, in a single reaction. We report the validation and application of this novel method using DNA samples derived from dried bloodspots (DBS) and whole blood of field isolates and clinical samples. We also use the parasite house-keeping gene (pfldh) to estimate P. falciparum parasite density to rule out low parasite density as a factor for false RDT negative results [5,13] when microscopic data is unavailable or if it is not reliable.
2. Methods
2.1. Plasmodium falciparum laboratory strains
Initial validation of the qPCR assay was performed using culture-adapted laboratory isolates with different pfhrp2 and pfhrp3 status, 3D7 (wildtype, West Africa origin), Dd2 (pfhrp2 deletion, Indochina origin), HB3 (pfhrp3 deletion, Honduras origin) were obtained from the Malaria Research Reference Reagent Repository (http://MR4.org). A culture-adapted isolate lacking both genes (3BD5, double deletion) was also obtained from Thomas Wellems (NIAID, US). Parasite cultures of 3D7, Dd2, HB3 and 3BD5 were tightly synchronized as ring stage trophozoites in vivo to simulate infected peripheral blood similar to previously used methods [16,17]. The WHO P. falciparum International standard (Pf INT), a reagent comprising lyophilised whole blood from a single hyperparasitaemic individual was obtained from NIBSC UK. Undiluted, this reagent represents a parasitaemia of 9.8% which is equivalent to 4.9 × 105 parasites per µl [16,18].
3. Clinical and field DNA samples
Clinical validation and application of the qPCR assay was performed using 462 DNA samples derived from both DBS and whole-blood samples. Parasite DNA was isolated from fifty DBS samples from suspected malaria patients in Eritrea; 299 samples from whole-blood collected in EDTA from symptomatic Tanzanian and Kenyan patients and anonymized 113 samples from UK malaria patients. Details of the study for the samples from Eritrea including study site, sample collection and IRB approvals have already been published [9,10]. Whole blood collected in EDTA from UK travellers with confirmed P. falciparum infections in 2018 was obtained from the Public Health England Malaria Reference Laboratory (MRL), London, UK. Samples from Kenya (Ahero) and Tanzania (Bagamoyo) were collected between Oct 2016 and Dec 2018 as part of a study of parasite clearance after treatment with artemisinin combination therapy. These samples from Kenya and Tanzania have aliquots of cryopreserved blood samples and were selected for the pfhrp2/3 deletion study to identify pfhrp2/3-deleted parasites for culture-adaptation.
4. DNA extraction
DNA was extracted from DBS from Eritrea and from whole blood from the MRL and from cultured laboratory isolates using a robotic DNA extraction system (Qiasymphony, QIAGEN, Germany), as previously described [18]. For the clinical samples collected in Kenya and Tanzania, 200 µl of whole-blood was extracted using the QIAamp Blood Mini Kit (Qiagen) into 200 µl of Buffer EB as per the manufacturer's instructions.
5. Multiplex qPCR development
5.1. Gene target selection and primer design
To design highly specific amplification primers that are conserved across global P. falciparum isolates, we carried out multiple alignments of pfhrp2 gene sequences from 1581 published P. falciparum genomes (MalariaGEN) from Africa, SE Asia and South America, and a similar alignment was also carried out for pfhrp3 (Figure S1). The DNA sequences of the genes were obtained from publicly available genomic data and the processing of the data has been described in our previous report [6]. We have also aligned the 3D7 DNA sequence of pfhrp2 (PF3D7_0831800) and pfhrp3 (PF3D7_1372200) to ensure that the conserved primers of the two genes do not cross-bind and are specific to pfhrp2 and pfhrp3 respectively (Figure S2). The DNA sequences of pfhrp2 and pfhrp3 were aligned using Geneious v. 10 (Biomatters, USA)
We used Plasmodium falciparum lactate dehydrogenase (pfldh, PF3D7_1324900), coded by a single-copy gene on chromosome 13, as a confirmatory gene for the presence and quality of parasite DNA as well as a target for measuring parasite density. We used previously published qPCR methods, with some modification of reaction conditions to amplify pfldh [13] and the human beta tubulin gene (HumTuBB) (Table S1) [19]. The latter was used both as an internal control and as a normalizer for measurement of parasite density and for detection of pfhrp2/3 deletions in polyclonal infections. All primers and probes were ordered from Eurofins Scientific (Germany).
5.2. Modification of pfhrp2 primer at the 3′ end
Due to limited availability of suitable conserved target sequence regions in pfhrp2 and pfhrp3 that are dissimilar between the isoform genes and to prevent non-specific cross-binding of the pfhrp2 primers to pfhrp3, we used a strategy altering nucleotides located at the 3′ end region (within the last 5 nucleotides) of both pfhrp2 primers (Table S1). In total, we designed six primers with different modifications at the 3′ end of the forward and reverse pfhrp2 primers, which were then tested empirically to identify primer pairs that delivered the best specificity while maintaining product yield (sensitivity).
5.3. Assay optimization
The multiplex qPCR assay designed in this study was optimized for primer and probe hybridization temperature; different primer and probe concentrations and different MgCl2 concentrations. All the optimization analyses were performed in triplicates in a RGQ rotor-gene (Qiagen, Germany).
The optimized final reaction conditions were performed in a final volume of 25 µl containing 1.6X NH4 buffer (Bioline); 4 mM MgCl2 (Bioline); 800 nM dNTPs (Bioline), 200 nM of pfhrp2 primers, 200 nM pfhrp3 primers, 120 nM of pfldh, 120 nM HumTuBB primers, 120 nM of pfhrp2 probe, 120 nM pfhrp3 probe, 80 nM of pfldh probe and 80 nM HumTuBB probe; 2 units of biotaq polymerase and 5 µl of extracted DNA (the lowest concentration used was two parasites per 5 µl). The optimal thermocycling conditions selected were 3 min at 95 °C, followed by 45 cycles of 15 s at 95 °C; 30 s at 54 °C and 30 s at 72 °C.
5.4. PCR efficiency, linear dynamic range and limit of detection
We conducted assay performance analysis based on the MIQE guidelines [20]. The efficiency of primer and probe combinations for each gene and linear dynamics of the qPCR assays were evaluated using seven 4-fold dilutions of Pf INT (from 12,500 to 3 parasites per µl). The last dilution series (3 parasites per µl) was then further diluted 2-fold (from 3 to 0.38 parasites per µl) to determine the measured limit of detection (LOD). The three laboratory strains (Dd2, HB3 and 3BD5) with known pfhrp2/3 status were included in the evaluation of linear dynamics to examine the effect of DNA concentration on the cross reactivity of the primers.
5.5. Assay precision, analytical sensitivity and specificity
Performance of the multiplex qPCR pfhrp2/3 assay was evaluated by measuring coefficient of variation across the seven four-fold dilution series of Pf INT. The specificity and sensitivity as well as the robustness of the assay was evaluated by testing the lowest two concentrations of Pf INT replicates of eight and 20 P. falciparum-negative whole blood samples in three different experiments.
5.6. Detection of parasites with pfhrp2 and pfhrp3 deletion hidden in polyclonal infections
To investigate whether the qPCR assay could detect pfhrp2/3-deleted parasites hidden in polyclonal infections robustly and accurately, we generated pairwise mixtures of known P. falciparum genotypes (Dd2 and Pf INT, HB3 and Pf INT, and 3BD5 and Pf INT) at different ratios i.e. 1:1, 5:1, 10:1, 100:1, 1000:1, 10,000:1, 1:100,000, 1:10,000, 1:1000, 1:100, 1:10, 1:5, 1:1. We estimated the abundance of pfhrp2/3 deletion genotypes in the mix relative to the whole parasite biomass as measured by relative quantification normalized to pfldh (measures the DNA of all strains) and humTuBB genes. The qPCR assay was also used to detect pfhrp2/3 deletions in patient samples with known polyclonal infection in patient samples, as determined by a peviously published high resolution melting qPCR assay [16].
5.7. Relative quantification using the pfldh gene
We used pfldh as a parasite target and HumTuBB as a normalizer to estimate relative parasite density of each sample. The lowest parasite density (parasite per µl) with a coefficient of variation of less than 35% was considered the limit of quantification for the assay [21]. We determined the performance of the pfldh qPCR in the multiplex assay by comparing parasite densities determined by a published pgmet duplex qPCR assay using samples from Eritrea [19].
5.8. Application to DNA from diverse field samples
The validated qPCR assay was then applied to DNA extracted from diverse field samples. Samples with Cq values lower than the Cq value of the limit of detection of individual genes are determined to be negative for the gene. Samples with HumTuBB positive and pfldh positive but negative for pfhrp2 or pfhrp3 are determined to be pfhrp2-deleted and pfhrp3-deleted respectively. Samples with HumTuBB positive but pfldh negative are determined to be parasite negative. Finally, samples with HumTuBB negative are considered to be invalid and the DNA extraction and/or PCR experiment should be repeated.
6. Statistical analysis
For all amplification curve analyses, the quantification cycle (Cq) threshold was placed above the amplification curve of No Template Control (NTC) and any crossing point between the Cq threshold and the amplification curve was considered positive (Cq value) for the specific sample. To evaluate assay precision, we calculated the coefficient of variation (CV) of parasite density (parasite per µl) as follows: CV% = (standard deviation/mean) × 100. To determine the limit of detection (LOD), we calculated the percentage of positive samples and the lowest sample with more than 3 parasites per PCR and with ≥ 95% of replicate samples detected was considered LOD [20]. Similarly, to determine the lower limit of quantification (LOQ), we calculated the parasite density using delta method and the sample with lowest parasite density (parasite per µl) with a CV ≤35% was considered LOQ for the assay [21]. Throughout the manuscript CV refers to variation in parasite density (parasite per µl).
For estimation of relative abundance of pfhrp2/3 deletion in a mixed-strain infection we used delta relative quantification method [19] as follows:
Before calculating the relative abundance of pfhrp2/3 strains, the Cq threshold of the positive control (calibrator, Pf INT, 0.98% parasitaemia) was adjusted in each channel in such a way that the Cq value is similar in all parasite target genes.
For comparison of parasite density estimated by two different qPCR assays, we used STATA (v 15, USA) software to perform linear regression.
7. Ethics approval and consent to participate
Ethical approval for collection of samples was obtained from each local ethical committee in Eritrea (Eritrean MOH Research and Ethical Committees), Kenya (KEMRI IRB, 3293) and Tanzania (MUHAS IRB, DA.282/298/01). Ethical approval for the samples from MRL patients was obtained from NHS England Research Ethics Committee (18/LO/0738). The ethical approval for the laboratory work for the Eritrean and MRL samples was obtained from LSHTM Ethical Review Committee (#11979 and #14710, respectively).
8. Results
We followed the MIQE guidelines for optimization of the qPCR assay; for analysis and reporting of the data [20].
8.1. Primer and probe selection, and in silico analysis
Initial assessment of pfhrp3 primers across exons 1 and 2 showed cross reactivity with pfhrp2 target (Figure S3) and a new set of primers within a specific conserved region of exon2 of pfhrp3 was designed. After initial assessment of pfhrp2 and pfhrp3 primers for specificity and length of the probe, two sets of pfhrp3 primers and three sets of pfhrp2 primers, including primers with modifications at the 3′ end, were selected for testing (Table S1). For pfhrp2 primers, of the nine combinations used, the lowest Cq value was obtained when pfhrp2_F1 and pfhrp2_R2 (modification at 3′ end) were combined and were selected for further optimization (Table S2). Since one single mutation in the pfhrp2 forward primer was found in one sample in The Gambia and three samples in Ghana, we have nucleotide redundancy in the synthesis of pfhrp2_F1 primer to reflect these mutations (Table S2). The other pfhrp2 primer combinations either produced fluorescence signal in Dd2 (pfhrp2-deleted laboratory strain) due to cross binding to pfhrp3 or generated relatively higher (more unfavorable) Cq values in pfhrp2-positive lab strains (HB3 and Pf INT) compared to the selected primer combinations (Figure S4). Interestingly, a single nucleotide change (T to G) decreased the Cq value by 7 (30 to 23) while two nucleotide changes (T to G and T to G) decreased the Cq value by 2 (30 to 28) (Table S2).
9. Detection of pfhrp2 and pfhrp3 in laboratory strains
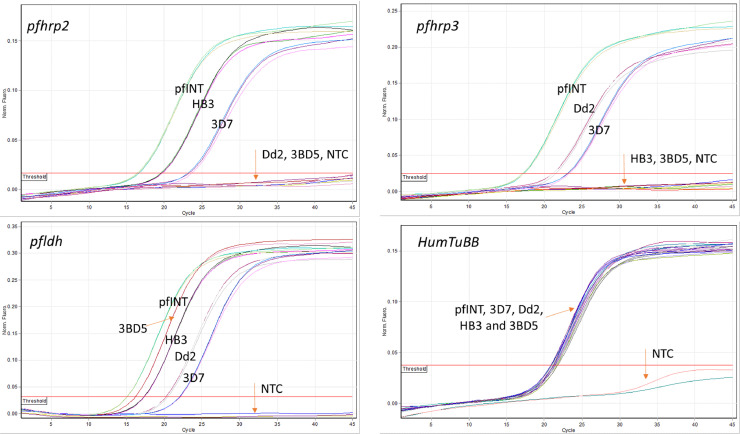
The qPCR assay correctly determined the pfhrp2 and pfhrp3 status of the four laboratory strains (on Dd2, HB3, 3BD5 and 3D7) and the Pf INT. No amplification of either pfhrp2 or pfhrp3 was observed in Dd2 and HB3, respectively, while 3BD5 produced no fluorescence signal in either the pfhrp2 or pfhrp3 channels (Fig. 1). 3D7 and Pf INT were pfhrp2 and pfhrp3 positive.
Fig. 1.
Amplification of four laboratory clones (3D7, Dd2, HB3 and 3BD5) and Pf INT in three parasite targets (pfhrp2, pfhrp3 and pfldh) and a human beta tubulin gene (HumTuBB). Laboratory clones 3D7 (wild type), Dd2 (pfhrp2 deletion), HB3 (hrp3 deletion), 3BD5 (both pfhrp2/3 deletion) and Pf INT (Plasmodium falciparum WHO International Standard, both pfhrp2/3 present) were amplified in triplicate targeting four different genes; pfhrp2, pfhrp3, pfldh and HumTuBB. The red horizontal line marks the threshold, the normalized fluorescence is measured on the y-axis and the number of cycles on the x-axis. Orange arrows point to different clones and the negative no template control (NTC).
10. Analytical sensitivity of the qPCR assay
The pfhrp2, pfhrp3 and pfldh qPCR assays allowed detection of three parasites per µl with a Cq standard deviation (SD) of 0.58, 0.47, 0.41, respectively (Table 1A), which corresponds to parasite density CVs of 9.4%, 9.3% and 8.9% of, respectively (Table 1B). The sample with dilution of the 1.5 parasite per µl showed a Cq SD of 0.80, 0.71 and 0.74, which corresponds to parasite density CVs of 32.6%, 26.1% and 28.1% respectively (Table S3). Though the assay also detected as low as 0.76 parasites per µl the SD value was very high (1.49, 1.37 and 1.48, respectively), and this corresponds to parasite density of CVs of 109%, 99% and 108% (Table S3). Therefore, the lowest parasite density that can be quantified with CV of 35% lies between 1.5 and 0.76 parasites per µl.
Table 1A.
Precision of the parasite target genes. Mean, standard deviation (SD) of quantification cycle (Cq) were calculated from amplifications of seven Pf INT 4-fold dilutions (in triplicate) with pfhrp2, pfhrp3 and pfldh assays.
| Mean Cq values | ||||||
|---|---|---|---|---|---|---|
| Parasite density (p/μl) |
pfhrp2 |
pfhrp3 |
Pfldh |
|||
| Mean | SD | Mean | SD | Mean | SD | |
| 12,500 | 23.18 | 0.06 | 23.46 | 0.02 | 22.44 | 0.04 |
| 3125 | 25.49 | 0.02 | 25.82 | 0.19 | 24.77 | 0.06 |
| 781 | 27.11 | 0.07 | 27.55 | 0.07 | 26.48 | 0.08 |
| 195 | 29.02 | 0.07 | 29.54 | 0.12 | 28.33 | 0.16 |
| 49 | 31.28 | 0.59 | 32.16 | 0.73 | 30.81 | 0.77 |
| 12 | 33.17 | 0.66 | 33.73 | 0.54 | 32.40 | 0.57 |
| 3 | 35.36 | 0.58 | 34.76 | 0.47 | 35.12 | 0.41 |
| Total variance | 28.95 | 0.15 | 29.31 | 0.18 | 28.32 | 0.17 |
Table 1B.
Precision of the parasite target genes. Mean, standard deviation (SD) of coefficient of variation (CV) parasite density were calculated from amplifications of seven Pf INT 4-fold dilutions (in triplicate) with pfhrp2, pfhrp3; and pfldh assays.
| Estimated parasite density (parasite per µl) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Parasite density (p/μl) | Mean | SD | CV | Mean | SD | CV | Mean | SD | CV |
| 12,500 | 11220.33 | 544.65 | 4.85 | 12137.31 | 776.08 | 6.39 | 13405.10 | 658.97 | 4.92 |
| 3125 | 2965.02 | 215.30 | 7.26 | 3098.08 | 196.15 | 6.33 | 3493.59 | 315.34 | 9.03 |
| 781 | 902.11 | 71.29 | 7.90 | 879.47 | 68.65 | 7.81 | 1003.27 | 80.72 | 8.05 |
| 195 | 197.70 | 16.65 | 8.42 | 181.92 | 13.88 | 7.63 | 229.74 | 22.57 | 9.82 |
| 49 | 47.96 | 5.81 | 8.43 | 34.23 | 2.81 | 8.20 | 47.41 | 3.52 | 7.42 |
| 12 | 14.23 | 1.28 | 8.99 | 12.76 | 0.93 | 7.29 | 17.35 | 1.30 | 7.48 |
| 3 | 2.24 | 0.21 | 9.40 | 4.49 | 0.42 | 9.29 | 1.90 | 0.17 | 8.88 |
| Total variance | 190.40 | 15.53 | 7.74 | 197.70 | 14.83 | 7.50 | 208.08 | 16.19 | 7.78 |
11. PCR efficiency and dynamic range
The efficiency of each qPCR was evaluated on 4-fold serial dilutions of Pf INT (from 12,500 to 3 parasite per µl) and each primer and probe combination resulted in similar PCR efficiency (Fig. 2) and this was evident by the Cq value generated by each combination. A 4-fold dilution of the Pf INT produced linear standard plots with 98%, 96% and 98% PCR amplification efficiency for pfhrp2, pfhrp3 and pfldh primer pairs respectively (Fig. 2). The coefficient of determination (R2) of the standard curve was 0.96–0.99 with a slope value of −3.370 to −3.429 for each assay.
Fig. 2.
Standard curve of 7 Pf INT samples diluted 4-fold starting from 12,500 parasites per µl. The three parasite assays (pfhrp2, pfhrp3 and pfldh) detected as low as 3 parasites per microliter with amplification efficiency of 98%, 97% and 96% and coefficient of determination (R2) −3.37, −3.39 and −3.43 respectively.
12. Performance of the qPCR assays
We assessed the precision of the assay by testing seven 4-fold dilution series of the Pf INT (starting concentration, 12,500 parasites per µl) in triplicate on three separate occasions. The mean SD for cycle quantification across the seven samples was 0.15, 0.18 and 0.17 for pfhrp2, pfhrp3 and pfldh target genes respectively (Table 1A), which corresponds to a calculated parasite density of CVs of 7.4%, 7.5% and 7.8% respectively (Table 1B). The sensitivity and specificity as well as the robustness of the assay was assessed using P. falciparum-negative blood samples and Pf INT (3 and 1.5 parasites per µl) in eight replicates on three different occasions. All the control samples were negative, while the Pf INT was positive indicating the absence of amplification inhibition and non-specific amplification (Table S4).
13. Detecting parasites with pfhrp2/3 deletions hidden in polyclonal infections
Both pfhrp2 and pfhrp3 qPCR assays showed good performance in detecting minor (as low as 20%; CI, 7.94–27.05) and major (as high as 99.99%; CI, 99.99–100) pfhrp2/3-deleted clones in the artificially mixed laboratory clones (Table S5). The pfhrp2/3 assays can also detect as low as 10% pfhrp2/3 deleted clones but the value lies within the confidence interval of replicate experiment of a single clone sample and therefore lacks confidence.
14. Workflow and throughput time
Performing the multiplex qPCR requires three steps: reaction set up of ~20 min, two hours for runtime and 30 min for analysis. Over all the estimated time was three hours for 72 reactions on the Rotorgene Q platform. In comparison, the estimated time for the conventional method, deploying several nested PCR assays in a 96-well plate format followed by electrophoresis is approximately 30 h. Time for DNA extraction is the same for both.
15. Validation and application of the qPCR assay on field samples
To determine the diagnostic sensitivity and specificity of the qPCR assay for detecting pfhrp2/3 deletions in field samples and to provide population estimates of deletion prevalence, we first investigated 50 DNA samples obtained from suspected P. falciparum patients from Eritrea whose pfhrp2/3 deletions were previously determined using the conventional nPCR method [10]. Results obtained from the qPCR assay were fully concordant with previously reported results from the same samples using the conventional nPCR method [10]. The qPCR assay correctly detected all the pfhrp2/3 positives (100% sensitivity for pfhrp2, n = 19, 95% CI = (82.4%, 100%); 100% sensitivity for pfhrp3, 95% CI= (66.4%; 100%), n = 9) and accurately determined the absence of pfhrp2/3 in the remaining samples (100% specificity for pfhrp2, n = 31, 95% CI=(88.8%,100%); 100% specificity for pfhrp3, n = 41, 95% CI=(91.4%,100%)) (Table 2). We then assessed samples obtained from clinical malaria patients from the MRL (n = 113), Kenya (n = 150) and Tanzania (n = 149). For the MRL samples, we selected 113 samples from countries with reported pfhrp2/3 deletion or high risk of emergence of pfhrp2/3 deletion [22]. The countries include Eritrea (n = 1), Ethiopia (n = 2), Kenya (n = 20), Tanzania (n = 16), Uganda (n = 27), Sudan (n = 29), South Sudan (n = 10), Djibouti (n = 1), Somalia (n = 3) and east Africa (unknown country, n = 2). Analysis of the pfhrp2/3 status of the MRL samples by qPCR showed 0.9%, 5.3% and 0.9% pfhrp2-/3+, pfhrp2+/3- and pfhrp2-/3- deletions respectively. One pfhrp2 deletion occurred in a UK traveler from Sudan and six pfhrp3 deletions occurred in UK travellers from Ethiopia (n = 1), Sudan (n = 1), South Sudan (n = 3) and Uganda (n = 1). There was evidence of pfhrp2/3 deletions in polyclonal infections in 3.5% of MRL samples in total (Table 2). Of the 149 whole blood samples collected from Tanzania, one sample (0.7%) carried a pfhrp2 deletions and another sample (0.7%) carried a pfhrp3 deletion while no deletions were observed in the 150 Kenyan samples. However, there were 5 samples (3.4%) carrying pfhrp2-deleted strains hidden in polyclonal infections in the Kenyan samples and one (0.7%) in the Tanzanian samples. Details of the deletions in each country are shown in Table 2.
Table 2.
Prevalence of pfhrp2 and pfhrp3 deletions including in polyclonal infections in clinical samples.
| DNA source | Country |
pfhrp2 and/or pfhrp3 deletions in samples |
pfhrp2/3 deletions in polyclonal infections |
||||
|---|---|---|---|---|---|---|---|
| pfhrp2 n(%) | pfhrp3 n(%) | pfhp2/3 n%) | pfhrp2 n(%) | pfhrp3 n(%) | pfhp2/3 n(%) | ||
| Eritrea | Eritrea (n = 50) | 0 | 10 (20) | 31 (62) | 3 (6) | 4 (8) | 0 |
| MRL | Djibouti (n = 1) | 0 | 0 | 0 | 0 | 0 | 0 |
| Kenya (n = 20) | 0 | 0 | 0 | 1 (5) | 1 (5) | 0 | |
| Eritrea (n = 1) | 0 | 0 | 0 | 0 | 0 | 1 (100) | |
| Ethiopia (n = 2) | 0 | 1 (50) | 0 | 0 | 0 | 0 | |
| Somalia (n = 3) | 0 | 0 | 0 | 0 | (2) 66.7 | 0 | |
| South Sudan (n = 12) | 0 | 3 (25) | 0 | 1 (10) | 0 | 0 | |
| Sudan (n = 29) | 1 (3.5) | 1 (3.5) | 1 (3.5) | 0 | 3 (10.3) | 0 | |
| Tanzania (n = 16) | 0 | 0 | 0 | 0 | 1 (6.3) | 0 | |
| Uganda (n = 27) | 0 | 1 (3.7) | 0 | 2 (7.4) | 3 (11.2) | 0 | |
| East Africa (n = 2) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kenya/Tanzania | Kenya (n = 150) | 0 | 0 | 0 | 5 (3.4) | 0 | 0 |
| Tanzania (n = 149) | 1 (0.7) | 1 (0.7) | 0 | 1 (0.7) | 0 | 0 | |
16. Estimation of parasite density using pfldh
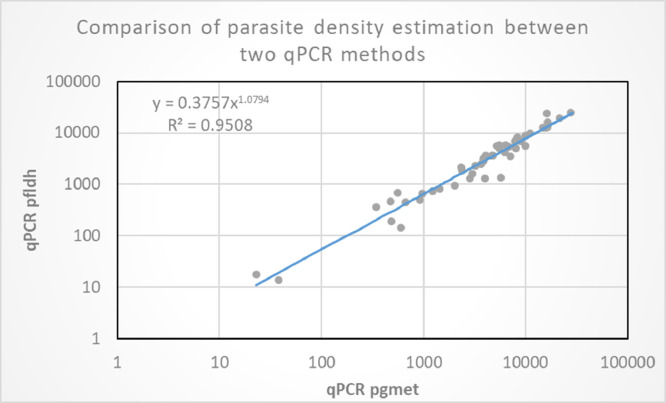
The pfldh gene was used as a parasite target for estimation of parasite density using relative quantification. After assessing its sensitivity, specificity and amplification efficiency the usefulness of the qPCR as an estimator of parasite density was evaluated by comparison to the previously published duplex pgmet qPCR assay [19]. We performed this comparison using 50 DNA samples from Eritrea and the pfldh qPCR showed high degree of correlation (R2=0.95) with the duplex pgmet qPCR (Fig. 3). The pgmet qPCR generated relatively higher parasite density compared to the pfldh qPCR and this is expected as the former targets multi-copy genes in the apicoplast genome, whereas the latter targets is a single-copy gene.
Fig. 3.
Comparing parasite density estimates produced by pfldh and pgmet qPCR methods using 50 Eritrean samples. The estimation was done using delta relative quantification methods in the presence of HumTuBB gene as a normalizer and Pf INT as a calibrator. The two qPCR methods showed strong agreement (R2 = 0.95). The value of X and Y are log of parasite per microliter.
17. Discussion
In this study, we describe the development, validation and application of a high-throughput multiplex qPCR assay for simultaneous determination of P. falciparum with pfhrp2 and pfhrp3 deletion genotypes in monoclonal and polyclonal infections, and estimation of parasite density. This highly sensitive and specific method enables accurate and robust assessment of parasite density between 1.5 and 0.76 parasites per µl with a CV of 35% [21]. The qPCR detection of each of the targets described here was achieved with high efficiency, R2 values greater than 0.96 and very low standard deviations among replicates of each dilution. An additional strength of the assay is the detection of pfhrp2/3 deletions at relative abundance as low as 20% and as high as 99% in mixed infections, simulated by mixture of cultured parasites. This is particularly relevant in moderate to high transmission settings, where such parasites have already emerged but can be masked in polyclonal infections. The assay can be used as a tool to monitor changes in frequency of deletions, providing potential early warning signs of emergence of P. falciparum with pfhrp2/3 deletions. This would allow appropriate measures to be taken to identify, respond and contain the spread of such parasites before they become sufficiently abundant to impact on case management and malaria control programs as has already occurred independently in South America and Eritrea [12,23].
The design of the multiplex qPCR assay provides several advantages over existing detection methods [5,14,24]. Firstly, the qPCR assay uses only one parasite target as a reference gene in a single assay to confirm the presence of DNA and to assess its quality while the nested PCR (nPCR) methods use three target genes in three different assays [5,25]. This reduces the cost and throughput time and simplifies the algorithm for interpreting results. Secondly, the parasite target gene (pfldh) used for DNA confirmation in the qPCR assay is a single copy gene, generating equivalent sensitivity to the pfhrp2/3 targets. When multi-copy genes (e.g., 18SrDNA and cytb) are used in the nPCR and other qPCR assays there is an increased risk of false-pfhrp2/3 deletion calls due to sensitivity differences with the single copy pfhrp2/3 genes [14,26]. It is difficult to evaluate the accuracy of pfhrp2/3 deletion calls in the literature, as most laboratories do not report the limit of detection of the nPCR methods used. Thirdly, the qPCR assay uses a human house-keeping gene as an internal control. Variability in DNA yield, or loss, introduced during sample collection, parasite DNA extraction or qPCR amplification can thus be corrected for, reducing the risk of false-pfhrp2/3 deletion calls. Using conventional methods, those samples negative by 18SrDNA may be excluded from further pfhrp2/3 deletion analysis as they are presumed parasite negatives, potentially underestimating the prevalence of pfhrp2/3 deletions by missing those samples where technical failure has caused this outcome. Fourthly, the multiplex qPCR assay accurately detects laboratory P. falciparum strains and clinical samples with pfhrp2/3 deletions when mixed as minor or major clones. The ability of the multiplex qPCR assay to detect pfhrp2/3 deletions in samples with low parasitaemia and in polyclonal infections is made possible due to three unique features: the choice of a single copy parasite gene (pfldh) for DNA quality confirmation; the inclusion of a human gene for normalization and the modification of the primers. Finally, the qPCR assay can also estimate relative parasite density using the human gene as a normalizer and Pf INT as a calibrator. This characteristic of the assay is useful because one of the criteria for confirming deletion of pfhrp2/3 is estimation of parasite density using microscopy to rule out low parasite density as a factor for lack of parasite target amplification [14]. However, microscopy is not always performed during community surveys, and quantification of parasite density using qPCR is required [5,13]. The qPCR assay not only detects P. falciparum parasites with pfhrp2/3 deletions but also simultaneously estimates parasite density in the same experiments, hence reducing time and cost.
Our study shows the presence of pfhrp2/3 deletions in infected UK travellers for the first time. Current UK guideline for malaria diagnosis is microscopic examination of thin and thick films, with RDT as a supplementary test when the microscopist is relatively inexperienced. Our findings suggest that RDT results should be interpreted cautiously, particularly when microscopic results are unreliable or unavailable. While pfhrp2/3 deletions were previously reported in Eritrea [10], Ethiopia [27] and Uganda [7] this is the first time such deletions are reported in Sudan and South Sudan, though a negative pfhrp2 result was reported in Sudan [28]. Interestingly, pfhrp2 and pfhrp3 deletions were also detected in polyclonal infections in Kenya, Eritrea, Somalia, South Sudan, Sudan and Uganda. This suggests the circulation of low frequency pfhrp2-deleted parasites in Somalia as minor strains in mixed infections. WHO recommends surveillance to determine the prevalence of pfhrp2/3 deletions in countries where sporadic reports of deletions occur and in neighbouring areas. If the prevalence of pfhrp2 gene deletions that cause false-negative HRP2-based RDT results in a representative sample is higher than 5%, HRP2-based RDTs should be replaced with an alternative P. falciparum diagnostic tool that is not exclusively reliant on detection of HRP2 [11]. If the prevalence is below 5% a repeat of the survey is recommended in 1–2 years and the detection of pfhrp2/3 deletions in polyclonal infections by the qPCR assay could be used to inform decisions about how soon to repeat the survey. For example, if the pfhrp2/3 deletions in polyclonal infections occur in medium to high transmission endemic settings, it may be preferable to survey during the dry season when the multiplicity of infection is lower and would allow accurate estimation of the pfhrp2/3 deletions.
There are several limitations to our assay and assays targeting deletions in general. The challenges of confirming the absence of a gene target require careful attention to lab workflow and DNA quality. First, the sensitivity of the qPCR assay demands careful laboratory workflows that prevent contamination. This is true of all qPCR assays but particularly important for discrimination of low-concentration deleted strains in polyclonal infections. Second, while the qPCR assay was carefully designed and optimized to avoid cross-binding, use of appropriate DNA controls is needed to monitor for unintentional amplification of pfhrp3 by pfhrp2 primers and vice versa. We recommend including at least two parasite negative controls (preferably Dd2 and HB3) for pfhrp2 and pfhrp3, respectively, in each experiment. The use of only a double-deleted strain (such as 3BD5) is not recommended. In addition, the qPCR assay targets only one additional single-copy parasite gene while the conventional methods for pfhrp2 and pfhrp3 genotyping have employed three independent parasite genes to ensure DNA quality and rule out DNA degradation [14]. Because the pfhrp2 and pfhrp3 qPCR amplicon lengths are shorter (98 bp and 84 bp, respectively) than typical amplicons generated by the conventional method (~300–800 bp), detection of these targets is expected to be more reliable. If the frequency of deletions is observed to be markedly increased near the limits of detection of the assay in a particular study, then confirmatory testing using a second gene target could be considered. Finally, due to limited conserved regions, the pfhrp2-specific primer covered known variant positions present in a minority of field samples in the MalariaGEN genome data. The qPCR assay was optimized taking into account the known sequence variants, and the use of nucleotide redundancies in the primer synthesis did not affect the yield in fluorescence signal (Cq value). The variants within our primer sequence are relatively fewer compared to the primer and probe sequences of other recently published qPCR assays [26,29], which we predict may be challenged by sequence variation (mutations, insertions and deletion) in the primer and probe sequences of pfhrp2 and pfhrp3 found in field samples from seven and nine countries respectively (Table S6).
In conclusion, our qPCR method for detection of pfhrp2/3 deletions is a robust alternative with several advantages over existing approaches. The qPCR assay has superior performance to existing methods in speed, cost and ease of interpretation in detecting pfhrp2/3-deleted P. falciparum parasites from DNA derived from whole blood or filter-paper bloodspots. Data from screening endemic country samples in returning travellers to the United Kingdom suggest that systematic surveillance of pfhrp2/3 deletions in Ethiopia, Sudan and South Sudan is warranted. Careful monitoring of pfhrp2/3 deletions in Somalia will also be required as the emergence of pfhrp2/pfhrp3-deleted parasites as a single-clone infection may soon occur. Based on the findings in this report and elsewhere in the literature, pfhrp2/3 deletions are present in 31 countries but the scale and scope is still not well elucidated and efforts to dramatically scale up surveillance are needed [15,30]. This qPCR assay can accurately and efficiently support surveillance efforts so that endemic countries have the data required to guide policy on RDT procurement and avert a serious public health threat.
Declaration of Competing Interest
JBP reports non-financial support in the form of in-kind donation of laboratory testing and reagents from Abbott Laboratories for studies of viral hepatitis and financial support from the World Health Organization. Other authors declare no conflict of interest.
Acknowledgments
Acknowledgment
We thank all the patients and staff who participated in this study. We thank Helena Stone for her support with the Malaria Reference Laboratory samples. We also thank Oksana Kharabora for assistance in the lab with the Kenyan and Tanzanian samples. We thank Qin Cheng (ADFMIDI/QIMR Berghofer) sharing with us the pfhrp2/3 data generated by conventional PCR to validate our qPCR assay.
Funding sources
This study was supported by Wellcome’s Institutional Strategic Support Fund to KBB (204928/Z/16/Z) and Department of Infection Biology's Rosemary Weir Research Prize to LG, NS and KBB. DN is funded by the PHE Malaria Reference Laboratory. RDK is supported by the DELTAS Africa Initiative grant # DEL-15-011 to THRiVE-2. Collection of samples from Kenya and Tanzania was supported by the National Institutes of Health (R01AI121558) to JJJ. The Malaria Reference Laboratory is funded by Public Health England (research contract awarded to CJS). The funders did not have any study design, data collection, data analysis, interpretation and writing of the report.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102757.
Appendix. Supplementary materials
References
- 1.WHO. World malaria report 2019. Geneva: World Health Organization. 2019.
- 2.WHO . Results of who product testing of malaria RDTs: round 8 (2016–2018) 2018. Malaria rapid diagnostic test performance. [Google Scholar]
- 3.Baker J., Gatton M.L., Peters J., Ho M.F., McCarthy J.S., Cheng Q. Transcription and expression of plasmodium falciparum histidine-rich proteins in different stages and strains: implications for rapid diagnostic tests. PLoS One. 2011;6(7):e22593. doi: 10.1371/journal.pone.0022593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N., Baker J., Andrews K.T., Gatton M.L., Bell D., Cheng Q. Effect of sequence variation in plasmodium falciparum histidine- rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. J Clin Microbiol. 2006;44(8):2773–2778. doi: 10.1128/JCM.02557-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beshir K.B., Sepulveda N., Bharmal J., Robinson A., Mwanguzi J., Busula A.O. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. 2017;7(1):14718. doi: 10.1038/s41598-017-15031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sepulveda N., Phelan J., Diez-Benavente E., Campino S., Clark T.G., Hopkins H. Global analysis of plasmodium falciparum histidine-rich protein-2 (pfhrp2) and pfhrp3 gene deletions using whole-genome sequencing data and meta-analysis. Infect Genet Evol. 2018;62:211–219. doi: 10.1016/j.meegid.2018.04.039. [DOI] [PubMed] [Google Scholar]
- 7.Thomson R., Beshir K.B., Cunningham J., Baiden F., Bharmal J., Bruxvoort K.J. pfhrp2 and pfhrp3 gene deletions that affect malaria rapid diagnostic tests for plasmodium falciparum: analysis of archived blood samples from 3 African countries. J Infect Dis. 2019;220(9):1444–1452. doi: 10.1093/infdis/jiz335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorado E.J., Okoth S.A., Montenegro L.M., Diaz G., Barnwell J.W., Udhayakumar V. Genetic characterisation of plasmodium falciparum isolates with deletion of the pfhrp2 and/or pfhrp3 genes in colombia: the amazon region, a challenge for malaria diagnosis and control. PLoS One. 2016;11(9) doi: 10.1371/journal.pone.0163137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berhane A., Russom M., Bahta I., Hagos F., Ghirmai M., Uqubay S. Rapid diagnostic tests failing to detect plasmodium falciparum infections in eritrea: an investigation of reported false negative RDT results. Malar J. 2017;16(1):105. doi: 10.1186/s12936-017-1752-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berhane A., Anderson K., Mihreteab S., Gresty K., Rogier E., Mohamed S. Major threat to malaria control programs by plasmodium falciparum lacking histidine-rich protein 2, eritrea. Emerg Infect Dis. 2018;24(3):462–470. doi: 10.3201/eid2403.171723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.WHO. Protocol for estimating the prevalence of pfhrp2/pfhrp3 gene deletions among symptomatic falciparum patients with false-negative RDT results. https://www.who.int/malaria/publications/atoz/hrp2-deletion-protocol/en/. Accessed 21 January 2020. 2018(WHO/CDS/GMP/2018.03).
- 12.Gamboa D., Ho M.F., Bendezu J., Torres K., Chiodini P.L., Barnwell J.W. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One. 2010;5(1):e8091. doi: 10.1371/journal.pone.0008091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parr J.B., Verity R., Doctor S.M., Janko M., Carey-Ewend K., Turman B.J. Pfhrp2-deleted plasmodium falciparum parasites in the democratic republic of the Congo: a national cross-sectional survey. J Infect Dis. 2017;216(1):36–44. doi: 10.1093/infdis/jiw538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Q., Gatton M.L., Barnwell J., Chiodini P., McCarthy J., Bell D. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. 2014;13:283. doi: 10.1186/1475-2875-13-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO. Response plan to pfhrp2 gene deletions. https://apps.who.int/iris/bitstream/handle/10665/325528/WHO-CDS-GMP-2019.02-eng.pdf. Accessed on 31 January 2020. 2019.
- 16.Beshir K.B., Diallo N., Sutherland C.J. Identifying recrudescent plasmodium falciparum in treated malaria patients by real-time PCR and high resolution melt analysis of genetic diversity. Sci Rep. 2018;8(1):10097. doi: 10.1038/s41598-018-28179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Schalkwyk D.A., Burrow R., Henriques G., Gadalla N.B., Beshir K.B., Hasford C. Culture-adapted plasmodium falciparum isolates from UK travellers: in vitro drug sensitivity, clonality and drug resistance markers. Malar J. 2013;12:320. doi: 10.1186/1475-2875-12-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson A., Busula A.O., Muwanguzi J.K., Powers S.J., Masiga D.K., Bousema T. Molecular quantification of plasmodium parasite density from the blood retained in used RDTs. Sci Rep. 2019;9(1):5107. doi: 10.1038/s41598-019-41438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beshir K.B., Hallett R.L., Eziefula A.C., Bailey R., Watson J., Wright S.G. Measuring the efficacy of anti-malarial drugs in vivo: quantitative PCR measurement of parasite clearance. Malar J. 2010;9:312. doi: 10.1186/1475-2875-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The miqe guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 21.Forootan A., Sjoback R., Bjorkman J., Sjogreen B., Linz L., Kubista M. Methods to determine limit of detection and limit of quantification in quantitative real-time PCR (qPCR) Biomol Detect Quantif. 2017;12:1–6. doi: 10.1016/j.bdq.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gatton M.L., Dunn J., Chaudhry A., Ciketic S., Cunningham J., Cheng Q. Use of pfhrp2-only RDTs rapidly select for pfhrp2-negative parasites with serious implications for malaria case management and control. J Infect Dis. 2017;215(7):1156–1166. doi: 10.1093/infdis/jix094. [DOI] [PubMed] [Google Scholar]
- 23.Berhane A M.S., Mohammed S., Embaye G H.F., Zehaie A., Chinorumba A C.J. Pfhrp2 detecting malaria rdts: alarming false negative results in eritrea. Proceedings of program and absracts of the 65th conference on American Soceity for tropical medicine and hygiene; Atlanta; 2016. p. 879. [Google Scholar]
- 24.Parr J.B., Anderson O., Juliano J.J., Meshnick S.R. Streamlined, PCR-based testing for pfhrp2- and pfhrp3-negative plasmodium falciparum. Malar J. 2018;17(1):137. doi: 10.1186/s12936-018-2287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar N., Pande V., Bhatt R.M., Shah N.K., Mishra N., Srivastava B. Genetic deletion of HRP2 and HRP3 in Indian plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125(1):119–121. doi: 10.1016/j.actatropica.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 26.Kreidenweiss A., Trauner F., Rodi M., Koehne E., Held J., Wyndorps L. Monitoring the threatened utility of malaria rapid diagnostic tests by novel high-throughput detection of plasmodium falciparum hrp2 and hrp3 deletions: a cross-sectional, diagnostic accuracy study. EBioMedicine. 2019;50:14–22. doi: 10.1016/j.ebiom.2019.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girma S., Cheaveau J., Mohon A.N., Marasinghe D., Legese R., Balasingam N. Prevalence and epidemiological characteristics of asymptomatic malaria based on ultrasensitive diagnostics: a cross-sectional study. Clin Infect Dis. 2019;69(6):1003–1010. doi: 10.1093/cid/ciy1005. [DOI] [PubMed] [Google Scholar]
- 28.Mussa A., Talib M., Mohamed Z., Hajissa K. Genetic diversity of plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of pfhrp2-based rapid diagnostic tests. BMC Res Notes. 2019;12(1):334. doi: 10.1186/s13104-019-4361-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schindler T., Deal A.C., Fink M., Guirou E., Moser K.A., Mwakasungula S.M. A multiplex qPCR approach for detection of pfhrp2 and pfhrp3 gene deletions in multiple strain infections of plasmodium falciparum. Sci Rep. 2019;9(1):13107. doi: 10.1038/s41598-019-49389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO. Malaria Threats Map: visualizing biological challenges to malaria control and elimination https://www.who.int/malaria/maps/threats-about/en/ Accessed on 31 January 2020. 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.