Short abstract
Objective
Circulating microRNAs (miRNAs) have promising potential as diagnostic or prognostic biomarkers for hepatocellular carcinoma (HCC). This study aimed to analyze the clinical significance of serum exosomal miR-320a expression in patients with HCC.
Methods
A total of 104 patients with HCC, 55 patients with chronic liver disease (CLD), and 50 healthy volunteers were enrolled. Serum exosomal miR-320a levels were measured by quantitative reverse-transcriptase polymerase chain reaction and compared among the groups. The relationships between exosomal miR-320a levels and clinicopathological factors in patients with HCC were also analyzed.
Results
Serum exosomal miR-320a levels were significantly lower in patients with HCC compared with patients with CLD and healthy controls. Receiver-operating characteristic curve analysis showed that serum exosomal miR-320a had good diagnostic value for distinguishing between HCC subjects and normal controls. Serum exosomal miR-320a levels were significantly elevated 1 month after surgery in patients with HCC. Moreover, serum exosomal miR-320a downregulation was strongly associated with positive lymph node metastasis, positive vein invasion, advanced TNM stage, and shorter survival. Serum exosomal miR-320a was confirmed as an independent prognostic marker for HCC.
Conclusions
Collectively, these results indicate that serum exosomal miR-320a might be a potential biomarker for the detection and prognosis of HCC.
Keywords: Serum level, exosomal miR-320a, hepatocellular carcinoma, biomarker, prognosis, diagnosis
Introduction
According to the Chinese National Cancer Center, liver cancer was the third most common cancer in China, with an estimated 410,000 new cases, and the second leading cause of cancer-related death, with an estimated 330,000 deaths in 2012.1 Hepatocellular carcinoma (HCC) accounts for 75% to 85% of all primary liver cancers, and its incidence and mortality have continued to increase in recent decades.2,3 Unfortunately, the asymptomatic nature of early HCC means that its survival rate remains extremely poor because most patients remain undiagnosed until a late stage.4,5 HCC has thus become a threat to global health, and identifying novel diagnostic and prognostic markers is critical for improving the prognosis of this malignancy.
MicroRNAs (miRNAs) are a group of small, highly conserved, non-coding single RNA molecules consisting of 20 to 24 nucleotides. MiRNAs regulate gene expression by post-translational binding to the 3′ untranslated region of their target mRNAs, leading to inhibition of translation or destabilization of the mRNAs.6,7 Accumulating evidence has implicated miRNAs in multiple physiological and pathological processes, including cell growth, apoptosis, differentiation, and migration.8,9 Many miRNAs have been reported to be aberrantly expressed in HCC. For instance, miR-146a-5p,10 miR-342-3p,11 and miR-2212 were significantly downregulated in HCC and were positively correlated with a favorable prognosis in patients with HCC, while miR-892a,13 miR-92a,14 and miR-65015 were overexpressed in HCC and associated with a poor prognosis, suggesting that these miRNAs might act as tumor suppressors or oncogenes, respectively, in HCC.
Exosomes are membrane vesicles with a maximum size of 150 nm that exist in urine, serum, plasma, breast milk, saliva, and other body fluids.16,17 Extracellular miRNAs in exosomes are relatively stable and may serve as noninvasive biomarkers in several types of cancer, including HCC. MiR-320a has been demonstrated to play a tumor suppressive role in HCC and downregulation of exosomal miR-320a in cancer-associated fibroblasts promoted HCC tumorigenesis, indicating that exosomal miR-320a levels secreted from the tumor microenvironment might be associated with the prognosis of HCC.20 However, to the best of our knowledge, expression levels of serum exosomal miR-320a and its potential clinical significance in HCC remain unclear. We therefore detected serum levels of exosomal miR-320a in patients with HCC and assessed their correlations with clinical features and survival in these patients.
Materials and methods
Study population
This study was reviewed and authorized by the Ethics Committee of Qingdao No.6 People’s Hospital and written informed consent was obtained from all participants. We enrolled patients with HCC or CLD who received treatment at Qingdao No.6 People’s Hospital from January 2012 to December 2013. The inclusion criteria for patients in HCC group were as follows: (1) pathologically confirmed HCC; (2) no treatment including chemotherapy, radiotherapy, transarterial chemoembolization, or ablation before collection of first blood samples; and (3) complete follow-up and clinical data. The exclusion criteria for HCC cases were as follows: (1) HCC not confirmed by histopathological examination; (2) patients who had received treatment before collection of the first blood sample; (3) and patients who rejected treatment, dropped out, or whose information was lost. CLD cases were confirmed by medical imaging, laboratory tests, and clinical symptoms. Patients with CLD who had received any treatment before collection of the first blood sample were excluded. Histological classification and clinical staging of HCC were based on the Barcelona Clinic Liver Cancer (BCLC) staging system. Healthy volunteers were enrolled as controls.
Overall survival (OS) was defined as the time from the day of diagnosis to the date of death or last follow-up. Disease-free survival (DFS) was defined as the time from the day of diagnosis to the date of relapse or last follow-up.
Up to 5 mL of peripheral blood was withdrawn from each participant before receiving any treatment and second blood samples were taken from HCC patients 1 month after surgery, centrifuged at 3000 ×g for 10 minutes within 2 hours, and the supernatant was stored at −80°C for further use. All blood samples were handled anonymously according to ethical and legal standards.
Exosomal RNA isolation and quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
Total exosomes were isolated using ExoQuick Exosome Precipitation Solution (System Biosciences, Mountain View, CA, USA) based on the manufacturer’s protocols. The exosomal pelleted fraction extracted from serum was resuspended in nuclease-free water. Total RNA was then extracted from the exosomes using a mirVana PARIS Kit (Ambion, Austin, TX, USA). During the RNA isolation step, 20 fmol of synthetic non-human cel-miR-39 was added to each sample. RNA concentration and purity were assessed using an Agilent Bioanalyzer 2100 with a Small RNA Chip (Agilent Technologies, Santa Clara, CA, USA). Reverse transcription was carried out with a TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Real-time PCR was performed on an ABI 7500 Fast Real-Time PCR System (Applied Biosystems) Each experiment was carried out in triplicate. Cel-miR-39 was used as a control, and the relative fold-change in serum exosomal miR-320a expression was calculated using the 2–ΔΔCt method.
Statistical analysis
Serum exosomal miR-320a expression levels were compared among the groups using the Mann–Whitney U test or Kruskal–Wallis test. The relationships between serum exosomal miR-320a expression and clinical features were evaluated by χ2 tests. The area under the receiver operating characteristic (ROC) curve (AUC) was analyzed to assess the diagnostic value of serum exosomal miR-320a. Cumulative OS and DFS rates were calculated by the Kaplan–Meier method and compared by log-rank tests. Univariate and multivariate Cox regression analyses were used to determine the factors related to OS. Statistical significance was defined as P < 0.05. All statistical analyses were performed using MedCalc 16.4.3 (MedCalc, Ostend, Belgium).
Results
Patients
We enrolled 104 patients with HCC, 55 with chronic liver disease (CLD), and 50 healthy controls. The clinical features of the participants are listed in Table 1. All HCC patients underwent surgery and also received similar adjuvant chemotherapy throughout the entire treatment period.
Table 1.
Demographic characteristics of the study subjects.
| HCC (n = 104) | CLD (n = 55) | Controls (n = 50) | |
|---|---|---|---|
| Age, years | |||
| <60 | 48 | 24 | 23 |
| ≥60 | 56 | 31 | 27 |
| Sex | |||
| Male | 77 | 38 | 32 |
| Female | 27 | 17 | 18 |
| Etiology | |||
| HCV | 6 | 8 | – |
| HBV | 68 | 39 | – |
| HBV + HCV | 14 | 6 | – |
| Alcohol | 12 | 2 | – |
| Autoimmune | 4 | – | – |
HCC, hepatocellular carcinoma; CLD, chronic liver disease; HCV, hepatitis C virus; HBV, hepatitis B virus.
Down-regulation of serum exosomal miR-320a in HCC patients
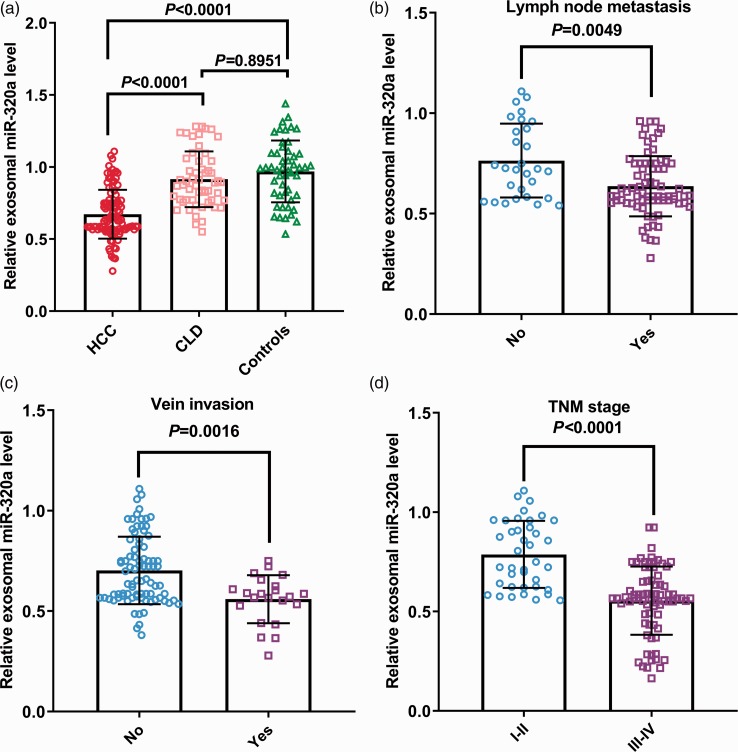
Serum exosomal miR-320a levels in patients with HCC or CLD and in healthy controls were detected by qRT-PCR. Serum exosomal miR-320a levels were significantly lower in patients with HCC compared with CLD patients and controls (both P < 0.0001) (Figure 1a). However, there was no significant difference between CLD patients and controls. Moreover, serum exosomal miR-320a expression levels were also significantly lower in patients with LNM compared with those without LNM (P = 0.0049) (Figure 1b) and in patients with vein invasion compared with those without vein invasion (P = 0.0016) (Figure 1c). Serum exosomal miR-320a levels were significantly higher in patients with early-stage (I/II) compared with advanced-stage HCC (III/IV) (P < 0.0001) (Figure 1d).
Figure 1.
(a) Serum exosomal miR-320a levels were significantly lower in patients with hepatocellular carcinoma (HCC). Among patients with HCC, serum exosomal miR-320a levels were significantly lower in patients with lymphatic metastasis (b) and distant metastasis (c) compared with those without, and in patients with advanced-stage compared with early-stage HCC (d).
Association between serum exosomal miR-320a and clinicopathological characteristics
Among the 104 patients with HCC, 49 subjects were assigned to the high-serum exosomal miR-320a expression group and the other 55 to the low-expression group based on the median serum exosomal miR-320a expression level. Low serum exosomal miR-320a expression was closely correlated with LNM (P = 0.0194), vein invasion (P = 0.0099), and TNM stage (P = 0.0005), but not with any other clinical parameters (Table 2).
Table 2.
Correlations between miR-320a expression and clinicopathological parameters in patients with hepatocellular carcinoma.
| Parameter | Cases | Serum exosomal miR-320a low | Serum exosomal miR-320a high | P |
|---|---|---|---|---|
| Age, years | 0.8084 | |||
| <60 | 48 | 26 | 22 | |
| ≥60 | 56 | 29 | 27 | |
| Sex | 0.7466 | |||
| Male | 77 | 40 | 37 | |
| Female | 27 | 15 | 12 | |
| AFP, ng/mL | 0.1433 | |||
| <20 | 37 | 16 | 21 | |
| ≥20 | 67 | 39 | 28 | |
| Differentiation | 0.1212 | |||
| Well | 19 | 7 | 12 | |
| Moderate/poor | 85 | 48 | 37 | |
| Liver cirrhosis | 0.1985 | |||
| No | 42 | 19 | 23 | |
| Yes | 62 | 36 | 26 | |
| Vein invasion | 0.0099 | |||
| No | 82 | 38 | 44 | |
| Yes | 22 | 17 | 5 | |
| TNM stage | 0.0005 | |||
| I–II | 39 | 12 | 27 | |
| III–IV | 65 | 43 | 22 | |
| Lymph node metastasis | 0.0194 | |||
| No | 29 | 10 | 19 | |
| Yes | 75 | 45 | 30 |
AFP, alfa-fetoprotein.
Diagnostic value of serum exosomal miR-320a in HCC and treatment response
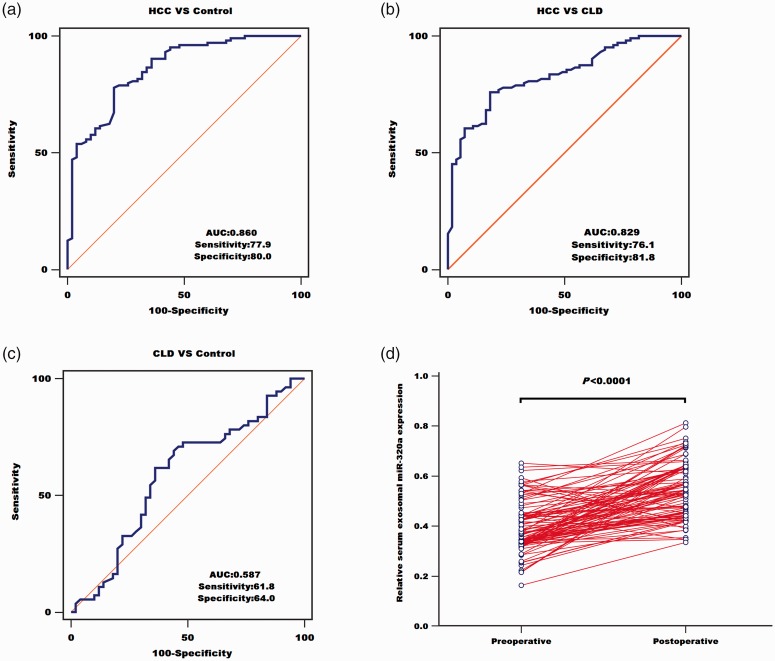
We also analyzed the diagnostic value of serum exosomal miR-320a. ROC curve analysis showed that the AUC value for serum exosomal miR-320a was 0.860 with a sensitivity of 77.9% and specificity of 80.0%, indicating that serum exosomal miR-320a could discriminate between patients with HCC and healthy subjects (Figure 2a). Serum exosomal miR-320a also yielded an AUC of 0.829 with 76.1% sensitivity and 81.8% specificity for differentiating between HCC and CLD patients (Figure 2b). However, its AUC for differentiating between CLD patients and controls was only 0.587 with 61.8% sensitivity and 64.0% specificity (Figure 2c). We also collected blood samples from all participants 1 month after their treatment, and measured the expression levels of serum exosomal miR-320a. As expected, serum exosomal miR-320a levels were significantly increased in postoperative compared with preoperative blood samples (P < 0.0001) (Figure 2d).
Figure 2.
Serum exosomal miR-320a was used to distinguish between patients with hepatocellular carcinoma (HCC) and healthy controls (a), between patients with HCC and those with chronic liver disease (CLD) (b), and between patients with CLD and healthy controls (c). (d) Serum exosomal miR-320a levels in patients with HCC were significantly elevated after treatment. AUC, area under the curve.
Prognostic value of serum exosomal miR-320a in HCC patients
Kaplan–Meier survival analysis revealed that patients in the low-serum exosomal miR-320a expression group had poorer OS than those in the high-expression group (P = 0.0032) (Figure 3a). Similarly, patients with higher serum exosomal miR-320a expression had significantly better DFS than those with lower expression (P = 0.0019) (Figure 3b).
Figure 3.
Kaplan–Meier curve estimates of the relationships between serum exosomal miR-320a and overall survival (a) and disease-free survival (b) in all patients with hepatocellular carcinoma.
Univariate and multivariate analyses showed that LNM (hazard ratio (HR) = 1.931, 95% confidence interval (CI) = 1.121–2.845, P = 0.023; HR = 2.678, 95% CI = 1.407–4.132, P = 0.011; respectively), vein invasion (HR = 2.163, 95% CI = 1.124–3.347, P = 0.015; HR = 2.817, 95% CI = 1.368–4.619, P = 0.010; respectively), TNM stage (HR = 2.595, 95% CI = 1.405–3.830, P = 0.012; HR = 3.258, 95% CI = 1.623–5.137, P = 0.004; respectively), and serum exosomal miR-320a (HR = 2.412, 95% CI = 1.337–3.714, P = 0.013; HR = 2.974, 95% CI = 1.562–4.628, P = 0.008; respectively) were strongly associated with OS, and these were recognized as independent prognostic factors in patients with HCC (Table 3).
Table 3.
Univariate and multivariate analyses of overall survival in 104 patients with hepatocellular carcinoma.
| Independent variable |
Univariate analysis |
Multivariate analysis |
||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P | HR | 95% CI | P | |
| Serum exosomal miR-320a | 2.412 | 1.337–3.714 | 0.013 | 2.974 | 1.562–4.628 | 0.008 |
| Vein invasion | 2.163 | 1.124–3.347 | 0.015 | 2.817 | 1.368–4.619 | 0.010 |
| Lymph node metastasis | 1.931 | 1.121–2.845 | 0.023 | 2.678 | 1.407–4.132 | 0.011 |
| TNM stage | 2.595 | 1.405–3.830 | 0.012 | 3.258 | 1.623–5.137 | 0.004 |
HR, hazard ratio; CI, confidence interval.
Discussion
The results of this study showed that serum exosomal miR-320a levels were greatly decreased in patients with HCC compared with patients with CLD and healthy controls. In addition, low serum exosomal miR-320a expression levels were significantly and positively linked to LNM, vein invasion, and advanced TNM stage. ROC analysis demonstrated that serum exosomal miR-320a had high sensitivity and specificity for the diagnosis of HCC. Serum exosomal miR-320a was also useful for monitoring treatment response. Moreover, serum exosomal miR-320a expression was frequently lower in HCC patients with poorer OS and DFS. Finally, univariate and multivariate analyses confirmed that serum exosomal miR-320a was an independent prognostic indicator in patients with HCC. Collectively, these results indicated that serum exosomal miR-320a might act as a tumor suppressor gene in HCC.
Consistent with the current results, previous studies19 found that miR-320a expression was reduced in HCC tissues and cells, while overexpression of miR-320a inhibited cancer cell proliferation and invasion in vitro by targeting c-Myc. Similarly, Lv et al.20 demonstrated that miR-320a expression was significantly downregulated in HCC tissues, and ectopic expression of miR-320a suppressed carcinogenesis in vitro by regulating HMGB1 expression. Moreover, miR-320a expression in cancer-associated fibroblast-derived exosomes was significantly downregulated compared with levels in para-cancerous fibroblasts,18 and in vitro and in vivo evidence also revealed that miR-320a overexpression inhibited carcinogenesis.18
MiR-320a has previously been reported in various tumor types. Overexpression of miR-320a significantly inhibited tumorigenesis in gastric cancer by regulating RAB14 and ADAM10.21,22 In addition, Fang et al.23 showed that plasma miR-320a levels were lower in patients with colorectal cancer (CRC) compared with normal controls, and were elevated in patients after surgical treatment. MiR-320a was frequently decreased in both CRC cell lines and colon cancer tissues.24 Restoration of miR-320a remarkably inhibited cell migration and invasion by targeting Rac124 and β-catenin.25 Interestingly, miR-320a expression was reduced in liver metastasis CRC tissues compared with primary cancerous tissues.26 MiR-320a was also decreased in multiple myeloma cell lines, and reduced miR-320a dramatically promoted cancer cell viability, inhibited apoptosis in vitro, and enhanced tumorigenicity in a mouse xenograft model.27 MiR-320a levels were downregulated in cancer specimens from patients with bladder transitional cell carcinoma.28 MiR-320a upregulation inhibited tumorigenicity by repressing the expression of integrin beta chain beta 3, and vice versa.28 Low expression of miR-320a was found in breast cancer tissue samples and cell lines, and its downregulation was associated with shortened survival and positive distant metastasis.29 Furthermore, ectopic expression of miR-320a dramatically suppressed the proliferation, migration, and invasion of breast cancer cells in vitro and tumor growth in vivo by silencing RAB14,30 RAB11A,31 or MTDH.32 Forced miR-320a expression led to reduced insulin-like growth factor 1 receptor (IGF-1R) and voltage-dependent anion-selective channel 1 levels in non-small cell lung cancer, and depletion of miR-320a significantly increased cell growth and invasion both in vitro and in vivo.33,34 MiR-320a levels were also decreased in cancerous glioma tissues and cell lines, and increased expression of miR-320a was inversely associated with tumor grade and patient survival. Additionally, in vitro and in vivo analyses revealed that miR-320a inhibited cancer cell proliferation and increased survival in a mouse model by degrading IGF-1R,35 staphylococcal nuclease domain-containing protein, and β-catenin.36 Qi et al. showed that miR-320a expression was greatly downregulated in nasopharyngeal carcinoma tissues and cell lines, and high miR-320a expression restrained cell growth, migration, and invasion in vivo by downregulating B lymphoma Mo-MLV insertion region 1 homolog protein.37 Similarly, miR-320a was also decreased in HCC tissues and cells lines and miR-320a overexpression suppressed tumor cell proliferation by regulating c-Myc.19 Overall, these results suggested that miR-320a exerted a suppressive role in different types of cancers.
This study was limited by its relatively small sample size, and further, large-scale prospective cohort studies are warranted to validate our findings. In addition, the molecular mechanism of exosomal miR-320a in HCC tumorigenesis also needs further investigation.
In conclusion, the current study demonstrated that serum exosomal miR-320a levels were reduced and were associated with an unfavorable prognosis in patients with HCC. Serum exosomal miR-320a might thus be a reliable marker for monitoring the progression of HCC.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
ORCID iD
Wenjing Dong https://orcid.org/0000-0003-1822-112X
References
- 1.Chen W, Zheng R, Zeng H, et al. The incidence and mortality of major cancers in China, 2012. Chin J Cancer 2016; 35: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Zucman-Rossi J, Pikarsky E, et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2016; 2: 16018. [DOI] [PubMed] [Google Scholar]
- 4.Kinoshita A, Onoda H, Fushiya N, et al. Staging systems for hepatocellular carcinoma: current status and future perspectives. World J Hepatol 2015; 7: 406–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carr BI. Hepatocellular carcinoma: current management and future trends. Gastroenterology 2004; 127: S218–S224. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001; 294: 862–864. [DOI] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11: 597–610. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Shen XJ, Zou Q, et al. Biological functions of microRNAs: a review. J Physiol Biochem 2011; 67: 129–139. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Ye ZH, Liang HW, et al. Down-regulation of miR-146a-5p and its potential targets in hepatocellular carcinoma validated by a TCGA- and GEO-based study. FEBS Open Bio 2017; 7: 504–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao Y, Zhang SG, Wang ZH, et al. Down-regulation of miR-342-3p in hepatocellular carcinoma tissues and its prognostic significance. Eur Rev Med Pharmacol Sci 2017; 21: 2098–2102. [PubMed] [Google Scholar]
- 12.Luo LJ, Zhang LP, Duan CY, et al. The inhibition role of miR-22 in hepatocellular carcinoma cell migration and invasion via targeting CD147. Cancer Cell Int 2017; 17: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia B, Tan L, Jin Z, et al. MiR-892a promotes hepatocellular carcinoma cells proliferation and invasion through targeting CD226. J Cell Biochem 2017; 118: 1489–1496. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Wu J, Xie C. MiR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting. Iran J Basic Med Sci 2017; 20: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng ZL, Li FJ, Gao F, et al. Upregulation of miR-650 is correlated with down-regulation of ING4 and progression of hepatocellular carcinoma. J Surg Oncol 2013; 107: 105–110. [DOI] [PubMed] [Google Scholar]
- 16.Lässer C. Exosomal RNA as biomarkers and the therapeutic potential of exosome vectors. Expert Opin Biol Ther 2012; 12: S189–S197. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Li C, Zhou T, et al. Role of exosomal proteins in cancer diagnosis. Mol Cancer 2017; 16: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Z, Li X, Sun W, et al. Loss of exosomal miR-320a from cancer-associated fibroblasts contributes to HCC proliferation and metastasis. Cancer Lett 2017; 397: 33–42. [DOI] [PubMed] [Google Scholar]
- 19.Xie F, Yuan Y, Xie L, et al. MiRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther 2017; 10: 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv G, Wu M, Wang M, et al. MiR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int 2017; 37: 1354–1364. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Liu H, Shao J, et al. MiR-320a serves as a negative regulator in the progression of gastric cancer by targeting RAB14. Mol Med Rep 2017; 16: 2652–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ge X, Cui H, Zhou Y, et al. MiR-320a modulates cell growth and chemosensitivity via regulating ADAM10 in gastric cancer. Mol Med Rep 2017; 16: 9664–9670. [DOI] [PubMed] [Google Scholar]
- 23.Fang Z, Tang J, Bai Y, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res 2015; 34: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Dong T, Zhou H, et al. MiR-320a suppresses colorectal cancer progression by targeting Rac1. Carcinogenesis 2014; 35: 886–895. [DOI] [PubMed] [Google Scholar]
- 25.Sun JY, Huang Y, Li JP, et al. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem Biophys Res Commun 2012; 420: 787–792. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, He X, Liu Y, et al. MicroRNA-320a inhibits tumor invasion by targeting neuropilin 1 and is associated with liver metastasis in colorectal cancer. Oncol Rep 2012; 27: 685–694. [DOI] [PubMed] [Google Scholar]
- 27.Lu Y, Wu D, Wang J, et al. MiR-320a regulates cell proliferation and apoptosis in multiple myeloma by targeting pre-B-cell leukemia transcription factor 3. Biochem Biophys Res Commun 2016; 473: 1315–1320. [DOI] [PubMed] [Google Scholar]
- 28.Shang C, Zhang H, Guo Y, et al. MiR-320a down-regulation mediates bladder carcinoma invasion by targeting ITGB3. Mol Biol Rep 2014; 41: 2521–2527. [DOI] [PubMed] [Google Scholar]
- 29.Yang H, Yu J, Wang L, et al. MiR-320a is an independent prognostic biomarker for invasive breast cancer. Oncol Lett 2014; 8: 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Wang L, Yang H, et al. Rab14 suppression mediated by miR-320a inhibits cell proliferation, migration and invasion in breast cancer. J Cancer 2016; 7: 2317–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Yang Z, Wang H, et al. MicroRNA-320a inhibits proliferation and invasion of breast cancer cells by targeting RAB11A. Am J Cancer Res 2015; 5: 2719–2729. [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, Wang JG, Zhang L, et al. MicroRNA-320a inhibits breast cancer metastasis by targeting metadherin. Oncotarget 2016; 7: 38612–38625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Shi C, Wang J, et al. MicroRNA-320a is downregulated in non-small cell lung cancer and suppresses tumor cell growth and invasion by directly targeting insulin-like growth factor 1 receptor. Oncol Lett 2017; 13: 3247–3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, Jiang G, Wang C, et al. Decreased expression of microRNA-320a promotes proliferation and invasion of non-small cell lung cancer cells by increasing VDAC1 expression. Oncotarget 2016; 7: 49470–49480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo T, Feng Y, Liu Q, et al. MicroRNA-320a suppresses in GBM patients and modulates glioma cell functions by targeting IGF-1R. Tumour Biol 2014; 35: 11269–11275. [DOI] [PubMed] [Google Scholar]
- 36.Li H, Yu L, Liu J, et al. MiR-320a functions as a suppressor for gliomas by targeting SND1 and β-catenin, and predicts the prognosis of patients. Oncotarget 2017; 8: 19723–19737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qi X, Li J, Zhou C, et al. MicroRNA-320a inhibits cell proliferation, migration and invasion by targeting BMI-1 in nasopharyngeal carcinoma. FEBS Lett 2014; 588: 3732–3738. [DOI] [PubMed] [Google Scholar]