Abstract
The biophysical environment of membrane phospholipids affects structure, function, and stability of membrane-bound proteins.1,2 Obesity can disrupt membrane lipids, and in particular, alter the activity of sarco/endoplasmic reticulum (ER/SR) Ca2+-ATPase (SERCA) to affect cellular metabolism.3–5 Recent evidence suggests that transport efficiency (Ca2+ uptake / ATP hydrolysis) of skeletal muscle SERCA can be uncoupled to increase energy expenditure and protect mice from diet-induced obesity.6,7 In isolated SR vesicles, membrane phospholipid composition is known to modulate SERCA efficiency.8–11 Here we show that skeletal muscle SR phospholipids can be altered to decrease SERCA efficiency and increase whole-body metabolic rate. The absence of skeletal muscle phosphatidylethanolamine (PE) methyltransferase (PEMT) promotes an increase in skeletal muscle and whole-body metabolic rate to protect mice from diet-induced obesity. The elevation in metabolic rate is caused by a decrease in SERCA Ca2+-transport efficiency, whereas mitochondrial uncoupling is unaffected. Our findings support the hypothesis that skeletal muscle energy efficiency can be reduced to promote protection from obesity.
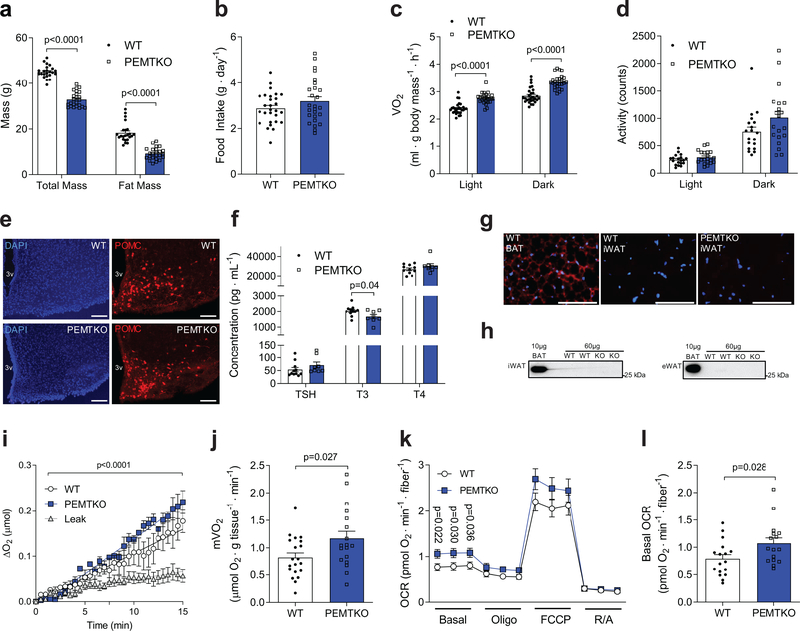
Genome-wide association data has identified PEMT polymorphism to be associated with obesity,12 and whole-body deletion of PEMT (PEMTKO) protects mice from high-fat diet (HFD)-induced weight gain.13 PEMTKO mice exhibit increased whole-body energy expenditure with no known mechanism.14 Raised at thermoneutrality (29°C) or room temperature, HFD-fed wild-type (WT) and PEMTKO mice were systematically evaluated. Consistent with previous reports, PEMTKO mice were protected from diet-induced weight gain (Fig. 1a, Supplementary Fig. 1a–e) without a decrease in food consumption (Fig. 1b), representing metabolic inefficiency (Supplementary Fig. 1f). Indirect calorimetry showed that PEMTKO mice demonstrated increased whole-body VO2 (Fig. 1c, Supplementary Fig. 1g&h) in the absence of increased spontaneous activity (Fig. 1d). Increased metabolic rate was not explained by differences in abundance of hypothalamic POMC or AgRP/NPY-expressing neurons (Fig. 1e, Supplementary Fig. 1i&j), or increases in thyroid hormones (Fig. 1f), ruling out these systemic mechanisms. A previous study showed no increase in UCP1 protein in brown adipose tissue,15 and we could not detect any UCP1 proteins in subcutaneous or visceral white adipose tissues (Fig. 1g&h), nor could we detect thermogenic reprograming in subcutaneous or visceral white adipose tissues (Supplementary Fig. 1k&l). Skeletal muscle is a major contributing tissue to total energy expenditure.12 Muscles from PEMTKO mice show a modest change in mitochondrial morphology without an increase in oxidative capacity.14 To evaluate the basal metabolic rate of skeletal muscle, we measured oxygen consumption of isolated extensor digitorum longus (EDL) muscles suspended ex vivo (mVO2).16 Strikingly, resting mVO2 was ~40% greater in PEMTKO mice compared to WT mice (Fig. 1i&j). To rule out a possibility that the differences in mVO2 came from differences in O2 transport gradient in the muscle, we measured O2 consumption in cultured intact muscle fibers from flexor digitorum brevis (FDB) muscle using a Seahorse platform. Basal O2 consumption rate in muscle fibers from PEMTKO mice was also ~40% greater compared to WT mice (Fig. 1k&l). Assessment of high-energy phosphates suggested that muscles deficient in PEMT may be in a state of accelerated ATP supply (Supplementary Fig. 1m&n). As muscle accounts for 20–30% of resting whole-body energy expenditure, it can be deduced that 8–12% out of the ~15% increase in whole-body energy expenditure (Fig. 1c) is accounted for by an increase in basal mVO2. The remaining difference is also likely largely accounted for by muscle energy expenditure when mice are active, making it somewhat unlikely that other tissues substantially contribute to greater whole-body metabolic rate in PEMTKO mice.
Figure 1.
PEMTKO mice exhibit increased muscle metabolic rate and are protected from diet-induced obesity. (a) Body mass and fat mass (n=23 for WT and 24 for PEMTKO), (b) food intake (n=28 for WT and 26 for PEMTKO), (c) whole-body VO2 (n=28 for WT and 26 for PEMTKO), and (d) activity (n=19 for WT and 20 for PEMTKO) of WT and PEMTKO mice after 8 weeks of HFD feeding. (e) Representative images of DAPI and POMC staining in arcuate nucleus of the hypothalamus from 3 mice per genotype, scale bar: 50 μm. (f) Thyroid stimulating hormone (TSH), triiodothyronine (T3), and thyroxine (T4) serum abundance (n=11 for WT and 8 for PEMTKO). (g) Representative immunocytochemical fluorescence images from two independent samples per genotype for DAPI (blue) and UCP1 (red) in WT brown adipose tissue (BAT), WT inguinal subcutaneous adipose tissue (iWAT), and PEMTKO iWAT, scale bar: 100 μm. (h) Representative western blots of UCP1 in iWAT and epidydymal adipose tissue (eWAT) (n=3 independent samples per genotype). (i) Representative O2 consumption tracing (n=7 for leak, 3 for WT, and 2 for PEMTKO) and (j) mean muscle oxygen consumption rate (mVO2, n=20 for WT and 18 for PEMTKO) in EDL muscles. (k, l) O2 consumption in FDB fibers (OCR: oxygen consumption rate, n=17 for WT and 16 for PEMTKO). Data are shown as the mean ± s.e.m. Statistical analyses in a,c,f,j,k,l were performed with an unpaired two-tailed t-test. Statistical analyses in i were performed with a two-way ANOVA.
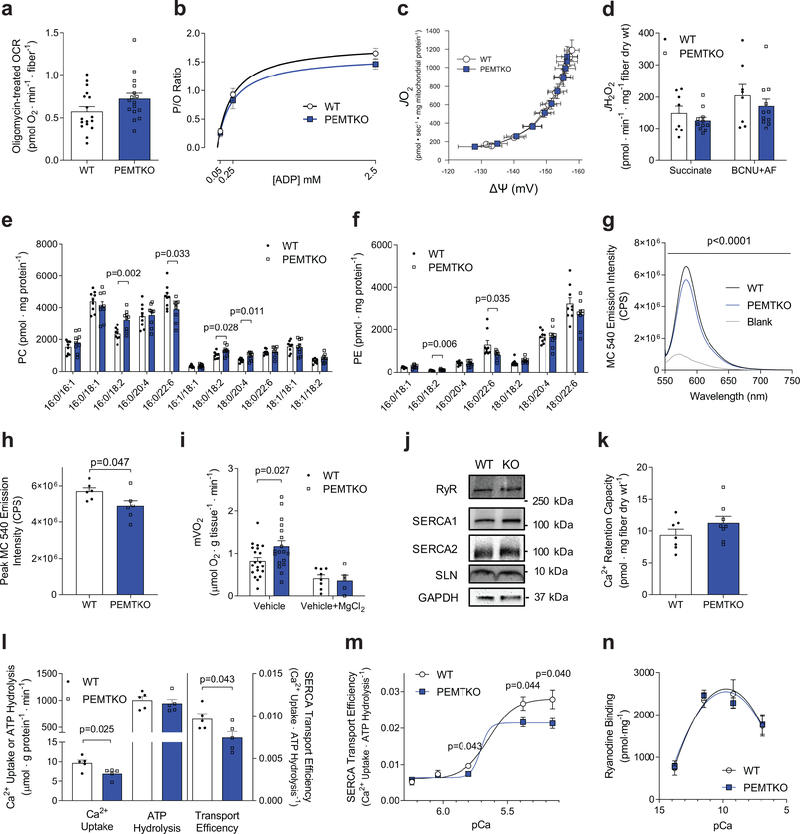
What is the molecular mechanism by which absence of PEMT increases skeletal muscle energy expenditure? Skeletal muscle mitochondria can be proton-uncoupled to be energetically inefficient which can protect mice from diet-induced obesity.17,18 Oligomycin-treated oxygen consumption rates (proton leak-dependent respiration) were not different between WT and PEMTKO fibers (Fig. 2a), but we performed high-resolution respirometry and fluorometry experiments to more carefully phenotype mitochondria. Nevertheless, respiratory efficiency of mitochondria (ATP produced / O2 reduced, or P/O ratio; Fig. 2b, Supplementary Fig. 2a–c), proton leak measured at multiple membrane potentials (Fig. 2c), or electron leak estimated with superoxide production (Fig. 2d, Supplementary Fig. 2d) were not different between WT and PEMTKO mice. There was a modest increase in glutathione buffering capacity (Supplementary Fig. 2e–g) in muscles from PEMTKO mice, suggesting that these mitochondria may be more adept to oxidative challenge. Consistent with previous reports,14 we did not observe any differences in skeletal muscle respiratory capacity (Supplementary Fig. 2h&i) or mitochondrial content (Supplementary Fig. 2j&k). It is important to note that cellular work (ΔGATP), not capacity, drives the metabolic rate. Thus, it is unsurprising that the increase in endogenous activity of mitochondrial oxidative phosphorylation occurs independent of an increase in oxidative capacity. Together, these data suggest that alteration in skeletal muscle mitochondrial efficiency or capacity does not explain the increased metabolic rate in PEMT-deficient muscles.
Figure 2.
Ca2+ transport inefficiency explains the increased muscle metabolic rate in PEMTKO muscle. (a) O2 consumption of FDB fibers after oligomycin exposure (n=17 for WT and 16 for PEMTKO). (b) Mitochondrial ATP production per oxygen reduced (P/O ratio, n=9 for WT and 11 for PEMTKO), (c) proton leak (n=9 for WT and 11 for PEMTKO), (d) electron leak as estimated by H2O2 production in permeabilized gastrocnemius fiber bundles (n=8,11). SR (e) phosphatidylcholine (PC) species and (f) phosphatidylethanolamine (PE) species (n=9 per genotype). (g) SR phospholipid headgroup packing determined by MC540 emission spectrum and (h) MC540 peak emission (n=6 per genotype). (i) Muscle respiration under resting (vehicle, data from Fig. 1j; n=20 for WT and 18 for PEMTKO) and Mg2+-stimulated (vehicle+MgCl2, n=8 for WT and 5 for PEMTKO) conditions. (j) Protein abundance of RyR (n=8 for WT and 7 for PEMTKO), SERCA1 (n=4 for WT and 4 for KO), SERCA2 (n=8 for WT and 7 for KO), SLN (n=4 for WT and 3 for PEMTKO), and GAPDH. (k) Mitochondrial Ca2+ retention capacity (n=7 for WT and 8 for PEMTKO). (l) SERCA-dependent Ca2+ transport, ATP hydrolysis, and transport efficiency (Ca2+/ATP hydrolysis) at pCa 5.8 (n=5 per genotype). (m) SERCA-dependent transport efficiency across multiple pCa (n=5 for WT and 6 for PEMTKO at pCa 6.24 and 6.04; n=5 for WT and 5 for KO at pCa 5.8; n=10 for WT and 9 for KO at pCa 5.38; n=12 for WT and 11 for KO at pCa 5.15). (n) Ryanodine receptor activity across multiple pCa (n=4 per pCa for each genotype). All results are from 8 week HFD-fed mice. Data are shown as the mean ± s.e.m., with exception of g which is shown as mean. Statistical analyses in e,f,h,i,l,m were performed with an unpaired two-tailed t-test. Statistical analyses in g were performed with a two-way ANOVA.
The lack of robust phenotype in skeletal muscle mitochondria is not surprising given that PEMT is located in ER,19 and PEMT deletion does not affect skeletal muscle mitochondrial membrane phospholipid composition.14 Instead, absence of PEMT affected phospholipid composition of muscle SR (Fig. 2e&f, Supplementary Fig. 2l&m) and increased the membrane headgroup packing of SR phospholipids (Fig. 2g&h), suggesting that PEMT may affect SR-resident ATPases. SERCA is a Ca2+ reuptake ion channel that derives its Ca2+ pumping energy from ATP hydrolysis. Indeed, SERCA-dependent ATP hydrolysis accounts for the majority of resting metabolic rate in muscle.20 Previous studies suggest that SR Ca2+ uncoupling, like H+ uncoupling in mitochondria, can increase energy expenditure.6,21 SERCA transport efficiency (Ca2+ uptake/ATP hydrolysis, or Ca2+/P ratio) is affected by the magnitude of Ca2+ gradient, SERCA uncoupling peptide sarcolipin (SLN),6,7,22 as well as membrane phospholipid composition.8–11 To evaluate whether the elevated mVO2 in PEMTKO mice is due to SERCA, we indirectly inhibited SERCA by incubating EDL muscles in the presence of Mg2+. Mg2+ ions block the ryanodine receptor (RyR) thereby preventing cytosolic Ca2+ release, ultimately leaving SERCA with no substrate to transport. Incubation with 10 mM of MgCl2 eliminated the increase in mVO2 of PEMTKO compared to WT mice, suggesting that the increase in basal mVO2 is dependent on SR calcium cycling (Fig. 2i).16 The difference was not explained by changes in protein abundance for RyR, SERCA1, or SERCA2 proteins (Fig. 2j, Supplementary Fig. 2n), protein and/or mRNA levels of known SERCA uncoupling proteins (Fig. 2j, Supplementary Fig. 2o), or mitochondrial calcium handling (Fig. 2k). To more definitively attribute elevated energy expenditure to SERCA, we quantified its transfer efficiency in SR vesicles isolated from skeletal muscle. Across multiple concentrations of free Ca2+, rates for Ca2+ uptake and ATP hydrolysis were quantified in the absence and presence of thapsigargin to determine SERCA-dependent and –independent rates (Fig. 2l&m, Supplementary Fig. 3a–l). Ca2+/P ratio was significantly lower in SR vesicles from PEMTKO compared to WT mice, with greater difference when cytosolic Ca2+ was higher (Fig. 2m). Alternatively, RyR channel could leak to induce thermogenesis.21,23 However, RyR activity was not different between WT and PEMTKO muscles (Fig. 2n), suggesting that this mechanism is unlikely to contribute to elevated basal energy expenditure in PEMTKO muscle. Together, these data implicate SERCA transport inefficiency, and not Ca2+-leak, to be the cellular process contributing toward elevated muscle energy expenditure in PEMTKO mice.
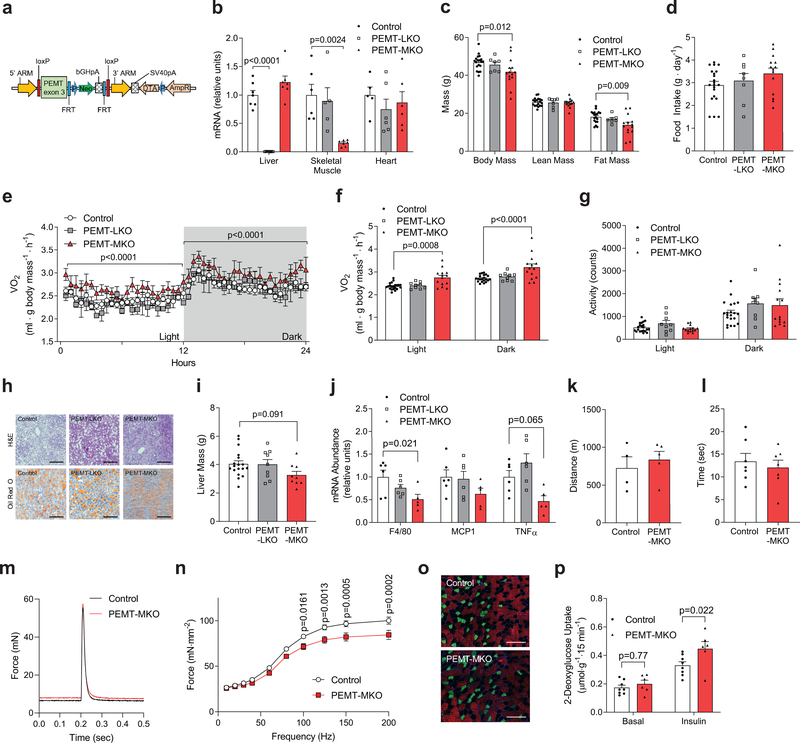
The liver is the predominant systemic source of phospholipids where these molecules are exported to peripheral tissues via VLDL. While PEMT is expressed in multiple tissues, liver contains the highest amount of PEMT among all cell types.19 Thus, it was unclear whether the muscle phenotype observed in whole-body PEMTKO mice was due to absence of PEMT in liver, or due to intrinsic loss of PEMT in skeletal muscle.24 First, we assessed PEMT mRNA expression in muscle from lean and obese humans as well as mice fed standard chow and HFD. Strikingly, obesity coincided with elevated PEMT expression in skeletal muscle (Supplementary Fig. 4a&b). These findings are consistent with observations that increased metabolic rate in PEMTKO mice are present only when mice are fed HFD.13 Next, in order to examine the tissue-specific effect of PEMT deletion, we generated a mouse strain with its Pemt gene flanked by loxP sites on the exon 3 (PEMTcKO+/+ or control mice, Fig. 3a). We then crossed these mice with Alb-Cre mice or tamoxifen-inducible HSA-MerCreMer mice to generate mice with liver-specific (PEMT-LKO) or skeletal muscle-specific (PEMT-MKO) deletion of PEMT (Supplementary Fig. 4c). Both strains were born with normal Mendelian ratio with no apparent defects. Tissue specificity of PEMT deletion was confirmed with quantitative PCR (Fig. 3b, Supplementary Fig. 4d). When challenged with HFD, control and PEMT-LKO mice equally gained lean and fat mass (Fig. 3c, Supplementary Fig. 4e–j). In contrast, skeletal muscle-specific deletion of PEMT protected mice from diet-induced obesity (Fig. 3c, Supplementary Fig. 4e–g,k&l) that was not explained by a difference in food intake (Fig. 3d, Supplementary Fig. 4h). Similar to whole-body PEMTKO mice,15 PEMT-MKO mice had increased whole-body metabolic rate (Fig 3e&f, Supplementary Fig. 4m), without an increase in physical activity (Fig. 3g). Nevertheless, PEMT-MKO mice were not more cold tolerant compared to controls (Supplementary Fig. 4n). To more definitely exclude liver PEMT in the hypermetabolic phenotype of whole-body PEMTKO mice, we re-expressed PEMT in liver of these mice with AAV8-TBG-PEMT infection. Consistent with findings in PEMT-LKO mice, liver PEMT had no effect on body weight, energy expenditure, activity, or food intake (Supplementary Fig. 4o–r). Together, increased metabolic rate in whole-body PEMTKO mice is not due to the lack of PEMT in liver, but rather due to the loss of PEMT in skeletal muscle. It is noteworthy that unlike PEMTKO13 or PEMT-LKO mice, PEMT-MKO mice did not develop hepatic steatosis or inflammation (Fig. 3h–j).13
Figure 3.
PEMT-MKO mice are protected from diet-induced obesity. (a) A targeting vector used to generate mice with conditional knockout for PEMT. (b) mRNA expression of PEMT in liver (n= 7 per genotype), skeletal muscle (tibialis anterior; n=6 for control, 5 for PEMT-LKO, and 7 for PEMT-MKO), and heart (n=5 for control, 6 for PEMT-LKO, and 6 for PEMT-MKO). (c) Body mass, lean mass, and fat mass after 8 weeks of HFD feeding (n=19 for control, 7 for PEMT-LKO, and 14 for PEMT-MKO). (d) Food intake (n=21 for control, 8 for PEMT-LKO, and 14 for PEMT-MKO), (e) representative 24 hour whole-body VO2 tracing (n= 8 for control, 2 for PEMT-LKO, and 6 for PEMT-MKO), (f) mean whole-body VO2 (n=21 for control, 9 for PEMT-LKO, and 14 for PEMT-MKO), and (g) activity (n=21 for control, 9 for PEMT-LKO, and 14 for PEMT-MKO) at 8 weeks of HFD feeding. (h) Representative H&E and Oil Red O staining in liver, scale bar: 200 μm (n=2 independent samples per genotype for H&E and 4 independent samples for Oil Red O). (i) Liver mass (n=18 for control, 9 for PEMT-LKO, and 9 for PEMT-MKO) and (j) mRNA levels of F4/80, MCP1, and TNFα from HFD fed mice (n=6 for control, 6 for PEMT-LKO, and 5 for PEMT-MKO). (k) Treadmill endurance tolerance test (n=4 for control and 5 for PEMT-MKO) and (l) inverted screen hang time (n=7 per genotype). (m) Representative twitch force tracing (n=10 per genotype) and (n) force-frequency curve of tetanic contractions in EDL muscles (n=10 per genotype). (o) Representative immunofluorescence images of muscle fiber-type, scale bar: 200 μm (Green: MHC IIa. Red: IIb. No color: IIx. n=5 per genotype). (p) 2-deoxyglucose uptake in soleus muscles (n=8 for control and 6 for PEMT-MKO). Data are shown as the mean ± s.e.m., with the exception of m which is a representative trace. All results are from 8 week HFD-fed mice. Statistical analyses in b,c,i,j were performed with an one-way ANOVA with Dunnett’s multiple comparisons test. Statistical analyses in e,f,n,p were performed with a two-way ANOVA and multiple comparisons were performed using Tukey’s (f) or Sidak’s (n,p) multiple comparisons tests.
We also characterized the consequence of PEMT deletion to skeletal muscle force-generating capacity with a series of in vivo and ex vivo tests. PEMT-MKO mice did not display deficiencies in their ability to run or grip strength (Fig 3k&l). We also characterized muscle contractile function with electrically-stimulated force production ex vivo. There was no difference in twitch force production (Fig. 3m, Supplementary Fig. 5a). Muscles from PEMT-MKO mice exhibited a slight reduction in tetanic force production (Fig. 3n, Supplementary Fig. 5b). Nonetheless, muscles from PEMT-MKO mice did not demonstrate greater fatigue (Supplementary Fig. 5c), suggesting that reduced force-generating capacity in PEMT-MKO muscles may come from its inability to effectively cycle Ca2+ in and out of SR, rather than deficiency in ATP production. There were no differences in the SDH staining, fiber-type, mitochondrial content, or ultra-structure of the mitochondria (Fig. 3o, Supplementary Fig 5d–h). Importantly, in agreement with decreased adiposity, muscles from PEMT-MKO mice were exquisitely insulin-sensitive (Fig. 3p).
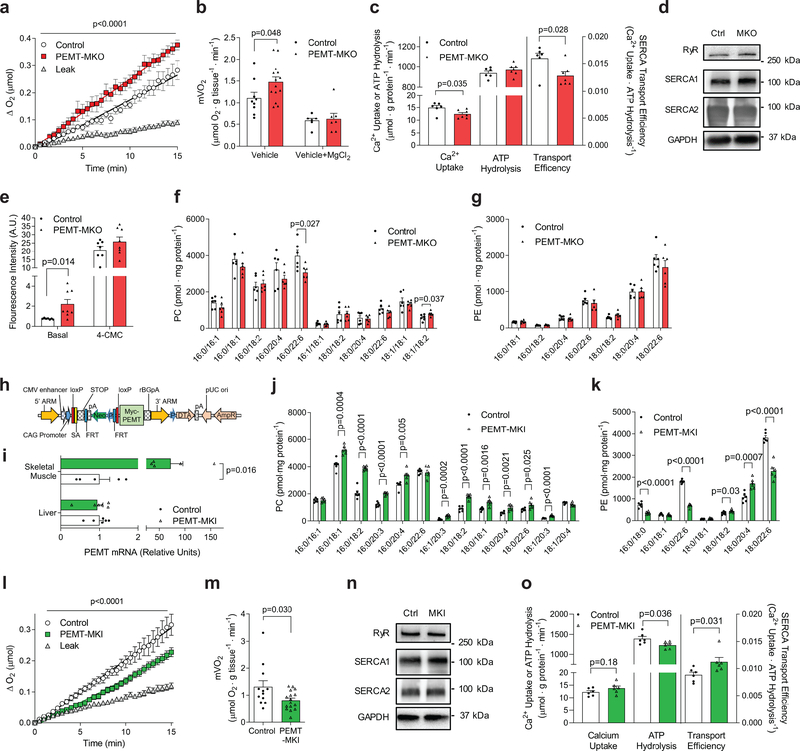
To confirm that increased whole-body energy expenditure in PEMT-MKO mice was due to an increase in skeletal muscle metabolic rate, we measured mVO2 in skeletal muscle from HFD-fed PEMT-MKO and control mice. mVO2 from PEMT-MKO mice were ~45% greater compared to control mice (Fig. 4a&b). Repeating these experiments in the presence of Mg2+ ions abrogated the difference in mVO2 (Fig. 4b), suggesting that like muscle from PEMTKO mice the energetic demand of SR Ca2+-flux is responsible for increased metabolic rate in skeletal muscle. Consistent with our findings in the whole-body PEMT knockout mice, Ca2+/P ratio was reduced in isolated SR vesicles from PEMT-MKO compared to control mice (Fig. 4c, Supplementary Fig. 6a&b). The phenotype was not explained by different protein or mRNA abundance for RyR, SERCA1 or SERCA2 in skeletal muscle (Fig. 4d, Supplementary Fig. 6c). Meanwhile, in cultured FDB fibers, basal cytosolic calcium concentration was greater in PEMT-MKO mice compared to control (Fig. 4e), likely reflecting SERCA inefficiency. In contrast, there was no difference in 4-CMC-stimulated Ca2+ release, suggesting that SR Ca2+ reserve is not different between these mice (Fig. 4e). Mass spectrometry analyses revealed differences in SR phospholipid composition between PEMT-MKO and control muscles with a notable decrease in 16:0/22:6-PC (primary product of PEMT25) (Fig. 4f&g, Supplementary Fig. 6d&e). Absence of PEMT did not alter UCP1 levels in brown or white adipose tissues (Supplementary Fig. 6f). Together, these data suggest that PE methylation in skeletal muscle modulate SR phospholipid composition, which in turn alters the energetic efficiency of SERCA ion pump.
Figure 4.
Skeletal muscle PEMT alters SR phospholipids to regulate muscle oxygen consumption. (a) Representative O2 consumption tracing (n=15 for leak, 7 for control, and 8 for PEMT-MKO) and (b) mean mVO2 under resting (vehicle; n= 9 for control and 14 for PEMT-MKO) or RyR-inhibited (vehicle+MgCl2; n= 6 for control and 7 for PEMT-MKO) conditions in EDL muscles from HFD-fed control and PEMT-MKO mice. (c) SERCA-dependent Ca2+ transport, ATP hydrolysis, and transport efficiency at pCa 5.8 (n=6 for control and 7 for PEMT-MKO). (d) Protein abundance of RyR, SERCA1, SERCA2, and GAPDH in soleus muscles from control and PEMT-MKO mice (n=4 per genotype). (e) Fluo-4 fluorescence intensity in basal and 2 mM 4-CMC-stimulated FDB fibers (n=7 for control and 8 for PEMT-MKO). (f, g) SR PC and PE in gastrocnemius muscles from control and PEMT-MKO mice (n=6 per genotype). (h) A targeting vector used to generate mice with conditional knock-in for PEMT. (i) PEMT mRNA levels in liver (n= 6 per genotype) and gastrocnemius muscles (n= 5 per genotype) from control and PEMT-MKI mice. (j, k) SR PC and PE in gastrocnemius muscles from control and PEMT-MKI mice (n= 6 per genotype). (l) Representative O2 consumption tracings (n=31 for leak, 12 for control, and 16 for PEMT-MKI) and (m) mean mVO2 (n=12 for control and 16 for PEMT-MKI) in EDL muscles from HFD-fed control and PEMT-MKI mice. (n) Protein abundance of RyR, SERCA1, SERCA2, and GAPDH (n= 8 per genotype). (o) SERCA-dependent Ca2+ transport, ATP hydrolysis, and transport efficiency at pCa 5.15 (n=6 per genotype). Data are shown as the mean ± s.e.m. All results are from 8 week HFD-fed mice. Statistical analyses in b,c,e,f,i,j,k,m,o were performed with an unpaired two-tailed t-test. Statistical analyses in a,l were performed with a two-way ANOVA.
As obesity coincided with an increase in muscle PEMT expression, we examined the effect of increased muscle PEMT expression on skeletal muscle metabolic rate. For these studies, mice with skeletal muscle-specific overexpression of PEMT were generated. First, we synthesized mice with conditional knock-in of PEMT (PEMTcKI+/+), which was accomplished by inserting a Pemt cDNA with a preceding stop codon that was flanked with loxP sites into a Rosa26 locus (Fig. 4h). These mice were crossed with HSA-MerCreMer+/− mice which produced PEMT-MKI (PEMTcKI+/−, HSA-MerCreMer+/−) and control (PEMTcKI+/−, HSA-MerCreMer−/−) mice. Skeletal muscle-specific overexpression of PEMT was confirmed by qPCR (Fig. 4i) which was sufficient to increase SR PC and decrease PE (Fig. 4j&k, Supplementary Fig 7a&b). Consistent with our observation that PEMT deletion increased mVO2, PEMT overexpression reduced mVO2 by ~40% (Fig. 4l&m). This occurred in the absence of changes in in SR Ca2+ handling proteins (Fig. 4n, Supplementary Fig. 6c). Consistent with data from PEMT deletion, muscles from PEMT-MKI demonstrated increased SERCA transport efficiency (Fig. 4o, Supplementary Fig. 6d&e). Despite exhibiting lower mVO2 and improved SERCA-transport efficiency, PEMT-MKI and control mice did not differ in whole-body energy expenditure, activity, food intake, or propensity for weight gain on chow or HFD diet (Supplementary Fig. 6f–r). It is unclear why the lower mVO2 in PEMT-MKI mice was not reflected in lower whole-body VO2. We did not detect greater UCP1 proteins in brown and white adipose tissues (Supplementary Fig. 6s). One possibility is that PEMT-MKI mice compensate for reduced resting mVO2 by increasing muscle shivering or small locomotion not detected by our motion sensor. Nevertheless, these data demonstrate that increased PE methylation is sufficient to decrease skeletal muscle metabolic rate.
Obesity is a major public health problem with at least 2.1 billion overweight people worldwide. Strategies to reduce energy efficiency, especially by means of mitochondrial uncoupling in brown and white adipose tissues,26 have attracted much attention as mechanisms that increase energy expenditure and decrease propensity for weight gain. While studies on UCP1-dependent thermogenesis in adipose tissues yield a strikingly robust phenotype in mouse models, it is yet unclear how targeting this mechanism would increase energy expenditure in humans. Meanwhile, skeletal muscle represents an organ with substantial contribution for whole-body energy expenditure.27 Exercise and shivering are two examples in which an increase in skeletal muscle energy expenditure effectively increases whole-body energy expenditure. Skeletal muscle tissues are also anatomically ideally situated for thermogenesis, away from vital organs and well-vascularized to dissipate heat. The current study uncovered a mechanism by which phospholipids alter the energy efficiency of skeletal muscle SERCA to affect propensity for obesity. In humans, skeletal muscle phospholipid metabolism becomes highly affected with obesity or exercise.28,29 We speculate that these changes may be responsible for alterations in skeletal muscle energy efficiency that are thought to occur with weight fluctuation or exercise training.30
Methods
Animal models
C57BL/6J mice (WT) were used as controls for whole-body PEMT knockout mice (PEMTKO).31 Using a traditional homologous-recombination method, PEMT-conditional knockout (PEMTcKO+/+) mice were generated by flanking Pemt exon 3 with loxP sites. PEMTcKO+/+ mice were then crossed with HSA-MerCreMer32 mice to generate mice with tamoxifen-inducible skeletal muscle-specific knockout of PEMT (PEMT-MKO: PEMTcKO+/+, HSA-MerCreMer+/−) or with Alb1-Cre33 mice to generate mice with liver-specific knockout of PEMT (PEMT-LKO: PEMTcKO+/+, Alb1-Cre+/−), all on a C57BL/6J background. For both PEMT-MKO and PEMT-LKO, PEMTcKO+/+, Cre−/− mice were used as littermate controls. PEMT conditional knock-in (PEMTcKI+/+) mice were generated by inserting myc-tagged human Pemt cDNA with a preceding stop codon that was flanked with loxP sites into a Rosa26 locus. PEMTcKI+/+ mice were crossed with HSA-MerCreMer mice to generate skeletal muscle specific knock-in of PEMT (PEMT-MKI: PEMTcKI+/+, HSA-MerCreMer+/−) and littermate controls (PEMTcKI+/+, HSA-MerCreMer−/−), all on a C57BL/6J background. For some experiments, Cre control mice (PEMTcKO−/− or PEMTcKI−/− with HSA-MerCreMer+/− or Alb1-Cre+/−) or tamoxifen-untreated control mice (PEMTcKO+/+, HSA-MerCreMer+/− or PEMTcKI+/−, HSA-MerCreMer+/− with no tamoxifen), which displayed no difference in phenotype compared to the control mice. At 10 weeks of age, PEMT-MKO, PEMT-MKI, and respective controls were injected with tamoxifen 5 consecutive days at a dose of 7.5 μg•g−1. Tamoxifen-treated mice were given a two-week washout before beginning high fat diet (HFD, 42% fat, Envigo TD.88137) feeding. PEMTKO, PEMT-LKO, and their respective control mice began HFD feeding at 10–12 weeks of age. All mice underwent 8 weeks of HFD feeding and were fasted 4 hours prior to terminal experiments and tissue collection. All mice were born with normal Mendelian ratios. Both male and female mice were studied with no difference in phenotype. All protocols were approved by Institutional Animal Care and Use Committees at East Carolina University and University of Utah.
AAV8 infection
PEMTKO mice were intravenously injected with 4×1011 GC of adeno-associated virus 8 (AAV8) expressing either green fluorescent protein (eGFP) or phosphatidylethanolamine methyltransferase (PEMT) regulated by thyroxine-binding globulin (AAV8-TBG-eGFP or AAV8-TBG-PEMT) promoter to drive liver-specific expression of eGFP or PEMT in PEMTKO mice (Vector Biolabs, Malvern, PA, USA). C57BL/6J mice were injected with AAV8-TBG-eGFP. Two weeks after viral injection, mice were fed with HFD for 8 weeks.
Ca2+ fluorescence
Flexor digitorum brevis (FDB) muscles were digested in Krebs-Henseleit Buffer (KHB) containing 2 mg/mL collagenase type II (Worthington Biochemical, Lakewood, NJ, USA) for two hours at 37°C. Digested FDB muscles were then transferred to KHB without collagenase or CaCl2 and titrated to separate them into individual muscle fibers. Fibers were plated on laminin coated 96-well plate and allowed to adhere for >30 minutes before experimentation. Fibers were used within 5 hours of dissection. Muscle fibers were loaded with 5 μM Fluo-4-AM (F14201, Invitrogen) and Pluronic F-127 (P3000MP, Invitrogen) in the dark for 30 minutes at room temperature. Fibers were then washed in dye-free solution supplemented with 25 μM N-benzyl-p-toluene sulfonamide (BTS, AAJ64910MA, Fisher Scientific) for 20 minutes to block myosin ATPase. Loaded fibers were imaged using EVOS FL microscope (Life Technologies Corp., Carlsbad, CA, USA). To stimulate Ca2+ release from the SR, fibers were injected with 2 mM 4-chloro-m-cresol (4-CMC), a ryanodine receptor agonist. Basal fluorescent measurements occurred in the absence of 4-CMC. Image J was used to trace individual muscle fibers, subtract background, and quantify fluorescence.
Cold tolerance test
Mice received subscapular injection of a wireless temperature transponder (IPTT-300, Bio Medic Data Systems, Seaford, DE, USA). One week following transponder injection, mice were placed in a 4°C room for eight hours with access to food and water. Body temperatures were recorded hourly.
Electron microscopy
Freshly dissected extensor digitorum longus (EDL) was cut into ~2 mm pieces and processed by the Electron Microscopy Core at University of Utah. Muscle pieces were submerged in fixative solution (1% glutaraldehyde, 2.5% paraformaldehyde, 100 mM cacodylate buffer pH 7.4, 6 mM CaCl2, 4.8% sucrose) and stored at 4°C overnight. Samples then underwent 3×10 minute washes in 100 mM cacodylate buffer (pH 7.4) prior to secondary fixation (2% osmium tetroxide) for 1 hour at room temperature. After secondary fixation, samples were subsequently rinsed for 5 minutes in cacodylate buffer and distilled H2O, followed by prestaining with saturated uranyl acetate for 1 hour at room temperature. After prestaining, samples were dehydrated with graded ethanol series (2 × 15 minutes each: 30%, 50%, 70%, 95%; then 3 × 20 minutes each: 100%) and acetone (3 × 15 minutes), and were infiltrated with EPON epoxy resin (5 hours 30%, overnight 70%, 3 × 2 hour 40 minute 100%, 100% fresh for embed). Samples were then polymerized for 48 hours at 60°C. Ultracut was performed using Leica UC 6 ultratome with sections at 70 nm thickness and mounted on 200 mesh copper grids. The grids with the sections were stained for 20 minutes with saturated uranyl acetate and 10 minutes with lead citrate. Sections were examined at an accelerating voltage of 120 kV in a JEOL-1400 Plus Transmission Electron Microscope (JEOL, Japan), equipped with CCD Gatan camera.
Histochemistry
Coronal brain sections were taken from WT and PEMTKO mice after a 8-week HFD feeding and immunohistochemistry for POMC (Phoenix Pharmaceuticals, Burlingame, CA, USA) and DAPI (4′,6-diamidino-2-phenylindole, 1 mg ml−1, Sigma Aldrich).34,35 Liver samples were frozen in Tissue-Tek O.C.T. compound and 10 μm sections were stained with Oil-red O staining or paraffin embedded and 5 μm stained with hematoxylin and eosin (H&E). Samples were imaged on Axio Scan Z.1 (Zeiss). Brown adipose (BAT) and subcutaneous inguinal white adipose tissues (iWAT) were embedded in paraffin and sectioned to 5 μm. Sections were deparaffinized and underwent antigen retrieval prior to overnight incubation with primary antibody for UCP1 (ab23481, Abcam) and subsequent secondary antibody incubation (Alexa Fluor 594, A32740, Invitrogen).
Membrane headgroup packing
Sarcoplasmic reticulum (SR)-enriched fraction was isolated from quadriceps muscle as described below (under SERCA-dependent Ca2+-uptake and ATPase Activity).4 30 μg of SR protein was added to 900 μL of HBSS (Gibco, 14025076) for a final volume of 1000 μL. In the cuvette, the sample was incubated at 37°C and stirred at 750 RPM. MC540 was added at a final concentration of 200 nM and samples were incubated for 10 minutes prior to excitation at 495 nm and a spectral scan from 550 to 750 nm. Blank measurements were recorded as no sample with MC540 probe at 10 minutes of incubation at 37°C. For peak intensity, the greatest emission value was subtracted from value of blank measurement for respective wavelength.
Metabolic phenotyping
TSE Calorimetry System (Chesterfield, MO) or Columbus Instruments Lab Animal Monitoring System (CLAMS) were used to measure VO2, RER, food intake, and physical activity. Mice were individually caged and acclimated for over 24 hours in the TSE Calorimetry System prior to collecting data. Body composition was determined using EchoMRI-700 (Echo Medical Systems, Houston, MA, USA) or Bruker Minispec NMR (Bruker, Germany). Metabolic efficiency was calculated by dividing the total energy intake over 8 weeks of HFD feeding energy intake by an increase in body mass or fat mass over the same period of time. The total energy intake was derived from caloric content of the HFD and the rate of its consumption.
Mitochondrial analysis
Skeletal muscle fiber bundles or mitochondria isolated from hindlimb muscles were analyzed by high-resolution respirometry (Oroboros O2k oxygraphs) and fluorometry (Horiba Fluoromax 3 fluorometers) to quantify ATP production per O2 reduction (P/O ratio),36 proton leak,18 H2O2 production,37 and Ca2+ retention.38 Substrate-dependent oxygen consumption was stimulated by concentrations of: 50 μM palmitoyl-CoA with 5 mM carnitine, 0.5 mM malate, 4 mM ADP, 5 mM pyruvate, 10 mM glutamate, 10 mM succinate, 10 μM cytochrome c, and 0.5 μM FCCP. Reduced glutathione (GSH) and glutathione disulfide (GSSG) were measured using high performance liquid chromatography (HPLC) from gastrocnemius homogenate.39
Muscle fiber-typing
Frozen, OCT-embedded hindlimb muscle samples (tibialis anterior, mixed fiber-type) were sectioned at 10 μm using a cryostat (Microtome Plus@). Following 1h blocking with M.O.M mouse IgG blocking (Vector, Cat#MKB-2213), myofiber sections were incubated for 1 h with concentrated BA.D5, SC.71, and BF.F3 (all 1:100; Developmental Studies Hybridoma Bank, University of Iowa) and laminin (1:200; Sigma, Cat#L9393) in 2.5% normal horse serum. To visualize laminin (for fiber border), myosin heavy chain I (MHC I), myosin heavy chain IIa (MHC IIa), and myosin heavy chain IIb (MHC IIb), slides were incubated for 1h with the following secondary antibodies: AMCA (1:250 Vector, Cat#CI-1000), Alexa Fluor 647 (1:250; Invitrogen (Carlsbad, CA), Cat#A21242), Alexa Fluor 488 (1:500; Invitrogen, Cat#A21121) and Alexa Fluor 555 (1:500; Invitrogen, Cat#A21426), respectively. Negative stained fibers were considered myosin heavy chain IIx (MHC IIx). After staining, slides were coverslipped with mounting media (Vector, Cat#H-1000). Stained slides were imaged with a fully automated wide field light microscope (Nikon, Nikon Corp.; Tokyo, Japan) with 10X objective lens. Images were captured using high sensitivity Andor Clara CCD (Belfast, UK).
Muscle force measurements ex vivo
Extensor digitorum muscles (EDL) were excised and suspended ex vivo in a tissue bath containing KHB + 8 mM glucose at 37°C. EDL were chosen as it can be sutured tendon to tendon and be utilized for highly-reproducible ex vivo measurements for force-generating capacity. Muscles were adjusted to optimal resting tension prior to recording twitch, force frequency, or fatigue measurements. Twitch and force frequency measurements were performed16 and fatigue measurements were assessed by 0.2 ms electrical pulse every 0.5 s for 2 minutes. All measurements were recorded using Aurora Scientific Dynamic Muscle Control software (DMCv5.5) and data were analyzed using Aurora Scientific Muscle Analysis software (DMAv5.3; Aurora Scientific, Aurora, ON, Canada).
Muscle function in vivo
High fat diet fed control and PEMT-MKO mice performed a low-intensity treadmill running test40 and inverted screen test.41 For treadmill running, fed mice were run for 1 h at 10 m/min, followed by an increase of 2 m/min every 15 min until exhaustion. For the inverted screen test, mice were placed on a screen that was then inverted. Mice were evaluated for their time to hang onto the screen. The test was repeated three times and the mean value for the top two times were reported.
Muscle 2-deoxyglucose uptake
Mice were fasted for four hours before anesthetization and excision of contralateral soleus muscles. Soleus muscles were chosen due to their highly-reproducible ex vivo measurements for insulin-stimulated glucose uptake. Muscles were initially incubated in a recovery buffer (KHB with 0.1% BSA, 8 mM glucose, and 2 mM mannitol) at 37 °C for 10 minutes. Following this step muscles were then moved to pre-incubation buffer (KHB with 0.1% BSA, 2 mM sodium pyruvate, and 6 mM mannitol) ± 50 mU/mL insulin for 15 minutes. After pre-incubation muscles were placed in incubation buffer (KHB with 0.1% BSA, 9 mM [14C]mannitol, 1 mM [3H]2-deoxyglucose) ± 50 mU/mL insulin for 15 minutes. Contralateral muscles were used for basal or insulin stimulated measurements. Immediately at the completion of the final 15-minute period, muscles were quickly blotted on ice-cold filter paper, snap-frozen, and stored at −80 °C until analyzed with liquid scintillation counting.4
Muscle oxygen consumption
For the measurement of oxygen consumption in isolated whole muscle tissues (mVO2), EDL muscles were excised and suspended ex vivo at a resting tension in a sealed tissue bath (Aurora Scientific, Inc. Model: 801C) containing O2-saturated KHB (-CaCl2) + 8 mM glucose at 37°C.16 Change in O2 saturation was measured with an MI-730 Dip-type O2 sensor (Microelectrode). mVO2 was calculated by subtracting the rate of O2 decline in the absence of muscle (O2 leak) from the rate of O2 decline in the presence of muscle. The measurements were taken both under resting and calcium-flux inhibited conditions (+MgCl2 10 mM). Seahorse platform was used for the measurements of oxygen consumption in isolated muscle fibers (OCR). Flexor digitorum brevis (FDB) muscles were excised and separated into single fibers as described above in Ca2+ fluorescence. Single fibers were plated onto a coated 24-well Seahorse plate and analyzed utilizing a Mito Stress Test kit (Agilent).42
Phospholipidomic analysis
Lipids were isolated using methyl-tert-butyl ether protocol.43 Lipids were reconstituted in methanol:toluene (9:1). Lipid extracts are separated on an Acquity UPLC CSH C18 1.7 μm 2.1 × 50 mm column maintained at 60 °C connected to an Agilent HiP 1290 Sampler, Agilent 1290 Infinity pump, equipped with an Agilent 1290 Flex Cube and Agilent 6490 triple quadrupole (QqQ) mass spectrometer. Results from LC-MS experiments were collected using Agilent Mass Hunter Workstation and analyzed using the software package Agilent Mass Hunter Quant B.07.00. Lipids are quantitated based on peak area ratios to the internal standards (330710, Avanti Polar Lipids, Inc.) added to the samples prior to extraction.
SERCA-dependent Ca2+-uptake and ATPase activity
Quadriceps muscle was homogenized [300 mM sucrose, 20 mM HEPES pH 7.4, Halt protease (78430)] and underwent differential centrifugation (10 min @ 1300g, 20 min @ 20000g, 2 hr 15 min @ 180000g)4 to pellet an SR-enriched fraction, which was resuspended in homogenization buffer. SR-fraction was quantified using BCA protein assay (Pierce, 23225) and 10 μg of SR protein was used in each replicate for SERCA-dependent Ca2+-uptake and ATPase activity assay. SERCA-dependent Ca2+ uptake and ATPase activity assays were performed in buffer containing 60 mM HEPES, 200 mM KCl, 15 mM MgCl2, 10 mM NaN3, 1 mM EGTA, and 0.005% Triton-X with varying concentrations of free calcium and with or without 15 μM thapsigargin. SERCA-dependent measures are calculated by taking the difference of values without thapsigargin to values with thapsigargin [Total (without thapsigargin) – SERCA independent (with thapsigargin) = SERCA-dependent]. Ca2+ uptake assay buffer additionally contained 5 mM of (COOK)2. ATPase activity assay buffer additionally contained 10 mM PEP, 1.5 mM NADH, 2.4–4 units of pyruvate kinase/ 3.6–5.6 units lactate dehydrogenases enzymes (Sigma, P0294).
Calcium uptake assay22 was started by the addition of 4 mM ATP and 45CaCl2 (Perkin Elmer, NEZ013001MC) to assay buffer with sample. After incubation for 15 minutes at 37°C with 300 RPM rotation, assay was quenched with the addition of 150 mM KCl and 1 mM LaCl3 and placed on ice. Samples were then filtered on to a 0.22 μm PES membrane filter (Millipore, GPWP02500), rinsed 3 × 5 mL PBS, and processed for scintillation counting. SERCA ATPase activity assay44 was performed on a 96-well plate reader. The assay was initiated by the addition of 4 mM ATP and the absorbance at 340 nm was recorded every 60 seconds for 30 minutes at 37°C. SERCA transport efficiency was determined by the ratio of SERCA-dependent Ca2+ uptake to SERCA-dependent ATPase hydrolysis.
Succinate dehydrogenase (SDH) staining
Muscle sections (see Muscle fiber-typing) were incubated in freshly-made SDH staining solution [0.367 mM nitrotetrazolium blue (N6876, Sigma) and 60 mM succinate acid disodium (224731, Sigma) in 1x PBS pH 7.4 (10010, Gibco)]. Sections were incubated at 37°C for 1 hour. Sections were rinsed in 30%, 60%, and 30% Acetone for 1 minute, followed by a three rinses with distilled H2O. Sections were then mounted (H-1000, Vector Laboratories) and imaged at 10x using Evos FL light microscope (Life Technologies Corp., CA, USA).
Multiplex analysis
Serum T3, T4, and TSH were measured with Milliplex MAP (RTHYMAG-30K, EMD Millipore, Billerica, MA).
Nucleotide analysis
Soleus muscles were freeze-clamped immediately post-dissection. Nucleotides were extracted using perchloric acid and concentrations of ATP, ADP, AMP, and IMP were determined by ultra-performance liquid chromatography.45
Quantitative PCR
Total RNA was extracted from tissues using TRIzol (Invitrogen). One μg of RNA was reverse transcribed using iScript cDNA synthesis kit (BioRad). PCR reactions were performed with a Life Technologies QuantStudio 12K Flex instrument using SYBR Green PCR Master Mix. Assays were performed in duplicate and all results were normalized to ribosomal protein L32 mRNA levels, which were unchanged between controls and respective experimental groups. All values were normalized to the mean of the control group. Primers were based on sequences in public databases.
Ryanodine binding assay
The protocol for ryanodine binding was adapted from a previously described method.46 Quadriceps muscle was homogenized and processed for an SR-enriched fraction (see SERCA-dependent Ca2+-uptake and ATPase Activity). 20 μg of SR-enriched sample was added to ryanodine binding solution with varying calcium concentrations [300 mM NaCl, 50 mM MOPS pH 7.2, 1 mM EDTA, 0.01% BSA, 0.1% CHAPS, 5 nM 3H-Ryanodine (Perkin Elmer, NET950250UC)]. Samples were run with and without 20 μM ryanodine to determine 3H-ryanodine binding. Samples were run in triplicate for 17 hours at 25°C and were rotated at 300 RPM. Samples were then filtered on a 0.22 μm PES membrane filter (Millipore, GPWP02500), washed 3 × 5 mL with PBS, and processed for scintillation counting.
Western blotting
Muscle homogenates were probed for proteins of mitochondrial oxidative phosphorylation (ab110413, Abcam), RyR (sc-13942, Santa Cruz), SERCA1 (ab2819, Abcam), SERCA2 (ab3625, Abcam), SLN (ABT13, Millipore), and GAPDH (2118, Cell Signaling). Brown adipose tissue (BAT), epidydymal white adipose (eWAT), or inguinal subcutaneous white adipose tissue (iWAT) were blotted for UCP1 (UCP11-A, Alpha Diagnostic). Additionally, BAT was blotted for COXIV (4844S, Cell Signaling) or Citrate Synthase (ab96600, Abcam).
Statistical analyses
Data are presented as means ± s.e.m. Statistical analyses were performed using GraphPad Prism 7.03 or SigmaPlot 8.0 software. Independent samples t-tests were used to compare two groups. For two-by-two comparisons, two-way ANOVA analyses were performed (main effect of genotype shown with an asterisk over a horizontal line) followed by appropriate post-hoc tests corrected for multiple comparisons. For all tests P<0.05 was considered statistically significant.
Reporting Summary
Additional information on experimental design is available in the Nature Research Reporting Summary.
Data Availability
Data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This research is supported by NIH DK107397, DK109888 (to K.F.), DK110656 (to P.D.N.), DK112826, DK108833 (to W.L.H.), DK115824, DK116450 (to S.A.S.), DK103930 (to C.J.V.), AR066660 (to E.E.S.), AR070200 (to J.J.B), HL129362 (to T.E.R.), and DK091317 (to T.S.T.), P&F funding from P30 DK020579 at Washington University in St. Louis (to K.F.), American Heart Association 18PRE33960491 (to A.R.P.V.) and 19PRE34380991 (to J.M.J.), Larry H. & Gail Miller Family Foundation (to P.J.F. and to C.J.V.), and Uehara Memorial Foundation (to H.E.). University of Utah Metabolomics Core Facility is supported by S10 OD016232, S10 OD021505, and U54 DK110858.
Footnotes
Competing Interests
The authors declare no competing financial interests.
References
- 1.Laganowsky A et al. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510, 172–175, doi: 10.1038/nature13419 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta K et al. The role of interfacial lipids in stabilizing membrane protein oligomers. Nature 541, 421–424, doi: 10.1038/nature20820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu S et al. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531, doi: 10.1038/nature09968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funai K et al. Muscle lipogenesis balances insulin sensitivity and strength through calcium signaling. Journal of Clinical Investigation 123, 1229–1240, doi: 10.1172/JCI65726 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Funai K et al. Skeletal Muscle Phospholipid Metabolism Regulates Insulin Sensitivity and Contractile Function. Diabetes 65, 1–37, doi: 10.2337/db15-0659 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bal NC et al. Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nature Medicine 18, 1575–1579, doi: 10.1038/nm.2897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurya SK et al. Sarcolipin is a key determinant of basal metabolic rate and its overexpression enhances energy expenditure and resistance against diet induced obesity. Journal of Biological Chemistry 290, jbc.M115.636878, doi: 10.1074/jbc.M115.636878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caffrey M & Feigenson GW Fluorescence quenching in model membranes. 3. Relationship between calcium adenosinetriphosphatase enzyme activity and the affinity of the protein for phosphatidylcholines with different acyl chain characteristics. Biochemistry 20, 1949–1961, doi: 10.1021/bi00510a034 (1981). [DOI] [PubMed] [Google Scholar]
- 9.Navarro J, Toivio-Kinnucan M & Racker E Effect of lipid composition on the calcium/adenosine 5’-triphosphate coupling ratio of the Ca2+-ATPase of sarcoplasmic reticulum. Biochemistry 23, 130–135 (1984). [DOI] [PubMed] [Google Scholar]
- 10.Lervik A, Bresme F, Kjelstrup S & Rubi JM On the thermodynamic efficiency of Ca2+-ATPase molecular machines. Biophysical Journal 103, 1218–1226, doi: 10.1016/j.bpj.2012.07.057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fajardo VA et al. Dietary docosahexaenoic acid supplementation reduces SERCA Ca2+ transport efficiency in rat skeletal muscle. Chemistry and Physics of Lipids 187, 56–61, doi: 10.1016/j.chemphyslip.2015.03.001 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Sharma NK, Langberg KA, Mondal AK & Das SK Phospholipid Biosynthesis Genes and Susceptibility to Obesity: Analysis of Expression and Polymorphisms. PLoS ONE 8, doi: 10.1371/journal.pone.0065303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs RL et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. Journal of Biological Chemistry 285, 22403–22413, doi: 10.1074/jbc.M110.108514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasseva G et al. Lack of phosphatidylethanolamine N-methyltransferase in mice does not promote fatty acid oxidation in skeletal muscle. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1861, 119–129, doi: 10.1016/j.bbalip.2015.11.008 (2016). [DOI] [PubMed] [Google Scholar]
- 15.Gao X et al. Insufficient glucose supply is linked to hypothermia upon cold exposure in mice lacking phosphatidylethanolamine N-methyltransferase. Journal of lipid research 56, 1701–1710, doi: 10.1194/jlr.M059287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrara PJ, Verkerke ARP, Brault JJ & Funai K Hypothermia Decreases O2 Cost for Ex Vivo Contraction in Mouse Skeletal Muscle. Med Sci Sports Exerc 50, 2015–2023, doi: 10.1249/MSS.0000000000001673 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B et al. Skeletal muscle respiratory uncoupling prevents diet-induced obesity and insulin resistance in mice. Nat Med 6, 1115–1120, doi: 10.1038/80450 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Fisher-Wellman, Kelsey H et al. Pyruvate dehydrogenase complex and nicotinamide nucleotide transhydrogenase constitute an energy-consuming redox circuit. Biochemical Journal 467, 271–280, doi: 10.1042/BJ20141447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Z, Vance JE, Chen MH, Voelker DR & Vance DE Cloning and expression of a novel phosphatidylethanolamine N-methyltransferase. A specific biochemical and cytological marker for a unique membrane fraction in rat liver. The Journal of biological chemistry 268, 16655–16663 (1993). [PubMed] [Google Scholar]
- 20.Smith IC, Bombardier E, Vigna C & Tupling a. R. ATP Consumption by Sarcoplasmic Reticulum Ca2+ Pumps Accounts for 40–50% of Resting Metabolic Rate in Mouse Fast and Slow Twitch Skeletal Muscle. PLoS ONE 8, 1–11, doi: 10.1371/journal.pone.0068924 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda K et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nature medicine 23, 1454–1465, doi: 10.1038/nm.4429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paran CW et al. Reduced efficiency of sarcolipin-dependent respiration in myocytes from humans with severe obesity. Obesity (Silver Spring) 23, 1440–1449, doi: 10.1002/oby.21123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durham WJ et al. RyR1 S-Nitrosylation Underlies Environmental Heat Stroke and Sudden Death in Y522S RyR1 Knockin Mice. Cell 133, 53–65, doi: 10.1016/j.cell.2008.02.042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shields DJ, Agellon LB & Vance DE Structure, expression profile and alternative processing of the human phosphatidylethanolamine N-methyltransferase (PEMT) gene1. Biochimica et Biophysica Acta - Molecular and Cell Biology of Lipids 1532, 105–114, doi: 10.1016/S1388-1981(01)00122-6 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Samborski RW, Ridgway ND & Vance DE Metabolism of molecular species of phosphatidylethanolamine and phosphatidylcholine in rat hepatocytes during prolonged inhibition of phosphatidylethanolamine N-methyltransferase. Journal of lipid research 34, 125–137 (1993). [PubMed] [Google Scholar]
- 26.Kajimura S, Spiegelman BM & Seale P Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell metabolism 22, 546–559, doi: 10.1016/j.cmet.2015.09.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zurlo F, Larson K, Bogardus C & Ravussin E Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86, 1423–1427, doi: 10.1172/JCI114857 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S et al. Skeletal muscle phosphatidylcholine and phosphatidylethanolamine respond to exercise and influence insulin sensitivity in men. Scientific reports 8, 6531, doi: 10.1038/s41598-018-24976-x (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newsom SA et al. Skeletal muscle phosphatidylcholine and phosphatidylethanolamine are related to insulin sensitivity and respond to acute exercise in humans. Journal of applied physiology 120, 1355–1363, doi: 10.1152/japplphysiol.00664.2015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravussin Y, Leibel RL & Ferrante AW Jr. A missing link in body weight homeostasis: the catabolic signal of the overfed state. Cell metabolism 20, 565–572, doi: 10.1016/j.cmet.2014.09.002 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walkey CJ, Donohue LR, Bronson R, Agellon LB & Vance DE Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proceedings of the National Academy of Sciences of the United States of America 94, 12880–12885, doi: 10.1073/pnas.94.24.12880 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCarthy JJ, Srikuea R, Kirby TJ, Peterson C. a. & Esser K a. Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skeletal Muscle 2, 8, doi: 10.1186/2044-5040-2-8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yakar S et al. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proceedings of the National Academy of Sciences 96, 7324–7329, doi: 10.1073/pnas.96.13.7324 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H et al. Rho-kinase regulates energy balance by targeting hypothalamic leptin receptor signaling. Nature Neuroscience 15, 1391–1398, doi: 10.1038/nn.3207 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laing BT, Do K, Matsubara T & Wert DW Voluntary exercise improves hypothalamic and metabolic function in obese mice. doi: 10.1530/JOE-15-0510 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Lark DS et al. Direct real-time quantification of mitochondrial oxidative phosphorylation efficiency in permeabilized skeletal muscle myofibers. American Journal of Physiology - Cell Physiology 311, C239–C245, doi: 10.1152/ajpcell.00124.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JM et al. Targeted overexpression of catalase to mitochondria does not prevent cardioskeletal myopathy in Barth syndrome. Journal of Molecular and Cellular Cardiology 121, 94–102, doi: 10.1016/j.yjmcc.2018.07.001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilliam LAA et al. Free Radical Biology and Medicine The anticancer agent doxorubicin disrupts mitochondrial energy metabolism and redox balance in skeletal muscle. Free Radical Biology and Medicine 65, 988–996, doi: 10.1016/j.freeradbiomed.2013.08.191 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferdaoussi M et al. Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional b cells Find the latest version : Isocitrate-to-SENP1 signaling amplifies insulin secretion and rescues dysfunctional β cells. 125, 3847–3860, doi: 10.1172/JCI82498.tive (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gan Z et al. The nuclear receptor PPARβ/δ programs muscle glucose metabolism in cooperation with AMPK and MEF2. Cold Spring Harbor Laboratory Press, 2619–2630, doi: 10.1101/gad.178434.111.4 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zechner C et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metabolism 12, 633–642, doi: 10.1016/j.cmet.2010.11.008 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuh RA, Jackson KC, Khairallah RJ, Ward CW & Spangenburg EE Measuring mitochondrial respiration in intact single muscle fibers. American journal of physiology. Regulatory, integrative and comparative physiology 302, R712–719, doi: 10.1152/ajpregu.00229.2011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A & Schwudke D methods Lipid extraction by methyl- tert -butyl ether for high-throughput lipidomics. 49, 1137–1146, doi: 10.1194/jlr.D700041-JLR200 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simonides WS & van Hardeveld C An assay for sarcoplasmic reticulum Ca2(+)-ATPase activity in muscle homogenates. Analytical biochemistry 191, 321–331, doi: 10.1016/0003-2697(90)90226-Y (1990). [DOI] [PubMed] [Google Scholar]
- 45.Brault JJ, Pizzimenti NM, Dentel JN & Wiseman RW Selective inhibition of ATPase activity during contraction alters the activation of p38 MAP kinase isoforms in skeletal muscle. Journal of Cellular Biochemistry 114, 1445–1455, doi: 10.1002/jcb.24486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aracena-Parks P et al. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. Journal of Biological Chemistry 281, 40354–40368, doi: 10.1074/jbc.M600876200 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that support the findings of this study are available from the corresponding author upon reasonable request.