Abstract
Context
The H19 long noncoding RNA (lncRNA) belongs to a highly conserved, imprinted gene cluster involved in embryonic development and growth control. We previously described a novel mechanism whereby the Anti-mullerian hormone (Amh) appears to be regulated by H19. However, the relationship between circulating H19 and markers of ovarian reserve including AMH not been investigated.
Objective
To determine whether H19 expression is altered in women with decreased ovarian reserve.
Design
Experimental study.
Setting
Yale School of Medicine (New Haven, USA) and Gazi University School of Medicine (Ankara, Turkey).
Patients or other participants
A total of 141 women undergoing infertility evaluation and treatment.
Intervention
Collection of discarded blood samples and cumulus cells at the time of baseline infertility evaluation and transvaginal oocyte retrieval, respectively.
Main outcome measure
Serum and cumulus cell H19 expression.
Results
Women with diminished ovarian reserve (as determined by AMH) had significantly lower serum H19 expression levels as compared to controls (p < 0.01). Serum H19 was moderately positively correlated with serum AMH. H19 expression was increased 3.7-fold in cumulus cells of IVF patients who demonstrated a high response to gonadotropins, compared to low responders (p < 0.05).
Conclusion
In this study, we show that downregulation of H19 in serum and cumulus cells is closely associated with decreased ovarian reserve, as measured by decreased AMH levels and reduced oocyte yield at oocyte retrieval. Further study with expanded sample sizes is necessary to determine whether H19 may be of use as a novel biomarker for diminished ovarian reserve.
Keywords: H19, Noncoding RNA, ncRNA, Ovarian reserve, AMH
Precis
Downregulation of H19 in serum and cumulus cells is associated with decreased ovarian reserve, as measured by decreased AMH levels and reduced oocyte yield at oocyte retrieval.
Introduction
The problem of ovarian aging has considerable impact on public health, and presently no viable preventative or treatment options are available. With age, a drastic decline in the quantity of follicles and oocytes (the “ovarian reserve”), occurs [1], leading to decreased female fecundity and infertility. Historical data, as well as studies of women undergoing fertility treatment, have shown that female fertility begins to decline as early as age 30 [2, 3]. Women are increasingly delaying childbearing [4]. In 2014, over 30% of first births occurred in women over 30 [4]. Thus, more and more women are attempting to conceive in their later reproductive years, when fertility is already in decline [2]. Indeed, birth rates in the U.S. are at a record low [5]. Adding to the complexity of this challenge is the fact that fecundity varies even among women of similar age groups [6], and some women experience idiopathic accelerated follicle loss and early menopause. Why follicle growth and development go so drastically awry in some women is poorly understood, and many gaps remain in our understanding of the processes regulating the recruitment and growth of ovarian follicles.
To understand the factors underpinning these developments, it is essential to define the processes regulating the normal physiology of ovarian aging. In reproductive age women, the pool of primordial follicles is continuously depleted through cyclic recruitment. This relentless loss of follicles leads to a condition known as diminished ovarian reserve (DOR). Women with DOR are at increased risk for infertility and poor ovarian response to ovarian stimulation during in vitro fertilization (IVF) cycles [7].
We previously described a novel mechanism whereby follicular recruitment appears to be regulated in part by the long noncoding RNA H19. H19 is highly conserved [8], expressed during embryogenesis and repressed in most adult tissues, with the exception of ovary, uterus, skeletal muscle and heart [9–13]. We showed that the ovaries of female H19 knockout mice exhibit accelerated follicular recruitment and atresia and a more rapid decline in fertility as compared to WT mice [14] We also observed that H19KO mice produce less anti-Mullerian hormone (AMH) as compared to WT mice, a phenotype that is recapitulated in vitro in granulosa cells after H19 knockdown [14]. AMH regulates the size of the follicular pool by inhibiting the initial recruitment of primordial follicles into the growing pool and modulating the sensitivity of growing follicles to follicle stimulating hormone, and is critical in the regulation of follicle numbers [15–17]. Mice lacking AMH exhibit early follicular recruitment and loss [16–18], and AMH sequence variants in humans have been associated with premature ovarian insufficiency (POI) [19].
AMH is an informative marker for the assessment of ovarian reserve, but its regulation remains poorly understood. Additionally, AMH is only one of several available markers of oocyte quantity. Poor ovarian reserve can be defined by multiple factors, including age, poor oocyte yield after IVF, and abnormal ovarian reserve testing markers including FSH7. A potential link between H19 and markers of ovarian reserve in women, including AMH, has not yet been investigated. Given the relationship between H19 and follicular recruitment we observed in mice, we sought to determine whether decreased circulating and follicular H19 expression is linked to poor ovarian reserve, as determined by clinical and biochemical markers, (including but not limited to AMH), in a population of women with infertility.
Materials and methods
Collection of patient samples
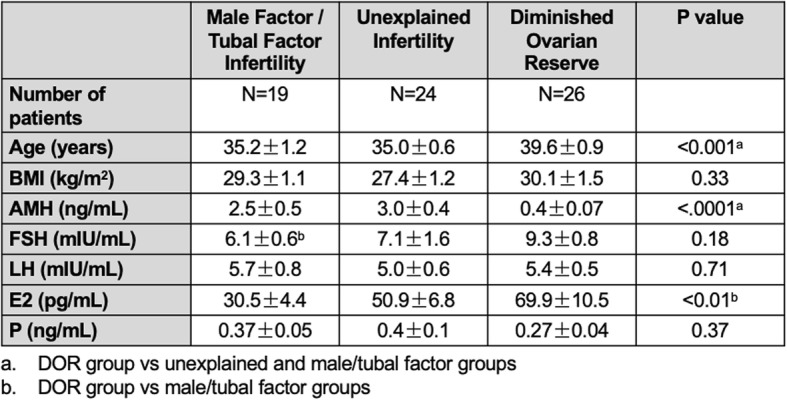
In order to evaluate circulating H19 expression and correlation with ovarian reserve in infertile women, discarded blood samples (2 mL) were collected from 69 patients undergoing evaluation for infertility at the Yale Fertility Center (New Haven, CT) (Table 1). Samples were collected in the early follicular phase (day 2–4 of the menstrual cycle). Serum was separated by centrifugation for 10 m at 12000 rpm at 4 °C and stored at − 80 °C until RNA extraction [20–22]. Patients were excluded if any of the following diagnoses were present: Cushing’s syndrome, hyperprolactinemia, adrenal hyperplasia, acromegaly, hypothalamic amenorrhea, hypothyroidism, or diabetes mellitus. Patients were divided into three groups: controls (women diagnosed with male and tubal factor infertility, n = 19), women with unexplained infertility (n = 24), and women with diminished ovarian reserve (DOR, defined as AMH < 1.1 ng/mL and antral follicle count < 5, n = 26). This study was approved by the Yale University Institutional Review Board (IRB protocol # 1606017946).
Table 1.
Descriptive statistics for patients from which serum samples were collected. Results of comparisons between baseline characteristics among the three groups are given. a.DOR group vs unexplained and male/tubal factor groups. b DOR group vs male/tubal factor groups
We also sought to determine whether H19 is detectable in cumulus cells of women undergoing IVF with oocyte retrieval, and whether expression correlates with ovarian reserve as measured by response to gonadotropins. For these studies, we utilized pooled cumulus cells collected from a total of 72 consecutive cycles in women undergoing infertility treatment with IVF-ICSI at Gazi University School of Medicine IVF Center (Ankara, Turkey) (Table 2). Causes of infertility included diminished ovarian reserve, male factor, tubal factor, anovulation, endometriosis, and unexplained infertility, or a combination of these factors. For patients undergoing agonist cycles, treatment was initiated by GnRH agonists during the luteal phase of the preceding cycle. Stimulation with gonadotropins was initiated after downregulation had been achieved (estradiol level < 50 pg/ml in the absence of ovarian cysts on transvaginal sonography). For patients undergoing antagonist cycles, treatment with gonadotropins was initiated on cycle day 3 when serum progesterone was < 1 ng/ml and transvaginal sonography confirmed absence of ovarian cysts. A GnRH antagonist was added for pituitary suppression after five days of gonadotropin stimulation. Stimulation protocols included 150–300 IU/day of gonadotropins, either recombinant (GonalF, Merck) or in combination with human menopausal gonadotropin (hMG, Menopur, Ferring). Patients received hCG (Ovidrel 250 μg, Merck) when two or more follicles > 18 mm in diameter were present with an adequate estradiol response. Oocytes were collected 36 h after hCG injection. Retrieved cumulus-oocyte complexes (COCs) were placed in culture medium (G-MOPS, VitroLife) and cumulus cells were dissected from the oocyte mechanically in the absence of hyaluronidase. Cumulus cells from each patient were pooled in a single eppendorf tube. Samples were washed twice with 0.5 ml 1X Phosphate Buffered Saline (PBS) and centrifuged at 1000 x g for 1 min with the supernatant removed after each wash. After the final wash, the cell pellet was resuspended in 50 μl SideStep™ Lysis and Stabilization Buffer (Stratagene, La Jolla, CA). Samples were stored at − 80 °C until analysis.
Table 2.
Descriptive statistics for patients from which cumulus cells were collected. Results of comparisons between baseline characteristics among the three groups are given
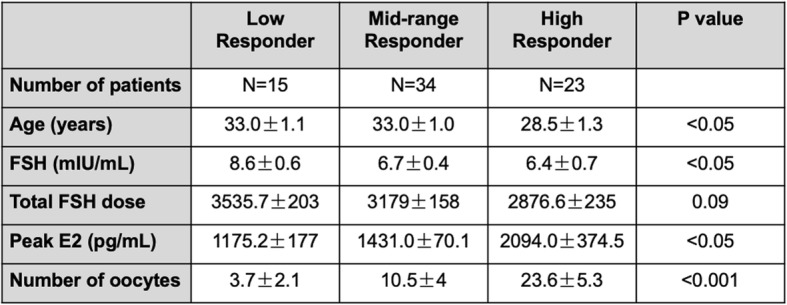
For study analysis, patients were stratified by percentiles based on the number of retrieved oocytes. The number of oocytes included in the first quartile, which correspond to percentile values lower than 25%, was considered as indication of poor response (PR) to controlled ovarian stimulation. Women producing more oocytes than the 75th % of their age group were considered high responders (HR) to controlled ovarian stimulation. Women whose oocyte production fell between 25-75th % of their age group were considered normal responders (NR). This study was approved by the Gazi University Institutional Review Board committee (IRB protocol #131/11.05.2011).
Materials
Primers for H19 and β-actin were purchased from Real Time Primers (Elkins Park, PA, USA). The Norgen Plasma/Serum RNA Extraction Kit was purchased from Norgen Biotek (Ontario, CA). The Qiagen miRNeasy Mini Kit and RNeasy MinElute Cleanup Kits were purchased from Qiagen (Germantown, MD, USA). SYBRGreen was purchased from BioRad Laboratories (Hercules, CA, USA).
RNA extraction, cDNA synthesis and RT-PCR
RNA extraction from stored blood samples was performed using the Norgen Plasma/Serum RNA Purification Mini Kit (Thorold, Ontario, Canada). To purify RNA from cumulus cells, the Qiagen miRNeasy Mini Kit and RNeasy MinElute Cleanup Kits were used according to the manufacturer’s instructions. RNA quantity and purity were determined using an ultraviolet spectrophotometer. Reverse transcription was carried out using the BioRad iScript cDNA synthesis kit (Hercules, CA, USA) in a 20ul reaction containing 0.5μg of total RNA. For cDNA synthesis, the reaction mixtures were incubated at 16 °C for 30 min, at 42 °C for 30 min, and at then 85 °C for 5 min. The cDNA was stored at − 80 °C until qPCR. qPCR was performed in a 25ul reaction containing 1.5ul of cDNA using Bio-Rad QSYBRGreen in a Bio-Rad iCycler. PCR was performed by initial denaturation at 95 °C for 5 min followed by 40 cycles of 30 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. All SYBR green runs had dissociation curves to predict potential primer-dimers. Specificity was verified by melting curve analysis and agarose gel electrophoresis. The threshold cycle (Ct) values of each sample were used in the post-PCR data analysis, and the delta-delta Ct method was used to calculate mRNA expression. H19 levels were normalized and expressed as fold change relative to that of β-actin. The mean level of H19 expression in the control (tubal/male) factor group was set to 1, and H19 expression in the other two groups was normalized to this control group. All SYBR green runs had dissociation curves to predict potential primer-dimers. Specificity was verified by melting curve analysis. The PCR primers for the indicated genes are listed below.
Human H19 forward: 5′-GCACCTTGGACATCTGGAGT.
Human H19 reverse: 5′-TTCTTTCCAGCCCTAGCTCA.
β-actin forward: AAGAGCTATGAGCTGCCTGA.
β-actin reverse: TACGGATGTCAACGTCACAC.
Statistical analysis
Descriptive statistics for all patients were analyzed with one-way ANOVA combined with Bonferroni post hoc analysis. To compare H19 expression levels, one-way ANOVA was performed with Bonferroni post hoc analysis. Association between serum H19 expression and AMH levels was measured using Pearson’s correlation coefficient analysis. All P values were considered to be statistically significant when p < 0.05. Statistical analysis was performed using GraphPad (Chicago, IL).
Results
H19 expression in serum
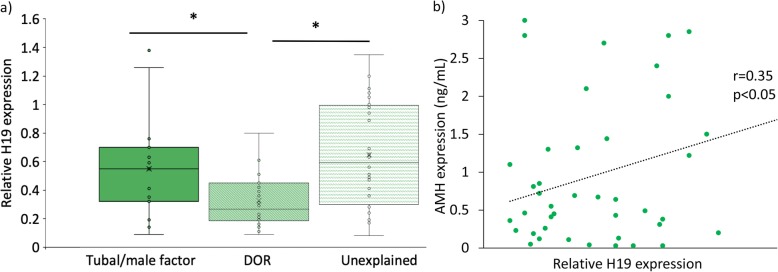
Women with DOR had significantly lower serum H19 expression level as compared to women with tubal factor infertility and women with unexplained infertility (Fig. 1a, p < 0.05). As expected, women with DOR also had significantly lower serum AMH levels 0.4 ng/mL for DOR vs 2.5 ng/mL for tubal factor and 3.0 ng/mL for unexplained infertility, p < 0.001). A moderate correlation was observed between serum H19 expression and AMH (Fig. 1b, p < 0.05).
Fig. 1.
Serum H19 expression in decreased in women with DOR. a) Expression level of H19 in women with DOR is presented as fold change relative to women without DOR (i.e. unexplained infertility and male/tubal factor). Relative H19 expression was decreased by half in women with DOR compared to women with male/tubal factor infertility, and by nearly 60% as compared to women with unexplained infertility (p < 0.05). B) A moderate positive correlation between H19 expression and serum AMH levels (r = 0.35) was observed; p < 0.05
H19 expression in cumulus cells
H19 expression was significantly decreased (4.6 fold) in cumulus cells of IVF patients who demonstrated a poor response to gonadotropins compared to high responders (Fig. 2, p < 0.05). Women who were poor responders also had higher cycle day 3 FSH levels (8.6 mIU/mL vs 6.4 mIU/mL, p < 0.05) and lower peak E2 during IVF stimulation (1175.2 pg/mL vs 2094 pg/mL, p < 0.05). Total gonadotropin dose did not differ between the two groups.
Fig. 2.
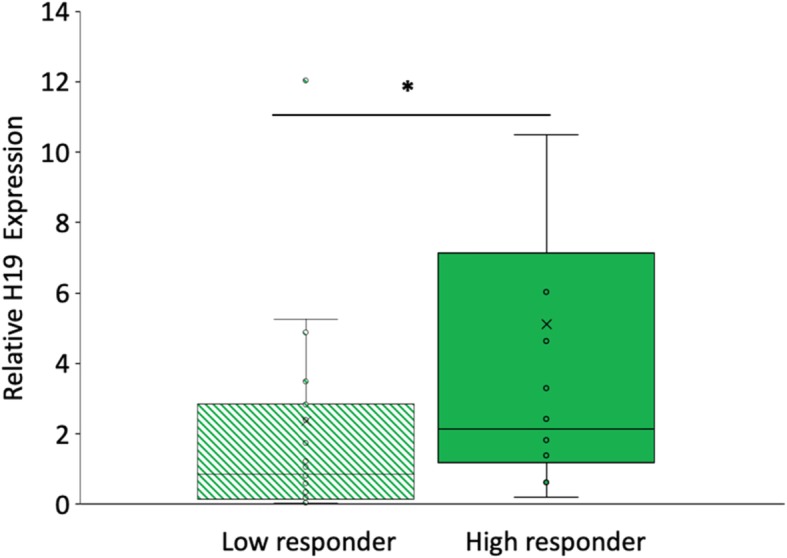
Serum H19 expression in women who are poor responders to IVF treatment. Expression level of H19 in women who were high and low responders to gonadotropins is presented as fold change relative to controls (women who had a normal / midrange gonadotropin response). Relative H19 expression was significantly decreased in “low responder” women (p < 0.05)
Discussion
In this study, we show that in women with evidence of decreased ovarian reserve, circulating and ovarian levels of H19 are decreased. We observed that women with diminished ovarian reserve, as evidenced by a poor response to gonadotropins and a low oocyte yield after in vitro fertilization, also exhibited decreased cumulus cell H19 expression. We also found that serum H19 was lower in women with decreased serum AMH levels (a proxy for ovarian reserve which is predictive of low oocyte yield at retrieval [23]). We also observed a moderate, statistically significant correlation between decreased serum H19 expression and decreased AMH. Altered expression profiles of microRNAs, another class of noncoding RNAs, has been linked with poor ovarian reserve in plasma and serum [24, 25], whole ovaries [26], follicular fluid [27], granulosa cells [28–30], and oocytes [31], and two studies have reported differential expression of piwi-interacting RNAs in GCs and oocytes from women with DOR [31, 32]. However, little is known about the role of long noncoding RNAs such as H19 in ovarian aging.
More work is needed to determine whether H19 may be useful as a marker of ovarian reserve alone or in combination with other markers, or as a prognostic marker for follicle loss remains to be determined. Currently available methods of determining ovarian reserve have their drawbacks. The antral follicle count, typically performed via transvaginal ultrasound, is invasive, uncomfortable, and subject to inter-observer variability. An early follicular phase FSH has poor sensitivity for predicting failure to conceive, and requires careful timing in order to be accurate. Estradiol has poor intra- and inter-cycle reliability and is useful only in conjunction with FSH [33]. Serum AMH has emerged as a widely used measure of ovarian reserve based on its consistency within and between menstrual cycles. AMH is useful as a predictor of ovarian response in controlled ovarian hyperstimulation cycles. However, the PPV in young women is poor, and AMH does not appear to predict time to pregnancy in non-infertile women [34]. Combining the different currently available ovarian reserve testing modalities does not appear to improve predictive ability over the use of a single test [35]. Importantly, all available tests are considered diagnostic rather than prognostic; we still seek the “holy grail” of a predictive test for follicle loss.
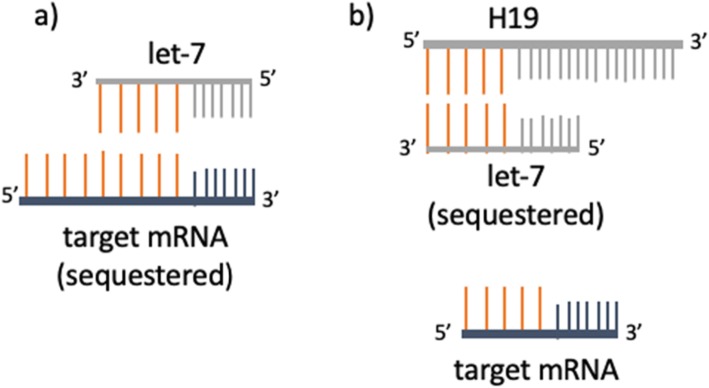
As a mechanistic explanation for how H19 might regulate ovarian reserve, we have shown that in mice, loss of H19 leads to accelerated follicular recruitment with increased spontaneous development of secondary, preantral, and antral follicles [14]. We also described a novel mechanism by which H19 appears to regulate AMH. AMH is a critical “gatekeeper” which inhibits the activation of dormant ovarian follicles and slows the growth of maturing follicles [15–17]. Our previous work demonstrated low AMH, increased follicular recruitment and subfertility in H19 knockout mice, suggesting a potential role for H19 in the regulation of anti-Müllerian hormone AMH [14]. A possible mechanism for this can be found in a microRNA (miRNA) intermediary, let-7, which functions as a negative regulator of target genes [36]. We have shown that H19 acts as a molecular “sponge” for let-7, binding and modulating its availability [36] (Fig. 3). The Amh mRNA contains putative binding sites for let-7 [14], and let-7 transfection leads to decreased Amh expression in GCs [14], supporting Amh as a novel let-7 target. Previous studies have demonstrated that let-7 expression is increased in GCs from women with poor ovarian reserve [30] and in plasma from women with premature ovarian insufficiency, a condition marked by early loss of ovarian follicles [25]. Taken together, these studies suggest a plausible ncRNA-mediated mechanism for AMH regulation by H19, via let-7, and points to a potential role for H19 in ovarian aging by identifying a direct link between ncRNAs and follicular and oocyte quantity.
Fig. 3.
H19 acts as a molecular “sponge” for let-7. (a) The microRNA let-7 functions as a negative regulator of target genes [36]. (b) We have shown that H19 acts as a molecular “sponge” for let-7, binding and modulating its availability [36]. The Amh mRNA contains putative binding sites for let-7 [14], and let-7 transfection leads to decreased Amh expression in GCs [14], supporting Amh as a novel let-7 target and representing a novel ncRNA mediated mechanism by which the bioavailability of Amh can be regulated by H19
This work raises the crucial question of whether H19 has a role in the regulation of ovarian aging. As expected, in order to examine whether H19 is related to markers of ovarian reserve, our studies included a study population of women who were older than controls. One question is whether the decreased H19 expression we have seen in these populations is correlated with aging itself. To our knowledge, no data exists regarding whether age-related changes in H19 expression occur with time in reproductive tissues. Loss of H19 has been linked to increased expression of markers of cellular aging and loss of cellular quiescence in hematopoietic stem cells and endothelial cells [37–39]. More work is necessary to clarify this question. However, even if the differences in H19 expression observed between populations are a result of age, the idea that H19 may represent a biological “rheostat” regulating the aging reproductive endocrine system beyond fertility is an intriguing one, especially given that ovarian aging has implications beyond fertility, including the serious health consequences of menopause such as vasomotor symptoms, osteoporosis, and cardiovascular disease.
The strengths of our study include the use of different testing modalities (serum and cumulus cells). Noncoding RNAs, including miRNAs and lncRNAs, can be collected from tissues as well as bodily fluids including plasma, serum, and urine [40, 41]. While tissue-specific changes in H19 expression have been investigated as potential biomarkers in other conditions including breast [42] cancer, this is the first report of H19 expression in cumulus cells, which are easily accessible at the time of oocyte retrieval. We also report a correlation between serum H19 and AMH, a marker of follicular response during IVF23. Circulating H19 has been explored as a diagnostic and prognostic biomarker for other conditions including coronary artery disease [43], multiple myeloma [44], and cancers including bladder and gastric cancer [22, 40, 41, 45, 46]. In the reproductive tract, circulating H19 is higher in women with polycystic ovary syndrome (PCOS) compared to controls [47], and H19 has been identified as a potential diagnostic and prognostic marker for epithelial ovarian [48] and cervical cancer [49]. However, a link between circulating H19 and markers of ovarian reserve has not previously been described. Moreover, our sample groups include a diverse population of women at a range of ages and ethnic backgrounds, lending generalizability to our results. Our study is limited by the small sample size and the fact that limited demographic data and no AMH was available from the population of women from whom cumulus cells were obtained. However, while the finding of low H19 in women with poor ovarian reserve may be correlative and requires further study, our findings regarding Amh and let-7 lend mechanistic plausibility to H19 as a causative factor. Lastly, it is well established that imprinted genes, including H19, are susceptible to alteration by assisted reproductive technologies [50–52]. However, our findings of low H19 in women with DOR are consistent across women undergoing controlled ovarian stimulation (COH) and those who presented prior to initiating COH. Additionally, our findings are consistent across different testing modalities (serum and cumulus cells).
In conclusion, our work suggests for the first time that circulating and intrafollicular H19 levels may be altered in women with diminished ovarian reserve. H19 may have an important role as a master regulator of ovarian reserve markers, particularly AMH, and further elucidation of the role of H19 in the ovarian aging process will enhance our understanding of normal folliculogenesis as well as the pathogenesis of diminished ovarian reserve. Further studies should be conducted to determine whether the association we have observed holds true in younger women with diminished ovarian reserve. In the future, H19 may also prove useful as a novel diagnostic biomarker for ovarian reserve and premature ovarian insufficiency, and has the potential to improve our diagnosis of DOR especially in situations where current ovarian reserve testing has proven inadequate.
Acknowledgements
The authors gratefully acknowledge support received from the grant K12 HD000849, awarded to the Reproductive Scientist Development Program (RSDP) by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the American Society for Reproductive Medicine as part of the RSDP, the Bennack-Polan Foundation grant, the NIH Loan Repayment Program, the Milstein Medical Asian American partnership Foundation (MMAAPF) and the Research Team of Female Reproductive Health and Fertility Preservation (201612065).
Abbreviations
- AMH
Anti-mullerian hormone
- COH
Controlled ovarian hyperstimulation
- DOR
Diminished ovarian reserve
- E2
Estradiol
- FSH
Follicle stimulating hormone
- GC
Granulosa cell
- hCG
Human chorionic gonadotropin
- hMG
Human menopausal gonadotropin
- IVF-ICSI
In vitro fertilization – intracytoplasmic sperm injection
- lncRNA
Long noncoding RNA
- miRNA
microRNA
- ncRNA
noncoding RNA
- PPV
Positive predictive value
Authors’ contributions
A.K. and X.X. contributed to study design and interpretation of data. X.X., Y.C., and M.B. performed the experiments. X.X. performed statistical analysis and prepared the figures and tables. All authors contributed to manuscript drafting. The author(s) read and approved the final manuscript.
Availability of data and materials
Data supporting findings are presented within the manuscript.
Ethics approval and consent to participate
These studies were approved by the Yale University Institutional Review Board (IRB protocol # 1606017946) and the Gazi University Institutional Review Board committee (IRB protocol #131/11.05.2011).
Consent for publication
All authors consent to publication of this study.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xi Xia, Email: xixia1126@hotmail.com.
Martina S. Burn, Email: martina.burn@yale.edu
Yong Chen, Email: fjmucy@hotmail.com.
Cengiz Karakaya, Email: karakayac@gazi.edu.tr.
Amanda Kallen, Email: Amanda.kallen@yale.edu.
References
- 1.Kallen A, Polotsky AJ, Johnson J. Untapped reserves: controlling primordial follicle growth activation. Trends Mol Med 2018;24(3):319–331. PMID: 29452791. [DOI] [PMC free article] [PubMed]
- 2.The American College of Obstetricians and Gynecologists Committee on Gynecologic Practice. Committee Opinion #589: Female Age-Related Fertility Decline. PMID: 26942393.
- 3.Schwartz D, Mayaux M. Results of artificial insemination in 2193 Nulliparrous women with Azoospermic husbands. N Engl J Med. 1982;306(7):404–6. 3141811. [DOI] [PubMed]
- 4.Mathews TJ, Hamilton BE. Mean Age of Mothers is on the Rise: United States, 2000–2014. NCHS Data Brief [Internet]. 2016;(232):1–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26828319 PMID: 26828319. [PubMed]
- 5.Martin JA, Brady MPH, Hamilton E, Osterman MJK, Driscoll AK, Mathews TJ. Births: Final Data for 2015. CDC Natl Vital Stat Reports [Internet]. 2017;66(1). Available from: http://www.cdc.gov/nchs/data_access/Vitalstatsonline.htm. [PubMed]
- 6.Chandra A, Copen C. Infertility and Impaired Fecundity in the United States , 1982–2010 : Data from the National Survey of family growth. CDC Natl Heal Stat Rep. 2013:67. [PubMed]
- 7.Ferraretti AP, La Marca A, Fauser BCJM, Tarlatzis B, Nargund G, Gianaroli L. ESHRE consensus on the definition of ‘poor response to ovarian stimulation for in vitro fertilization: the Bologna criteria. Hum Reprod. 2011;26(7):1616–1624. doi: 10.1093/humrep/der092. [DOI] [PubMed] [Google Scholar]
- 8.Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. BioEssays. 2010;32(6):473–480. PMID: 20486133. [DOI] [PubMed]
- 9.Ariel I, Weinstein D, Voutilainen R, Schneider T, Lustig-Yariv O, De-Groot N, Hochberg A. The expression of the imprinted gene H19 in the human female reproductive organs. Diagnostic Mol Pathol 1997;61(1):17–25. PMID: 9028733. [DOI] [PubMed]
- 10.Rachmilewitz J, Goshen R, Ariel I, Schneider T, de Groot N, Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett 1992;309(1):25–28. PMID: 1709450. [DOI] [PubMed]
- 11.Khatib H, Schutzkus V. The expression profile of the H19 gene in cattle. Mamm Genome 2006;17(9):991–996. PMID: 16964441. [DOI] [PubMed]
- 12.Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991;351(6322):153–155. PMID: 1709450. [DOI] [PubMed]
- 13.Ripoche MA, Kress C, Poirier F, Dandolo L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 1997;11(12):1596–1604. PMID: 9203585. [DOI] [PubMed]
- 14.Qin C, Xia X, Fan Y, Jiang Y, Chen Y, Zhang N, Uslu B, Johnson J, Kallen AN. A novel, noncoding-RNA-mediated, post-transcriptional mechanism of anti-Mullerian hormone regulation by the H19/let-7 axis†. Biol Reprod 2018;0(August):1–11. PMID: 30137224. [DOI] [PMC free article] [PubMed]
- 15.Broekmans FJ, Soules MR, Fauser BC. Ovarian aging: mechanisms and clinical consequences. Endocr Rev 2009;30(5):465–493. PMID: 19589949. [DOI] [PubMed]
- 16.Durlinger ALL, Kramer P, Karels B A S, Jong FHDE, Uilenbroek J a NTHJ, Grootegoed JA, Themmen APN, di Clemente N, Ghaffari S, Pepinsky RB, Pieau C, Josso N, Cate RL, Vigier B. Control of primordial Follilce recruitment by AMH in mouse ovary. Endocrinology. 1999;140(12):5789–5796. PMID: 1319894. [DOI] [PubMed]
- 17.Durlinger AL, Visser JA, Themmen AP. Regulation of ovarian function: the role of anti-Mullerian hormone. Reproduction. 2002;124(5):601–609. PMID: 12416998. [DOI] [PubMed]
- 18.Durlinger ALL, Gruijters MJG, Kramer P, Karels B, Kumar TR, Matzuk MM, Rose UM, De Jong FH, Uilenbroek JTJ, Grootegoed JA, Themmen APN. AMH attenuates the effects of FSH on follicle development in the mouse ovary. Endocrinology [Internet]. 2001 [cited 2017 Jul 17];142(11):4891–4899. PMID: 11606457. [DOI] [PubMed]
- 19.Alvaro Mercadal B, Imbert R, Demeestere I, Gervy C, De Leener A, Englert Y, Costagliola S, Delbaere A. AMH mutations with reduced in vitro bioactivity are related to premature ovarian insufficiency. Hum Reprod 2015;30(5):1196–1202. PMID: 25750103. [DOI] [PubMed]
- 20.Wang J, Zhao H, Fan Z, Li G, Ma Q, Tao Z, Wang R, Feng J, Luo Y. Long noncoding RNA H19 promotes Neuroinflammation in ischemic stroke by driving histone Deacetylase 1–dependent M1 microglial polarization. Stroke. 2017;48(8):2211–2221. PMID: 28630232. [DOI] [PubMed]
- 21.Wu Y, Wang Y-Q, Weng W-W, Zhang Q-Y, Yang X-Q, Gan H-L, Yang Y-S, Zhang P-P, Sun M-H, Xu M-D, Wang C-F. A serum-circulating long noncoding RNA signature can discriminate between patients with clear cell renal cell carcinoma and healthy controls. Oncogenesis. 2016;5(2):e192. PMID: 26878386. [DOI] [PMC free article] [PubMed]
- 22.Wang J, Yang K, Yuan W, Gao Z. Determination of serum Exosomal H19 as a noninvasive biomarker for bladder Cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–9316. doi: 10.12659/MSM.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borges E, Braga DPAF, Setti A, Figueira R. de C, Iaconelli a. the predictive value of serum concentrations of anti-Müllerian hormone for oocyte quality, fertilization, and implantation. J Bras Reprod Assist. 2017;21(3):176–182. doi: 10.5935/1518-0557.20170035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong L, Peng S, Li Y, Fang Y, Wang Q, Klausen C, Yin C, Wang S, Leung PCK, Yang X. MiR-106a increases Granulosa cell viability and is Downregulated in women with diminished ovarian reserve. J Clin Endocrinol Metab. 2018;103(6):2157–66. [DOI] [PubMed]
- 25.Dang Y, Zhao S, Qin Y, Han T, Li W, Chen ZJ. MicroRNA-22-3p is down-regulated in the plasma of Han Chinese patients with premature ovarian failure. Fertil Steril. Elsevier Inc. 2015;103(3):802–807.e1. doi: 10.1016/j.fertnstert.2014.12.106. [DOI] [PubMed] [Google Scholar]
- 26.Schneider A, Matkovich SJ, Victoria B, Spinel L, Bartke A, Golusinski P, Masternak MM. Changes of ovarian microRNA profile in long-living Ames dwarf mice during aging. PLoS One. 2017;12(1):1–19. doi: 10.1371/journal.pone.0169213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno JM, Núñez MJ, Quiñonero A, Martínez S, De La Orden M, Simón C, Pellicer A, Díaz-García C, Domínguez F. Follicular fluid and mural granulosa cells microRNA profiles vary in in vitro fertilization patients depending on their age and oocyte maturation stage. Fertil Steril. 2015;104(4):1037–1046.e1. doi: 10.1016/j.fertnstert.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Woo I, Christenson LK, Gunewardena S, Ingles SA, Thomas S, Ahmady A, Chung K, Bendikson K, Paulson R, McGinnis LK. Micro-RNAs involved in cellular proliferation have altered expression profiles in granulosa of young women with diminished ovarian reserve. J Assist Reprod Genet. 2018;35(10):1777–1786. doi: 10.1007/s10815-018-1239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo H, Han Y, Liu J, Zhang Y. Identification of microRNAs in granulosa cells from patients with different levels of ovarian reserve function and the potential regulatory function of miR-23a in granulosa cell apoptosis. Gene Elsevier. 2019;686(November 2018):250–260. doi: 10.1016/j.gene.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 30.Hong L, Peng S, Li Y, Fang Y, Wang Q, Klausen C, Yin C, Wang S, Leung PCK, Yang X. MiR-106a increases Granulosa cell viability and is Downregulated in women with diminished ovarian reserve. J Clin Endocrinol Metab. 2018;103(6):2157–2166. doi: 10.1210/jc.2017-02344. [DOI] [PubMed] [Google Scholar]
- 31.Barragán M, Pons J, Ferrer-Vaquer A, Cornet-Bartolomé D, Schweitzer A, Hubbard J, Auer H, Rodolosse A, Vassena R. The transcriptome of human oocytes is related to age and ovarian reserve. Mol Hum Reprod. 2017;23(8):535–548. doi: 10.1093/molehr/gax033. [DOI] [PubMed] [Google Scholar]
- 32.Chen D, Zhang Z, Chen B, Ji D, Hao Y, Zhou P, Wei Z, Cao Y. Altered microRNA and Piwi-interacting RNA profiles in cumulus cells from patients with diminished ovarian reserve. Biol Reprod. 2017;97(1):91–103. doi: 10.1093/biolre/iox062. [DOI] [PubMed] [Google Scholar]
- 33.Pfeifer S, Butts S, Dumesic D, Fossum G, Giudice L, Gracia C, La Barbera A, Odem R, Pisarska M, Rebar R, Richard R, Rosen M, Sandlow J, Vernon M, Widra E. Testing and interpreting measures of ovarian reserve: A committee opinion. Fertil Steril. American Society for Reproductive Medicine; 2015;103(3):e9–e17.
- 34.Steiner AZ, Pritchard D, Stanczyk FZ, Kesner JS, Meadows JW, Herring AH, Baird DD. Association between biomarkers of ovarian reserve and infertility among older women of reproductive age. Jama. 2017;318(14):1367–1376. doi: 10.1001/jama.2017.14588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pfeifer S, Butts S, Dumesic D, Fossum G, Giudice L, Gracia C, La Barbera A, Odem R, Pisarska M, Rebar R, Richard R, Rosen M, Sandlow J, Vernon M, Widra E. Testing and interpreting measures of ovarian reserve: A committee opinion. Fertil Steril [Internet]. American Society for Reproductive Medicine; 2015;103(3):e9–e17. Available from: 10.1016/j.fertnstert.2014.12.093.
- 36.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 LncRNA antagonizes Let-7 MicroRNAs. Mol Cell 2013;52(1):101–112. PMID: 24055342. [DOI] [PMC free article] [PubMed]
- 37.Venkatraman A, He XC, Thorvaldsen JL, Sugimura R, Perry JM, Tao F, Zhao M, Christenson MK, Sanchez R, Yu JY, Peng L, Haug JS, Paulson A, Li H, Zhong XB, Clemens TL, Bartolomei MS, Li L. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature. 2013;500(7462):345–349. PMID: 23863936. [DOI] [PMC free article] [PubMed]
- 38.Eisenberg I, Kotaja N, Goldman-wohl D, Imbar T. microRNA in Human Reproduction. microRNA Med Evid. 2015. p. 353–387. PMID: 26663190. [DOI] [PubMed]
- 39.Hofmann P, Sommer J, Theodorou K, Kirchhof L, Fischer A, Li Y, Perisic L, Hedin U, Maegdefessel L, Dimmeler S, Boon RA. Long non-coding RNA H19 regulates endothelial cell aging via inhibition of STAT3 signalling. Cardiovasc Res 2019;115(1):230–242. PMID: 30107531. [DOI] [PMC free article] [PubMed]
- 40.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 2015;5(May):1–10. PMID: 26096073. [DOI] [PMC free article] [PubMed]
- 41.Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric Cancer. J Clin Lab Anal 2016;30(6):1100–1105. PMID: 27184562. [DOI] [PMC free article] [PubMed]
- 42.Li Z, Li Y, Li Y, Ren K, Li X, Han X, Wang J. Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J Biochem Mol Toxicol 2017;31(e21933):1–9. PMID: 28544374. [DOI] [PubMed]
- 43.Xiong G, Jiang X, Song T. The overexpression of lncRNA H19 as a diagnostic marker for coronary artery disease. Rev Assoc Med Bras. 2019;65(2):110–117. doi: 10.1590/1806-9282.65.2.110. [DOI] [PubMed] [Google Scholar]
- 44.Pan Y, Chen H, Shen X, Wang X, Ju S, Lu M, Cong H. Serum level of long noncoding RNA H19 as a diagnostic biomarker of multiple myeloma. Clin Chim Acta [internet]. Elsevier; 2018;480(January):199–205. Available from: 10.1016/j.cca.2018.02.019. [DOI] [PubMed]
- 45.Ghaedi H, Mozaffari MAN, Salehi Z, Ghasemi H, Zadian SS, Alipoor S, Hadianpour S, Alipoor B. Co-expression profiling of plasma miRNAs and long noncoding RNAs in gastric cancer patients. Gene. 2019;687(October 2018):135–142. doi: 10.1016/j.gene.2018.11.034. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, He A, Liu B, Huang Z, Mei H. Potential role of lncRNA H19 as a Cancer biomarker in human cancers detection and diagnosis: a pooled analysis based on 1585 subjects. Biomed Res Int. 2019;2019:1–11. doi: 10.1155/2019/9056458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin L, Huang C, Yan X, Wang Y, Li Z, Wei X. Long non-coding RNA H19 is associated with polycystic ovary syndrome in Chinese women: a preliminary study. Endocr J. 2019. [DOI] [PubMed]
- 48.Lou Y, Jiang H, Cui Z, Wang X, Wang L, Han Y. Gene microarray analysis of lncRNA and mRNA expression profiles in patients with high-grade ovarian serous cancer. Int J Mol Med. 2018;42(1):91–104. doi: 10.3892/ijmm.2018.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin S, Gao Y, Yang Y, Zhang L, Zhang T, Yu J, Shi C. Identifying molecular markers of cervical Cancer based on competing endogenous RNA network analysis. Gynecol Obstet Investig. 2018:1–10. [DOI] [PubMed]
- 50.Sakian S, Louie K, Wong EC, Havelock J, Kashyap S, Rowe T, Taylor B, Ma S. Altered gene expression of H19 and IGF2 in placentas from ART pregnancies. Placenta Elsevier Ltd; 2015;36(10):1100–1105. PMID: 26386650. [DOI] [PubMed]
- 51.Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, Hu JF, Behr B, Giudice L, Hoffman AR. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod 2005;11(9):631–640. PMID: 16219628. [DOI] [PubMed]
- 52.Shi X, Ni Y, Zheng H, Chen S, Zhong M, Wu F, Xia R, Luo Y. Abnormal methylation patterns at the IGF2/H19 imprinting control region in phenotypically normal babies conceived by assisted reproductive technologies. Eur J Obstet Gynecol Reprod Biol 2011;158(1):52–55. PMID: 21555179. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data supporting findings are presented within the manuscript.