Abstract
Background
Since numerous pathological conditions are evoked by unwanted dendritic cell (DC) activity, therapeutic agents modulating DC functions are of great medical interest. In regenerative medicine, cellular secretomes have gained increasing attention and valuable immunomodulatory properties have been attributed to the secretome of γ-irradiated peripheral blood mononuclear cells (PBMCs). Potential effects of the PBMC secretome (PBMCsec) on key DC functions have not been elucidated so far.
Methods
We used a hapten-mediated murine model of contact hypersensitivity (CH) to study the effects of PBMCsec on DCs in vivo. Effects of PBMCsec on human DCs were investigated in monocyte-derived DCs (MoDC) and ex vivo skin cultures. DCs were phenotypically characterised by transcriptomics analyses and flow cytometry. DC function was evaluated by cytokine secretion, antigen uptake, PBMC proliferation and T-cell priming.
Findings
PBMCsec significantly alleviated tissue inflammation and cellular infiltration in hapten-sensitized mice. We found that PBMCsec abrogated differentiation of MoDCs, indicated by lower expression of classical DC markers CD1a, CD11c and MHC class II molecules. Furthermore, PBMCsec reduced DC maturation, antigen uptake, lipopolysaccharides-induced cytokine secretion, and DC-mediated immune cell proliferation. Moreover, MoDCs differentiated with PBMCsec displayed diminished ability to prime naïve CD4+T-cells into TH1 and TH2 cells. Furthermore, PBMCsec modulated the phenotype of DCs present in the skin in situ. Mechanistically, we identified lipids as the main biomolecule accountable for the observed immunomodulatory effects.
Interpretation
Together, our data describe DC-modulatory actions of lipids secreted by stressed PBMCs and suggest PBMCsec as a therapeutic option for treatment of DC-mediated inflammatory skin conditions.
Funding
This research project was supported by the Austrian Research Promotion Agency (Vienna, Austria; grant “APOSEC” 862068; 2015–2019) and the Vienna Business Agency (Vienna, Austria; grant “APOSEC to clinic” 2343727).
Keywords: Cellular secretome, Peripheral blood mononuclear cells, Dendritic cell-mediated inflammatory skin conditions, Contact hypersensitivity, Drug discovery, Immunomodulatory lipids
Nomenclature
- γ
gamma
- *
indicates statistical significance
- §
indicates statistical significance
- †
indicates statistical significance
- ALOX
arachinodate lipoxygenase
- ALOX12
arachidonate 12-lipoxygenase
- ALOX12B
arachidonate 12-lipoxygenase, 12R type
- ALOX12P2
arachidonate 12-lipoxygenase pseudogene 2
- ALOX12P2
arachidonate 12-lipoxygenase pseudogene 2
- ALOX15
arachidonate 15-lipoxygenase
- ALOX15B
arachidonate 15-lipoxygenase, type B
- ALOX5
arachidonate 5-lipoxygenase
- ALOX5AP
arachinodate lipoxygenase 5-activating protein
- ALOXE3
arachidonate lipoxygenase 3
- ANOVA
analysis of variance
- APC
antigen-presenting cell
- C57BL/6J
mouse inbred strain established at The Jackson Laboratory
- Ccl11
chemokine C—C motif ligand 11
- Ccl20
chemokine C—C motif ligand 20
- Ccl5
chemokine C—C motif ligand 5
- Ccl7
chemokine C—C motif ligand 7
- CD11c
cluster of differentiation 11c
- CD14
cluster of differentiation 14
- CD1a
cluster of differentiation 1a
- CD1b
cluster of differentiation 1b
- CD1c
cluster of differentiation 1c
- CD1d
cluster of differentiation 1d
- CD1e
cluster of differentiation 1e
- CD4
cluster of differentiation 4
- CD83
cluster of differentiation 83
- CH
contact hypersensitivity
- CLEC4A
C-Type Lectin Domain Family 4 Member A
- Cntf
ciliary neurotrophic factor
- Ctf1
cardiotrophin 1
- CSF1R
colony stimulating factor 1 receptor
- CSF2RA
colony stimulating factor 2 receptor alpha subunit
- CSF2RB
colony stimulating factor 2 receptor subunit beta
- CSF3R
colony stimulating factor 3 receptor
- Cxcl12
chemokine C-X-C motif ligand 12
- Cxcl9
chemokine C-X-C motif ligand 9
- DC
dendritic cell
- DEG
differentially expressed genes
- DFU
diabetic foot ulcer
- DMEM
dulbecco's modified eagle medium
- DNFB
dinitrofluorobenzene
- EC
endothelial cell
- ELISA
enzyme-linked immunosorbent assay
- EV
extracellular vesicle
- FB
fibroblast
- GEM
gel bead-in-emulsions
- GM-CSF
human granulocyte-macrophage colony-stimulating factor
- GMP
good manufacturing practice
- GO
gene ontology
- HETE
hydroxyeicosatetraenoic acid
- HI-FBS
heat-inactivated fetal bovine serum
- HLA
human leukocyte antigen
- HLA
human leukocyte antigen
- IFNg
interferon gamma
- IL-12p70
interleukin 12 p70
- IL13
interleukin 13
- IL-17
interleukin 17
- Il17a
interleukin 17a
- Il1b
interleukin 1 beta
- IL-23
interleukin 23
- IL-4
interleukin 4
- Il7
interleukin 7
- IPA
ingenuity pathway analysis
- ITGAX
integrin alpha X
- KC
keratinocyte
- LC
Langerhans cell
- LOXHD1
lipoxygenase homology domains 1
- LPS
lipopolysaccharides
- Lta
lymphotoxin alpha
- LTA4H
leukotriene A4 hydrolase
- Ltb
lymphotoxin beta
- LTB4R
leukotriene B4 receptor
- mannose R
mannose receptor.
- MHC
major histocompatibility complex
- MLR
mixed lymphocyte reaction
- MoDC
monocyte-derived dendritic cell
- Mstn
myostatin
- NO
nitric oxide
- Osm
oncostatin M
- OxPAPC
oxidized 1-palmitoyl-2-arachidonyl-sn- glycero‑3-phosphorylcholine
- PBMC
peripheral blood mononuclear cell
- PBMCsec
secretome obtained from γ-irradiated PBMCs
- PCA
principle component analysis
- PEG
polyethylene glycol
- PGE
prostaglandin E
- PTGER2
prostaglandin E receptor 2 (subtype EP2)
- PTGER4
prostaglandin E receptor 4 (subtype EP4)
- PTGES2
prostaglandin E synthase 2
- PTGES3
prostaglandin E synthase 3 (cytosolic)
- PTGIR
prostaglandin I2 (prostacyclin) receptor (IP)
- PTGS1
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase)
- PTGS2
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
- RMA
robust multi-array average
- RNA
ribonucleic acid
- RP11-10B2.1
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (PTGS1) pseudogene
- RPMI
Roswell Park Memorial Institute
- SAPC
1-stearoyl-2-arachidonoyl-sn‑glycero-phosphocholine
- SC
stem cell
- TH1 cell
type 1 T helper cell
- TH2 cell
type 2 T helper cell
- TLR7
toll-like receptor 7
- Tnf
tumor necrosis factor
- TNFa
tumor necrosis factor alpha
- Tnfrsf11b
tumor necrosis factor receptor superfamily member 11b
- UMAP
uniform Manifold Approximation and Projection
Research in context.
Evidence before this study
Several diseases are attributed to unwanted dendritic cell (DC) activity, including contact hypersensitivity (CH). Modulating DC activity is therefore of high clinical interest. Our previous studies showed strong anti-inflammatory effects of the secretome obtained from irradiated white blood cells (PBMCsec). Though PBMCsec was successfully tested in various clinical indications, such as myocardial infarction and neural damage, the potential of the secretome to treat DC-mediated diseases is still not known.
Added value of this study
In this study, we demonstrate that PBMCsec is able to alleviate CH symptoms in a CH mouse model and we identify mitigated antigen-presenting cell activity as the underlying cause, indicating valuable therapeutic potential of the secretome. PBMCsec strongly diminishes major DC functions, including cytokine secretion, antigen uptake, and T cell priming in human DCs in vitro and in skin ex vivo. We further show that lipids present in the secretome mainly account for the observed effects.
Implications of all the available evidence
Using in vivo, ex vivo, and in vitro approaches, this study provides data for the inhibition of key DC characteristics and functions by the secretome obtained from irradiated white blood cells. More specifically, we showed that lipids predominantly account for the anti-inflammatory effects of PBMCsec. Therefore, our results suggest the use of PBMCsec or secretome-derived lipids for treating DC-mediated inflammatory diseases.
Alt-text: Unlabelled box
1. Introduction
Dendritic cells (DCs) are professional antigen-presenting cells (APCs) orchestrating adaptive immune responses [1], [2], [3], [4]. The vast majority of DCs originate from bone marrow-resident DC precursor cells [5]. Alternatively, DCs can develop from monocytes under inflammatory or infectious conditions [6]. Langerhans cells are tissue-resident DCs of the skin and, though functionally similar to DCs, originate from distinct progenitor cells of the embryonal yolk sac and foetal liver [7]. Upon antigen exposure and pathogenic stimulus, DCs become mature, a process involving changes in expression of lymphocytic co-stimulatory molecules and in secretion of immunomodulatory cytokines [8], [9], [10], [11] first described by Schuler and Steinman in 1985 [12]. Cells lacking a co-stimulus can undergo a partial maturation, leading to homeostatic and tolerogenic DC maturation in steady state [8]. Mature DCs subsequently migrate into lymphoid organs, where naïve T cells are primed to differentiate into specific effector T cell subsets [1,2]. Though creating the indispensable linchpin between innate and adaptive immunity, DCs may adversely instigate the immune system and have been implicated in the pathomechanistic events of inflammatory skin conditions, allergic reactions, graft-versus-host-disease, and human immunodeficiency virus infection [13], [14], [15]. Consequently, tight control of DC function is of particular importance to evade unwanted immune responses and clinically modulating DC activity represents an attractive approach for various therapeutic interventions.
Allergic contact dermatitis, also known as contact hypersensitivity (CH), is an inflammatory skin disease with more than 20 percent of the general population suffering from hypersensitivity to at least one contact allergen [16] and whose prevalence is increasing [17,18]. Common irritant classes causing the characteristic symptoms of itching, erythema, and edema include metals, antibiotics, and preservatives [19]. Over the past decades, extensive research on CH pathology has contributed to a better understanding of the pathomechanistic immunologic events. Nonetheless, clinical treatment options remain limited to date, since the complex and multifaceted disease etiology represents a major obstacle for development of effective therapeutic agents. Murine CH represents a well-established model to study eczematous skin reactions, whereby sensitization and, after a brief intermission phase, elicitation of immune responses are provoked by topical application of low molecular weight chemicals, so called haptens [20]. Numerous cell types are involved in shaping the immunological responses leading to CH, including epidermal keratinocytes, T helper cells, memory and regulatory T cells, cutaneous DCs, mast cells, and neutrophils. Murine CH is a powerful model allowing the testing of immunosuppressive agents for treatment of allergic contact dermatitis [20].
Investigations on stem cell (SC)-based tissue regeneration have provided the medical community with encouraging pre-clinical results [21], and SC-based therapies have been considered a promising tool for regeneration of various injured tissues and organs [22], [23], [24]. Yet, pioneer clinical trials in humans failed to meet the high expectations [25,26]. Pursuing studies administering conditioned medium from mesenchymal SCs to injured cardiac tissues revealed that secreted factors, rather than SCs themselves, exert beneficial paracrine effects and account for most of the initial findings [27], [28], [29]. Our group showed that γ-irradiated peripheral blood mononuclear cells (PBMCs) represent an attractive, and in contrast to SCs, easily accessible and rich source for cellular secretomes with comparable action spectra, including cytoprotection and immunomodulation [30,31]. Recently, the importance of γ-irradiation-induced necroptosis for the pro-angiogenic actions of the PBMC secretome (PBMCsec) has been reported [32]. Versatile modes of action have already been elucidated [30,[33], [34], [35], [36]] and various clinical indications have been described for PBMCsec, including wound healing [36,37], acute myocardial infarction [30], autoimmune myocarditis [38], cerebral ischemia [39], and spinal cord injury [40]. A diverse spectrum of biomolecules, including lipids, proteins, and extracellular vesicles (EVs), are secreted by γ-irradiated PBMCs and have been shown to account for the observed effects [34,37]. Cellular secretomes represent a pleiotropic mix of biologically active substances and the exact composition of PBMCsec has been increasingly studied in the past years. Components of the medium used for PBMCsec manufacturing, such as albumin and cholesterol, have been determined [41] and proteins secreted by irradiated PBMCs have been quantified [30,41]. In addition, molecular composition and actions of extracellular vesicles present in PBMCsec have been extensively investigated [37] and different lipid species have been identified in PBMCsec [34]. Anti-inflammatory, cytoprotective, and pro-angiogenic activities have already been attributed to PBMCsec [36,42] and an effect of PBMCsec on T cell-mediated inflammation has been reported [38]. In spite of previous reports studying secretome obtained from PBMCsec and its effects on DC maturation and antigen uptake [38], potential immunomodulatory effects of PBMCsec on key DC functions and DC-mediated skin inflammation remain largely elusive to date. Since lipid species and lipid-metabolizing enzymes have been implicated in DC maturation and function [43] and presence of immunologically active (oxidized) lipid species in PBMCsec has been reported previously [34], we sought to determine the role of lipids secreted by stressed PBMCs in potential, PBMCsec-mediated, immunomodulatory effects on DCs.
2. Materials and methods
2.1. Ethics statement
To isolate DC precursor cells, leukoreduction system chambers (Trima Accel, Terumo BCT, Lakewood, CO, USA) were purchased from the Department of Blood Group Serology and Transfusion Medicine of the Medical University of Vienna (Vienna, Austria) and all volunteers provided written informed consent. Blood sample collection from volunteers was approved by the ethics committee of the Medical University of Vienna (vote number 1539/2017). PBMCs for PBMCsec manufacturing were obtained from voluntary blood donors. All donors gave written informed consent for blood withdrawal and for use of residual material for scientific purposes. Isolation of skin biopsies was approved by the ethics committee of the Medical University of Vienna (vote number 217/2010) and all donors provided written informed consent. Animal experiments were performed in accordance with guidelines of the Institutional Review Board of the Medical University of Vienna and the Austrian guidelines for the use and care of laboratory animals. Animal experiments were approved by the Austrian Federal Ministry of Education, Science and Research (Vienna, Austria; ethics vote number BMBWF-66.009/0037-V/3b/2018).
2.2. PBMC isolation and generation of PBMC secretome
Secretomes of PBMCs were produced in compliance with good manufacturing practice (GMP) by the Austrian Red Cross, Blood Transfusion Service for Upper Austria (Linz, Austria) as described [37]. Briefly, PBMCs were obtained by Ficoll-Paque PLUS (GE Healthcare, Chicago, IL, USA)-assisted density gradient centrifugation and exposed to 60 Gy Caesium 137 γ-irradiation (IBL 437C, Isotopen Diagnostik CIS GmbH, Dreieich, Germany). Cells were adjusted to a concentration of 2.5 × 107 cells/mL and cultured in phenol red-free CellGenix GMP DC medium (CellGenix GmbH, Freiburg, Germany) for 24 ± 2 h. Cells and cellular debris were removed by centrifugation and supernatants were passed through a 0.2 µm filter. For viral clearance, methylene blue treatment was performed as described [44]. Secretomes were lyophilized, terminally sterilized by high-dose γ-irradiation (Gammatron 1500, UKEM 60Co irradiator with a maximum capacity of 1.5 MCi), and cryopreserved. Lyophilisates were reconstituted in 0.9% NaCl (B. Braun Melsungen AG, Melsungen, Germany). All experiments were performed using secretomes produced under GMP with the following batches: A000918399086, A000918399095, and A000918399098, A000918399101, A000918399102, and A000918399105.
2.3. Fractionating PBMC secretome
Total lipids were purified according to the procedure described by Folch et al. with minor modifications [45]. In detail, one part reconstituted PBMCsec was combined with 9 parts 2:1 (vol/vol) chloroform-methanol mixture and excessively vortexed. The emulsion was acidified by adding 0.7 m formic acid (¼ the volume of CHCl3−MetOH) and homogenized by vigorous shaking. Phase separation was performed on ice for 30 min. The organic (lower) phase was collected and solvents were eliminated by rotary vacuum evaporation (475 mbar, 100 rpm, 60 °C water bath temperature). Proteins were isolated by polyethylene glycol (PEG) 4000-assisted precipitation (Sigma-Aldrich, St. Louis, MO, USA) with PEG-4000 addition at a final concentration of 30% (wt/vol). Solutions were incubated for 30 min on ice and proteins were isolated by centrifugation (8000 g, 10 min, 4 °C). EVs were obtained by ultracentrifugation as described previously (110,000 g for 2 h at 4 °C) [34,37]. Eventually, EVs, lipids, and proteins were reconstituted in CellGenix GMP DC medium in the same volume of secretome initially used for fractionation.
2.4. Generation of monocyte-derived dendritic cells (MoDCs)
PBMCs were isolated by density centrifugation as described above and DC precursor cells were enriched by adhesion on tissue culture-treated flasks [46]. After 1.5 h, cells were trypsinized (trypsin-EDTA 0.25%, phenol red; Gibco, Waltham, MA, USA) and seeded at a density of 4 × 104 cells/cm2. In a 6-well plate, 3.8 × 105 cells were plated in a total volume of 3 mL medium. MoDCs were generated in RPMI-1640 medium GlutaMAX (Gibco) supplemented with 10% (vol/vol) heat-inactivated foetal bovine serum (HI-FBS, Gibco), 25 ng/mL human interleukin 4 (IL-4, 5 × 106 units/mg), and 50 ng/mL human granulocyte-macrophage colony-stimulating factor (GM-CSF, 1 × 107 units/mg) (both PeproTech, Rocky Hill, NJ, USA) for 7 days. In parallel, MoDCs were differentiated in the presence of PBMCsec or fractions (designated MoDC-PBMCsec, MoDC-lipids, MoDC-proteins, and MoDC-EVs). CellGenix GMP DC medium used to culture PBMCs served as vehicle control (MoDC-vehicle). The effective dose was titrated to a final concentration equivalent to the secretome of 6.25 × 106 PBMCs/mL. During differentiation, reconstituted PBMCsec was present in a final amount of 25% (vol/vol). Fractions and medium vehicle were added accordingly. Supplemented medium, secretome, and fractions were freshly replenished on day 3 of differentiation.
2.5. Ex vivo treatment of human skin biopsies with PBMCsec
Six mm whole skin biopsies were obtained from three healthy female volunteers (30–45 years old) during abdominoplasty. Biopsies were incubated in Dulbecco's modified eagle medium (DMEM, Gibco) supplemented with penicillin, streptomycin, 10% (vol/vol) HI-FBS and 20% (vol/vol) vehicle or PBMCsec. In addition, 100 µL PBMCsec or vehicle were injected intradermally. After a 24 h-culture at 37 °C, viable cells were isolated and prepared for sequencing as described previously [47].
2.6. Single-cell RNA sequencing (scRNAseq) and bioinformatics analyses
Viable cell suspensions were loaded onto a Chromium Controller and Single Cell 3′ Library & Gel Bead Kit v2 (10x Genomics, Pleasanton, CA, USA) according to the manufacturer's protocol and as described previously [48]. In brief, gel bead-in-emulsions (GEMs) were generated by cell suspensions (up to 10,000 per sample) and gel beads containing reverse transcription reagents in a nanoliter-scale water-in-oil emulsion. cDNA in GEMs was generated using C1000 Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA), GEMs were broken, cDNA was isolated and washed. Libraries were sequenced using the Illumina HiSeq 3000/4000 platform (Illumina, Inc., San Diego, CA, USA) and the 75 bp paired-end configuration. GEM generation, cDNA synthesis, library preparation, and sequencing were performed by the Biomedical Sequencing Facility at the Research center for Molecular Medicine of the Austrian Academy of Sciences (Vienna, Austria). Pre-processing of the scRNAseq data was performed using Cell Ranger software (version 3.0.2., 10x Genomics). Raw sequencing files were demultiplexed using the Cell Ranger ‘mkfastq’-pipeline. Each sample was aligned to the human reference genome (GRCh38-1.2.0) using the Cellranger ‘count’-pipeline, and raw expression data were analysed by R (version 3.5.1., R Foundation for Statistical Computing, Vienna, Austria).
Secondary bioinformatics analysis was performed using the R-package ‘Seurat’ (Seurat v3, Satija Lab, New York University, New York City, NY, USA) [49,50]. Cells of all datasets were first analysed for their unique molecular identifier (UMI) and mitochondrial gene counts, and cells with low (<300) or high (>2500) UMI counts or high percentage of mitochondrial genes (>4%) were excluded from further analysis. Data were integrated in a standardized workflow as recommended by the developers of the “Seurat”-package, including data normalization, identification of variable genes, finding anchors for integration based on variable genes, integration of all datasets, scaling of data, principle component analysis (PCA) with JackStraw procedure, and unsupervised clustering with a resolution of 0.7 based on Uniform Manifold Approximation and Projection (UMAP) [51]. Cell types were identified based on a marker gene panel and on differentially expressed genes (DEGs) in every cluster. DEGs were calculated by Wilcoxon rank sum test with Bonferroni correction for adjusted p-values. As recommended by the “Seurat” developers, data in feature plots, violin plots, heat maps and trajectories demonstrating features that vary across conditions were displayed based on the “RNA”-count slot, and data reflecting the entire dataset were displayed based on the “integrated” dataset. Gene ontology (GO) networks based on DEGs were created using the ClueGO plugin [52] of Cytoscape (version 3.7.2, Institute for Systems Biology, Seattle, WA, USA) (EBI UniProt GOA) [53].
2.7. Hapten-induced murine contact hypersensitivity
1-Fluoro-2,4-dinitrobenzene (DNFB, Sigma-Aldrich) was used to induce inflammatory skin conditions in C57BL/6 J mice at the age of 12 weeks as described previously [54,55] with minor modifications. Twenty µL 0.25% (vol/vol) DNFB were topically administered in a 3:1 acetone-to-olive oil mixture on shaved back skins on days 0 and 1. Ears were treated daily with PBMCsec or lipids, while contralateral ears received vehicle medium for 6 consecutive days starting from day 0. To facilitate administration and substance absorbance by the tissue, treatment solutions were mixed 3:1 (vol/vol) in Ultrasicc oil/water emulsion base (Hecht-Pharma GmbH, Bremervoerde, Germany). Skin inflammation was elicited by challenging ears of sensitized mice with DNFB on day 7. Naïve controls were left untouched. Twenty-four hours post DNFB re-challenge, ear thickness was assessed using an electronic digital micrometer (0–25 mm, Marathon Management Inc., Wilsonville, OR, USA), measuring thickness of the outer two-thirds of the ear whilst avoiding skin folds at the ear base. Measurements were performed in triplicates. Tissues were harvested for RNA isolation as described below and histological assessment. Cytokine and chemokine expressions were assessed by RT2 Profiler PCR Array Mouse Cytokines and Chemokines (Qiagen, Redwood City, CA, USA) according to the manufacturer's instructions.
2.8. Allergen-independent skin inflammation
To induce allergen-independent skin inflammation, mice (C57BL/6 J, 12 weeks of age) were treated with imiquimod (Aldara 5% crème, Meda AB, AB, Solna, Sweden) [56]. For 6 consecutive days, ears were treated with PBMCsec or vehicle for paired comparison as described above. Six hours later, a total of 4.2 mg imiquimod (83.3 mg crème) per ear were topically applied. On day 7, mice were sacrificed and ear thickness was determined as described above. Ear tissues were isolated for histology.
2.9. Flow cytometry
Flow cytometric assessment was routinely performed on FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA) as recommended by the manufacturer. To assess DC differentiation, cluster of differentiation (CD) 1a and CD11c expressions were assessed using fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD1a antibody (clone HI149, 1:100; BD Biosciences) and phycoerythrin (PE)-conjugated mouse anti-human CD11c antibody (clone S-HCL-3, 1:100; BD Biosciences). CD14 expression was quantified by PE-conjugated mouse anti-human CD14 antibody (clone M5E2, 1:100; BD Biosciences). DC maturation was determined using FITC-conjugated mouse anti-human CD83 (clone HB15e, 1:100; BD Biosciences) and allophycocyanin (APC)-conjugated mouse anti-human leukocyte antigen (HLA)-DR (clone G46-6, 1:400; BD Biosciences). For data acquisition and analysis, CellQuest Pro (BD Biosciences) and FlowJo (FlowJo LLC, Ashland, OR, USA, version 10) softwares were used, respectively.
2.10. Transcriptomics and bioinformatics analyses
For transcriptomics, CD14+ monocytes were enriched from PBMCs by magnetic sorting using human CD14 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany) and autoMACS Pro-Separator (Miltenyi Biotec) as suggested by the manufacturer. Total RNA was isolated using peqGOLD TriFast (Qiagen) according to the manufacturer's recommendations. RNA cleanup was performed using RNeasy MinElute Cleanup Kit (Qiagen) and RNA quality was assessed by Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA, USA). Two-hundred ng RNA were used for gene expression analysis using Affymetrix human gene 2.1 ST arrays (Affymetrix Inc., Santa Clara, CA, USA). Hybridization and scanning were performed as suggested by the manufacturer. Custom chip description file was used for robust multi-array average (RMA) signal extraction and normalization. RMA values below 90 were considered non-detectable. GeneSpring Version 15.0 software (Agilent Technologies) was used for PCA. Venn diagrams were generated using the biovenn web application (http://www.biovenn.nl/index.php, accessed on September 14th, 2018). Web-based heatmapping was used to visualize results [57]. To identify regulated signaling pathways, genes displaying an average transcriptional upregulation of ≥ 1.5 or an average downregulation of ≤ 0.5 between PBMCsec versus medium controls were selected for Ingenuity Pathway Analysis (IPA, Qiagen) [58]. Biological processes associated with differentially regulated genes were identified by GO enrichment analysis using PANTHER classification system [59,60].
2.11. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA isolation, reverse transcription, and qPCR were performed as described previously [37,61]. Primer sequences to amplify respective genes from human cDNA libraries were designed using Primer3 Input (version 0.4.0) and synthesized by Microsynth AG (Balgach, Switzerland) [62,63]. HLA-DRA sense 5′ cgatcaccaatgtacctcca 3′, antisense 5′ cctgtggtgacaggttttcc 3′, HLA-DPB1 sense 5′ ggaacagccagaaggacatc 3′, antisense 5′ cattcaggaaccatcggact 3′, HLA-DQA1 sense 5′ cagctcagaacaccaactgc 3′, antisense 5′ aggcagtctccttcctctcc 3′, CD1A sense 5′ gccaggacatcgtcctctac 3′, antisense 5′ tctctcaccccaaaaggaga 3′, CD1B sense 5′ ctcaggctggttggatgatt 3′, antisense 5′ gctatgccctggatctcaaa 3′, CD1C sense 5′ agactcatggctgggacagt 3′, antisense 5′ ttccagaatgcagctcacag 3′.
2.12. Mixed lymphocyte reaction (MLR)
Maturation of MoDCs was induced by exposure to 100 endotoxin units (eu)/mL lipopolysaccharides (LPS) from Escherichia coli 0111:B4 (Sigma-Aldrich) on day 7 of differentiation [64,65]. During DC differentiation and maturation, vehicle, PBMCsec, or fractions were present. After incubation for 16 h, cells were washed thoroughly to remove secretome, fractions, and LPS. 8 × 104 mature MoDCs were co-incubated with 4 × 105 allogeneic, cell proliferation dye-stained PBMCs (eFluor 670, eBioscience; Thermo Fisher Scientific, Waltham, MA, USA). Proliferation was assessed by flow cytometry on day 4. Non-proliferating cells were defined as the cell population displaying the highest fluorescence intensity. Proliferating cells were discriminated in cells undergoing two and multiple cellular divisions, indicated by intermediate and low fluorescence intensities, respectively.
2.13. T cell priming co-cultures
To study DC-mediated priming of T cells into T helper cell subsets, naïve CD4+ T cells were enriched from PBMCs using human Naïve CD4+ T Cell Isolation Kit II (Miltenyi Biotec) as suggested by the manufacturer. On day 6, MoDC differentiated with secretome, lipids, or vehicle were maturated with LPS as described above, washed with Dulbecco's phosphate-buffered saline (DPBS, Gibco), and seeded in flat-bottom 96-well plates at a density of 1.3 × 105 cells/cm2. For each condition, allogeneic, naïve CD4+ T cells were added in a DC-to-T cell ratio of 1:20. Cells were co-cultured for 6 days in RPMI 1640 with medium changes on day 3. Supernatants were used for protein quantification of TH1 and TH2 cytokines.
2.14. Antigen uptake assay
Immature MoDC were incubated with dextran (FITC-conjugated with an average molecular weight of 10,000 Da, Sigma-Aldrich) for 6 h at 37 °C. Cells at 4 °C served as controls. Flow cytometry was performed as described above.
2.15. Protein quantification
Protein levels of secreted cytokines were determined by proteome profiler cytokine array (R&D Systems, Minneapolis, MN, USA) and quantified by ELISA (IFNγ and IL13, both R&D Systems; IL12p70 and TNFα, both eBioscience) as recommended by the manufacturers. 3.8 × 105 cells in 3 mL medium were used for protein quantification of LPS-treated samples. To determine cytokine concentrations during MLR, 8 × 104 MoDCs together with 4 × 105 allogeneic PBMCs were cultured in 200 µL medium. In T cell priming experiments, supernatants were obtained from 4 × 104 DCs and 8 × 105 naive T cells cultured in 200 µL medium. Chemiluminescence was detected by ChemiDoc XRS imaging systems (Bio-Rad Laboratories) using Image Lab software (version 5.2.1., Bio-Rad Laboratories). Mean gray values 8-bit gray-scale images were determined using the ImageJ measure tool (v1.50i, National Institutes of Health, Bethesda, MD, USA) [66]. Photometric measurements were performed by FLUOstar OPTIMA (BMG Labtech, Ortenburg, Germany) using OPTIMA software (version 2.20R2, BMG Labtech) and MARS Data Analysis Software (version 2.41, BMG Labtech).
2.16. Lipid quantification
Resolvins were quantified by ELISAs (R&D Systems as recommended by the manufacturer. Photometric assessment was performed as described above.
2.17. Statistical analyses
Sample size calculation was performed based on preliminary experimental data (online tool https://clincalc.com/stats/samplesize.aspx, α = 0.05, power = 80%) and data were collected at prospectively defined endpoints. No outliers were excluded. Experiments were repeated using different donors and animals, respectively, in at least triplicates. Animal studies were performed in matched case-control sets. For in vitro and ex vivo experiments, samples of the same donor were exposed to all conditions in parallel. Treatments were not administered blinded. Data were statistically evaluated using GraphPad Prism 6 software (GraphPad Software Inc., LA Jolla, CA, US). Ordinary one-way ANOVA and multiple comparison post hoc tests with Dunnett's or Sidak's correction were carried out as indicated. Two datasets were analysed by one-tailed student's t-test. Box plot diagrams were generated using first and third quartiles as boxes and bars as medians. Whiskers indicate minimal and maximal values, respectively. In bar diagrams, data are shown as arithmetic means ± standard error of the mean.
3. Results
3.1. PBMCsec mitigates hapten-induced tissue inflammation by reducing cellular infiltration to sensitized areas and by curbing local synthesis of pro-inflammatory immune mediators
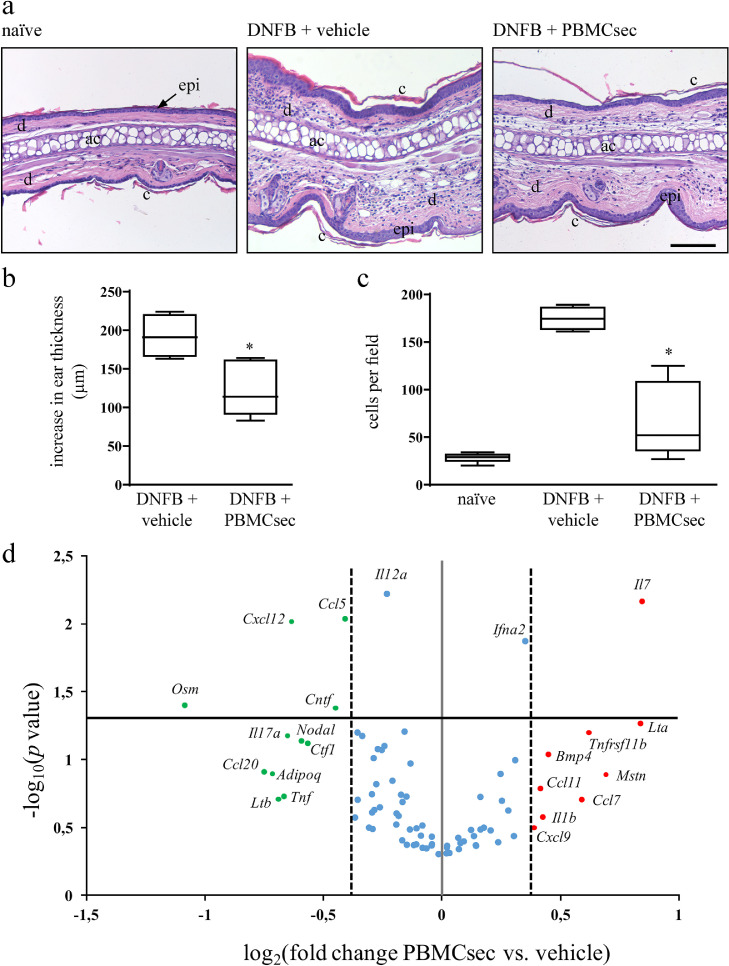
We first sought to investigate whether PBMCsec might affect characteristic events of skin inflammation. Using murine DNFB-induced CH as a model to study DC-mediated inflammatory conditions [20], we assessed potential effects of PBMCsec on CH-associated events. The degree of ear swelling, reflecting severity of immune response, was found remarkably reduced by PBMCsec 24 h post DNFB challenge (average 192.8 ± 11.2 µm increase in thickness of vehicle-treated ears versus 123.8 ± 14.6 µm increase in ear thickness with PBMCsec, p<.05 versus medium) (Fig. 1a & b). Histological investigation revealed diminished ear swelling of PBMCsec-treated ears compared to medium alone (Fig 1b), corroborating results obtained by micrometric assessment. In addition, total numbers of infiltrating immune cells per field were diminished by PBMCsec application compared to vehicle medium (68 ± 15.8 cells per field with PBMCsec versus 174.8 ± 5.03 cells/field with medium; p<.05) (Fig 1c).
Fig. 1.
PBMC secretome alleviates ear swelling in DNFB-induced CH and abolishes immune cell recruitment to challenged skin. (a) Hematoxylin/eosin-stained ears 24 h after DNFB re-challenge with vehicle or PBMCsec treatment. Naïve ears served as untreated controls. Representative micrographs of n = 5 mice per treatment condition are shown. ac, auricular cartilage; c, cornified epidermis; d, dermis; epi, epidermis. Scale bar, 200 µm. (b) Increase in ear thickness assessed by micrometer-assisted measurements 24 h post DNFB elicitation. * indicates p<.05 PBMCsec versus vehicle. N = 5 mice per group. Ordinary one-way ANOVA was performed. Dunnett's multiple comparison test was carried out to compare groups versus vehicle control. (c) Number of cells per field. * denotes p<.05 PBMCsec versus vehicle. N = 5 mice per group. Ordinary one-way ANOVA was performed. Dunnett's multiple comparison test was carried out to compare groups versus vehicle control. (d) qPCR analysis of genes encoding cytokines and chemokines in ears of DNFB-sensitized mice. Red and green dots indicate up- and down-regulated genes, respectively, in PBMCsec- versus medium-treated ears. Blue dots represent genes considered not differentially regulated. Dotted lines denote log2-transformed 1.3-fold regulations when comparing PBMCsec and medium. Solid gray line represents log2 transformation of PBMCsec / medium = 1. Solid black line indicates -log10 of p = .05. N = 5 mice per condition. p-values were calculated by one-tailed student's t-test with equal variances between the two samples.
As multiple cell types contribute to CH pathology, we sought to determine whether the anti-inflammatory effect of PBMCsec observed in DNFB-sensitized mice results from compromised function of APCs. Therefore, we tested PBMCsec in an allergen-independent mouse model of skin inflammation. Mouse ears repeatedly challenged with imiquimod, a toll-like receptor 7 (TLR7) agonist, were treated with secretome or vehicle and tissue inflammation was analysed. We observed that the characteristic imiquimod-induced symptoms of skin inflammation, including tissue swelling, stromal cell infiltration, thickening of the cornified layer, and epidermal keratinocyte hyperproliferation, were comparable between vehicle- and PBMCsec-treated ears (average 94.9 ± 11.3 µm increase in thickness in vehicle-treated ears and 69.3 ± 8.8 µm increase in thickness with PBMCsec, p>.05) (average 372.3 ± 25.8 cells per field with vehicle versus 349.4 ± 19.0 cells per field with PBMCsec, p>.05) (supplementary Fig S1). Mechanistically, these data suggest that the symptom-alleviating effect of PBMCsec in CH mice presumably depends on the action of PBMCsec on DCs.
As hapten exposure induces a pathomechanistic cascade causing vast immune cell activation and recruitment to challenged tissues, we profiled expression of selected cytokines and chemokines in ears of DNFB-sensitized mice and observed a strong immunomodulatory effect of PBMCsec. Twenty-four hours post DNFB-induced elicitation, numerous genes reportedly implicated in skin inflammation, such as Ccl5, Cxcl12, Osm, Cntf, Il17a, Ccl20, Adipoq, and Tnf, were weakly expressed with PBMCsec treatment compared to medium (p <0.05 for Ccl5, Cxcl12, Osm, and Cntf) (Fig 1d). Conceivably, these data suggest that PBMCsec alleviates symptoms of skin inflammation by affecting the activity of cutaneous APCs, by modulating cyto- and chemokine expression and, consequently, preventing exacerbated immune cell infiltration.
3.2. PBMCsec compromises classical phenotypic and secretory DC characteristics
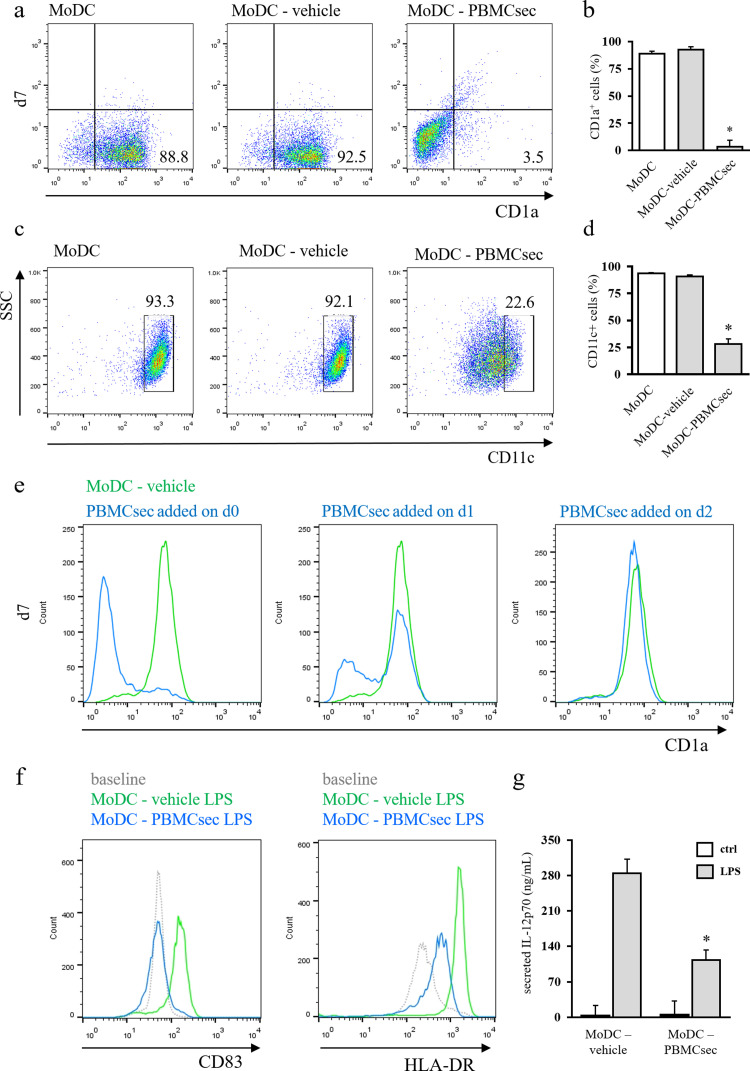
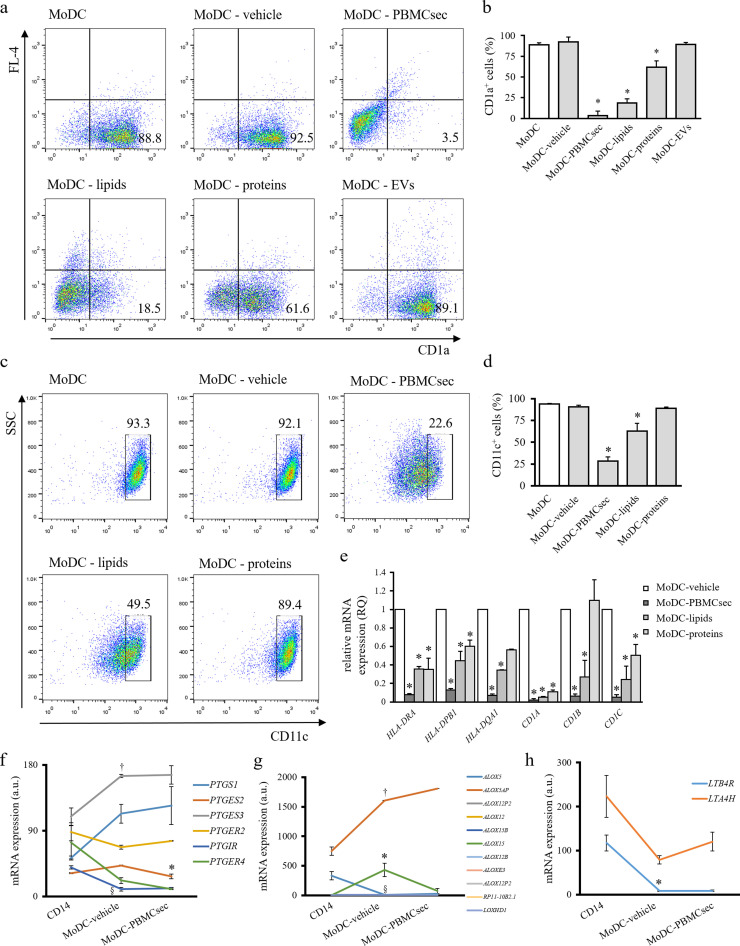
As pleiotropic immunomodulatory effects of PBMCsec are already known [38,67] and since we observed alleviated CH by PBMCsec-dependent DC functional modulation, we sought to determine whether PBMCsec affects the cytokine-driven differentiation of blood-derived monocytes into DCs in vitro. Therefore, monocytes were stimulated with IL-4 and GM-CSF in the presence of vehicle or PBMCsec and efficacy of cytokine-driven differentiation was evaluated by flow cytometric assessment of CD1a and CD11c expressions. After 7 days of differentiation, the majority of cells expressed CD1a with differentiation medium alone (MoDC, 87.7 ± 2.5% CD1a+ cells) and with addition of vehicle medium (MoDC-vehicle, 91.2 ± 2.6% CD1a+ cells; p>.05 vs MoDC) (Fig. 2a & b). By comparison, supplementation with secretome starting from day 0 largely abrogated CD1a expression (MoDC-PBMCsec 5.3 ± 5.7% CD1a+ cells on day 7, p<.05 versus medium control). In line with these data, CD11c expression was compromised by PBMCsec (93.3%, 92.1%, and 22.6% CD11c+ cells in MoDC, MoDC-vehicle, and MoDC-PBMCsec, respectively; p<.05 PBMCsec vs. vehicle) (Fig. 2c & d). We furthermore assessed expression of CD14, a marker for monocytes. While freshly isolated monocytes displayed high CD14 levels, CD14 expression was downregulated in both MoDC-vehicle and MoDC-PBMCsec (supplementary Fig S2).
Fig. 2.
PBMCsec prevents differentiation-associated marker expression and DC cytokine secretion. Representative dot plots of (a) CD1a and (c) CD11c expressions of MoDC differentiated in the presence of vehicle medium and PBMCsec on day 7. Numbers in (a) and (c) indicate frequency of cells. N = 5 and n = 3 donors in (a) and (c), respectively. Statistical analysis of (b) CD1a+ and (d) CD11c+ cells in MoDC-vehicle and MoDC-PBMCsec. * indicates p<.05 PBMCsec versus vehicle. N = 5 donors and n = 3 donors in (b) and (d), respectively. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed. (e) CD1a expression dynamics in the course of DC differentiation with PBMCsec addition at indicated time points. N = 3 donors. (f) Flow cytometric assessment of DC maturation marker expressions in LPS-challenged MoDC-vehicle and MoDC-PBMCsec. Representative histograms of n = 3 donors are shown. (g) Quantification of secreted IL-12p70 by ELISA. N = 3 donors. Asterisk denotes p<.05 LPS-challenged MoDC-PBMCsec versus LPS-treated MoDC-vehicle. Ordinary one-way ANOVA was performed and LPS-treated groups were compared by Sidak's multiple comparisons.
To assess dynamics of MoDC differentiation, PBMCsec was added at various time points and CD1a expression was evaluated over time. The majority of cells was found CD1a-positive starting from day 2 of differentiation (46.6%, 84.8%, and 80% CD1a+ cells on day 1, 2, and 3, respectively) (supplementary Fig S3). Addition of PBMCsec at different time points revealed that PBMCsec preserves the status of the time point when PBMCsec was added (12.9%, 58.2%, and 86.9% CD1a+ cells on day 7 with PBMCsec supplementation starting from day 0, 1, and 2, respectively) (Fig 2e).
We furthermore sought to investigate potential effects of PBMCsec on DC maturation. To this end, we assessed expression of CD83 and HLA-DR after LPS-induced maturation in vehicle- and PBMCsec-treated MoDCs. As shown in Fig 2f, addition of PBMCsec significantly reduced LPS-induced upregulation of CD83 and HLA-DR, indicating diminished DC maturation.
As release of immunomodulatory agents is a major function of DCs, we compared cytokine secretion properties of mature MoDCs-vehicle and MoDC-PBMCsec by protein array. In response to LPS, MoDC differentiated in the presence of secretome secreted fewer DC-relevant cytokines, including IL-12p70, IL-23, and TNFα (supplementary Fig S4). Data obtained by cytokine profiler were further corroborated by enzyme-linked immunosorbent assay (ELISA)-assisted quantification of secreted IL-12p70 (Fig 2g).
Together, these results show that PBMCsec attenuates expression of DC differentiation- and maturation-associated markers and abrogates release of DC-specific cytokines in response to antigen encounter.
3.3. PBMCsec suppresses antigen presentation and induces immunosuppressive and cytoprotective pathways in CD1a+ and CD11c+ human skin cells
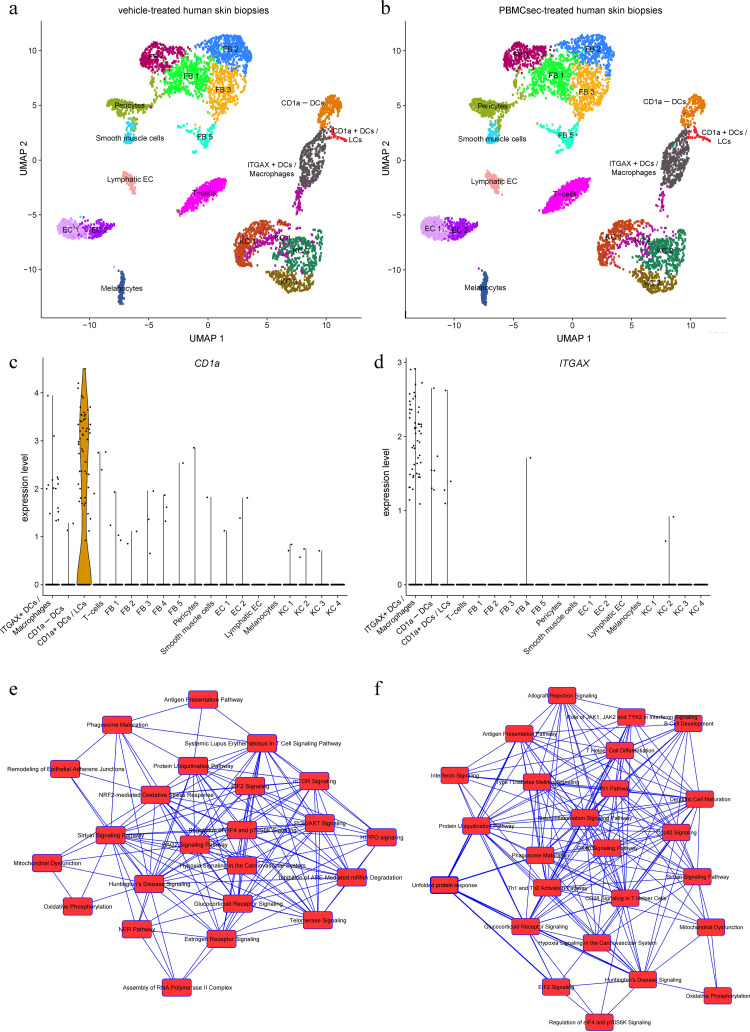
Our in vitro findings show that PBMCsec does not affect expression of DC markers once their expression was induced during differentiation (Fig 2e). To investigate whether PBMCsec is nevertheless able to functionally affect differentiated DCs present in human skin in situ, we performed scRNAseq of ex vivo treated skin biopsies. Using unsupervised clustering together with established cluster markers, we could identify all major cell types present in the human skin (Fig 3a). Compared to vehicle, PBMCsec treatment had no effect on the cellular composition of the skin samples i.e. no specific cell cluster was depleted or expanded by adding PBMCsec to human skin biopsies (Fig 3b). Differences on transcriptional level, however, were evident throughout all cell clusters (data not shown). Since we were interested in APCs, we then focused our analyses on CD1a+ DCs (i.e. Langerhans cells and a subset of dermal DCs) and ITGAX (CD11c)-expressing DCs (Fig. 3c & d) and selected genes downregulated by PBMCsec for GO functional classification by ClueGO Cytoscape. In CD1a+ cells, we found that, amongst others, biological processes associated with antigen presentation, intracellular transport, response to cytokines, and apoptotic processes were affected in PBMCsec-treated skin compared to vehicle (supplementary Table S1). Furthermore, PBMCsec negatively regulated genes involved in pro-inflammatory pathways and antigen receptor-mediated signaling in ITGAX+ cells (supplementary Table S2). Genes were further analysed by IPA. Genes downregulated in CD1a+ cells were, amongst others, associated with processes such as antigen presentation and phagosome maturation (Fig 3e), corroborating results obtained by Cytoscape-assisted functional annotations. Similarly, we detected downregulated signaling pathways for antigen presentation, DC maturation, interferon signaling, allograft rejection, and T-cell differentiation in CD11c+ cells in skin treated with PBMCsec (Fig 3f). In both cell populations, several diseases in the category of dermatological diseases and conditions, such as atopic dermatitis and drug-induced hypersensitivity syndrome, were annotated to PBMCsec-downregulated genes (supplementary Tables S3 & S4).
Fig. 3.
PBMCsec downregulates antigen presentation pathways in CD1a+ and CD11c+ human skin cells. Human skin biopsies were treated ex vivo with vehicle or PBMCsec and used for scRNAseq. Unsupervised clustering together with established cell markers allowed identification of the major cell types present in skin, such as epidermal keratinocytes, fibroblasts, melanocytes, and various immune cells. Clustering was performed for (a) vehicle- and (b) PBMCsec-treated human skin. EC, endothelial cells; FB, fibroblasts; KC, keratinocytes. Expression levels of (c) CD1a and (d) ITGAX in the respective, identified cell clusters. Heights of violin blots indicate expression levels, while widths represent numbers of cells. Points represent individual cells. Genes significantly downregulated by PBMCsec were determined in (e) CD1a+ and (f) ITGAX+ cells and used to identify canonical pathways associated with these genes using IPA. Biological processes (nodes) are connected by common genes. Differentially regulated genes were identified by Wilcoxon rank sum test with Bonferroni correction for adjusted p values. N = 3 donors.
Together, these results suggest that PBMCsec exerts anti-inflammatory and cytoprotective effects by modulating biological functions of skin-resident DCs.
3.4. PBMCsec modulates the transcriptional landscape of MODC
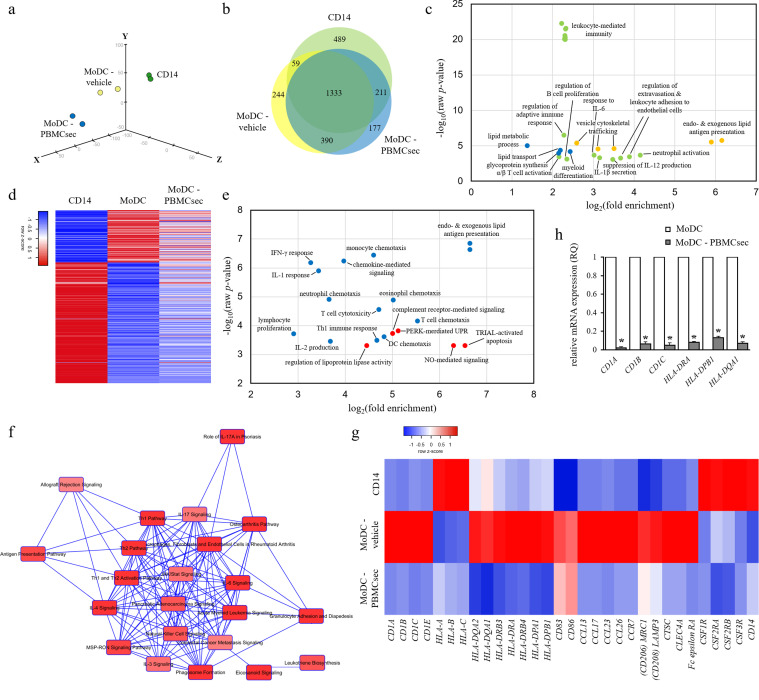
Since the PBMC secretome diminished expression of classical DC markers and reduced release of DC cytokines, we aimed to unravel genes and pathways differentially regulated by PBMCsec. To this end, we compared transcriptional profiles of MoDCs-vehicle and MoDCs differentiated in the presence of PBMCsec. Global gene expression profiles revealed few differences between groups (supplementary Fig S5), indicating scarce transcriptional changes of lineage-committed cells. PCA clustered MoDC-PBMCsec separately from CD14+ cells and MoDC (Fig 4a). Though PBMCsec prevents expression of classical DC differentiation markers, MoDC-PBMCsec displayed a distinct expression pattern compared to that of freshly isolated CD14+ cells, presumably due to the culture period of 7 days.
Fig. 4.
PBMCsec profoundly alters transcriptional profile of MoDC. (a) PCA of CD14+ cells and MoDCs differentiated in the presence of vehicle medium or PBMCsec. N = 2 donors per condition. (b) Genes commonly and exclusively expressed by CD14+ cells, MoDC-vehicle, and MoDC-PBMCsec are shown. Numbers indicate number of genes expressed in the respective sets. (c) GO terms associated with genes exclusively expressed in CD14+ cells (green), MoDC (yellow), and MoDC- PBMCsec (blue), respectively. Each dot represents one biological process. p-values and fold enrichments were used to depict GO terms and were calculated by PANTHER classification system. N = 2 donors per condition. (d) Genes expressed in CD14+ but not in MoDC and vice versa were compared to their expression in MoDC-PBMCsec. Z-scores were calculated by heatmapper.ca. N = 2 donors per condition. (e) GO terms of genes expressed in MoDC but not in MoDC-PBMCsec (blue) and vice versa (red). Each dot represents one biological process. Cut-off values of MoDC / MoDC-PBMCsec fold changes were set to 0.25 and 4, respectively, to identify differentially regulated genes between groups. p-values and fold enrichments were calculated by PANTHER classification system and used for visualization of the results. N = 2 donors per condition. (f) Canonical pathways activated in MoDC-vehicle but not in MoDC-PBMCsec were identified by IPA. Pathways (nodes) are connected by common genes. N = 2 donors per group. (g) Expression levels of genes related to DC function. Z-scores were calculated by heatmapper.ca. N = 2 donors per condition. (h) qPCR analysis of CD1A, CD1B, CD1C, HLA-DRA, HLA-DPB1, and HLA-DQA1 expressions in MoDC-vehicle and MoDC-PBMCsec. N = 3 donors per group. p-values were calculated by one-tailed student's t-test with equal variances between the two samples. Asterisks denote significant difference between MoDC and MoDC-PBMCsec.
Genes expressed in different subsets were analysed by web application-based, area-proportional venn diagrams (Fig 4b). To limit analysis to strongly expressed genes, an expression cut-off value of 90 was chosen. Relative complements, i.e. genes exclusively expressed in CD14+, MoDC, and MoDC-PBMCsec, respectively, are listed in supplementary Table S5. We subsequently used these genes to perform GO enrichment analysis using the PANTHER overrepresentation test to identify biological processes active in the respective cellular subgroups (Fig 4c). Processes associated with adaptive immune responses, B and T cell function, and leukocyte-mediated immunity were detected in CD14+ cells (489 genes), while events involving vesicle trafficking and antigen presentation were exclusively identified in differentiated MoDC (244 genes). Intriguingly, genes regulating lipid transport and lipid metabolic processes were found enriched in MoDC-PBMCsec (177 genes), suggesting a role for lipids in PBMCsec-mediated suppression of DC differentiation marker expression.
We next investigated genes most differentially expressed between CD14+ cells and MoDC, i.e. genes highly expressed in CD14+ cells but not expressed in differentiated MoDC and vice versa (Fig 4d). The expression of these most differentially expressed genes was strongly affected by PBMCsec (Fig 4d). When calculating GO terms for genes expressed in MoDC but not in MoDC-PBMCsec (supplementary Table S6), lipid antigen presentation, chemotaxis of various cells, and type 1 T helper cell (TH1) immune response were enriched (Fig 4e), suggesting an inhibitory effect of PBMCsec on these biological processes. Conversely, regulation of lipoprotein lipase activity and nitric oxide (NO)-mediated signaling were found strongly upregulated in MoDC-PBMCsec, while being inactive in MoDC. Similar results were obtained by IPA when canonical pathways overrepresented in MoDC-vehicle but inactive in MoDC-PBMCsec were analysed (Fig 4f & supplementary Table S7).
We then focused our analyses on specific genes associated with DC phenotype and functions. Among the mRNAs most differentially regulated between vehicle and PBMCsec, we found members of the CD1 and HLA family, antigen receptors and -processing molecules, cytokines and chemokines (Fig 4g). While CD1A, CD1B, CD1C, and CD1E were highly expressed in MoDCs with vehicle medium, PBMCsec prevented differentiation-associated upregulation of CD1 molecules, corroborating results obtained from CD1a flow cytometry. Similar tendencies were observed when studying HLA class II genes, while HLA class I genes were strongly expressed in CD14 cells but showed low levels in MoDC-vehicle and MoDC-PBMCsec. We furthermore investigated expression levels of genes essential for DCs (CD83, CD86, chemokine ligands and receptors, CD206, CD208, cathepsin C, CLEC4A, and Fc epsilon receptor, Fig 4g) and found regulatory effects of PBMCsec on these genes. Lastly, we analysed the expression of genes highly expressed in monocytes. Freshly isolated CD14+ cells strongly expressed colony-stimulating factor receptors (CSF1R, CSF2RA, CSF2RB, and CSF3R) and CD14, while we observed low expression of these monocyte marker molecules in both MoDC-vehicle and MoDC-PBMCsec. mRNA sequencing data were confirmed by RT-qPCR analysis, when expression of CD1A, CD1B, CD1C, HLA-DRA, HLA-DPB1, and HLA-DQA1 were low in DCs cultured with PBMCsec compared to MoDC-vehicle (Fig 4h). Taken together, these data show that PBMCsec remarkably modulates the transcriptional profile by regulating multiple gene expression patterns and signaling pathways related to key DC functions and DC differentiation.
3.5. Lipids secreted by γ-irradiated PBMCs predominantly account for immunomodulatory effects of PBMCsec on DCs
Since our GO enrichment analysis suggested a role for lipids in PBMCsec-mediated compromised DC phenotype and lipids are known to affect DC activity [68], [69], [70], we sought to determine whether lipids and/or other biomolecule(s) present in PBMC secretome were responsible for diminished DC characteristics after PBMCsec treatment. Therefore, the major secretome fractions proteins, lipids, and EVs were isolated from PBMCsec and individually added during DC differentiation. Hardly any adherent cells were detected and no major differences in cell morphology and numbers were observed when cells were exposed to the secretome or the different fractions (supplementary Fig S6). Intriguingly, lipids secreted by irradiated PBMCs effectively abrogated cytokine-induced differentiation of monocytes, comparably to the extent of PBMCsec (75.7 ± 2.6% and 83.4 ± 2.6% CD1a+ cells with differentiation medium alone and control medium, respectively; 13.98 ± 5.8% and 16.2 ± 4.9% CD1a+ with PBMCsec and lipids, respectively; both p<.0001 compared to control medium) (Fig. 5a & b). By comparison, secreted proteins and EVs displayed moderate and no effects on differentiation process, respectively (51.6 ± 7.8% CD1a+ cells with proteins, p<.5 versus control medium; 77.9 ± 5.2% CD1a+ cells with EVs, p>.05 compared to control medium). Interestingly, purified lipids showed lower activity than the whole cell secretome, suggesting that an interplay or additive effects of several fractions are necessary for the full immunomodulatory action of PBMCsec. Similar results were obtained when CD11c expression was assessed (Fig. 5c & d). In detail, CD11c levels were reduced from 93.7 ± 0.6 and 90.5 ± 1.5% positive cells in MoDC and MoDC-medium, respectively, to 28.3 ± 3.3, 62.7 ± 5.9, and 89.1 ± 1.9 CD11c+ with PBMCsec, lipids, and proteins, respectively. In addition, we performed qPCR analysis of our CD1 and HLA gene panel and detected strong inhibitory effects of PBMCsec and lipids on the expression of these genes, while proteins displayed diminished ability to prevent induction of DC differentiation-associated genes (Fig 5e). Taken together, these results indicate that lipids present in the secretome of γ-irradiated PBMC, at least to a large extent, account for the constrained phenotype observed in MoDC-PBMCsec.
Fig. 5.
Lipids secreted by irradiated PBMC abolish expression of DC differentiation-associated molecules. (a) CD1a and (c) CD11c expression of MoDCs differentiated with IL-4/GM-CSF in the presence of either control medium, PBMCsec, or indicated subfractions. Numbers in (a) and (c) indicate frequency of cells. N = 5 and n = 3 donors per group for (a) and (c), respectively. Statistical analyses of (b) CD1a and (d) CD11c expressions. Asterisks denote a significant reduction in the number of positively stained cells of respective groups compared to control vehicle. N = 5 donors and n = 3 donors in (b) and (d), respectively. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed. (e) qPCR analysis of CD1A, CD1B, CD1C, HLA-DRA, HLA-DPB1, and HLA-DQA1 expressions in MoDC-vehicle, MoDC-PBMCsec, MoDC-lipids, and MoDC-proteins. N = 3 donors per group. Ordinary one-way ANOVA was performed with Dunnett's multiple comparisons to compare groups versus vehicle. Asterisks indicate significant p-values versus vehicle. Expression values of genes associated with (f) prostaglandin synthesis, (g) arachidonate lipoxygenases, and (h) leukotriene-associated genes. N = 2 donors per group. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed. * in (f) indicates significantly lower PTGES2 expression of PBMCsec versus vehicle. † in (f) indicates significantly higher PTGES3 expression when comparing vehicle versus CD14. § in (f) indicates significantly lower PTGIR expression of vehicle versus CD14.* in (g) indicates significantly higher ALOX15 expression of vehicle versus CD14. † in (g) indicates significantly higher ALOX15AP expression when comparing vehicle versus CD14. § in (g) indicates significantly lower ALOX5 expression of vehicle versus CD14. * in (h) indicates significantly lower LTB4R expression in vehicle versus CD14.
To elucidate a potential underlying mechanism by which lipids abrogate expression of DC molecular marker, we assessed levels of genes reportedly involved in DC differentiation in our gene chip array and evaluated putative effects of PBMCsec on their expression. We first focused our analysis on genes involved in antioxidant mechanisms and genes encoding lipid-metabolizing enzymes. Expressions of nuclear factor erythroid 2-related factor 2 (NRF2) target genes and antioxidant genes remained largely unaffected by PBMCsec (supplementary Table S8). Only moderate effects of PBMCsec were detected when expressions of prostaglandin (PTG)-synthesizing enzymes and receptors were assessed (Fig 5f). Among arachinodate lipoxygenase genes (ALOX), ALOX15 was found remarkably downregulated by PBMCsec compared to vehicle medium (Fig 5g). Of note, ALOX5-activating protein (ALOX5AP) was strongly upregulated with secretome, while ALOX5 expression remained unaltered compared to vehicle (Fig 5g). Moreover, differentiation-associated downregulation of leukotriene A4 hydrolase LTA4H was impaired by PBMCsec, while LTB4R expression was not regulated by PBMC secretome (Fig 5h). These data suggest that PBMCsec prevents the DC differentiation-associated expression of ALOX15.
3.6. Lipids secreted by γ-irradiated PBMC compromise major MODC functions
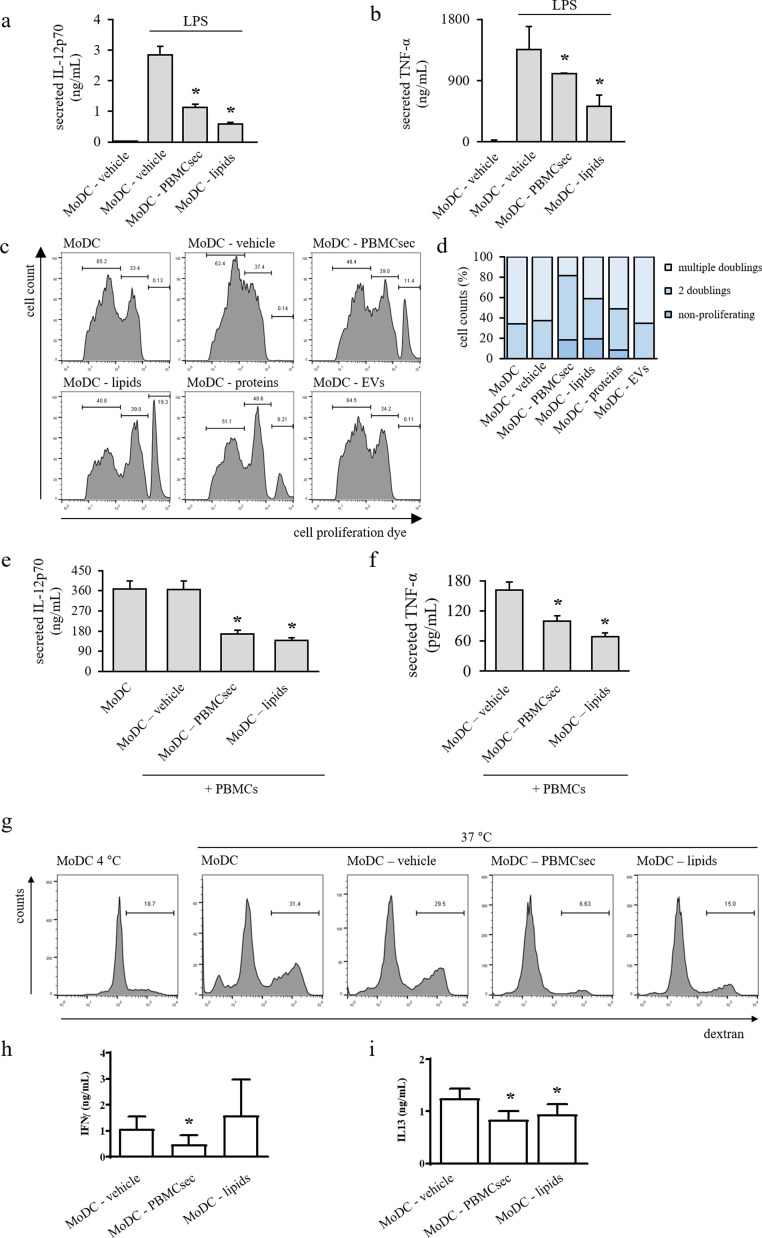
Based on the encouraging findings obtained from our analytical assessments, we furthermore sought to evaluate potency of MoDCs differentiated with PBMCsec and its fractions in a number of functional assays. First, we aimed to determine the effect of the lipid fraction of the secretome on LPS-induced release of DC cytokines. While LPS treatment caused vast secretion of IL-12p70 and TNF-α in MoDC-vehicle, we detected lower levels of respective cytokines with PBMCsec and lipids after LPS stimulation (Fig. 6a & b). Based on these results, we asked whether DC-mediated immune cell proliferation was compromised by PBMCsec. Therefore, DC maturation was induced by brief exposure to LPS and stimulatory properties of mature DCs were tested in an MLR with allogeneic, cell proliferation dye-labelled PBMCs (Fig. 6c & d). While few non-proliferating cells were detectable in MoDCs generated with differentiation medium alone, control medium, and EVs (0.12, 0.13, and 0.11%, respectively), PBMCsec and the lipid fraction, when added during MoDC differentiation, effectively prevented immune cell proliferation, indicated by 11.5 and 18.2% non-proliferating cells in MLR, respectively. We furthermore observed a moderate effect of PBMC-secreted proteins in cell proliferation-inducing abilities of MoDC (7.14% cell proliferation dyehigh cells with MoDC-proteins). Fewer cells having undergone two doublings were observed with differentiated MoDC, MoDC-vehicle, and MoDC-EVs compared to MoDC-PBMCsec, MoDC-lipids, and MoDC-proteins (27.4, 27.9, and 28.7% versus 34.01, 34.6, and 32.3%, respectively). Conversely, more cells with multiple doublings indicated by low proliferation dye signal were detected in MoDC with differentiation and control medium as well as with EVs (72.5, 71.6, and 71.2%, respectively), when compared to MoDC-PBMCsec, MoDC-lipids, and MoDC-proteins (54.4, 47.1, and 60.6%, respectively). Accordingly, we observed less secreted IL-12p70 and TNF-α during MLR with MoDC-PBMCsec compared to DCs differentiated with vehicle (Fig. 6e & f). Additionally, a functional dextran uptake assay revealed decreased antigen endocytosis by PBMCsec- and lipids-treated DCs compared to vehicle medium (Fig 6g).
Fig. 6.
PBMCsec and lipids functionally impair MoDCs. Amounts of (a) IL-12p70 and (b) TNF-α released by LPS-treated MoDC-PBMCsec and MoDC-lipids. * indicates p<.05 versus vehicle. N = 3 donors per group. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed. (c) Intensity of cell proliferation dye staining reflecting non-proliferating and proliferating cell populations in MLR. Numbers indicate frequencies per gate. Representative histograms of biological triplicates are shown. (d) Quantification of proliferating and non-proliferating cells. N = 3 donors per group. Concentrations of (e) secreted IL-12p70 and (f) secreted TNF-α during MLR with MoDC-medium, MoDC-PBMCsec, and MoDC-lipids. * denotes p<.05 versus vehicle. N = 3 donors per group. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed. (g) Dextran uptake of MoDC differentiated with control medium, PBMCsec, or lipids. Numbers indicate frequencies of cells per gate. One representative histogram of biological triplicates is shown. Secretion of (h) IFNγ and (i) IL-13 during co-culture of naïve T cell with MoDC differentiated in the presence of medium vehicle, PBMCsec, or lipids. * indicates p<.05 versus vehicle. N = 3 donors per group. Ordinary one-way ANOVA and Dunnett's multiple comparisons of all groups versus vehicle were performed.
A major role of mature DCs is the initiation of lymphocytic immune responses. Since we detected diminished expression of characteristic DC molecules and reduced secretion of pro-inflammatory mediators, we next sought to determine whether CD4+ T cell-priming activity of DCs was mitigated by PBMCsec and lipid treatment. To this end, mature MoDC differentiated with secretome, lipids, or vehicle were co-cultured with allogenic naïve, CD4+ T cells and release of TH1 and TH2 response-specific cytokines IFN-γ and IL13, respectively, was assessed. After 6 days of co-incubation, we found that secretion of both cytokines was diminished when MoDC were differentiated with PBMCsec or, to a lesser extent, with lipids compared to vehicle (Fig. 6h & i), indicating that the PBMC secretome delimits T cell-activating properties of DCs, presumably by compromising expression of DC marker molecules and release of inflammation-promoting cytokines. Since resolvins, lipid mediators with inflammation-resolving properties [71,72], are known to impair DC function, we quantified the presence of this lipid class in our secretome. As shown in supplementary Fig S7, PBMCsec indeed contains several resolvins, suggesting a potential role in the anti-inflammatory action of PBMCsec. Of note, all of the detected resolvins showed significantly lower levels in the secretome of non-irradiated PBMCs (not shown).
3.7. Lipids present in PBMCsec mitigate hapten-induced tissue swelling and reduce immune cell infiltration to sensitized areas
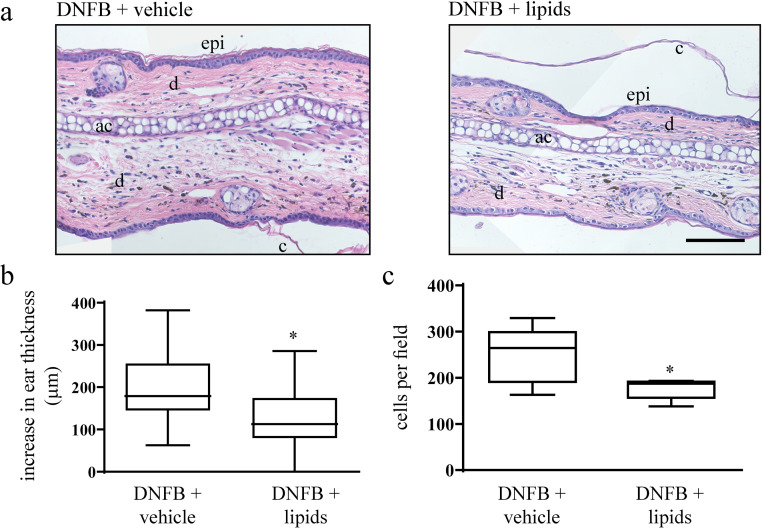
As we observed strong inhibitory effects of PBMCsec on murine CH in vivo and since we could identify lipid species as the predominant bioactive molecules mediating the observed anti-inflammatory effects of PBMCsec on DCs, we next asked whether lipids isolated from PBMCsec were capable of mitigating CH comparably to the extent of PBMCsec. To this end, we induced DNFB-mediated CH and topically applied PBMCsec-derived lipids. Treating ears of DNFB-sensitized mice with lipids caused reduced ear thickness (average 184.05 ± 20.5 µm increase in ear thickness with medium vehicle versus 130.3 ± 18.7 µm increase in ear thickness with lipids, p<.05) (Fig. 7a & b), comparable to, yet not as effective as the whole secretome (Fig. 1a & b). Moreover, amounts of infiltrating cells into the stroma were remarkably reduced by lipids compared to medium (176.6 ± 9.1 cells per field with lipids versus 248.6 ± 24.7 cells/field with medium; p<.05) (Fig 7c). In conclusion, our data suggest a strong therapeutic potential of lipids released by stressed PBMCs in inflammatory skin conditions.
Fig. 7.
Lipids secreted by stressed PBMCs alleviate symptoms of DNFB-induced CH. (a) Hematoxylin/eosin-stained ears 24 h after DNFB elicitation. Representative micrographs of n = 5 mice per treatment condition are shown. ac, auricular cartilage; c, cornified epidermis; d, dermis; epi, epidermis. Scale bar, 200 µm. (b) Increase in ear thickness determined by micrometric assessment 24 h post DNFB re-challenge. * denotes p<.05 versus vehicle. N = 5 mice per group. One-tailed student's t-test for paired samples was performed to compare PBMCsec versus vehicle. (c) Number of stromal cells per field. * indicates p<.05 versus vehicle. N = 5 mice per group. One-tailed student's t-test for paired samples was performed to compare PBMCsec versus vehicle.
4. Discussion
DCs, as sentinels of immunity, represent a clinically attractive target implicated in numerous disease aetiologies, including allograft rejection, autoimmune diseases, and inflammatory skin conditions. In the current study, we provide evidence that the secretome released by γ-irradiated PBMCs considerably affects the differentiation and function of MoDCs and differentiated DCs present in human skin. We were furthermore able to show that cells differentiated in the presence of PBMCsec showed significantly reduced cytokine secretion upon LPS stimulation. As a consequence, T cell-stimulatory capacities of PBMCsec-treated MoDC were dampened. In a recent study, Henry and colleagues demonstrated mitigated T cell activation as a result of reduced IL-12 secretion [73]. Apart from IL-12, we also found decreased IL-23 release in response to secretome treatment. Interestingly, DCs secreting IL-23 have been recently implicated in disease aetiology of atopic dermatitis, by polarizing CD4+ T cell subsets towards an inflammatory phenotype, eventually leading to epidermal thickening [74]. In line with these data, the reduced production of IL-12 and IL-23 by PBMCsec-treated DCs suggest a therapeutically beneficial effect of the secretome for inflamed skin conditions, such as CH.
Bioinformatics analysis of our transcriptomics data revealed that lipid transport and lipid metabolic processes were affected in MoDCs generated with addition of PBMCsec, thus corroborating a role for lipids in impaired DC differentiation. Further in vivo studies showed that indeed the lipid fraction of the secretome exerted strong effects in the CH mouse model. One explanation for our observation might be that oxidized phospholipids present in the secretome prevent binding and signaling of LPS, as it was recently demonstrated by Bochkov and colleagues [75]. In previous works of our group, we could show that PBMCsec contains a variety of non-oxidized, native phospholipids, phospholipids with hydroperoxides and hydroxides, lysophospholipid species, and phospholipids with carbonyl modifications [34,37]. In addition, we here show that different resolvins are present in PBMCsec, which have known immunomodulatory activity on DCs [72] and could therefore be involved in the observed effects of PBMCsec on DC function. Though oxidized phospholipids have been implicated in the maturation of DCs [43], we already observe a differentiation-impairing effect prior to maturation. Therefore, (oxidized) lipids most likely act independently of LPS signaling as described by Buchkov et al. in our settings. The exact underling mechanism by which PBMCsec-derived lipids exert their anti-inflammatory action, however, is still not known. Several modes of action are conceivable. Though effects of prostaglandins and leukotrienes on DCs have been reported previously [69,76,77], our chip data do not show differential regulation of prostaglandin- and leukotriene-associated genes by PBMCsec, suggesting a prostaglandin- and leukotriene-independent action of PBMCsec on DC function. Recently, the relevance of arachinodate 15-lipoxygenase in DC function [43] and dermal inflammation [78] was described. Rothe and colleagues reported enhanced DC maturation in the absence of ALOX15 evidenced by decreased levels of MHC class II molecules, costimulatory molecules, and activation markers. Moreover, diminished levels of the ALOX15 products PAPC—OH and PAPC—OOH were detected when ALOX15 was depleted. Conversely, these data could suggest that ALOX15-derived phospholipid oxidation products are required to attenuate DC maturation. However, our gene chip data revealed that PBMCsec abolished the differentiation-associated upregulation of ALOX15. Why DC differentiated in the presence of PBMCsec display impaired maturation in spite of low ALOX15 expression remains to be determined. Conceivably, exogenously added anti-inflammatory lipids present in PBMCsec, such as resolvins, might account for alleviated tissue inflammation while simultaneously interfering with the endogenous ALOX15 machinery of DCs. In this regard, it is worth mentioning that an Alox15-dependent mechanism of resolvin D2 production maintaining skin integrity by suppressing dermal inflammation has been recently described [78]. In addition, resolvin E1, a lipid mediator with inflammation-resolving properties, has been shown to impair DC motility, attenuate T cell priming, and prevent activation of effector T cells in murine contact hypersensitivity [78]. Interestingly, most of the previously mentioned works reported an immunosuppressive action via up-regulation of IL-10. In contrast, we could not detect IL-10 secretion by secretome-treated DCs in our cytokine profiler. In line with these data, Blueml demonstrated that the inhibitory effects of OxPAPC did not involve IL-10 [70], suggesting the presence of alternative mechanisms. Profiling the exact immunologically active lipid species secreted by irradiated PBMCs, e.g. by in-depth mass spectrometric analyses, and the lipid metabolism of secretome-treated DCs might deepen our understanding of lipid-mediated impaired DC differentiation.
Our bioinformatics analysis revealed significant differences between vehicle- and PBMCsec-treated human skin, suggesting diminished antigen presentation and processing and downregulated inflammatory processes. Contradictory data for both disease-promoting and regulatory functions of skin-resident DCs in CH have been repeatedly reported [79], [80], [81], [82]. In spite of ongoing debates about the exact involvements and contributions of different cell types to CH disease aetiology, we hypothesize that PBMCsec exerts anti-inflammatory effects on inflamed murine and human skin, at least partially, by modulating the immunogenic potential of skin-resident APCs. In line with this hypothesis, we observe only minor immunomodulatory actions of PBMCsec in an antigen-independent, TLR7-mediated mouse model of skin inflammation. Analysis of the local cytokine and chemokine milieu identified several immune mediators differentially expressed by PBMCsec, including IL-7, Ccl5, Cxcl12, oncostatin M (Osm), and adiponectin. The most relevant cytokines and chemokines and their roles in contact dermatitis have been described previously by Lee et al. [83]. Regulating the expressions of these molecules by PBMCsec might provide a potential mechanism by which PBMCsec contribute to alleviated CH symptoms. However, future studies will be necessary to elucidate the exact contribution of PBMCsec to changes of the local cytokine and chemokine milieu in inflammatory skin conditions.
In our experiments, PBMCsec was present during differentiation as well as during LPS-induced maturation. Effects of PBMCsec on differentiated DCs have been reported previously. Hoetzenecker et al. investigated the effect of PBMCsec on DC maturation and observed only minor effect on the expression of maturation markers and antigen uptake [38]. In contrast to this study, we observe diminished DC maturation when PBMCsec was added during differentiation and maturation. Moreover, antigen uptake was strongly reduced when DCs were differentiated in the presence of PBMCsec. Together, these data suggest that adding the secretome during differentiation rather than maturation is crucial for delimiting DC characteristics and functions.
Multiple cell types are involved in the induction of CH in the DNFB mouse model used in our study. Although we found only minor impact of the secretome in an antigen-independent model, we cannot completely rule out potential involvements of other cell types in the immunomodulatory action of PBMCsec. In addition to modulating APCs in the skin, PBMCsec might also modulate T cell activity, as effects on CD4+ T cell have been previously reported in an autoimmune myocarditis mouse model [38]. Delineating additional potential mechanism and other cell types modulated by PBMCsec treatment, e.g. by scRNAseq analyses, is subject of future investigations.
Safety and tolerability of topically applying PBMCsec has already been demonstrated in a phase I safety clinical trial (clinicaltrials.gov identifier: NCT02284360), where no therapy-related, serious adverse events were detected [84]. Furthermore, toxicological testing of intravenously administered PBMCsec was performed in two rodent models and subcutaneous tolerance was determined in minipigs [85]. In their study, Wuschko and colleagues observed no major toxicities or local signs of intolerance. Together, these data support the safe use of PBMCsec in various administration routes. Currently, an international, multi-center, randomized, double-blinded phase II clinical trial has been initiated to investigate the regenerative effects of topically applied PBMCsec in diabetic foot ulcer (DFU) with chronic non-healing wounds (EudraCT number 2018–001,653–27). This clinical trial will pave the way for future clinical administrations of PBMCsec in other inflammatory skin conditions.
Declaration of Competing Interest
The Medical University of Vienna has claimed financial interest and HJA holds patents related to this work (WO 2010/079,086 A1, WO 2010/070,105, PCT/EP2018/085,955, EPO 19,165,340.1, EPO application 19,219,342.3). All other authors declare no potential conflicts of interest.
Acknowledgments
Acknowledgments
We thank Dr. Hans Peter Haselsteiner for his belief in this private-public partnership to augment patients’ health. We thank Heidemarie Rossiter, BSc for critical review of the manuscript and Ing. Bahar Golabi and Barbara Lengauer for excellent technical assistance.
Funding sources
This research project was supported by the Austrian Research Promotion Agency (Vienna, Austria; Grant “APOSEC” 862068; 2015–2019) and the Vienna Business Agency (Vienna, Austria; Grant “APOSEC to clinic” 2343727). The funding sources had no involvement in study design, data collection, analysis, or interpretation, in writing of the report or in the decision to submit the manuscript for publication. The corresponding authors had full access to all data in the study and had final responsibility for the decision to submit for publication.
Author contributions
MM, HJA, and ML conceived and planned the experiments. ML, DC, LN, AG, AP, and VV performed the experiments. ML, MM, HJA, FG, and AG contributed to the interpretations of the results. ML and MM wrote the original manuscript. All authors reviewed the final manuscript.
Data availability
Raw data of single cell RNA sequencing and transcriptomics are available upon request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102774.
Contributor Information
Hendrik J. Ankersmit, Email: hendrik.ankersmit@meduniwien.ac.at.
Michael Mildner, Email: michael.mildner@meduniwien.ac.at.
Appendix. Supplementary materials
References
- 1.Inaba K., Young J.W., Steinman R.M. Direct activation of CD8+ cytotoxic T lymphocytes by dendritic cells. J Exp Med. 1987;166(1):182–194. doi: 10.1084/jem.166.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Inaba K., Metlay J.P., Crowley M.T., Steinman R.M. Dendritic cells pulsed with protein antigens in vitro can prime antigen-specific, MHC-restricted T cells in situ. J Exp Med. 1990;172(2):631–640. doi: 10.1084/jem.172.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinman R.M. Decisions about dendritic cells: past, present, and future. Annu Rev Immunol. 2012;30:1–22. doi: 10.1146/annurev-immunol-100311-102839. [DOI] [PubMed] [Google Scholar]
- 4.Collin M., Bigley V. Human dendritic cell subsets: an update. Immunology. 2018;154(1):3–20. doi: 10.1111/imm.12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K. In vivo analysis of dendritic cell development and homeostasis. Science. 2009;324(5925):392–397. doi: 10.1126/science.1170540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lutz M.B., Strobl H., Schuler G., Romani N. GM-CSF Monocyte-Derived Cells and Langerhans Cells As Part of the Dendritic Cell Family. Front Immunol. 2017;8:1388. doi: 10.3389/fimmu.2017.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeffel G., Wang Y., Greter M., See P., Teo P., Malleret B. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209(6):1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalod M., Chelbi R., Malissen B., Lawrence T. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J. 2014;33(10):1104–1116. doi: 10.1002/embj.201488027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bender A., Sapp M., Schuler G., Steinman R.M., Bhardwaj N. Improved methods for the generation of dendritic cells from nonproliferating progenitors in human blood. J Immunol Methods. 1996;196(2):121–135. doi: 10.1016/0022-1759(96)00079-8. [DOI] [PubMed] [Google Scholar]
- 10.Cella M., Scheidegger D., Palmer-Lehmann K., Lane P., Lanzavecchia A., Alber G. Ligation of CD40 on dendritic cells triggers production of high levels of interleukin-12 and enhances T cell stimulatory capacity: T-T help via APC activation. J Exp Med. 1996;184(2):747–752. doi: 10.1084/jem.184.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou L.J., Tedder T.F. CD14+ blood monocytes can differentiate into functionally mature CD83+ dendritic cells. Proc Natl Acad Sci U S A. 1996;93(6):2588–2592. doi: 10.1073/pnas.93.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuler G., Steinman R.M. Murine epidermal Langerhans cells mature into potent immunostimulatory dendritic cells in vitro. J Exp Med. 1985;161(3):526–546. doi: 10.1084/jem.161.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavrois M., Neidleman J., Kreisberg J.F., Greene W.C. In vitro derived dendritic cells trans-infect CD4 T cells primarily with surface-bound HIV-1 virions. PLoS Pathog. 2007;3(1):e4. doi: 10.1371/journal.ppat.0030004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu C.C., Di Meglio P., Nestle F.O. Harnessing dendritic cells in inflammatory skin diseases. Semin Immunol. 2011;23(1):28–41. doi: 10.1016/j.smim.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffner U.A., Maeda Y., Cooke K.R., Reddy P., Ordemann R., Liu C. Host dendritic cells alone are sufficient to initiate acute graft-versus-host disease. J Immunol. 2004;172(12):7393–7398. doi: 10.4049/jimmunol.172.12.7393. [DOI] [PubMed] [Google Scholar]
- 16.Peiser M., Tralau T., Heidler J., Api A.M., Arts J.H., Basketter D.A. Allergic contact dermatitis: epidemiology, molecular mechanisms, in vitro methods and regulatory aspects. Current knowledge assembled at an international workshop at BfR, Germany. Cell Mol Life Sci. 2012;69(5):763–781. doi: 10.1007/s00018-011-0846-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buttazzo S., Prodi A., Fortina A.B., Corradin M.T., Larese Filon F. Sensitization to Rubber Accelerators in Northeastern Italy: the Triveneto Patch Test Database. Dermatitis. 2016;27(4):222–226. doi: 10.1097/DER.0000000000000205. [DOI] [PubMed] [Google Scholar]
- 18.Montgomery R., Stocks S.J., Wilkinson S.M. Contact allergy resulting from the use of acrylate nails is increasing in both users and those who are occupationally exposed. Contact Derm. 2016;74(2):120–122. doi: 10.1111/cod.12497. [DOI] [PubMed] [Google Scholar]
- 19.de Groot A.C. New Contact Allergens: 2008 to 2015. Dermatitis. 2015;26(5):199–215. doi: 10.1097/DER.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 20.Gaspari A.A., Katz S.I., Martin S.F. Contact Hypersensitivity. Curr Protoc Immunol. 2016;113:1–7. doi: 10.1002/0471142735.im0402s113. 4.2. [DOI] [PubMed] [Google Scholar]
- 21.Orlic D., Kajstura J., Chimenti S., Jakoniuk I., Anderson S.M., Li B. Bone marrow cells regenerate infarcted myocardium. Nature. 2001;410(6829):701–705. doi: 10.1038/35070587. [DOI] [PubMed] [Google Scholar]
- 22.Rama P., Matuska S., Paganoni G., Spinelli A., De Luca M., Pellegrini G. Limbal stem-cell therapy and long-term corneal regeneration. N Engl J Med. 2010;363(2):147–155. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- 23.Bartunek J., Behfar A., Dolatabadi D., Vanderheyden M., Ostojic M., Dens J. Cardiopoietic stem cell therapy in heart failure: the C-CURE (Cardiopoietic stem Cell therapy in heart failURE) multicenter randomized trial with lineage-specified biologics. J Am Coll Cardiol. 2013;61(23):2329–2338. doi: 10.1016/j.jacc.2013.02.071. [DOI] [PubMed] [Google Scholar]
- 24.Kuo T.K., Hung S.P., Chuang C.H., Chen C.T., Shih Y.R., Fang S.C. Stem cell therapy for liver disease: parameters governing the success of using bone marrow mesenchymal stem cells. Gastroenterology. 2008;134(7):2111–2121. doi: 10.1053/j.gastro.2008.03.015. 21.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behfar A., Crespo-Diaz R., Terzic A., Gersh B.J. Cell therapy for cardiac repair–lessons from clinical trials. Nat Rev Cardiol. 2014;11(4):232–246. doi: 10.1038/nrcardio.2014.9. [DOI] [PubMed] [Google Scholar]
- 26.Gyongyosi M., Wojakowski W., Lemarchand P., Lunde K., Tendera M., Bartunek J. Meta-Analysis of Cell-based CaRdiac stUdiEs (ACCRUE) in patients with acute myocardial infarction based on individual patient data. Circ Res. 2015;116(8):1346–1360. doi: 10.1161/CIRCRESAHA.116.304346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gnecchi M., He H., Liang O.D., Melo L.G., Morello F., Mu H. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat Med. 2005;11(4):367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- 28.Gnecchi M., He H., Noiseux N., Liang O.D., Zhang L., Morello F. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20(6):661–669. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- 29.Mirotsou M., Zhang Z., Deb A., Zhang L., Gnecchi M., Noiseux N. Secreted frizzled related protein 2 (Sfrp2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc Natl Acad Sci U S A. 2007;104(5):1643–1648. doi: 10.1073/pnas.0610024104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lichtenauer M., Mildner M., Hoetzenecker K., Zimmermann M., Podesser B.K., Sipos W. Secretome of apoptotic peripheral blood cells (APOSEC) confers cytoprotection to cardiomyocytes and inhibits tissue remodelling after acute myocardial infarction: a preclinical study. Basic Res Cardiol. 2011;106(6):1283–1297. doi: 10.1007/s00395-011-0224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavo N., Zimmermann M., Pils D., Mildner M., Petrasi Z., Petnehazy O. Long-acting beneficial effect of percutaneously intramyocardially delivered secretome of apoptotic peripheral blood cells on porcine chronic ischemic left ventricular dysfunction. Biomaterials. 2014;35(11):3541–3550. doi: 10.1016/j.biomaterials.2013.12.071. [DOI] [PubMed] [Google Scholar]
- 32.Simader E., Beer L., Laggner M., Vorstandlechner V., Gugerell A., Erb M. Tissue-regenerative potential of the secretome of gamma-irradiated peripheral blood mononuclear cells is mediated via TNFRSF1B-induced necroptosis. Cell Death Dis. 2019;10(10):729. doi: 10.1038/s41419-019-1974-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beer L., Mildner M., Gyöngyösi M., Ankersmit H.J. Peripheral blood mononuclear cell secretome for tissue repair. Apoptosis. 2016;21(12):1336–1353. doi: 10.1007/s10495-016-1292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beer L., Zimmermann M., Mitterbauer A., Ellinger A., Gruber F., Narzt M.S. Analysis of the Secretome of Apoptotic Peripheral Blood Mononuclear Cells: impact of Released Proteins and Exosomes for Tissue Regeneration. Sci Rep. 2015;5:16662. doi: 10.1038/srep16662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasiri M.M., Beer L., Nemec L., Gruber F., Pietkiewicz S., Haider T. Dying blood mononuclear cell secretome exerts antimicrobial activity. Eur J Clin Invest. 2016;46(10):853–863. doi: 10.1111/eci.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacker S., Mittermayr R., Nickl S., Haider T., Lebherz-Eichinger D., Beer L. Paracrine Factors from Irradiated Peripheral Blood Mononuclear Cells Improve Skin Regeneration and Angiogenesis in a Porcine Burn Model. Sci Rep. 2016;6:25168. doi: 10.1038/srep25168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner T., Traxler D., Simader E., Beer L., Narzt M.S., Gruber F. Different pro-angiogenic potential of gamma-irradiated PBMC-derived secretome and its subfractions. Sci Rep. 2018;8(1):18016. doi: 10.1038/s41598-018-36928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoetzenecker K., Zimmermann M., Hoetzenecker W., Schweiger T., Kollmann D., Mildner M. Mononuclear cell secretome protects from experimental autoimmune myocarditis. Eur Heart J. 2015;36(11):676–685. doi: 10.1093/eurheartj/ehs459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altmann P., Mildner M., Haider T., Traxler D., Beer L., Ristl R. Secretomes of apoptotic mononuclear cells ameliorate neurological damage in rats with focal ischemia. F1000Res. 2014;3:131. doi: 10.12688/f1000research.4219.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haider T., Hoftberger R., Ruger B., Mildner M., Blumer R., Mitterbauer A. The secretome of apoptotic human peripheral blood mononuclear cells attenuates secondary damage following spinal cord injury in rats. Exp Neurol. 2015;267:230–242. doi: 10.1016/j.expneurol.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 41.Laggner M., Gugerell A., Bachmann C., Hofbauer H., Vorstandlechner V., Seibold M. Reproducibility of GMP-compliant production of therapeutic stressed peripheral blood mononuclear cell-derived secretomes, a novel class of biological medicinal products. Stem Cell Res Ther. 2020;11(1):9. doi: 10.1186/s13287-019-1524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mildner M., Hacker S., Haider T., Gschwandtner M., Werba G., Barresi C. Secretome of peripheral blood mononuclear cells enhances wound healing. PLoS ONE. 2013;8(3):e60103. doi: 10.1371/journal.pone.0060103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothe T., Gruber F., Uderhardt S., Ipseiz N., Rossner S., Oskolkova O. 12/15-Lipoxygenase-mediated enzymatic lipid oxidation regulates DC maturation and function. J Clin Invest. 2015;125(5):1944–1954. doi: 10.1172/JCI78490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gugerell A., Sorgenfrey D., Laggner M., Raimann J., Peterbauer A., Bormann D. Viral safety of APOSEC. Blood Transfus. 2019 doi: 10.2450/2019.0249-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 46.Bennett W.E., Cohn Z.A. The isolation and selected properties of blood monocytes. J Exp Med. 1966;123(1):145–160. doi: 10.1084/jem.123.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vorstandlechner V., Laggner M., Kalinina P., Haslik W., Radtke C., Shaw L. Deciphering the functional heterogeneity of skin fibroblasts using single-cell RNA sequencing. FASEB J. 2020 doi: 10.1096/fj.201902001RR. [DOI] [PubMed] [Google Scholar]
- 48.Rendeiro A.F., Krausgruber T., Fortelny N., Zhao F., Penz T., Farlik M. Chromatin mapping and single-cell immune profiling define the temporal dynamics of ibrutinib drug response in chronic lymphocytic leukemia. bioRxiv. 2019 doi: 10.1038/s41467-019-14081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd Comprehensive Integration of Single-Cell Data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becht E., McInnes L., Healy J., Dutertre C.A., Kwok I.W.H., Ng L.G. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 52.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lotia S., Montojo J., Dong Y., Bader G.D., Pico A.R. Cytoscape app store. Bioinformatics. 2013;29(10):1350–1351. doi: 10.1093/bioinformatics/btt138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ayush O., Lee C.H., Kim H.K., Im S.Y., Cho B.H., Lee H.K. Glutamine suppresses DNFB-induced contact dermatitis by deactivating p38 mitogen-activated protein kinase via induction of MAPK phosphatase-1. J Invest Dermatol. 2013;133(3):723–731. doi: 10.1038/jid.2012.373. [DOI] [PubMed] [Google Scholar]
- 55.Tomobe Y.I., Morizawa K., Tsuchida M., Hibino H., Nakano Y., Tanaka Y. Dietary docosahexaenoic acid suppresses inflammation and immunoresponses in contact hypersensitivity reaction in mice. Lipids. 2000;35(1):61–69. doi: 10.1007/s11745-000-0495-0. [DOI] [PubMed] [Google Scholar]
- 56.van der Fits L., Mourits S., Voerman J.S., Kant M., Boon L., Laman J.D. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182(9):5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 57.Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song X., Narzt M.S., Nagelreiter I.M., Hohensinner P., Terlecki-Zaniewicz L., Tschachler E. Autophagy deficient keratinocytes display increased DNA damage, senescence and aberrant lipid composition after oxidative stress in vitro and in vivo. Redox Biol. 2017;11:219–230. doi: 10.1016/j.redox.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47(D1):D419–DD26. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sukseree S., Chen Y.T., Laggner M., Gruber F., Petit V., Nagelreiter I.M. Tyrosinase-cre-mediated deletion of the autophagy gene Atg7 leads to accumulation of the RPE65 variant M450 in the retinal pigment epithelium of C57BL/6 Mice. PLoS ONE. 2016;11(8) doi: 10.1371/journal.pone.0161640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 63.Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lloberas N., Rama I., Llaudó I., Torras J., Cerezo G., Cassis L. Dendritic cells phenotype fitting under hypoxia or lipopolysaccharide; adenosine 5′-triphosphate-binding cassette transporters far beyond an efflux pump. Clin Exp Immunol. 2013;172(3):444–454. doi: 10.1111/cei.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindén O., Dohlsten M., Boketoft A., Hedlund G., Sjögren H.O. Catalase and lipopolysaccharide enhance proliferation in the rat mixed lymphocyte reaction. Scand J Immunol. 1987;26(3):223–228. doi: 10.1111/j.1365-3083.1987.tb02255.x. [DOI] [PubMed] [Google Scholar]
- 66.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ankersmit H.J., Hoetzenecker K., Dietl W., Soleiman A., Horvat R., Wolfsberger M. Irradiated cultured apoptotic peripheral blood mononuclear cells regenerate infarcted myocardium. Eur J Clin Invest. 2009;39(6):445–456. doi: 10.1111/j.1365-2362.2009.02111.x. [DOI] [PubMed] [Google Scholar]
- 68.Zeyda M., Säemann M.D., Stuhlmeier K.M., Mascher D.G., Nowotny P.N., Zlabinger G.J. Polyunsaturated fatty acids block dendritic cell activation and function independently of NF-kappaB activation. J Biol Chem. 2005;280(14):14293–14301. doi: 10.1074/jbc.M410000200. [DOI] [PubMed] [Google Scholar]
- 69.Jozefowski S., Biedroń R., Bobek M., Marcinkiewicz J. Leukotrienes modulate cytokine release from dendritic cells. Immunology. 2005;116(4):418–428. doi: 10.1111/j.1365-2567.2005.02241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blüml S., Kirchberger S., Bochkov V.N., Krönke G., Stuhlmeier K., Majdic O. Oxidized phospholipids negatively regulate dendritic cell maturation induced by TLRs and CD40. J Immunol. 2005;175(1):501–508. doi: 10.4049/jimmunol.175.1.501. [DOI] [PubMed] [Google Scholar]
- 71.Buckley C.D., Gilroy D.W., Serhan C.N. Proresolving lipid mediators and mechanisms in the resolution of acute inflammation. Immunity. 2014;40(3):315–327. doi: 10.1016/j.immuni.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sawada Y., Honda T., Hanakawa S., Nakamizo S., Murata T., Ueharaguchi-Tanada Y. Resolvin E1 inhibits dendritic cell migration in the skin and attenuates contact hypersensitivity responses. J Exp Med. 2015;212(11):1921–1930. doi: 10.1084/jem.20150381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Henry C.J., Ornelles D.A., Mitchell L.M., Brzoza-Lewis K.L., Hiltbold E.M. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol. 2008;181(12):8576–8584. doi: 10.4049/jimmunol.181.12.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoon J., Leyva-Castillo J.M., Wang G., Galand C., Oyoshi M.K., Kumar L. IL-23 induced in keratinocytes by endogenous TLR4 ligands polarizes dendritic cells to drive IL-22 responses to skin immunization. J Exp Med. 2016;213(10):2147–2166. doi: 10.1084/jem.20150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bochkov V.N., Kadl A., Huber J., Gruber F., Binder B.R., Leitinger N. Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature. 2002;419(6902):77–81. doi: 10.1038/nature01023. [DOI] [PubMed] [Google Scholar]
- 76.Harizi H., Juzan M., Pitard V., Moreau J.F., Gualde N. Cyclooxygenase-2-issued prostaglandin e(2) enhances the production of endogenous IL-10, which down-regulates dendritic cell functions. J Immunol. 2002;168(5):2255–2263. doi: 10.4049/jimmunol.168.5.2255. [DOI] [PubMed] [Google Scholar]
- 77.Harizi H., Grosset C., Gualde N. Prostaglandin E2 modulates dendritic cell function via EP2 and EP4 receptor subtypes. J Leukoc Biol. 2003;73(6):756–763. doi: 10.1189/jlb.1002483. [DOI] [PubMed] [Google Scholar]
- 78.Kim S.N., Akindehin S., Kwon H.J., Son Y.H., Saha A., Jung Y.S. Anti-inflammatory role of 15-lipoxygenase contributes to the maintenance of skin integrity in mice. Sci Rep. 2018;8(1):8856. doi: 10.1038/s41598-018-27221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Igyarto B.Z., Jenison M.C., Dudda J.C., Roers A., Müller W., Koni P.A. Langerhans cells suppress contact hypersensitivity responses via cognate CD4 interaction and langerhans cell-derived IL-10. J Immunol. 2009;183(8):5085–5093. doi: 10.4049/jimmunol.0901884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaplan D.H., Jenison M.C., Saeland S., Shlomchik W.D., Shlomchik M.J. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23(6):611–620. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 81.Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 2005;22(5):643–654. doi: 10.1016/j.immuni.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 82.Wang L., Bursch L.S., Kissenpfennig A., Malissen B., Jameson S.C., Hogquist K.A. Langerin expressing cells promote skin immune responses under defined conditions. J Immunol. 2008;180(7):4722–4727. doi: 10.4049/jimmunol.180.7.4722. [DOI] [PubMed] [Google Scholar]
- 83.Lee H.Y., Stieger M., Yawalkar N., Kakeda M. Cytokines and chemokines in irritant contact dermatitis. Mediators Inflamm. 2013;2013 doi: 10.1155/2013/916497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simader E., Traxler D., Kasiri M.M., Hofbauer H., Wolzt M., Glogner C. Safety and tolerability of topically administered autologous, apoptotic PBMC secretome (APOSEC) in dermal wounds: a randomized Phase 1 trial (MARSYAS I) Sci Rep. 2017;7(1):6216. doi: 10.1038/s41598-017-06223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wuschko S., Gugerell A., Chabicovsky M., Hofbauer H., Laggner M., Erb M. Toxicological testing of allogeneic secretome derived from peripheral mononuclear cells (APOSEC): a novel cell-free therapeutic agent in skin disease. Sci Rep. 2019;9(1):5598. doi: 10.1038/s41598-019-42057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data of single cell RNA sequencing and transcriptomics are available upon request.