Abstract
Background and Aims
Recent findings indicate that Nod factor signalling is tightly interconnected with phytohormonal regulation that affects the development of nodules. Since the mechanisms of this interaction are still far from understood, here the distribution of cytokinin and auxin in pea (Pisum sativum) nodules was investigated. In addition, the effect of certain mutations blocking rhizobial infection and subsequent plant cell and bacteroid differentiation on cytokinin distribution in nodules was analysed.
Methods
Patterns of cytokinin and auxin in pea nodules were profiled using both responsive genetic constructs and antibodies.
Key Results
In wild-type nodules, cytokinins were found in the meristem, infection zone and apical part of the nitrogen fixation zone, whereas auxin localization was restricted to the meristem and peripheral tissues. We found significantly altered cytokinin distribution in sym33 and sym40 pea mutants defective in IPD3/CYCLOPS and EFD transcription factors, respectively. In the sym33 mutants impaired in bacterial accommodation and subsequent nodule differentiation, cytokinin localization was mostly limited to the meristem. In addition, we found significantly decreased expression of LOG1 and A-type RR11 as well as KNOX3 and NIN genes in the sym33 mutants, which correlated with low cellular cytokinin levels. In the sym40 mutant, cytokinins were detected in the nodule infection zone but, in contrast to the wild type, they were absent in infection droplets.
Conclusions
In conclusion, our findings suggest that enhanced cytokinin accumulation during the late stages of symbiosis development may be associated with bacterial penetration into the plant cells and subsequent plant cell and bacteroid differentiation.
Keywords: Bacterial penetration, bacteroid differentiation, cytokinins, auxin, KNOX3 transcription factor, NIN transcription factor, immunolocalization, composite plants, pea mutants, plant cell differentiation
INTRODUCTION
Legumes develop a structurally and functionally unique organ, the nodule, in response to inoculation with nitrogen-fixing bacteria collectively called rhizobia. De novo morphogenesis of nodules is induced by rhizobial signalling molecules, the Nod factors (Denarie et al., 1996), as well as microRNA (miRNA) (Simon et al., 2009), tRNA-derived small RNA fragments (tRFs) (Ren et al., 2019) and small rDNA-derived RNA (srRNA) (Jin et al., 2020); although similar to other processes associated with the development of new tissues and organs, nodule formation is also controlled by endogenous plant regulators. Multiple findings indicate that Nod factor signalling is tightly interconnected with changes in phytohormone synthesis, accumulation and perception that affect the infection and development of nodules.
Cytokinin and auxin are essential for regulating both rhizobial infection and nodule organogenesis (Thimann, 1936; Libbenga and Harkes, 1973; Cooper and Long, 1994; Mathesius et al., 1998; Rightmyer and Long, 2011; Demina et al., 2019). According to previous reports, the formation of auxin response maxima in various legumes precedes the initiation of nodule formation, and, most probably, results from a reduction in polar auxin transport or, alternatively, an auxin biosynthesis stimulation (Mathesius et al., 1998; Boot et al., 1999; Pacios-Bras et al., 2003; Huo et al., 2006; Suzaki et al., 2012; Ng et al., 2015; Schiessl et al., 2019). During early nodulation stages, some LAX (LIKE-AUX1) genes encoding auxin importers were found to be highly expressed in the developing primordia in Medicago truncatula (de Billy et al., 2001; Roy et al., 2017), suggesting that auxin plays an important role in initiating and maintaining cell proliferation to form the nodule primordia (Mathesius et al., 1998; Suzaki et al., 2012). In addition, auxin is involved in the regulation of infection thread progression (Breakspear et al., 2014; Laplaze et al., 2015).
The key role of cytokinins in nodule initiation was demonstrated by the phenotype of M. truncatula and Lotus japonicus mutants cre1 (cytokinin response1) and hit1 (hyperinfected1) affecting the cytokinin receptors MtCRE1 and LjLHK1 (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Plet et al., 2011). The loss-of-function mutants cre1 and hit1 are defective in nodule formation (Gonzalez-Rizzo et al., 2006; Murray et al., 2007). In contrast, in the snf2 (spontaneous nodule formation2) mutant carrying a gain-of-function mutation in LjLHK1, the development of nodule-like structures was observed in the absence of rhizobia (Tirichine et al., 2007). Similarly, in M. truncatula plants expressing MtCRE1 with an amino acid replacement in the CHASE domain (L267F), spontaneous nodules appeared (Ovchinnikova et al., 2011). It has been shown that the cytokinin pathway may regulate certain flavonoids acting on polar auxin transport through its effect on PIN-formed (PIN) auxin efflux carriers, that influences auxin accumulation in cortical cells during the early stages of nodulation (Plet et al., 2011; Ng et al., 2015). Moreover in L. japonicus and M. truncatula, cytokinin signalling also positively regulates auxin accumulation, probably by regulating genes involved in auxin biosynthesis (Suzaki et al., 2012; Schiessl et al., 2019).
In accordance with the suggested involvement of cytokinins in the regulation of early stages of nodulation, the cytokinin biosynthesis genes ISOPENTENYL TRANSFERASE (IPT) and LONELY GUY (LOG) are up-regulated during these stages. LjIPT2 and LjIPT4 in L. japonicus and their homologues MtIPT2 (Medtr4g117330) and MtIPT4 (Medtr2g022140) in M. truncatula as well as PsIPT1 and PsIPT2 in Pisum sativum are quickly induced in response to rhizobial inoculation or Nod factor treatment followed by cytokinin accumulation in roots (van Zeijl et al., 2015; Jardinaud et al., 2016; Dolgikh et al., 2017; Reid et al., 2017). However, in addition to their early activation, other genes are strongly up-regulated during the late stages of nodulation, such as LjIPT1 and LjIPT3 in L. japonicus, or MtIPT1 (Medtr1g110590) and MtIPT3 (Medtr1g072540) in M. truncatula and their homologues PsIPT4 and PsIPT3 in P. sativum, respectively, as well as MtLOG1, MtLOG2, PsLOG1 and PsLOG2 (Chen et al., 2014; Mortier et al., 2014; Azarakhsh et al., 2015; Dolgikh et al., 2017; Reid et al., 2017). Moreover, knock down of LjIPT3 or MtLOG1 and MtLOG2 by RNA interference leads to decreased nodulation (Chen et al., 2014; Mortier et al., 2014). This suggests that cytokinin signalling activated in plant cells during the late stages of symbiosis development fulfils specific functions that remain to be elucidated. In contrast, the cytokinins produced by rhizobia are capable of influencing the efficiency of symbiosis but do not affect the formation of nodules (Kisiala et al., 2013).
Genes involved in cytokinin signalling such as those encoding the B- and A-type response regulators (B- and A-type RRs) were shown to be activated in the rhizodermis and cortical cells during nodule initiation (Gonzalez-Rizzo et al., 2006; Lohar et al., 2006; Vernié et al., 2008; Ariel et al., 2012; Held et al., 2014; van Zeijl et al., 2015; Liu et al., 2018). Recent data showed that cytokinins tightly interplay with several transcriptional regulators of nodulation such as NSP2, NIN and EFD (Vernié et al., 2008; Ariel et al., 2012; Liu et al., 2018). It has been found that the transcription factor for cytokinin signalling, the B-type RR, activates NSP2, a key regulator of nodulation (Ariel et al., 2012). Analysis of mutants showed that the response to exogenously applied cytokinins depended on NSP2 and NIN, affecting nodule development (Heckmann et al., 2011). Several putative cytokinin response elements have been found in the NIN promoter region and shown to be important to regulate NIN expression in the pericycle during formation of nodule primordia (Liu et al., 2018). Another transcription factor, EFD, directly regulates the A-type RR, restricting the action of cytokinin (Vernié et al., 2008). Since EFD plays a positive role in nodule differentiation, the cytokinins may be involved in the regulation of the late stages of nodulation associated with rhizobial infection and subsequent plant cell and bacteroid differentiation. This suggests cross-talk between the regulators of cytokinin- and Nod factor-activated signalling, which requires further investigation.
To elucidate the specific functions of cytokinin and auxin signalling during the late stages of symbiosis development, we analysed the cytokinin and auxin dynamics in wild-type pea plants and Fix– mutants defective in the genes encoding key transcription factors IPD3/CYCLOPS (sym33 mutants) and EFD (sym40 mutant). The mutants in the sym33 gene were impaired in the infection process due to the absence of bacterial release from the infection threads (Tsyganov et al., 1998; Voroshilova et al., 2009; Tsyganova et al., 2019). The sym40 mutant had nodules with abnormal histological organization, hypertrophied infection droplets and abnormal bacteroids (Tsyganov et al., 1998). The results revealed that the cytokinin distribution was significantly altered in these mutants impaired in the late stages of nodule development. To further elucidate the link between the cytokinin response and localization and abnormalities in the late stages of nodulation, we also assessed the cytokinin distribution in pea sym31 and sym26 mutants defective in plant and bacteroid differentiation (Borisov et al., 1997; Serova et al., 2018).
MATERIALS AND METHODS
Plant material, bacterial strains and plant growth conditions
Pea (Pisum sativum L.) SGE and Sprint-2 wild-type lines and derived mutant lines having nodule development arrest at different stages were used in this study (Table 1). To perform gene expression studies, surface-sterilized seeds were transferred to Petri dishes with 1 % water agar and germinated in the dark at 23 °C for a 4–5 d period. Following germination, plants were transferred into pots with vermiculite and grown in a growth chamber at 21 °C and 60 % humidity in a 16 h/8 h light/dark cycle. For immunolocalization analysis, three independent experiments were performed. Surface-sterilized seeds were transferred into vermiculite supplemented with nutrient solution without nitrogen (Fåhraeus, 1957). In all experiments, the seedlings were inoculated with Rhizobium leguminosarum bv. viciae strain 3841 (Wang et al., 1982). Growth conditions for plants and bacteria were as described previously (Ivanova et al., 2015). Nodules were harvested 2 and 4 weeks after inoculation. For each variant, 10–15 nodules from different plants were analysed.
Table 1.
Plant material used in the study
| Lines | Phenotype | References |
|---|---|---|
| SGE | Wild type | Kosterin and Rozov (1993); Tsyganov et al. (1998) |
| SGEFix−-2 (sym33-3) | ‘Locked’ infection threads inside the nodule, occasional bacterial release | Tsyganov et al. (1994, 1998, 2011); Voroshilova et al. (2001, 2009 ) |
| SGEFix−-5 (sym33-2) | ‘Locked’ infection threads inside the nodule, no bacterial release | Ovchinnikova et al. (2011); Tsyganov et al. (2013); Tsyganova et al. (2019) |
| SGEFix−-1 (sym40) | Hypertrophied infection droplets and infection threads, abnormal bacteroids | Tsyganov et al. (1994, 1998); Voroshilova et al. (2009) |
| SGEFix−-3 (sym26) | Premature degradation of symbiotic structures | Tsyganov et al. (2000); Serova et al. (2018) |
| Sprint-2 | Wild type | Borisov et al., (1994, 1997) |
| Sprint-2Fix−(sym31) | Undifferentiated bacteroids, several bacteroids are surrounded by a common symbiosome membrane | Borisov et al. (1994, 1997) |
RNA isolation and cDNA synthesis
Pea nodules were harvested 2 and 3 weeks after inoculation and frozen in liquid nitrogen. Total RNA was isolated from approx. 50–100 mg of tissue per sample using the TriZol reagent (Thermo Fisher Scientific, USA) as previously described (Azarakhsh et al., 2015). RNA (1–2.5 μg) was used as template for cDNA synthesis with RevertAid Reverse Transcriptase (Thermo Fisher Scientific) for 1 h at 42 °C followed by 5 min at 95 °C. Aliquots of cDNA were diluted 1:10 for subsequent quantitative PCR analysis.
Quantitative reverse transcription–PCR (qRT–PCR) analysis
The qRT–PCR analysis was performed on a CFX-96 real-time PCR detection system with a C1000 thermal cycler (Bio-Rad Laboratories, USA). Relative expression was normalized against constitutively expressed Ubiquitin and Actin genes from pea. Each PCR was carried out with SYBR Green intercalating dye in a total volume of 10 µL. Reactions were conducted in triplicate and then averaged. Cycle threshold (Ct) values were obtained using the accompanying software, with data analysed according to the 2–ΔΔCT method (Livak and Schmittgen, 2001). PCR primers were designed using the Vector NTI program (Thermo Fisher Scientific), with all primer sequences employed in expression analysis listed in Supplementary data Table S1. Each experiment was repeated a minimum of three times using independent biological samples (n = 3–4).
Agrobacterium rhizogenes-mediated plant transformation
For plant transformation, 4- to 5-day-old pea seedlings were transferred in sterile dark plastic boxes with Jensen’s medium to light conditions and incubated for 3–4 d. Seedlings were cut at the hypocotyl region and transformed with freshly grown Agrobacterium rhizogenes strain ARqua 1 (Quandt et al., 1993) carrying an appropriate plasmid. DR5::GFP-NLS (NLS = nuclear localization signal; Ilina et al., 2018) and MtRR4::GUS (Plet et al., 2011) were used for transformation. Plants were placed in plastic vessels (Duchefa, The Netherlands) on Jensen’s agar, with the area exposed by the cut covered with wet cotton wool and aluminium foil (Leppyanen et al., 2019). The seedlings were co-cultivated with A. rhizogenes for a period of 10–14 d at 21 °C (16 h/8 h light/darkness), transferred to Emergence medium containing 150 mg mL–1 cefotaxime and incubated for 3–4 d. Before transferring into pots with vermiculite saturated with Jensen’s medium containing 1.5 mm NH4NO3, emerging roots were analysed under the fluorescent stereomicroscope on a SteREO Discovery, V8 (Zeiss, Germany). Transgenic roots were selected by visualization of DsRED or GFP (green fluorescent protein) expression. The primordia and nodules which appeared were used for GFP and β-glucuronidase (GUS) localization. To localize GFP, the roots with primordia and nodules were fixed in freshly prepared fixative solution and sectioned as previously described (Ilina et al., 2018). For GUS staining, the material was fixed in freshly prepared 3 % paraformaldehyde, 0.1 % Tween-20 in phosphate-buffered saline (PBS; pH 7.4) under vacuum (−0.9 bar; ME 1C vacuum pump, Vacuubrand, Germany) for 3 min, three times at 10 min intervals. GUS staining was performed overnight at 37 °C. Imaging and analysis of sections of DR5::GFP-NLS-transformed nodules were carried out using the laser scanning confocal system LSM 510 META equipped with ZEN 2009 software (Zeiss). Sections of GUS-stained nodules were examined on an Axio Imager.Z1 (Zeiss) microscope, and were imaged with an Axiocam 506 colour (Zeiss) digital microscope camera. Two independent experiments were performed for each variant. A total of 10–15 nodules from different transgenic roots (n = 8–10) were used for analysis in each variant.
Trans-zeatin riboside immunolocalization and confocal laser scanning microscopy
For immunolocalization of trans-zeatin riboside, collected nodules were fixed and sectioned as described by Kitaeva et al. (2016) with minor changes. Material was fixed in freshly prepared fixative solution (3 % paraformaldehyde, 0.5 % glutaraldehyde, 0.1 % Tween-20, 0.1 % Triton X-100) in 1/3 MTSB (50 mM PIPES, 5 mM MgSO4·7H2O, 5 mM EGTA, pH 6.9) under a vacuum (−0.9 bar; ME 1C vacuum pump, Vacuubrand) for 7 min, three times at 15 min intervals. Then samples were incubated in the fixative solution overnight at 4 °C, followed by rinsing in 1/3 MTSB three times. Nodules were mounted into 3 % agarose blocks and longitudinally sectioned (50 μm) using a HM650V microtome with a vibrating blade (Microm, Germany). Sections were incubated in blocking solutions, first in 5 % bovine serum albumin (BSA), 0.5 % goat serum and 0.2 % cold fish gelatin in MTSB for 30 min at 28 °C, followed by 2 mg mL−1 acetylated BSA (BSA-C) in Tris-buffered saline (TBS; 50 mm Tris–HCl, 150 mm NaCl, pH 7.5) for 30 min at 28 °C. Next, sections were incubated overnight at 4 °C in 1 % BSA in TBS with primary rabbit anti-trans-zeatin riboside IgG or rabbit anti-N6-isopentenyladenosine IgG (Agrisera, Sweden) at 1:100 dilution. Sections were washed in TBS ten times for 10 min then incubated in 5 % BSA in TBS for 25 min at 28 °C. Sections were then incubated in 1 % BSA in TBS with secondary goat anti-rabbit IgG Alexa Fluor 488 (Thermo Fisher Scientific) at 1:750 dilution for 90 min at 28 °C. In order to visualize nuclei and bacteria, sections were then washed three times for 10 min in TBS and stained with propidium iodide (0.5 μg mL−1) for 8 min. Sections were then washed twice in TBS for 10 min and mounted under coverslips in ProLong Gold antifade reagent (Thermo Fisher Scientific).
As a positive control for the primary antibody specificity, nodules were incubated in 40 mM trans-zeatin riboside (Sigma-Aldrich, USA), 0.1 % Tween-20 and 0.1 % Triton X-100 in 1/3 MTSB under vacuum (−0.9 bar, three times for 20 min, at 15 min intervals) before fixation (Supplementary data Fig. S1A–C). As a blank control, nodules were fixed in FAA (3.8 % paraformaldehyde and 3.8 % acetic acid in 70 % ethanol) solution (Supplementary data Fig. S1D–F). To ensure binding specificity of secondary antibodies with primary antibodies, a negative control was performed with anti-trans-zeatin riboside antibody omitted during the immunolocalization protocol (Supplementary data Fig. S1G–I). Analysis of sections was carried out using the laser scanning confocal systems LSM 510 META or LSM 780 (Zeiss), equipped with ZEN 2009 or ZEN 2.3pro software (Zeiss), respectively.
Immunogold labelling and transmission electron microscopy
The immunogold labelling procedure was described previously (Tsyganova et al., 2009). Sections were treated with the primary rabbit anti-trans-zeatin riboside IgG antibody (Agrisera) diluted 1:25 in 0.1 % BSA-C in PBS (2.48 g L–1 NaH2PO4, 21.36 g L–1 Na2HPO4, 87.66 g L–1 NaCl, pH 7.2) at 4 °C overnight. After rinsing four times in 0.1 % BSA-C in PBS, samples were incubated with a 10 nm gold-conjugated secondary antibody goat anti-rat IgG (Amersham International, UK), diluted 1:50 in 0.1 % BSA-C in PBS for 4 h at 37 °C. The grids containing the sections were counterstained with 2 % aqueous uranyl acetate for 1 h followed by Reynolds’ lead citrate for 1 min. Nodule tissues were examined under a transmission electron microscope, JEM-1400 (Jeol, Japan), at 80 kV. Images were obtained using a Veleta side-mounted CCD camera (Olympus, Germany).
RESULTS
Сytokinin and auxin response patterns in wild-type pea nodules
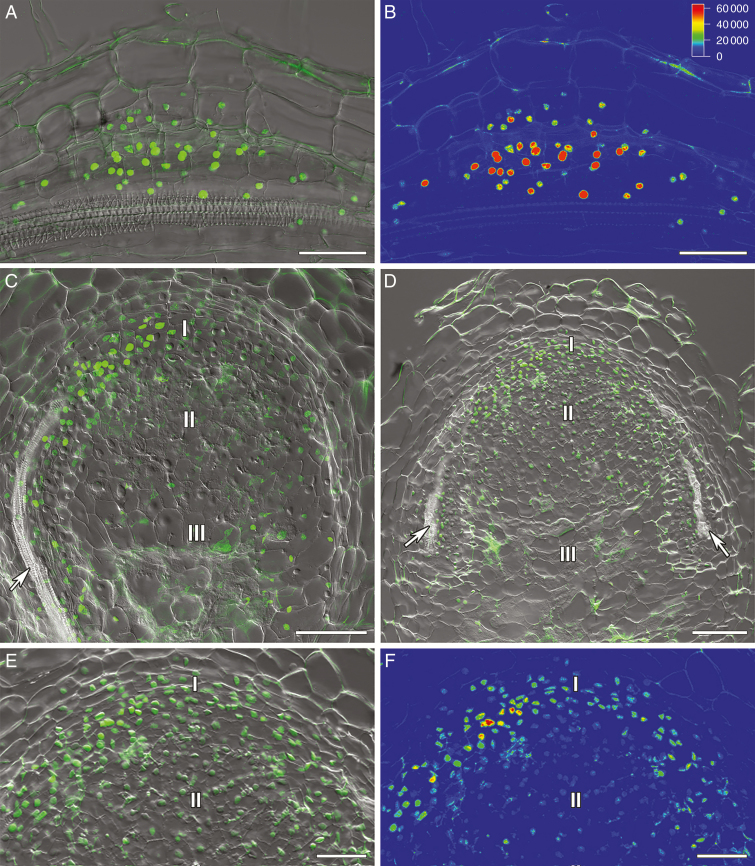
The pattern of cytokinin and auxin distribution was previously investigated in wild-type nodules of model legumes M. truncatula and L. japonicus, and more recently in soybean (Glycine max) (Plet et al., 2011; Suzaki et al., 2012; Held et al., 2014; Fisher et al., 2018). However, there was only limited information about cytokinin and auxin localization in pea nodules. To study the importance of auxin and cytokinin in the development of nodules in pea, composite plants carrying genetic constructs with auxin- and cytokinin-responsive promoters and reporter genes DR5::GFP-NLS (Ilina et al., 2018) and pMtRR4::GUS having four 12 bp RR-binding sites (RRBSs) (Ariel et al., 2012) were examined. In pea SGE wild-type plants containing the DR5::GFP-NLS construct, the auxin response maxima were observed during the early stages of nodule development in dividing cells of the pericycle, endoderm and inner cortex of the root (Fig. 1A, B). In 2-week-old nodules, the response to auxin was localized in the meristem, bordering cells around vascular bundles and peripheral tissues (Fig. 1C; Supplementary data Fig. S2A, B). A similar pattern was found in 3-week-old nodules (Fig. 1D–F; Supplementary data Fig. S2C, D), indicating the possible role of auxin in maintaining the meristem activity and the processes occurring in the vasculature.
Fig. 1.
Visualization of auxin response maxima in the nodule primordia and nodules of composite SGE pea wild-type plants containing the DR5::GFP-NLS construct. (A, B) Nodule primordium. (C) Two-week-old nodule. (D–F) Three-week-old nodules: (D) general view; (E, F) meristem. (A, C, D, E) Merge of differential interference contrast and green channel used for the GFP-NLS signal in green. (B, F) The heatmap shows colour-coded fluorescence signal intensities for the green signal channel; the quantification scale is the same for B and F images. I, meristem zone; II, infection zone; III, nitrogen fixation zone; arrows indicate vascular bundles. Scale bars are 50 µm (A, B, E, F) and 100 µm (C, D).
Analysis of transgenic roots containing the pMtRR4::GUS construct revealed the response to cytokinins in the primordia and emerging nodules (Fig. 2A, B). In the 2-week-old pea nodules, the GUS reporter was detected in the meristem, the infection zone and the apical part of the nitrogen fixation zone (Fig. 2C, D). These observations suggested that the cytokinins participate not only in maintaining the meristem activity in nodules, but also in controlling the late stages of the nodule development associated with the infection process and the differentiation of nodule tissue. Since our analysis indicated that cytokinins could be important for regulation of the late stages of symbiosis development, we performed a more detailed analysis of the cytokinin distribution in pea mutants defective in bacterial penetration into plant cells and subsequent tissue differentiation.
Fig. 2.
Localization of RR4 expression in pea nodule primordia and 2-week-old nodules expressing a pMtRR4::GUS genetic construct. (A, B) Nodule primordia; pMtRR4::GUS expression is visible in the pericycle and inner cortical cells of composite SGE pea wild-type plants. (C, D) Two-week-old pea nodules. Cytokinin response is stimulated in nodule primordia and in the meristem, infection zone and apical part of the nitrogen fixation zone of 2-week-old pea nodules. I, meristem zone; II, infection zone; III, nitrogen fixation zone; arrows indicate vascular bundles. Scale bars are 50 µm (A, B), 100 µm (C) and 75 µm (D).
Cytokinin response patterns in sym33-2 and sym33-3 mutants defective in the gene encoding the IPD3/CYCLOPS transcription factor
The transcription factor IPD3/CYCLOPS encoded by the Sym33 gene in pea may be involved in infection development and intracellular bacterial accommodation (Tsyganov et al., 1998; Ovchinnikova et al., 2011; Tsyganova et al., 2019), as was shown in model legumes (Messinese et al., 2007; Yano et al., 2008). In addition, IPD3/CYCLOPS is also required for transactivation of NIN transcription factor, which may be important for regulation of organogenesis (Singh et al., 2014). Moreover, recent studies in pea forming the indeterminate nodule type allowed the discovery of a strong mutant allele, sym33-4, that failed to develop nodules (Zhernakov et al., 2019). The pattern of cytokinin distribution in ipd3/cyclops mutants of legumes with the indeterminate nodule type has not been studied in detail. To investigate the cytokinin distribution in sym33-2 and sym33-3 mutants, the analysis was performed in composite pea plants. We found that the cytokinin distribution was significantly altered in the transgenic sym33-2 and sym33-3 mutants containing pMtRR4::GUS.
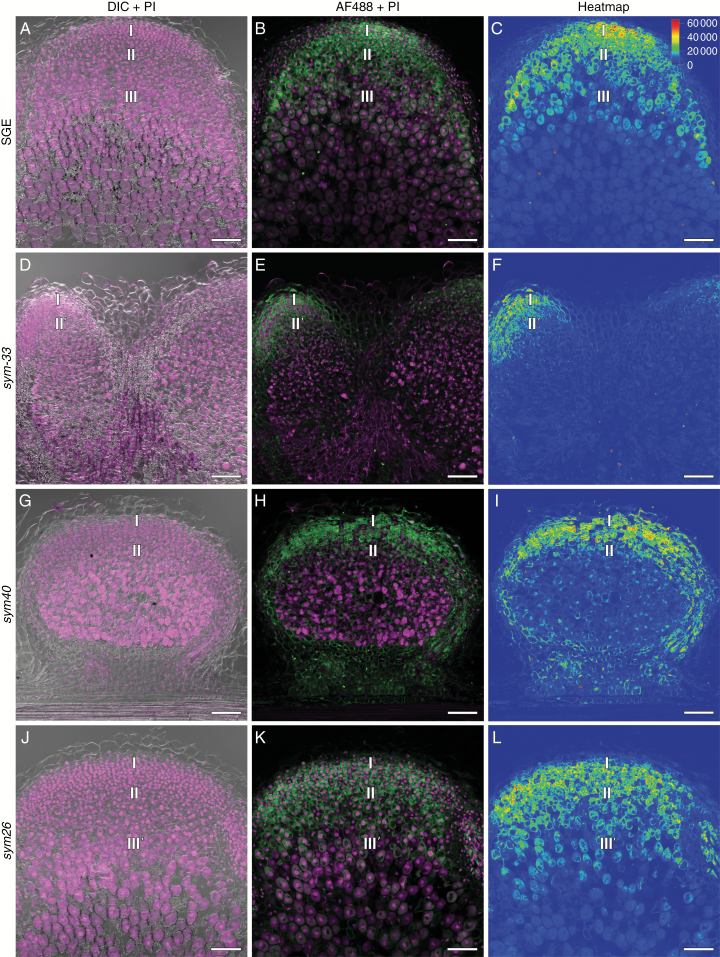
The distribution of the cytokinin response was reduced in the sym33-2 and sym33-3 mutants as compared with that in the wild-type line SGE (Fig. 3A). In 2-week-old nodules of sym33-2 and sym33-3, the GUS reporter was detectable only in vascular tissues, and weak staining was associated with the meristem (Fig. 3B, C). Thus, the changes in the biosynthesis and distribution of cytokinins in the pea sym33-2 and sym33-3 mutants may be related to the disturbances in the development of symbiosis. In mutants impaired in the Sym33 gene, due to the impaired infection process and the absence of release of bacteria from the infection threads (Tsyganov et al., 1998; Voroshilova et al., 2009; Tsyganova et al., 2019), the differentiation of nodule tissue probably does not occur. Thus, cytokinins could be involved in differentiation control during the late stages of symbiosis development. At the same time, it could not be excluded that the level of bacteroid differentiation may affect cytokinin accumulation in the infected cells of nodules.
Fig. 3.
Localization of RR4 expression in pea nodules of the wild type and mutants expressing the pMtRR4::GUS construct 2 weeks after inoculation. (A) Two-week-old SGE pea nodules. (B) sym33-3 mature nodule. pMtRR4::GUS expression is detected in peripheral tissues including vascular bundles. (C) sym33-2 mature nodule. (D) sym40. I, meristem zone; II, infection zone; II', infection thread propagation zone; III, nitrogen fixation zone; arrows indicate vascular bundles. Scale bars are 100 µm (A, B, D) and 50 µm (C).
Distribution of cytokinins in the sym40 mutant defective in the gene encoding the EFD transcription factor
The pea Sym40 gene is orthologous to the M. truncatula EFD gene (Nemankin, 2011). Analysis of mutants showed that the transcription factor EFD is required for the formation of functional nodules and essential for nodule differentiation in M. truncatula and pea (Tsyganov et al., 1998; Vernié et al., 2008; Voroshilova et al., 2009). In addition, because the M. truncatula efd mutants and pea sym40 mutant had more nodules, along with more frequent infection threads in the rhizodermis and root cortex, the EFD transcription factor may exert negative nodulation control (Vernié et al., 2008; Voroshilova et al., 2009).
In the transgenic sym40 mutant, containing the pMtRR4::GUS construct, we detected a cytokinin response in the central part of the nodules where the release of bacteria occurred (Fig. 3D). This part can be described as the infection zone of the mutant, although it significantly differed from the nodules of the wild-type line SGE. We concluded that the cytokinin accumulation in the central part of the nodule and the release of bacteria into the plant cells may be interconnected. The disturbances of differentiation processes in the sym40 mutant (Voroshilova et al., 2009) indicated that the cytokinins could play a role in regulating plant cell differentiation.
Immunolocalization of trans-zeatin riboside and N6-isopentenyladenosine in the wild type and mutants blocked at different stages of nodule development
To further study the role of cytokinins in the differentiation of nodules and rhizobial cells, we assessed immunolocalization of trans-zeatin riboside and N6-isopentenyladenosine, transport forms of cytokinins, in nodules of the wild type and the corresponding sym33-3 and sym40 mutants defective in the genes encoding the IPD3/CYCLOPS and EFD transcription factors.
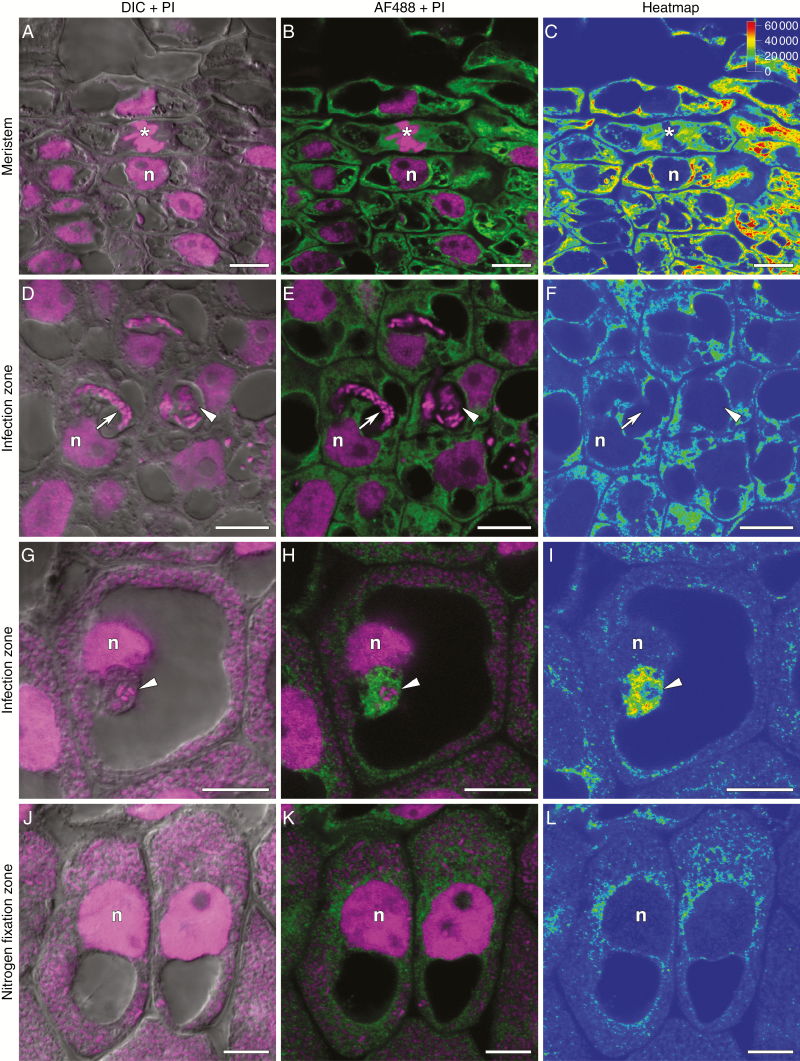
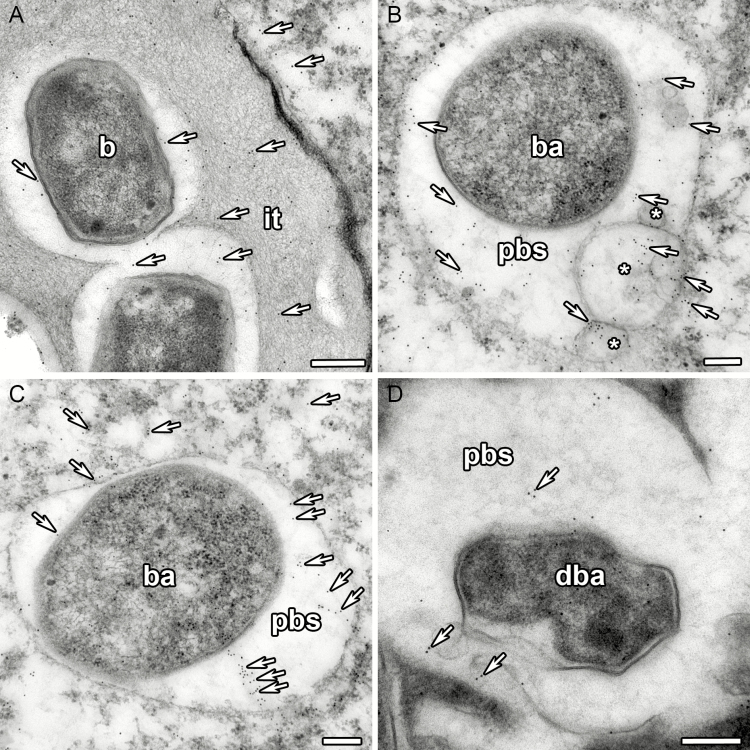
In 2-week-old nodules of the wild-type line SGE (Fig. 4A–C), the anti-trans-zeatin riboside antibody signal was detected in the meristem (Fig. 5A–C), the infection zone and the proximal part of the nitrogen fixation zone. A similar pattern was observed for N6-isopentenyladenosine in the 2-week-old nodules of the SGE wild type (Supplementary data Fig. S3A–C, G–I). These results were consistent with the data obtained with the cytokinin-responsive construct. In the early infection zone, trans-zeatin riboside was found in the plant cell cytoplasm but was absent in the infection threads and infection droplets (Fig. 5D–F). However, in the late infection zone and the proximal part of the nitrogen fixation zone, trans-zeatin riboside was present in the infection threads and infection droplets (Fig. 5G–I). The immunogold labelling of trans-zeatin riboside detected gold particles in the matrix of infection threads and in the exopolysaccharide capsule of bacteria (Fig. 6A). In these zones, the signal for trans-zeatin riboside was associated with the symbiosomes (Fig. 5D–L). The immunogold labelling confirmed the large amount of trans-zeatin riboside in symbiosomes, both in the peribacteroid space and within the bacteroids (Fig. 6C). Vesicles labelled with gold particles were associated with symbiosomes (Fig. 6B). In all zones, the signal for trans-zeatin riboside was not detected in the nuclei. In 4-week-old nodules of the SGE line (Supplementary data Fig. S4A–C), the signal intensity was much lower, especially in the nitrogen fixation zone. The immunogold labelling showed low amounts of trans-zeatin riboside in senescent symbiosomes (Fig. 6D).
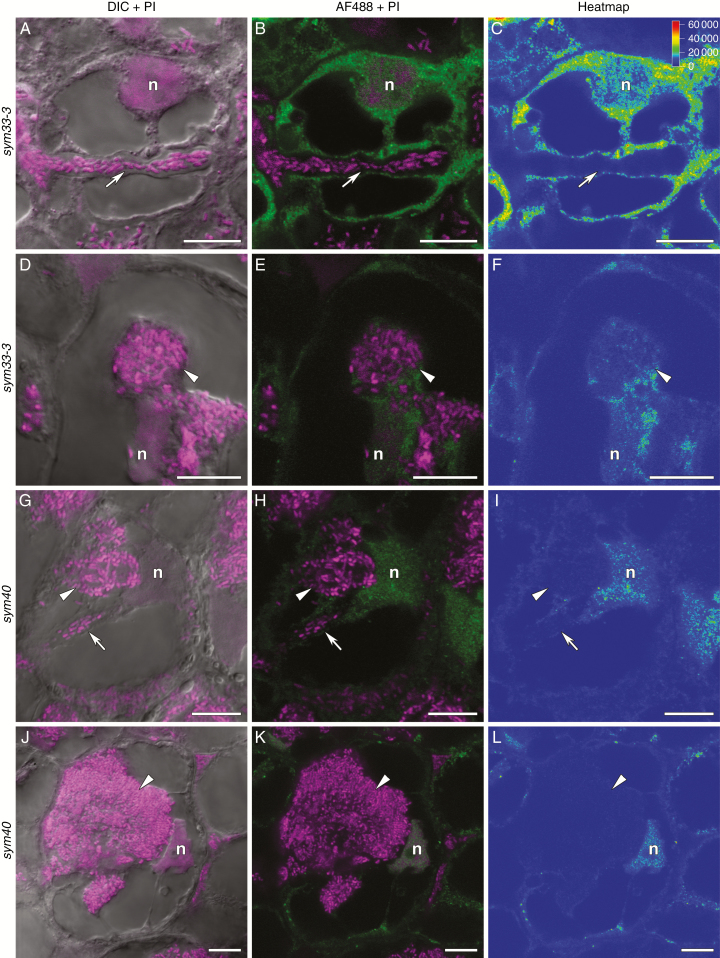
Fig. 4.
Immunolocalization of trans-zeatin riboside in the 2-week-old nodules of pea SGE wild type and corresponding mutants, general view. (A–C) SGE. (D–F) sym33-3. (G–I) sym40. (J–L) sym26. (A, D, G, J) Merge of differential interference contrast (DIC) and magenta channel. (B, E, H, K) Merge of green and magenta channels. A single optical section is presented: trans-zeatin riboside in green and DNA (bacteria and nuclei) in magenta. (C, F, I, L) The heatmap shows colour-coded fluorescence signal intensities for the green signal channel; the quantification scale is the same for all images. PI, propidium iodide; AF488, Alexa Fluor 488. I, meristem zone; II, infection zone; II', infection thread propagation zone; III, nitrogen fixation zone; III', zone corresponding to the nitrogen fixation zone of wild-type nodules. Scale bars are 100 µm.
Fig. 5.
Immunolocalization of trans-zeatin riboside in the 2-week-old nodules of pea SGE wild type. (A–C) Meristem. (D–F) Cells from the early infection zone. (G–I) Cells from the late infection zone. (J–L) Cells from the nitrogen fixation zone. (A, D, G, J) Merge of differential interference contrast (DIC) and magenta channel. (B, E, H, K) Merge of green and magenta channels. A single optical section is presented: trans-zeatin riboside in green and DNA (bacteria and nuclei) in magenta. (C, F, I, L) The heatmap shows colour-coded fluorescence signal intensities for the green signal channel; the quantification scale is the same for all images. PI, propidium iodide; AF488, Alexa Fluor 488. n, nucleus; *, mitosis; arrows indicate infection threads; arrowheads indicate infection droplets. Scale bars are 10 µm.
Fig. 6.
Immunogold localization of trans-zeatin riboside in the 2-week-old nodules of pea SGE wild type. (A) Infection thread. (B) Symbiosome with vesicles labelled with gold particles. (C) Mature symbiosome. (D) Degrading symbiosome. it, infection thread; b, bacterium; ba, bacteroid; pbs, peribacteroid space; dba, degrading bacteroid; *, vesicle; arrows indicate gold particles. Scale bars are 250 nm (A) and 200 nm (B–D).
Two-week-old nodules of the sym33-3 mutant forming nodules with ‘locked’ infection threads (Tsyganov et al., 1998) exhibited a maximal amount of trans-zeatin riboside in the meristem (Fig. 4D–F). The trans-zeatin riboside was absent in the ‘locked’ infection threads but present in the nuclei and cytoplasm of colonized cells (Fig. 7A–C) and in the infection droplets (Fig. 7D–F), which are occasionally formed (Tsyganov et al., 2011). A similar pattern was observed for N6-isopentenyladenosine (Supplementary data Fig. S3D–F, J–L). In 4-week-old nodules, the signal for trans-zeatin riboside was limited to the meristem (Supplementary data Fig. S4D–F).
Fig. 7.
Immunolocalization of trans-zeatin riboside in 2-week-old nodules of pea mutants sym33-3 (A–F) and sym40 (G–L). (A–C) Colonized cell with a ‘locked’ infection thread. (D–F) Colonized cell with an infection droplet. (G–I) Recently infected cell with an infection droplet. (J–L) Colonized cell with a hypertrophied infection droplet. (A, D, G, J) Merge of differential interference contrast (DIC) and magenta channel. (B, E, H, K) Merge of green and magenta channels. A single optical section is presented: trans-zeatin riboside in green and DNA (bacteria and nuclei) in magenta. (C, F, I, L) The heatmap shows colour-coded fluorescence signal intensities for the green signal channel; the quantification scale is the same for all images. PI, propidium iodide; AF488, Alexa Fluor 488. n, nucleus; arrows indicate infection threads; arrowheads indicate infection droplets. Scale bars are 10 µm.
Although the signal distribution patterns of trans-zeatin riboside and N6-isopentenyladenosine were similar in the nodules of SGE wild type and sym33-3 mutants, trans-zeatin riboside had a higher labelling intensity than N6-isopentenyladenosine. Thus, only trans-zeatin riboside labelling was used in the analysis of other genotypes.
In 2-week-old nodules of the sym40 mutant forming nodules with abnormal histological organization, hypertrophied infection droplets and abnormal bacteroids (Tsyganov et al., 1998), the maximum signal for trans-zeatin riboside was detected in the meristem (Fig. 4G–I) and in infected cells (Figs 4G–I and 7G–I). In contrast to wild-type nodules, the nuclei had accumulated large amounts of trans-zeatin riboside (Fig. 7G–L), while the hypertrophied infection droplets were free from the signal (Fig. 7G–L). In 4-week-old nodules of the sym40 mutant, the signal intensity for trans-zeatin riboside was reduced (Supplementary data Fig. S4G–I) as in the wild-type nodules.
Along with the sym33-3 and sym40 ineffective mutants, two other mutants, the sym26 mutant forming nodules with morphologically differentiated bacteroids and premature degradation of symbiotic structures (Serova et al., 2018) and the sym31 mutant forming nodules with undifferentiated bacteroids (Borisov et al., 1997), were used. In the 2-week-old nodules of the sym26 mutant that typically form nodules with morphologically differentiated bacteroids undergoing premature degradation (Serova et al., 2018), the distribution of trans-zeatin riboside was similar to that in the wild-type nodules (Fig. 4J–L). However, in mutant nodules, the respective signals were detected in the nuclei (Fig. 8G–L). In 4-week-old nodules, the trans-zeatin riboside level was dramatically reduced (Supplementary Fig. S4J–L). These results further confirmed that cytokinins are involved in the infection process in mature nodules and in the nodule tissue and bacteroid differentiation.
Fig. 8.
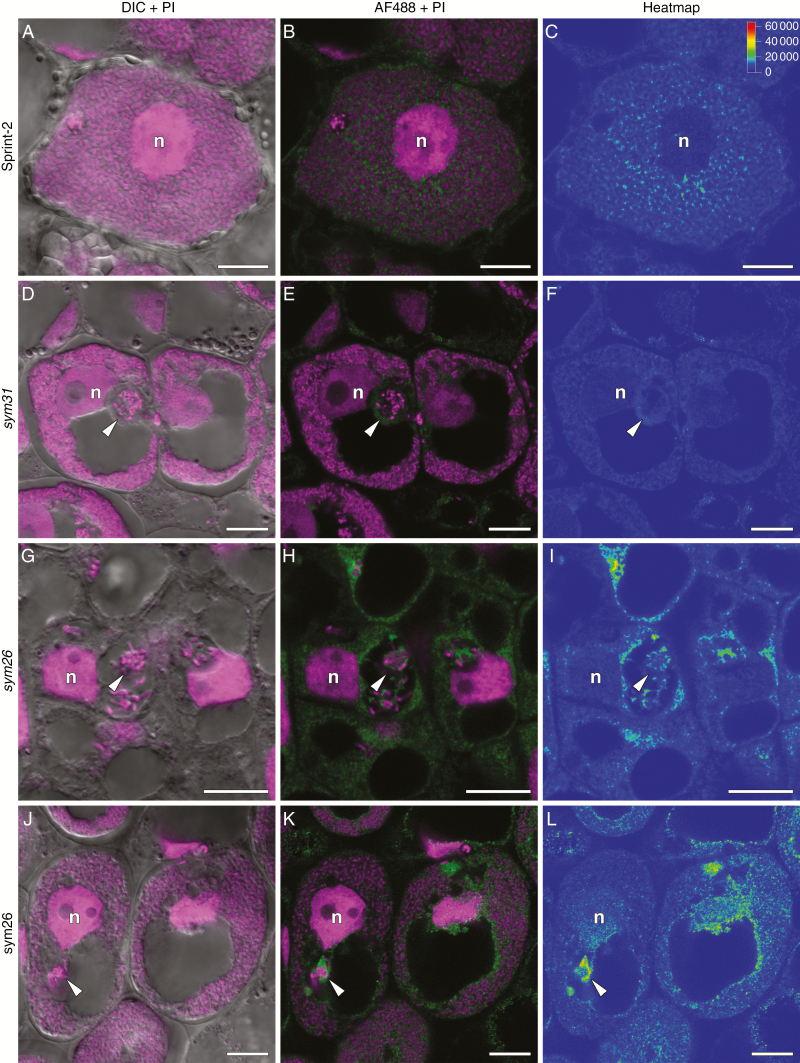
Immunolocalization of trans-zeatin riboside in 2-week-old nodules of pea Sprint-2 wild type (A–C), corresponding sym31 mutant (D–F) and sym26 mutant (G–L). (A–C) Infected cell from the nitrogen fixation zone. (D–F) Infected cell from the zone corresponding to the nitrogen fixation zone. (G–I) Recently infected cell with an infection thread and droplet. (J–L) Infected cell from the zone corresponding to the nitrogen fixation zone. (A, D, G, J) Merge of differential interference contrast (DIC) and magenta channel. (B, E, H, K) Merge of green and magenta channels. A single optical section is presented: trans-zeatin riboside in green and DNA (bacteria and nuclei) in magenta. (C, F, I, L) The heatmap shows colour-coded fluorescence signal intensities for the green signal channel; the quantification scale is the same for all images. PI, propidium iodide; AF488, Alexa Fluor 488. n, nucleus; arrowheads indicate infection droplets. Scale bars are 10 µm.
In the 2-week-old nodules of sym31 mutants that typically form nodules with undifferentiated bacteroids (Borisov et al., 1997), the strongest trans-zeatin riboside signal was observed in the meristem and the infection zone, but the signal intensity was decreased in the zone that corresponded to the nitrogen fixation zone in the wild type (Supplementary data Fig. S5D–F). In the infected cells of sym31 mutants, the signal intensity was significantly lower (Fig. 8D–F) than that in the Sprint-2 wild type (Fig. 8A–C). In 4-week-old nodules, the trans-zeatin riboside level was sharply decreased (Supplementary data Fig. S5J–L).
Expression of cytokinin metabolic genes ISOPENTENYL TRANSFERASE (IPT) and LONELY GUY (LOG) in pea wild type and mutants
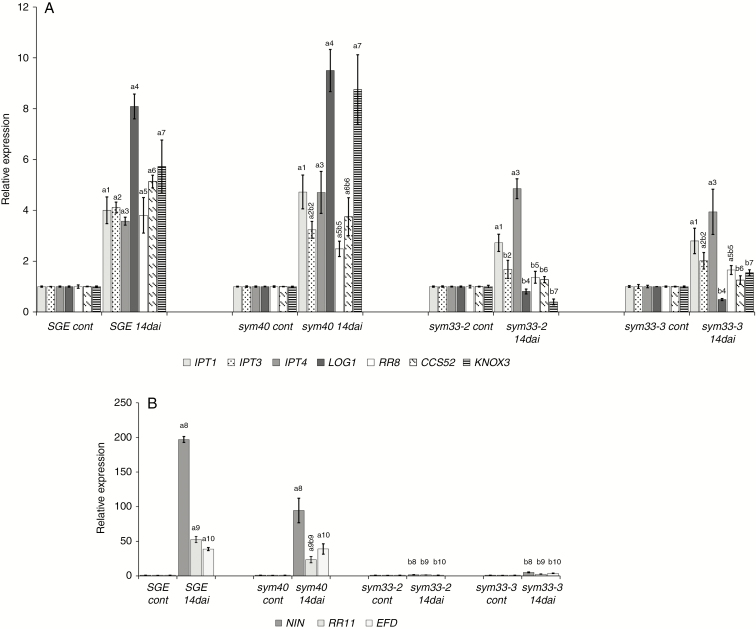
To elucidate the basis for the changes in the hormone level in developing nodules, we profiled the expression of plant genes controlling the metabolism of these phytohormones, as well as the primary response. In accordance with our previous results for other pea cultivars, the expression levels of LOG1, IPT1, IPT3, IPT4 and A-type response regulators RR8 and RR11 were up-regulated noticeably in the nodules of pea SGE wild type (Azarakhsh et al., 2015; Dolgikh et al., 2017). In contrast, in the 2-week-old nodules of sym33-2 and sym33-3 mutants, the expression levels of LOG1 and RR11 were significantly reduced (Fig. 9A, B). The expression levels of IPT3 and RR8 were also decreased as compared with those in the wild-type nodules, while the levels of IPT1 and IPT4 did not change significantly. A similar pattern was found in the mature 3-week-old nodules of these mutants (Supplementary data Fig. S6). In mutants defective in the sym33 gene, the nodules remained uninfected because they contain ‘locked’ infection threads without bacterial release into the cytoplasm of the plant cells (Tsyganov et al., 1998; Tsyganova et al., 2019). These observations suggested that stimulation of cytokinin biosynthesis and additional activation of the cytokinin receptor in pea nodules may be associated with the release of bacteria from infection threads and subsequent plant cell differentiation (Fig. 9A, B).
Fig. 9.
Expression pattern of cytokinin metabolic and regulatory genes IPT1, IPT3, IPT4, LOG1, RR8, CCS52A and KNOX3 (A), and NIN, RR11 and EFD (B) in pea wild type and sym33-2, sym33-3 and sym40 mutants 2 weeks after inoculation (14 dai). Transcript levels of genes were normalized against pea Ubiquitin and Actin genes. For each gene, the transcript level in non-inoculated roots of the wild type or mutants was set to 1 (control), and the level in nodules of the inoculated wild type or mutants was calculated relative to the control values. The graphs show the results of three independent experiments. The error bars represent the s.e.m. of three repeats. To conduct one biological repeat, the fragments of non-inoculated main roots or nodules from 3–4 plants were collected and used to isolate RNA. Pairwise comparisons were made using Dunn’s test, and the CLD algorithm was performed to summarize the sets of groups that differed significantly for each gene (groups are shown with different letters on the histogram). Kruskal–Wallis one-way analysis of variance showed significant differences in expression of genes IPT3 [H(d.f. = 3) = 9.59, P = 0.022], LOG1 [H(d.f. = 3) = 10.17, P = 0.017], RR8 [H(d.f. = 3) = 10.42, P = 0.015], KNOX3 [H(d.f. = 3) = 11.18, P = 0.011], NIN [H(d.f. = 3) = 11.36, P = 0.0099], EFD [H(d.f. = 3) = 9.56, P = 0.023] and RR11 [H(d.f. = 3) = 10.51, P = 0.015] depending on the type of plant.
To test this assumption, we also conducted an analysis of the sym40 pea mutant, in which the infection process and differentiation were impaired. In the nodules of this mutant, the hypertrophic infection threads were formed but the release of bacteria from the infection threads occurred with a delay (Tsyganov et al., 1998; Voroshilova et al., 2009). In contrast to the sym33-2 and sym33-3 mutants, an increase of LOG1, IPT3, RR8 and RR11 gene expression was detected in the nodules of the sym40 mutant (Fig. 9A, B), where the infection of the nodules occurred, but subsequent stages of plant cell and bacteroid differentiation were impaired. This indicated a link between the activation of these genes and certain stages of nodule development.
The balance between cell proliferation and differentiation may be controlled by cell cycle regulators. The transition of cells to endoreduplication may be linked to the expression of CCS52A (Takahashi et al., 2013). In contrast to the wild type and the sym40 mutant, we were not able to detect an increase in the level of CCS52A in the nodules of sym33-2 and sym33-3 mutants (Fig. 9A, B).
It has been shown that the KNOX3 transcription factor may be involved in the upregulation of LOG and IPT genes in pea (Azarakhsh et al., 2015). In addition, the NIN transcription factor is known to induce the expression of the cytokinin receptor CRE1 in cortical cells (Vernié et al., 2015). In our experiments, the expression of KNOX3 and NIN was significantly stimulated in the 2-week-old nodules of the SGE wild type and sym40 mutant, but not in sym33-2 and sym33-3 mutants (Fig. 9A, B). The decreased level of KNOX3 expression in sym33 mutants was also found previously (Dolgikh et al., 2019). These observations suggested that the KNOX3 and NIN transcription factors were involved in promoting the accumulation of cytokinins in pea nodules.
Previous studies in legume plants have shown that the transcription factor EFD is required for the formation of functional nodules and essential for nodule differentiation (Vernié et al., 2008). In our experiments, we were not able to detect an increased EFD level in the nodules of sym33-2 and sym33-3 mutants in contrast to the wild type (Fig. 9A, B). These observations suggested that activation of EFD in the wild type may be associated with the subsequent nodule differentiation.
DISCUSSION
In addition to studying the well-established function of cytokinin and auxin in the initiation of nodule formation (for a recent review, see Gamas et al., 2017), in this study, we have investigated the effect of these phytohormones on other aspects of nodulation. In 2- and 3-week-old pea nodules, the response to auxin was localized in the meristem, bordering cells around vascular bundles and peripheral tissues. Similar localization patterns have been reported in the nodules of other legumes, such as L. japonicus, Trifolium repens and G. max (Mathesius et al., 1998; Suzaki et al., 2012; Fisher et al., 2018). This pattern may indicate a critical role for auxin in maintaining the meristem and functioning of the vasculature. A previous report indicated that 2- and 3-week-old pea nodules had maximal cytokinin levels in the meristem and infection zone, and 4-week-old nodules maintained a similar level of cytokinins in the meristem but showed significantly reduced levels in the infection zone (Syōno et al., 1976). Owing to the correlation between the decrease in cytokinin levels with age of the nodules and the decrease in their meristematic activity, it has been suggested that cytokinins affect nodule morphogenesis by regulating the mitotic activity of the nodule meristem (Syōno et al., 1976). In our experiments, we used reporter constructs and assessed the immunolocalization of trans-zeatin riboside and N6-isopentenyladenosine in 2- and 4-week-old wild-type pea nodules and found that the cytokinin distribution profile resembled the previously described pattern (Fig. 10). We propose that in addition to the localization of cytokinins in the meristem of wild-type nodules, their presence in the infection zone and apical part of the nitrogen fixation zone supports a potential role for cytokinins in the control of intracellular bacterial accommodation and tissue differentiation. The first experimental evidence for this assumption was obtained using M. truncatula, a legume with a similar nodulation type, demonstrating that the M. truncatula cre1 mutant defective in cytokinin perception displayed significantly disturbed zonation and tissue differentiation in rarely appearing nodules (Plet et al., 2011). In Arachis hypogaea, switching off AhHK1 encoding cytokinin receptor histidine-kinase1 led to the formation of undifferentiated nodules associated with the proliferation of infected cells (Kundu and DasGupta, 2017). To study the potential role of cytokinins in nodulation regulation, we performed a detailed analysis using pea ineffective mutants impaired in the genes encoding IPD3/CYCLOPS and EFD transcription factors.
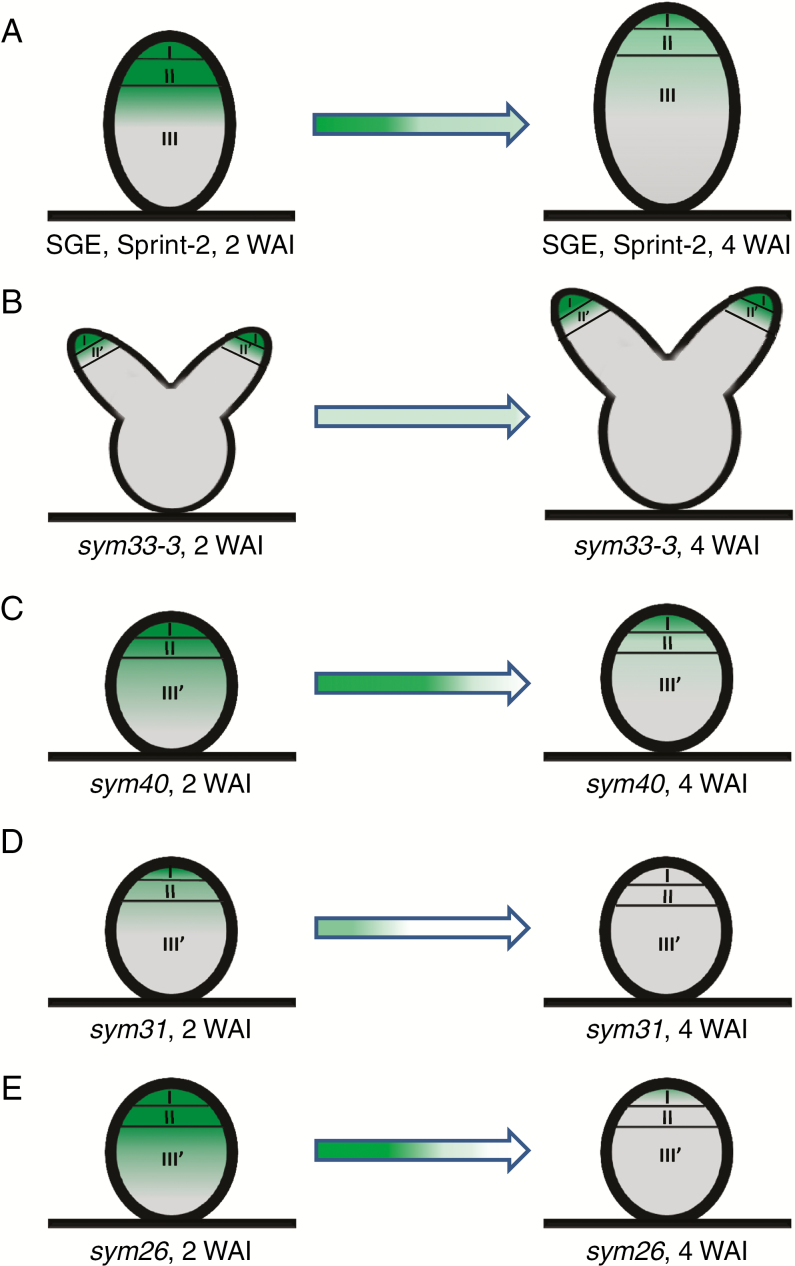
Fig. 10.
Scheme of cytokinin localization in nodules of the wild type (A) and mutants sym33-3 (B), sym40 (C), sym31 (D) and sym26 (E) at 2 and 4 weeks after inoculation (WAI). Zones of the nodule are designated by Roman numerals: I, meristem; II, infection zone; II', infection thread propagation zone; III, fixation zone; and III', zone corresponding to the nitrogen fixation zone in the wild type. A decrease in the colour intensity of the arrow indicates a reduction in cytokinin levels.
Altered patterns of cytokinin response and immunolocalization in sym33 and sym40 mutants defective in IPD3/CYCLOPS and EFD transcription factors, respectively, indicate their involvement in the control of the late stages of symbiosis
In this study, we found altered patterns of cytokinin response and immunolocalization in sym33 and sym40 mutants defective in IPD3/CYCLOPS and EFD transcription factors, respectively. In sym33 mutants, the cytokinin localization area was mostly limited to the meristem, whereas cytokinins were absent in the central part of non-infected nodules (Fig. 10B). In contrast, in the sym40 mutant, cytokinins were detected in the infection zone, where the bacteria are released into the cytoplasm of plant cells (Fig. 10C). However, the bacterial release into plant cells was delayed in this mutant. Therefore, the enhanced cytokinin accumulation during the late stages of symbiosis development may be linked to bacterial penetration into the plant cells and subsequent plant cell differentiation. The interconnection between cytokinin regulation and bacterial release has been previously suggested (Held et al., 2014). Indeed, in the L. japonicus lhk1 mutant, the delayed formation of rare nodules was completely abolished by this mutation that prevented bacterial entry (Held et al., 2014).
In legumes with indeterminate nodule type, the primary cytokinin accumulation occurred in the rhizodermis (root hairs) and later in the cells of developing primordia (Lohar et al., 2004; Op den Camp et al., 2011; Jardinaud et al., 2016). The sym33 mutants developed a reduced number of nodule primordia due to an impaired infection process, and these nodule primordia remained uninfected because the infection threads grew only in the outer cortex and did not penetrate the primordia (Voroshilova et al., 2009). In contrast, co-ordinated infection thread development and nodule organogenesis occurred in the sym40 mutant, and infection threads penetrated the nodule primordia (Voroshilova et al., 2009). These observations indicated a co-ordinated signal exchange between the rhizodermis and the cortical layers during nodule development in the sym40 mutant but not in the sym33 mutants.
Role of the NIN and KNOX3 transcription factors in the regulation of late stages of nodulation
Potential candidates for regulators that perform such non-cell-autonomous functions are transcription factors that are activated in response to Nod factor exposure in the rhizodermis and may be translocated into cortical cells. Indeed, in the nodules of the sym33 mutants, the expression levels of genes encoding the NIN and KNOX3 transcription factors were significantly reduced. Owing to the involvement of IPD3/CYCLOPS in the transcription activation of NIN (Singh et al., 2014) and its potential translocation into the cortical cells (Vernié et al., 2015), the mutations in the Sym33 gene may interrupt a co-ordinated signal exchange between tissue layers. In this study, we also showed a possible link between the IPD3/CYCLOPS transcription factor and KNOX3 activation. As previously reported, the KNOX transcription factors perform non-cell-autonomous functions in plants (Hake et al., 2004). This suggests that KNOX3 may play a critical role in the signal exchange between the rhizodermis and the cortical layers during the early stages of symbiosis development. However, this suggestion needs to be verified in further investigations.
The lack of cytokinins in the central part of nodules in the sym33 mutants defective in bacterial accommodation demonstrates that regulators controlled by the IPD3/CYCLOPS transcription factor may be involved in signal transduction that triggers secondary cytokinin accumulation in nodules. Previously, the involvement of the KNOX3 transcription factor in activating genes required for cytokinin biosynthesis regulation has been confirmed in pea and M. truncatula (Azarakhsh et al., 2015; Di Giacomo et al., 2017). Based on this finding, we used pea mutants to examine the expression of crucial genes controlling cytokinin biosynthesis and response. Decreased expression levels of LOG1 and IPT3, along with those of RR8 and RR11, in the sym33 mutants may represent further evidence that KNOX3 acts during the late stages of nodule development and in the active nodule. It is interesting to note that the NIN transcription factor can stimulate CRE1 expression in cortical cells during nodule formation (Vernié et al., 2015). In addition, the NIN expression may be dependent on the cytokinins, because cytokinin response elements have been found in the NIN promoter region (Liu et al., 2018). Therefore, activation of KNOX3 and NIN expression during the late nodulation stages may be linked to cytokinin accumulation and perception in the infection zone and apical part of the nitrogen fixation zone. At the current stage of research, the analysis of infection-impaired pea mutants revealed the relationship among NIN and KNOX3 activation, bacterial release and cytokinin accumulation in nodules. The bacterial release and continued Nod factor production in bacteroids may be important for cytokinin biosynthesis stimulation. However, this assumption requires further investigation. Which processes can be potentially regulated by these cytokinins?
Influence of cytokinins on plant cell differentiation
Recent studies identified legume mutants defective in the genes necessary to control endoreduplication, which is related to the process of tissue differentiation during nodule formation (Suzaki et al., 2014; Yoon et al., 2014). Since the transition of cells to endoreduplication can be directly controlled by cytokinins, as demonstrated in arabidopsis (Takahashi et al., 2013), the cytokinins in legumes may affect tissue differentiation during nodule development. To test this hypothesis, we performed an expression analysis of CCS52A in sym33 and sym40 mutants. We failed to detect CCS52A upregulation in sym33 mutants, suggesting a link between cytokinins and endoreduplication control. Some CCS52A expression was detected in the sym40 mutant but it was not as pronounced as that in the wild-type pea, indicating an association with the tissue differentiation defects in the mutant (Voroshilova et al., 2009). The impaired differentiation in sym33 mutants corresponded to a reduced expression level of the gene encoding the EFD transcription factor that acts during the late stages of nodule development. This implies that regulation of plant tissue differentiation involving cytokinins, CCS52A and EFD is required for the formation of functional nitrogen-fixing nodules.
The underlying mechanism for the participation of phytohormones in cell differentiation in nodule tissues has yet to be elucidated, but a detailed analysis of this regulation has been performed on the roots of the model plant arabidopsis. Importantly, cytokinins are involved in regulating the root system architecture. In the parental root meristem, cytokinin signalling ensures the balance between cell proliferation and differentiation (Ioio et al., 2007; Moubayidin et al., 2010). Cytokinins control the cell differentiation rate by negatively regulating the positive effect of auxin on cell proliferation. In the arabidopsis main root meristem, cytokinins, via the receptor AHK3 and B-type response regulators ARR1 and ARR12, directly activate the transcription of SHY2/IAA3, a member of the Aux/IAA family of auxin signalling repressors (Ioio et al., 2007, 2008). Since SHY2/IAA3 negatively regulates PIN expression, its activation results in limiting auxin transport and allows cell differentiation in the root transition zone (Ioio et al., 2008). In addition, the cytokinin-activated B-type response regulator ARR2 directly upregulates the expression of CCS52A1, which encodes an activator of an E3 ubiquitin ligase, also described as the anaphase-promoting complex/cyclosome, mediating degradation of cell cycle regulators. It co-ordinates root growth by promoting endoreduplication and restricting cell proliferation in the root meristem.
Furthermore, cytokinins act as negative regulators of the lateral root initiation (Laplaze et al., 2007), and the downregulation of their accumulation allowed the specification of lateral root founder cells (Bielach et al., 2012). In the pericycle cells between the existing lateral root primordia, cytokinins negatively control lateral root initiation by reducing the expression of the auxin efflux carrier PIN, thus preventing the establishment of the auxin maximum (Del Bianco et al., 2013). Similar mechanisms may be involved in regulating nodule formation. If synthesized under Nod factor control, cytokinins negatively regulate PIN expression in the root cortex, which enhances local auxin accumulation (Plet et al., 2011). Therefore, cytokinin-dependent regulation of the auxin response maximum occurs during the initiation of symbiosis development that stimulates local cell proliferation, which suggests a common mechanism for regulating the development of roots and nodules. However, the exact role of cytokinins during the late stages of nodulation associated with plant cell and bacteroid differentiation remains to be elucidated.
Effect of sub-cellular localization of cytokinins
We observed an unusually strong accumulation of trans-zeatin riboside in the nuclei of sym33-3, sym40 and sym26 mutants, whereas the trans-zeatin riboside signal was almost absent in the nuclei of cells from different histological nodule zones in the SGE and Sprint-2 wild-type lines, as well as in the sym31 mutants. This indicated that sym33-3, sym40 and sym26 affected the cellular transport of trans-zeatin riboside. Previously, different cytokinin forms have been detected in the nuclei of somatic and zygotic embryos of Tilia cordata (Kärkönen and Simola, 1999) and somatic embryos of Dactylis glomerata (Ivanova et al., 1994). However, the precise function of cytokinins in the nuclei merits further investigation.
Cytokinins negatively regulate infection thread and infection droplet development in nodules
In contrast to the positive effect that cytokinins exert on nodule organogenesis, they have a negative impact on rhizobial infection, as demonstrated by the Ljlhk1-1 mutant with a manifested hyperinfection phenotype (Murray et al., 2007). In L. japonicus plants with a mutated ckx3 gene encoding cytokinin oxidase, the number of infection threads was lower than that in wild-type plants; interestingly, treatment with the ethylene synthesis inhibitor aminoethoxyvinylglycine restored the number of infections. This observation indicates that cytokinins negatively affect the early stages of infection by ethylene signalling (Reid et al., 2016). In our work, we demonstrated the absence of detectable amounts of trans-zeatin riboside in young infection threads in the nodules of the SGE wild type and in the ‘locked’ infection threads in sym33-3 mutants. This cytokinin was also absent in young infection droplets in the wild-type nodules. However, in mature infection droplets, enhanced levels of trans-zeatin riboside were observed in wild-type nodules and in sym33-3 and sym26 mutant nodules. Surprisingly, trans-zeatin riboside was not detectable in hypertrophied infection droplets formed in nodules of the sym40 mutant. The observed trans-zeatin riboside distribution may indicate that in mature nodules, some cytokinin forms are also involved in the negative regulation of infection, limiting infection thread growth and, specifically, the development of infection droplets. It should be noted that in the nodules of sym33-2 and sym33-3 mutants, the infection thread is highly ramified (Voroshilova et al., 2009; Tsyganova et al., 2019), probably because of decreased cytokinin levels and the absence of a negative cytokinin effect on infection thread growth.
Cytokinins promote bacteroid differentiation
It has been recently demonstrated in A. hypogaea with aeschynomenoid-type determinate nodules that gene silencing by AhHK1-RNAi dramatically inhibited the ability of bacteria to differentiate to bacteroids (Kundu and DasGupta, 2017). In this study, we used pea mutants accommodating different degrees of bacteroid differentiation. In the sym31 mutants forming undifferentiated bacteroids (Borisov et al., 1997), there was a significant decline in the signal intensity of the trans-zeatin riboside label associated with symbiosomes (Fig. 10D). However, in the sym26 mutants forming nodules with morphologically differentiated bacteroids (Serova et al., 2018), the signal intensity of the trans-zeatin riboside label associated with bacteroids resembled that in the SGE wild-type line (Fig. 10E). Hence, our results indicate a possible positive regulation of bacteroid differentiation by cytokinins in indeterminate nodules. Alternatively, the level of bacteroid differentiation may affect cytokinin accumulation in the infected cells of nodules. Thus, future experiments should examine the inter-relationships between both processes during nodulation.
Cytokinins negatively regulate nitrogen fixation in nodules
Another potential function of cytokinins in nodule formation could include controlling nitrogen fixation. It has been shown that cytokinin receptor inactivity results in reduced nodule formation and decreased nitrogen fixation in M. truncatula cre1 mutants (Boivin et al., 2016), whereas the use of kinetin in chickpea plants increased nitrogen fixation (Fatima et al., 2008). However, the elevated cytokinin content in L. japonicus ckx3-2 mutants, as well as in wild-type plants treated with the synthetic cytokinin 6-benzylaminopurine, enhanced the sensitivity of efficient nodule formation to nitrates, which demonstrates a negative effect of cytokinins on nitrogen fixation (Reid et al., 2016). Our analysis detected a sharp reduction in the trans-zeatin riboside level in the nitrogen fixation zone of 4-week-old nodules during the active nitrogen fixation stage (Fig. 10A), which appears to correlate well with the negative effects of cytokinins on nitrogen fixation.
Thus, cytokinins may play a multifunctional role during the late stages of nodule development and the active nitrogen fixation stage. Our analysis using the ipd3/cyclops and efd pea mutants assessed the potential effect of cytokinins on plant cell and bacteroid differentiation, infection thread development, and nodule function.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following.
Figure S1: trans-zeatin riboside immunolocalization in control 2-week-old pea nodules.
Figure S2: visualization of auxin response maxima in the nodules of composite SGE pea wild-type plants containing the DR5::GFP-NLS construct.
Figure S3: N6-isopentenyladenosine immunolocalization in the 2-week-old pea nodules of SGE wild type and sym33-3 mutants.
Figure S4: trans-zeatin riboside immunolocalization in the 4-week-old pea nodules of SGE wild type and corresponding mutants.
Figure S5: trans-zeatin riboside immunolocalization in pea nodules of Sprint-2 wild type and sym31 mutants.
Figure S6: expression pattern of cytokinin metabolic and regulatory genes in pea wild type and sym33-2, sym33-3 and sym40 mutants 3 weeks after inoculation.
Table S1: list of primers used in this study.
ACKNOWLEDGEMENTS
We are very grateful to Dr Florian Frugier (IPS2, France) for providing the pMtRR4::GUS construct. The research was performed using equipment of the Core Centrum ‘Genomic Technologies, Proteomics and Cell Biology’ in ARRIAM, the Core Facilities Center ‘Cell and Molecular Technologies in Plant Science’ at the Komarov Botanical Institute RAS (Saint Petersburg, Russia) and Research Resource Centre ‘Molecular and Cell Technologies’ at the Saint Petersburg State University. The authors declare they have no competing interests.
FUNDING
This work was financially supported by the Russian Science Foundation (grant 16-16-10043 for analysis of hormone distribution in composite pea plants, grant 16-16-10035 for immunolocalization of cytokinins in root nodules).
LITERATURE CITED
- Ariel F, Brault-Hernandez M, Laffont C, et al. . 2012. Two direct targets of cytokinin signaling regulate symbiotic nodulation in Medicago truncatula. The Plant Cell 24: 3838–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azarakhsh M, Kirienko AN, Zhukov VA, Lebedeva MA, Dolgikh EA, Lutova LA. 2015. KNOTTED1-LIKE HOMEOBOX 3: a new regulator of symbiotic nodule development. Journal of Experimental Botany 66: 7181–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielach A, Podlešáková K, Marhavý P, et al. . 2012. Spatiotemporal regulation of lateral root organogenesis in Arabidopsis by cytokinin. The Plant Cell 24: 3967–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Billy F, Grosjean C, May S, Bennett M, Cullimore JV. 2001. Expression studies on AUX1-like genes in Medicago truncatula suggest that auxin is required at two steps in early nodule development. Molecular Plant-Microbe Interactions 14: 267–277. [DOI] [PubMed] [Google Scholar]
- Boivin S, Fonouni-Farde C, Frugier F. 2016. How auxin and cytokinin phytohormones modulate root microbe interactions. Frontiers in Plant Science 7: 1240. doi: 10.3389/fpls.2016.01240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot KJ, van Brussel AA, Tak T, Spaink HP, Kijne JW. 1999. Lipochitin oligosaccharides from Rhizobium leguminosarum bv. viciae reduce auxin transport capacity in Vicia sativa subsp. nigra roots. Molecular Plant-Microbe Interactions 12: 839–844. [Google Scholar]
- Borisov AY, Rozov S, Tsyganov V, et al. . 1994. Identification of symbiotic genes in pea (Pisum sativum L.) by means of experimental mutagenesis. Genetika (Russian Federation) 30: 1484–1494. [Google Scholar]
- Borisov AY, Rozov SM, Tsyganov VE, Morzhina EV, Lebsky VK, Tikhonovich IA. 1997. Sequential functioning of Sym-13 and Sym-31, two genes affecting symbiosome development in root nodules of pea (Pisum sativum L). Molecular and General Genetics 254: 592–598. [DOI] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, et al. . 2014. The root hair ‘infectome’ of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. The Plant Cell 26: 4680–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chen W, Li X, et al. . 2014. Knockdown of LjIPT3 influences nodule development in Lotus japonicus. Plant & Cell Physiology 55: 183–193. [DOI] [PubMed] [Google Scholar]
- Cooper JB, Long SR. 1994. Morphogenetic rescue of Rhizobium meliloti nodulation mutants by trans-zeatin secretion. The Plant Cell 6: 215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bianco M, Giustini L, Sabatini S. 2013. Spatiotemporal changes in the role of cytokinin during root development. New Phytologist 199: 324–338. [DOI] [PubMed] [Google Scholar]
- Demina IV, Maity PJ, Nagchowdhury A, et al. . 2019. Accumulation of and response to auxins in roots and nodules of the actinorhizal plant Datisca glomerata compared to the model legume Medicago truncatula. Frontiers in Plant Science 10: 1085. doi: 10.3389/fpls.2019.01085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denarie J, Debelle F, Prome J-C. 1996. Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annual Review of Biochemistry 65: 503–535. [DOI] [PubMed] [Google Scholar]
- Di Giacomo E, Laffont C, Sciarra F, Iannelli MA, Frugier F, Frugis G. 2017. KNAT3/4/5-like class 2 KNOX transcription factors are involved in Medicago truncatula symbiotic nodule organ development. New Phytologist 213: 822–837. [DOI] [PubMed] [Google Scholar]
- Dolgikh EA, Shaposhnikov AI, Dolgikh AV, et al. . 2017. Identification of Pisum sativum L. cytokinin and auxin metabolic and signaling genes, and an analysis of their role in symbiotic nodule development. International Journal of Plant Physiology and Biochemistry 9: 22–35. [Google Scholar]
- Dolgikh AV, Kirienko AN, Tikhonovich IA, Foo E, Dolgikh EA. 2019. The DELLA proteins influence the expression of cytokinin biosynthesis and response genes during nodulation. Frontiers in Plant Science 10: 432. doi: 10.3389/fpls.2019.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus G. 1957. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. Journal of general microbiology 16: 374–381. [DOI] [PubMed] [Google Scholar]
- Fatima Z, Bano A, Sial R, Aslam M. 2008. Response of chickpea to plant growth regulators on nitrogen fixation and yield. Pakistan Journal of Botany 40: 2005–2013. [Google Scholar]
- Fisher J, Gaillard P, Fellbaum CR, Subramanian S, Smith S. 2018. Quantitative 3D imaging of cell level auxin and cytokinin response ratios in soybean roots and nodules. Plant, Cell & Environment 41: 2080–2092. [DOI] [PubMed] [Google Scholar]
- Gamas P, Brault M, Jardinaud M-F, Frugier F. 2017. Cytokinins in symbiotic nodulation: when, where, what for? Trends in Plant Science 22: 792–802. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S, Crespi M, Frugier F. 2006. The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. The Plant Cell 18: 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S, Smith HMS, Holtan H, Magnani E, Mele G, Ramirez J. 2004. The role of KNOX genes in plant development. Annual Review of Cell and Developmental Biology 20: 125–151. [DOI] [PubMed] [Google Scholar]
- Heckmann AB, Sandal N, Bek AS, et al. . 2011. Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Molecular Plant-Microbe Interactions 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M, Hou H, Miri M, et al. . 2014. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. The Plant Cell 26: 678–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X, Schnabel E, Hughes K, Frugoli J. 2006. RNAi phenotypes and the localization of a protein::GUS fusion imply a role for Medicago truncatula PIN genes in nodulation. Journal of Plant Growth Regulation 25: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilina EL, Kiryushkin AS, Semenova VA, Demchenko NP, Pawlowski K, Demchenko KN. 2018. Lateral root initiation and formation within the parental root meristem of Cucurbita pepo: is auxin a key player? Annals of Botany 122: 873–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioio RD, Linhares FS, Scacchi E, et al. . 2007. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Current Biology 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Ioio RD, Nakamura K, Moubayidin L, et al. . 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- Ivanova KA, Tsyganova AV, Brewin NJ, Tikhonovich IA, Tsyganov VE. 2015. Induction of host defences by Rhizobium during ineffective nodulation of pea (Pisum sativum L.) carrying symbiotically defective mutations sym40 (PsEFD), sym33 (PsIPD3/PsCYCLOPS) and sym42. Protoplasma 252: 1505–1517. [DOI] [PubMed] [Google Scholar]
- Ivanova MI, Todorov IT, Atanassova L, Dewitte W, Onckelen HAV. 1994. Co-localization of cytokinins with proteins related to cell proliferation in developing somatic embryos of Dactylis glomerata L. Journal of Experimental Botany 45: 1009–1017. [Google Scholar]
- Jardinaud M-F, Boivin S, Rodde N, et al. . 2016. A laser dissection-RNAseq analysis highlights the activation of cytokinin pathways by Nod factors in the Medicago truncatula root epidermis. Plant Physiology 171: 2256–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D, Meng X, Wang Y, Wang J, Zhao Y, Chen M. 2020. Expression and regulation of small RNAs in the plant–microorganism symbioses in Medicago truncatula. In: de Bruijn FJ, ed. The model legume Medicago truncatula. Hoboken: John Wiley and Sons, 919–922. [Google Scholar]
- Kärkönen A, Simola LK. 1999. Localization of cytokinins in somatic and zygotic embryos of Tilia cordata using immunocytochemistry. Physiologia Plantarum 105: 355–365. [Google Scholar]
- Kisiala A, Laffont C, Emery RJN, Frugier F. 2013. Bioactive cytokinins are selectively secreted by Sinorhizobium meliloti nodulating and nonnodulating strains. Molecular Plant-Microbe Interactions 26: 1225–1231. [DOI] [PubMed] [Google Scholar]
- Kitaeva AB, Demchenko KN, Tikhonovich IA, Timmers ACJ, Tsyganov VE. 2016. Comparative analysis of the tubulin cytoskeleton organization in nodules of Medicago truncatula and Pisum sativum: bacterial release and bacteroid positioning correlate with characteristic microtubule rearrangements. New Phytologist 210: 168–183. [DOI] [PubMed] [Google Scholar]
- Kosterin OE, Rozov SM. 1993. Mapping of the new mutation blb and the problem of integrity of linkage group I. Pisum Genetics 25: 27–31. [Google Scholar]
- Kundu A, DasGupta M. 2017. Silencing of putative cytokinin receptor histidine kinase1 inhibits both inception and differentiation of root nodules in Arachis hypogae. Molecular Plant-Microbe Interactions 31: 187–199. [DOI] [PubMed] [Google Scholar]
- Laplaze L, Benkova E, Casimiro I, et al. . 2007. Cytokinins act directly on lateral root founder cells to inhibit root initiation. The Plant Cell 19: 3889–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplaze L, Lucas M, Champion A. 2015. Rhizobial root hair infection requires auxin signaling. Trends in Plant Science 20: 332–334. [DOI] [PubMed] [Google Scholar]
- Leppyanen IV, Kirienko AN, Dolgikh EA. 2019. Agrobacterium rhizogenes-mediated transformation of Pisum sativum L. roots as a tool for studying the mycorrhizal and root nodule symbioses. PeerJ 7: e6552. doi: 10.7717/peerj.6552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbenga KR, Harkes PAA. 1973. Initial proliferation of cortical cells in the formation of root nodules in Pisum sativum L. Planta 114: 17–28. [DOI] [PubMed] [Google Scholar]
- Liu J, Rutten L, Limpens E, et al. . 2018. A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. The Plant Cell 31: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Lohar DP, Schaff JE, Laskey JG, Kieber JJ, Bilyeu KD, Bird DM. 2004. Cytokinins play opposite roles in lateral root formation, and nematode and rhizobial symbioses. The Plant Journal 38: 203–214. [DOI] [PubMed] [Google Scholar]
- Lohar DP, Sharopova N, Endre G, et al. . 2006. Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiology 140: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathesius U, Schlaman HRM, Spaink HP, Of Sautter C, Rolfe BG, Djordjevic MA. 1998. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. The Plant Journal 14: 23–34. [DOI] [PubMed] [Google Scholar]
- Messinese E, Mun J-H, Yeun LH, et al. . 2007. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Molecular Plant-Microbe Interactions 20: 912–921. [DOI] [PubMed] [Google Scholar]
- Mortier V, Wasson A, Jaworek P, et al. . 2014. Role of LONELY GUY genes in indeterminate nodulation on Medicago truncatula. New Phytologist 202: 582–593. [DOI] [PubMed] [Google Scholar]
- Moubayidin L, Perilli S, Ioio RD, Di Mambro R, Costantino P, Sabatini S. 2010. The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Current Biology 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K. 2007. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Nemankin N. 2011. Analysis of pea (Pisum sativum L.) genetic system, controlling development of arbuscular mycorrhiza and nitrogen-fixing symbiosis. PhD thesis (in Russian), Saint-Petersburg State University, Russia. [Google Scholar]
- Ng JLP, Hassan S, Truong TT, et al. . 2015. Flavonoids and auxin transport inhibitors rescue symbiotic nodulation in the Medicago truncatula cytokinin perception mutant cre1. The Plant Cell 27: 2210–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp RHM, De Mita S, Lillo A, et al. . 2011. A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiology 157: 2013–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikova E, Journet E-P, Chabaud M, et al. . 2011. IPD3 controls the formation of nitrogen-fixing symbiosomes in pea and Medicago spp. Molecular Plant-Microbe Interactions 24: 1333–1344. [DOI] [PubMed] [Google Scholar]
- Pacios-Bras C, Schlaman HRM, Boot K, et al. . 2003. Auxin distribution in Lotus japonicus during root nodule development. Plant Molecular Biology 52: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Plet J, Wasson A, Ariel F, et al. . 2011. MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. The Plant Journal 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Quandt HJ, Pühler A, Broer I. 1993. Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. Molecular Plant-Microbe Interactions 6: 699–706. [Google Scholar]
- Reid DE, Heckmann AB, Novák O, Kelly S, Stougaard J. 2016. CYTOKININ OXIDASE/DEHYDROGENASE3 maintains cytokinin homeostasis during root and nodule development in Lotus japonicus. Plant Physiology 170: 1060–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DE, Nadzieja M, Novak O, Heckmann AB, Sandal N, Stougaard J. 2017. Cytokinin biosynthesis promotes cortical cell responses during nodule development. Plant Physiology 175: 361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Wang X, Duan J, Ma J. 2019. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365: 919–922. [DOI] [PubMed] [Google Scholar]
- Rightmyer AP, Long SR. 2011. Pseudonodule formation by wild-type and symbiotic mutant Medicago truncatula in response to auxin transport inhibitors. Molecular Plant-Microbe Interactions 24: 1372–1384. [DOI] [PubMed] [Google Scholar]
- Roy S, Robson FC, Lilley JLS, et al. . 2017. MtLAX2, a functional homologue of the auxin importer AtAUX1, is required for nodule organogenesis. Plant Physiology 174: 326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, et al. . 2019. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Current Biology 29: 3657–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serova TA, Tsyganova AV, Tsyganov VE. 2018. Early nodule senescence is activated in symbiotic mutants of pea (Pisum sativum L.) forming ineffective nodules blocked at different nodule developmental stages. Protoplasma 255: 1443–1459. [DOI] [PubMed] [Google Scholar]
- Simon SA, Meyers BC, Sherrier DJ. 2009. MicroRNAs in the rhizobia legume symbiosis. Plant Physiology 151: 1002–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Katzer K, Lambert J, Cerri M, Parniske M. 2014. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host and Microbe 15: 139–152. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. 2012. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 139: 3997–4006. [DOI] [PubMed] [Google Scholar]
- Suzaki T, Ito M, Yoro E, et al. . 2014. Endoreduplication-mediated initiation of symbiotic organ development in Lotus japonicus. Development 141: 2441–2445. [DOI] [PubMed] [Google Scholar]
- Syōno K, Newcomb W, Torrey JG. 1976. Cytokinin production in relation to the development of pea root nodules. Canadian Journal of Botany 54: 2155–2162. [Google Scholar]
- Takahashi N, Kajihara T, Okamura C, et al. . 2013. Cytokinins control endocycle onset by promoting the expression of an APC/C activator in Arabidopsis roots. Current Biology 23: 1812–1817. [DOI] [PubMed] [Google Scholar]
- Thimann KV. 1936. On the physiology of the formation of nodules on legume roots. Proceedings of the National Academy of Sciences, USA 22: 511–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirichine L, Sandal N, Madsen LH, et al. . 2007. A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107. [DOI] [PubMed] [Google Scholar]
- Tsyganov VE, Borisov AY, Rozov SM, Tikhonovich IA. 1994. New symbiotic mutants of pea obtained after mutagenesis of laboratory line SGE. Pisum Genetics 26: 36–37. [Google Scholar]
- Tsyganov VE, Morzhina EV, Stefanov SY, Borisov AY, Lebsky VK, Tikhonovich IA. 1998. The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function. Molecular and General Genetics 259: 491–503. [DOI] [PubMed] [Google Scholar]
- Tsyganov VE, Voroshilova VA, Borisov AY, Tikhonovich IA, Rozov SM. 2000. Four more symbiotic mutants obtained using EMS mutagenesis of line SGE. Pisum Genetics 32: 63. [Google Scholar]
- Tsyganov VE, Seliverstova E, Voroshilova V, et al. . 2011. Double mutant analysis of sequential functioning of pea (Pisum sativum L.) genes Sym13, Sym33, and Sym40 during symbiotic nodule development. Russian Journal of Genetics: Applied Research 1: 343. [Google Scholar]
- Tsyganov VE, Voroshilova V, Rozov S, Borisov AY, Tikhonovich I. 2013. A new series of pea symbiotic mutants induced in the line SGE. Russian Journal of Genetics: Applied Research 3: 156–162. [Google Scholar]
- Tsyganova AV, Tsyganov VE, Findlay КC, Borisov AY, Tikhonovich IA, Brewin NG. 2009. Distribution of legume arabinogalactanprotein-extensin (AGPE) glycoproteins in symbiotically defective pea mutants with abnormal infection threads. Cell and Tissue Biology 51: 93–102. [PubMed] [Google Scholar]
- Tsyganova AV, Ivanova KA, Tsyganov VE. 2019. Histological and ultrastructural nodule organization of the pea (Pisum sativum) mutant SGEFix–5 in the Sym33 gene encoding the transcription factor PsCYCLOPS/PsIPD3. Ecological genetics 17: 65–70. [Google Scholar]
- Vernié T, Moreau S, de Billy F, et al. . 2008. EFD is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. The Plant Cell 20: 2696–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernié T, Kim J, Frances L, et al. . 2015. The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. The Plant Cell 27: 3410–3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voroshilova VA, Boesten B, Tsyganov VE, Borisov AY, Tikhonovich IA, Priefer UB. 2001. Effect of mutations in Pisum sativum L. genes blocking different stages of nodule development on the expression of late symbiotic genes in Rhizobium leguminosarum bv. viciae. Molecular Plant-Microbe Interactions 14: 471–476. [DOI] [PubMed] [Google Scholar]
- Voroshilova VA, Demchenko KN, Brewin NJ, Borisov AY, Tikhonovich IA. 2009. Initiation of a legume nodule with an indeterminate meristem involves proliferating host cells that harbour infection threads. New Phytologist 181: 913–923. [DOI] [PubMed] [Google Scholar]
- Wang TL, Wood EA, Brewin NJ. 1982. Growth regulators, Rhizobium and nodulation in peas. Planta 155: 350–355. [DOI] [PubMed] [Google Scholar]
- Yano K, Yoshida S, Müller J, et al. . 2008. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proceedings of the National Academy of Sciences, USA 105: 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HJ, Hossain MS, Held M, et al. . 2014. Lotus japonicus SUNERGOS1 encodes a predicted subunit A of a DNA topoisomerase VI that is required for nodule differentiation and accommodation of rhizobial infection. The Plant Journal 78: 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A, den Camp RHO, Deinum EE, et al. . 2015. Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Molecular Plant 8: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Zhernakov AI, Shtark OY, Kulaeva OA, et al. . 2019. Mapping-by-sequencing using NGS-based 3′-MACE-Seq reveals a new mutant allele of the essential nodulation gene Sym33 (IPD3) in pea (Pisum sativum L.). PeerJ 7: e6662. doi: 10.7717/peerj.6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.