Abstract
This study establishes the applicability of imine-based dynamic combinatorial chemistry to discover non-covalent ligands for RNA targets. We elucidate properties underlying the reactivity of arylamines and demonstrate target-guided amplification of tight binders in an amiloride-based dynamic library.
Graphical Abstract

RNA molecules are increasingly recognized and pursued as novel targets that would expand the scope of “druggable” space, and several promising RNA-binding small molecules have been discovered.1–4 Successful approaches to RNA ligand discovery have included scaffold-based synthesis, screening of general and RNA-biased libraries, sequence- and structure-based design, ensemble-based virtual screening, modular assembly of multivalent ligands, fragment-based screening, and dynamic combinatorial chemistry.2, 5–24
While successful in specific cases, many of these approaches suffer from technical challenges that limit generalizability. In the case of microarray-based screening, potential ligands can be missed if surface-immobilization greatly affects their binding to RNA. Screening methods that involve labeling RNA with a fluorophore are limited by the potential impact of the label on the RNA conformational landscape. Indicator displacement assays provide a label-free solution-based alternative, however, these assays are limited to small RNA constructs. Computational prediction of ligands based on the RNA secondary structure motifs,25 while promising for some systems, has not been developed for RNAs with highly folded binding pockets. Finally, structure-based design is difficult due to inherent challenges with solving three-dimensional structures of large complex RNAs at high resolution and the importance of dynamics in recognition, though advances are rapidly being made.26
Dynamic combinatorial chemistry (DCC), on the other hand, is a uniquely poised approach as it allows tandem in situ synthesis and screening of a large diverse library of ligands27–29 and has the potential to allow targeting of RNAs with a wide range of complexity without a priori knowledge of RNA structure or dynamics. Miller and co-workers demonstrated the utility of this approach by using disulfide exchange to generate a dynamic resin-bound library of 11,325 members that has led to the selection of bioactive ligands for RNA targets in HIV and myotonic dystrophy.19 One limitation of this method has been the use of disulfide chemistry, which requires replacement of the exchanging bond with a bio-isostere.30
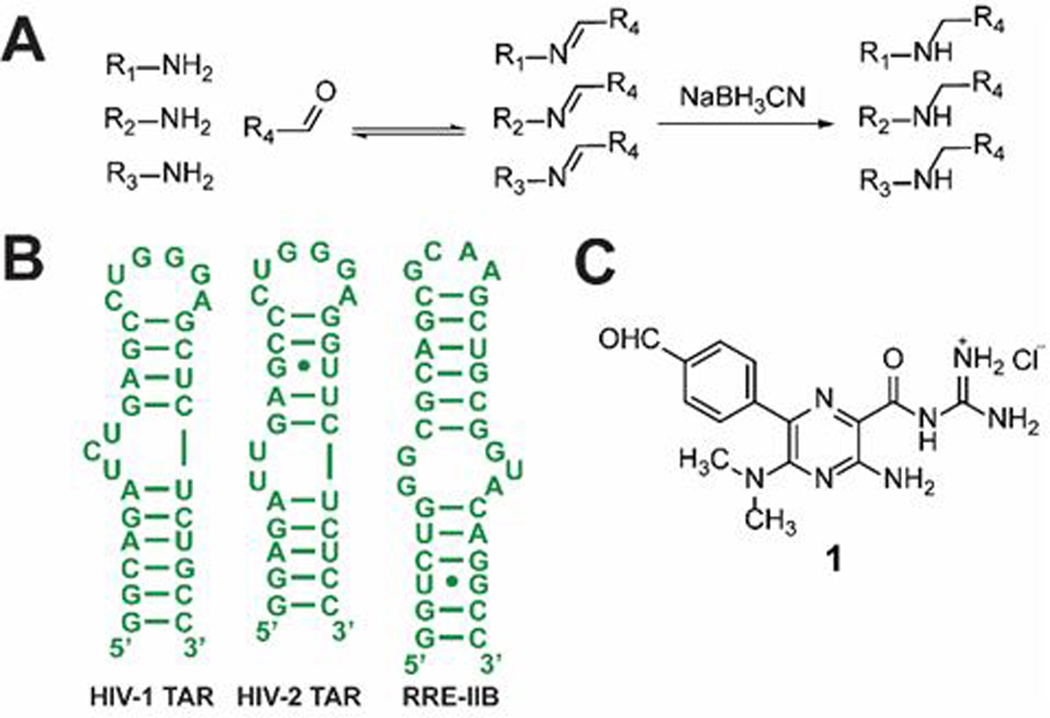
To develop a complementary method that addresses some of the limitations of current approaches to RNA-ligand discovery, we worked within the area of template-guided ligand selection by DCC. We expanded on work previously done in this area by introducing imine exchange chemistry (Fig. 1A).31 Imine exchange also allows access to diverse ligands as researchers can utilize the large selection of amines and aldehydes or ketones that are commercially available.32 Rayner and co-workers used imine-based DCC to conjugate nucleosides at the 3’ end of an RNA construct, supporting the compatibility of imine-based DCC with RNA.33 Recently, Dash and co-workers showed the promise of imine-based DCC for nucleic acids by targeting G-Quadruplex DNA.34 However, the target in this work was conjugated on a magnetic nanoplatform, a format that has potential to affect ligand binding by altering the conformational landscape of the target, especially in the case of RNA that has a highly dynamic nature.35 Our efforts to adapt imine-based DCC for RNA targets were thus focused on developing a general, solution-based method that applies to many different systems.
Fig. 1.
Model system for DCC studies. (A) Representation of imine exchange. Addition of a reducing agent locks the imine library into stable secondary amines; (B) Structures of RNA constructs; (C) Aldehyde analogue of the amiloride scaffold.
To determine the applicability of solution imine-based DCC to RNA systems we sought to identify the types of amines amenable to DCC under certain conditions, the amplification factors that can be expected, and how the relative amplification factors relate to the relative binding affinities of the library members. As a model system, we used an aldehyde analogue of an amiloride-based scaffold shown to be tunable for differential binding to multiple RNAs (Fig. 1C).36, 37 A tunable scaffold is particularly desirable for these studies because it has the potential to generate both tight and weak binders. Three RNA constructs were chosen for this study (Fig. 1B): the HIV-1 transactivation response element (TAR) for which the amiloride scaffold was initially optimized, the related RNA HIV-2 TAR, and stem IIB of the HIV-1 Rev response element (RRE-IIB).38 These RNAs make an excellent model system for RNA-small molecule interactions as they have been extensively studied in efforts to discover novel anti-HIV therapeutics, and their biological roles are well understood.
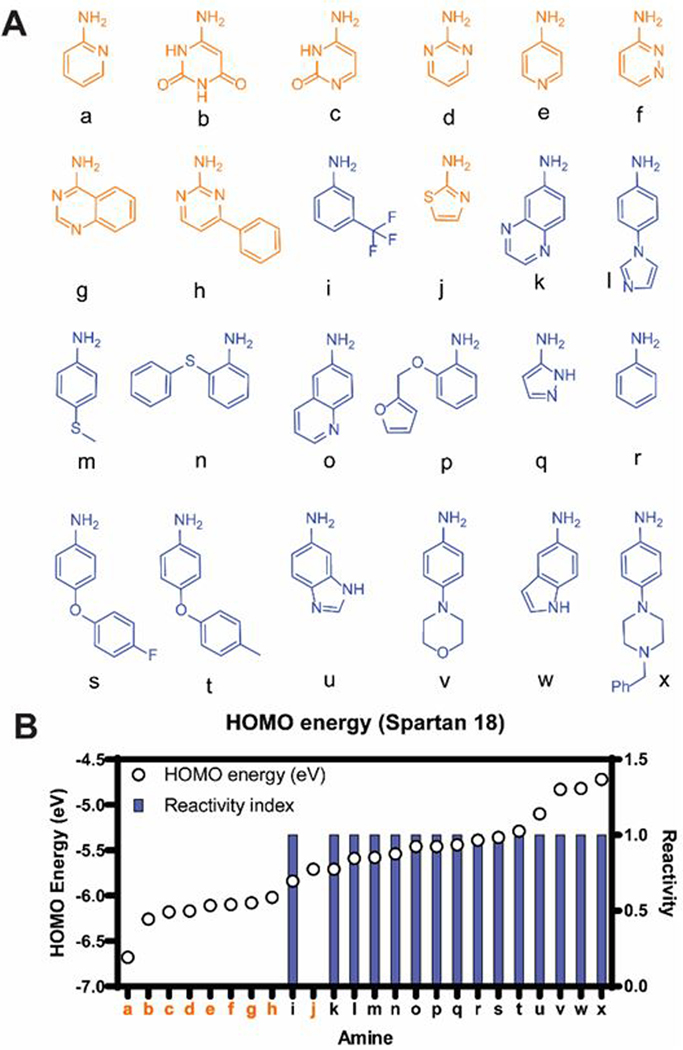
Our studies began by selecting amine building blocks suited for a rapid DCC-based method. We chose to first focus on aromatic amines because RNA ligands (not based on aminoglycosides) tend to have fewer sp3 centers compared to protein ligands.39, 40 Twenty-four commercially available aromatic amines were selected and tested for imine formation in buffer at pH 6.3 (Fig. 2A and S1). Since our DCC procedure would include the addition of a reducing agent (NaBH3CN) to convert imine library members into secondary amines that are stable to water-based analysis methods,27 amine reactivity was tested under these conditions using reductive alkylation as a proxy for imine formation (Fig. 3A). Fifteen amines showed high reactivity (>90% aldehyde consumption), while the remaining amines showed no reactivity (Fig. 2). To understand the basis of this differential reactivity, we compared the calculated HOMO energies of the amines and found that the reactive amines generally had higher HOMO energies than the unreactive ones (Fig. 2B), while no trend was observed with pKa values (Fig. S2). This result is consistent with the study by Lehn and co-workers which showed that incorporation of amine HOMO energies improved modeling of the stability of imines in aqueous media while pKa alone was poorly correlated to the imine formation constant.41 Of note, all the unreactive amines contain a nitrogen atom in the same ring carrying the NH2 group (Fig. 2). For these amines, the poor reactivity may derive from diminished electron density around the NH2 group as suggested by the preferential protonation of the ring nitrogen.42 The correlation of the energy prediction to experimental reactivity facilitates library design as it allows for the filtering of commercial amines and the purchase of only those expected to be reactive.
Fig. 2.
Selection of amine subunits. (A) Structures of amines tested for reactivity (orange: unreactive; blue: reactive); (B) Relationship between amine reactivity and their HOMO energies. For reactive amines (reactivity index = 1), HPLC analysis showed >90% consumption of aldehyde and presence of the secondary amine. For unreactive amines (reactivity index = 0), no secondary amine formation was observed. See Fig. S1 for reaction conditions.
Fig. 3.
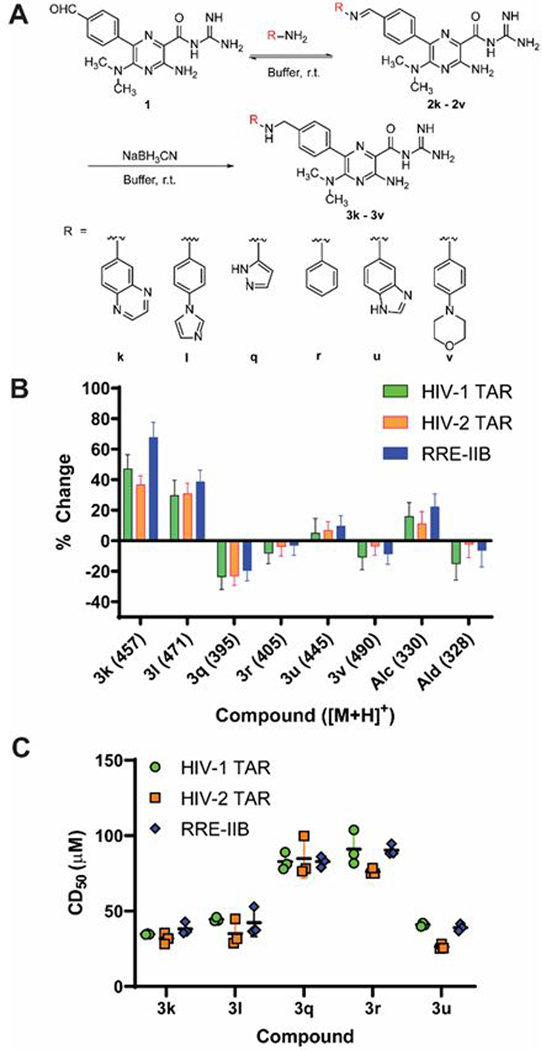
DCC and TOPRO-1 assay with HIV-1 TAR, HIV-2 TAR and RRE-IIB. (A) DCC reaction scheme. The aldehyde and amines were incubated in the presence or absence of RNA in buffer (20 mM BisTris, 25 mM NaCl, 1 mM MgCl2, 0.1 mM EDTA, pH = 6.3, 5% v/v DMSO). (B) Percent change of the peak area of each compound in the presence of RNA relative to the no-RNA control reaction (average ± standard deviation of three independent experiments). (C) TOPRO-1 displacement assay of library members with HIV-1 TAR, HIV-2 TAR and RRE-IIB. CD50: Competitive dosage for 50% reduction in fluorescence signal. Error bars: standard deviation.
Of the 15 reactive amines, six were chosen for the DCC proof of concept study. Seven amines were excluded based on anticipated low buffer solubility of the resulting amiloride analogues (Fig. S3). Given the chemical similarity between amines k and o and between amines u and w, we decided to use only one amine in each pair. In both cases, the analogue with higher nitrogen count (k and u) was chosen to increase the chances of solubility.
DCC experiments with HIV-1 TAR, HIV-2 TAR and RRE-IIB were performed by incubating the six amines, the amiloride aldehyde, and the RNA in buffer (pH = 6.3) at ambient temperature (~21 ˚C) for 20 hours (Fig. 3A). Dimethylsulfoxide (DMSO) was used at 5% v/v to enhance ligand solubility while maintaining biologically-relevant conditions given the known susceptibility of RNA conformations to high DMSO content.43 A reducing agent (NaBH3CN) was added to convert imine library members into secondary amines that are stable to water-based analysis methods (See ESI for detailed procedures).27 Negative control reactions (no RNA) were run in parallel under the same conditions. The slightly acidic conditions were chosen because we observed that reductive amination was significantly higher yielding at pH 6.3 compared to pH 6.9 and pH 7.4 (Fig. S4). After incubation, the reactions were analysed by electrospray ionization mass spectrometry (ESI-MS) with single ion monitoring, which provides high sensitivity and resolution compared to commonly HPLC analysis. The peak area of each compound in RNA-containing reactions was compared to that in the negative control to give a measure of how its abundance changes in the presence of RNA.
As shown in Fig. 3B, differential amplification and suppression were observed. Compounds 3k and 3l were amplified in the presence of RNA, suggesting that these compounds bound more strongly than the other library members to each RNA. On the other hand, compound 3q shows marked suppression, followed by compounds 3r and 3v. These compounds are expected to have the lowest affinity for the RNAs amongst all library members. Lastly, compound 3u shows slight amplification, suggesting a moderate binding affinity compared to 3k and 3l. An unexpected increase in alcohol by-product from aldehyde reduction was also observed in the presence of RNA.
To test whether the observed amplification pattern indeed corresponded to binding affinity, the library members were independently synthesized and evaluated for binding to HIV-1 TAR, HIV-2 TAR and RRE-IIB using the TOPRO-1 fluorescent indicator displacement assay.44 Compounds 3k, 3l and 3u displaced TOPRO-1 twice as strongly as compounds 3q and 3r to all three RNAs, consistent with the higher amplification in DCC (Fig. 3C, Table S8). A CD50 value could not be recorded for 3v since the binding was too weak for convergence during curve fitting (Fig. S10–S12), consistent with suppression in DCC. Both the aldehyde and alcohol were substantially weak binders, with the aldehyde showing somewhat stronger displacement at high concentrations compared to the alcohol (Fig. S10–S12). Therefore, the increase in alcohol by-product in DCC may be a result of decreased reaction efficiency of the aldehyde with the amines when it is bound to RNA.45 These results show that imine-based DCC can effectively discriminate between binders and non-binders for a given RNA target and that the secondary amines resulting from the incorporation of a reducing agent still generally reflect the behaviour of the underlying dynamic library of imines. We will note, however, that the binding affinities of library members did not perfectly correlate to the DCC amplification pattern. These discrepancies likely reflect the difficulty of discriminating between highly similar compounds within a narrow range of affinities (26–91 µM; Table S8).
To further verify these results, we used surface plasmon resonance (SPR) as an orthogonal assay to test the binding of compounds 3k–3u to HIV-1 TAR. Similar to the TOPRO-1 displacement assay, compounds 3k, 3l, and 3u bound considerably stronger than 3q (Fig. S13), further supporting that amplification in imine-based DCC corresponds to stronger binding of the library member.
In this work, we establish the applicability of imine-based DCC to RNA systems. We showed that the method is versatile, as amplification of binders was observed for multiple RNAs with structure and sequence diversity. Of note, this method is highly sensitive as differentiation was observed among compounds with highly similar binding affinities (Fig. 3C). Interestingly, the three compounds with higher affinity (3k, 3l and 3u) were derived from amines containing two aromatic rings, supporting the potential for identification of properties advantageous for RNA-binding using imine-based DCC. Furthermore, the use of highly reactive amines enables considerably faster identification of ligands compared to previous imine-based DCC studies.27 Studies to understand conditions under which heteroaromatic amines and alkylamines can be used are underway as well as the expansion of the library to increase chances of selectivity and begin applying imine-based DCC to large complex RNAs. In particular, we are investigating the use of imine-based DCC to expedite the discovery of multivalent ligands through dialdehyde or diamine linkers. We expect this methodology to be a valuable addition to current approaches of targeting RNA particularly because it does not require large library synthesis or knowledge of high-resolution structure.
Supplementary Material
Acknowledgments
We would like to thank Dr. Brittany Morgan (University of Michigan) and Jordan Forte (Wake Forest School of Medicine) for their input in the initial selection of amines, Dr. Peter Silinski (Director, Analytical Services, Duke University) for optimizing the ESI-MS method used for DCC reaction analysis and Dr. Brian Watts (Manager of Duke DHVI BIA Core) for assistance with SPR experiments. We thank Duke University undergraduate students Noelia Boldizsar and Oluwafikemi Faleye for assistance in synthesizing library members, Shantal Jayawickreme for synthesizing RRE-IIB RNA, and members of the Hargrove Lab for valuable discussion. The authors acknowledge financial support from Duke University, the U.S. NIH (U54 AI150470), the NSF Career Award (CAREER 1750375) and the Prostate Cancer Foundation Young Investigator Award. A.U.J. acknowledges additional support from the Lews Siegel Fellowship from the Pratt School of Engineering, Duke University.
Footnotes
† Electronic Supplementary Information (ESI) available: The methods employed, additional figures and tables, spectroscopic and chromatographic data, and binding curves. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Morgan BS, Forte JE and Hargrove AE, Nucleic Acids Res., 2018, 46, 8025–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sztuba-Solinska J, Chavez-Calvillo G. and Cline SE, Bioorg. Med. Chem., 2019, 27, 2149–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hermann T, Wiley Interdisciplinary Reviews: RNA, 2016, DOI: 10.1002/wrna.1373, doi: 10.1002/wrna.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connelly CM, Moon MH and Schneekloth JS Jr., Cell Chem. Biol., 2016, 23, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang L, Watkins D, Jin Y, Gong C, King A, Washington AZ, Green KD, Garneau-Tsodikova S, Oyelere AK and Arya DP, ACS Chem. Biol., 2015, 10, 1278–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratmeyer L, Zapp ML, Green MR, Vinayak R, Kumar A, Boykin DW and Wilson WD, Biochemistry, 1996, 35, 13689–13696. [DOI] [PubMed] [Google Scholar]
- 7.Patwardhan NN, Ganser LR, Kapral GJ, Eubanks CS, Lee J, Sathyamoorthy B, Al-Hashimi HM and Hargrove AE, MedChemComm, 2017, 8, 1022–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donlic A, Morgan BS, Xu JL, Liu A, Roble C Jr. and Hargrove AE, Angew. Chem., Int. Ed., 2018, 57, 13242–13247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abulwerdi FA, Shortridge MD, Sztuba-Solinska J, Wilson R, Le Grice SF, Varani G. and Schneekloth JS Jr., J. Med. Chem., 2016, 59, 11148–11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rynearson KD, Charrette B, Gabriel C, Moreno J, Boerneke MA, Dibrov SM and Hermann T, Bioorg. Med. Chem. Lett., 2014, 24, 3521–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sztuba-Solinska J, Shenoy SR, Gareiss P, Krumpe LR, Le Grice SF, O’Keefe BR and Schneekloth JS Jr., J. Am. Chem. Soc., 2014, 136, 8402–8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorenz DA, Song JM and Garner AL, Bioconjug. Chem., 2015, 26, 19–23. [DOI] [PubMed] [Google Scholar]
- 13.Rzuczek SG, Southern MR and Disney MD, ACS Chem. Biol., 2015, 10, 2706–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costales MG, Childs-Disney JL and Disney MD, in Therapeutics RNA, ed. Garner AL, Springer International Publishing, Cham, 2018, DOI: 10.1007/7355_2016_21, pp. 1–16. [DOI] [Google Scholar]
- 15.Zhou Y, Gregor VE, Sun Z, Ayida BK, Winters GC, Murphy D, Simonsen KB, Vourloumis D, Fish S, Froelich JM, Wall D. and Hermann T, Antimicrob. Agents Chemother., 2005, 49, 4942–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganser LR, Lee J, Rangadurai A, Merriman DK, Kelly ML, Kansal AD, Sathyamoorthy B. and Al-Hashimi HM, Nat. Struct. Mol. Biol., 2018, 25, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee J, Bai Y, Chembazhi UV, Peng S, Yum K, Luu LM, Hagler LD, Serrano JF, Chan HYE, Kalsotra A. and Zimmerman SC, Proc. Natl. Acad. Sci. U. S. A., 2019, DOI: 10.1073/pnas.1820827116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran T, Childs-Disney JL, Liu B, Guan L, Rzuczek S. and Disney MD, ACS Chem. Biol., 2014, 9, 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller BL, in RNA Therapeutics, ed. Garner AL, Springer International Publishing, Cham, 2018, DOI: 10.1007/7355_2016_23, pp. 17–45. [DOI] [Google Scholar]
- 20.Moumné R, Catala M, Larue V, Micouin L. and Tisné C, Biochimie, 2012, 94, 1607–1619. [DOI] [PubMed] [Google Scholar]
- 21.Garavis M, Lopez-Mendez B, Somoza A, Oyarzabal J, Dalvit C, Villasante A, Campos-Olivas R. and Gonzalez C, ACS Chem. Biol, 2014, 9, 1559–1566. [DOI] [PubMed] [Google Scholar]
- 22.Warner KD, Homan P, Weeks KM, Smith AG, Abell C. and Ferre-D’Amare AR, Chem. Biol., 2014, 21, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jamali H, Khan HA, Stringer JR, Chowdhury S. and Ellman JA, J. Am. Chem. Soc., 2015, 137, 3616–3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Connelly CM, Abulwerdi FA and Schneekloth JS Jr., Methods Mol. Biol., 2017, 1518, 157–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M. and Childs-Disney JL, ACS Chem. Biol., 2016, DOI: 10.1021/acschembio.6b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang K, Li S, Kappel K, Pintilie G, Su Z, Mou T-C, Schmid MF, Das R. and Chiu W, Nat. Comm., 2019, 10, 5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbett PT, Leclaire J, Vial L, West KR, Wietor J-L, Sanders JKM and Otto S, Chem. Rev., 2006, 106, 3652–3711. [DOI] [PubMed] [Google Scholar]
- 28.Ramström O. and Lehn J-M, Nat. Rev. Drug Discovery, 2002, 1, 26–36. [DOI] [PubMed] [Google Scholar]
- 29.Ni C, Zha D, Ye H, Hai Y, Zhou Y, Anslyn EV and You L, Angew. Chem., Int. Ed., 2018, 57, 1300–1305. [DOI] [PubMed] [Google Scholar]
- 30.Ofori LO, Hoskins J, Nakamori M, Thornton CA and Miller BL, Nucleic Acids Res., 2012, 40, 6380–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belowich ME and Stoddart JF, Chem. Soc. Rev., 2012, 41, 2003–2024. [DOI] [PubMed] [Google Scholar]
- 32.Kalliokoski T, ACS Comb. Sci., 2015, 17, 600–607. [DOI] [PubMed] [Google Scholar]
- 33.Bugaut A, Toulme JJ and Rayner B, Angew. Chem. Int. Ed. Engl., 2004, 43, 3144–3147. [DOI] [PubMed] [Google Scholar]
- 34.Jana S, Panda D, Saha P, Pantos̨ GD and Dash J, J. Med. Chem., 2019, 62, 762–773. [DOI] [PubMed] [Google Scholar]
- 35.Mustoe AM, Brooks CL and Al-Hashimi HM, Annu. Rev. Biochem., 2014, 83, 441–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patwardhan NN, Cai Z, Umuhire Juru A. and Hargrove AE, Org. Biomol. Chem., 2019, 17, 9313–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patwardhan NN, Cai Z, Newson CN and Hargrove AE, Org. Biomol. Chem., 2019, 17, 1778–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Grice SF, Curr. Top. Microbiol. Immunol., 2015, 389, 147–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan BS, Forte JE, Culver RN, Zhang Y. and Hargrove AE, Angew. Chem., Int. Ed., 2017, 56, 13498–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morgan BS, Sanaba BG, Donlic A, Karloff DB, Forte JE, Zhang Y. and Hargrove AE, ACS Chem. Biol., 2019, 14, 2691–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godoy-Alcántar C, Yatsimirsky AK and Lehn JM, J. Phys. Org. Chem., 2005, 18, 979–985. [Google Scholar]
- 42.Konishi H, Kato H. and Yonezawa T, Theor. Chim. Acta, 1970, 19, 71–82. [Google Scholar]
- 43.Lee J, Vogt CE, McBrairty M. and Al-Hashimi HM, Anal. Chem., 2013, 85, 9692–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wicks SL and Hargrove AE, Methods, 2019, 167, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huc I. and Lehn J-M, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 2106–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.