Abstract
Background and aims
Camelina (Camelina sativa, Brassicaceae) has attracted interest in recent years as a novel oilseed crop, and an increasing number of studies have sought to enhance camelina’s yield potential or to modify the composition of its oil. The ability of camelina to cross-hybridize with its wild relative, C. microcarpa, is of interest as a potential source of genetic variability for the crop.
Methods
Manual crosses were performed between the crop C. sativa and its wild relative C. microcarpa; F1 and F2 progenies were obtained. Cytology was used to study meiosis in the parents and F1s and to evaluate pollen viability. Flow cytometry was used to estimate nuclear DNA amounts and fatty acid methyl ester analysis was used to evaluate the lipid composition of F3 seeds.
Key Results
The F1 plants obtained by interspecific crossing presented severe abnormalities at meiosis and low pollen viability, and produced very few F2 seeds. The F2s presented diverse phenotypes and in some cases severe developmental abnormalities. Many F2s were aneuploid. The F2s produced highly variable numbers of F3 seeds, and certain F3 seeds presented atypical lipid profiles.
Conclusions
Considering the meiotic abnormalities observed and the probability of aneuploidy in the F2 plants, the C. microcarpa accessions used in this study would be difficult to use as sources of genetic variability for the crop.
Keywords: Camelina sativa, Camelina microcarpa, interspecific outcrossing, gene flow, aberrant meiosis, interspecific hybrid, aneuploidy
INTRODUCTION
In recent decades, increased interest in developing more sustainable agriculture has led to renewed interest in novel crops. Among these, one of the most promising is camelina (Camelina sativa), an oilseed crop in the family Brassicaceae that is closely related to the model plant species Arabidopsis thaliana. Archaeological remains of camelina from 6000 BC have been found in Armenia (Hovsepyan and Willcox, 2008), and there is clear evidence of the cultivated species in France and elsewhere in Western Europe starting in the late Bronze Age (Toulemonde, 2010). Camelina continued to be an important oilseed crop in Northern Europe until the end of the 19th century, after which it was progressively replaced by others, such as oilseed rape.
As has been presented in depth in recent review articles (Eynck and Falk, 2013; Vollmann and Eynck, 2015; Faure and Tepfer, 2016; Berti et al., 2016; Sainger et al., 2017), camelina has numerous attractive features that explain the renewed interest: the oil extracted from its seeds has several potential uses that could grow in importance, either for food, for biofuel or for other industrial uses, and the residual seed-cake can be used in animal feed. Furthermore, the plant is remarkably robust, growing well on marginal lands with little or no inputs required, and is resistant to major pathogens and pests of other Brassicaceae crops, such as oilseed rape. Because camelina has been a neglected crop for the past century, the available cultivars have not undergone extensive improvement using modern plant breeding strategies. In recent years, several laboratories have presented evidence suggesting that the genetic diversity present in current cultivars is relatively narrow (Vollmann et al., 2005; Ghamkhar et al., 2010; Manca et al., 2013; Galasso et al., 2015; Singh et al., 2015; Kurasiak-Popowska et al., 2018; Luo et al., 2019), which greatly diminishes the prospects for crop improvement.
There are (at least) two possible solutions for this concern. First, numerous laboratories have recently created camelina genotypes expressing transgenes that modify the composition of its oil reserves (for reviews see Berti et al., 2016; Faure and Tepfer, 2016; Sainger et al., 2017), and CRISPR-Cas9 genome-edited camelina lines with enhanced oil composition have also been created (Jiang et al., 2017; Morineau et al., 2017). The release of transgenic/edited camelina varieties faces regulatory restrictions, however.
As an alternative, the use of wild genetic resources is an interesting path to explore. Within the genus Camelina, C. microcarpa is of particular interest, since many accessions have a genome structure that is similar to that of the crop (Séguin-Swartz et al., 2013; Martin et al., 2017, 2019; Mandáková et al., 2019). Camelina sativa and certain accessions of C. microcarpa have an allohexaploid genome (Kagale et al., 2014), composed of three closely related sub-genomes (2n = 6x = 40). A very recent paper has demonstrated that C. sativa and C. microcarpa show the same global chromosome organization (Mandakova et al., 2019), supporting the view that C. microcarpa could be the parental species of the crop (Brock et al., 2018). It is also of note that crosses between Canadian accessions of hexaploid C. microcarpa can yield viable, partly fertile F1 progeny in C. microcarpa × C. sativa crosses (Séguin-Swartz et al., 2013; Martin et al., 2019). Crosses between C. sativa and C. microcarpa are thus of interest from a plant breeder’s perspective, as they provide the opportunity of using the crop’s closest wild relatives as sources of diversity. This relies on the fitness of F1 hybrids and succeeding generations, which has not been evaluated previously.
The objective of this study was to evaluate the viability of F1 hybrids and succeeding generations. We first show that C. sativa × C. microcarpa F1 plants had profoundly disturbed meiosis and low pollen viability, but that a few F2 seeds could nonetheless be obtained. The F2 individuals displayed a range of developmental abnormalities, and evaluation of nuclear DNA amounts suggests that many, if not all, were aneuploids. Nonetheless, certain F2 individuals produced abundant F3 seeds, demonstrating that the barriers to hybridization between C. sativa and hexaploid C. microcarpa, although strong, are not totally impermeable. As a proof of concept, we also show that certain F3 seeds presented atypical lipid profiles, which opens potential avenues for plant breeding.
MATERIALS AND METHODS
Plant materials, growing conditions and interspecific crosses
DsRed F, a homozygous line of Camelina sativa ‘Céline’ expressing a DsRed transgene has been described (Julié-Galau et al., 2014). Camelina microcarpa populations have been described across Europe and Western and Central Asia (Calasan et al., 2019). In France, C. microcarpa is present in scattered populations primarily in the southern and eastern regions of the country (https://inpn.mnhn.fr/espece/cd_nom/87577). Seeds of two accessions of C. microcarpa were obtained from the Conservatoire Botanique du Bassin Parisien – Muséum National d’Histoire Naturelle (CBNP:MNHN): 03CF1063, referred to as Guillestre, and temporary accession 2487, referred to as Fleurey. Except when vernalization was applied, plants were grown in the greenhouse with a 16-h daylength, 25 °C day and 22 °C night temperature, and 40% relative humidity. Plants that were vernalized were first grown for 1 month as above, then transferred to a greenhouse maintained above 5 °C for 1 month, then returned to normal conditions.
For manual crosses, flowers of the female parent were emasculated before anthesis to avoid selfing, and were pollinated manually with the pollen of the male parent.
Cytology
Male meiotic spreads for 4′,6-diamidino-2-phenylindole (DAPI) staining were prepared as described for Arabidopsis in Chelysheva et al. (2013) from buds fixed in Carnoy’s fixative (absolute ethanol:acetic acid, 3:1, v/v). Mature pollen grain viability was estimated as described by Alexander (1969).
Flow cytometry
Relative nuclear DNA amounts were determined with a CyFlo Space flow cytometer as described by Marie and Brown (1993). For each plant genotype, three young leaf samples were analysed. This entailed chopping with a fresh razor blade in Galbraith’s chopping buffer (Galbraith et al., 1983), addition of propidium iodide, and analysis according to the manufacturer’s protocol. The position of the different ploidy peaks of hexaploid C. sativa ‘Céline’ had been determined previously relative to diploid Arabidopsis thaliana and tetraploid Capsella bursa-pastoris (Julié-Galau et al., 2014), and thus served as internal reference.
Analysis of seed fatty acid composition
Fatty acid methyl esters (FAMEs) were extracted from single seeds, and then analysed by GC–MS according to Li et al. (2006). For each sample, a single seed was put in a glass tube to which was added methanol (1 mL), toluene (0.3 mL) and sulphuric acid (25 µL). The tubes were closed and warmed at 80 °C for 90 min. After cooling, hexane (0.5 mL) and water (1.5 mL) were added. The tubes were shaken and then centrifuged for 5 min at 1469 g. The hexane phase (50 µL) was transferred to a GC vial for injection. The samples were injected into an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. The polar column was a BPX70 (SGE) (30 m). Oven temperature ramp was 16 °C min−1 from 70 °C to 160 °C, then 4 °C min−1 up to 240 °C and 10 °C min−1 up to 260 °C for 4 min (run length 31.6 min). Helium constant flow rate was 1.52 mL min−1. Temperatures were as follows: injector 250 °C, transfer line 290 °C, source 250 °C and quadrupole 150 °C.
RESULTS
Our work is based on previous observations that C. sativa and hexaploid C. microcarpa are cross-compatible (Séguin-Swartz et al., 2013; Martin et al., 2019); therefore, we have not re-assessed the frequency with which F1 hybrids can be produced. In this study, we used the previously described line of C. sativa ‘Céline’ expressing a constitutive DsRed transgene (Julié-Galau et al., 2014) as pollen donor to produce C. microcarpa × C. sativa hybrids. The presence of the DsRed marker gene facilitates confirmation that the F1 seeds produced were genuine hybrids. Two French C. microcarpa accessions were used: CBNBP-MNHN accession 03CF1063 from Guillestre, originating ~50 km west of Gap, and CBNBP-MNHN temporary accession 2487, collected at Fleurey-sur-Ouche, ~10 km west of Dijon. Neither required stratification for germination, and both required vernalization to induce timely flowering, but would eventually flower in the greenhouse if maintained under non-vernalizing conditions for several months. Plants of the two C. microcarpa accessions were morphologically indistinguishable and behaved in the same manner in crosses with C. sativa, so further analyses of progeny were focused on those from crosses with the Guillestre accession, except for the meiosis studies, in which both accessions were used.
Similar to what has been reported with several Canadian hexaploid C. microcarpa accessions (Séguin-Swartz et al., 2013; Martin et al., 2019), for both French C. microcarpa accessions, manual pollination of emasculated flowers with pollen from DsRed C. sativa readily yielded DsRed-positive F1 seeds, and the reciprocal crosses were also successful, as judged by the phenotype of the putative C. sativa × C. microcarpa F1 plants.
Characterization of C. microcarpa × C. sativa F1 hybrids
Six F1 hybrids were grown in the greenhouse under long-day conditions. In the early vegetative phase, they remained in a rosette, as did the C. microcarpa parent, whereas the stems of the C. sativa elongated without a rosette phase (Fig. 1A). Later, after 2 months of growth, the F1 hybrids had bolted and began to flower, but the C. microcarpa parents remained in a rosette (Fig. 1B). Induction of flowering in the hybrids was therefore intermediate between the two parents.
Fig. 1.
Plants of cultivated camelina, its wild relative C. microcarpa and the F1 hybrid. (Left to right) C. sativa ‘Céline’, F1 hybrid C. microcarpa × C. sativa and C. microcarpa Guillestre, 1 (A) and 2 (B) months after sowing. Plants were grown in 14-cm pots.
Although in the early phases of flowering the F1 hybrids appeared to be entirely sterile, at later stages a few seeds were produced. When pollen viability was evaluated by Alexander staining, nearly all pollen grains of the parental species were viable, as expected, but only a small percentage of the pollen of the F1 hybrids (<3 %) was viable (Table 1). These values are in the low range relative to those reported by Séguin-Swartz et al. (2013) and Martin et al. (2019), who reported pollen viabilities in F1 hybrids that ranged from 1.3 to 45 %. The low pollen viability was likely the cause of the very low ability of the F1 plants to produce self-progeny, since they produced only 30–50 seeds per plant. This low seed productivity was also reflected in the weight of the seeds per plant (Table 2).
Table 1.
Alexander staining of pollen of C. sativa, C. microcarpa and F1 hybrids
| Stain positive | Stain negative | Percentage stain positive | |
|---|---|---|---|
| C. sativa DsRed F | 245 | 2 | 99.2 |
| C. microcarpa Guillestre | 122 | 0 | 100 |
| C. microcarpa Guillestre × C. sativa DsRed F | 6 | 255 | 2.3 |
| C. sativa DsRed F × C. microcarpa Guillestre | 8 | 426 | 1.8 |
Viability of freshly harvested pollen grains was evaluated by Alexander staining.
Table 2.
Weight of seeds produced by individual C. microcarpa × C. sativa F1 and F2 plants
| Sample | Seed weight (g) |
|---|---|
| C. sativa DsRed F | 12.00 |
| 15.21 | |
| C. microcarpa Guillestre | 3.95 |
| 5.90 | |
| F 1 of C. microcarpa Guillestre × C. sativa DsRed F | |
| F1-A | 0.03 |
| F1-B | 0.01 |
| F 2 of C. microcarpa Guillestre × C. sativa DsRed F | |
| F2-1 | < 0.01 |
| F2-2 | 0 |
| F2-3 | 2.68 |
| F2-4 | 0.47 |
| F2-5 | 0.54 |
| F2-6 | 0.03 |
| F2-7 | 0.05 |
| F2-8 | 0 |
| F2-9 | 0 |
| F2-10 | 0.01 |
| F2-11 | 0.07 |
| F2-12 | 0 |
| F2-13 | 2.14 |
| F2-14 | 0.13 |
| F2-15 | 0 |
| F2-16 | < 0.01 |
| F2-17 | 3.88 |
| F2-18 | < 0.01 |
| F2-19 | 0 |
| F2 -20 | 0.30 |
| F2-21 | < 0.01 |
| F2-22 | 0 |
| F2-23 | 0 |
| F2-24 | 0 |
| F2-25 | 0 |
| F2-26 | 0.19 |
| F2-27 | 0.26 |
| F2-28 | 2.01 |
| F2-29 | 0.39 |
| F2-30 | 0 |
| F2-31 | 0 |
| F2-32 | 0 |
| F2-33 | 0 |
| F2-34 | 0 |
| F2-35 | 0 |
| F2-36 | 0 |
| F2-37 | 0 |
| F2-38 | 1.07 |
| F2-39 | 1.04 |
| F2-40 | 0.66 |
| F2-41 | < 0.01 |
Meiotic defects in F1 hybrids between C. sativa and C. microcarpa
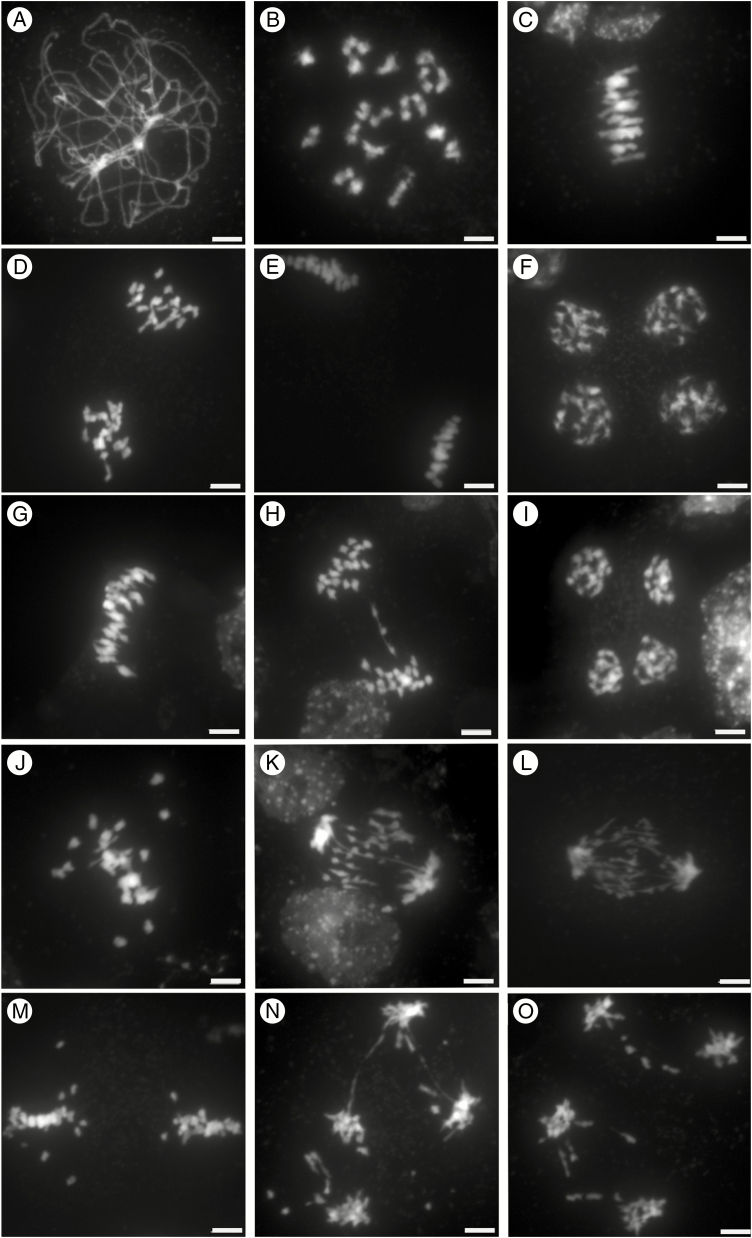
The strong sterility observed in the F1 hybrids prompted us to investigate male meiosis by staining pollen mother-cell chromosome spreads with DAPI. As a prerequisite, we first characterized male meiosis in C. sativa, which had not been assessed previously. We observed all the conspicuous landmarks of canonical meiotic progression. At pachytene, pairs of homologous chromosomes formed a ribbon-like structure that resulted from their intimate association via the synaptonemal complex (Fig. 2A). The synaptonemal complex then disassembled, and chromosomes condensed until discrete separate bivalents became visible at diakinesis (Fig. 2B). In all cells (n = 12), about 20 bivalents were discernible, while no obvious multivalents (association of more than two chromosomes) were observed. At metaphase I, bivalents were under tension, with homologous centromeres directed towards opposite poles; at this stage, the homologues were still connected with one another by chiasmata, the cytological manifestation of meiotic crossovers (n = 10; Fig. 2C). At anaphase I the homologues separated, leading to the formation of dyads corresponding to two groups of 20 chromosomes (Fig. 2D). The chromosomes then partially decondensed before the second meiotic division took place. At metaphase II, the individual chromosomes, each consisting of two chromatids, aligned on the metaphase II spindle (Fig. 2E). At anaphase II, individual chromatids migrated to opposite spindle poles, forming four groups of 20 chromatids at telophase II (Fig. 2F), which ultimately gave rise to a tetrad of four microspores (n = 4). All these results indicate that meiosis is very regular in C. sativa, which, despite being a hexaploid, displays diploid meiotic behaviour, at least cytologically.
Fig. 2.
Meiosis in C. sativa, C. microcarpa and their F1 hybrid. DAPI staining of male meiotic cells in C. sativa ‘Céline’ (A–F), C. microcarpa Fleurey (G–I) and C. microcarpa Fleurey × C. sativa ‘Céline’ F1 hybrids (J–O). Pachytene (A); diakinesis (B); metaphase I (C,G, J); anaphase I (D, H, K, L); metaphase II (E, M), early anaphase II (N,O); telophase II (F, I). Scale bar = 5 μm.
Male meiosis in C. microcarpa was very similar, although a few meiotic defects were observed (Fig. 2G–I; Supplementary Data Fig. S1A–C). Briefly, as in C. sativa, chromosomes first condensed, synapsed and recombined during the first meiotic division. Again, about 20 bivalents could be visualized at metaphase I (Fig. 2G; n = 15 in C. microcarpa Fleurey). Occasional chromosomal abnormalities appeared at anaphase I, with some laggards and/or fragments in some cells of C. microcarpa Fleurey (n = 6; Fig. 2H). The second division then separated sister chromatids and resulted in four pools of chromosomes, each consisting of a single chromatid (Fig. 2I; Supplementary Data Fig. S1C).
The situation in C. sativa × C. microcarpa Fleurey F1 hybrids was very different, illustrating a wide range of meiotic defects. Numerous univalents (i.e. chromosomes that failed to form crossovers) were observed in all metaphase I plates (n = 41), scattered around the spindle equator (Fig. 2J). Anaphase I (n = 24) and telophase I were also very irregular, showing the presence of numerous laggards (Fig. 2K) and bridges (Fig. 2L) in some cells. In particular, we observed at least one clear instance in which the bridge was very close to one or two fragments (Fig. 2K), suggesting, if confirmed, that a crossover may have occurred within a paracentric inversion. The second division followed in the usual way, except for the presence of fragments in some ‘dyads’ and (individual) chromatids unaligned on the metaphase II plate or lagging between the two groups of separating chromatids (Fig. 2M). Bridges connecting different groups of individualized chromatids were also observed at anaphase II/telophase II, suggesting sister-chromatid breakage and rejoining at an earlier stage (n = 7; Fig. 2N, O). Similar, although less numerous, defects were observed during meiosis of C. microcarpa Guillestre × C. sativa: univalents at diakinesis (Supplementary Data Fig. S1D) and metaphase I (n = 93; Supplementary Data Fig. S1E, F), laggards and bridges at anaphase I and anaphase II (n = 4; Supplementary Data Fig. S1G, H) and unbalanced tetrads (Supplementary Data Fig. S1I).
Characterization of F2 generation plants
As stated above, and despite the conspicuous abnormalities observed during male meiosis of F1 hybrids between C. sativa and C. microcarpa, 20–50 F2 seeds were eventually produced by self-fertilization of the F1s. The F2 progeny of two C. microcarpa Guillestre × C. sativa F1 hybrids were grown in the greenhouse, the first (42 seeds) without vernalization, the second (41 seeds) with a 1-month vernalization period. As shown in Fig. 3A, B, in the non-vernalized group the F2 plants displayed highly diverse phenotypes, with marked differences in leaf morphology, stem elongation, and the ability to enter reproductive phase. In some F2 individuals, flowering was only moderately delayed relative to C. sativa, whereas others did not flower after several months in the absence of vernalization. Leaf shape and the presence/absence of teeth on the leaf margin were also extremely variable. Since there was no evident correlation between the diverse morphological parameters, no attempt to quantify them was made. Two F2 individuals displayed remarkably abnormal floral development: in plant F2-B5 the sepals were not shed, and developed into elongated leaf-like appendages (Fig. 3D); in plant F2-E7 all floral organs were leaflike (Fig. 3E). Both plants were entirely sterile.
Fig. 3.
Phenotype of F2 plants. (A) A control C. sativa plant (left) and four representative F2 individuals are shown 2 months after sowing, without vernalization. (B) Two-month-old F2 plants seen from above. (C) A normal inflorescence is shown in comparison with abnormal floral development of (D) plant F2-B5 and (E) plant F2-E7, shown 3 months after sowing.
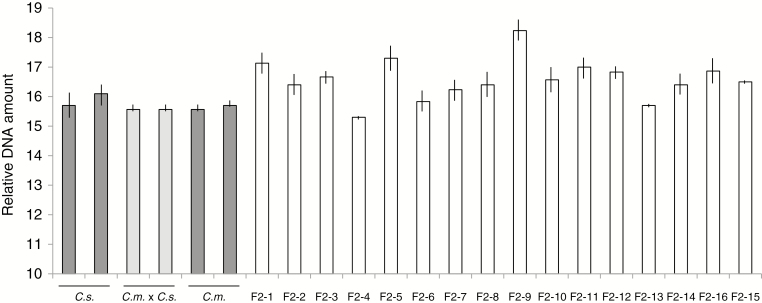
Considering the abnormal meiosis observed in pollen formation and the transgressive segregation of developmental features, it was of interest to evaluate the nuclear DNA amounts of F2 progeny. This was carried out on the second series of F2 plants, which had been vernalized. As was expected, since it was already known that C. sativa and C. microcarpa Guillestre are both hexaploid, the DNA amounts of the two parental species and their F1 progeny were equivalent (Fig. 4). By contrast, the DNA amounts of F2 individuals were quite diverse, and several had distinctly higher nuclear DNA amounts than the parental species or the F1 hybrids, the extreme case being F2-9, in which the nuclear DNA amount was more than 10% greater than the highest parental value; these results strongly suggest that many F2s were aneuploid to varying degrees.
Fig. 4.
Flow cytometry of nuclear DNA amounts. Two individuals of the parental C. sativa and C. microcarpa parents (dark grey bars), two F1C. microcarpa × C. sativa hybrids (light grey bars) and 16 F2 individuals (white bars) are shown. Relative DNA amount refers to the position of peaks on the flow cytometer readout. The analysis was carried out in triplicate; mean ± s.e.m. values are shown. C.s., C. sativa; C.m. × C.s., F1 hybrid; C.m., C. microcarpa Guillestre.
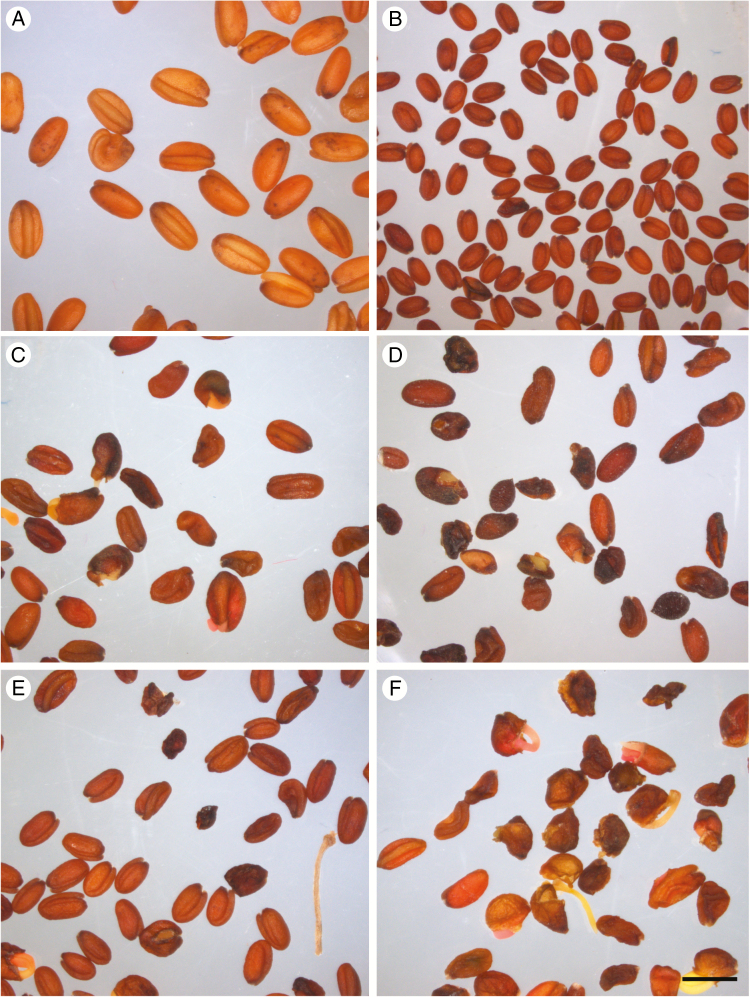
When vernalized, all F2 plants were able to flower, but their fertility was highly variable. As shown in Table 2, approximately half of them were sterile or nearly so (fewer than ten seeds were harvested from plants indicated <0.01 g). However, four F2 individuals produced abundant seeds (F2-3, F2-13, F2-17 and F2-28), but F3 seed quality was highly heterogeneous, with many seeds showing diverse abnormalities, such as abortion, incomplete filling and premature germination (Fig. 5). The viability of F3 seeds was not evaluated.
Fig. 5.
Seeds produced by C. sativa, C. microcarpa Guillestre and F1 and F2 progeny. Seeds produced by self-pollinated C. sativa (A), C. microcarpa (B), F1 hybrids F1-A (C) and F1-B (D), and F2 hybrids F2-3 (E) and F2-13 (F). Scale bar = 2 mm.
Lipid composition of F3 seeds
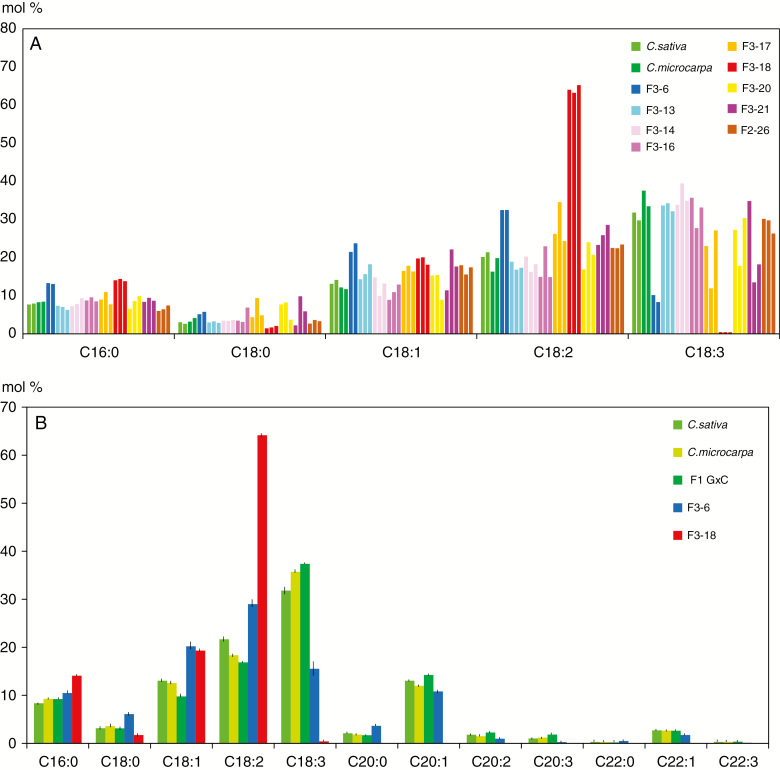
A major objective of research in camelina is modification of the seed oil composition, so it was of interest to evaluate whether the lipid composition of the seeds produced by hybrid progenies was as profoundly modified as the F2 plant phenotypes. Since each seed within an F3 population is likely to be genetically distinct, in particular since many F2s were probably aneuploid, the fatty acid composition of three seeds produced by each F2 plant was analysed individually; representative results are shown in Fig. 6A. The fatty acid profiles of the parental species and the F1 hybrids were essentially identical, with C18:3 > C18:2 > C18:1 = 20:1 > C16:0. As expected, the fatty acid composition within many F3 populations was quite heterogeneous, suggesting that genes controlling fatty acid composition were segregating within these populations. One F3 population, F3-18, was clearly different, with all three seeds showing greatly increased levels of C18:2, and with C18:3 correspondingly reduced to near zero. Since the F2-18 plant produced only three seeds, its progeny could not be analysed further. The three F3-6 seeds displayed a similar but less drastic change in C18:2 and C18:3, and since F2-6 produced sufficient seed, more seeds of the F3-6 population were analysed (Fig. 6B). These results confirm that the fatty acid composition of the F3-6 seeds is distinct from that of the controls, with an increased level of C18:2 and decreased C18:3.
Fig. 6.
Relative fatty acid profiles of F2 and F3 hybrid seeds. (A) Major fatty acid profile of individual seeds of nine F3 populations and controls was analysed by GC–MS. For each population, two or three seeds were analysed. Each bar represents a single seed. (B) Mean values (± s.e.m.) obtained with a greater number of individual seeds. F1 G×C = F2 seeds produced by the cross C. microcarpa × C. sativa. For all samples n = 9, except F1 G×C (n = 4) and F3-18 (n = 3).
Discussion
Camelina is a re-emerging crop that shows limited genetic diversity (Vollmann et al., 2005; Ghamkhar et al., 2010; Manca et al., 2013; Galasso et al., 2015; Singh et al., 2015; Kurasiak-Popowska et al., 2018; Luo et al., 2019). Our aim in this study was to explore whether C. microcarpa could be used as a source of useful genes to widen the genetic diversity of C. sativa. Our results suggest that this approach is not as simple as it looked.
Our results first confirmed previous findings that C. sativa and C. microcarpa are sexually compatible (Séguin-Swartz et al., 2013; Martin et al., 2019). We observed that the F1 hybrids, unremarkably, displayed a morphological phenotype intermediate between that of the two parental species during the vegetative phase, and the lipid composition of the F1 seeds was equivalent to that of the parents. Transgressive phenotypes were also observed for some traits, such as the concentrations of certain primary metabolites following virus infections, which were markedly higher in the F1 hybrids than in both parental species (Chesnais et al., 2019).
We also observed that F1C. microcarpa × C. sativa hybrids showed very reduced fertility. This is very likely due to the frequent and diverse meiotic abnormalities we observed in these plants, in particular the presence of univalents, bridges and fragments in many cells. Likewise, observation of bridges at anaphase I/telophase I or anaphase II/telophase II may be indicative of chromosome and/or sister-chromatid breakage and rejoining (Fig. 2; Supplementary Data Fig. S1). Although further studies are required to analyse the exact nature and mechanisms associated with the observed meiotic defects (e.g. Friebe et al., 2005), their wide occurrence does not support the assumption that C. sativa was domesticated from C. microcarpa (Brock et al., 2018), or that it was from C. microcarpa genotypes that would be quite different from those used in this study. Considering the meiotic abnormalities we observed in the C. microcarpa × C. sativa F1s, it is also surprising that Mandáková et al. (2019) observed no differences in chromosome structure between C. sativa and the C. microcarpa accession they studied. This apparent discrepancy raises the question whether different C. microcarpa accessions may in fact have different chromosome organizations, some that are sativa-like (e.g. ones in which the F1s have higher fertility), and others with more divergent structure. In this connection, we observed varying degrees of meiotic defects in the two hybrids we characterized. It was also observed that certain Canadian C. microcarpa accessions yielded F1s with distinctly higher levels of pollen viability (up to 45 %) in crosses with C. sativa (Séguin-Swartz et al., 2013; Martin et al., 2019). We do not know whether these hybrids show a more regular meiosis compared with those characterized here, but these genotypes might be better candidates for introgressing genes from C. microcarpa into the crop. Furthermore, these results suggest that the likelihood of introgression of transgenes from genetically modified camelina would be lower with the C. microcarpa accessions studied here compared with accessions with higher F1 fertility (Séguin-Swartz et al., 2013; Martin et al., 2019).
Surprisingly, and in spite of the low F1 fertility, some F2 plants can be obtained, which makes it possible move on to the next generations. Notably, we identified four F2 individuals (out of 41) that produced a significant number of seeds, which suggests, rather surprisingly, that later generations of self-progeny could be readily obtainable. We also observed lipid profiles that were unlike those of the parental species in the progeny of certain F2s. This suggests that interspecific hybrids could indeed be a useful source of novel traits in camelina. However, considering the number of meiotic abnormalities observed in the F1 progenies and the aneuploidy of many F2 individuals, returning to the normal genome structure of cultivated camelina through a series of backcrosses may remain an arduous process.
Altogether our results demonstrate that C. sativa and C. microcarpa may be less related to one another than previously thought, or that there is a great diversity of genome structure within C. microcarpa. In terms of pre-breeding (gene introgression from C. microcarpa into cultivated camelina), our results suggest that, at least for C. microcarpa genotypes similar to those studied here, this would not be a project to be engaged in lightly.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: meiosis in C. microcarpa Guillestre and its F1 hybrid with C. sativa ‘Céline’.
Funding
The Institut Jean-Pierre Bourgin benefits from the support of Saclay Plant Sciences-SPS (ANR-17-EUR-0007).
Acknowledgments
We thank Anne-Marie Chèvre, Jean-Denis Faure and Fabien Nogué for helpful comments on the manuscript. Seeds of both C. microcarpa accessions were kindly provided by the Seed Bank of the Conservatoire Botanique du Bassin Parisien – Muséum National d’Histoire Naturelle (CPNBP-MNHN). This work has benefited from the support of the Institut Jean-Pierre Bourgin’s Plant Observatory technological platforms. M.T. conceived the overall project. M.T. performed the crosses, plant observations and flow cytometry. Cytology was performed by A.H. with guidance from E.J. FAMEs were analysed by F.T. M.T wrote the manuscript, with input from the other authors, notably E.J.
LITERATURE CITED
- Alexander M. 1969. Differential staining of aborted and nonaborted pollen. Biotechnic and Histochemistry 44: 117–122. [DOI] [PubMed] [Google Scholar]
- Berti M, Gesch R, Eynck C, Anderson J, Cermak S. 2016. Camelina uses, genetics, genomics, production, and management. Industrial Crops and Products 94: 690–710. [Google Scholar]
- Brock JR, Dönmez AA, Beilstein MA, Olsen KM. 2018. Phylogenetics of Camelina Crantz. (Brassicaceae) and insight on the origin of gold-of-pleasure (Camelina sativa). Molecular Phylogenetics and Evolution 127: 834–842. [DOI] [PubMed] [Google Scholar]
- Calasan AZ, Seregin AP, Hurka H, Hofford NP, Neuffer B. 2019. The Eurasian steppe belt in time and space: phylogeny and historical biogeography of the false flax (Camelina Crantz, Camelineae, Brassicaceae). Flora. doi: 10.1016/j.flora.2019.151477. [DOI] [Google Scholar]
- Chelysheva L, Grandont L, Grelon M. 2013. Immunolocalization of meiotic proteins in Brassicaceae: method 1. Methods in Molecular Biology 990: 93–101. [DOI] [PubMed] [Google Scholar]
- Chesnais Q, Mauck KE, Bogaert F, et al. 2019. Virus effects of plant quality and vector behavior are species specific and do not depend on host physiological phenotype. Journal of Pest Science. doi: 10.1007/s10340-019-01082-z. [DOI] [Google Scholar]
- Eynck C, Falk KC. 2013. Camelina (Camelina sativa). In: Singh BP, ed. Biofuel crops: production, physiology and genetics. Fort Valley, GA: : CAB International, 369–391. [Google Scholar]
- Faure J-D, Tepfer M. 2016. Camelina, a Swiss knife for plant lipid biotechnology. Oilseeds Fats Crops Lipids 23. doi: 10.1051/ocl/2016023. [DOI] [Google Scholar]
- Friebe B, Zhang P, Linc G, Gill BS. 2005. Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenetic and Genome Research 109: 293–297. [DOI] [PubMed] [Google Scholar]
- Galasso I, Manca A, Braglia L, Ponzoni E, Breviario D. 2015. Genomic fingerprinting of Camelina species using cTBP as molecular marker. American Journal of Plant Sciences 6: 1184–1200. [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Sharma DP, Firoozabady E. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 2290: 1049–1051. [DOI] [PubMed] [Google Scholar]
- Ghamkhar K, Croser J, Aryamanesh N, Campbell M, Kon’kova N, Francis C. 2010. Camelina (Camelina sativa (L.) Crantz) as an alternative oilseed: molecular and ecogeographic analyses. Genome 53: 558–567. [DOI] [PubMed] [Google Scholar]
- Hovsepyan R, Willcox G. 2008. The earliest finds of cultivated plants in Armenia: evidence from charred remains and crop processing residues in pisé from the Neolithic settlements of Aratasche and Aknashen. Vegetation History and Archeobotany 17 (Suppl 1): 63–71. [Google Scholar]
- Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP. 2017. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnology Journal 15: 648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julié-Galau S, Bellec Y, Faure J-D, Tepfer M. 2014. Evaluation of the potential for interspecific hybridization between Camelina sativa and related wild Brassicaceae in anticipation of field trials of GM camelina. Transgenic Research 23: 67–74 [DOI] [PubMed] [Google Scholar]
- Kagale S, Koh C, Nixon J, et al. 2014. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nature Communications 5: 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasiak-Popowska D, Tomkowiak A, Czlopinska M, Bocianowski J, Weigt D, Nawracala J. 2018. Analysis of yield and genetic similarity of Polish and Ukrainian Camelina sativa genotypes. Industrial Crops and Products 123: 667–675. [Google Scholar]
- Li Y, Beisson F, Pollard M, Ohlrogge J. 2006. Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915. [DOI] [PubMed] [Google Scholar]
- Luo Z, Brock J, Dyer JM, Kutchan T, et al. 2019. Genetic diversity and population structure of a Camelina sativa spring panel. Frontiers in Plant Science 10: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manca A, Pecchia P, Mapelli S, Masella P, Galasso I. 2013. Evaluation of genetic diversity in a Camelina sativa (L.) Crantz collection using microsatellite markers and biochemical traits. Genetic Resources and Crop Evolution 60: 1223–1236. [Google Scholar]
- Mandáková T, Pouch M, Brock JR, Al-Shehbaz IA, Lysak MA. 2019. Origin and evolution of diploid and allopolyploid Camelina genomes were accompanied by chromosome shattering. Plant Cell 31: 2596–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Brown SC. 1993. A cytometric exercise in plant DNA histograms, with 2C values for 70 species. Biology of the Cell 78: 41–51. [DOI] [PubMed] [Google Scholar]
- Martin SL, Smith TW, James T, Shalabi F, Kron P Sauder CA. 2017. An update on the Canadian range, abundance and ploidy of Camelina spp. (Brassicaceae) east of the Rocky Mountains. Botany 95: 405–417. [Google Scholar]
- Martin SL, Lujan-Toro BE, Sauder CA, James T, Ohadi S, Hall LM. 2019. Hybridization rate and hybrid fitness for Camelina microcarpa Andrz. ex DC (♀) and Camelina sativa (L.) Crantz (Brassicaceae) (♂). Evolutionary Applications 12: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morineau C, Bellec Y, Tellier F, et al. 2017. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploidy Camelina sativa. Plant Biotechnology Journal 15: 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainger M, Jaiwal A, Sainger PA, Chaudary D, Jaiwal R, Jaiwal PK. 2017. Advances in genetic improvement of Camelina sativa for biofuel and industrial bio-products. Renewable and Sustainable Energy Reviews 68: 623–637. [Google Scholar]
- Séguin-Swartz G, Nettleton JA, Sauder C, Warwick SI, Gugel RK. 2013. Hybridization between Camelina sativa (L.) Crantz (false flax) and North American Camelina species. Plant Breeding 132: 390–396. [Google Scholar]
- Singh R, Bollina V, Higgins EE, et al. 2015. Single-nucleotide polymorphism identification and genotyping in Camelina sativa. Molecular Breeding 35: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulemonde F. 2010. Camelina sativa, l’or végétal du Bronze et du Fer. Anthropobotanica 1.1: 3–14. [Google Scholar]
- Vollmann J, Eynck C. 2015. Camelina as a sustainable oilseed crop: contributions of plant breeding and genetic engineering. Biotechnology Journal 10: 525–535. [DOI] [PubMed] [Google Scholar]
- Vollmann J, Grausgruber H, Stift G, Dryzhyruk V, Lelley T. 2005. Genetic diversity in camelina germplasm as revealed by seed quality characteristics and RAPD polymorphism. Plant Breeding 124: 446–453. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.