Abstract
Background
Multiple factors contribute to the etiology of addiction, including genetics, sex, and a number of addiction-related behavioral traits. One behavioral trait where individuals assign incentive salience to food stimuli (“sign-trackers”, ST) are more impulsive compared to those that do not (“goal-trackers”, GT), as well as more sensitive to drugs and drug stimuli. Furthermore, this GT/ST phenotype predicts differences in other behavioral measures. Recent studies have implicated the gut microbiota as a key regulator of brain and behavior, and have shown that many microbiota-associated changes occur in a sex-dependent manner. However, few studies have examined how the microbiome might influence addiction-related behaviors. To this end, we sought to determine if gut microbiome composition was correlated with addiction-related behaviors determined by the GT/ST phenotype.
Methods
Outbred male (N=101) and female (N=101) heterogeneous stock rats underwent a series of behavioral tests measuring impulsivity, attention, reward-learning, incentive salience, and locomotor response. Cecal microbiome composition was estimated using 16S rRNA gene amplicon sequencing. Behavior and microbiome were characterized and correlated with behavioral phenotypes. Robust sex differences were observed in both behavior and microbiome; further analyses were conducted within sex using the pre-established goal/sign-tracking (GT/ST) phenotype and partial least squares differential analysis (PLS-DA) clustered behavioral phenotype.
Results
Overall microbiome composition was not associated to the GT/ST phenotype. However, microbial alpha diversity was significantly decreased in female STs. On the other hand, a measure of impulsivity had many significant correlations to microbiome in both males and females. Several measures of impulsivity were correlated with the genus Barnesiella in females. Female STs had notable correlations between microbiome and attentional deficient. In both males and females, many measures were correlated with the bacterial families Ruminocococcaceae and Lachnospiraceae.
Conclusions
These data demonstrate correlations between several addiction-related behaviors and the microbiome specific to sex.
Keywords: Addiction, Sign-tracker, Microbiome, Gut-brain axis, Sex
Research in context.
Evidence before this study
There is growing evidence supporting a role for the trillions of bacteria within the gut in many aspects of brain function and behaviour. However, there has been limited studies focused on the relationship between microbiota composition and key behaviours associated to increased risk of addiction. There are two well established behavioral characteristics predictive of addiction, where subjects assign motivation for reward to the actual reward, or motivation for reward based on external cues indicative of the reward. This difference in reward learning is called goal-tracking (actual reward) and sign-tracking (external cues of reward). Research has shown that rodents that sign-track are more impulsive and more predisposed to drug seeking. The relationship between goal-tracking and sign-tracking and microbiota composition is unknown. Moreover, whether sex differences occur in terms in microbiota responses in addiction is unclear.
Added value of this study
This study takes advantage of intensive behavioural characterization of rats coupled with microbiome profiling. Furthermore, the study focused on this predisposed predictive behavioural phenotype (goal-tracking and sign-training) in relation to the gut-microbiome and implicated clear sex differences in the response.
Implications of all the available evidence
Findings from this study show that the microbiome is an important aspect of behavioural regulation. Moreover, it emphasises that further research into psychological disorders and addiction need to account for sex differences. For treatments to be developed for drug addiction, more attention needs to be given to the inherent behavioral differences between males and females.
Alt-text: Unlabelled box
1. Introduction
Addiction is a complex disorder; many factors modulate addiction severity and treatment efficacy. Extensive research has identified socioeconomic status [37], childhood trauma [27,84], genetics [70], sex [11], and psychological comorbidities as factors that contribute to addiction risk and severity [36]. Due to the complex etiology of addiction, treatment is a trial and error process, frequently requiring a combination of therapies [38]. More research is necessary to investigate how genetic and environmental factors contribute to addiction. Additionally, new factors such as the effect of diet, microbiome, and social interventions require greater attention. In particular, manipulation of the gut microbiome offers an intriguing target for new addiction treatments [91].
The gut microbiota consists of a myriad of bacteria, archaea, fungi, and viruses colonizing the host gastrointestinal tract and influencing many host systems, such as metabolism [8,78], immune function [47,53], hypothalamic-pituitary-adrenal axis response, and brain [22,42,62,87]. The gut microbiota is defined as the community of microbes residing in the intestines; the microbiome is the genomic DNA from all microbes in that community. In the context of affective disorders, both animal and human studies have strongly linked the composition of the gut microbiota to the behavioral aspects of these disorders [12,15,22,23,48,100]. Furthermore, microbiota has been linked to psychiatric disorders that involve an array of behavioral abnormalities, such as autism spectrum disorder [35,42,61,86]. Previous research has indicated a link between gut microbiome, addiction, and drugs of abuse [50,56,63,69,71,81,98].
Drugs of abuse activate ‘the reward pathway’, which includes cortical innervations in the ventral tegmental area (VTA), striatum, and prefrontal cortex (PFC). Behavioral measures that have been developed in rodents to explore this pathway, include: locomotor response to novelty [28], measures of impulsivity [75], attention [33], and reward-stimulus learning [29]. In the case of reward-stimulus learning, the unconditioned stimuli (USs) and the conditioned stimuli (CSs) can activate this reward pathway [83]. Individual differences in the activation of this pathway promote differential behavioral responsivity to CSs [28]; "sign-tracking" rats (STs) approach CSs more than their "goal-tracking" counterparts (GTs) [31,64]. Additionally, STs and GTs are differentially sensitive to the motivational properties of several abused drugs [80,94,96,97]. Furthermore, STs are more impulsive as measured by tests of impulsive action and choice [49,58,66,92]. To conclude, this GT/ST phenotype predicts other behavioral phenotypes [68]. Interestingly, sign-tracking behavior is more frequent in female than male rats [49,73]. Impulsivity has also been associated with increases in addiction-related behavior, as well as attention deficit disorder in both rodents [34,49] and humans [79].
The relationship between sex hormones, microbiome and behavior is gaining attention [2,17,44,45,82]. During development factors like genetics and hormones, contribute to sexual dimorphism and associated to sex-differences in the microbiome [3,44,45]. Clinical and pre-clinical research has shown differences in microbiota composition associated with altered metabolism of essential vitamins and nutrients from the diet, with increased metabolic function seen in females compared to males [2,13,101]. Worldwide, illicit drug use is significantly lower in females compared to males [95]. However, females self-administer drugs more readily than males and are more vulnerable to addiction to a variety of licit and illicit drugs [11,99].
This study, to our knowledge, is the first to investigate sex-specific relationships between addiction-related behavioral measures and the microbiome in a large dataset. The pre-established GT/ST phenotype was characterized for this study, as it predicts differences in other addiction-related behavioral measures. A total of 54 behavioral measures associated with addiction were used to determine a behavioral phenotype to group subjects by. These results are presented alongside a clustered behavioral phenotype constrained by goal/sign-tracker phenotype. Differences in behavioral measures and microbiome were characterized for each phenotype, within sex.
2. Methods
2.1. Animals
Male and female heterogenous stock (HS) rats were bred at Medical College of Wisconsin and then shipped to the University at Buffalo for behavioral testing. This National Institutes of Health (NIH)-derived outbred rat colony shows broad phenotypic and genotypic variation [88], making it an ideal choice for the study of individuals differences. Rats (N=202) were housed in pairs in plastic cages (42.5 cm × 22.5 cm × 19.25 cm); males and females housed in the same room in alternating cages in testing order. In the event of odd numbers of rats, rats were housed individually. Animals were kept in reversed 12-h light/dark cycle and housed in controlled temperature and humidity conditions. Lights were on in the colony room from 19:00 to 07:00 hours. Behavioral testing occurred 6 days/week between of 08:30 and 12:30 hours during the dark phase of the light cycle. Food (#8604, Harlan Inc., Indianapolis, IN) was available ad libitum in the home cage. Animals were treated in compliance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and the experiments were conducted in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the University at Buffalo, The State University of New York.
2.2. Behavioral tests
Behavioral testing was carried out on rats beginning at 63 days of age (see Table 1). All behavioral tests were conducted in the dark phase of the light cycle. Epochs for behavioral tests consisted of consecutive 3-minute intervals across a test session.
Table 1.
Study design flow chart – Age in days (first column), Procedure (second column).
| Age | Procedure |
|---|---|
| 0-21 | Rats reared in Medical College of Wisconson |
| 21 | Rats arrive in Buffalo, NY (21 days old). |
| 60 | Rats housed until they are young adults (60 days of age) |
| 63 | Locomotor Response to Novelty |
| 86 | Light Reinforcement |
| 129 | Choice Reaction Time Task |
| 154 | Delay Discounting |
| 171 | Pavlovian Conditioned Approach (1+5 days), then Conditioned Reinforcement (1 day) |
| 185 | Cocaine Cue Preference |
| 200 | Rats sacrificed; cecal samples collected |
2.2.1. Locomotor response to novelty (Loco)
To assess locomotor response to novelty, rats were placed in a 24 x 45 cm clean plastic standard laboratory cage. A Hamilton Kinder motor monitor frame contained infrared photo detectors which measured locomotor activity by beam breaks. Rats were placed into the test cages for one hour. Only the first 18 min of the 1 h test session were used for analysis. Each rat was tested only once. Measures used for this test included total locomotor activity (Loco.Activity), epoch with the greatest activity (Loco.MaxAct), total distance travelled (Loco.Distance), epoch with the greatest distance travelled (Loco.MaxDist), time in center (Loco.Center), total number of rears (Loco.Rear), epoch with the greatest number of rears (Loco.MaxRear).
2.2.2. Light reinforcement (LR)
In-house constructed operant chambers were used for testing (see Supp. Methods Fig 2). The visual stimulus (VS) reinforcer used in the experiment was the onset of the light located in the middle of the back wall of the test chamber. Onset of the VS reinforcer produced an illuminance of 68 lx, as measured from the center of the test chamber. The VS reinforcer was illuminated for 5 s each time it was presented. Each test chamber was housed in a Coleman Cooler (Model # 3000000187), which blocked external audiovisual sources of stimulation. The pre-exposure phase consisted of six 18 min sessions. Light reinforcement testing took place immediately after pre-exposure testing with one day off in-between. The light reinforcement phase consisted of six 18 min sessions. During light reinforcement testing, rats were placed in dark experimental chambers and snout pokes into the aperture designated as ‘active’ resulted in 5 s illumination according to a variable interval 1 min (VI1) schedule of reinforcement. Measures from light reinforcement testing included total number of light reinforcers (LR.Reinforcers), total number of InActive responses during test (LR.InActive), total number of Active and InActive responses during test (LR.Total), epoch with greatest total responses (LR.TotMax) and epoch with greatest InActive responses (LR.InActMax).
Fig. 2.
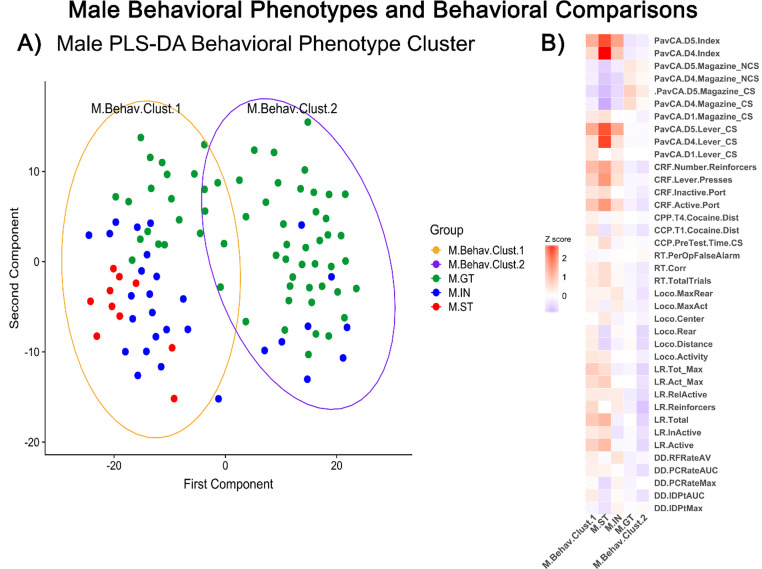
Male behavioral phenotypes and behavioral comparisons – (A) Behavioral Phenotype Cluster visualized in principal coordinate analysis (PCA) of PLS-DA clustered behavioral measures. Each dot represents an individual rat, distance from one dot to another represents overall differences in behavioral measures.Goal-trackers (GT) colored green, intermediate (IN) blue, sign-trackers (ST) red. (B) Z score indicate increases (red) or decreases (blue) in behavioral measures by behavioral cluster group and goal/sign-tracker phenotype group compared to entire male population. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2.3. Choice reaction time task (RT)
Locally constructed experimental chambers were used for the choice reaction time task. The test panel had two water dispensers located on either side of a centrally located snout-poke hole (see Supp. Methods Fig. 3). The water dispenser and stimulus lights were arranged so that they were level with the rat's eyes when the rat's snout interrupted an infrared beam in the center snout-poke hole. Rats were placed into the test cages for 18 minutes for each test session. Rats initiated trials by holding their snout in the center snout hole until the left stimulus light was turned on (hold time). Once the hold time criterion was reached and the imperative stimulus was presented, the rat had 3 s to respond by removing its snout and inserting it into the left feeder hole (reaction time), or the trial ended and the trial was counted as an omission. If the rat made a correct response, the rat received a water reinforcer (30 μl) and the trial ended. A false alarm was recorded when the rat pulled its snout out of the center hole to respond to the left water feeder hole prior to the onset of the imperative stimulus. Premature initiations are defined similar to a false alarm, except that the rat pulls out of the center snout poke hole before the imperative stimulus occurs and then puts its snout back into the center hole without going to the left water feeder hole. The final 3 test sessions were used for analysis. Measures for this test included total number of correct responses (RT.Corr), mean reaction time (RT.MeanRT), per opportunity (trial) premature initiations (RT.PerOPInit), per opportunity false alarms (RT.POFA), and total omissions (RT.Omissions).
Fig. 3.
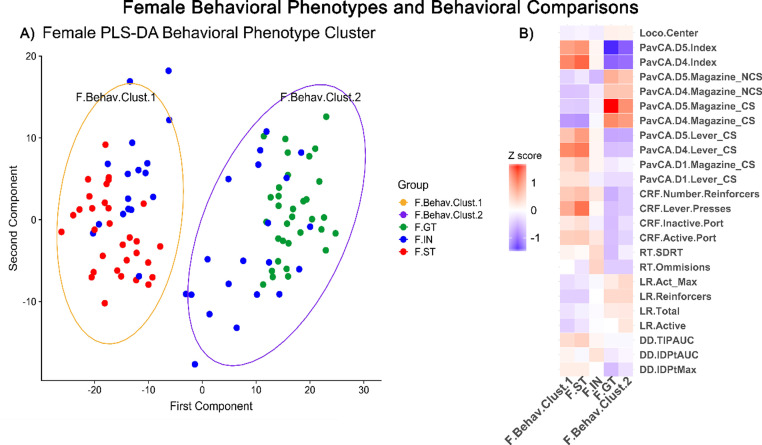
Female behavioral phenotypes and behavioral comparisons – (A) Behavioral Phenotype Cluster visualized in principal coordinate analysis (PCA) of PLS-DA clustered behavioral measures. Each dot represents an individual rat, distance from one dot to another represents overall differences in behavioral measures. Goal-trackers (GT) colored green, intermediate (IN) blue, sign-trackers (ST) red. (B) Z score indicate increases (red) or decreases (blue) in behavioral measures by behavioral cluster group and goal/sign-tracker phenotype group compared to entire female population. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2.4. Delay discounting (DD)
Delay discounting was measured using a sequential patch depletion procedure. This procedure mimics naturally occurring choice problems confronting animals while foraging in resource scarce environments (i.e., travel delays and patch depletion). Water was restricted and only made available for 20 minutes following testing. Behavior was measured in in-house constructed operant chambers (see Supp. Methods Fig. 2). In the laboratory patch depletion procedure rats drank water at both the left and center water feeders. Rats received successively smaller amounts of water every 4 s by remaining at the same feeder. The amount of water was initially 150 µL and was then decreased by 20% after each delivery from the same feeder. The rats could reset the amount of water to the initial maximum of 150 µL by switching to the alternative water feeder. However, changing to a new patch resulted in a delay to activation of the new feeder (travel time delay). During the delay, water was not available at either feeder. A change in patch was indicated by a snout poke into the alternative non-active feeder (patch). The indifference point (IDPt) was defined as the amount of water available at the current feeder (or patch) when the rat chose to switch to the new patch. Test sessions lasted for 10 minutes or until the rats consumed a cumulative total 5 ml of water, which ever occurred first. The area under the curve (AUC) was calculated for successive session measures. Behavioral measures from this test included indifference point area maximum (DD.IDPMax) and under the curve (DD.IDPtAUC), patch change rate area under the curve (DD.PCRateAUC), and average reinforcer rate (DD.RFRate).
2.2.5. Pavlovian conditioned approach (PavCA) and conditioned reinforcement (CRF)
To examine individual differences in the propensity to attribute incentive salience to reward cues, HS rats were first exposed to a Pavlovian conditioning paradigm, wherein a cue (lever) was repeatedly paired with the presentation of a reward (food). Before animals underwent the standard PavCA procedure, they received ~25 banana-flavored food pellets (Envigo, #F0059) in their homecages for 2 days. They then underwent one day of magazine training, during which they were placed into Med-associates conditioning chambers, and food pellets were delivered on a VI 30 s (1–60 s) schedule. For the subsequent 5 PavCA conditioning days, rats received 25 CS to US conditioning trials, presented on a VI 90 s (30–150 s) schedule. During each trial, an 8 s presentation of an illuminated lever CS preceded the delivery of a food pellet. On the day following the final session of PavCA, we performed conditioned reinforcement (CRF) in which rats were able to nose poke for presentations of the lever CS. All variables were derived from lever presses and magazine entries, including latencies and probability. Measures for PavCA included index scores on days 4 and 5 [see [64] for a description of this index]; briefly, scores ranged from -1 to 1, with negative numbers indicating magazine directed responses (goal-tracking), and positive numbers indicating lever-CS directed responses (sign-tracking). CRF measures used were total number of lever presses (CRF.LeverPresses), total number of active nose-poke port entries (CRF.ActivePort) and total number of CS lever presentations (CRF.Reinforcers).
2.2.6. Cocaine cue preference (CCP)
To examine the individual differences in approach to a cocaine-associated cue, HS rats were tested for their response to a cocaine-paired tactile cue [see [65] for details]. Briefly, testing chambers were constructed with black acrylic walls (47.5 cm length x 15.5 cm width x 30 cm height) with black spray-painted textured floors that were either stainless steel rods (“grid”) or perforated steel (“hole”), or half grid and half hole. After one day of habituation to the chamber and one "pre-test" day to determine grid-hole preferences, rats were given four conditioning trials. Each trial was two days, one saline-paired day and one cocaine-paired day. On saline-paired days, rats were given an injection of saline (i.p.) and placed into a chamber with either a uniform grid or hole floor (whichever was their preferred floor). On the cocaine-paired day, rats are given an injection of cocaine (10 mg/mL; Nat. Inst. Of Drug Abuse, Bethesda, MD) and placed in a chamber with the opposite uniform floor (i.e., non-preferred floor). On the last "post-test" day, rats were given a saline injection, and presented with both cocaine- and saline-paired floors. The time spent on each floor was analyzed to determine cocaine cue preference. The primary dependent measures are change in time spent on the cocaine-paired floor from pre-test to post-test (CCP.PreTest.Time.CS - CCP.PostTest.Time.CS = CCP.dtCS). Secondary measures included cocaine change in locomotor activity on trails 1 and 4 (CCP.T1.Cocaine.Dist CCP.T4.Cocaine.Dist).
2.3. Cecal microbiome collection and sequencing
Cecum was collected and snap-frozen on dry ice. Protocols for 16S rRNA microbiome sequencing were used as previously described [71]. Briefly, cecal contents from frozen cecum (stored at -80ºC) were extracted under a sterile hood. The QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) was used to extract bacterial DNA from cecal contents using the manufacturer's handbook (Second Edition 2012). Samples were prepared for 16S sequencing using the Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA, USA), as described in the Illumina 16S library preparation workflow. 16S bacterial rRNA gene was amplified using primers targeting the V3-V4 hypervariable region (Forward: 5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; Reverse: 5′GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC) (Sigma Aldrich Ireland ltd., Wicklow, Ireland). The Illumina V3–V4 primers were selected for their high coverage (94.5% bacteria) while remaining in the amplicon size necessary for sequencing to sequence at 2 × 250 bp [51]. 16S rRNA amplicons were sequenced on the Illumina MiSeq platform, multiplexed on 4 separate runs (~50 samples per run) (Teagasc, Moorepark, Ireland). No negative control was used in processing of DNA or sequencing.
2.4. Microbiome sequence processing
All sequences in FASTQ files format were filtered using PRINSEQ. Sequences with length less than 150 nucleotides or with low quality at the 3’ end were removed. Paired-end reads with a minimum overlap of 20 base-pairs were joined using FASTQ-join and analyzed with QIIME (Quantitative Insights Into Microbial Ecology, v1.9.1). Sequence quality was checked and chimeras removed, remaining sequences were clustered into Operational Taxonomic Units (OTUs ; 97% identity level) using USEARCH (Version 7.0-64bit). The average number of high-quality sequences generated per sample was 153,561 ± 84,269 SD. Taxonomy was assigned to OTUs using Silva version 123. Alpha diversity (Observed, Chao1, Shannon, Simpson) indices were calculated with QIIME. No negative control was used in processing of DNA or sequencing.
2.5. Statistical analysis
Data was analyzed in R (v3.3.3) and RStudio (v1.0.136). Plots were generated in R using ggplot2 package (v2.2.1). All testing was corrected for multiple comparisons using the qvalue R package (v2.18.0). For OTU correlations to behavior, q-value confidence interval was set to 0.15. For within-sex correlations, the q-value of 0.15 was accepted due to the exploratory nature of this study. In this dataset, q=0.15 indicates that of the 30 reported significant correlations within sex, only 4-5 of them may be false positives. For all other analyses, q-value was set to 0.05.
2.5.1. Behavioral analysis
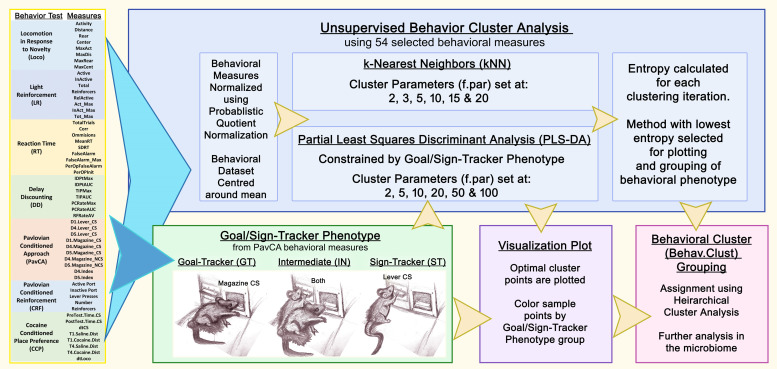
A total of 54 behavioral measures were selected for analysis based on relevance to addiction. Behavioral differences were assessed by behavioral cluster and goal/sign-tracking phenotype within sex using Kruskal-Wallis and Wilcoxon test. Behavioral measures significantly different by group were plotted by z-score to visualize trends between grouping phenotypes. Factor analysis was performed to test influence of weight and age at time of dissection, generation, and goal/sign-tracker phenotype using the ADONIS (PERMANOVA) function of the vegan (2.4-3) R package.
2.5.1.1. Sign-tracker/goal-tracker classification
Rats were classified as sign/goal-trackers based on PavCA Index Score (= [PavCA Score (Day 4) +PavCA Score (Day 5)]/2; see [64] for details [64]. Subjects were classified as sign-tracker (ST) (PavCA Index Score between +0.5 and +1), goal-tracker (GT) (PavCA Index Score between -0.5 and -1), and intermediate (IN) (PavCA Index Score between -0.49 and 0.49), based on the classification method previously described [64].
2.5.1.2. Behavioral cluster analysis
All behavioral measures (total=54) were used for cluster analysis using the KODAMA package (v1.4). Behavioral data within sex was normalized using probabilistic quotient normalization and centered to zero. Data was clustered using PLS-DA (partial least squares differential analysis) grouped by goal/sign-tracking phenotype and using multiple levels of cross-validation. Entropy was tested on point values for each cross-validation level to find optimal cluster. For both the female and male behavioral data sets, the lowest entropy cluster was found with PLS-DA set to parameter (f.par) 100. Hierarchical cluster analysis was performed to select optimal number of clusters and to assign samples to cluster. Results were then plotted using PCA and colored based goal/sign-tracking phenotype.
2.5.2. Microbiome analysis
Microbiome was assessed by behavioral cluster and goal/sign-tracking phenotype within sex. Kruskal-Wallis and Wilcoxon test were used to assess statistical significance in alpha diversity indices and taxonomic comparisons between groups. Beta diversity was calculated using Euclidian distance visualized and analyzed using the vegan community ecology package (v2.4-3). Adonis (PERMANOVA) function from vegan assessed beta diversity significance by generation, behavioral phenotype, sign/goal-tracker phenotype (GT, ST, IN), age and weight at time of dissection. For Spearman correlations and taxonomic comparisons, Phylum, Family, Genus, and OTUs (Operational Taxonomic Units) were filtered by median >0.01%. For Spearman correlations this filtering resulted in sequences only present in >50% of samples and normalized to relative abundance. Correlations were performed between OTU-level bacterial abundance and behavioral measures, subset by grouping phenotype within sex. For correlation analysis, all behavioral measures were classified into 3 major categories: reward-learning, impulsivity, and locomotion (Suppl. Table 1A). For Spearman correlations within sex, the qvalue R package (v2.60) was used to select false discovery rate. Due to the exploratory nature of this study, the q-value of 0.15 was accepted as it allowed reporting of interesting trends while still maintaining a low rate of false positives (4–5 total).
3. Results
3.1. Behavioral cluster analysis
Preliminary analysis showed that all non-PavCA behavioral measures clustered by the goal/sign-tracking phenotype (GT/ST) (Fig. S1). Thus, we sought to evaluate the goal/sign-tracking phenotype, within sex, alongside a novel group cluster constrained by this pre-established grouping. Partial least squared differential analysis (PLS-DA) of all 54 behavioral measures (Fig. 1) within sex show a separation along the x-axis (Figs. 2A and 3A). In both males and females, the sign-tracking phenotype clusters in the negative (left) side of the x-axis (Figs. 2A and 3A). Behavioral clusters that contained the sign-tracking phenotype were labeled F.Behav.Clust.1 for female sup-population and M.Behav.Clust.1 for males. Samples clustering to the right of the x-intercept were labeled F.Behav.Clust.2 and M.Behav.Clust.2, for females and males, respectively.
Fig. 1.
Flow Diagram of how behavioral measures are used to create goal/sign-tracking phenotype and behavioral cluster phenotype.
3.2. Behavioral differences by behavioral cluster within sex
Factor analysis of 54 behavioral measures revealed that weight at the end of the experiment was the only significant contributing factor in both females (R2 = 0.036, p = 0.028) and males (R2=0.037, p = 0.018). Goal/sign-tracker phenotype, generation, and age at time of dissection were not significant factors (p > 0.05) contributing to variations in all behavioral measures.
All 54 behavior measures were tested within sex by goal/sign-tracker phenotype (GT, IN, ST) and behavioral cluster (Behav.Clust.1, Behav.Clust.2). Females tested by goal/sign-tracking phenotype, 15 behavioral measures were significantly different (Kruskal-Wallis, p < 0.05), with 13 of these measures due to differences in GTs compared to STs (Wilcoxon, p < 0.01). In males tested by goal/sign-tracking phenotype, 11 behavioral measures were significantly different (Kruskal-Wallis, p < 0.001), with 8 of these measures due to differences in GTs compared to STs (Wilcoxon, p < 0.001). Behavioral measure comparisons by behavioral cluster revealed 18 significant measures in females and 28 significant measures in males (Wilcoxon, p < 0.05). Z-score was used to visualize behavioral comparisons by groups, behavioral cluster and GT/ST phenotype, within sex (Figs. 2B and 3B). See supplementary material for behavioral comparisons between sex (Supp. Section 1.2 and Fig. 1B).
3.2.1. Group differences in Pavlovian conditioned reinforcement (CRF)
Within both sexes and grouping phenotypes, significant differences were seen in measures for nose pokes in reinforcing port (CRF.Active.Port) and number of lever presses (CRF.Lever.Presses). In both male and female, significant differences in CRF.Active.Port were seen in the goal-tracking (GT) phenotype compared to sign-tracking (ST) (Wilcoxon, p < 0.001) and behavioral cluster grouping (Behav.Clust.1 vs. Behav.Clust.2) (Wilcoxon, p < 0.001). Differences in CRF.Lever.Presses were explained by GT compared to ST group (Wilcoxon, p < 0.001) and behavior cluster (Wilcoxon, p < 0.001), in both sexes. Additionally, the number of nose pokes into active port for presentation of lever reinforcer (CRF.Number.Reinforcers) was significantly different in both males and females by GT compared to ST phenotype (Wilcoxon, p < 0.001).
3.2.2. Group differences in cocaine cue preference (CCP)
Only the male behavioral cluster showed significant differences in cocaine-induced locomotor activity in Trial 1 measured by distance travelled (CCP.T1.Cocaine.Dist) (Wilcoxon, p < 0.001). This difference was explained by the increases in M.Behav.Clust.1 compared to M.Behav.Clust.2 (Fig. 2B).
3.2.3. Group differences in choice reaction time task (RT)
Reaction time (RT) measures of standard deviation of reaction time per epoch (RT.SDRT) and number of failed responses defined as omissions (RT.Omissions) were significantly different by female goal/sign-tracking phenotype. For both measures, this result was explained by increases in IN group compared to GT (Wilcoxon, p < 0.01) (Fig. 3B).
3.2.4. Group differences in locomotor response to novelty (Loco)
Similar to measures from CCP, M.Behav.Clust.1 had significant increases in distance travelled (Loco.Distance) compared to M.Behav.Clust.2 (Wilcoxon, p<0.01). This was also seen in comparisons between male behavioral clusters in measures of exploratory rearing and maximum rears (Loco.Rear and Loco.MaxRear) (Wilcoxon, p<0.05), locomotor activity and maximum activity (Loco.Activity and Loco.MaxAct, p<0.05).
3.2.5. Group differences in light reinforcement (LR)
Measures from the LR task were significantly different by behavioral cluster in both males and females. Total number of light onset reinforcers (LR.Reinforcers), total active and inactive responses (LR.Total), total active responses (LR.Active), and maximum active responses in an epoch (LR.Act_ Max) were significantly different (Wilcoxon, p<0.01). These measures were decreased in F.Behav.Clust.1 compared to F.Behav.Clust.2, while they were increased in M.Behav.Clust.1 compared to M.Behav.Clust.2 (Fig. 2B & 3B).
3.2.6. Group differences in delay discounting (DD)
In female behavioral cluster, F.Behav.Clust.1 had significant increases in change time in patch with experimenter imposed delay across epochs (DD.TIPAUC) compared to F.Behav.Clust.2 (Wilcoxon, p < 0.05) (Fig. 3B). In the male behavioral cluster, M.Behav.Clust.1 was significantly increased in measures of patch change rate area under the curve (DD.PCRateAUC) and indifference point area under the curve (DD.IDPtAUC) compared to M.Behav.Clust.2 (Wilcoxon, p < 0.01) (Fig. 2B).
3.3. Microbiome diversity
In males, microbiome beta diversity was significant by generation (R2=0.039, p < 0.001) and age at the end of the experiment (R2=0.021, p < 0.001). Females were only significant by generation (R2=0.019, p<0.001). No significant differences were seen for weight, sign/goal-tracker phenotype, or behavioral cluster in either males or females.
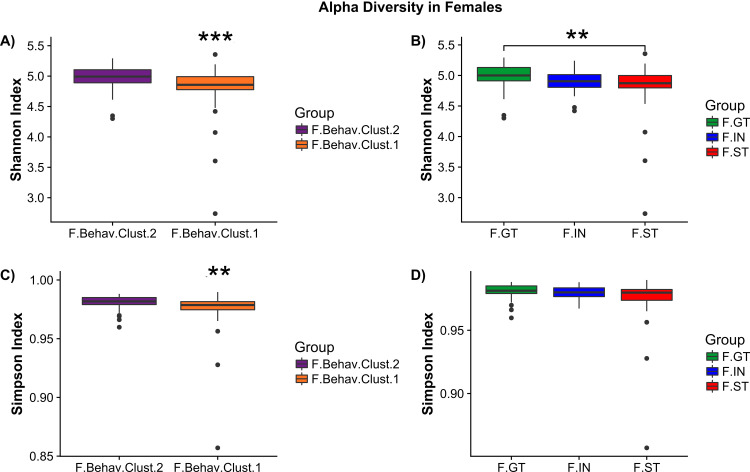
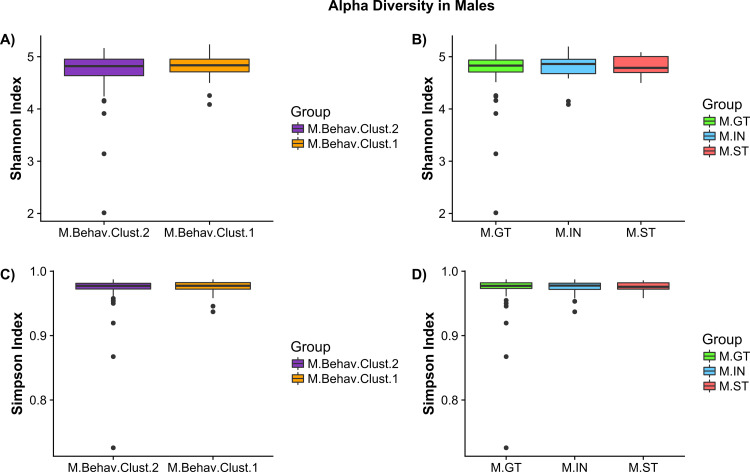
Alpha diversity analysis by female behavioral cluster revealed significant decreases in Shannon (Wilcoxson, p < 0.001) and Simpson (Wilcoxson, p<0.001) index measures in F.Behav.Clust.1 compared to F.Behav.Clust.2 (Fig. 5A and 5C). Females ST group also had significant reductions in Shannon measure of alpha diversity compared to GT (Fig. 5B) There were no significant differences in alpha diversity measures in males (Fig. 4).
Fig. 5.
Female alpha diversity by phenotype group – (A) Shannon index measure of alpha diversity by behavioral cluster (F.Behav.Clust.1 = orange, F.Behav.Clust.2 = purple). (B) Shannon index by goal/sign-tracking phenotype: goal-tracker (F.GT = green), intermediate (F.IN = blue), and sign-tracker (F.ST = red). (C) Simpson index measures of female alpha diversity by behavioral cluster. (D) Simpson index of female goal/sign-tracker phenotype. Asterisks indicate significance: ‘***’ p<0.001, ‘**’ p<0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Male alpha diversity by phenotype group – (A) Shannon index measure of alpha diversity by behavioral cluster (M.Behav.Clust.1 = orange, M.Behav.Clust.2 = purple). (B) Shannon index by goal/sign-tracking phenotype: goal-tracker (M.GT = green), intermediate (M.IN = blue), and sign-tracker (M.ST = red). (C) Simpson index measures of male alpha diversity by behavioral cluster. (D) Simpson index of male goal/sign-tracker phenotype. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Bacterial differences by groups within sex
No significant differences were seen in either phenotype group by sex at the Phylum, Family, Genus, or OTU level after FDR correction. In the female behavioral cluster, genus-level Blautia (Wilcoxson, q=0.062) and OTU-level Papillibacter OTU_128 and Blautia OTU_160 approached significance (Wilcoxson, q = 0.055). All other results had an FDR q-value greater than 0.15.
3.5. OTU level bacterial correlations to behavior
3.5.1. Correlations by Male and Female Goal/Sign-Tracker Phenotype
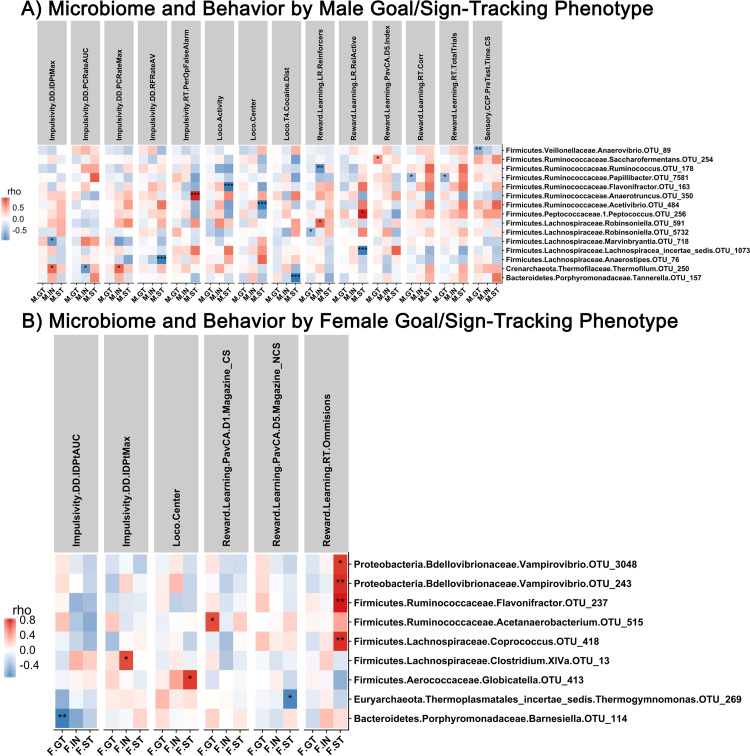
In males, behaviors correlated to refined OTUs by GT/ST phenotype. The strongest correlation was between cocaine induced locomotor activity during trial 4 and Tanneralla OTU_157 (Spearman, rho=-0.976, p < 0.001). Further correlations were seen in OTU-level bacteria belonging to family Lachnispiroceae and Ruminococcaceae in measures of reward learning (LR, PavCA, RT), in reaction time and light reinforcement measures, locomotion and impulsivity measures from delay discounting (Spearman, |rho|>0.722, p < 0.001). An OTU-level bacteria from genus Thermofilum significantly correlated to impulsivity measures only in the male intermediate (IN) phenotype (Spearman, |rho|>0.709, p < 0.001). Overall, the strongest correlations were seen in the male STs (Spearman, |rho|>0.939, p < 0.001) (Fig. 6A).
Fig. 6.
Correlation analysis in male and female goal/sign-tracking phenotype. (A) Correlations between OTU-level bacteria and behavior in male goal/sign-tracking phenotype. (B) Correlations between OTU-level bacteria and behavior in female goal/sign-tracking phenotype. Positive correlations indicated in red, negative correlations indicated in blue. Significance that passes FDR indicated by asterisk: ‘***’ q<0.05, ‘**’ q<0.10, ‘*’ q<0.15. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In females, less significant correlations between microbiome and behavior by GT/ST phenotype were seen. A total of 9 significant correlations were observed, of these 6 were significant correlations within ST phenotype. A strong trend was seen in ST females with correlations between reaction time omissions and specific OTUs assigned to genera Vampirovibrio, Coprococcus, and Flavinofractor (Spearman, rho>0.729, p < 0.001). Significant correlations were also found between impulsivity measures and OTU bacteria assigned to genera Clostridium XIVa and Barnesiella (Spearman, rho>0.682, p<0.001) (Fig. 6B).
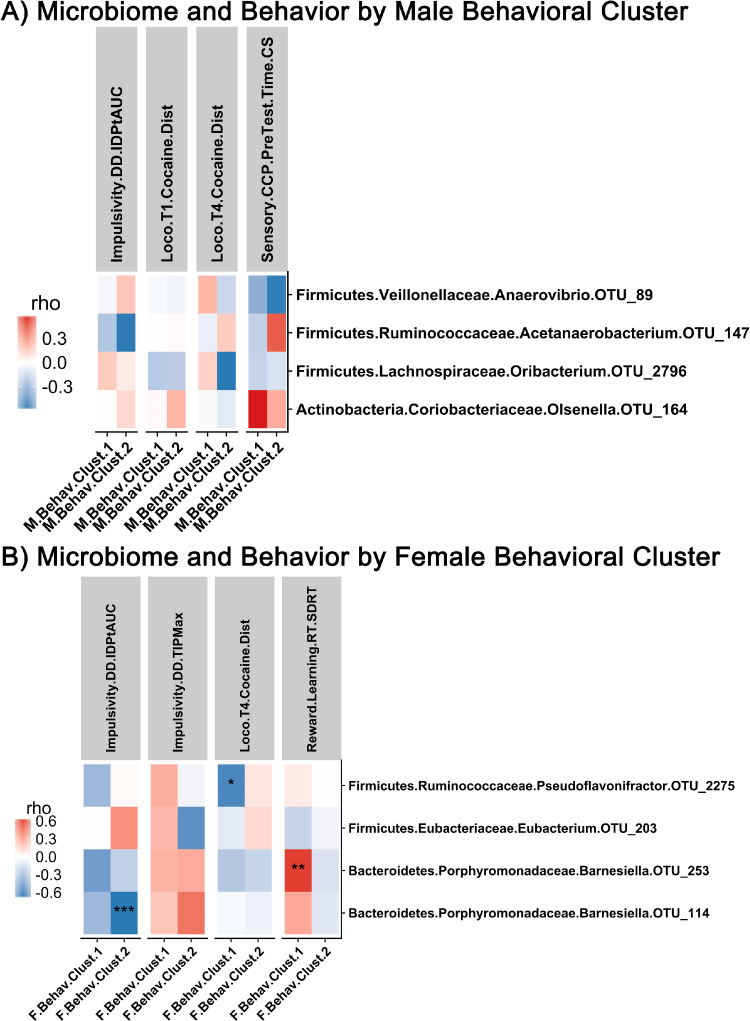
3.5.2. Correlations by male and female behavioral cluster
No significant correlations were seen between microbiome and behavior when analyzed by male behavioral cluster (Fig. 7A). In females, impulsivity measures and attention measures were significantly correlated with OTUs assigned to genus Barnesiella (Spearman, |rho|>0.620, p<0.001) (Fig. 7B).
Fig. 7.
Correlation analysis in male and female behavioral clusters. (A) Correlation between OTU-level bacteria and behavior in male behavioral cluster phenotype. (B) Correlation between OTU-level bacteria and behavior in female behavioral cluster phenotype. Positive correlations indicated in red, negative correlations indicated in blue. Significance that passes FDR indicated by asterisk: ‘***’ q<0.05, ‘**’ q<0.10, ‘*’ q<0.15. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5.3. Correlations by male and female
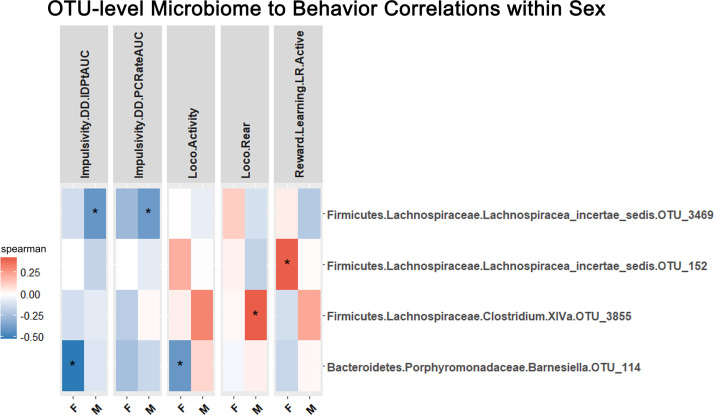
Spearman correlations were performed within the entire male and female sub-populations between behaviors and OTU-level bacteria (N=101, Males and Females) (Fig. 8). Impulsivity measures of indifference point area under the curve and patch change rate area under the curve was significantly correlated with Lachnospiracaea_incertae_sedis OTU_3469 (Impulsivity.DD.IDPtAUC and Impulsivity.DD.PCRateAUC, rho<-0.393, p < 0.001) in males. In females, indifference point area under the curve and locomotor activity were negatively correlated to Barnesiella OTU_114 (Impulsivity.DD.IDPtAUC and Loco.Activity, rho<-0.407, p < 0.001). In males, exploratory rearing was positively correlated to Clostridium XlVa OTU_3855; additionally, males had many OTU correlations to sensory preference for CPP floor before testing (see Supp. Materials). A positive correlation between reward learning and Lachnospiracea incertae sedis OTU_152 was observed in females (Reward.Learning.LR.Active, rho=0.407, p < 0.001) (Fig. 8).
Fig. 8.
Correlation analysis by sex. Correlations between OTU-level bacteria and behavior in entire female and male populations. Positive correlations indicated in red, negative correlations indicated in blue. Significance that passes FDR indicated by asterisk: ‘***’ q<0.05, ‘**’ q<0.10, ‘*’ q<0.15. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Growing evidence suggests that understanding the complex relationship between addiction-related behaviors and microbiome is important in illuminating factors associated with addiction vulnerability. Overall microbiome composition, as measured by beta-diversity, was not associated to the goal/sign-tracking (GT/ST) phenotype, and also not in Random Forest analysis (Supp. Material, Section 1.1.1.2). However, correlations revealed novel sex-dependent links between addiction-associated behavior and microbiome. In females, operational taxonomic units (OTUs) from bacteria Barnesiella repeatedly correlated with behavioral measures of impulsivity, measured by indifference point area under the curve in delay discounting task (DD.IDPtAUC) (Figs. 7 and 8). Moreover, alpha diversity was reduced in female behavioral phenotypes associated with increased addiction vulnerability (Fig. 5). Female PLS-DA cluster phenotype (F.Behav.Clust.1 and F.Behav.Clust.2, Fig. 3A) revealed that behavioral differences followed trends similar to GT/ST phenotype. Most behavioral measures in F.Behav.Clust.1 aligned with sign-trackers (STs) and F.Behav.Clust 2 similar to goal-tracking (GTs) phenotype (Fig. 3B). In males, fewer trends were seen in both microbiome and behavior. All groups had significant correlations between OTU-level bacteria, predominantly belonging to families Ruminococcaceae and Lachnospiraceae, and behavioral measures of impulsivity, attention, reward-learning, and locomotor response to novelty.
Sign-tracking (ST) is defined as the propensity to imbue a conditioned stimulus with incentive salience [30,64]. STs are more responsive to both food and drug CSs than GTs and tend to engage in addiction-related behaviors. Increased ST behavior is positively correlated to increased dopamine levels in the nucleus accumbens during amphetamine self-administration [92]. Additionally, in selectively bred high/low response (HR/LR) rats, both male and female HR rats sign-track while LR counterparts goal-track. HR females have a greater propensity to self-administer cocaine than LR females and HR & LR male rats [25]. In line with previous work [49,73], this study had more females assigned to the addictive-associated ST phenotype, compared to males.
There is contradictory data in humans with regard to gender differences in the incidence of addiction. However, research of both humans and other animal species indicates that females may, in some cases, display a greater propensity for addiction. A human genome-wide-association study (GWAS) showed females having greater impulsive behavior, as measured by delay discounting [79]. Furthermore, GWAS studies suggest that opioid dependence is linked to sex specific single nucleotide polymorphisms [99]. In Cloninger's typology, Type II alcoholism has an early onset, a genetic propensity, and more common in males, while Type I has a late onset and is seen in both males and females [20]. Differences in hormones underlie some sex differences in addiction-related behavior. Furthermore, intensity of drug usage and drug withdrawal vary depending on menstrual cycle. In the striatum, dopamine levels are higher during estrus in rats, when estradiol is elevated. Estradiol is known to increase positive affect, locomotor sensitization, and acquisition of self-administration to psychomotor stimulants in female rats [10].
Measures of impulsivity, attention, reward-learning, and locomotor response to novelty were significantly different by behavioral phenotypes and correlated to the microbiome. In delay discounting, the indifference point/area under the curve measure (DD.IDPtAUC) has previously been linked to addiction in humans [76]. This impulsivity measure correlated to OTUs assigned to the family Lachnospiraceae in males and OTUs assigned to the genus Barnesiella in females. The divergence in gut bacteria correlated to impulsivity, indicates that sex-specific commensal bacteria may have similar effects on behavior. In males and females, measures of Pavlovian Conditioned Approach (PavCA) were significantly correlated with bacterial OTUs in family Ruminococcaceae. In males, reaction time (RT) measures correlated to bacterial OTUs in family Ruminococcaceae. Intriguingly, female STs revealed strong correlations between reaction time attention measure (RT.Omissions) and OTUs in family Ruminococcaceae, including genus Flavonifrator, and family Lachnospiraceae, as well as two OTUs in genus Vampirovibrio, which preys on algae, and is capable of competitively replacing taxa in the microbiome [4,89]. Significant correlations were seen in light reinforcement (LR) and locomotor response (Loco) measures and bacterial OTUs in the family Ruminococcaceae, including the genus Flavonifrator, and Lachnospiraceae in the male GT/ST phenotype. In the present study, taxonomic differences between bacterial composition in GT/IN/ST phenotypes did not reach significance. However, the observed correlations repeatedly associated to Ruminococcaceae align with previous addiction research showing reductions in Ruminococcaceae in opioid users [1], alcohol consumption [16,57], and alcohol addiction severity [56]. Research investigating alcohol consumption and microbiome has also previously reported changes in Lachnospiraceae in mice receiving oral and vapor alcohol [16,71] and alcohol addiction severity in humans [56]. Our recent work on the microbiome and addiction phenotype has illuminated intriguing trends in bacteria from the families Ruminococcaceae and Lachnospiraceae correlating to dopamine receptor expression in the striatum and measures of anxiety, novelty induced locomotor activity, impulsivity, and compulsive alcohol seeking in male rats [43].
Sex and/or gender are significant factors to consider when investigating the microbiome-gut-brain axis [44,45]. Previous research reveals sex-associated microbiota differences are strongly linked to metabolic processes [2,24,60,101] and influence brain and behavior [19,40]. Segregation of male and female rats by same sex co-housing may confound and/or inflate sex differences observed, especially in light of coprophagic behavior in rats - an environmental factor that may influence microbiota more than host genetics [77]. However, in a study of howler monkeys the microbiome continued to be distinct between sex despite shared environment and parent [2]. In this study, co-housed rats showed clear sex-differences in percent occurrence of shared GT/IN/ST phenotypes (55% in males vs. 31% in females, co-housed) compared to the ST phenotype only (12% in males vs. 29% in females, co-housed) (Supp. Table 2). Clearly, further investigation is necessary to elucidate the connections between sex and microbiome composition as well as factors such as genotype [70] and cagemates [9]. These studies should also include fecal transplantation across different phenotypes.
Females associated with an ST phenotype had decreased alpha diversity compared to GT (Fig. 5). No difference was seen in alpha diversity between GT/IN/ST phenotypes in males (Fig. 4). Reduced alpha diversity is commonly associated with poor health, often the result of common factors, with reductions reported in mental health conditions including stress and depression [5,48]. Reduced alpha diversity has also been shown in chronic-intermittent vapor ethanol exposure [71] and antibiotic depletion is linked to increased locomotor response to cocaine [50]. Alterations in Ruminococcaceae and Lachnospiraceae have been characterized in many health conditions. Importantly, reduced intestinal permeability, liver damage, and inflammation [1,5,6,16,54,55,57,72] have been linked to alterations in these two family-level bacteria in conditions of psychological stress [7,26], depression [18,46,67], autism [35,42], and dopaminergic-mediated disorders [56,72]. Many novel bacteria were identified in this study in relation to addictive phenotype and microbiome. OTUs associated with genus Flavonifractor correlated with locomotor activity in males and RT omissions in females. Increased abundance of Flavonifractor is linked to major depressive disorder [46] and bipolar disease [21]; the mechanism of how Flavonifractor effects the brain is not well known, though it is believed to cause oxidative stress and inflammation. Furthermore, OTUs associated to Barnesiella were repeatedly correlated to behaviors in females, particularly impulsivity measures. In a human study comparing microbiota composition of alcohol drinkers and non-drinkers, a top differentiated OTU increased with alcohol consumption belonged to an uncharacterized Barnesiella [52]. Genus Barnesiella has also previously been characterized as preventing colonization of infection bacteria [90, 93], thus reducing inflammatory profile in the gastrointestinal tract. In this study, we see two novel taxa which have been characterized as replacing taxa in the microbiome, Barnesiella and Vampirovibrio. These novel taxa were also related to measures of attention and impulsivity. Previous research in humans reported a negative relationship between Clostridium XIVa and depression in females [18].
A limitation of this study is the lack of negative controls for DNA extraction, library preparation, and sequencing. Due to lack of negative controls, low-abundance bacteria significantly correlating with behavioral measures should be interpreted with caution. An example of this is OTU_250 assigned to genus Thermofilum (Silva reference database); this low-abundance bacteria showed significant correlations with impulsivity behavior in the male IN phenotype. However, further investigation into this unusual taxon revealed significant differences in batches from DNA processing and sequencing (Supp. Figs. 8–10). Furthermore, taxonomic assignment of the OTU_250 sequence to alternative reference databases were inconclusive (Supp. Material, Section 1.1.1.15). Further investigation is necessary to determine if these identified bacteria preferentially colonize certain sexes, the mechanism for the preference, in addition to how these certain bacteria are influencing brain and behavior. Furthermore, the impact of immune function must be investigated in these behavioral phenotypes associated with specific bacteria [74]. Research has shown that gut bacteria impact neuroimmunology [14,85], and neuroimmunology has been linked to addiction [41].
The current studies are also limited by the fact that single timepoints were taken and that 16S-based sequencing only allows for genus-level specificity to be unmasked. Further studies should also focus on functional metabolomic analysis. Future studies should also examine how the neurobiological substrates of the behavioral changes observed are regulated by the microbiome. Indeed, the gut microbiome has been shown to regulate cortical morphology and neurotransmitter expression, notably in the prefrontal cortex, which is involved in impulsivity behaviors [39,40,59]. Moreover, investigations into the role of the vagus nerve as a conduit of signals from the gut to the brain are also warranted [15,32].
5. Conclusion
This is the first time, to our knowledge, that extensive characterization of within sex addiction-phenotype and behavioral measures have been associated to the microbiome. Overall microbiome composition was not capable of predicting addiction phenotype. The most robust findings in this study indicate that microbiome is associated with locomotor response, reward-stimulus learning, impulsivity and attention. Notably, impulsivity measures repeatedly correlated to certain bacteria in males and females. This novel work implies that sex as a factor must be considered in both behavior and microbiome research. Further investigation is necessary to elucidate factors that contribute to sex differences in the microbiome, and how these differences influence other addiction-related measures like drug-self administration and relapse.
Declaration of Competing Interest
JFC & TGD are in receipt of research funding from 4D‐Pharma, Mead Johnson, Nutricia, and Cremo. Timothy Dinan has been an invited speaker at meetings organized by Servier, Lundbeck, Janssen, and AstraZeneca. John Cryan has been an invited speaker at meetings organized by Mead Johnsen, Alkermes, and Janssen.
Acknowledgments
Funding sources: National Institutes for Drug Abuse P50DA037844, Science Foundation Ireland 12/RC/2273. AB was supported by a fellowship from the Wellcome Trust (105941/Z/14/Z). None of the funders played a role in study design, writing, or interpretation.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102769.
Appendix. Supplementary materials
References
- 1.Acharya C., Betrapally N.S., Gillevet P.M., Sterling R.K., Akbarali H., White M.B., Ganapathy D., Fagan A., Sikaroodi M., Bajaj J.S. Chronic opioid use is associated with altered gut microbiota and predicts readmissions in patients with cirrhosis. Aliment Pharmacol Ther. 2017;45:319–331. doi: 10.1111/apt.13858. [DOI] [PubMed] [Google Scholar]
- 2.Amato K.R., Leigh S.R., Kent A., Mackie R.I., Yeoman C.J., Stumpf R.M., Wilson B.A., Nelson K.E., White B.A., Garber P.A. The role of gut microbes in satisfying the nutritional demands of adult and juvenile wild, black howler monkeys (Alouatta Pigra) Am J Phys Anthropol. 2014;155:652–664. doi: 10.1002/ajpa.22621. [DOI] [PubMed] [Google Scholar]
- 3.Audet M.C. Stress-induced disturbances along the gut microbiota-immune-brain axis and implications for mental health: does sex matter? Front Neuroendocrinol. 2019;54 doi: 10.1016/j.yfrne.2019.100772. [DOI] [PubMed] [Google Scholar]
- 4.Baer, M.L., Williams, H.N., 2015. Vampirovibrio, Bergey's Manual of Systematics of Archaea and Bacteria, pp. 1-2.
- 5.Bailey M.T., Dowd S.E., Galley J.D., Hufnagle A.R., Allen R.G., Lyte M. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajaj J.S., Hylemon P.B., Ridlon J.M., Heuman D.M., Daita K., White M.B., Monteith P., Noble N.A., Sikaroodi M., Gillevet P.M. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675–G685. doi: 10.1152/ajpgi.00152.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bangsgaard Bendtsen K.M., Krych L., Sorensen D.B., Pang W., Nielsen D.S., Josefsen K., Hansen L.H., Sorensen S.J., Hansen A.K. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One. 2012;7:e46231. doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barton W., Penney N.C., Cronin O., Garcia-Perez I., Molloy M.G., Holmes E., Shanahan F., Cotter P.D., O'Sullivan O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut. 2018;67:625–633. doi: 10.1136/gutjnl-2016-313627. [DOI] [PubMed] [Google Scholar]
- 9.Baud A., Mulligan M.K., Casale F.P., Ingels J.F., Bohl C.J., Callebert J., Launay J.M., Krohn J., Legarra A., Williams R.W., Stegle O. Genetic variation in the social environment contributes to health and disease. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker H.C., Lopez M.F., Doremus-Fitzwater T.L. Effects of stress on alcohol drinking: a review of animal studies. Psychopharmacol (Berl) 2011;218:131–156. doi: 10.1007/s00213-011-2443-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker J.B., Hu M. Sex differences in drug abuse. Front Neuroendocrinol. 2008;29:36–47. doi: 10.1016/j.yfrne.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bercik P., Verdu E.F., Foster J.A., Macri J., Potter M., Huang X., Malinowski P., Jackson W., Blennerhassett P., Neufeld K.A., Lu J., Khan W.I., Corthesy-Theulaz I., Cherbut C., Bergonzelli G.E., Collins S.M. Chronic gastrointestinal inflammation induces anxiety-like behavior and alters central nervous system biochemistry in mice. Gastroenterology. 2010;139:2102–2112. doi: 10.1053/j.gastro.2010.06.063. e2101. [DOI] [PubMed] [Google Scholar]
- 13.Bolnick D.I., Snowberg L.K., Hirsch P.E., Lauber C.L., Org E., Parks B., Lusis A.J., Knight R., Caporaso J.G., Svanback R. Individual diet has sex-dependent effects on vertebrate gut microbiota. Nat Commun. 2014;5:4500. doi: 10.1038/ncomms5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braniste V., Al-Asmakh M., Kowal C., Anuar F., Abbaspour A., Toth M., Korecka A., Bakocevic N., Ng L.G., Kundu P., Gulyas B., Halldin C., Hultenby K., Nilsson H., Hebert H., Volpe B.T., Diamond B., Pettersson S. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3009759. 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bravo J.A., Forsythe P., Chew M.V., Escaravage E., Savignac H.M., Dinan T.G., Bienenstock J., Cryan J.F. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bull-Otterson L., Feng W., Kirpich I., Wang Y., Qin X., Liu Y., Gobejishvili L., Joshi-Barve S., Ayvaz T., Petrosino J., Kong M., Barker D., McClain C., Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. doi: 10.1371/journal.pone.0053028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chaban B., Links M.G., Jayaprakash T.P., Wagner E.C., Bourque D.K., Lohn Z., Albert A.Y., van Schalkwyk J., Reid G., Hemmingsen S.M. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:23. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J.J., Zheng P., Liu Y.Y., Zhong X.G., Wang H.Y., Guo Y.J., Xie P. Sex differences in gut microbiota in patients with major depressive disorder. Neuropsychiatr Dis Treat. 2018;14:647–655. doi: 10.2147/NDT.S159322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke G., Grenham S., Scully P., Fitzgerald P., Moloney R.D., Shanahan F., Dinan T.G., Cryan J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry. 2013;18:666–673. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 20.Cloninger C.R., Sigvardsson S., Bohman M. Type I and type II alcoholism: an update. Alcohol Res Health. 1996;20:18. [PMC free article] [PubMed] [Google Scholar]
- 21.Coello K., Hansen T.H., Sorensen N., Munkholm K., Vedel Kessing L., Pedersen O., Vinberg M. Gut microbiota composition in patients with newly diagnosed bipolar disorder and their unaffected first-degree relatives. Brain Behav Immun. 2019;75:112–118. doi: 10.1016/j.bbi.2018.09.026. [DOI] [PubMed] [Google Scholar]
- 22.Collins S.M., Kassam Z., Bercik P. The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol. 2013;16:240–245. doi: 10.1016/j.mib.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V., Guzzetta K.E., Jaggar M., Long-Smith C.M., Lyte J.M., Martin J.A., Molinero-Perez A., Moloney G., Morelli E., Morillas E., O'Connor R., Cruz-Pereira J.S., Peterson V.L., Rea K., Ritz N.L., Sherwin E., Spichak S., Teichman E.M., van de Wouw M., Ventura-Silva A.P., Wallace-Fitzsimons S.E., Hyland N., Clarke G., Dinan T.G. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- 24.Davey K.J., O'Mahony S.M., Schellekens H., O'Sullivan O., Bienenstock J., Cotter P.D., Dinan T.G., Cryan J.F. Gender-dependent consequences of chronic olanzapine in the rat: effects on body weight, inflammatory, metabolic and microbiota parameters. Psychopharmacol (Berl) 2012;221:155–169. doi: 10.1007/s00213-011-2555-2. [DOI] [PubMed] [Google Scholar]
- 25.Davis B.A., Clinton S.M., Akil H., Becker J.B. The effects of novelty-seeking phenotypes and sex differences on acquisition of cocaine self-administration in selectively bred high-responder and low-responder rats. Pharmacol Biochem Behav. 2008;90:331–338. doi: 10.1016/j.pbb.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunphy-Doherty F., O'Mahony S.M., Peterson V.L., O'Sullivan O., Crispie F., Cotter P.D., Wigmore P., King M.V., Cryan J.F., Fone K.C.F. Post-weaning social isolation of rats leads to long-term disruption of the gut microbiota-immune-brain axis. Brain Behav Immun. 2018;68:261–273. doi: 10.1016/j.bbi.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Enoch M.A., Hodgkinson C.A., Yuan Q., Shen P.H., Goldman D., Roy A. The influence of GABRA2, childhood trauma, and their interaction on alcohol, heroin, and cocaine dependence. Biol Psychiatry. 2010;67:20–27. doi: 10.1016/j.biopsych.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flagel S.B., Clark J.J., Robinson T.E., Mayo L., Czuj A., Willuhn I., Akers C.A., Clinton S.M., Phillips P.E., Akil H. A selective role for dopamine in stimulus-reward learning. Nature. 2011;469:53–57. doi: 10.1038/nature09588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flagel S.B., Robinson T.E. Neurobiological basis of individual variation in stimulus-reward learning. Curr Opin Behav Sci. 2017;13:178–185. doi: 10.1016/j.cobeha.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flagel S.B., Watson S.J., Akil H., Robinson T.E. Individual differences in the attribution of incentive salience to a reward-related cue: influence on cocaine sensitization. Behav Brain Res. 2008;186:48–56. doi: 10.1016/j.bbr.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flagel S.B., Watson S.J., Robinson T.E., Akil H. Individual differences in the propensity to approach signals vs goals promote different adaptations in the dopamine system of rats. Psychopharmacol (Berl) 2007;191:599–607. doi: 10.1007/s00213-006-0535-8. [DOI] [PubMed] [Google Scholar]
- 32.Fulling C., Dinan T.G., Cryan J.F. Gut microbe to brain signaling: what happens in vagus. Neuron. 2019;101:998–1002. doi: 10.1016/j.neuron.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Gancarz A.M., Robble M.A., Kausch M.A., Lloyd D.R., Richards J.B. Association between locomotor response to novelty and light reinforcement: sensory reinforcement as a rodent model of sensation seeking. Behav Brain Res. 2012;230:380–388. doi: 10.1016/j.bbr.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gancarz A.M., San George M.A., Ashrafioun L., Richards J.B. Locomotor activity in a novel environment predicts both responding for a visual stimulus and self-administration of a low dose of methamphetamine in rats. Behav Process. 2011;86:295–304. doi: 10.1016/j.beproc.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golubeva A.V., Joyce S.A., Moloney G., Burokas A., Sherwin E., Arboleya S., Flynn I., Khochanskiy D., Moya-Perez A., Peterson V., Rea K., Murphy K., Makarova O., Buravkov S., Hyland N.P., Stanton C., Clarke G., Gahan C.G.M., Dinan T.G., Cryan J.F. Microbiota-related changes in bile acid & tryptophan metabolism are associated with gastrointestinal dysfunction in a mouse model of autism. EBioMedicine. 2017;24:166–178. doi: 10.1016/j.ebiom.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant B.F., Goldstein R.B., Saha T.D., Chou S.P., Jung J., Zhang H., Pickering R.P., Ruan W.J., Smith S.M., Huang B., Hasin D.S. Epidemiology of DSM-5 alcohol use disorder: results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72:757–766. doi: 10.1001/jamapsychiatry.2015.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heilig M., Epstein D.H., Nader M.A., Shaham Y. Time to connect: bringing social context into addiction neuroscience. Nat Rev Neurosci. 2016;17:592–599. doi: 10.1038/nrn.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heilig M., Goldman D., Berrettini W., O'Brien C.P. Pharmacogenetic approaches to the treatment of alcohol addiction. Nat Rev Neuroscsi. 2011;12:670–684. doi: 10.1038/nrn3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoban A.E., Stilling R.M., G M.M., Moloney R.D., Shanahan F., Dinan T.G., Cryan J.F., Clarke G. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome. 2017;5:102. doi: 10.1186/s40168-017-0321-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoban A.E., Stilling R.M., Ryan F.J., Shanahan F., Dinan T.G., Claesson M.J., Clarke G., Cryan J.F. Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry. 2016;6:e774. doi: 10.1038/tp.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofford R.S., Russo S.J., Kiraly D.D. Neuroimmune mechanisms of psychostimulant and opioid use disorders. Eur J Neurosci. 2019;50:2562–2573. doi: 10.1111/ejn.14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiao E.Y., McBride S.W., Hsien S., Sharon G., Hyde E.R., McCue T., Codelli J.A., Chow J., Reisman S.E., Petrosino J.F., Patterson P.H., Mazmanian S.K. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jadhav K.S., Peterson V.L., Halfon O., Ahern G., Fouhy F., Stanton C., Dinan T.G., Cryan J.F., Boutrel B. Gut microbiome correlates with altered striatal dopamine receptor expression in a model of compulsive alcohol seeking. Neuropharmacology. 2018;141:249–259. doi: 10.1016/j.neuropharm.2018.08.026. [DOI] [PubMed] [Google Scholar]
- 44.Jaggar M., Rea K., Spichak S., Dinan T.G., Cryan J.F. You've got male: sex and the microbiota-gut-brain axis across the lifespan. Front Neuroendocrinol. 2020;56 doi: 10.1016/j.yfrne.2019.100815. [DOI] [PubMed] [Google Scholar]
- 45.Jasarevic E., Morrison K.E., Bale T.L. Sex differences in the gut microbiome-brain axis across the lifespan. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang H., Ling Z., Zhang Y., Mao H., Ma Z., Yin Y., Wang W., Tang W., Tan Z., Shi J., Li L., Ruan B. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly J.R., Borre Y., C O.B., Patterson E., El Aidy S., Deane J., Kennedy P.J., Beers S., Scott K., Moloney G., Hoban A.E., Scott L., Fitzgerald P., Ross P., Stanton C., Clarke G., Cryan J.F., Dinan T.G. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–118. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 49.King C.P., Palmer A.A., Woods L.C., Hawk L.W., Richards J.B., Meyer P.J. Premature responding is associated with approach to a food cue in male and female heterogeneous stock rats. Psychopharmacol (Berl) 2016;233:2593–2605. doi: 10.1007/s00213-016-4306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiraly D.D., Walker D.M., Calipari E.S., Labonte B., Issler O., Pena C.J., Ribeiro E.A., Russo S.J., Nestler E.J. Alterations of the host microbiome affect behavioral responses to cocaine. Sci Rep. 2016;6:35455. doi: 10.1038/srep35455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., Glockner F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kosnicki K.L., Penprase J.C., Cintora P., Torres P.J., Harris G.L., Brasser S.M., Kelley S.T. Effects of moderate, voluntary ethanol consumption on the rat and human gut microbiome. Addict Biol. 2019;24:617–630. doi: 10.1111/adb.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kundu P., Blacher E., Elinav E., Pettersson S. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 54.Leclercq S., De Saeger C., Delzenne N., de Timary P., Starkel P. Role of inflammatory pathways, blood mononuclear cells, and gut-derived bacterial products in alcohol dependence. Biol Psychiatry. 2014;76:725–733. doi: 10.1016/j.biopsych.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 55.Leclercq S., de Timary P., Delzenne N.M., Starkel P. The link between inflammation, bugs, the intestine and the brain in alcohol dependence. Transl Psychiatry. 2017;7:e1048. doi: 10.1038/tp.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leclercq S., Matamoros S., Cani P.D., Neyrinck A.M., Jamar F., Starkel P., Windey K., Tremaroli V., Backhed F., Verbeke K., de Timary P., Delzenne N.M. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 2014;111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Llopis M., Cassard A.M., Wrzosek L., Boschat L., Bruneau A., Ferrere G., Puchois V., Martin J.C., Lepage P., Le Roy T., Lefevre L., Langelier B., Cailleux F., Gonzalez-Castro A.M., Rabot S., Gaudin F., Agostini H., Prevot S., Berrebi D., Ciocan D., Jousse C., Naveau S., Gerard P., Perlemuter G. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830–839. doi: 10.1136/gutjnl-2015-310585. [DOI] [PubMed] [Google Scholar]
- 58.Lovic V., Saunders B.T., Yager L.M., Robinson T.E. Rats prone to attribute incentive salience to reward cues are also prone to impulsive action. Behav Brain Res. 2011;223:255–261. doi: 10.1016/j.bbr.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luczynski P., Whelan S.O., O'Sullivan C., Clarke G., Shanahan F., Dinan T.G., Cryan J.F. Adult microbiota-deficient mice have distinct dendritic morphological changes: differential effects in the amygdala and hippocampus. Eur J Neurosci. 2016;44:2654–2666. doi: 10.1111/ejn.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Markle J.G., Frank D.N., Mortin-Toth S., Robertson C.E., Feazel L.M., Rolle-Kampczyk U., von Bergen M., McCoy K.D., Macpherson A.J., Danska J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 61.Mayer E.A., Padua D., Tillisch K. Altered brain-gut axis in autism: comorbidity or causative mechanisms? Bioessays. 2014;36:933–939. doi: 10.1002/bies.201400075. [DOI] [PubMed] [Google Scholar]
- 62.Mayer E.A., Tillisch K., Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meckel K.R., Kiraly D.D. A potential role for the gut microbiome in substance use disorders. Psychopharmacol (Berl) 2019;236:1513–1530. doi: 10.1007/s00213-019-05232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer P.J., Lovic V., Saunders B.T., Yager L.M., Flagel S.B., Morrow J.D., Robinson T.E. Quantifying individual variation in the propensity to attribute incentive salience to reward cues. PLoS One. 2012;7:e38987. doi: 10.1371/journal.pone.0038987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meyer P.J., Ma S.T., Robinson T.E. A cocaine cue is more preferred and evokes more frequency-modulated 50-kHz ultrasonic vocalizations in rats prone to attribute incentive salience to a food cue. Psychopharmacol (Berl) 2012;219:999–1009. doi: 10.1007/s00213-011-2429-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Meyer P.J., Tripi J.A. Sign-tracking, response inhibition, and drug-induced vocalizations. In: Tomie A, Morrow JD, editors. Sign-tracking and drug addiction. Maize Publishing; Ann Arbor, MI: 2018. [Google Scholar]
- 67.Naseribafrouei A., Hestad K., Avershina E., Sekelja M., Linlokken A., Wilson R., Rudi K. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26:1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 68.Nasser H.M., Chen Y.W., Fiscella K., Calu D.J. Individual variability in behavioral flexibility predicts sign-tracking tendency. Front Behav Neurosci. 2015;9:289. doi: 10.3389/fnbeh.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ning T., Gong X., Xie L., Ma B. Gut Microbiota Analysis in Rats with Methamphetamine-Induced Conditioned Place Preference. Front Microbiol. 2017;8:1620. doi: 10.3389/fmicb.2017.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palmer A.A., de Wit H. Translational genetic approaches to substance use disorders: bridging the gap between mice and humans. Hum Genet. 2012;131:931–939. doi: 10.1007/s00439-011-1123-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peterson V.L., Jury N.J., Cabrera-Rubio R., Draper L.A., Crispie F., Cotter P.D., Dinan T.G., Holmes A., Cryan J.F. Drunk bugs: chronic vapour alcohol exposure induces marked changes in the gut microbiome in mice. Behav Brain Res. 2017;323:172–176. doi: 10.1016/j.bbr.2017.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petrov V.A., Saltykova I.V., Zhukova I.A., Alifirova V.M., Zhukova N.G., Dorofeeva Y.B., Tyakht A.V., Kovarsky B.A., Alekseev D.G., Kostryukova E.S., Mironova Y.S., Izhboldina O.P., Nikitina M.A., Perevozchikova T.V., Fait E.A., Babenko V.V., Vakhitova M.T., Govorun V.M., Sazonov A.E. Analysis of gut microbiota in patients with Parkinson's disease. Bull Exp Biol Med. 2017;162:734–737. doi: 10.1007/s10517-017-3700-7. [DOI] [PubMed] [Google Scholar]
- 73.Pitchers K.K., Flagel S.B., O'Donnell E.G., Woods L.C., Sarter M., Robinson T.E. Individual variation in the propensity to attribute incentive salience to a food cue: influence of sex. Behav Brain Res. 2015;278:462–469. doi: 10.1016/j.bbr.2014.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rea K., Dinan T.G., Cryan J.F. The microbiome: a key regulator of stress and neuroinflammation. Neurobiol Stress. 2016;4:23–33. doi: 10.1016/j.ynstr.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reynolds B., Ortengren A., Richards J.B., de Wit H. Dimensions of impulsive behavior: Personality and behavioral measures. Personal Individ Differ. 2006;40:305–315. [Google Scholar]
- 76.Reynolds B., Richards J.B., Horn K., Karraker K. Delay discounting and probability discounting as related to cigarette smoking status in adults. Behav Process. 2004;65:35–42. doi: 10.1016/s0376-6357(03)00109-8. [DOI] [PubMed] [Google Scholar]
- 77.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., Costea P.I., Godneva A., Kalka I.N., Bar N., Shilo S., Lador D., Vila A.V., Zmora N., Pevsner-Fischer M., Israeli D., Kosower N., Malka G., Wolf B.C., Avnit-Sagi T., Lotan-Pompan M., Weinberger A., Halpern Z., Carmi S., Fu J., Wijmenga C., Zhernakova A., Elinav E., Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 78.Ryan P.M., Patterson E., Kent R.M., Stack H., O'Connor P.M., Murphy K., Peterson V.L., Mandal R., Wishart D.S., Dinan T.G., Cryan J.F., Seeley R.J., Stanton C., Ross R.P. Recombinant incretin-secreting microbe improves metabolic dysfunction in high-fat diet fed rodents. Sci Rep. 2017;7:13523. doi: 10.1038/s41598-017-14010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanchez-Roige S., Fontanillas P., Elson S.L., Pandit A., Schmidt E.M., Foerster J.R., Abecasis G.R., Gray J.C., de Wit H., Davis L.K., MacKillop J., Palmer A.A., Me Research T. Genome-wide association study of delay discounting in 23,217 adult research participants of European ancestry. Nat Neurosci. 2018;21:16–18. doi: 10.1038/s41593-017-0032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saunders B.T., Robinson T.E. A cocaine cue acts as an incentive stimulus in some but not others: implications for addiction. Biol Psychiatry. 2010;67:730–736. doi: 10.1016/j.biopsych.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheperjans F., Pekkonen E., Kaakkola S., Auvinen P. Linking smoking, coffee, urate, and parkinson's disease - a role for gut microbiota? J Parkinsons Dis. 2015;5:255–262. doi: 10.3233/JPD-150557. [DOI] [PubMed] [Google Scholar]
- 82.Schnorr S.L., Candela M., Rampelli S., Centanni M., Consolandi C., Basaglia G., Turroni S., Biagi E., Peano C., Severgnini M., Fiori J., Gotti R., De Bellis G., Luiselli D., Brigidi P., Mabulla A., Marlowe F., Henry A.G., Crittenden A.N. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schultz W., Dayan P., Montague P.R. A neural substrate of prediction and reward. Science. 1997;275:1593–1599. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- 84.Schwandt M.L., Heilig M., George D.T., Hommer D., Ramchandani V.A. Childhood trauma in alcohol dependence: vulnerability and relative resilience. Alcohol. 2017;60 [Google Scholar]
- 85.Scott K.A., Ida M., Peterson V.L., Prenderville J.A., Moloney G.M., Izumo T., Murphy K., Murphy A., Ross R.P., Stanton C., Dinan T.G., Cryan J.F. Revisiting Metchnikoff: Age-related alterations in microbiota-gut-brain axis in the mouse. Brain Behav Immun. 2017;65:20–32. doi: 10.1016/j.bbi.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 86.Sherwin E., Bordenstein S.R., Quinn J.L., Dinan T.G., Cryan J.F. Microbiota and the social brain. Science. 2019;366:6465. doi: 10.1126/science.aar2016. [DOI] [PubMed] [Google Scholar]
- 87.Sherwin E., Dinan T.G., Cryan J.F. Recent developments in understanding the role of the gut microbiota in brain health and disease. Ann N Y Acad Sci. 2018;1420:5–25. doi: 10.1111/nyas.13416. [DOI] [PubMed] [Google Scholar]
- 88.Solberg Woods L.C., Palmer A.A. Using Heterogeneous Stocks for Fine-Mapping Genetically Complex Traits. Methods Mol Biol. 2019;2018:233–247. doi: 10.1007/978-1-4939-9581-3_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Soo R.M., Woodcroft B.J., Parks D.H., Tyson G.W., Hugenholtz P. Back from the dead; the curious tale of the predatory cyanobacterium Vampirovibrio chlorellavorus. PeerJ. 2015;3:e968. doi: 10.7717/peerj.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinway S.N., Biggs M.B., Loughran T.P., Jr., Papin J.A., Albert R. Inference of Network Dynamics and Metabolic Interactions in the Gut Microbiome. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Temko J.E., Bouhlal S., Farokhnia M., Lee M.R., Cryan J.F., Leggio L. The Microbiota, the Gut and the Brain in Eating and Alcohol Use Disorders: A 'Menage a Trois'? Alcohol Alcohol. 2017;52:403–413. doi: 10.1093/alcalc/agx024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tomie A., Grimes K.L., Pohorecky L.A. Behavioral characteristics and neurobiological substrates shared by Pavlovian sign-tracking and drug abuse. Brain Res Rev. 2008;58:121–135. doi: 10.1016/j.brainresrev.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ubeda C., Bucci V., Caballero S., Djukovic A., Toussaint N.C., Equinda M., Lipuma L., Ling L., Gobourne A., No D., Taur Y., Jenq R.R., van den Brink M.R., Xavier J.B., Pamer E.G. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Versaggi C.L., King C.P., Meyer P.J. The tendency to sign-track predicts cue-induced reinstatement during nicotine self-administration, and is enhanced by nicotine but not ethanol. Psychopharmacol (Berl) 2016;233:2985–2997. doi: 10.1007/s00213-016-4341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.World Health Organization . United Nations Offices on Drugs and Crime (UNODC); New York: 2012. World Drug Report. [Google Scholar]
- 96.Yager L.M., Pitchers K.K., Flagel S.B., Robinson T.E. Individual variation in the motivational and neurobiological effects of an opioid cue. Neuropsychopharmacology. 2015;40:1269–1277. doi: 10.1038/npp.2014.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yager L.M., Robinson T.E. Individual variation in the motivational properties of a nicotine cue: sign-trackers vs. goal-trackers. Psychopharmacol (Berl) 2015;232:3149–3160. doi: 10.1007/s00213-015-3962-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yan A.W., Fouts D.E., Brandl J., Starkel P., Torralba M., Schott E., Tsukamoto H., Nelson K.E., Brenner D.A., Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96–105. doi: 10.1002/hep.24018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang B.Z., Han S., Kranzler H.R., Palmer A.A., Gelernter J. Sex-specific linkage scans in opioid dependence. Am J Med Genet B Neuropsychiatr Genet. 2017;174:261–268. doi: 10.1002/ajmg.b.32507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng P., Zeng B., Zhou C., Liu M., Fang Z., Xu X., Zeng L., Chen J., Fan S., Du X., Zhang X., Yang D., Yang Y., Meng H., Li W., Melgiri N.D., Licinio J., Wei H., Xie P. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host's metabolism. Mol Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 101.Zhernakova A., Kurilshikov A., Bonder M.J., Tigchelaar E.F., Schirmer M., Vatanen T., Mujagic Z., Vila A.V., Falony G., Vieira-Silva S. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.