Abstract
The first outbreak of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occurred in Wuhan, Hubei Province, China, in late 2019. The subsequent COVID-19 pandemic rapidly affected the health and economy of the world. The global approach to the pandemic was to isolate populations to reduce the spread of this deadly virus while vaccines began to be developed. In March 2020, the first phase I clinical trial of a novel lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine, mRNA-1273, which encodes the spike protein (S protein) of SARS-CoV-2, began in the United States (US). The production of mRNA-based vaccines is a promising recent development in the production of vaccines. However, there remain significant challenges in the development and testing of vaccines as rapidly as possible to control COVID-19, which requires international collaboration. This review aims to describe the background to the rationale for the development of mRNA-based SARS-CoV-2 vaccines and the current status of the mRNA-1273 vaccine.
MeSH Keywords: Clinical Trial; COVID-19; Pandemics; RNA, Messenger; Spike Glycoprotein, Coronavirus; Vaccines
Background
In late 2019, in Wuhan, Hubei Province, China, an outbreak occurred of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This outbreak was followed by the release of the first situation report from the World Health Organization (WHO) on January 20, 2020, and since then, a global pandemic has resulted in an increasing number of deaths [1–3]. The pandemic has resulted in uncertainty regarding when or whether the pandemic will end, and whether COVID-19 will follow the same clinical course as severe acute respiratory syndrome (SARS) did 17 years ago. However, the current view is that this novel coronavirus infection will persist, and without a treatment, a vaccine is urgently required [4].
Review of the epidemiology of the SARS epidemic of 2003 shows that it disappeared suddenly without resulting in a pandemic, and this was also the reason for the lack of development of a SARS vaccine [5]. However, COVID-19 is different from SARS based on its ability to spread globally. Currently, antiviral pharmaceuticals and immunotherapies are two principal therapeutic approaches for COVID-19 [6], whereas vaccines are still considered the most promising way to eradicate this virus [7]. In general, DNA-based and protein-based vaccines have been the major approaches for the development of stable and effective vaccines with less innate immunogenicity [8,9]. However, mRNA-based vaccines have become a potentially preventive approach, as they are safer, more efficient, and easier to manufacture [10]. Due to the rapid spread of COVID-19, mRNA-based vaccine development seems to be an approach to prevent infection and to prevent a second wave of this pandemic.
Typically, at least 12 to 18 months is required to develop a new vaccine, using current processes for vaccine development, clinical trials, and regulatory approval [11]. Therefore, it is unlikely that an effective vaccine will be developed in time to control the current pandemic. However, on March 16 2020, the first phase I clinical trial of a novel lipid nanoparticle (LNP)-encapsulated mRNA-based vaccine, mRNA-1273, which encodes the spike protein (S protein) of SARS-CoV-2, began in the United States (US), conducted by Moderna and the Vaccine Research Center (VRC) of the National Institute of Allergy and Infectious Diseases (NIAID) [12,13]. The first patient enrolled in this phase 1 study was a 43-year-old woman at the Kaiser Permanente Washington Health Research Institute in Seattle, Washington, USA [12]. The phase I study of mRNA-1273 is being conducted to evaluate the safety and immunogenicity, with the following trials currently being planned to evaluate the efficacy and effective dose of the new mRNA vaccine [13]. This review aims to describe the background to the rationale for the development of mRNA-based SARS-CoV-2 vaccines and the current status of the mRNA-1273 vaccine.
Biochemical and Molecular Roadmap of SARS-CoV-2
The terminology for the RNA virus that causes COVID-19, SARS-CoV-2, has been established by the International Committee on Taxonomy of Viruses (ICTV) [14], due to its extensive homology with the 2003 SARS coronavirus. The SARS-CoV-2 coronavirus belongs to the subfamily of Coronavirinae, with a genomic structure of (+)ss-RNA of 30 kb in length that includes a 5′-cap structure and 3′-poly-A tail [15]. From the viral RNA, polyprotein 1a/1ab (pp1a/pp1ab) is synthesized in the host to form 16 non-structural proteins (nsps) that organize the replication-transcription complex (RTC) in double-membrane vesicles (DMVs). The nRTC synthesizes a set of minus-strand subgenomic RNAs (sgRNAs) discontinuously [16]. Between open reading frames (ORFs), transcription terminates, and then a subsequent acquisition of a leader RNA occurs. During this process, subgenomic mRNAs need these sgRNAs as the templates [14,17]. At least six ORFs exist for a typical CoV, including SARS-CoV-2.
The first ORFs (ORF1a/b) with over 65% of the whole genome length encode 16 nsps. Of note, two polypeptides (pp1a and pp1ab) come from a −1 frameshift between ORF1a and ORF1b. For the other ORFs on the 35% of the genome close to the 3′-terminus encode at least four main structural proteins, including spike (S), membrane (M), envelope (E), and nucleocapsid (N). All these structural and non-structural proteins are translated from the sgRNAs [14,16,17]. Currently, more than 200 complete and partial genome sequences of SARS-CoV-2 have been decoded and deposited in the Global Initiative on Sharing All Influenza Data (GISAID) database (https://www.gisaid.org/) and in the National Institutes of Health (NIH) GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/?term=covid-19).
Phylogenetic analysis showed that SARS-CoV-2 was closely related to two SARS-like coronaviruses present in bats, including bat-SL-CoVZC45 and bat-SL-CoVZXC21, with 88% identity, and showed 79% homology with SARS-CoV, and 50% with MERS-CoV [18]. However, homology modeling disclosed that SARS-CoV-2 had a similar RBD structure to that of SARS-CoV, despite amino acid variation at some key residues [19,20]. These findings suggest that SARS-CoV-2 emerged from a single animal source within a short period [21]. However, because the sequence similarity between SARS-CoV-2 and its close relatives bat-SL-CoVZC45 and bat-SL-CoVZXC21 is less than 90%, the bat-derived viruses may not be the direct origins of SARS-CoV-2.
Engineering mRNA-Based SARS-CoV-2 Vaccines
Vaccines based on the cytoplasmic expression of chimeric mRNAs containing curated ORF viral sequences possess great potential to translate directly in the cytoplasm and block chromosomal integration [22]. Once injected, the delivered mRNA can be processed by immune cells and start to produce targeted protein directly through translation, of which is being followed by activating other immune cells to recognize the newly produced viral protein to make antibodies.
Two types of RNA vaccine exist against infectious pathogens, or non-replicating mRNA vaccine and self-amplifying or replicon RNA vaccine [10,23]. Due to different delivery methods, non-replicating mRNA vaccines are further divided as ex vivo loading of dendritic cell and direct in vivo injection into various anatomical sites [10]. The penetration of the lipid membrane barrier is the first step for exogenous mRNA to reach the cytoplasm before the translation of functional protein happen [24]. Also, the uptake mechanisms of mRNA vaccines show cell specificity [25], and the physicochemical properties of the mRNA may significantly influence its cellular delivery and organ distribution [26]. All these factors must be considered when designing an effective mRNA-based vaccine. Even so, an mRNA vaccine is still considered the most promising candidate because it can be scaled rapidly, which can save time when the rapidly spreading COVID-19 emerged and started to infect millions of people worldwide [7,27].
As a (+)ss-RNA virus, SARS-CoV-2 possesses self-amplifying RNA that can realize extreme RNA replication in the cytosol [28]. This finding supports the role of mRNA-based vaccine development. However, the safety and efficacy of mRNA vaccines for use in humans remain unknown. The hypothetical benefits of mRNA vaccine seem strong, whereas limitations such as the delivery and stability issues related to RNA degradation, and the safety concerns due to immunogenicity hinder its development [29]. The results from the phase I trial of the mRNA-1273 vaccine are awaited [13].
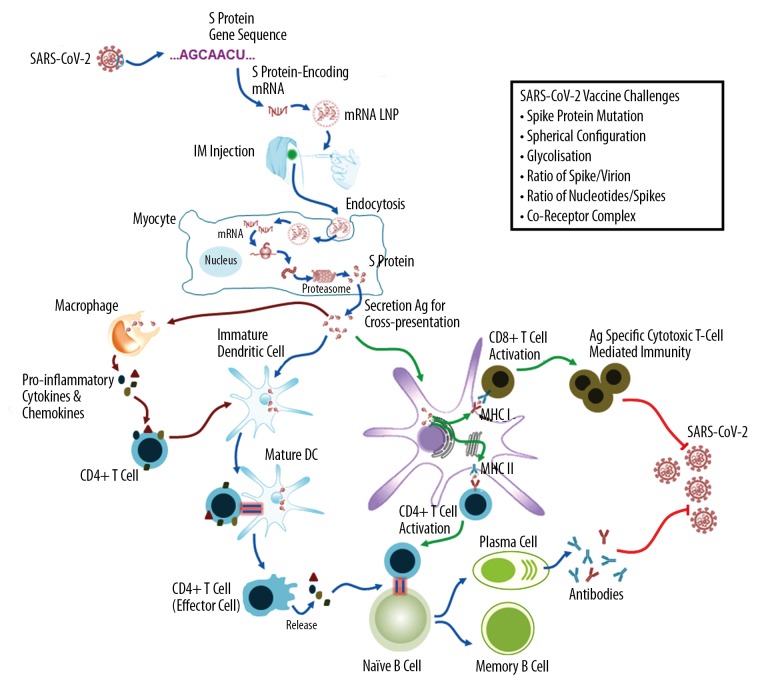
The mRNA-based vaccines actively induce activation of both B cell responses and T cell cytotoxicity. First, the mRNA vaccines use the mRNA sequence of the target protein that recombine in vitro, rather than the sequence of the target antibody. Subsequently, the recombinant target protein mRNA sequence is carried by LNPs and enters the somatic cytoplasm to achieve direct translation and encoding the target protein. When the target protein is released from the host cell, the antigen-presenting cells will quickly capture and process the heterologous protein. Then, the presentation of MHC I and MHC II on the surface of the antigen-presenting cell membrane [30]. This step is important for the subsequent activation of B cells and T cells and is also the key to the humoral and cytotoxic response. Figure 1 is a schematic representation of mRNA-based active immunization.
Figure 1.
Schematic diagram of the mRNA-based vaccine targeted to the spike protein (S protein) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The mRNA-based vaccine targeted to the S protein of SARS-CoV-2 works by active immunization. This technique will not use part of the virus but only recombine mRNA of the S protein in vitro according to the gene sequence, which is coated with lipid nanoparticles for effective delivery. Once injected into the muscle, the myocytes take up the lipid nanoparticle (LNPs) and then release the mRNAs into the cytoplasm for translation into the S proteins. These endogenously synthesized S proteins will be secreted to activate both humoral and cellular immune responses. S protein – spike protein; IM – intramuscular, LNP – lipid nanoparticle; DC – dendritic cell; MHC – major histocompatibility complex; Ag – antigen.
Targeting the SARS-CoV-2 S Protein Sequence in mRNA Vaccine Development
Finding the most suitable target site for SARS-CoV-2 vaccine development is extremely important. The spike glycoprotein (S protein) is now a key target for vaccine development, therapeutic antibody generation, and the clinical diagnosis of COVID-19.
SARS-CoV-2 enters the host cell by using highly glycosylated homotrimeric S protein to achieve fusion with cell membranes through its structural changes. This process includes: the S1 subunit binds to the host cell receptor, which triggers trimeric instability that is followed by the separation of the S1 subunit from the S2 subunit to form a highly stable fusion structure [19–21]. To access host cell receptors, RBD in the S1 subunit undergoes hinge-like conformational changes to hide or expose key sites for receptor binding, which is very similar to SARS-CoV [19–21]. This high homology of RBD suggests that the COVID-19 virus shares the same host cell receptor ACE2 as SARS-CoV [19–21].
Although there are similarities, COVID-19 has its own characteristics. The most significant change is the RRAR amino acid sequence with a S1/S2 protease cleavage site, which is consistent with the characteristics of a Furin recognition site. This common phenomenon occurs more frequently in influenza viruses rather than in SARS viruses that only have a single arginine [31]. Also, SARS-CoV-2 and RaTG13 S proteins have 29 amino acid residues that differ, 17 of which are located at the RBD site. The RBD of SARS-CoV-2 is much closer to the center of the trimeric S protein. One of the three RBDs in the S protein will spiral upwards to form a spatial conformation that helps the virus bind to the host receptor ACE2 easily, which suggests that SARS-CoV-2 would be more infectious than SARS [32]. A cross-reactivity test of RBD of SARS-CoV-2 was performed using the RBD monoclonal antibody of SARS, and it was found that this antibody did not cross-react with SARS-CoV-2 [33]. These results provide an important structural biological basis for designing vaccines more accurately and discovering antiviral drugs.
S protein helps the virus to enter target cells, but this endocytosis simultaneously depends on both the binding of S protein to membrane ACE2 receptors and the initiative activation of S protein by cellular proteases [34]. Therefore, a vaccine against S protein provides an approach for preventing the proliferation and spread of SARS-CoV-2. The vaccine can prevent the initial activation of the S protein by blocking the S protein binding to ACE2. SARS-CoV-2 infects cells in a transmembrane serine protease 2 (TMPRSS2) protein-coding gene-independent manner, and cathepsins B and L directly activate S protein in cells that do not express TMPRSS2, and SARS-CoV-2 infects cells in a TMPRSS2-dependent manner. Recently, Hoffmann et al. showed that the use of TMPRSS2 inhibitors significantly blocked the entry of SARS-CoV-2 into cell lines expressing TMPRSS2 and that promoting TMPRSS2 expression antagonized this inhibition [35]. Therefore, if the antibody titer of S protein is high enough to prevent the virus being engulfed into endosomes or undergoing fusion at the host cell surface, then virus infection associated with S protein activation and the intracellular release of the virus particles will be effectively inhibited.
Given the bioavailability of ACE2 as a key determinant of the spread and infectious capacity of SARS-CoV-2, if enough S protein antibodies are formed in the body, then two therapeutic effects may occur. Firstly, the S protein-antibody complex will be quickly cleared by the immune system, which will subsequently result in clearance of the virus itself. Secondly, ACE2 bioavailability will be significantly reduced, which helps mitigate the spread and infectivity of the virus [36]. Although empirical data from both SARS-CoV and SARS-CoV-2 research have identified ACE2 as the primary portal for cellular entry of these two viruses [19–21], the probability of a membrane-associated co-receptor complex remains. Therefore, targeting of ACE2/SARS-CoV-2 binding may not be the rate-limiting step for inhibiting viral infection.
Even with hopes of an mRNA-based vaccine targeting the S protein, it is important to note that the stoichiometric relationship between S protein and the immune response occurs in several ways. First, the intrinsic ratio of outer surface S protein per SARS-CoV-2 virion (s/v) may predetermine the preference of induced IgG in immunological responses. The inherent ratio of nucleotides to the S protein (n/s) may determine whether there are redundant S proteins when IgG effectively neutralizes the S protein, which reduces the immune efficacy of IgG. Therefore, both ratios will finally determine how high the antibody titer should be and how many vaccine boosters are needed so that it can effectively neutralize the virus without concerns about the existence of redundant S protein complexes that may effectively remove IgG antibodies to leave unblocked S protein available for processing and binding to ACE2. Although several previous studies have investigated this finding in human immunodeficiency viruse (HIV) infection and concluded that low spike density with wide spacing of the envelope spikes is unhelpful for the activation of B cells [37,38], this remains unclear for SARS-CoV-2, and it is a challenging factor for vaccine development against SARS-CoV-2.
An important consideration relates to mutations in the S protein that may affect the degree of viral pathogenesis [39]. Direct evidence of functionally meaningful S protein mutations affects the S protein SARS-CoV-2 and the host cell ACE2, which appears to mediate a higher binding affinity when compared with SARS-CoV [40]. The reason for this increased S protein/ACE2 affinity in SARS-CoV-2 (−15.7 Kcal/mol) versus that of SARS-CoV (−14.1 Kcal/mol) is mainly attributed to three main factors [40]. First, the emergence of two loops around the RBDs in SARS-CoV-2 may promote its chemical interaction with ACE2 by increasing the number of atoms needed. Second, more amino acid residues in the S protein of SARS-CoV-2 determine a higher number of protein-protein contacts between S protein and ACE2. Third, a longer capping loop in the S protein of SARS-CoV-2 favors its interaction with the receptor. Therefore, it is difficult to ensure that the new vaccine that is targeted to S protein can be used long term or in the near future, it there is the possibility that the vaccine will not be effective because of mutations in the S protein.
Potential Advantages and Limitations of mRNA-based SARS-CoV-2 Vaccines
For protein-based immunization, repeated vaccine administrations due to the clearance and indication may be required to maintain a therapeutically effective level [41]. Furthermore, some non-specific effects may result from these typical vaccines, such as cross-reactive TCRs and antibodies, bystander activation of pre-existing effector or memory cells, and classical cell-mediated immunity may function as either beneficial or unfavorable to the public health of the population [42]. Also, although there have been previous studies on the safety of live attenuated or killed vaccines, and the safety of these vaccines used in clinics is very reliable, the potential for inducing infection still exists [43].
There are four main safety and efficacy advantages of the use of mRNA-based antiviral vaccines over traditional approaches. First, mRNA-based antiviral vaccines minimize the potential risk of infection and insertion-induced mutagenesis due to natural degradation of mRNA in the cellular microenvironment [44]. Second, the immunogen’s high efficacy due to engineered mRNA structural modifications improves its stability and translation efficacy. Third, the high potency of mRNA-based vaccines capable of generating potent antiviral neutralizing immunoglobulins with only one or two low-dose immunizations [23,45] may induce strong immune responses by activating both CD8+ and CD4+ T cells [46]. Fourth, engineered mRNA production facilitates large-scale production of sufficient vaccine doses required to treat mass populations [29,47]. All these factors make the mRNA vaccine more suitable for a rapid response to the emerging COVID-19 pandemic.
Although these beneficial features of mRNA vaccines provide some hope for the development of the first clinically applicable SARS-CoV-2 mRNA vaccine, recent reports regarding rare cases of moderate or severe reactions for different mRNA vaccines have raised concerns about safety and immunogenicity, including in the primary outcome findings of the phase I trial on mRNA-1273 [20,48]. Therefore, it is important to clearly understand the potential risks of this type of mRNA-based vaccine, which include local and systemic inflammatory responses, the biodistribution and persistence of the induced immunogen expression, possible development of autoreactive antibodies and toxic effects of any non-native nucleotides and delivery system components [48–50].
Glycosylation of viral envelopes is a very common biological phenomenon. Glycosylation refers to the process in which proteins or lipids are linked to sugar chains by the action of enzymes. Glycosylation is one of the important forms of post-translational modification of proteins and is an essential way to regulate protein localization, function, persistence, and diversity [51,52]. For viruses, their replication and invasion into host cells are closely associated with the glycosylation of their structural proteins [53]. The main purpose of viral vaccine development is to evoke an immune response to fight the virus. However, the high glycosylation of proteins on the surface of the viral envelope works like invisible camouflage, which can help the virus to successfully escape the detection of the human immune system and achieve the purpose of survival [54,55]. Therefore, the higher the degree of glycosylation, the greater the probability that the virus will escape the immune response and the lower the success rate of vaccine development.
Therefore, overcoming the highly glycosylated viral envelope is another challenge of SARS-CoV-2 vaccine development. SARS-CoV-2 includes highly glycosylated spherical particles and has a large structure containing at least 66 N-linked glycan sites, of which 54 sites show similarity with SARS-CoV [56]. In contrast, the glycan sites of HIV are between three times and six times that of the influenza virus, which is one of the major reasons why an HIV vaccine has not been successfully developed at this time [57]. However, SARS-CoV-2 has more than twice the glycan sites of HIV [58]. This unusual degree of glycosylation means that SARS-CoV-2 may mutate quickly, making the development of a vaccine extremely difficult. However, vaccine development failures caused by glycosylation remains significant, because vaccine design is based on the traditional inactivated or attenuated live vaccines, and different traditional vaccines may induce various glycosylation patterns of antibodies that subsequently affect the protective role of the antibodies [59]. However, the use of mRNA-based vaccine technology and targeting only the S protein, rather than the entire virus particle, may lead to the human immune system producing S protein antibodies without the influence of viral glycosylation.
Perspectives
The successful delivery of oligonucleotide-based therapies [60] has provided new opportunities to develop mRNA-based vaccines. Given their beneficial properties, which include lack of persistence, lack of integration into the genome, the absence of induction of autoantibodies, the ability to produce mRNA vaccines in large quantities, and their high purity [61], an mRNA-based vaccine has become a hopeful choice for the development of a SARS-CoV-2 vaccine. Currently, no clinically applicable vaccine against COVID-19 is available, and even one of the first vaccines, mRNA-1273, is undergoing a phase I safety and immunogenicity trial [13]. The recently developed encapsulated RNA delivery and self-amplifying RNA technique assists in the ease of use of the mRNA vaccine with an increased success rate. However, several issues still need to be clarified for mRNA-based vaccines. Concerning the worldwide spread of COVID-19, the development of an effective and safe vaccine in time has become the most promising way to control the current viral pandemic. If currently available technology can be used to develop a SARS-CoV-2 vaccine, which may be clinically applicable, this will give hope for controlling the COVID-19 pandemic.
However, further research and development should continue to ensure that an effective SARS-CoV-2 vaccine is developed. There are several challenges to address as new formulations are required to stabilize the mRNA vaccine at higher temperatures during its distribution. A more suitable packaging formula for the mRNA vaccine within the nanoparticles is required to ensure vaccine stability. The best way should be sought to anchor the RNase inhibitor into the mRNA vaccine co-formulation and to develop new ways to purify or remove the mRNA vaccine-related reaction components. Studies are still required to investigate the mechanisms for reducing the innate immune response to the delivered mRNA vaccine. Also, novel in vivo delivery methods requires development to maximize the uptake efficiency of the mRNA vaccine, and to determine which immune signaling pathways are the most effective in response to the exogenous mRNA vaccine.
In addition to the described technical difficulties related to mRNA vaccines, the difficulties associated with vaccine development need to be resolved. Even though clinical trials of some vaccines have begun [13], there remain several technical issues that affect vaccine development and efficacy that require further study. First, strict bioinformatics analysis of the probability of a membrane-associated co-receptor complex is required to find the rate-limiting pattern of antibodies that affect ACE2/SARS-CoV-2 binding. The stoichiometric relationship between the S protein and the strength of the immune response requires studies on the intrinsic ratio of the outer surface S protein per SARS-CoV-2 virion (s/v) and nucleotides to the S protein (n/s). The precise function of viral self-defense proteins and the spherical configuration of the virion require further research, as does the mutation probability of the S protein that affects viral tropism and receptor affinity. There is also the potential hindrance of the high glycosylated state of the viral envelope to vaccine development. Therefore, further in-depth studies are needed to elucidate the structure and corresponding physiological and immunological properties of the S protein and also other structural proteins that have a potential role in vaccine development, including mRNA based vaccines.
Conclusions
This review aimed to describe the background to the rationale for the development of mRNA-based SARS-CoV-2 vaccines and the current status of the mRNA-1273 vaccine. Although mRNA vaccines are commencing human clinical trials, due to the rapid global spread of this new viral pandemic, it may not be possible to develop a safe and effective vaccine for SARS-CoV-2 in time to prevent the increasing number of deaths due to this novel RNA virus.
Footnotes
Source of support: Self financing
References
- 1.Guo YR, Cao QD, Hong ZS, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Mil Med Res. 2020;7(1):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (WHO) Novel Coronavirus (2019-nCoV) Situation Report-1. Jan 21, 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200121-sitrep-1-2019-ncov.pdf?sfvrsn=20a99c10_4.
- 3.World Health Organization (WHO) Coronavirus (COVID-19) https://covid19.who.int.
- 4.BASE Medicine Task Force. COVID-19: Facts and recommendations from A to Z. Sci Insigt. 2020;33(1):138–58. [Google Scholar]
- 5.Zumla A, Chan JF, Azhar EI, et al. Coronaviruses – drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–47. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong L, Hu S, Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov Ther. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 7.Prompetchara E, Ketloy C, Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac J Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 8.Porter KR, Raviprakash K. DNA vaccine delivery and improved immunogenicity. Curr Issues Mol Biol. 2017;22:129–38. doi: 10.21775/cimb.022.129. [DOI] [PubMed] [Google Scholar]
- 9.Lazo L, Valdes I, Guillén G, et al. Aiming at the heart: The capsid protein of dengue virus as a vaccine candidate. Expert Rev Vaccines. 2019;18(2):161–73. doi: 10.1080/14760584.2019.1574575. [DOI] [PubMed] [Google Scholar]
- 10.Iavarone C, O’hagan DT, Yu D, et al. Mechanism of action of mRNA-based vaccines. Expert Rev Vaccines. 2017;16(9):871–81. doi: 10.1080/14760584.2017.1355245. [DOI] [PubMed] [Google Scholar]
- 11.Verch T, Trausch JJ, Shank-Retzlaff M. Principles of vaccine potency assays. Bioanalysis. 2018;10(3):163–80. doi: 10.4155/bio-2017-0176. [DOI] [PubMed] [Google Scholar]
- 12.Pharmaceutical Business Review. Moderna doses first patient with mRNA-1273 in coronavirus vaccine trial. https://www.pharmaceutical-business-review.com/news/moderna-mrna-1273-coronavirus-trial/
- 13.Safety and immunogenicity study of 2019-nCoV Vaccine (mRNA-1273) to prevent SARS-CoV-2 Infection. ClinicalTrials.gov Identifier: NCT04283461. https://clinicaltrials.gov/ct2/show/NCT04283461.
- 14.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–44. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Gayle AA, Wilder-Smith A, Rocklöv J. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2) doi: 10.1093/jtm/taaa021. pii: taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarghampoor F, Azarpira N, Khatami SR, et al. Improved translation efficiency of therapeutic mRNA. Gene. 2019;707:231–38. doi: 10.1016/j.gene.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 17.World Health Organization (WHO) Naming the coronavirus disease (COVID-19) and the virus that causes it. World Health Organization; https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it. [Google Scholar]
- 18.Sun P, Lu X, Xu C, et al. Understanding of COVID-19 based on current evidence. J Med Virol. 2020 doi: 10.1002/jmv.25722. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Lin D, Sun X, et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 doi: 10.1126/science.abb3405. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan R, Zhang Y, Li Y, et al. Structural basis for the recognition of the SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–48. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen KG, Rambaut A, Lipkin WI, et al. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–52. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch S, Jasny E, Schmidt KE, Petsch B. New vaccine technologies to combat outbreak situations. Front Immunol. 2018;9:1963. doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardi N, Weissman D. Nucleoside modified mRNA vaccines for infectious diseases. Methods Mol Biol. 2017;1499:109–21. doi: 10.1007/978-1-4939-6481-9_6. [DOI] [PubMed] [Google Scholar]
- 24.Midoux P, Pichon C. Lipid-based mRNA vaccine delivery systems. Expert Rev Vaccines. 2015;14(2):221–34. doi: 10.1586/14760584.2015.986104. [DOI] [PubMed] [Google Scholar]
- 25.Schlake T, Thess A, Fotin-Mleczek M, Kallen KJ. Developing mRNA-vaccine technologies. RNA Biol. 2019;9(11):1319–30. doi: 10.4161/rna.22269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kramps T, Elbers K. Introduction to RNA Vaccines. Methods Mol Biol. 2017;1499:1–11. doi: 10.1007/978-1-4939-6481-9_1. [DOI] [PubMed] [Google Scholar]
- 27.Yuen KS, Ye ZW, Fung SY, et al. SARS-CoV-2 and COVID-19: The most important research questions. Cell Biosci. 2020;10:40. doi: 10.1186/s13578-020-00404-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cascella M, Rajnik M, Cuomo A, et al. StatPearls. Treasure Island (FL): StatPearls Publishing; 2020. Jan, Features, evaluation and treatment coronavirus (COVID-19) https://www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed] [Google Scholar]
- 29.Zarghampoor F, Azarpira N, Khatami SR, et al. Improved translation efficiency of therapeutic mRNA. Gene. 2019;707:231–38. doi: 10.1016/j.gene.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Reichmuth AM, Oberli MA, Jaklenec A, et al. mRNA vaccine delivery using lipid nanoparticles. Ther Deliv. 2016;7(5):319–34. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus 2019) – recent trends. Eur Rev Med Pharmacol Sci. 2020;24(4):2006–11. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 33.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–63. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Fang L, Yang Z, et al. Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB J. 2019;33(12):14575–87. doi: 10.1096/fj.201901624RR. [DOI] [PubMed] [Google Scholar]
- 35.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dhama K, Sharun K, Tiwari R, et al. COVID-19, an emerging coronavirus infection: Advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020 doi: 10.1080/21645515.2020.1735227. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klasse PJ. Modeling how many envelope glycoprotein trimers per virion participate in human immunodeficiency virus infectivity and its neutralization by antibody. Virology. 2007;369(2):245–62. doi: 10.1016/j.virol.2007.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magnus C, Rusert P, Bonhoeffer S, et al. Estimating the stoichiometry of human immunodeficiency virus entry. J Virol. 2009;83(3):1523–31. doi: 10.1128/JVI.01764-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shang J, Wan Y, Liu C, et al. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16(3):e1008392. doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortega JT, Serrano ML, Pujol FH, Rangel HR. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J. 2020;19:410–17. doi: 10.17179/excli2020-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenberg Y, Sack M, Montefiori D, et al. Pharmacokinetics and immunogenicity of broadly neutralizing HIV monoclonal antibodies in macaques. PLoS One. 2015;10(3):e0120451. doi: 10.1371/journal.pone.0120451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uthayakumar D, Paris S, Chapat L, et al. Non-specific effects of vaccines illustrated through the BCG example: From observations to demonstrations. Front Immunol. 2018;9:2869. doi: 10.3389/fimmu.2018.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamboj M, Sepkowitz KA. Risk of transmission associated with live attenuated vaccines given to healthy persons caring for or residing with an immunocompromised patient. Infect Control Hosp Epidemiol. 2007;28(6):702–7. doi: 10.1086/517952. [DOI] [PubMed] [Google Scholar]
- 44.Lim B, Lee K. Stability of the osmoregulated promoter-derived proP mRNA is posttranscriptionally regulated by RNase III in Escherichia coli. J Bacteriol. 2015;197(7):1297–305. doi: 10.1128/JB.02460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlake T, Thess A, Thran M, Jordan I. mRNA as novel technology for passive immunotherapy. Cell Mol Life Sci. 2019;76(2):301–28. doi: 10.1007/s00018-018-2935-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knights AJ, Nuber N, Thomson CW, et al. Modified tumour antigen-encoding mRNA facilitates the analysis of naturally occurring and vaccine-induced CD4 and CD8 T cells in cancer patients. Cancer Immunol Immunother. 2009;58(3):325–38. doi: 10.1007/s00262-008-0556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohto T, Konishi M, Tanaka H, et al. Inhibition of the inflammatory pathway enhances both the in vitro and in vivo transfection activity of exogenous in vitro-transcribed mRNAs delivered by lipid nanoparticles. Biol Pharm Bull. 2019;42(2):299–302. doi: 10.1248/bpb.b18-00783. [DOI] [PubMed] [Google Scholar]
- 48.Peck KM, Lauring AS. Complexities of viral mutation rates. J Virol. 2018;92(14):e01031–17. doi: 10.1128/JVI.01031-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pepini T, Pulichino AM, Carsillo T, et al. Induction of an IFN-mediated antiviral response by a self-amplifying RNA vaccine: Implications for vaccine design. J Immunol. 2017;198:4012–24. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–36. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 51.van den Boogert MAW, Rader DJ, Holleboom AG. New insights into the role of glycosylation in lipoprotein metabolism. Curr Opin Lipidol. 2017;28(6):502–6. doi: 10.1097/MOL.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 52.Breloy I, Hanisch FG. Functional roles of O-glycosylation. Molecules. 2018;23(12):3063. doi: 10.3390/molecules23123063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe Y, Bowden TA, Wilson IA, Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj. 2019;1863(10):1480–97. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schulze IT. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J Infect Dis. 1997;176(Suppl 1):S24–28. doi: 10.1086/514170. [DOI] [PubMed] [Google Scholar]
- 55.Vigerust DJ, Shepherd VL. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007;15(5):211–18. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vankadari N, Wilce JA. Emerging WuHan (COVID-19) coronavirus: Glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. 2020;9(1):601–4. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Behrens AJ, Struwe WB, Crispin M. Glycosylation profiling to evaluate glycoprotein immunogens against HIV-1. Expert Rev Proteomics. 2017;14(10):881–90. doi: 10.1080/14789450.2017.1376658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe Y, Allen JD, Wrapp D, et al. Site-specific analysis of the SARS-CoV-2 glycan shield. BioRxiv. doi: 10.1126/science.abb9983. 2020.03.26.010322. https://www.biorxiv.org/content/10.1101/2020.03.26.010322v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehta N, Alter G. Opportunities to exploit antibody glycosylation in vaccination. Future Virol. 2017;12:325–28. [Google Scholar]
- 60.Lundin KE, Gissberg O, Smith CI. Oligonucleotide therapies: The past and the present. Hum Gene Ther. 2015;26(8):475–85. doi: 10.1089/hum.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pascolo S. Vaccination with messenger RNA (mRNA) Handb Exp Pharmacol. 2008;183:221–35. doi: 10.1007/978-3-540-72167-3_11. [DOI] [PubMed] [Google Scholar]