Abstract
There is a major clinical need for new therapies for treatment of chronic itch. Many of the molecular components involved in itch neuro-transmission are known including the neuropeptide NPPB, a transmitter required for normal itch responses to multiple pruritogens in mice. Here we investigated the potential for a novel strategy for treatment of itch that involves the inhibition of the NPPB-receptor, NPR1. Since there are no available effective human NPR1 (hNPR1) antagonists, we performed a high-throughput cell-based screen and identified 15 small molecule hNPR1 inhibitors. Using in vitro assays, we demonstrated that these compounds specifically inhibit hNPR1 and murine NPR1 (mNPR1). In vivo, NPR1 antagonism attenuated behavioral responses to both, acute and chronic itch challenged mice. Together our results suggest that inhibiting NPR1 might be an effective strategy for treating acute and chronic itch.
One sentence summary:
Natriuretic peptide receptor 1 plays a main role in itch and its inhibition with small molecules has therapeutic effects in mouse models of acute and chronic itch
Introduction
Itch is an unpleasant sensation associated with skin irritation which elicits the strong urge to scratch. Whereas most itch has a relatively low prevalence and is easily managed (1), chronic itch has a major negative impact on quality of life and current treatments are largely ineffective (2, 3). The underlying pathophysiological mechanisms for chronic itch are poorly understood (4, 5). For this reason, clinically chronic itch is categorized based on its apparent origin with dermatological, systemic, neurological, somatoform, and mixed pruritus (6). Itch stimuli themselves are thought to be detected in the skin by dedicated sensory neurons which innervate the skin and express G protein-coupled, toll-like, and interleukin receptors (5, 7–11). While the repertoire of known receptors is large, the number of itch cell-types which detect itch stimuli is small. The itch-sensory neurons comprise two distinct populations, those expressing the mas-related G protein-coupled receptor A3 (Mrgpra3) and those expressing the neuropeptide natriuretic polypeptide b (Nppb) (12, 13). Both these classes of neurons transmit itch through a common spinal cord circuit dependent on NPPB (14, 15). Additionally, sensory neuron-derived NPPB has recently been suggested to drive inflammation in different forms of dermatitis, both in humans and mice, thereby enhancing pruritus (16).
Work from our laboratory identified the spinal cord receptor for NPPB, natriuretic peptide receptor 1 (NPR1) as a potential target for the treatment of itch (13). We demonstrated that elimination of NPPB as well as the ablation of spinal interneurons expressing Npr1 profoundly attenuated scratching responses to many pruritogens in mice (13). These results indicate that NPPB is a critical component required for the excitation of spinal cord Npr1-expressing interneurons. The NPR1 receptor belongs to a small guanylate cyclase (GC) family of receptor proteins which are specifically activated by natriuretic peptides (NP) (17). NPR1 binds both NPPB and NPPA with high affinity, and has a much lower affinity for the third NP, NPPC (17). Three cyclic peptide analogues of NPPB have been reported to inhibit NPR1 (18–21). However, one of these, A-71915, was reported to be ineffective at blocking itch in mice. Specifically, A-71915, when administered intrathecally, was unable to attenuate acute itch (22). This result suggests that inhibition of spinal cord NPR1 may not be an effective method for relieving itch. To investigate this further, we characterized the properties of A-71915 and found that the compound is a strong partial agonist not a neutral antagonist of murine NPR1 (mNPR1), suggesting a possible explanation for its lack of potency in relieving itch. Using high-throughput screening (HTS) we identified human NPR1 (hNPR1) antagonists and show that one of these compounds can relieve itch in vivo in mouse models of itch.
Results
Similar expression of NPPB in human and mouse DRG neurons
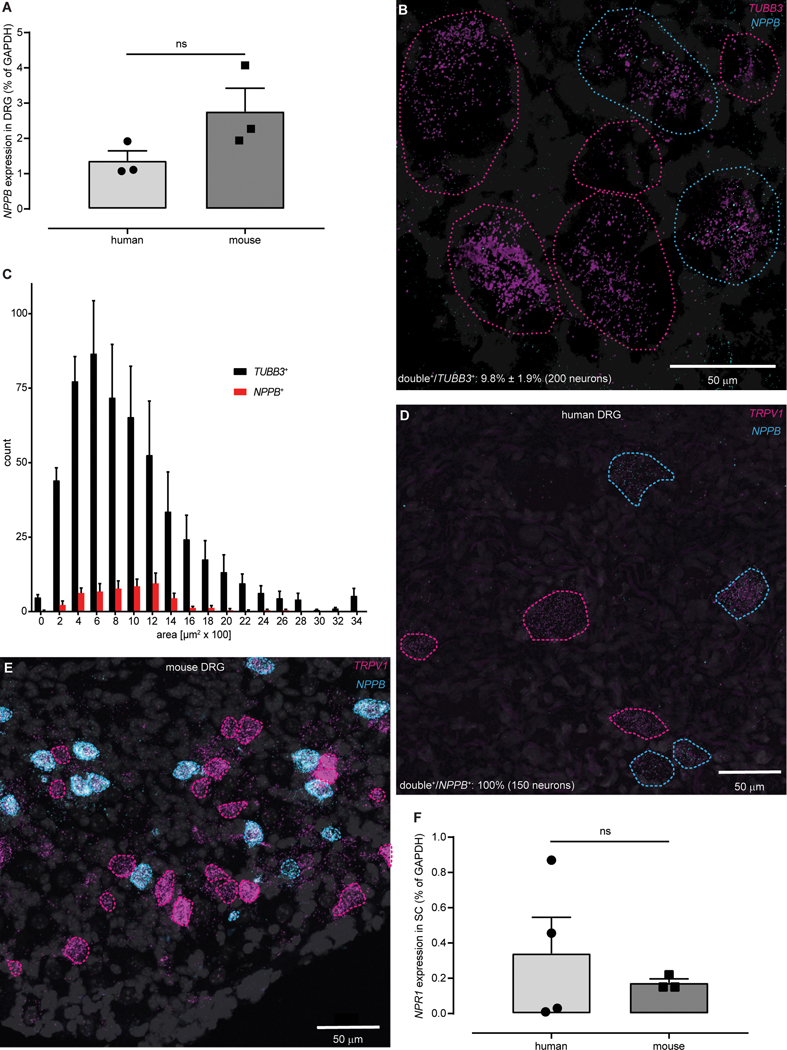
Research on the basic mechanism for itch has been predominantly performed in the mouse animal model (4). The molecules for many pathways in sensory neurons in humans and mice have been assumed to be similar to each other, although in some instances the molecules may not be the same (23–25). Therefore, as a starting point, to motivate the investigation of NPPB as a target for development of itch therapies, we examined if human dorsal root ganglia (DRG) express NPPB. We detected, by qPCR, similar amounts of NPPB transcripts in cDNA from human and mouse DRG (Fig. 1A). In concordance with this result, using double label in situ hybridization (ISH), we found that the numbers of NPPB-neurons (9.8% ± 1.9%; 200 of 2086 neurons, n = 4 donors) in human DRG are approximately the same as previously reported in mice (26) and human NPPB-neurons likewise have small to medium diameter (Fig. 1B,C). In mice, Nppb is co-expressed with TrpV1 (13). Therefore, if NPPB has a comparable role in humans then, it should have a similar distribution in human DRG. ISH revealed, similar to mouse (26), that all NPPB-positive human DRG neurons co-express TRPV1 (150 neurons, n = 4 donors) and that these neurons are a fraction of the TRPV1-neurons (Fig. 1D,E). In addition, for NPPB to function in a similar way in humans to that reported in mice (13) then, its receptor NPR1 should be expressed in spinal cord. To test this, we compared expression of human and mouse Npr1 in spinal cord using qPCR and found that NPR1 is expressed in similar amounts (Fig. 1F). Together these results suggest that mouse and human itch neurotransmission likely use the same signaling molecules.
Fig. 1: The NPPB-NPR1 itch signaling pathway is conserved between mice and humans.
(A) qPCR-based quantification of expression did not show a significant difference in amounts of NPPB transcripts between human and mouse DRG (P = 0.1241, unpaired t-test, n = 3). (B) Representative double ISH images of a field of human DRG with neurons stained for NPPB (cyan) and TUBB3 (magenta). NPPB-positive and NPPB-negative neurons are outlined with cyan and magenta dots respectively, DAPI counterstain is displayed in grey. (C) Quantification of the soma-size of NPPB- (red) compared to TUBB3-stained (black) neurons (n = 4). Representative double ISH images, of fields of human (D) and mouse (E) DRG, reveal that NPPB (cyan) and TRPV1 (magenta) are co-expressed. In human and mouse DRG, NPPB is expressed in a subset of TRPV1-neurons (cyan-dotted profiles), single-labeled TRPV1-neurons are indicated with magenta-dotted profiles. (F) qPCR-based quantification of expression did not show a significant difference in amounts of NPR1 transcripts between human and mouse spinal cord (ns P > 0.9999, Mann-Whitney, n = 4 (human) 3 (mouse)).
A-71915, a mixed NPR1 antagonist/agonist
When the gene for Nppb is eliminated in knockout mice or Npr1-expressing spinal neurons are ablated, itch responses are attenuated, suggesting that NPR1 signaling is critical for itch responses (13). However, it was reported that A-71915, a relatively potent hNPR1 antagonist (18), does not block acute itch responses (22). An explanation for this may be that A-71915 does not effectively block itch in vivo. As the pharmacodynamic interactions of A-71915 with mNPR1 have not been tested, we developed a method to measure inhibition of mNPR1 by A-71915.
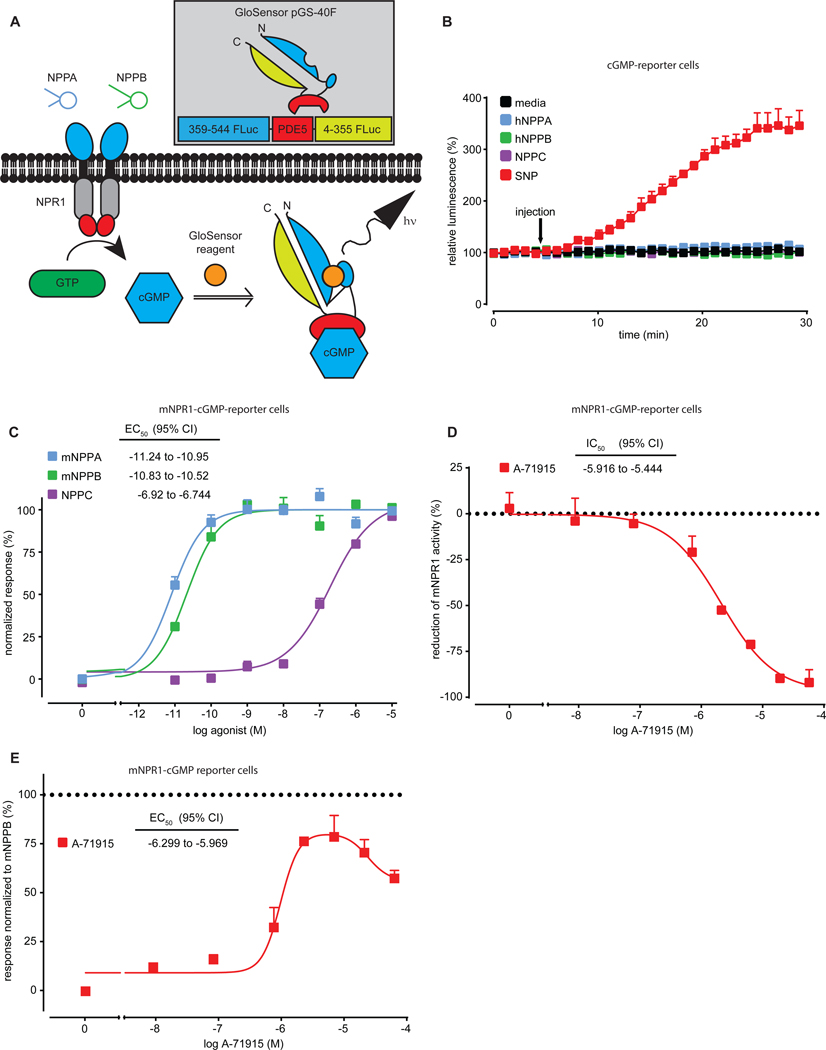
Given that the NPR1 receptor is a ligand dependent GC (17), we reasoned that monitoring agonist-induced changes of intracellular cyclic guanosine monophosphate (cGMP) amounts would be a straight forward way to examine NPR1 inhibition. To measure the production of cGMP by NPR1, we utilized a circular permutated Firefly luciferase molecule which was functionally linked to the cGMP binding domain of human phosphodiesterase 5 (GloSensor technology pGS-40F, (27)). This reporter together with a luciferase substrate (GloSensor reagent) and a sensitive method for detection of luciferase activity (light emission) permitted us, in real-time, to measure cGMP in cells (Fig. 2A). We expressed the cGMP-sensor in human embryonic kidney 293 (HEK-293) cells and examined luciferase activity after stimulation with the NO-donor sodium nitroprusside (SNP), which activates ubiquitously expressed soluble GC (Fig. 2B). As expected, SNP generated a long-lasting increase in luciferase activity. In contrast, stimulation of cGMP reporter cells with hNPPA, hNPPB and NPPC did not increase cGMP (Fig. 2B), indicating that HEK-293 cells do not express detectable endogenous NP receptors. Next, we transiently expressed mNpr1 and cGMP-sensor in HEK-293 cells and examined receptor activation. As expected, mNPPA, mNPPB and NPPC dose-dependently increased reporter activity and had stimulation potencies similar to those previously reported (Fig. 2C) (17) and, as previously reported for hNPR1 (18), A-71915 inhibited NP-induced mNPR1 activity (Figure 2D). In addition, our results showed that A-71915, in the absence of NP, evoked a dose-dependent activation of mNPR1, indicating that, instead of being a neutral antagonist as described for hNPR1 (18), A-71915 acted as a partial agonist (Fig. 2E).
Fig. 2: A-71915 is a partial agonist of mNPR1.
(A) Schematic depicting our strategy to measure NPR1 activity. NPR1 is stimulated by NP to increase synthesis of cGMP, in turn, increased cGMP alters the conformation of a PDE5-Firefly luciferase-based sensor (cGMP-sensor) which results in hydrolysis of GloSensor reagent and production of light. (B) Time course experiments quantifying luminescence of HEK-293 cells transiently expressing cGMP-sensor stimulated with the soluble GC activator SNP (333 μM; red), media (black), hNPPA (blue), hNPPB (green), and NPPC (purple)(10 nM each). (C) Quantification of activity of HEK-293 cells transiently expressing mNPR1 and cGMP-sensor stimulated with mNPPB (green), mNPPA (blue) and NPPC (purple). (D) Quantification of inhibition of mNPR1-cGMP-sensor cells with A-71915 (5 minutes after addition of A-71915, cells were treated with 1 nM mNPPB). (E) Quantification of mNPR1-cGMP-sensor cells shows partial agonist activity for A-71915. Data represent means SEM of triplicate (B) or duplicate (C-E) measurements.
Identification of novel hNPR1 antagonists
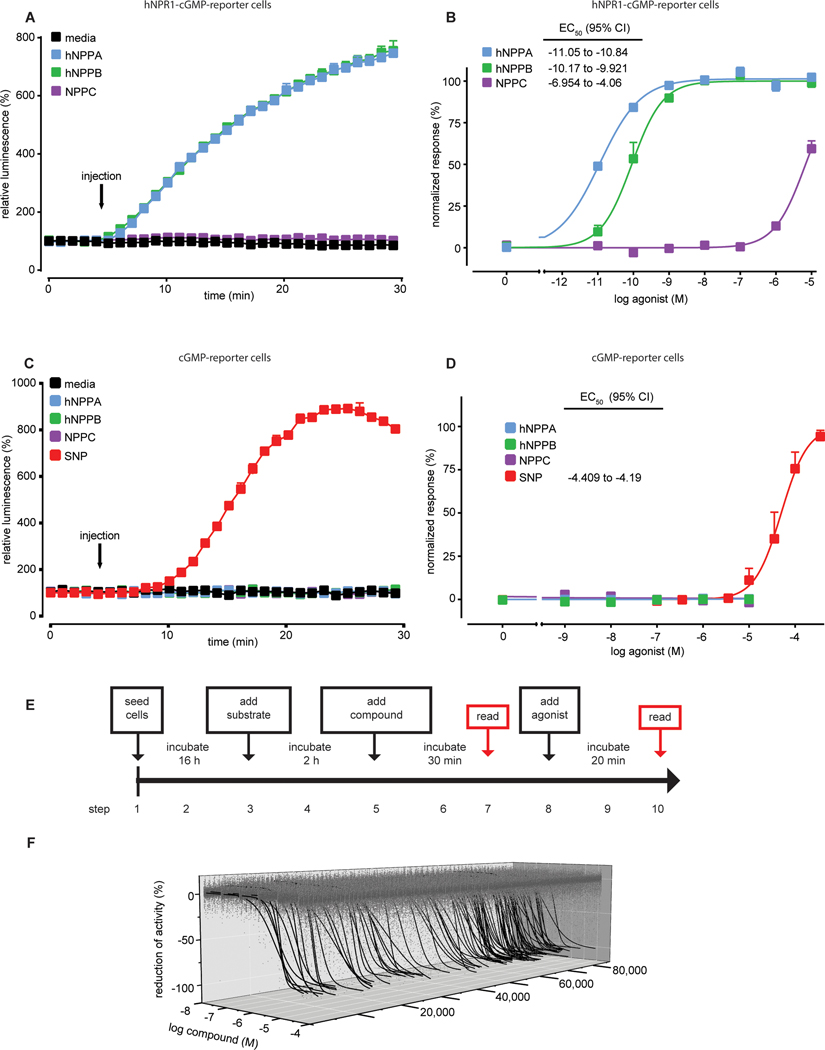
Next, we searched for molecules with good antagonistic properties to investigate if inhibition of NPR1 is a viable approach for alleviating itch. We used a quantitative high-throughput screening (qHTS) method to identify candidate molecules able to inhibit NPR1 from a large chemical library. With a few modifications, we used the same cell-based approach to perform qHTS as we used to determine the properties of A-71915. To increase sensitivity and allow miniaturization to a 1536-well format, we developed a stable cGMP-sensor cell-line expressing hNRP1, a control cell-line with stable expression of cGMP-sensor, and a hNPR2 cell-line (fig. S1). hNPR1-cGMP-reporter cells responded to hNPPA, hNPPB and NPPC with appropriate potencies (Fig. 3A,B), whereas cGMP-reporter cells lacked responses to hNPPA, hNPPB and NPPC but were strongly activated by SNP in a dose-dependent fashion (Fig. 3C,D). To enable assessment of the inhibition of agonist-induced hNPR1 activity and effects of compounds on basal hNPR1 activity, we performed reads prior to addition and 30 minutes after application of agonist (Fig. 3E). To improve identification of active compounds and to assist in ranking compounds, we also conducted primary screens at six log concentrations (28). Our final hNPR1 sensor assay was highly consistent; screening of the LOPAC1280 (1280 compounds) and BU-CMD (1838 compounds) libraries gave Z’ factor scores of 0.73 ± 0.07 and 0.72 ± 0.2, respectively. Our assay also had a low hit rate with these small libraries of compounds and none of the molecules identified as inhibitors from these small libraries passed counter-screens (data file S1).
Fig. 3: Cell-based screen identifies candidate small molecule inhibitors of hNPR1.
(A and C) Time course experiments quantifying luminescence of stable cell lines expressing pGS-40F and hNPR1 (A) and pGS-40F alone (C) stimulated by media (black), hNPPA (blue), hNPPB (green), NPPC (purple) (10 nM each) and SNP (333 μM, red). (B and D) Quantification of activity of HEK-hNPR1-cGMP-sensor cells (B) and HEK-cGMP-sensor cells (D) with hNPPA (blue), hNPPB (green), NPPC (purple), and SNP (red). (E) Schematic depicts the time course of our qHTS assay. (F) Representative 3-axis plot of concentration-response curve profiles for compounds from the Genesis chemical library; 519,417 concentration response values are displayed in grey (1574 out-lie values were not plotted). Out of the 3.9% active compounds, 105 compounds with greatest efficacy (maximum antagonism > 90%) are displayed (black traces). Curves were fit using a four-parameter logistic regression. Data represent means SEM of duplicate (A-C) or triplicate (D) measurements.
For the main qHTS, we utilized an automated robotic system (29) and employed the National Center of Advancing Translational Sciences Genesis library which consists of a chemically and structurally diverse set of small molecules suited for rapid chemical modification (86,437 compounds screened, data file S2). Compounds which inhibited activity (Fig. 3F) were identified using automated software (28). The overall hit rate of our screen was 3.9%. These candidate compounds were prioritized based on their potency, efficacy, and structural relationships, and from all positive compounds 1,408 were judged to be strong candidates for further study (see methods for details).
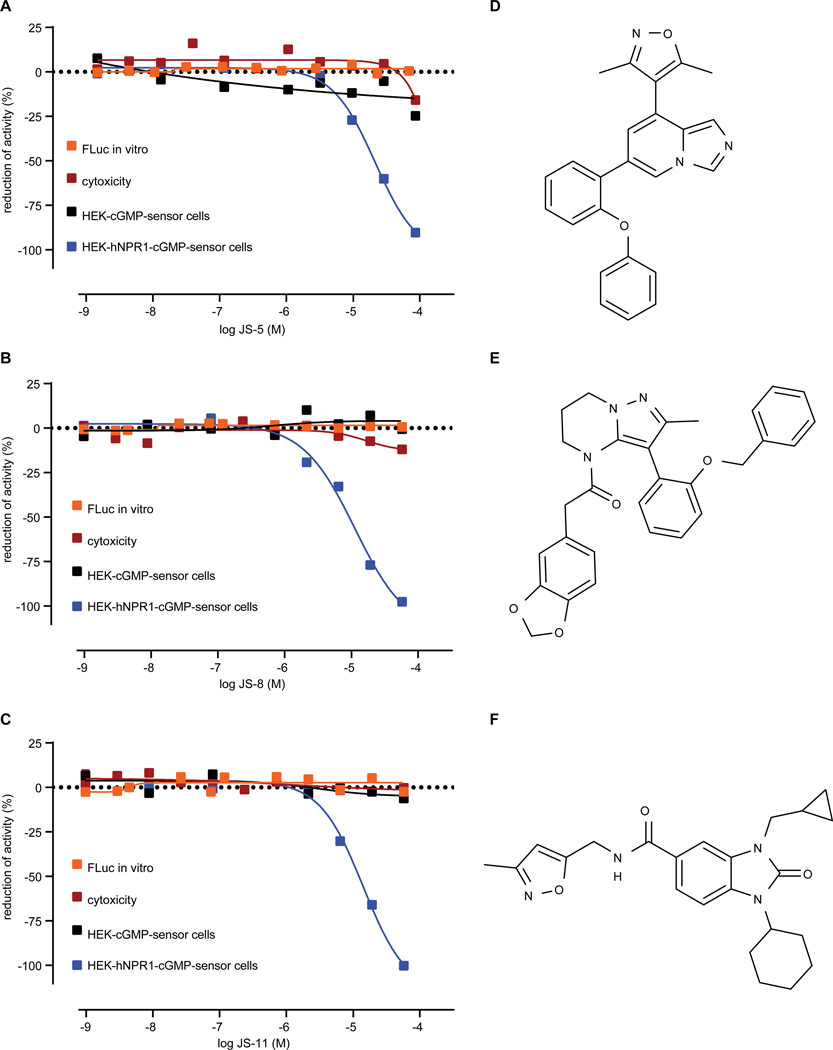
Although we identified compounds based on their inhibition, these compounds might be interfering with components of the assay instead of directly inhibiting hNPR1. Therefore, we employed overlapping strategies to eliminate false positives and identify bona-fide hNPR1 inhibitors. First, we repeated at 8 concentrations, qHTS assays on all selected compounds confirming all the selected compounds from our primary screen. Second, we subtracted compounds which interfered directly with luciferase, or molecules which were cytotoxic ((30), see methods). Third, to eliminate compounds that directly block the activity of the cGMP-sensor or sequester GloSensor reagent, we tested for the inhibition of luciferase activity upon activation of soluble GC in HEK-cGMP-sensor cells (using SNP). These counter-screens eliminated most of the initial candidate compounds (data file S3). Only 15 candidates remained after counter-screens were completed (Table S1,2). Figure 4 shows three of these candidates which potently inhibit hNPR1 activity. These compounds inhibited hNPR2 and hNPR1 with similar potency (fig. S2). The compounds JS-5, JS-8, and JS-11 show structural similarity, suggesting a possible common mode of action, and potentially a shared binding site for this class of antagonists. To examine cross-reactivity of hNPR1 antagonists further, we investigated if JS-11 could inhibit the cyclase domain of the structurally similar adenylate cyclases (ACs) family of enzymes (31). Endogenously expressed in HEK-293 cells, ACs were stimulated with forskolin and amounts of cyclic adenosine monophosphate (cAMP) were measured using a cAMP biosensor (GloSensor technology pGS-22F, (27)), using a similar approach to the one we employed to measure cGMP. Despite using a dose-range of JS-11 which completely inhibited NP-induced hNPR1 activity, ACs were not inhibited (fig. S3). In addition, a screen of the SafetyScreen44 panel of off-target G protein-coupled receptors, transmitter transporters, ion channels, nuclear receptors, and enzymes with clinical reference (32) only exposed inhibition of CCKAR and HTR2A, two receptors without clearly defined roles in itch sensation (Table S3). Therefore, JS-11 is likely a fairly selective antagonist of NPR1.
Fig. 4: Candidate inhibitors attenuate specifically hNPR1 activity.
Quantification of inhibition of hNPR1 activity (blue squares), Firefly luciferase activity (orange squares), SNP-induced activity (black squares), and cytotoxicity for HEK-hNPR1-cGMP-sensor cells (red squares) by JS-5 (A), JS-8 (B), and JS-11 (C). Data were collected from qHTS assays. (D-F) Chemical structures of JS-5 (D), JS-8 (E), and JS-11 (F).
Validation of candidate hNPR1 inhibitors
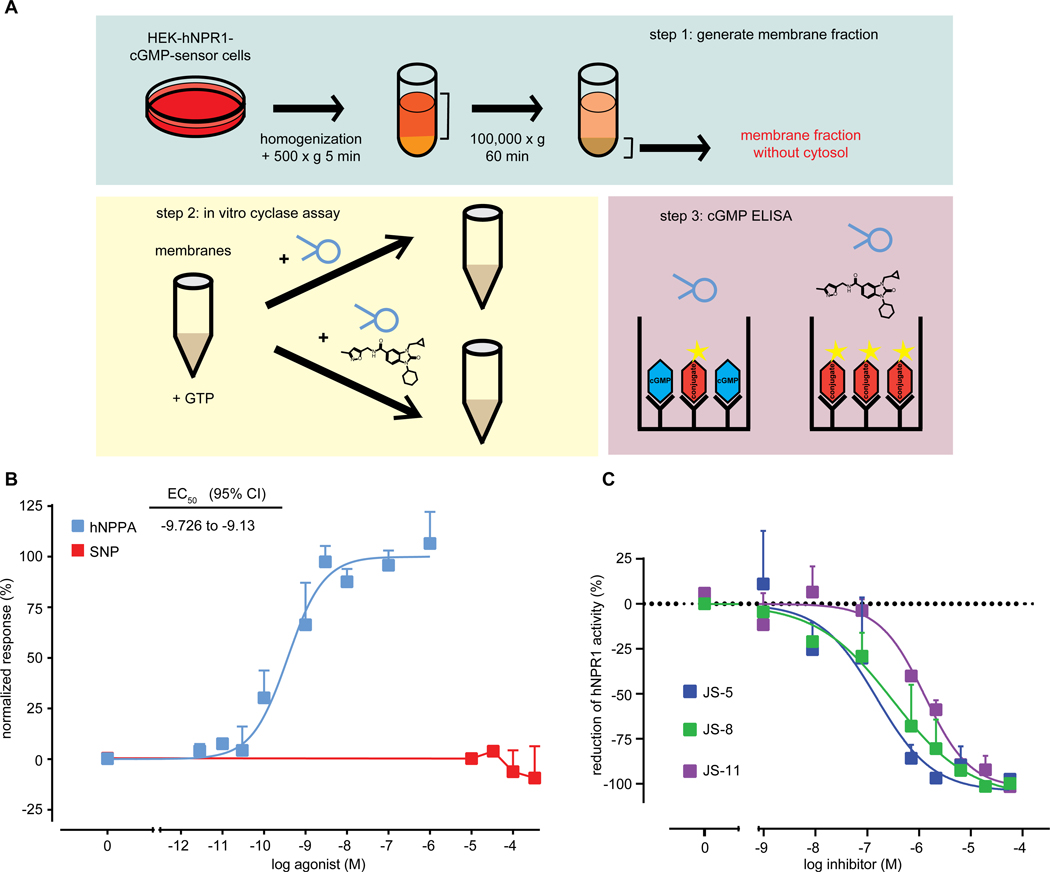
To study candidate compounds further, we selected 12 molecules from those identified initially which we could obtain in large amounts and at high purity and we developed an independent strategy to confirm their direct inhibition of hNPR1. The approach we used was to directly determine cGMP production by stimulated hNPR1 in an in vitro assay (Fig. 5A). As expected, we found that the activation of hNPR1 with hNPPA increased cGMP amounts, while activation of membranes with SNP did not (Fig. 5B). All tested hNPR1 antagonists blocked cGMP production by hNPR1 (Table S4, Fig. 5C). The recorded potencies of inhibition in the in vitro and cell-based assays, when corrected for differences in assay sensitivity, were very similar (Table S5).
Fig. 5: Cell-free membrane cyclase assay confirms that candidate compounds are specific antagonists of hNPR1.
(A) Schematic depicts our strategy to measure hNPR1 activity with an in vitro assay. A crude membrane fraction was prepared from HEK-hNPR1-cGMP-sensor cells. Incubation of hNPR1 membranes with GTP and NP results in production of cGMP and cGMP was measured using an ELISA test. (B) Quantifications of cGMP production by hNPR1 membranes, stimulated by hNPPA (blue) and SNP (red). (C) Quantification of inhibition of hNPPA-stimulated (1 nM) hNPR1 activity by JS-5 (blue), JS-8 (green), or JS-11 (purple). Data represent means SEM for triplicate (B, hNPPA), duplicate (B, SNP), and duplicate measurements (C).
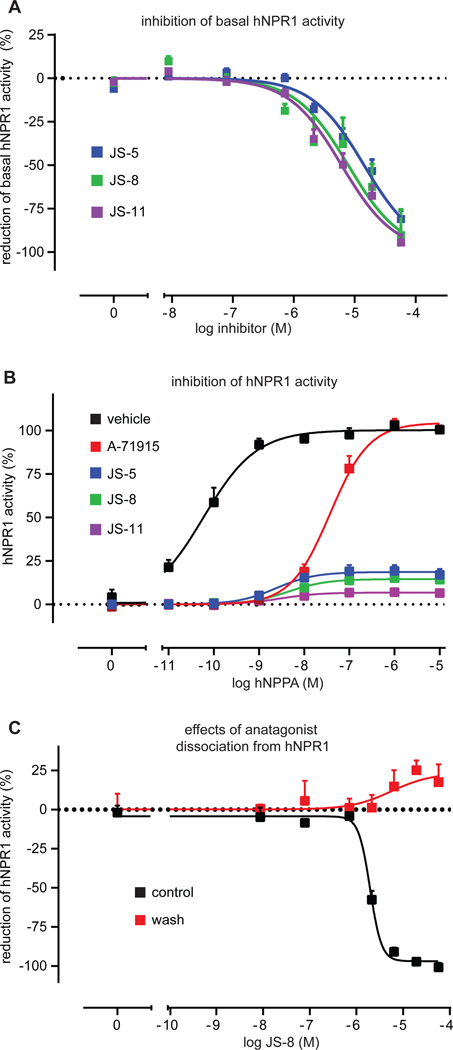
We found that both, the basal and agonist-induced hNPR1 activity were inhibited by antagonists (Fig. 6A). Since A-71915 does not inhibit basal hNPR1 activity, this suggested to us that the antagonists we identified inhibit hNPR1 activity via a different mechanism. To explore this further, we measured, with fixed concentrations of A-71915, JS-5, JS-8, and JS-11, receptor activity to increasing concentrations of hNPPA. Whereas A-71915 induced a right-shift in hNPPA potency without any effect on maximal efficacy, our antagonists reduced maximal responses even at extremely high hNPPA concentrations (105-fold higher than EC50) (Fig. 6B). This apparent lack of effect of increased agonist concentrations could either indicate a non-competitive inhibition of hNPR1, or it could be explained by slow dissociation of antagonists from hNPR1. The latter explanation would mean that washing hNPR1 membranes, after incubation with antagonist, should have little effect on receptor inhibition. To examine this possibility, we added the antagonist JS-8 and measured inhibition of hNPPA-induced hNPR1 activation before and after washing. We found that even a single 5-minute washing step was sufficient to completely recover hNPR1 activity (Fig. 6C), suggesting that the antagonists we identified have fast dissociation rates. Therefore, the antagonists we identified probably block hNPR1 via a non-competitive mechanism.
Fig. 6: hNPR1 antagonists inhibit receptor activity through a non-competitive mechanism.
(A) Quantification of the inhibition of basal hNPR1 activity by antagonists, JS-5 (blue), JS-8 (green), and JS-11 (purple). (B) Quantification of hNPR1 activity to increasing concentrations of hNPPA in the presence of a fixed concentration of antagonists (5 μM); JS-5 (blue), JS-8 (green), JS-11 (purple), A-71915 (red), and saline (black). (C) Quantification of antagonist dissociation from hNPR1. HEK-hNPR1-cGMP-sensor cells were treated with JS-8 and where either given a 5-minute washing step (red), or we not treated (black). Next, cells were stimulated with hNPPA (60 pM) to test if JS-8 dissociates rapidly from hNPR1. Data represent means SEM of triplicate (A, C) and duplicate (B) measurements.
Inhibition of itch in vivo by NPR1 antagonists
As we finally wanted to test if inhibition of NPR1 can alleviate itch in mouse models, we next examined the potencies of inhibition of compounds on mNPR1 (Table S6). Comparison of inhibition of hNPR1 and mNPR1 revealed, when corrected for differences in assay sensitivity, that most of the identified compounds had similar inhibitory properties at mouse and human receptors (Table S5), suggesting that they might block behavioral responses to itch-inducing agents in vivo in mice.
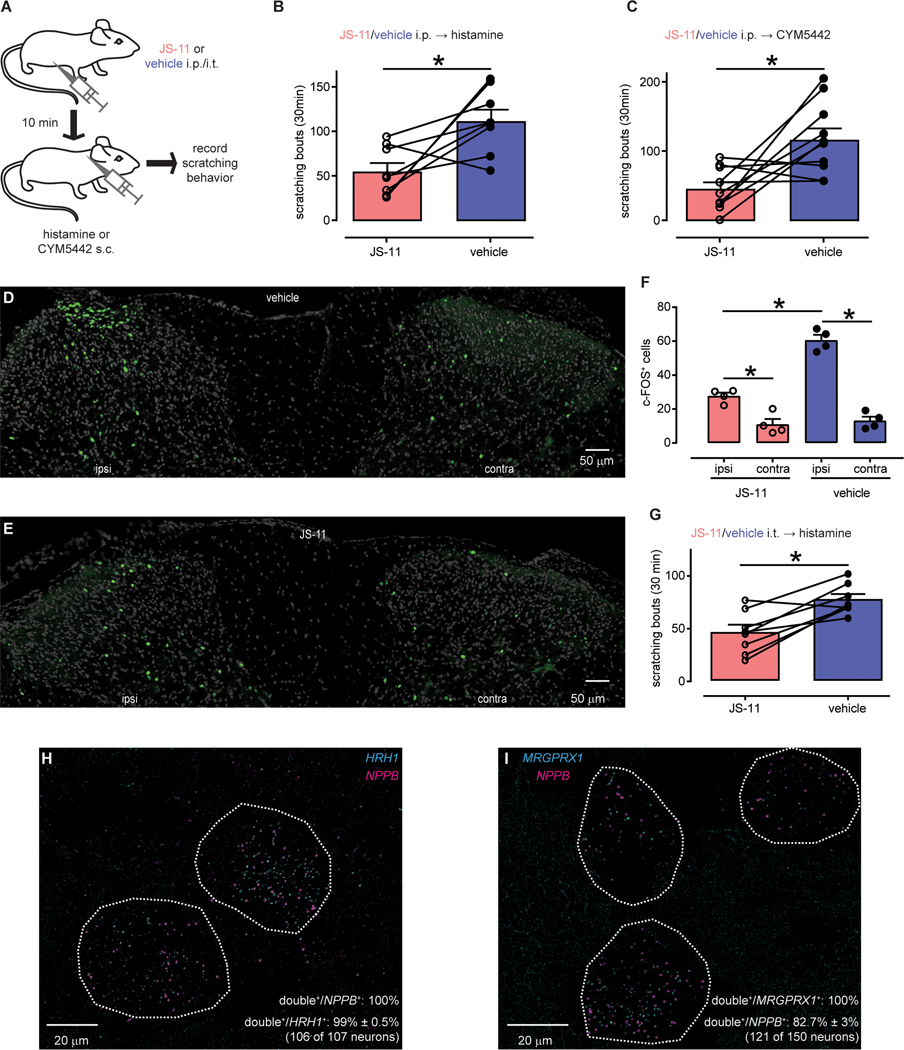
Since NPR1 signaling has been suggested to be critical for acute itch (13), we first sought to investigate the ability of NPR1 antagonists to inhibit acute itch responses. We determined that compound JS-11 might be suited to block itch in vivo because it has a relatively high water solubility and membrane permeability and a reasonable half-life (Table S6). Next, we examined the pharmacokinetic properties of JS-11 in vivo. We found that, after intraperitoneal delivery (5 mg/kg), JS-11 concentrations decayed with first-order kinetics in plasma and central nervous system (CNS) tissues with half-lifes of approximately 30 minutes and JS-11 readily crossed the blood-brain-barrier (33) (Figure S4). Amounts of JS-11 in the CNS reached concentrations that should, based on the calculated Ki, inhibit mNPR1. JS-11 did not have major untoward effects on mouse behavior when administered intraperitoneally (163 μg, ~7.5 mg/kg), did not alter spontaneous locomotor activity, and did not change rotarod performance (fig. S5A–C). Next, we tested whether scratching responses induced by intradermal injections of histamine were attenuated by JS-11 (Fig. 7A). Treatment with JS-11 reduced scratching responses to histamine by more than a half (Fig. 7B + fig. S5D). To corroborate this result, we assayed if JS-11 can inhibit itch-responses induced by a second agent that elicits scratching in mice, CYM5442, which activates the sphingosine-1-phosphate receptor 1 (26). Our results showed that JS-11 attenuates CYM5442-induced scratching (Fig. 7C + fig. S5E).
Fig. 7: NPR1 antagonist inhibits acute itch-behavior.
(A) Schematic depicting the strategy employed to test effects of JS-11 in a mouse model of acute itch. (B-C) Quantification of scratching responses to histamine (B, n = 8) and CYM5442 (C, n = 10) (B, *P = 0.0221; C, *P = 0.0128, paired t-test). Mice were intraperitoneally injected with 163 μg JS-11, or vehicle and 10 minutes later injected into the nape of the neck with pruritogens (100 μg, histamine and 8.9 μg, CYM5442). (D-E) Representative images of c-FOS immunostaining in the spinal cord following intradermal calf injection of histamine (100 μg) and prior administration of JS-11 (163 μg) or vehicle (20% DMSO). (F) Quantification of the number of c-FOS-positive neurons (average from 6 sections for each animal; n = 4 mice per treatment). Significant differences were assessed using 1-way ANOVA and Sidak’s multiple comparisons post-hoc test. JS-11 reduced the number of spinal c-FOS-positive neurons ipsilateral (ipsi) to the histamine injection (*P < 0.0001) without affecting basal activity on the contralateral (contra) side (ns P = 0.9953). Histamine significantly increased numbers of c-FOS neurons ipsilateral to the injection side in both treatment groups (JS-11: *P = 0.0058; vehicle: *P < 0.0001). (G) Quantification of the effect of intrathecal delivery of JS-11 (16.3 μg) and vehicle (20% DMSO) on numbers of scratching bouts to histamine (100 μg into the nape of the neck). Itch responses were significantly reduced by administration of JS-11 (*P = 0.0030, paired t-test, n = 8). (H-I) Representative double ISH images of human DRG sections revealed neurons stained for NPPB (magenta H and I) and HRH1 (H, cyan) and MRGPRX1 (I, cyan). Neurons positive for NPPB and itch-receptors are highlighted with white-dotted profiles.
Previously we showed that itch can be attenuated by ablation of spinal cord Npr1-neurons (13). To examine if JS-11 inhibits itch through a spinal cord pathway, we examined pruritogen-activated c-FOS expression in spinal cord neurons (34). Corroborating that JS-11 likely inhibits this pathway, JS-11 substantially reduced the number of c-FOS-positive neurons to intradermal histamine challenge (Fig. 7D–F). In addition, intrathecal delivery of JS-11 (16.3 μg, ~0.75 mg/kg) strongly reduced scratch responses of histamine-challenged mice (Fig. 7G + fig. S5F).
For NPR1 antagonists to have translatable potential, NPPB should also be required for human itch. While it is not formally possible to directly test this requirement in humans (at this stage), a precondition for NPPB to have a role in human itch neurotransmission would be that it should be expressed in the appropriate sensory neurons. We showed that NPPB is expressed in a subset of human TRPV1-sensory neurons (Fig. 1), but we wondered if it is also co-expressed with known itch receptors. Therefore, we performed double label ISH on human DRG sections with NPPB and the histamine receptor HRH1 (35) and chloroquine receptor MRGPRX1 (12, 36). We found that HRH1 is largely co-expressed with NPPB (Fig. 7H; 99 ± 0.5% of HRH1-neurons express NPPB (106 of 107 neurons, n = 4 donors); all NPPB-neurons express HRH1) and that all MRGPRX1-positve cells co-express NPPB (Fig. 7I; 121 MRGPRX1-neurons express NPPB, n = 4 donors), although some NPPB-expressing cells were MRGPRX1-negative (17.3 ± 3%; 29 neurons; n = 4 DRG donors).
A potential concern with NPR1 antagonists is the known vasodilatory effects of these receptors (37), which could potentially produce unwanted cardiovascular effects. To investigate this possible side-effect, we examined effects of acute intraperitoneal injection of JS-11 on blood pressure and heart rate. Figure S6 shows that, except for a slight change in the kinetics of a transient (< 5 minutes) drop in blood pressure directly after injection, we could not detect drug-induced changes in either blood pressure or heart rate.
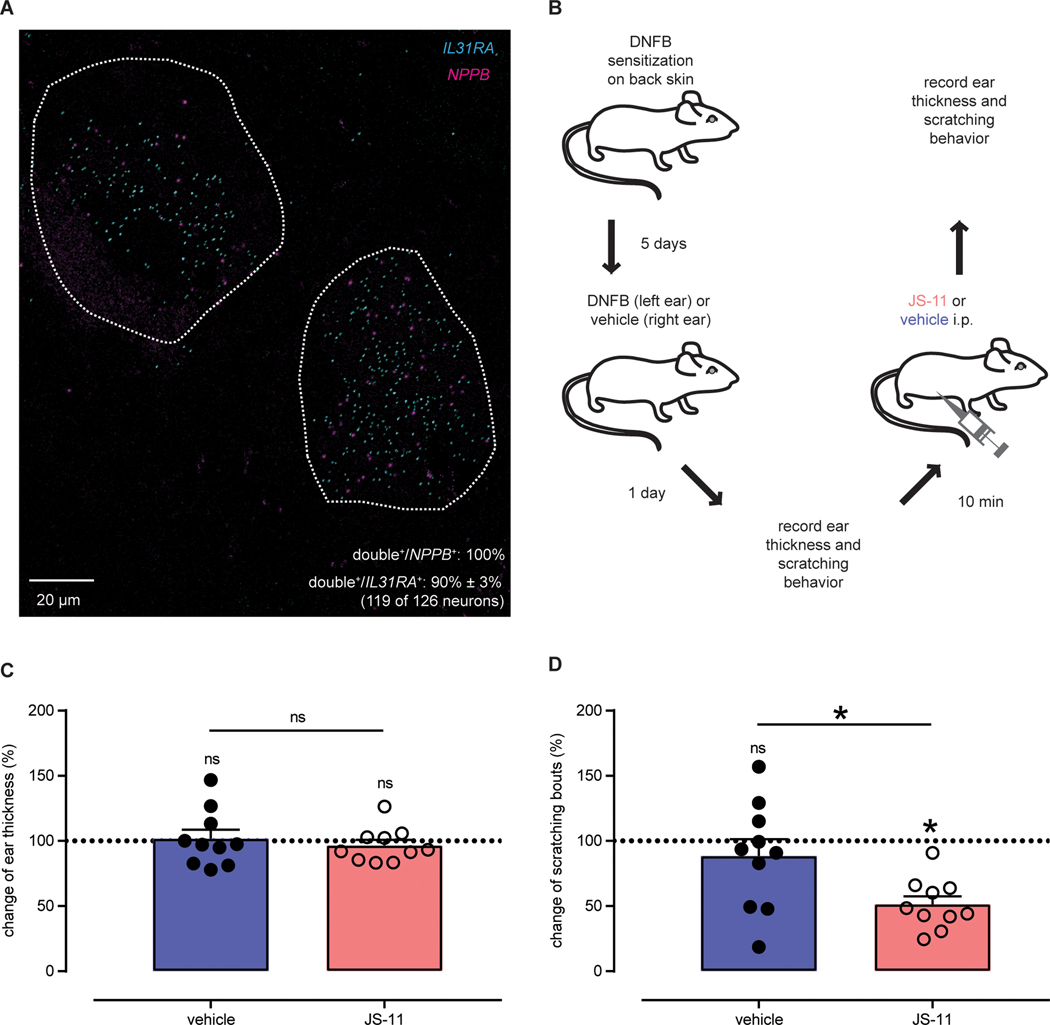
Clinically, most acute itch can be controlled with anti-histamines (1) however, anti-histamines lack effectiveness against persistent forms of itch (2, 3). In different skin disorders associated with persistent itch, including atopic and contact dermatitis, T cell-derived interleukin 31 (IL31) has been postulated to be a mediator for disease progression and correlated with disease severity in humans (38–41). Therefore, we examined the expression of NPPB with IL31RA. Like for HRH1 and MRGPRX1 itch receptors, ISH revealed that most of the human NPPB-neurons co-express IL31RA (Fig. 8A; 90 ± 3% of IL31RA-neurons express NPPB (119 of 126, n = 4 donors); all NPPB-neurons express IL31RA). To further investigate the significance of this co-expression in vivo, we tested the potential for blocking IL31-mediated itch (42) with NPR1 antagonist in a model of contact hypersensitivity (Fig. 8B). If NPPB is an important mediator of this type of itch, then we anticipated that NPR1 antagonist would attenuate scratching. In this model, scratching but not skin inflammation is dependent on IL31 (42). In line with this notion, as assessed by measuring ear thickness after hapten challenge, acute blockade of NPR1 with JS-11 had no effect on skin inflammation (Fig. 8C). By contrast, JS-11 attenuated hapten-induced scratching responses by about a half (Fig. 8D + fig. S7). This result indicates that ongoing peripheral drive contributes to pruritus in a model of persistent inflammatory dermatitis and suggests that antagonism of NPR1 might be an approach to treat chronic itch.
Fig. 8: NPR1 antagonism inhibits itching in a mouse model of contact dermatitis.
(A) Representative double ISH image of a human DRG section reveals that NPPB (magenta) and IL31RA (cyan) are co-expressed. Neurons co-expressing NPPB and IL31RA are highlighted with white-dotted profiles. (B) Schematic depicts the experimental strategy to examine the effects of JS-11 on a mouse model of contact hypersensitivity-induced itch. (C) Quantification of the effects of JS-11 treatment on contact dermatitis-induced changes in ear thickness. There were no significant differences between JS-11 (pink, 163 μg) and vehicle (blue, 20% DMSO) groups (n=10). Ear thickness was analyzed using one-sample t-test against a theoretical mean of 100% (vehicle: ns P = 0.8020 and JS-11: ns P = 0.4384) and differences between treatment groups were assessed using unpaired t-test (ns P = 0.5283). (D) Quantification of the effects of JS-11 treatment on contact dermatitis-induced changes in scratching responses. Itch-behavior was significantly reduced by administration of JS-11 (pink, 163 μg) compared to vehicle (blue, 20% DMSO) (n = 10). Scratching responses were analyzed using one-sample t-test against a theoretical mean of 100% (vehicle: ns P = 0.3951 and JS-11: *P < 0.0001) and differences between treatment groups were assessed using unpaired t-test (*P = 0.0193).
Discussion
Chronic itch is a major concern for large numbers of people, particularly because most available treatments are, at best, only partially effective (3). Scratching associated with itch is often the principle relief for patients and this leads long term to severe skin damage. In addition, these skin abrasions are disfiguring, can lead to infections, and this itch scratch cycle seriously reduces quality of life for sufferers. The underlying molecular and cellular mechanisms for itch have started to be understood in mice and other model organisms (4, 7, 43). In turn, this has led to the possibility of targeting, for the development of new treatments, key signaling molecules. For instance, small molecule antagonists for gastrin releasing peptide receptor, a neurotransmitter receptor required for itch (44), have been developed (45, 46). Here, in proof-of-principle studies, we examined whether inhibition of the signaling through another crucial neurotransmitter, NPPB, might be a viable method to reduce itch.
The initial rationale for choosing NPPB as a target for treatment of itch was based on knowledge of its critical importance for acute itch responses to multiple itch inducing compounds in mice (13). In addition, the expression of Nppb in the skin was shown to be dramatically increased in models of itch (16, 47, 48) as well as in patients diagnosed with atopic dermatitis or psoriasis (16, 49), suggestive of involvement of NPPB in persistent itch. Furthermore, in renal failure, a condition known for a high incidence of itch (61), blood NPPB concentrations were found to be elevated in human subjects and correlated with itch ratings (50). We show that NPPB is expressed in a subset of TRPV1-expressing human DRG neurons and is co-expressed with the itch receptors HR1R and MRGPRX1 (36, 51). Additionally, we find that IL31RA, a key player for the development of inflammatory skin diseases associated with chronic itch (38–41), is co-expressed with NPPB in human DRG neurons. For the most part, this expression pattern and other characteristics of human NPPB-neurons are reminiscent of those in mouse, suggesting that NPPB probably plays a similar function in humans and mice (13). Our data also point to an interesting distinction between mice and humans; while in mice Mrgpra3 and Mrgprc11, receptors that share some pharmacological similarities with the human MRGPRX1 (25), are not co-expressed with Nppb (15), MRGPRX1 and NPPB are co-expressed in humans.
Given that NPPB is a potential target for development of treatments of itch, we searched for potent NPR1 antagonists. Initially, we tested the previously identified NPR1 inhibitor A-71915 and found that it is a strong partial agonist, not a neutral antagonist of mNPR1. Since A-71915 is structurally related to NP (18), it likely competes with NP for the same binding site. This domain is the least conserved region of murine and hNPR1, potentially explaining this species-specific difference in pharmacodynamics. The fact that A-71915 is not a neutral antagonist but is instead a strong partial agonist, suggests a potential explanation for the lack of efficacy of A-71915 on itch behavior in vivo (22). Also, A-71915 may have very short half-life in humans (52–55). For these reasons, we embarked on HTS to identify novel NPR1 antagonists which can better inhibit NPR1 activity. Our HTS identified 15 new hNPR1 antagonists. Although these molecules efficiently inhibited both, human and mNPR1, they also inhibited hNPR2 and, at least for JS-11, inhibited CCKAR and HTR2A. In future studies, this information, may be helpful in identifying more specific NPR1 antagonists. Pharmacological analysis of antagonists revealed that they likely act as non-competitive inhibitors of NPR1. Similar but oppositional allosteric effects on NPR1 activity have been described before (56, 57). In particular, binding of a chloride ion to a region of the extracellular domain that is not directly involved in ligand binding favored NPPA binding to NPR1 (57) and binding of adenosine triphosphate to the intracellular kinase homology domain increased NPPA-induced NPR1 activation (56). Although the exact molecular mechanism of NPR1 inhibition by our antagonists is unknown, it might involve interference with one, or more steps of the sequential ligand-induced NPR1 activation cascade. Upon binding of NP to the orthosteric binding site, that is built asymmetrically by two NPR1 molecules, a twisting motion is thought to be induced (58). This, in turn, leads to activation of the intracellular GC domain (58). However, since structural data is lacking for the intracellular domains of NPR1 (59, 60), it is unclear which exact structural reconfigurations of the NPR1 intracellular domains are involved. We note that there are some common structural elements to the inhibitors we identified. For example, the nitrogen-containing indene cores in JS-5, −8 and −11 may embody structural features of adenosine or guanosine triphosphate sufficient for occupancy of the nucleotide-binding site(s) conserved in NPR1 and NPR2, thus explaining the non-competitive nature of NP antagonism and equivalent NPR subtype-selectivity observed. However, our findings that ACs are not inhibited by JS-11, suggest that these antagonists are not broad cyclase inhibitors (31).
In the present study, we found that blocking NPR1 is not only effective in attenuating acute itch but also reduced scratching in a model for persistent inflammatory dermatitis that is associated with chronic itch. At this point, we cannot exclude the possibility that the itch-reducing effects of JS-11 may, at least in part, be caused by inhibition of off-targets. However, this is very unlikely as NPR2, CCKAR, and HTR2A have not been associated with histamine- or CYM5442-induced itch or with spontaneous itch in persistent inflammatory dermatitis. Based on previous reports of the mechanism by which NPPB acts (13), we propose that the site of action of JS-11 is the Npr1-expressing interneurons in the spinal cord. Consistent with this, JS-11 readily crosses the blood-brain-barrier and reaches concentrations that, based on our in vitro characterization, predict mNPR1 inhibition. Also, we found that intrathecal delivery of JS-11 phenocopied the same itch-attenuating effects of systemic administration and pruritogen-induced activation of spinal dorsal horn neurons was likewise reduced by JS-11.
As well as being expressed in spinal interneurons, NPR1 is expressed in other tissues, including the kidney and vasculature (37), raising concerns about potential unwanted side effects of NPR1 inhibition. We measured cardiovascular effects of JS-11 after acute delivery as a test of preclinical safety. In this acute setting, we did not find major effects on blood pressure or heart rate. This lack of effect may be because the JS-11 dose we employed did not suppress cardiovascular effects of NPPA, the most potent agonist of NPR1, as efficiently as the pruritic effects of NPPB. Future studies are needed to systematically examine all possible potential side effects of NPR1 antagonists, especially under chronic NPR1 inhibition. However, NPR1 inhibition may still be practical in renal failure patients because one of the major sites of action of NP is the kidney. Since uremic itch affects a large percentage of kidney failure patients and there are few effective treatments, an anti-itch drug would considerably improve the quality of life for this growing patient population (61).
There are limitations of translatability to the clinic of NPR1 antagonism and employing JS-11, to alleviate itch. The investigational inhibitor JS-11 is not suitable for use in the clinic because of its relatively low affinity, issues of cross-reactivity, and insufficient physicochemical properties. Further, NPR1 inhibition may remain problematic because of unwanted on-site effects. In addition, pre-clinical tests in large animals that include a thorough mechanistic investigation of JS-11 effects need to be conducted. However, although there are additional steps and hurdles before NPR1 antagonists might be used for the treatment of itch, our data suggest that NPR1 might be a potential target for treating acute and chronic itch.
Materials and Methods
Study Design
The primary research objective was to determine if itch could be reduced by antagonism of the NPPB receptor NPR1. All other hypotheses were related to this objective. The research subjects and units of investigation were cell-culture cells, DRG and spinal cord tissue from human donors and mice in controlled laboratory experiments. Animals were randomly assigned to two groups and the experimenter was not blinded. Sample sizes for in vitro and cell-based assays were those used by other labs in the field. For animal experiments, sample sizes were based on experience and were of a size generally used in the itch field. Data from two mice (Fig. 7B) were excluded, as these mice did not complete testing because they were euthanized for reasons unconnected with the experiment.
Statistical Analysis
All data is shown as the mean ± standard error of the mean and statistical analyses were performed with Prism 7.0. To determine if samples were normally distributed D’Agostino & Pearson (N > 7) or Shapiro-Wilk (N < 7) tests were performed. Differences between mean values of two groups were analyzed using unpaired or paired two-tailed t-test or two-tailed Mann-Whitney or Wilcoxon test. Mean differences of more than two groups were analyzed with 1-way ANOVA with Sidak’s multiple comparisons post-hoc test or Friedman test with Dunn’s multiple comparisons post-hoc test. Differences in scratching time-course experiments were analyzed with 2-way ANOVA and Sidak’s multiple comparisons post-hoc test. Differences were considered significant for p < 0.05 *. Exact p-values and definition and number of replicates are given in the respective figure legend.
Supplementary Material
Fig. S1. Generation of HEK-hNPR2-cGMP-sensor cells.
Fig. S2. hNPR1 inhibitors also block hNPR2.
Fig. S3. JS-11 does not inhibit ACs.
Fig. S4. In vivo pharmacokinetics of JS-11.
Fig. S5. Effects of NPR1 antagonist on general motor behavior and itch responses.
Fig. S6. JS-11 does not cause extensive cardiovascular side effects.
Fig. S7. JS-11 inhibits itching in a mouse model of contact dermatitis.
Fig. S8. General plate map layout for qHTS.
Fig. S9. qHTS concentration response curves for JS-1 through JS-15.
Table S1. Summary of qHTS and counter-screens.
Table S2. Corroboration of hNPR1 inhibition in screening assay.
Table S3. SafteyScreen44-dependent test for off-targets of JS-11.
Table S4. Inhibition of hNPR1 in membrane cyclase assay.
Table S5. Ki values for hNPR1 and mNPR1.
Table S6. In vitro pharmacokinetics of JS-11 and JS-8
Table S7. hNPR1 antagonists also inhibit mNPR1.
Table S8. PubChem AIDs deposited and used for this study.
Table S9. Clinical information of human DRG donors.
Data file S1. LOPAC pilot screening data.
Data file S2. Genesis primary qHTS screening data.
Data file S3. qHTS follow-up screening data.
Data file S4. Raw data.
Acknowledgments
We thank C. Klumpp-Thomas for automated screening support, the NCGC compound management team for sample preparation, Drs. X. Xu, P. Shah and A. Wang for PK studies, and R. MacArthur for informatics assistance. We thank A. Noguchi (NHLBI-phenotyping core) for measuring cardiovascular studies and Dr. C. Xiao (NIDDK) for performing locomotor measurements. Human tissue was obtained from the NIH NeuroBioBank at the University of Maryland, Baltimore, MD.
Funding
This work was supported by the intramural research program of the National Institute of Dental and Craniofacial Research, National Institutes of Health, project ZIADE000721–18 (MAH) and National Center for Advancing Translational Sciences, National Institutes of Health, project 1ZIATR000053–04 (JI).
Footnotes
Competing interests
The NIH have a pending patent application (Compositions and methods for the inhibition of pruritus, No. 62/581,420) that covers part of the work described in this manuscript (H.J.S., P.D., J.I., and M.A.H.).
Data and materials availability
All the data for this study is present in the main text or in the Supplementary Materials. In addition, pharmacological screening data has been deposited with PubChem (see Table S8).
References
- 1.Dalgard F, Svensson A, Holm JO, Sundby J, Self-reported skin morbidity in Oslo. Associations with sociodemographic factors among adults in a cross-sectional study. Br J Dermatol 151, 452–457 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Kini SP, DeLong LK, Veledar E, McKenzie-Brown AM, Schaufele M, Chen SC, The impact of pruritus on quality of life: the skin equivalent of pain. Arch Dermatol 147, 1153–1156 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Yosipovitch G, Greaves MW, Schmelz M, Itch. Lancet 361, 690–694 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Bautista DM, Wilson SR, Hoon MA, Why we scratch an itch: the molecules, cells and circuits of itch. Nature neuroscience 17, 175–182 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikoma A, Steinhoff M, Stander S, Yosipovitch G, Schmelz M, The neurobiology of itch. Nat Rev Neurosci 7, 535–547 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Stander S, Weisshaar E, Mettang T, Szepietowski JC, Carstens E, Ikoma A, Bergasa NV, Gieler U, Misery L, Wallengren J, Darsow U, Streit M, Metze D, Luger TA, Greaves MW, Schmelz M, Yosipovitch G, Bernhard JD, Clinical classification of itch: a position paper of the International Forum for the Study of Itch. Acta Derm Venereol 87, 291–294 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Han L, Dong X, Itch mechanisms and circuits. Annu Rev Biophys 43, 331–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azimi E, Xia J, Lerner EA, Peripheral Mechanisms of Itch. Curr Probl Dermatol 50, 18–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azimi E, Reddy VB, Pereira PJS, Talbot S, Woolf CJ, Lerner EA, Substance P activates Mas-related G protein-coupled receptors to induce itch. J Allergy Clin Immunol 140, 447–453 e443 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson SR, The L, Batia LM, Beattie K, Katibah GE, McClain SP, Pellegrino M, Estandian DM, Bautista DM, The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell 155, 285–295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNeil B, Dong X, Peripheral mechanisms of itch. Neurosci Bull 28, 100–110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, Kim A, Ru F, Guan Y, Weng HJ, Geng Y, Undem BJ, Kollarik M, Chen ZF, Anderson DJ, Dong X, Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell 139, 1353–1365 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra SK, Hoon MA, The cells and circuitry for itch responses in mice. Science (New York, N.Y 340, 968–971 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, Li Z, McNeil B, He S, Guan Y, Xiao B, Lamotte RH, Dong X, A subpopulation of nociceptors specifically linked to itch. Nature neuroscience 16, 174–182 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Polgar E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, Zeilhofer HU, Watanabe M, Riddell JS, Todd AJ, Hoon MA, Circuit dissection of the role of somatostatin in itch and pain. Nature neuroscience 21, 707–716 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng J, Moriyama M, Feld M, Buddenkotte J, Buhl T, Szollosi A, Zhang J, Miller P, Ghetti A, Fischer M, Reeh PW, Shan C, Wang J, Steinhoff M, New mechanism underlying IL-31-induced atopic dermatitis. J Allergy Clin Immunol 141, 1677–1689 e1678 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Alexander SP, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, Peters JA, Harmar AJ, Collaborators C, The Concise Guide to PHARMACOLOGY 2013/14: catalytic receptors. British journal of pharmacology 170, 1676–1705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delporte C, Winand J, Poloczek P, Von Geldern T, Christophe J, Discovery of a potent atrial natriuretic peptide antagonist for ANPA receptors in the human neuroblastoma NB-OK-1 cell line. European journal of pharmacology 224, 183–188 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Kambayashi Y, Nakajima S, Ueda M, Inouye K, A dicarba analog of beta-atrial natriuretic peptide (beta-ANP) inhibits guanosine 3’,5’-cyclic monophosphate production induced by alpha-ANP in cultured rat vascular smooth muscle cells. FEBS letters 248, 28–34 (1989). [DOI] [PubMed] [Google Scholar]
- 20.Weber W, Fischli W, Hochuli E, Kupfer E, Weibel EK, Anantin--a peptide antagonist of the atrial natriuretic factor (ANF). I. Producing organism, fermentation, isolation and biological activity. J Antibiot (Tokyo) 44, 164–171 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Wyss DF, Lahm HW, Manneberg M, Labhardt AM, Anantin--a peptide antagonist of the atrial natriuretic factor (ANF). II. Determination of the primary sequence by NMR on the basis of proton assignments. J Antibiot (Tokyo) 44, 172–180 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Kiguchi N, Sukhtankar DD, Ding H, Tanaka K, Kishioka S, Peters CM, Ko MC, Spinal Functions of B-Type Natriuretic Peptide, Gastrin-Releasing Peptide, and Their Cognate Receptors for Regulating Itch in Mice. The Journal of pharmacology and experimental therapeutics 356, 596–603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ, A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell 106, 619–632 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Lembo PM, Grazzini E, Groblewski T, O’Donnell D, Roy MO, Zhang J, Hoffert C, Cao J, Schmidt R, Pelletier M, Labarre M, Gosselin M, Fortin Y, Banville D, Shen SH, Strom P, Payza K, Dray A, Walker P, Ahmad S, Proenkephalin A gene products activate a new family of sensory neuron--specific GPCRs. Nature neuroscience 5, 201–209 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Solinski HJ, Gudermann T, Breit A, Pharmacology and Signaling of MAS-Related G Protein-Coupled Receptors. Pharmacological reviews 66, 570–597 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Solinski HJ, Kriegbaum MC, Tseng PY, Earnest TW, Gu X, Barik A, Chesler AT, Hoon MA, Nppb-neurons are sensors of mast cell-induced itch. Cell Rep 26, 3561–3573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan F, Binkowski BF, Butler BL, Stecha PF, Lewis MK, Wood KV, Novel genetically encoded biosensors using firefly luciferase. ACS Chem Biol 3, 346–351 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP, Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proceedings of the National Academy of Sciences of the United States of America 103, 11473–11478 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michael S, Auld D, Klumpp C, Jadhav A, Zheng W, Thorne N, Austin CP, Inglese J, Simeonov A, A robotic platform for quantitative high-throughput screening. Assay Drug Dev Technol 6, 637–657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang SW, Lopez-Anido C, MacArthur R, Svaren J, Inglese J, Identification of drug modulators targeting gene-dosage disease CMT1A. ACS Chem Biol 7, 1205–1213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sunahara RK, Beuve A, Tesmer JJ, Sprang SR, Garbers DL, Gilman AG, Exchange of substrate and inhibitor specificities between adenylyl and guanylyl cyclases. The Journal of biological chemistry 273, 16332–16338 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Bowes J, Brown AJ, Hamon J, Jarolimek W, Sridhar A, Waldron G, Whitebread S, Reducing safety-related drug attrition: the use of in vitro pharmacological profiling. Nature reviews. Drug discovery 11, 909–922 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Hitchcock SA, Pennington LD, Structure-brain exposure relationships. Journal of medicinal chemistry 49, 7559–7583 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Bell AM, Gutierrez-Mecinas M, Polgar E, Todd AJ, Spinal neurons that contain gastrin-releasing peptide seldom express Fos or phosphorylate extracellular signal-regulated kinases in response to intradermal chloroquine. Mol Pain 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shim WS, Oh U, Histamine-induced itch and its relationship with pain. Mol Pain 4, 29 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sikand P, Dong X, Lamotte RH, BAM8–22 Peptide Produces Itch and Nociceptive Sensations in Humans Independent of Histamine Release. J Neurosci 31, 7563–7567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potter LR, Abbey-Hosch S, Dickey DM, Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev 27, 47–72 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Cevikbas F, Wang X, Akiyama T, Kempkes C, Savinko T, Antal A, Kukova G, Buhl T, Ikoma A, Buddenkotte J, Soumelis V, Feld M, Alenius H, Dillon SR, Carstens E, Homey B, Basbaum A, Steinhoff M, A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J Allergy Clin Immunol 133, 448–460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weidinger S, Novak N, Atopic dermatitis. Lancet 387, 1109–1122 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Neis MM, Peters B, Dreuw A, Wenzel J, Bieber T, Mauch C, Krieg T, Stanzel S, Heinrich PC, Merk HF, Bosio A, Baron JM, Hermanns HM, Enhanced expression levels of IL-31 correlate with IL-4 and IL-13 in atopic and allergic contact dermatitis. J Allergy Clin Immunol 118, 930–937 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Rabenhorst A, Hartmann K, Interleukin-31: a novel diagnostic marker of allergic diseases. Curr Allergy Asthma Rep 14, 423 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Takamori A, Nambu A, Sato K, Yamaguchi S, Matsuda K, Numata T, Sugawara T, Yoshizaki T, Arae K, Morita H, Matsumoto K, Sudo K, Okumura K, Kitaura J, Matsuda H, Nakae S, IL-31 is crucial for induction of pruritus, but not inflammation, in contact hypersensitivity. Sci Rep 8, 6639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoon MA, Molecular dissection of itch. Current opinion in neurobiology 34, 61–66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun YG, Chen ZF, A gastrin-releasing peptide receptor mediates the itch sensation in the spinal cord. Nature 448, 700–703 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Liu XY, Liu ZC, Sun YG, Ross M, Kim S, Tsai FF, Li QF, Jeffry J, Kim JY, Loh HH, Chen ZF, Unidirectional cross-activation of GRPR by MOR1D uncouples itch and analgesia induced by opioids. Cell 147, 447–458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun YG, Zhao ZQ, Meng XL, Yin J, Liu XY, Chen ZF, Cellular basis of itch sensation. Science (New York, N.Y 325, 1531–1534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ewald DA, Noda S, Oliva M, Litman T, Nakajima S, Li X, Xu H, Workman CT, Scheipers P, Svitacheva N, Labuda T, Krueger JG, Suarez-Farinas M, Kabashima K, Guttman-Yassky E, Major differences between human atopic dermatitis and murine models, as determined by using global transcriptomic profiling. J Allergy Clin Immunol 139, 562–571 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Liu B, Tai Y, Achanta S, Kaelberer MM, Caceres AI, Shao X, Fang J, Jordt SE, IL-33/ST2 signaling excites sensory neurons and mediates itch response in a mouse model of poison ivy contact allergy. Proceedings of the National Academy of Sciences of the United States of America 113, E7572–E7579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nattkemper LA, Tey HL, Valdes-Rodriguez R, Lee H, Mollanazar NK, Albornoz C, Sanders KM, Yosipovitch G, The Genetics of Chronic Itch: Gene Expression in the Skin of Patients with Atopic Dermatitis and Psoriasis with Severe Itch. J Invest Dermatol 138, 1311–1317 (2018). [DOI] [PubMed] [Google Scholar]
- 50.Shimizu Y, Sonoda A, Nogi C, Ogushi Y, Kanda R, Yamaguchi S, Nohara N, Aoki T, Yamada K, Nakata J, Io H, Kurusu A, Hamada C, Horikoshi S, Tomino Y, B-type (brain) natriuretic peptide and pruritus in hemodialysis patients. Int J Nephrol Renovasc Dis 7, 329–335 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies MG, Greaves MW, Sensory responses of human skin to synthetic histamine analogues and histamine. British journal of clinical pharmacology 9, 461–465 (1980). [PMC free article] [PubMed] [Google Scholar]
- 52.Hunt PJ, Richards AM, Espiner EA, Nicholls MG, Yandle TG, Bioactivity and metabolism of C-type natriuretic peptide in normal man. J Clin Endocrinol Metab 78, 1428–1435 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Mukoyama M, Nakao K, Hosoda K, Suga S, Saito Y, Ogawa Y, Shirakami G, Jougasaki M, Obata K, Yasue H, et al. , Brain natriuretic peptide as a novel cardiac hormone in humans. Evidence for an exquisite dual natriuretic peptide system, atrial natriuretic peptide and brain natriuretic peptide. The Journal of clinical investigation 87, 1402–1412 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakao K, Sugawara A, Morii N, Sakamoto M, Yamada T, Itoh H, Shiono S, Saito Y, Nishimura K, Ban T, et al. , The pharmacokinetics of alpha-human atrial natriuretic polypeptide in healthy subjects. Eur J Clin Pharmacol 31, 101–103 (1986). [DOI] [PubMed] [Google Scholar]
- 55.Yandle TG, Richards AM, Nicholls MG, Cuneo R, Espiner EA, Livesey JH, Metabolic clearance rate and plasma half life of alpha-human atrial natriuretic peptide in man. Life sciences 38, 1827–1833 (1986). [DOI] [PubMed] [Google Scholar]
- 56.Antos LK, Abbey-Hosch SE, Flora DR, Potter LR, ATP-independent activation of natriuretic peptide receptors. The Journal of biological chemistry 280, 26928–26932 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Misono KS, Atrial natriuretic factor binding to its receptor is dependent on chloride concentration: A possible feedback-control mechanism in renal salt regulation. Circ Res 86, 1135–1139 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Ogawa H, Qiu Y, Ogata CM, Misono KS, Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction. The Journal of biological chemistry 279, 28625–28631 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Ogawa H, Qiu Y, Philo JS, Arakawa T, Ogata CM, Misono KS, Reversibly bound chloride in the atrial natriuretic peptide receptor hormone-binding domain: possible allosteric regulation and a conserved structural motif for the chloride-binding site. Protein Sci 19, 544–557 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van den Akker F, Zhang X, Miyagi M, Huo X, Misono KS, Yee VC, Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor. Nature 406, 101–104 (2000). [DOI] [PubMed] [Google Scholar]
- 61.Combs SA, Teixeira JP, Germain MJ, Pruritus in Kidney Disease. Semin Nephrol 35, 383–391 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, Caponigro G, Hieronymus H, Murray RR, Salehi-Ashtiani K, Hill DE, Vidal M, Zhao JJ, Yang X, Alkan O, Kim S, Harris JL, Wilson CJ, Myer VE, Finan PM, Root DE, Roberts TM, Golub T, Flaherty KT, Dummer R, Weber BL, Sellers WR, Schlegel R, Wargo JA, Hahn WC, Garraway LA, COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mishra SK, Holzman S, Hoon MA, A nociceptive signaling role for neuromedin B. J Neurosci 32, 8686–8695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Southall NT, Jadhav A, Huang R, Nguyen T, Wang Y, Enabling the Large-Scale Analysis of Quantitative High-Throughput Screening Data Handbook of Drug Screening Second Edition, 442–464 (2009). [Google Scholar]
- 65.Yasgar A, Shinn P, Jadhav A, Auld D, Michael S, Zheng W, Austin CP, Inglese J, Simeonov A, Compound Management for Quantitative High-Throughput Screening. JALA Charlottesv Va 13, 79–89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thorne N, Shen M, Lea WA, Simeonov A, Lovell S, Auld DS, Inglese J, Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem Biol 19, 1060–1072 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avdeef A, Strafford M, Block E, Balogh MP, Chambliss W, Khan I, Drug absorption in vitro model: filter-immobilized artificial membranes. 2. Studies of the permeability properties of lactones in Piper methysticum Forst. Eur J Pharm Sci 14, 271–280 (2001). [DOI] [PubMed] [Google Scholar]
- 68.Shah P, Kerns E, Nguyen DT, Obach RS, Wang AQ, Zakharov A, McKew J, Simeonov A, Hop CE, Xu X, An Automated High-Throughput Metabolic Stability Assay Using an Integrated High-Resolution Accurate Mass Method and Automated Data Analysis Software. Drug Metab Dispos 44, 1653–1661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra SK, Tisel SM, Orestes P, Bhangoo SK, Hoon MA, TRPV1-lineage neurons are required for thermal sensation. EMBO J 30, 582–593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones BJ, Roberts DJ, The quantiative measurement of motor inco-ordination in naive mice using an acelerating rotarod. J Pharm Pharmacol 20, 302–304 (1968). [DOI] [PubMed] [Google Scholar]
- 71.Kim SM, Eisner C, Faulhaber-Walter R, Mizel D, Wall SM, Briggs JP, Schnermann J, Salt sensitivity of blood pressure in NKCC1-deficient mice. Am J Physiol Renal Physiol 295, F1230–1238 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawashima Y, Kameo K, Kato M, Hasegawa M, Tomisawa K, Hatayama K, Hirono S, Moriguchi I, Structure-activity studies of 3-benzoylpropionic acid derivatives suppressing adjuvant arthritis. Chem Pharm Bull (Tokyo) 40, 774–777 (1992). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Generation of HEK-hNPR2-cGMP-sensor cells.
Fig. S2. hNPR1 inhibitors also block hNPR2.
Fig. S3. JS-11 does not inhibit ACs.
Fig. S4. In vivo pharmacokinetics of JS-11.
Fig. S5. Effects of NPR1 antagonist on general motor behavior and itch responses.
Fig. S6. JS-11 does not cause extensive cardiovascular side effects.
Fig. S7. JS-11 inhibits itching in a mouse model of contact dermatitis.
Fig. S8. General plate map layout for qHTS.
Fig. S9. qHTS concentration response curves for JS-1 through JS-15.
Table S1. Summary of qHTS and counter-screens.
Table S2. Corroboration of hNPR1 inhibition in screening assay.
Table S3. SafteyScreen44-dependent test for off-targets of JS-11.
Table S4. Inhibition of hNPR1 in membrane cyclase assay.
Table S5. Ki values for hNPR1 and mNPR1.
Table S6. In vitro pharmacokinetics of JS-11 and JS-8
Table S7. hNPR1 antagonists also inhibit mNPR1.
Table S8. PubChem AIDs deposited and used for this study.
Table S9. Clinical information of human DRG donors.
Data file S1. LOPAC pilot screening data.
Data file S2. Genesis primary qHTS screening data.
Data file S3. qHTS follow-up screening data.
Data file S4. Raw data.